Abstract
Alphavirus-based replicon vector systems (family Togaviridae) have been developed as expression vectors with demonstrated potential in vaccine development against both infectious diseases and cancer. The single-cycle nature of virus-like replicon particles (VRP), generated by supplying the structural proteins from separate replicable helper RNAs, is an attractive safety component of these systems. MicroRNAs (miRNAs) have emerged as important cellular RNA regulation elements. Recently, miRNAs have been employed as a mechanism to attenuate or restrict cellular tropism of replication-competent viruses, such as oncolytic adenoviruses, vesicular stomatitis virus, and picornaviruses as well as nonreplicating lentiviral and adenoviral vectors. Here, we describe the incorporation of miRNA-specific target sequences into replicable alphavirus helper RNAs that are used in trans to provide the structural proteins required for VRP production. VRP were found to be efficiently produced using miRNA-targeted helper RNAs if miRNA-specific inhibitors were introduced into cells during VRP production. In the absence of such inhibitors, cellular miRNAs were capable of downregulating helper RNA replication in vitro. When miRNA targets were incorporated into a replicon RNA, cellular miRNAs were capable of downregulating replicon RNA replication upon delivery of VRP into animals, demonstrating activity in vivo. These data provide the first example of miRNA-specific repression of alphavirus replicon and helper RNA replication and demonstrate the feasibility of miRNA targeting of expression vector helper functions that are provided in trans.
MicroRNAs (miRNAs) are small RNAs of approximately 21 nucleotides (nt) in length that have been identified in cells and are associated with regulation of mRNA translation and stability (46, 49, 50). The first miRNA described (lin-4) was shown to control the translation of a cellular mRNA (lin-14) involved in the timing of Caenorhabditis elegans larval development (51, 76). Subsequently, numerous miRNAs that control cellular activities, such as proliferation, cell death, fat metabolism, neuronal patterning in nematodes, hematopoietic differentiation, and plant development, have been identified (4, 11, 16, 17, 21, 36, 60). These small regulatory RNAs have been identified in a wide range of animals, including mammals, fish, worms, and flies (1, 2, 18, 33, 42, 47, 48, 56, 57, 59). The miRNA sequences are transcribed from specific miRNA genes, as independent transcriptional units, throughout the genome (47, 49, 53) or may be produced in coordination with intron processing of specific mRNAs (66, 78).
The cellular production of miRNAs begins with transcription of large precursor primary miRNAs which are processed by a nuclear RNase III-like enzyme, Drosha (52, 73). The large precursor RNAs are termed pri-miRNAs, and the smaller Drosha-processed species, termed pre-miRNAs, are exported from the nucleus by Exportin 5 (77). The pre-miRNA, exported from the nucleus, is then processed in the cytoplasm by another RNase III-like enzyme, called Dicer, into a mature miRNA (27, 35, 41). The mature miRNA is then transferred to the RNA-induced silencing complex (RISC), which guides the miRNA to its target RNA (80). The 5′-most 7 to 8 nt (specifically nt 2 to 8) of the miRNA (sometimes referred to as the seed sequence) are involved in Watson-Crick base pairing with nucleotides in the 3′ untranslated region (UTR) of the target mRNA (54). If the base pairing is perfect, the target mRNA is cleaved by the RISC endonuclease activity. Alternatively, if the base pairing is imperfect, the target mRNA becomes translationally inactive, and protein expression is affected without mRNA degradation (reviewed in reference 6).
A replicon vector system has been derived from an attenuated strain of Venezuelan equine encephalitis virus (VEEV) (family Togaviridae; genus Alphavirus). From the positive-sense ∼11-kb genomic VEEV RNA, four nonstructural proteins (nsP1 to nsP4) are translated and function to replicate a full-length negative-sense RNA. The negative-sense RNA serves as a template for the replication of additional genomic RNA and, for synthesis of a subgenomic mRNA (26S mRNA), produced in 10-fold molar excess compared to genomic RNA, which directs the synthesis of the VEEV structural proteins (reviewed in reference 71). The structural proteins are translated initially as a polyprotein that is cotranslationally and posttranslationally cleaved to release the capsid protein and the envelope glycoproteins (E1 and E2). Because VEEV is a positive-sense RNA virus, full-length cDNA clones can be used to generate RNA transcripts that, when introduced into susceptible cells, will initiate a complete virus replication cycle and generate infectious virus. Similarly, foreign genes can be inserted in place of the VEEV structural protein gene region, and an RNA transcript from such a plasmid, when introduced into cells, will replicate and express the heterologous genes (64). This self-amplifying replicon RNA will direct the synthesis of large amounts of the foreign gene product within the cell, typically reaching levels of 15 to 20% of total cell protein (62). Because the replicon RNA does not contain the structural genes for VEEV, it is a single-cycle, propagation-defective RNA and replicates only within the cell into which it is introduced. The replicon RNA can be packaged into virus-like replicon particles (VRP) by supplying the structural protein genes of VEEV in trans (62). Replicon RNA is packaged into VRP when cells are cotransfected with replicon RNA and helpers encoding the capsid and envelope genes, which together encode the full complement of VEEV structural proteins. Replicable RNA helper transcripts can be provided by transfection (8, 10, 23, 24, 55, 62, 70, 74) or as pol II transcripts from stably transfected packaging cell lines (61).
Alphaviruses have a known propensity for nonhomologous recombination (29, 31, 63). A significant advance in reducing the probability of generating replication-competent virus (RCV) was described when the structural protein genes were separated onto two different RNA helpers (23, 62, 70). Early “split helper” RNA designs contain the 5′ and 3′ sequences required for replication as well as an alphavirus 26S subgenomic promoter that normally controls production of the alphavirus structural protein mRNA. Recently, second-generation split helpers have been designed, where the 26S promoter has been removed from the helper RNAs (Δ26S helpers) (37). Removal of the 26S promoter from the helper RNAs further reduces the probability of functional recombination events between helper RNAs and the replicon RNA, as multiple, precise, nonhomologous recombinations are required (37).
As described above, the probability of functional recombination between the replicon and helpers is low when using a two-helper-RNA system; however, some potential remains, and there are other theoretical ways for RNA combinations to arise that may not reconstitute a full genome and yet have limited capacity to be passaged. One possibility is that one or both of the helpers could form single recombinants with the replicon RNA or be packaged into the same or separate particles. Multiple infection of the same cell could then provide all of the genetic sequences required to initiate subsequent cycles. It has been demonstrated previously that a helper RNA coding for a reporter protein could be copackaged into Sindbis replicon particle preparations (58). Volkova et al. demonstrated that a tripartite VRP preparation similar to that described above could be maintained through serial cell culture passage if the nonstructural genes nsP1 to nsP3 were present on capsid and glycoprotein (GP) helper RNAs (74). For these reasons, we have designed “suicide” helper RNAs that function only in the cells used to package VRP but not in other cells. Because it is clear that the miRNAs can control cellular mRNA translation and/or stability and miRNAs are ubiquitous (active in animals ranging from mammals to flies), we have designed alphavirus helper systems that use these elements to target the helper RNAs in environments in which they are not intended to function, such as in VRP-vaccinated individuals. Here, we describe the incorporation of ubiquitous cellular miRNA target sequences into capsid and GP helper RNAs and demonstrate the miRNA-specific inhibition of helper replication in vitro. Furthermore, we have incorporated miRNA targets into replicon vector RNAs and demonstrate that naturally occurring cellular miRNAs are able to target these RNAs and significantly inhibit their ability to replicate both in vitro and in vivo.
MATERIALS AND METHODS
Construction of helpers containing 3′ plus-strand miRNA target sequences.
This set of helpers was designed to have the target for cellular miRNA action be present on the positive-strand RNA message produced during helper replication. A DNA fragment that coded for the reverse complement (RC) sequence of six different miRNAs aligned in tandem was synthesized. The miRNA target sequences are specific for let-7, lin-4, miR-101, miR-155, miR-17, and miR-19, in that order counting from the 5′ end (Table 1). The miRNA targets are represented by the identifiers RC1 through RC6 as indicated in Table 1. A unique SphI restriction site was engineered before the 5′-most miRNA target sequence (RC1), a unique EcoRV restriction site was engineered between the third (RC3) and fourth (RC4) miRNA target sequences, and a unique PmeI restriction site was engineered after the last miRNA (RC6) target sequence. In addition, immediately downstream of the miRNA target sequences, 63 bp corresponding to the VEE 3′ UTR, 55 bp corresponding to a poly(A) stretch, and 8 bp corresponding to a unique NotI restriction site were also synthesized. The 281-bp fragment was digested with SphI and NotI restriction enzymes and then ligated into Δ26S capsid (dHcap6-m1) and Δ26S GP (dHgp6-m1) helper plasmids linearized with the same two enzymes. The dHcap6-m1 and dHgp6-m1 helpers have been described previously (37). The resulting helper plasmids were designated dHcap6-m1 3′ RC1-6-plus and dHgp6-m1 3′ RC1-6-plus (Fig. 1 A).
TABLE 1.
Sequence of miRNA targets incorporated into alphavirus RNAs
| miRNA name | Target identifier | RC sequence of target (5′-3′) |
|---|---|---|
| let-7 | RC1 | AACTATACAACCTACTACCTCA |
| lin-4 | RC2 | ACACAGTCGAAGGTCTCAGGGA |
| miR-101 | RC3 | CTTCAGTTATCACAGTACTGTA |
| miR-155 | RC4 | CCCCTATCACGATTAGCATTAA |
| miR-17 | RC5 | ACTACCTGCACTGTAAGCACTTTG |
| miR-19 | RC6 | TCAGTTTTGCATAGATTTGCACA |
| miR-143 | RC7 | GAGCTACAGTGCTTCATCTCA |
| miR-181 | RC9 | ACTCACCGACAGCGTTGAATGTT |
| miR-124 | RC10 | GGCATTCACCGCGTGCCTTA |
| miR-133 | RC12 | CAGCTGGTTGAAGGGGACCAAA |
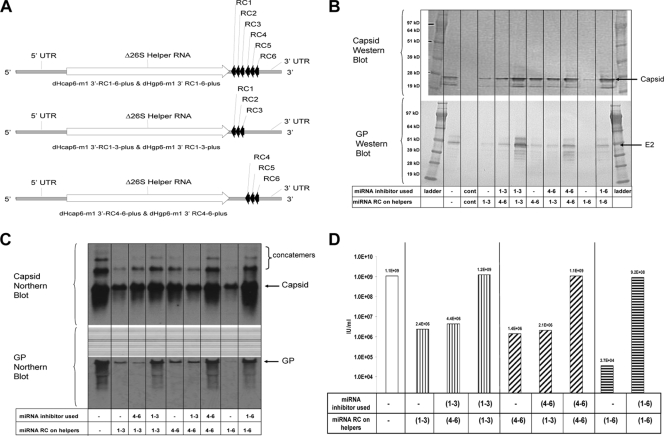
FIG. 1.
Experiments to demonstrate miRNA activity on helper RNAs containing matched target complements (3′ UTR positive-strand RNA). (A) Schematic of the Δ26S miRNA-targeted helper RNAs. (B) Capsid- and GP-specific Western blot analysis of cell protein lysates collected 18 h postelectroporation with miRNA-targeted helper RNAs and a replicon RNA in the absence or presence of unmatched or matched complements of miRNA inhibitors. (C) Capsid- and GP-specific Northern blot analysis of total cellular RNA collected 18 h postelectroporation with miRNA-targeted helper RNAs and a replicon RNA in the absence or presence of unmatched or matched complements of miRNA inhibitors. (D) IFA titer of VRP generated with miRNA-targeted helper RNAs and a replicon RNA in the absence or presence of unmatched or matched complements of miRNA inhibitors. Cont, negative control Vero cell lysates.
Helper plasmids coding for the first three (let-7, lin-4, and miR-101; i.e., RC1 to -3) or the last three (miR-155, miR-17, and miR-19; i.e., RC4 to -6) miRNA target sequences were constructed. Helpers with the first three miRNA targets (RC1 to -3) were constructed by digesting dHcap6-m1 3′ RC1-6-plus and dHgp6-m1 3′ RC1-6-plus helpers with EcoRV and NotI restriction enzymes to remove the miRNA targets RC4 to -6 and the VEE 3′ UTR. The VEE 3′ UTR was replaced by digesting dHcap6-m1 with SphI, treating the DNA with T4 DNA polymerase to produce a blunt end, and then digesting the DNA further with NotI to release a 122-bp fragment. The 122-bp fragment was then ligated into the EcoRV/NotI-digested capsid and GP helpers described above, generating helpers designated dHcap6-m1 3′ RC1-3-plus and dHgp6-m1 3′ RC1-3-plus (Fig. 1A).
Helpers coding for the last three miRNA targets (RC4 to -6) were constructed by digesting dHcap6-m1 3′ RC1-6-plus and dHgp6-m1 3′ RC1-6-plus helpers with EcoRV and RsrII restriction enzymes to remove the capsid or GP genes and the miRNA targets RC1 to -3. The capsid and GP genes were replaced by digesting dHcap6-m1 or dHgp6-m1 DNA with SphI, treating the DNAs with T4 DNA polymerase to produce a blunt end, and then digesting the DNA further with RsrII to release the VEE structural protein genes. The RsrII/SphI(T4) capsid and GP gene fragments were gel purified and then ligated into the RsrII/EcoRV-digested miRNA RC4 to -6 plasmid backbones described above, generating helpers designated dHcap6-m1 3′ RC4-6-plus and dHgp6-m1 3′ RC4-6-plus (Fig. 1A).
Construction of capsid helper containing 3′ minus-strand miRNA target sequences.
Additional helpers were constructed that provide the target sequences for cellular miRNA action on the minus-strand replicative intermediate RNA. A forward primer specific to the miR-19 (RC6) sequence was engineered to code for a unique SphI restriction site. A reverse primer specific for let-7 (RC1) was engineered with a unique PmeI restriction site. PCR amplification was carried out on dHcap6-m1 3′ RC1-6-plus DNA with these primers, resulting in a 155-bp product. The PCR product was digested with SphI and PmeI restriction enzymes and ligated into SphI and PmeI linearized dHcap6-m1 3′ RC1-6-plus. The resultant capsid helper (dHcap6-m1 3′-RC1-6-minus) generates the miRNA target sequences on the minus-strand replicative intermediate RNA (see Fig. 3A).
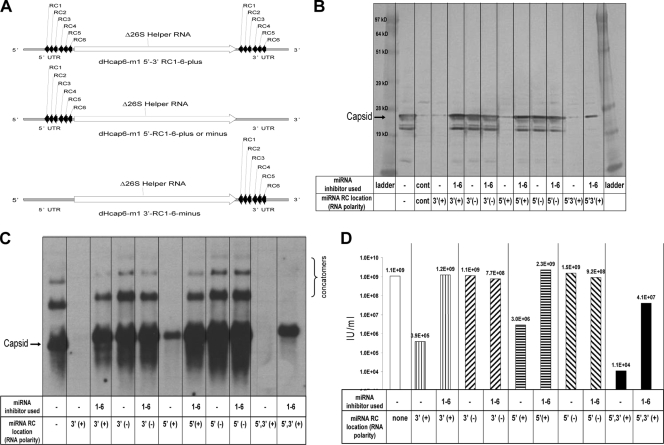
FIG. 3.
Experiments to determine the location requirement of miRNA targets on capsid helper RNAs (5′ and/or 3′ UTR location on positive- and negative-strand RNA). (A) Schematic of the location of miRNA targets on Δ26S capsid helper RNAs. (B) Capsid-specific Western blot analysis of cell protein lysates collected 18 h postelectroporation with miRNA-targeted capsid helper RNAs and a replicon RNA in the absence or presence of matched complements of miRNA inhibitors. (C) Capsid-specific Northern blot analysis of total cellular RNA collected 18 h postelectroporation with miRNA-targeted capsid helper RNAs and a replicon RNA in the absence or presence of matched complements of miRNA inhibitors. (D) IFA titer of VRP generated with miRNA-targeted capsid helper RNAs and a replicon RNA in the absence or presence of matched complements of miRNA inhibitors. Cont, negative control Vero cell lysates.
Construction of helpers containing 5′ plus- and minus-strand miRNA target sequences.
Capsid helpers were constructed that contain the miRNA target sequences (RC1 to -6) in the 5′ UTR such that the targets would be present on either the plus- or minus-strand RNA. A unique RsrII restriction site is present immediately 5′ to the structural gene in the dHcap6-m1 helper plasmid, and it was used to generate these constructs. Forward and reverse primers engineered to code for RsrII restriction sites were designed to amplify the miRNA RC1-6 cassette. The 155-bp PCR product was digested with the RsrII restriction enzyme and ligated into dHcap6-m1 DNA linearized with RsrII enzyme. The orientation of the miRNA RC1-6 PCR product was determined by sequence analysis, and clones with the miRNA RC1-6 fragment in only the reverse orientation (minus-strand RNA) were obtained. These clones were designated dHcap6-m1 5′-RC1-6-minus (see Fig. 3A). To generate a capsid helper with the miRNA RC1 to -6 targets 5′ of the structural gene on the plus-strand RNA, additional steps were required. First, the 5′ RsrII restriction site that was introduced when the minus-strand construct was produced was mutated to a unique BglII restriction site by using site-directed mutagenesis (Stratagene, La Jolla, CA), resulting in a helper designated dHcap6-m1 5′BglII-RC1-6-minus. Next, primers were engineered to amplify the plus-strand miRNA RC1 to -6 target sequence with a 5′ BglII restriction site and a 3′ RsrII restriction site. The ca. 147-bp product of amplification was digested with BglII and RsrII restriction enzymes and gel purified. The dHcap6-m1 5′BglII-RC1-6-minus DNA was then digested with BglII and RsrII to remove the minus-strand-oriented miRNA targets, and the plus-strand miRNA target fragment was cloned in its place. The helper that expressed the miRNA targets on the 5′ plus-strand RNA was designated dHcap6-m1 5′-RC1-6-plus (see Fig. 3A).
Construction of helpers containing 5′ and 3′ plus-strand miRNA target sequences.
A capsid helper was constructed that coded for miRNA RC1 to -6 targets in both the 5′ and 3′ UTR positions on the plus-strand RNA template. To accomplish this, the dHcap6-m1 5′-RC1-6-plus helper DNA was digested with SphI and NotI restriction enzymes to release the wild-type 3′ UTR sequence. The dHcap6-m1 3′ RC1-6-plus helper DNA was digested with SphI and NotI restriction enzymes, and the 3′ UTR fragment (ca. 270 bp) containing the miRNA RC1 to -6 targets was gel purified. The linearized dHcap6-m1 5′-RC-plus DNA was ligated with the 3′ UTR miRNA RC1-6 fragment, and the resulting helper was designated dHcap6-m1 5′-3′-RC1-6-plus DNA (see Fig. 3A).
Construction of helpers containing individual miRNA targets repeated six times on the 3′ UTR plus strand.
Capsid helpers coding for individual miRNA targets repeated six times, on the positive-strand RNA message produced during helper replication, were also constructed. Six copies of each individual miRNA target were chosen because that is the total number of miRNA targets tested in the RC1 to -6 constructs, and the length of the inserted sequence in the 3′ UTR would also be maintained in the new constructs relative to the RC1 to -6 constructs. DNA fragments (BlueHeron, Inc., Bothell, WA) that coded for the RC sequence of six copies of each individual miRNA aligned in tandem were de novo synthesized. The identifiers for the respective 6-mer miRNA fragments were as follows: RC1x6, RC2x6, RC3x6, RC4x6, RC5x6, and RC6x6. A unique SphI restriction site was engineered before the first miRNA target sequence, and a unique PmeI restriction site was engineered after the last copy of the miRNA target sequence. The respective miRNA RCx6 sequence fragments were digested with SphI and PmeI restriction enzymes and then ligated individually in place of the miRNA RC1-6 sequence fragment found in dHcap6-m1 3′ RC1-6-plus by digesting the helper with SphI and PmeI restriction enzymes.
Construction of miRNA-targeted replicon and TC-83 infectious clone vectors.
The replicon expressing the influenza A/Wyoming hemagglutinin (HA) gene, pERK/383/EV71/A(Wyo)/HA, has been described previously (34). The miRNA target sequences for RC1 to -6 were digested from the dHcap6-m1 3′ RC1-6-plus helper by using restriction enzymes SphI and NotI. The ca. 270-bp fragment released after digestion was isolated and ligated into the 3′ UTR of the pERK/383/EV71/A(Wyo)/HA replicon linearized with SphI and NotI restriction enzymes, generating a replicon designated pERK/383/EV71/A(Wyo)/HA RC1-6-plus. Replicon vectors expressing the chloramphenicol acetyltransferase (CAT) reporter gene that contained miRNA targets located in the 3′ UTR were constructed. The miRNA targets were cloned into a CAT replicon vector as tandem single copies (RC1 to -6) or as six tandem copies of the same miRNA target (RC1x6, RC2x6, RC3x6, RC4x6, RC5x6, and RC6x6). The replicon vector, pERK/342/EMCV/CAT, used for all constructions has been described previously (38). The miRNA target sequences for RC1x6 to RC6x6 were digested from their respective dHcap6-m1RC helpers (described above) by using restriction enzymes SphI and NotI. The miRNA target fragments released after digestion were then isolated and ligated into the pERK/342/EMCV/CAT replicon vector linearized with the same restriction enzymes.
Plasmid pVE/IC-92, coding for the full-length genome of TC-83 virus, was obtained from the Centers for Disease Control and Prevention (43). A DNA fragment that coded for the miRNA targets RC1-RC5 as well as the target sequences for miR-181 (56), miR-124, miR-143, and miR-133 was synthesized (48). The additional sequences were introduced to add tissue-specific miRNA targets to complement the more general targets represented by RC1 to RC5. The synthesized DNA fragment was engineered with unique 5′ SphI and 3′ PmeI restriction endonuclease sites. The synthesized DNA fragment was digested with SphI and PmeI restriction enzymes and ligated into plasmid pVE/IC-92 linearized with the same restriction enzymes such that the miRNA targets would be present on the positive-strand 3′ UTR of the TC-83 full-length genome RNA. The resulting plasmid was termed pTC-83 mi.
RNA transcription, electroporation, and VRP or TC-83 virus production.
Capped replicon, TC-83, and helper RNAs were in vitro transcribed using a T7 RiboMax kit (Promega, Madison, WI) by following the manufacturer's instructions. RNAs were purified using RNeasy purification columns (Qiagen, Valencia, CA) by following the manufacturer's instructions. The procedures used for producing VRP vaccines were based on modifications of published methods (62). Vero cells (8 × 107) suspended in 0.6 ml phosphate-buffered saline (PBS; Gibco) were electroporated with 30 μg of replicon RNA, 20 μg of capsid helper RNA, and 60 μg of GP helper RNA by using a Bio-Rad gene pulser (Bio-Rad, Hercules, CA). TC-83 virus was generated by electroporating 30 μg of infectious clone RNA into Vero cells as described above. When miRNA-targeted RNAs were used to generate VRP or virus, miRNA inhibitors (9 nmol of each inhibitor) were included in the electroporation. The miRNA inhibitors were 2′-O-methyl-modified RNA oligonucleotides (Eurogentec, San Diego, CA) with the RC sequence to the target miRNA (sequences shown in Table 1). Cells were pulsed four times with the electroporator set at 580 V and 25 μF. Electroporated cell suspensions were seeded into roller bottles containing serum-free medium and incubated at 37°C in 5% CO2 for 16 to 24 h. VRP or TC-83 virus was harvested from culture fluids, and the infectious titer of the preparation was measured by antigen-specific immunofluorescence assay (IFA) or plaque assay, respectively. VRP preparations were tested in a cytopathic effect (CPE) assay to ensure the absence of detectable RCV (38). VRP generated with a replicon vector containing miRNA targets were titrated on Vero cells electroporated with miRNA inhibitors to allow the replicon vector to replicate unhindered by the cellular miRNAs. VRP generated with a replicon vector containing miRNA targets were also analyzed by quantitative reverse transcription-PCR (RT-PCR) to determine genome equivalents for each preparation. VRP were formulated with human serum albumin (1%) and stored at −80°C until used.
VRP concentration, expressed as infectious units (IU) per ml, was determined by an IFA in which serial dilutions of VRP were added to Vero cell monolayers in 48-well plates, cultured overnight, fixed with acetone, and reacted with VEEV nsP2-specific goat polyclonal antibodies followed by fluorescein isothiocyanate-labeled secondary antibody to enumerate positive cells.
Titers of virus generated with the TC-83 mi infectious clone RNA were determined by plaque assay on Vero cells electroporated with miRNA inhibitors to allow replication to occur unhindered by cellular miRNAs. In parallel, mock-electroporated Vero cells were used to determine virus plaque titers in cells that did not receive miRNA inhibitors. TC-83 and TC-83 mi virus preparations were diluted serially, added to Vero cell monolayers, and incubated for 1 h, and then the cells were overlaid with 2 ml of a 1.2% Avicel (FMC, Rockland, ME), 2% fetal bovine serum (FBS), and 1× minimal essential medium (MEM; Gibco) solution. Plates were incubated at 37°C in 5% CO2 for 48 h. After incubation, virus titers were determined by fixing the monolayers with an acetone:methanol (1:1) solution containing 5% crystal violet and by enumerating the plaques.
CAT ELISA and Western blot analysis.
CAT expression was quantified by enzyme-linked immunosorbent assay (ELISA) using miRNA-targeted, CAT replicon VRP-infected cell lysates and a commercially available CAT ELISA kit (Boehringer Mannheim, Indianapolis, IN), by following the manufacturer's instructions. All cell types analyzed were infected with VRP at a multiplicity of infection (MOI) of 1 based on the number of genome copies rather than the number of IU. The cell lysates were produced 18 h postinfection, and total protein concentration was determined for each sample by using a bicinchoninic acid (BCA) protein kit (Pierce, Rockford, IL). The methods used to generate Vero cell lysates and carry out Western blot analysis have been described previously (32). VEEV-specific polyclonal goat anti-capsid or goat anti-GP antibodies were used to detect structural protein expression by Western blot analysis.
RT-qPCR analysis of VRP.
To determine the number of genome equivalents present in each miRNA-targeted CAT replicon VRP preparation, a standard one-step quantitative RT-PCR (RT-qPCR) protocol was performed on an Applied Biosystems 7500 fast real-time PCR sequence detection system (Applied Biosystems, Foster City, CA). Amplification was detected by means of a fluorogenic probe designed to anneal to a region of the nsP2 gene on the replicon between the two primers. A 5′ reporter dye (6-carboxyfluorescein [FAM]) and a 3′ quencher dye (BHQ-1) were attached to the probe. Proximity of the reporter and quencher dyes resulted in the suppression of reporter fluorescence prior to amplification. Upon successful amplification of the target region, the 5′ exonuclease activity of DNA polymerase released the reporter dye from the hybridized probe, resulting in a fluorescent signal. Purified VEE replicon RNA was used to generate a standard curve in the assay, and the fluorescent signal of each VRP sample was measured not to exceed 40 PCR cycles and compared to the fluorescent signal of the standards to determine genome equivalents.
Northern analysis.
Northern analysis was carried out on total cellular RNA collected from VRP-infected cells. Total cellular RNA was collected from the cells 18 h postinfection by using SV Total RNA isolation columns (Promega, Madison, WI) by following the manufacturer's instructions. The RNAs were quantified by UV absorption, and 3 μg of each were run on 1% glyoxal agarose gels before being transferred to a BrightStar Plus membrane (Ambion, Inc., Austin, TX) by passive transfer. The RNAs were UV cross-linked to the membrane, blocked with UltraHyb (Ambion) solution for 1 h at 45°C, and probed overnight with UltraHyb solution containing biotinylated RNA probes specific for nsP2, capsid, or GP genes at 68°C. After overnight hybridization, the blot was processed for chemiluminescent RNA detection by using a BrightStar BioDetect kit (Ambion) by following the manufacturer's instructions and visualization after exposure to autoradiograph film (Kodak, Rochester, NY).
Vaccination of mice and sample collection.
Animal protocols were reviewed and approved by the Integrated Laboratory Systems (Research Triangle Park, NC) Institutional Care and Use Committee. Female, 6- to 8-week-old BALB/c mice (Charles River Laboratory, Kingston, NY) were immunized with 1 × 106 IU of VRP expressing the influenza A/Wyoming hemagglutinin (HA) gene that were produced using combinations of miRNA-targeted helpers and replicon vectors. Mice were immunized at 0 and 3 weeks by subcutaneous (SC) injection into the rear footpad. Influenza virus HA-specific antibody and T-cell responses were monitored 1 week after each immunization. Blood samples were collected by retro-orbital bleeds for all groups. Eight mice from each group were sacrificed 1 week after each immunization, and splenocytes were collected from individual animals for T-cell analysis.
HA ELISA.
For HA ELISA analysis, dilutions of sera from VRP-vaccinated mice were made with PBS containing 1% bovine serum albumin (BSA) and 0.05% Tween 20, beginning with 1:40 followed by 2-fold dilutions up to 1:2,560. Prebleed samples were diluted to only 1:40. ELISA plates (Nunc, Rochester, NY), which had been coated with recombinant A/Wyoming HA antigen (50 ng/well) (Protein Sciences Inc., Meriden, CT) in carbonate/bicarbonate buffer (Sigma-Aldrich, St. Louis, MO) overnight at 4°C, were incubated with 200 μl/well of blocking buffer (PBS, 3% BSA) at 30°C for 1 h. Blocked plates were washed three times with 200 μl of PBS. Fifty microliters of diluted sera was added to the plates in duplicate and incubated at 30°C for 1 h. Plates were washed three times with 200 μl PBS. One hundred microliters of alkaline phosphatase-conjugated anti-mouse polyvalent immunoglobulins (IgG, IgA, IgM) (Sigma) diluted in blocking buffer (1:500) was added to each well. Plates with secondary antibody were incubated for 1 h at room temperature and then washed three times with 200 μl PBS. Plates were developed using Fast p-nitrophenyl phosphate tablet sets (Sigma) and read at a wavelength of 405 nm. An optical density at 405 nm (OD405) of 0.2 or greater was considered positive.
HA ELISPOT assay.
Splenocytes were isolated from individual animals and HA-specific gamma interferon enzyme-linked immunospot (ELISPOT) assays were performed to determine the number of antigen-specific cytokine-secreting T cells. This procedure has been described previously (65).
Statistical analyses.
Mean HA-specific ELISPOT spot-forming cell (SFC) counts and mean reciprocal HA-specific ELISA titers were analyzed using Student's t test and one-way analysis of variance software associated with the GraphPad Prism 5.01 program (GraphPad Software, San Diego, CA).
RESULTS
Experiments to demonstrate functional miRNA activity on helper RNAs.
Experiments were conducted to determine whether eukaryotic miRNAs could be used to reduce alphavirus helper replication and expression within cells by placing miRNA target sequences in the 5′ or 3′ UTRs of these RNAs. Six miRNA target sequences that represent evolutionarily conserved or ubiquitous eukaryotic miRNAs were selected (7, 22, 69). The miRNA targets used in these studies are listed in Table 1.
Figure 1 is a representative example of the initial experiments designed to determine whether a complement of six miRNA targets (let-7, lin-4, miR-101, miR-155, miR-17, and miR-19 target RC1 to -6) or subsets of these targets (let-7, lin-4, and miR-101 target RC1 to -3, or miR-155, miR-17, and miR-19 target RC4 to -6) would be able to control helper replication when located in the 3′ UTR of the plus-strand RNA (Fig. 1A). Vero cells were electroporated with a replicon vector combined with capsid and GP helpers with matching complements of miRNA targets on each helper. Specifically, replicon RNA was combined with capsid and GP helpers with miRNA targets RC1 to -6 or with both helpers carrying the miRNA RC1 to -3 targets or with both helpers carrying the miRNA RC4 to -6 targets. Each of the helper and replicon combinations were electroporated into Vero cells in the presence or absence of miRNA inhibitors (2′-O-methylated RNA oligonucleotides) specific for the complete complement of miRNA targets present on the helpers. In addition, the helper RNA combinations with miRNA targets RC1 to -3 and RC4 to -6 were also combined with unmatched miRNA inhibitor combinations to demonstrate the specificity of the inhibitors used. The electroporated cells were seeded into medium and incubated for 18 h, and samples were collected to examine helper-specific protein expression (Western blot), helper-specific RNA replication (Northern blot), and VRP yield. Western blot analysis indicated that expression of capsid and GP was reduced in the absence of miRNA inhibitors specific for the miRNA targets present on the helpers (Fig. 1B). The difference in protein expression was particularly evident for miRNA-targeted GP helpers in the absence of specific miRNA inhibitors. Northern blot analysis indicated that capsid and GP helper replication levels mirrored the respective helper protein expression in the presence or absence of the matched miRNA inhibitors (Fig. 1C). Interestingly, capsid-reactive RNAs larger than the unit-length helper RNA were detected by Northern blot analysis. Preliminary sequence analysis of the capsid-reactive RNAs that are larger than unit-length RNAs indicates that they are nonfunctional capsid concatemers (data not shown). A self-priming copy-back mechanism of RNA synthesis was proposed for generation of a similar RNA species identified by Hardy and Rice after an analysis of the importance of the 3′ conserved sequence element for Sindbis virus replication (30). The molecular characterization of the concatemer RNAs noted by Northern blotting is the source of ongoing research in our group, and results from those experiments will be reported elsewhere. The relative capsid and GP expression and helper RNA replication levels were reflected in the dramatic differences observed in VRP yields. Cultures that did not receive the appropriate miRNA inhibitors (specific for the miRNA targets on the helpers) produced 3 to 4.5 orders of magnitude less VRP (Fig. 1D).
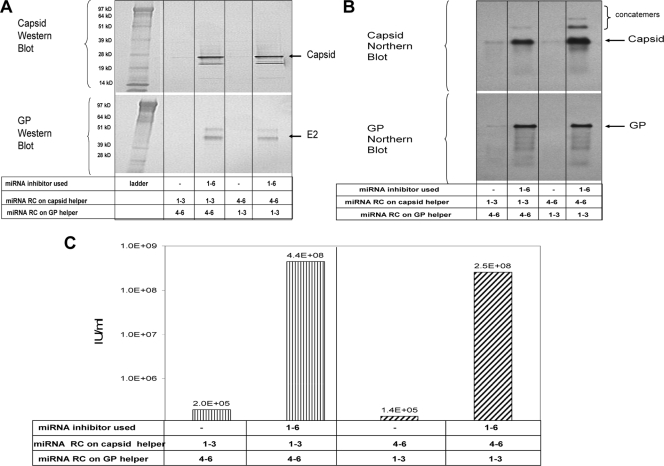
The experiment described above matched the miRNA targets on each helper. Figure 2 is a representative example of experiments conducted to determine whether different combinations of miRNA targets could be present on the helpers and still rescue VRP yields when the appropriate combination of miRNA inhibitors was supplied. Combinations of a capsid helper with RC1 to -3 targets and a GP helper with RC4 to -6 (and vice versa) were tested in the presence or absence of a mixture of all six miRNA inhibitors. Western blot analysis indicated that expression of capsid and GP was reduced in the absence of miRNA inhibitors specific for the miRNA targets present on the helpers (Fig. 2A). Northern blot analysis indicated that capsid and GP helper replication levels mirrored the respective helper protein expression in the presence or absence of the matched miRNA inhibitors (Fig. 2B). As found in the experiment with matched miRNA targeted helpers, the relative capsid and GP expression and helper RNA replication levels predicted the VRP yields produced. VRP yields were 3 orders of magnitude lower in cells that did not receive the miRNA inhibitors than in those that did (Fig. 2C).
FIG. 2.
Experiments to demonstrate miRNA activity on helper RNAs containing unmatched target complements (3′ UTR positive-strand RNA). (A) Capsid- and GP-specific Western blot analysis of cell protein lysates collected 18 h postelectroporation with miRNA-targeted helper RNAs and a replicon RNA in the absence or presence of matched complements of miRNA inhibitors. (B) Capsid- and GP-specific Northern blot analysis of total cellular RNA collected 18 h postelectroporation with miRNA targeted helper RNAs and a replicon RNA in the absence or presence of matched complements of miRNA inhibitors. (C) IFA titer of VRP generated with miRNA-targeted helper RNAs and a replicon RNA in the absence or presence of matched complements of miRNA inhibitors.
Experiments to determine location requirement of miRNA targets on helper RNAs.
To determine whether the miRNA targets RC1 to -6 could function in locations other than the 3′ UTR of the plus-strand capsid helper RNA, additional capsid helpers that moved the miRNA RC1 to -6 targets to the 5′ UTR of the capsid helper RNA on both the plus-strand and minus-strand RNA templates for replication were constructed. The miRNA RC1 to -6 targets were also engineered in the 3′ UTR on the minus-strand RNA template. In addition, a helper that coded for miRNA targets on the plus-strand RNA both 5′ and 3′ of the capsid gene was constructed. A schematic representation of the miRNA-targeted capsid helpers is shown in Fig. 3 A. Figure 3 is a representative example of the experiments that were conducted with a combination of replicon RNA, unmodified Δ26S GP helper RNA, and one of the miRNA RC1 to -6 targeted capsid helper RNAs in the presence and absence of miRNA inhibitors specific for the miRNA targets present on the helpers. The electroporated cells were seeded into medium and incubated overnight. After incubation (∼18 h), samples were again collected to examine capsid helper-specific protein expression, RNA replication, and VRP yield. The results of capsid-specific Western and Northern blot analysis are shown in Fig. 3B and C, respectively. The VRP yields produced using the different miRNA-targeted capsid helpers are shown in Fig. 3D. Both Western and Northern blot analyses indicated that the miRNA targets function most effectively when on the plus-strand helper RNA. Helper expression and replication were significantly reduced when the plus-strand RNA was targeted and miRNA inhibitors were not supplied at the time of RNA electroporation of cells. In addition, miRNA targets in either a 5′ or 3′ location relative to the capsid gene or in both the 5′ and 3′ locations simultaneously were effective at reducing expression and replication of the helper RNA in the absence of miRNA inhibitor addition. As noted in previous experiments, VRP yields were 3 orders of magnitude lower in cells that did not receive the miRNA inhibitors than in those that did (Fig. 3D).
Experiments to determine contribution of individual miRNA targets on helper RNA replication.
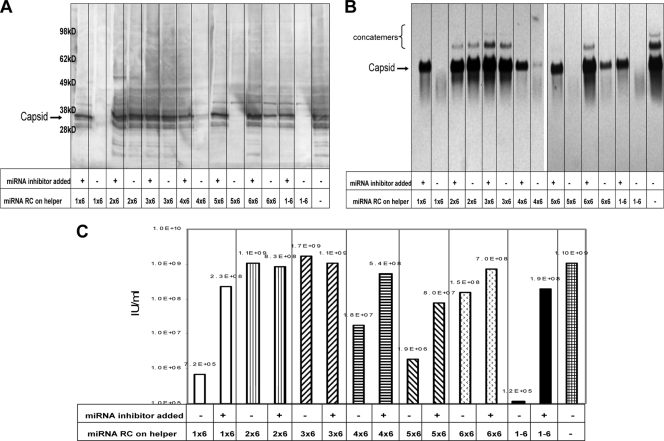
To determine what effect each miRNA target was contributing to the overall control of replication in Vero cells, six copies of individual target miRNA sequences were cloned into the 3′ UTR of the capsid helper plus-strand RNA. Figure 4 is a representative example of experiments using these capsid helper RNAs and unmodified Δ26S GP helper RNA combined with replicon RNA and electroporated into Vero cells in the presence or absence of miRNA inhibitors specific for the miRNA targets present on the capsid helper. The electroporated cells were seeded into medium and incubated overnight. After incubation (∼18 h), samples were collected to examine capsid-specific protein expression, RNA replication, and VRP yield. The results of capsid-specific Western and Northern blot analyses are shown in Fig. 4A and B, respectively. These results suggest that cellular miRNAs specific for let-7 (RC-1), miR-155 (RC-4), miR-17 (RC-5), and miR-19 (RC-6) are capable of downregulating helper expression and replication (in the absence of specific miRNA inhibitor) and that miRNAs specific for lin-4 (RC-2) and miR-101 (RC-3) are less effective at controlling helper expression/replication in Vero cells. The VRP yields produced using the different miRNA-targeted capsid helpers are shown in Fig. 4C. The capsid helper containing all six miRNA targets (RC1 to -6) demonstrated the largest difference in VRP yields in the presence and absence of miRNA inhibitor (3 orders of magnitude), suggesting an additive effect of the miRNA targets on helper replication. The 6-mer of miRNA let-7 (RC1x6) target demonstrated the next largest effect on VRP yield (∼2.5 orders of magnitude), with miR-155 (RC4x6) and miR-17 (RC5x6) both demonstrating ∼1.5 orders of magnitude differences in VRP yield. The helper with six copies of the miR-19 target (RC6x6) produced ∼0.5 log less VRP yield, while lin-4 (RC2x6) and miR-101 (RC3x6) targets demonstrated little difference in VRP yield in Vero cells (Fig. 4C).
FIG. 4.
Experiments to determine the contribution of individual miRNA targets on capsid helper RNA replication in Vero cells (six individual miRNA copies in tandem on the 3′ UTR positive-strand RNA). (A) Capsid-specific Western blot analysis of cell protein lysates collected 18 h postelectroporation with miRNA-targeted capsid helper RNAs and a replicon RNA in the absence or presence of matched complements of miRNA inhibitors. (B) Capsid-specific Northern blot analysis of total cellular RNA collected 18 h postelectroporation with miRNA-targeted capsid helper RNAs and a replicon RNA in the absence or presence of matched complements of miRNA inhibitors. (C) IFA titer of VRP generated with miRNA-targeted capsid helper RNAs and a replicon RNA in the absence or presence of matched complements of miRNA inhibitors.
Experiments to determine cell type specificity of miRNA activity on alphavirus RNA replication.
All of the previous experiments were conducted in Vero cells. To determine whether the miRNA targets we have tested in Vero cells have the same effect in other cell types, replicon vectors that express the CAT reporter gene were constructed with six copies of each individual miRNA target. VRP preparations were prepared by electroporating Vero cells, in the presence of the relevant miRNA inhibitor, with each of the miRNA-targeted CAT replicon RNAs combined with unmodified Δ26S capsid and Δ26S GP helper RNAs. To eliminate difficulty in determining accurate Vero cell IU titers of these VRP preparations (due to the effect of the miRNAs present in Vero cells reducing the effective IU titer), replicon genome equivalents (GE) were determined by quantitative reverse transcriptase PCR. The genome equivalent titers were then used to control the MOI for each of the cell types analyzed. A summary of the relative miRNA activities detected in each cell type is shown in Table 2 and is based on the percentage of CAT expressed from each of the miRNA-targeted replicons relative to an unmodified CAT replicon.
TABLE 2.
Relative miRNA activity in different cell lines
| Cell type | Relative % CAT (miRNA) replicon vs CAT (control) replicon expression in indicated cell linel |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human |
Hamster |
NHP kidney Veroi | Bovine turbinate BTj | Chicken embryo CEFk | |||||||
| Muscle |
Glioblastoma MO59Kd | Lung MRC-5e | Hepatocyte PHHf | Kidney BHKg | Ovary CHOh | ||||||
| HEL-299a | RDb | SkMc | |||||||||
| CAT (control) | 100 (0.6) | 100 (0.1) | 100 (0.1) | 100 (0.9) | 100 (0.5) | 100 (0.0) | 100 (0.0) | 100 (0.1) | 100 (0.0) | 100 (0.8) | 100 (0.1) |
| CAT (RC1-6) | 0 (0.6) | 5 (1.4) | 2 (0.3) | 0 (0.0) | 1 (1.7) | NT | 1 (0.6) | 0 (0.0) | 0 (0.0) | 0 (1.0) | 1 (0.1) |
| CAT (RC1x6) | 0 (0.0) | 20 (7.7) | 1 (0.4) | 0 (0.0) | 0 (0.3) | 0 (0.0) | 0 (0.1) | 0 (0.1) | 0 (0.0) | 0 (1.0) | 0 (0.0) |
| CAT (RC2x6) | 3 (0.4) | 20 (8.9) | 11 (2.1) | 15 (6.8) | 4 (1.5) | 11 (8.4) | 31 (9.3) | 1 (1.1) | 42 (11.0) | 8 (6.8) | 3 (0.1) |
| CAT (RC3x6) | 26 (8.2) | 41 (25.1) | 38 (11.8) | 50 (13.5) | 49 (13.3) | 0 (0.0) | 86 (19.5) | 36 (25.9) | 37 (11.3) | 50 (14.8) | 55 (8.2) |
| CAT (RC4x6) | 13 (2.0) | 52 (18.0) | 42 (9.6) | 46 (7.5) | 57 (20.9) | 32 (30.7) | 81 (15.6) | 66 (32.4) | 11 (2.4) | 55 (15.9) | 75 (9.5) |
| CAT (RC5x6) | 7 (1.2) | 0 (0.2) | 14 (3.7) | 6 (2.7) | 35 (13.0) | 0 (0.0) | 13 (7.0) | 0 (0.2) | 1 (0.9) | 18 (6.8) | 0 (0.0) |
| CAT (RC6x6) | 11 (1.2) | 8 (3.3) | 30 (5.1) | 28 (8.7) | 45 (18.2) | 0 (0.0) | 60 (15.0) | 8 (3.9) | 14 (2.9) | 32 (13.9) | 1 (0.1) |
ATCC CCL-137.
ATCC CCL-136.
Primary human skeletal muscle (Lonza Walkersville, Inc., Walkersville, MD).
ATCC CRL-2365.
ATCC CCL-171.
PHH (primary human hepatocytes, ADMET Technologies, Durham, NC).
ATCC CCL-10.
ATCC CCL-61.
ATCC CCL-81.
ATCC CRL-1390.
Primary chicken embryo fibroblast cells.
CAT protein expression measured in duplicate in two separate experiments. All percentages are based on the CAT replicon expression level (with no miRNA targets) for the respective cell type set at 100%. Numbers in parentheses represent 1 standard deviation. NT, not tested; NHP, nonhuman primate.
VRP generated with a CAT replicon carrying all six of the miRNA targets (CAT 1 to 6) expressed very little CAT protein in any cell type tested. Similarly, minimal CAT protein could be detected in cells infected with VRP containing a replicon RNA carrying the let-7 miRNA target (CAT RC1x6). These results indicate that the let-7 miRNA is present in all of the cell types tested. CAT protein levels detected in cells infected with the other individual miRNA targeted CAT replicons indicated a range of protein expression. These data show that each different cell type had a different functional level of active miRNAs available to control the replication of replicon RNA.
Experiments to determine the effect of miRNA targets on propagation of TC-83 virus in Vero cells.
A minimum of two separate recombination events between a replicon RNA and miRNA-targeted helper RNAs is required to reconstitute a functional genome and generate an RCV. The miRNA targets engineered into the 3′ UTR of the helper RNAs are likely to be maintained after the second recombination event required to generate RCV because the miRNA targets are located in close proximity to the conserved 3′ sequence required for replication of all alphaviruses. To determine the effect that miRNA targets would have on propagation of an RCV generated by the unlikely recombination events described above, we engineered miRNA targets into the 3′ UTR of an infectious clone of the VEEV vaccine strain, TC-83 (termed pTC-83 mi). TC-83 mi virus was produced by electroporating Vero cells with RNA transcribed in vitro from the TC-83 mi plasmid DNA. The TC-83 mi in vitro-transcribed RNA was combined with miRNA inhibitors prior to electroporation of Vero cells to allow unhindered genome replication and production of virus. The TC-83 mi virus was then titrated by plaque assay in Vero cells. The Vero cells used in the plaque assays were electroporated with or without miRNA inhibitors and then seeded in parallel into six-well plates prior to setting up virus titrations; if the miRNA targets were effective at reducing the replication of the viral genome, the titer of the TC-83 mi virus preparation should be lower on cells that did not receive miRNA inhibitor prior to infection than on those cells that did receive miRNA inhibitor. In parallel, wild-type TC-83 virus was titrated on Vero cells treated as described above to determine whether the presence of miRNA inhibitors in the cells would affect plaque titer of a virus that did not carry miRNA targets in the 3′ UTR of its genome. The mean virus titers of three separate plaque assay experiments are represented in Table 3. TC-83 mi virus demonstrated a >500-fold lower effective plaque titer on Vero cells that had not been treated with miRNA inhibitors relative to that on Vero cells that had been treated with miRNA inhibitors (Student's t test; P < 0.001). Importantly, wild-type TC-83 virus demonstrated no significant difference in plaque titer on either type of Vero cell preparation (Student's t test; P = 0.75). These data indicate that the presence of miRNA inhibitors in the Vero cells did not influence virus plaque titer unless the virus genome carried miRNA targets and that, in the absence of miRNA inhibitors, the replication of TC-83 mi virus was significantly reduced compared to when miRNA inhibitors were present.
TABLE 3.
Effect of miRNA targets on plaque titer of TC-83 mi virus
| Virus | No. of PFU/ml determined on Vero cellsa |
Fold change in PFU titer with miRNA inhibitor treatment | |
|---|---|---|---|
| No miRNA inhibitor | Plus miRNA inhibitor | ||
| TC-83 | 5.1 × 108 (1.0 × 108) | 5.3 × 108 (1.2 × 107) | +1.04 |
| TC-83 mi | 1.6 × 106 (5.3 × 105) | 1.1 × 109 (1.6 × 108) | +687.5 |
Values shown in parentheses represent 1 standard deviation.
Experiments to determine in vivo miRNA activity on alphavirus RNA replication.
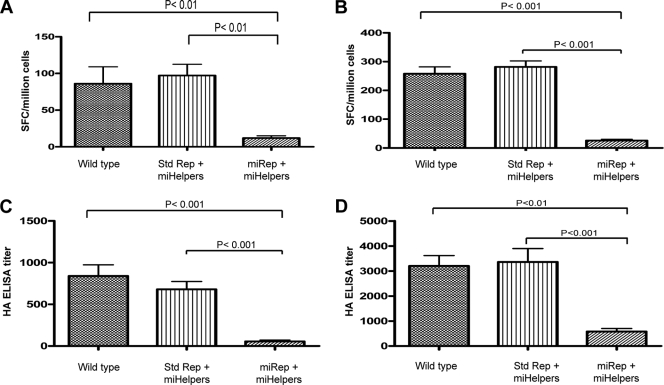
The data described above indicate that miRNA-targeted helper, replicon, and TC-83 virus genomic RNAs can be controlled by the action of cellular miRNAs of cells in culture (in vitro). To determine whether the same miRNA control demonstrated in vitro could be demonstrated in vivo, a number of VRP preparations were produced and tested in BALB/c mice. A summary of the helper and replicon RNA combinations used to generate VRP is shown in Table 4. All VRP preparations produced with RNAs containing miRNA targets were generated by coelectroporation of miRNA inhibitors. Groups of 16 mice were immunized with 1 × 106 IU of each of the VRP. In addition, each VRP preparation was analyzed by RT-qPCR to ensure that each group of animals received the same number of genome equivalents. Seven days after the priming dose, half of the mice (n = 8) were sacrificed, the splenocytes were collected, and HA-specific gamma interferon ELISPOT analysis was conducted. The results of the 7-day prime HA-specific ELISPOT analysis are shown in Fig. 5 A. There were significantly fewer SFCs in the group immunized with VRP generated with miRNA-targeted replicon RNA than in groups immunized with VRP generated with unmodified RNAs or with VRP generated with both capsid and GP miRNA-targeted helpers. Three weeks after the priming dose, the remaining mice in each group were boosted with their respective HA VRP. Seven days after the boost, the remaining animals were sacrificed, and the splenocytes were collected for HA-specific gamma interferon ELISPOT analysis. The postboost HA-specific ELISPOT analysis results are shown in Fig. 5B. After the boost, the number of SFCs detected in the group immunized with VRP generated using miRNA-targeted replicon was significantly lower than those detected in all of the other VRP-immunized groups. Furthermore, HA-specific ELISA of both postprime (Fig. 5C) and postboost (Fig. 5D) serum samples demonstrated that animals immunized with VRP generated with a miRNA-targeted replicon had significantly lower titers than those in all other VRP-immunized groups. These data demonstrate that the miRNA-targeted replicon VRP were significantly inhibited from inducing an HA-specific cellular or humoral immune response. Moreover, the data suggest that the added miRNA targets were biologically active in vivo and addition of miRNA targets within the replicon RNA resulted in intracellular control of the targeted RNA.
TABLE 4.
Combinations of replicon and Δ26S helper RNAs used to generate VRP for mouse study
| Group identifier | Combination with: |
||
|---|---|---|---|
| Replicon RNA | Capsid helper | GP helper | |
| Wild type | pERK/383/EV71/HA | dHcap6-m1 | dHgp6-m1 |
| Std Rep + miHelpers | pERK/383/EV71/HA | dHcap6-m1 3′ RC1-6-plus | dHgp6-m1 3′ RC1-6-plus |
| miRep + miHelpers | pERK/383/EV71/HA 3′ RC1-6-plus | dHcap6-m1 3′ RC1-6-plus | dHgp6-m1 3′ RC1-6-plus |
FIG. 5.
Experiments to determine in vivo miRNA activity on alphavirus RNA replication. Groups of 16 mice were immunized with each VRP at 0 and 3 weeks. The mean number of HA-specific gamma interferon cytokine-secreting T cells/1 × 106 splenocytes 1 week after the prime (A) and 1 week after the boost (B) are represented. Error bar represents 1 standard error. The mean anti-HA specific ELISA titers 1 week after the prime (C) and 1 week after the boost (D) are represented. Error bar represents 1 standard error.
DISCUSSION
Here, we have demonstrated that sequences that represent the targets for cellular miRNAs can be exploited to downregulate alphavirus helper, replicon, and TC-83 genomic RNA replication in vitro and alphavirus replicon RNA in vivo. miRNA targets incorporated into viral genomes have been shown to restrict replication in specific cell types, imparting a level of cellular tropism control to the engineered organism (5, 12-15, 20, 25, 28, 39, 40, 72). The miRNA tropism control has been demonstrated in RCVs (5, 15, 20, 25, 28, 39, 40) as well as in gene expression vectors (12-14, 72). Examples of miRNA-modified gene expression vectors are the most relevant comparison for the alphavirus replicon system described here. Brown et al. demonstrated that engineering targets for mir-142-3p, a hematopoietic specific miRNA, into the 3′ UTR of a lentiviral vector significantly reduced hematopoietic cell expression of the lentiviral transgene both in vitro and in vivo (12-14). An E1 and E3 gene-deleted adenovirus (Ad) vector expressing the herpes simplex virus thymidine kinase (HSVtk) gene has been engineered to contain targets for miR-122a in the 3′ UTR of the expressed HSVtk gene (72). The miR-122a-modified Ad HSVtk vector demonstrated significantly less hepatotoxicity in ganciclovir-treated mice than in mice treated similarly with an Ad vector that was not targeted with the liver-specific miRNA target. These examples demonstrate that the miRNA target-defined, tissue-specific control of transgene expression from replication-defective viral vectors is possible. The examples described above incorporate miRNA targets in cis within the genome of the viral expression vector.
The approach we have adopted places the miRNA targets on RNAs that work in trans to provide helper functions. The expression of alphavirus structural proteins, from helper RNAs in this vector system, is not intended to occur in any cells other than those used to produce VRP. By targeting the helper RNAs with miRNAs that are evolutionarily conserved and ubiquitous, it is anticipated that these RNAs would be degraded in a wide range of cell types. So, in contrast to tissue-specific, highly defined control, our approach is to enlist miRNA control from as diverse a range of cell populations as possible. Examination of the capacity of individual miRNA targets to inhibit replicon expression of CAT protein in different cell lines (11 cell lines in total) demonstrated a range of miRNA control in each. The range of miRNA activity noted in the cell lines tested demonstrates the advantage of introducing multiple different miRNA targets into helper RNAs in order to maximize inhibition in vivo.
We have shown that the miRNA target sequences can be located either 5′ or 3′ of the structural gene on the helper RNAs. Importantly, the miRNA targets were effective only if present on the positive-strand RNA, as miRNA targets found on the negative-strand templates for helper replication had little negative effect on replication. These data are in agreement with those of others with respect to functional miRNA target location on RNAs (5, 15, 20, 26, 28, 39, 40, 79). It is possible that the miRNA targets located on the negative-strand RNA are not accessible to the cytoplasmic RISC because this RNA is associated with replication complexes on cytoplasmic vacuoles (44, 45, 67, 68). Unlike the negative-strand replication intermediate RNA, the positive-strand genomic RNAs are found in the cytoplasm available for translation or encapsidation by nucleocapsid protein. Because of their location, the cytoplasmic positive-strand RNAs are available for miRNA-mediated RISC degradation.
We noted that when the same complement of miRNA targets were present on both helper RNAs, the level of replication inhibition was not as great as when the helpers each carried different miRNA targets. This is most readily noted by inspection of the relative amounts of capsid RNA concatemers detected by Northern blot analysis. In the absence of miRNA inhibitors, capsid RNA concatemers were detected when the same complement of miRNA targets was present on both helper RNAs (Fig. 1C), while concatemers were nearly undetectable when the miRNA complement was not matched on each helper RNA (Fig. 2B). Furthermore, although low, VRP yields when produced in the absence of miRNA inhibitor were more than 10 times higher if the miRNA targets were matching on both helpers than if the miRNA targets were different on each helper RNA (compare VRP titers in Fig. 1D and 2C). It is possible that having the same complement of miRNA targets on both helpers actually produces an miRNA sponge effect (19). That is, the targeted helper RNAs function to compete for the cytoplasmic stores of cellular miRNAs, subtly relieving the inhibition that would be realized if each helper RNA had a different complement of miRNA targets.
The VRP yields produced with miRNA-targeted helper RNAs in the absence of miRNA inhibitor were reduced by 3 to 4 orders of magnitude if compared to VRP yields when miRNA inhibitors were present. Interestingly, although miRNA-targeted helper RNA replication was reduced in the absence of miRNA inhibitor, it was not completely arrested (Fig. 1C). It is likely that the miRNA complement within Vero cells is capable of keeping helper replication below a threshold for efficient VRP production but not at a level that completely inhibits VRP generation. This suggests that even subtle effects on helper RNA replication, due to cellular miRNA control of targeted helpers, can reduce VRP production by orders of magnitude. The individual miRNA targets on the helper RNAs did not contribute equally to the inhibition noted in Vero cells, where three miRNA targets (let-7, miR-155, and miR-17) demonstrated the largest effect. The other three miRNA targets (miR-19, lin-4, and miR-101) demonstrated significant inhibitory potential when tested in other cell types. These observations support our goal of including miRNA targets that are active in a large range of cell types. There is also an advantage to having miRNA targets on the helper RNAs that are not active in the Vero cells used to produce VRP vaccines but that are active in multiple other cell types, because miRNA inhibitors for these targets are not required during VRP production.
The purpose of incorporating miRNA targets on helper RNAs was to control their replication in cells outside those used to generate VRP. Alphaviruses have been shown to undergo nonhomologous recombination (29, 31, 63), and the helper RNAs are a source for these events. RCV may be generated by recombination of helper RNAs with replicon RNA or copackaging of helper RNA (24, 31, 63, 75). The probability of functional recombination between the replicon and helpers is low when using a two-helper RNA system (8, 10, 23, 24, 55, 62, 70, 74). The probability of generating functional recombinant RNAs has been lowered even further by implementing alphavirus helper RNAs that lack the 26S promoter (37). However, there are still theoretical ways for RNA combinations to arise that do not reconstitute a full genome yet have limited capacity to be passaged (58, 74). Replicon RNA recombination with miRNA-targeted helper RNA would likely result in incorporation of the miRNA targets into the 3′ UTR of the newly formed recombinant RNA. This is because the miRNA target sequences are engineered immediately upstream of the conserved 3′ sequences required for replication of the helper RNAs. The presence of the miRNA targets would then lead to the degradation of the recombinant replicon RNA by the miRNA complement found within cells. This is supported, in vitro, by the reduction in CAT reporter gene expression noted from 3′ UTR miRNA-targeted CAT-replicon vectors and by the significant reduction in plaque titer noted from a 3′ UTR miRNA-targeted TC-83 virus in Vero cells. Furthermore, significant reduction in both cellular and humoral responses induced in mice, by VRP containing an miRNA-targeted replicon RNA, provides in vivo support for cellular miRNA-specific control of these targeted RNAs. These data provide evidence that the miRNA complement resident in the mouse cells infected by these miRNA modified replicon VRP can inhibit RNA replication.
This is the first demonstration of the use of miRNA targets on an alphavirus replicon system. Alphavirus replicons are powerful expression vectors that are used widely for research purposes (reviewed in references 3 and 64). Furthermore, alphavirus replicons have been used successfully in vaccine development and have recently shown great promise in human clinical trials (9). Incorporation of miRNA targets into the helper component of vector systems offers another barrier to reconstitution of a functional genome by making any recombinant RNA susceptible to the miRNA regulatory system present in host cells.
Acknowledgments
This work was supported in part by NIH/NIAID grants 1 UC1 AI067183-01 (K.I.K.) and 5 U01 AI056438-03 (J.S.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Published ahead of print on 26 May 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Ambros, V. 2003. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113:673-676. [DOI] [PubMed] [Google Scholar]
- 2.Aravin, A. A., M. Lagos-Quintana, A. Yalcin, M. Zavolan, D. Marks, B. Snyder, T. Gaasterland, J. Meyer, and T. Tuschl. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5:337-350. [DOI] [PubMed] [Google Scholar]
- 3.Atkins, G. J., M. N. Fleeton, and B. J. Sheahan. 2008. Therapeutic and prophylactic applications of alphavirus vectors. Expert Rev. Mol. Med. 10:e33. [DOI] [PubMed] [Google Scholar]
- 4.Aukerman, M. J., and H. Sakai. 2003. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15:2730-2741. Epub 2003 Oct 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes, D., M. Kunitomi, M. Vignuzzi, K. Saksela, and R. Andino. 2008. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe 4:239-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 7.Baskerville, S., and D. P. Bartel. 2005. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11:241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berglund, P., M. Sjoberg, H. Garoff, G. J. Atkins, B. J. Sheahan, and P. Liljestrom. 1993. Semliki Forest virus expression system: production of conditionally infectious recombinant particles. Biotechnology (NY) 11:916-920. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein, D. I., E. R. Reap, K. Katen, A. Watson, K. Smith, P. Norberg, R. A. Olmsted, A. Hoeper, J. Morris, S. Negri, M. F. Maughan, and J. D. Chulay. 2009. Randomized, double-blind, phase 1 trial of an alphavirus replicon vaccine for cytomegalovirus in CMV seronegative adult volunteers. Vaccine 28:484-493. [DOI] [PubMed] [Google Scholar]
- 10.Bredenbeek, P. J., I. Frolov, C. M. Rice, and S. Schlesinger. 1993. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 67:6439-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennecke, J., D. R. Hipfner, A. Stark, R. B. Russell, and S. M. Cohen. 2003. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113:25-36. [DOI] [PubMed] [Google Scholar]
- 12.Brown, B. D., A. Cantore, A. Annoni, L. S. Sergi, A. Lombardo, P. Della Valle, A. D'Angelo, and L. Naldini. 2007. A microRNA-regulated lentiviral vector mediates stable correction of hemophilia B mice. Blood 110:4144-4152. Epub 2007 Aug 28. [DOI] [PubMed] [Google Scholar]
- 13.Brown, B. D., B. Gentner, A. Cantore, S. Colleoni, M. Amendola, A. Zingale, A. Baccarini, G. Lazzari, C. Galli, and L. Naldini. 2007. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 25:1457-1467. [DOI] [PubMed] [Google Scholar]
- 14.Brown, B. D., M. A. Venneri, A. Zingale, L. Sergi, and L. Naldini. 2006. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat. Med. 12:585-591. Epub 2006 Apr 23. [DOI] [PubMed] [Google Scholar]
- 15.Cawood, R., H. H. Chen, F. Carroll, M. Bazan-Peregrino, N. van Rooijen, and L. W. Seymour. 2009. Use of tissue-specific microRNA to control pathology of wild-type adenovirus without attenuation of its ability to kill cancer cells. PLoS Pathog. 5:e1000440. Epub 2009 May 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, C. Z., L. Li, H. F. Lodish, and D. P. Bartel. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83-86. Epub 2003 Dec 4. [DOI] [PubMed] [Google Scholar]
- 17.Chen, X. 2004. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303:2022-2025. Epub 2003 Jul 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dostie, J., Z. Mourelatos, M. Yang, A. Sharma, and G. Dreyfuss. 2003. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 9:180-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebert, M. S., J. R. Neilson, and P. A. Sharp. 2007. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4:721-726. Epub 2007 Aug 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edge, R. E., T. J. Falls, C. W. Brown, B. D. Lichty, H. Atkins, and J. C. Bell. 2008. A let-7 microRNA-sensitive vesicular stomatitis virus demonstrates tumor-specific replication. Mol. Ther. 16:1437-1443. Epub 2008 Jun 17. [DOI] [PubMed] [Google Scholar]
- 21.Emery, J. F., S. K. Floyd, J. Alvarez, Y. Eshed, N. P. Hawker, A. Izhaki, S. F. Baum, and J. L. Bowman. 2003. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13:1768-1774. [DOI] [PubMed] [Google Scholar]
- 22.Farh, K. K., A. Grimson, C. Jan, B. P. Lewis, W. K. Johnston, L. P. Lim, C. B. Burge, and D. P. Bartel. 2005. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 310:1817-1821. Epub 2005 Nov 24. [DOI] [PubMed] [Google Scholar]
- 23.Frolov, I., E. Frolova, and S. Schlesinger. 1997. Sindbis virus replicons and Sindbis virus: assembly of chimeras and of particles deficient in virus RNA. J. Virol. 71:2819-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geigenmüller-Gnirke, U., B. Weiss, R. Wright, and S. Schlesinger. 1991. Complementation between Sindbis viral RNAs produces infectious particles with a bipartite genome. Proc. Natl. Acad. Sci. U. S. A. 88:3253-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gitlin, L., and R. Andino. 2003. Nucleic acid-based immune system: the antiviral potential of mammalian RNA silencing. J. Virol. 77:7159-7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottwein, E., X. Cai, and B. R. Cullen. 2006. A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J. Virol. 80:5321-5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grishok, A., A. E. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D. L. Baillie, A. Fire, G. Ruvkun, and C. C. Mello. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106:23-34. [DOI] [PubMed] [Google Scholar]
- 28.Gu, S., L. Jin, F. Zhang, P. Sarnow, and M. A. Kay. 2009. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat. Struct. Mol. Biol. 16:144-150. Epub 2009 Feb 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hajjou, M., K. R. Hill, S. V. Subramaniam, J. Y. Hu, and R. Raju. 1996. Nonhomologous RNA-RNA recombination events at the 3′ nontranslated region of the Sindbis virus genome: hot spots and utilization of nonviral sequences. J. Virol. 70:5153-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardy, R. W., and C. M. Rice. 2005. Requirements at the 3′ end of the Sindbis virus genome for efficient synthesis of minus-strand RNA. J. Virol. 79:4630-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill, K. R., M. Hajjou, J. Y. Hu, and R. Raju. 1997. RNA-RNA recombination in Sindbis virus: roles of the 3′ conserved motif, poly(A) tail, and nonviral sequences of template RNAs in polymerase recognition and template switching. J. Virol. 71:2693-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hooper, J. W., A. M. Ferro, J. W. Golden, P. Silvera, J. Dudek, K. Alterson, M. Custer, B. Rivers, J. Morris, G. Owens, J. F. Smith, and K. I. Kamrud. 2009. Molecular smallpox vaccine delivered by alphavirus replicons elicits protective immunity in mice and non-human primates. Vaccine 28:494-511. Epub 2009 Oct 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houbaviy, H. B., M. F. Murray, and P. A. Sharp. 2003. Embryonic stem cell-specific microRNAs. Dev. Cell 5:351-358. [DOI] [PubMed] [Google Scholar]
- 34.Hubby, B., T. Talarico, M. Maughan, E. A. Reap, P. Berglund, K. I. Kamrud, L. Copp, W. Lewis, C. Cecil, P. Norberg, J. Wagner, A. Watson, S. Negri, B. K. Burnett, A. Graham, J. F. Smith, and J. D. Chulay. 2007. Development and preclinical evaluation of an alphavirus replicon vaccine for influenza. Vaccine 25:8180-8189. Epub 2007 Oct 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutvágner, G., J. McLachlan, A. E. Pasquinelli, E. Balint, T. Tuschl, and P. D. Zamore. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293:834-838. Epub 2001 Jul 12. [DOI] [PubMed] [Google Scholar]
- 36.Johnston, R. J., and O. Hobert. 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426:845-849. Epub 2003 Dec 14. [DOI] [PubMed] [Google Scholar]
- 37.Kamrud, K. I., K. Alterson, M. Custer, J. Dudek, C. Goodman, G. Owens, and J. F. Smith. 2010. Development and characterization of promoterless helper RNAs for production of alphavirus replicon particles. J. Gen. Virol. [Epub ahead of print.] doi: 10.1099/vir.0.020081-0. [DOI] [PMC free article] [PubMed]
- 38.Kamrud, K. I., M. Custer, J. M. Dudek, G. Owens, K. D. Alterson, J. S. Lee, J. L. Groebner, and J. F. Smith. 2007. Alphavirus replicon approach to promoterless analysis of IRES elements. Virology 360:376-387. Epub 2006 Dec 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelly, E. J., E. M. Hadac, S. Greiner, and S. J. Russell. 2008. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat. Med. 14:1278-1283. Epub 2008 Oct 26. [DOI] [PubMed] [Google Scholar]
- 40.Kelly, E. J., R. Nace, G. N. Barber, and S. J. Russell. 2010. Attenuation of vesicular stomatitis virus encephalitis through microRNA targeting. J. Virol. 84:1550-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, J., A. Krichevsky, Y. Grad, G. D. Hayes, K. S. Kosik, G. M. Church, and G. Ruvkun. 2004. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl. Acad. Sci. U. S. A. 101:360-365. Epub 2003 Dec 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kinney, R. M., G. J. Chang, K. R. Tsuchiya, J. M. Sneider, J. T. Roehrig, T. M. Woodward, and D. W. Trent. 1993. Attenuation of Venezuelan equine encephalitis virus strain TC-83 is encoded by the 5′-noncoding region and the E2 envelope glycoprotein. J. Virol. 67:1269-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kujala, P., A. Ikaheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kaariainen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lagos-Quintana, M., R. Rauhut, W. Lendeckel, and T. Tuschl. 2001. Identification of novel genes coding for small expressed RNAs. Science 294:853-858. [DOI] [PubMed] [Google Scholar]
- 47.Lagos-Quintana, M., R. Rauhut, J. Meyer, A. Borkhardt, and T. Tuschl. 2003. New microRNAs from mouse and human. RNA 9:175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lagos-Quintana, M., R. Rauhut, A. Yalcin, J. Meyer, W. Lendeckel, and T. Tuschl. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12:735-739. [DOI] [PubMed] [Google Scholar]
- 49.Lau, N. C., L. P. Lim, E. G. Weinstein, and D. P. Bartel. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294:858-862. [DOI] [PubMed] [Google Scholar]
- 50.Lee, R. C., and V. Ambros. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294:862-864. [DOI] [PubMed] [Google Scholar]
- 51.Lee, R. C., R. L. Feinbaum, and V. Ambros. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843-854. [DOI] [PubMed] [Google Scholar]
- 52.Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Radmark, S. Kim, and V. N. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415-419. [DOI] [PubMed] [Google Scholar]
- 53.Lee, Y., K. Jeon, J. T. Lee, S. Kim, and V. N. Kim. 2002. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 21:4663-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lewis, B. P., C. B. Burge, and D. P. Bartel. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15-20. [DOI] [PubMed] [Google Scholar]
- 55.Liljestrom, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology (NY) 9:1356-1361. [DOI] [PubMed] [Google Scholar]
- 56.Lim, L. P., M. E. Glasner, S. Yekta, C. B. Burge, and D. P. Bartel. 2003. Vertebrate microRNA genes. Science 299:1540. [DOI] [PubMed] [Google Scholar]
- 57.Lim, L. P., N. C. Lau, E. G. Weinstein, A. Abdelhakim, S. Yekta, M. W. Rhoades, C. B. Burge, and D. P. Bartel. 2003. The microRNAs of Caenorhabditis elegans. Genes Dev. 17:991-1008. Epub 2003 Apr 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu, X., and J. Silver. 2001. Transmission of replication-defective Sindbis helper vectors encoding capsid and envelope proteins. J. Virol. Methods 91:59-65. [DOI] [PubMed] [Google Scholar]
- 59.Mourelatos, Z., J. Dostie, S. Paushkin, A. Sharma, B. Charroux, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16:720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palatnik, J. F., E. Allen, X. Wu, C. Schommer, R. Schwab, J. C. Carrington, and D. Weigel. 2003. Control of leaf morphogenesis by microRNAs. Nature 425:257-263. Epub 2003 Aug 20. [DOI] [PubMed] [Google Scholar]
- 61.Polo, J. M., B. A. Belli, D. A. Driver, I. Frolov, S. Sherrill, M. J. Hariharan, K. Townsend, S. Perri, S. J. Mento, D. J. Jolly, S. M. Chang, S. Schlesinger, and T. W. Dubensky, Jr. 1999. Stable alphavirus packaging cell lines for Sindbis virus and Semliki Forest virus-derived vectors. Proc. Natl. Acad. Sci. U. S. A. 96:4598-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pushko, P., M. Parker, G. V. Ludwig, N. L. Davis, R. E. Johnston, and J. F. Smith. 1997. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology 239:389-401. [DOI] [PubMed] [Google Scholar]
- 63.Raju, R., S. V. Subramaniam, and M. Hajjou. 1995. Genesis of Sindbis virus by in vivo recombination of nonreplicative RNA precursors. J. Virol. 69:7391-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rayner, J. O., S. A. Dryga, and K. I. Kamrud. 2002. Alphavirus vectors and vaccination. Rev. Med. Virol. 12:279-296. [DOI] [PubMed] [Google Scholar]
- 65.Reap, E. A., S. A. Dryga, J. Morris, B. Rivers, P. K. Norberg, R. A. Olmsted, and J. D. Chulay. 2007. Cellular and humoral immune responses to alphavirus replicon vaccines expressing cytomegalovirus pp65, IE1, and gB proteins. Clin. Vaccine Immunol. 14:748-755. Epub 2007 Apr 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodriguez, A., S. Griffiths-Jones, J. L. Ashurst, and A. Bradley. 2004. Identification of mammalian microRNA host genes and transcription units. Genome Res. 14:1902-1910. Epub 2004 Sep 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salonen, A., T. Ahola, and L. Kaariainen. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salonen, A., L. Vasiljeva, A. Merits, J. Magden, E. Jokitalo, and L. Kaariainen. 2003. Properly folded nonstructural polyprotein directs the Semliki Forest virus replication complex to the endosomal compartment. J. Virol. 77:1691-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sempere, L. F., S. Freemantle, I. Pitha-Rowe, E. Moss, E. Dmitrovsky, and V. Ambros. 2004. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5:R13. Epub 2004 Feb 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smerdou, C., and P. Liljestrom. 1999. Two-helper RNA system for production of recombinant Semliki Forest virus particles. J. Virol. 73:1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suzuki, T., F. Sakurai, S. Nakamura, E. Kouyama, K. Kawabata, M. Kondoh, K. Yagi, and H. Mizuguchi. 2008. miR-122a-regulated expression of a suicide gene prevents hepatotoxicity without altering antitumor effects in suicide gene therapy. Mol. Ther. 16:1719-1726. Epub 2008 Jul 29. [DOI] [PubMed] [Google Scholar]
- 73.Tomari, Y., and P. D. Zamore. 2005. MicroRNA biogenesis: Drosha can't cut it without a partner. Curr. Biol. 15:R61-R64. [DOI] [PubMed] [Google Scholar]
- 74.Volkova, E., R. Gorchakov, and I. Frolov. 2006. The efficient packaging of Venezuelan equine encephalitis virus-specific RNAs into viral particles is determined by nsP1-3 synthesis. Virology 344:315-327. Epub 2005 Oct 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weiss, B. G., and S. Schlesinger. 1991. Recombination between Sindbis virus RNAs. J. Virol. 65:4017-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wightman, B., I. Ha, and G. Ruvkun. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75:855-862. [DOI] [PubMed] [Google Scholar]
- 77.Yi, R., Y. Qin, I. G. Macara, and B. R. Cullen. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17:3011-3016. Epub 2003 Dec 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ying, S. Y., and S. L. Lin. 2005. Intronic microRNAs. Biochem. Biophys. Res. Commun. 326:515-520. [DOI] [PubMed] [Google Scholar]
- 79.Ylösmaki, E., T. Hakkarainen, A. Hemminki, T. Visakorpi, R. Andino, and K. Saksela. 2008. Generation of a conditionally replicating adenovirus based on targeted destruction of E1A mRNA by a cell type-specific microRNA. J. Virol. 82:11009-11015. Epub 2008 Sep 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zamore, P. D., T. Tuschl, P. A. Sharp, and D. P. Bartel. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25-33. [DOI] [PubMed] [Google Scholar]