Abstract
Branched-chain amino acids (BCAAs) are synthesized in plants from branched-chain keto acids, but their metabolism is not completely understood. The interface of BCAA metabolism lies with branched-chain aminotransferases (BCAT) that catalyze both the last anabolic step and the first catabolic step. In this study, six BCAT genes from the cultivated tomato (Solanum lycopersicum) were identified and characterized. SlBCAT1, -2, -3, and -4 are expressed in multiple plant tissues, while SlBCAT5 and -6 were undetectable. SlBCAT1 and -2 are located in the mitochondria, SlBCAT3 and -4 are located in chloroplasts, while SlBCAT5 and -6 are located in the cytosol and vacuole, respectively. SlBCAT1, -2, -3, and -4 were able to restore growth of Escherichia coli BCAA auxotrophic cells, but SlBCAT1 and -2 were less effective than SlBCAT3 and -4 in growth restoration. All enzymes were active in the forward (BCAA synthesis) and reverse (branched-chain keto acid synthesis) reactions. SlBCAT3 and -4 exhibited a preference for the forward reaction, while SlBCAT1 and -2 were more active in the reverse reaction. While overexpression of SlBCAT1 or -3 in tomato fruit did not significantly alter amino acid levels, an expression quantitative trait locus on chromosome 3, associated with substantially higher expression of Solanum pennellii BCAT4, did significantly increase BCAA levels. Conversely, antisense-mediated reduction of SlBCAT1 resulted in higher levels of BCAAs. Together, these results support a model in which the mitochondrial SlBCAT1 and -2 function in BCAA catabolism while the chloroplastic SlBCAT3 and -4 function in BCAA synthesis.
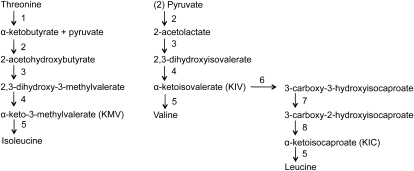
The branched-chain amino acids (BCAAs) Leu, Ile, and Val are primary metabolites synthesized in plants and are essential nutrients in animals. They are synthesized from Thr or pyruvate in plastids (Schulze-Siebert et al., 1984; Hagelstein et al., 1997). Thr feeds into the Ile pathway and pyruvate into the Val pathway, after which the same four enzymatic steps are shared to form these two amino acids. Leu is synthesized by a branch of the Val pathway starting with 3-methyl-2-oxobutanoic acid in four enzymatic steps (Holmberg and Petersen, 1988; Kohlhaw, 2003; Fig. 1).
Figure 1.
Synthetic pathways to BCAAs in plants. 1, Thr deaminase. 2, Acetolactate synthase. 3, Acetolactate isomeroreductase. 4, Dihydroxy acid dehydratase. 5, BCAT. 6, 2-Isopropylmalate synthase. 7, Isopropylmalate isomerase. 8, Isopropylmalate dehydrogenase.
Although synthesis of BCAAs is well characterized in plants, regulation of catabolism is not completely understood. Catabolism is believed to be initiated in mitochondria, where the branched-chain keto acid (BCKA) dehydrogenase complex is located (Taylor et al., 2004). The primary fates of BCAAs in plant cells are peptide elongation, glutamate recycling, Glc- and Suc-linked branched-chain esters, branched-chain fatty acid synthesis, and respiration through the synthesis of tricarboxylic acid cycle intermediates (Kandra et al., 1990; Walters and Steffens, 1990; Kroumova et al., 1994; Daschner et al., 1999; Li et al., 2003; Beck et al., 2004; Taylor et al., 2004; Engqvist et al., 2009). BCAA catabolism likely has other functions in plant metabolism. For example, Gu et al. (2010) showed that a mutation in isovaleryl-CoA dehydrogenase, an enzyme in the BCAA catabolic pathway, influences the metabolism of many unrelated compounds in Arabidopsis (Arabidopsis thaliana) seeds, including 12 amino acids.
Branched-chain aminotransferase (BCAT) enzymes are at the interface of BCAA synthesis and catabolism, reversibly catalyzing the interconversion of BCAAs to BCKAs. Leu is converted to 4-methyl-2-oxopentanoic acid (KIC), Ile to 3-methyl-2-oxopentanoic acid (KMV), and Val to 3-methyl-2-oxobutanoic acid (KIV). BCATs have been studied in only a few plant species. In spinach (Spinacia oleracea), there are two known BCATs, one with a higher affinity toward KIV and the other with a higher affinity toward KIC and KMV, indicating substrate preference (Binder et al., 2007). In Arabidopsis, there are six BCATs. AtBCAT1 localizes to mitochondria and is thought to be active primarily in catabolism. AtBCAT2, -3, and -5 localize to chloroplasts, suggesting roles in BCAA synthesis (Diebold et al., 2002). AtBCAT4 is cytosolic (Schuster et al., 2006), and the location of AtBCAT6 is suggested to be cytosolic because of its lack of a defined target peptide sequence (Diebold et al., 2002). Complementation analysis in BCAT-deficient yeast strains confirmed the functions of AtBCAT1, -2, -3, -5, and -6 but not AtBCAT4 (Diebold et al., 2002). AtBCAT1 is the most likely candidate for initiating BCAA breakdown (Schuster and Binder, 2005), although AtBCAT5 has also been found in mitochondrial fractions (Binder et al., 2007). AtBCAT1 catabolizes all BCAAs in almost all tissue types, and its affinity is greatest in the order Ile > Leu > Val. AtBCAT2 expression is observed only in flowers and is elevated under stress, while AtBCAT6 is expressed in flowers and siliques. Expression of the other AtBCATs is not as tissue specific (Liepman and Olsen, 2004). Two studies in Arabidopsis showed that both the chloroplastic AtBCAT3 and the cytosolic AtBCAT4 also participate in Met chain elongation and the production of aliphatic glucosinolates (Schuster et al., 2006; Knill et al., 2008). In another example of diverse BCAT function, a Nicotiana benthamiana chloroplastic BCAT was implicated in transcriptional regulation of KNOX genes that affect levels of gibberellins. This enzyme was also able to restore the growth of a BCAT-deficient yeast and was expressed highly in young leaves, suggesting that it has a primary role in BCAA synthesis (Gao et al., 2009). Together, these studies show that BCATs have functions beyond amino acid metabolism, making it important to understand the characteristics of each enzyme form.
This study focuses on characterization of the tomato (Solanum lycopersicum) BCAT gene family, their enzymatic properties, and their role in BCAA metabolism. Our results provide insight into the specific functions of BCAT isoforms in tomato. We show that tomato BCATs are diverse in subcellular location, substrate preference, and expression. Finally, we provide evidence that different BCAT alleles influence BCAA content in fruit.
RESULTS
Cloning of SlBCAT cDNAs
In order to better understand the dynamics of BCAA metabolism, we identified six unique tomato sequences potentially encoding BCAT enzymes in the SOL Genomics Network tomato EST database (http://solgenomics.net/index.pl; Mueller et al., 2005). Full-length cDNAs of each gene were cloned and sequenced. The unigene SGN-U569828 (SlBCAT1; 45 members) has the most ESTs of all putative SlBCATs, while the unigene SGN-U569952 (SlBCAT3; 27 members) has the second highest, both far surpassing the numbers of ESTs of the other putative SlBCATs. Phylogenetic analysis of all putative SlBCATs and comparisons with Arabidopsis BCATs (Diebold et al., 2002) revealed that SlBCAT1 is most similar to the AtBCAT2 and AtBCAT1 genes from Arabidopsis. The unigene SGN-U569830 (SlBCAT2; seven members) is most similar to AtBCAT3. SlBCAT3 and the unigene SGN-U569953 (SlBCAT4; seven members) are highly similar to each other and most similar to AtBCAT5. The unigenes SGN-U569831 (SlBCAT5; five members) and SGN-U569829 (SlBCAT6; two members) are most similar to AtBCAT2 and most similar to each other within the putative SlBCATs (Supplemental Figs. S1 and S2).
Expression of SlBCATs
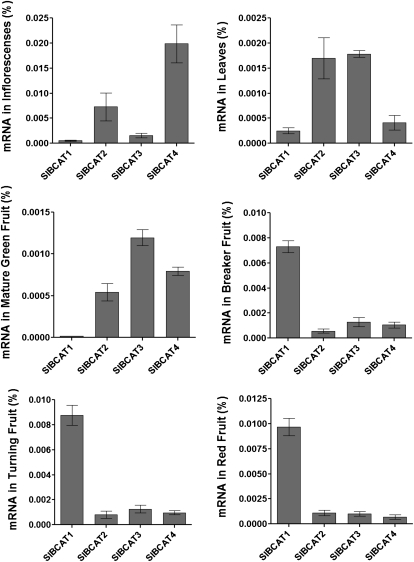
To gain a better understanding of the different roles of each SlBCAT family member, expression analysis was performed by quantitative reverse transcription (qRT)-PCR on all six SlBCAT cDNAs. Tissues tested were young leaves, inflorescences at 1 d post anthesis (dpa), and mature green, breaker, turning, and red ripe fruit stages (Fig. 2). Expression of SlBCAT1 is higher in ripening and red fruit than all other SlBCATs, is very low in leaves and inflorescences, and is undetectable in green fruit. SlBCAT2 is expressed in all tissues at similar levels except for inflorescences, where it is much more highly expressed. SlBCAT3 is expressed in all tissues and is most highly expressed in leaves. Expression of SlBCAT4 is highest in inflorescences but relatively low in all other tissues compared with the other SlBCATs. No SlBCAT5 and SlBCAT6 transcripts were detected in any of the tissues tested.
Figure 2.
Quantification of SlBCAT RNA in different tissue types. Analysis was performed on three biological and three technical replicates for each sample. Values represent percentage of total mRNA per sample ± sd, calculated from a standard curve for each gene. Note differences in y axes. Expression of SlBCAT5 and SlBCAT6 was below the limit of detection.
BCAA Levels in Various Tissues of Tomato
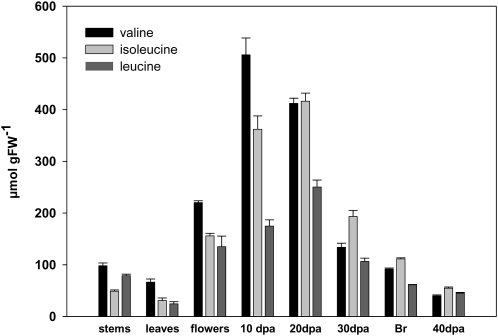
We next determined the levels of Val, Ile, and Leu in leaf tissue, flowers, and various stages of fruit (10 dpa, 20 dpa, 30 dpa, breaker, and 40 dpa). Stem and leaf tissue contained relatively low levels of Val, Leu, and Ile, which ranged between 24.4 and 98.2 μmol g−1 fresh weight, with considerably higher levels of Val than Leu or Ile (Fig. 3). Floral tissues contained approximately twice the content of all three amino acids as leaves. The relatively high expression of SlBCAT2 and SlBCAT4 in inflorescences may be related to the higher concentration of BCAAs in this tissue than in leaves. Young fruits (10 dpa) displayed three to five times the content observed in leaves. Indeed, the levels of Val peaked at this time point, whereas those of Ile and Leu increased until 20 dpa before declining considerably, with all three amino acids being present at contents similar to, or lower than, those observed in leaves by 40 dpa. These data on fruit development are consistent with those reported previously (Carrari et al., 2006).
Figure 3.
HPLC profile of BCAA content in different tissues of tomato. Br, Breaker stage fruit; FW, fresh weight.
Subcellular Localization of SlBCATs
Plant organelles have specific functions that can change during cell and organ development, as is the case with ripening tomato fruit. Therefore, the subcellular locations of metabolic enzymes can be important in predicting function. All six SlBCAT cDNAs were cloned with a C-terminal E-GFP gene fusion, expressed in N. benthamiana leaf protoplasts, and analyzed with confocal microscopy (Fig. 4).
Figure 4.
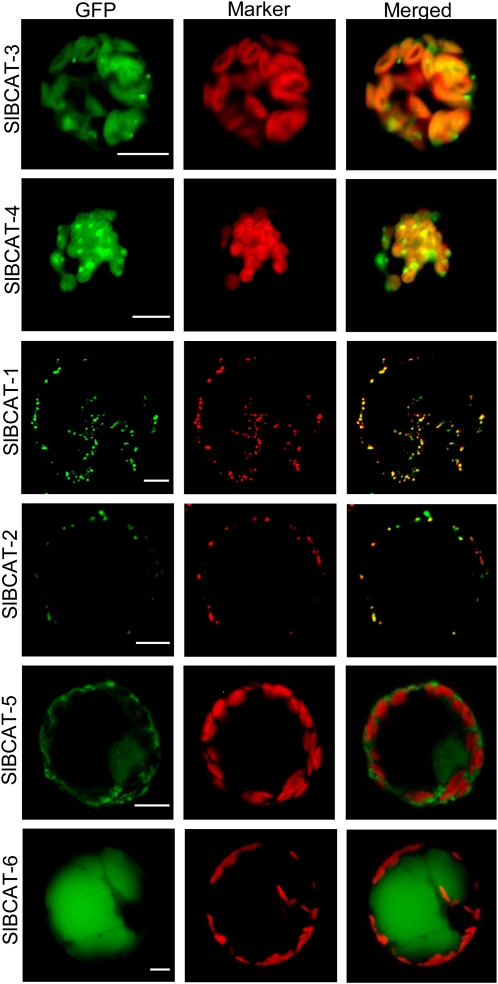
Subcellular localization of SlBCAT proteins. Each cDNA was fused to E-GFP at the C terminus and expressed in N. benthamiana leaf protoplasts. The left column shows GFP fluorescence, the middle column shows marker fluorescence, and the right column shows merging of GFP and marker. Chlorophyll autofluorescence was used to show the presence of chloroplasts for SlBCAT3, -4, -5, and -6. MitoTracker Orange dye was used to show mitochondria for SlBCAT1 and -2. Bars = 10 μm.
SlBCAT3 and SlBCAT4 were localized to plastids, consistent with localization algorithm software and homology with the chloroplast-localized AtBCAT proteins. SlBCAT1 and SlBCAT2 were localized to mitochondria, consistent with localization prediction software outputs. Mitochondrial localization was confirmed with MitoTracker Orange stain. SlBCAT5 appeared to be localized to the cytosol, consistent with the lack of an N-terminal targeting signal. SlBCAT6 appeared to be localized to the vacuole, based on the E-GFP signal filling the majority of the space inside the protoplasts, typical of vacuoles in leaf cells.
The mitochondria and chloroplast locations of SlBCAT1 to -4 suggest that each may have specific functions in BCAA catabolism and anabolism, respectively. Similarly, the cytoplasmic and vacuolar locations of SlBCAT5 and SlBCAT6, respectively, suggest unique metabolic functions for these two enzymes.
Functional Verification by Complementation
In order to demonstrate BCAT function in vivo, a complementation assay was performed in Escherichia coli. The E. coli genome contains one BCAT gene, ilvE. A second gene, tyrB, encoding an aromatic amino acid aminotransferase (EC 2.6.1.57), can partially restore BCAT activity in ΔilvE cells (Gelfand and Steinberg, 1977; Powell and Morrison, 1978; Vartak et al., 1991). The knockout strains for each gene were obtained from the Keio Collection (Baba et al., 2006), and a double knockout strain was constructed. The strain ΔilvE/ΔtyrB completely lacks BCAT activity and does not grow on medium lacking all BCAAs.
SlBCAT1, SlBCAT2, SlBCAT3, and SlBCAT4 were cloned into the E. coli expression vector pBAD24 under control of the Pbad promoter (Guzman et al., 1995) and transformed into the ΔilvE/ΔtyrB strain. Protein gel blots of cell extracts confirmed that BCAT concentrations did not vary greatly (data not shown). The growth rates of all four lines in minimal medium lacking all amino acids were compared with the wild type (Table I). SlBCAT3 and SlBCAT4 were significantly better at restoring growth (52% and 39% of the wild-type rate, respectively) than SlBCAT1 and SlBCAT2 (18% and 11% of the wild-type rate, respectively). The more effective restoration of growth by the chloroplastic SlBCAT3 and SlBCAT4 supports the hypothesis that they are the major BCAA-synthesizing enzymes in tomato. The relatively reduced growth of cells expressing SlBCAT1 and SlBCAT2 suggests that these mitochondrial enzymes are less efficient in the forward direction.
Table I. Growth of E. coli BCAT knockouts complemented with SlBCATs.
Slope is measured as change in OD600 per hour. All strains were grown on minimal medium lacking amino acids.
| Variable | Wild Type | Knockout | SlBCAT3 | SlBCAT4 | SlBCAT1 | SlBCAT2 |
| Slope | 0.159 | 0.0004 | 0.083 | 0.062 | 0.029 | 0.017 |
Enzyme Activity of SlBCATs
In order to determine the kinetic properties of each SlBCAT, proteins were expressed in E. coli cells and purified. Enzyme assays were performed with each recombinant SlBCAT in both the forward (amino acid-forming) and reverse (amino acid-degrading) directions. Table II shows the Km, Vmax, Kcat, and Kcat/Km values with all six branched-chain substrates. All SlBCATs functioned in both forward and reverse directions with all six substrates. SlBCAT3 has the highest efficiency in the forward direction than the reverse, having higher affinity for the BCKAs than BCAAs. SlBCAT4 exhibits a slight preference in the forward direction for KMV and Ile in the reverse direction. Like SlBCAT3, its most closely related form, SlBCAT4 is most efficient in the forward direction, consistent with the role of chloroplastic BCATs in BCAA synthesis in tomato. SlBCAT1 showed relatively low efficiency in the forward direction and much higher efficiency on Leu and Ile in the reverse direction. This preference, together with its mitochondrial location, supports a primarily catabolic function. SlBCAT2, also located in mitochondria, has a much higher affinity for the BCAAs than the corresponding BCKAs, similar to SlBCAT1, suggesting that it also principally functions in BCAA catabolism. SlBCAT5 was most efficient in the forward direction, with highest affinities towards KMV and KIC. SlBCAT6 was relatively highly efficient in both the forward and reverse directions, with highest efficiency when providing KIV as substrate.
Table II. Kinetic parameters of SlBCATs.
Activities of purified recombinant SlBCAT proteins on all BCAA and BCKA substrates. Km is presented as average ± se. Km data were obtained using GraphPad Prism5 software. Other parameters were obtained by calculations listed in “Materials and Methods.” Substrate indicates substrate used in the assay, and Enzyme indicates SlBCAT isoform used in the assay.
| Substrate | Enzyme | Km | Vmax | Kcat | Kcat/Km |
| mm | nkat mg−1 | s−1 | μm−1 s−1 | ||
| KIC | |||||
| 1 | 7.09 ± 0.92 | 0.5 | 11.7 | 0.002 | |
| 2 | 7.90 ± 0.80 | 3.5 | 84.8 | 0.011 | |
| 3 | 0.35 ± 0.06 | 1.1 | 28.1 | 0.080 | |
| 4 | 0.41 ± 0.02 | 1.4 | 35.0 | 0.085 | |
| 5 | 0.34 ± 0.06 | 2.2 | 54.6 | 0.160 | |
| 6 | 0.22 ± 0.02 | 1.2 | 28.7 | 0.130 | |
| KMV | |||||
| 1 | 11.65 ± 1.89 | 0.7 | 16.3 | 0.001 | |
| 2 | 12.40 ± 0.90 | 2.8 | 67.6 | 0.006 | |
| 3 | 0.19 ± 0.02 | 1.0 | 23.6 | 0.120 | |
| 4 | 0.14 ± 0.01 | 0.8 | 18.4 | 0.131 | |
| 5 | 0.19 ± 0.02 | 1.0 | 23.6 | 0.120 | |
| 6 | 0.16 ± 0.02 | 1.0 | 22.7 | 0.140 | |
| KIV | |||||
| 1 | 5.57 ± 0.75 | 1.0 | 23.0 | 0.004 | |
| 2 | 5.50 ± 0.60 | 3.5 | 84.3 | 0.015 | |
| 3 | 0.65 ± 0.07 | 1.9 | 46.5 | 0.070 | |
| 4 | 0.37 ± 0.02 | 2.5 | 60.6 | 0.164 | |
| 5 | 1.20 ± 0.10 | 0.9 | 22.1 | 0.020 | |
| 6 | 0.15 ± 0.01 | 4.6 | 109.8 | 0.730 | |
| Leu | |||||
| 1 | 0.56 ± 0.04 | 1.6 | 39.1 | 0.070 | |
| 2 | 0.20 ± 0.02 | 0.3 | 8.1 | 0.040 | |
| 3 | 2.70 ± 0.30 | 4.8 | 121.0 | 0.045 | |
| 4 | 0.57 ± 0.03 | 0.7 | 17.9 | 0.031 | |
| 5 | 1.80 ± 0.10 | 4.7 | 118.0 | 0.066 | |
| 6 | 0.21 ± 0.02 | 0.6 | 15.7 | 0.075 | |
| Ile | |||||
| 1 | 0.67 ± 0.09 | 1.6 | 40.8 | 0.061 | |
| 2 | 0.31 ± 0.02 | 0.3 | 7.6 | 0.025 | |
| 3 | 4.90 ± 0.90 | 6.9 | 174.0 | 0.036 | |
| 4 | 0.43 ± 0.03 | 0.8 | 20.0 | 0.047 | |
| 5 | 3.20 ± 0.20 | 6.5 | 163.0 | 0.051 | |
| 6 | 0.34 ± 0.03 | 0.8 | 20.0 | 0.059 | |
| Val | |||||
| 1 | 1.00 ± 0.10 | 2.0 | 50.5 | 0.050 | |
| 2 | 1.40 ± 0.50 | 0.2 | 3.8 | 0.003 | |
| 3 | 2.00 ± 0.20 | 4.4 | 111.0 | 0.056 | |
| 4 | 1.40 ± 0.10 | 0.8 | 20.6 | 0.015 | |
| 5 | 2.60 ± 0.20 | 4.9 | 123.0 | 0.047 | |
| 6 | 1.20 ± 0.10 | 1.0 | 24.0 | 0.020 |
Taken together, the data are consistent with the chloroplastic enzymes SlBCAT3 and -4 functioning principally in BCAA anabolism and the mitochondrial enzymes SlBCAT1 and -2 functioning principally in BCAA catabolism. The functions of SlBCAT5 and -6 are not apparent.
SlBCAT Transgenic Analysis
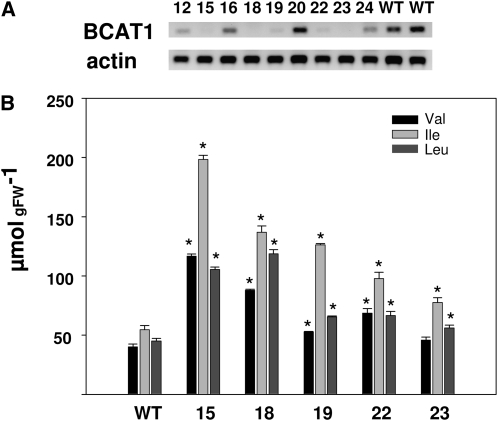
In order to further evaluate the functions of BCAT enzymes in vivo, transgenic plants overexpressing or underexpressing BCAT cDNAs were generated. A 1,138-bp SlBCAT1 fragment was cloned in the antisense orientation into the transformation vector pK2WG7 under the control of the cauliflower mosaic virus promoter. Nine selected lines were amplified in tissue culture, and six plants per line were grown in the greenhouse. No apparent phenotype was observed in any of these lines. Evaluation of the relative levels of SlBCAT1 gene expression in leaves of 6-week-old plants is presented in Figure 5A. Having identified lines with reduced expression of SlBCAT1, we determined the levels of amino acids in red ripe pericarp fruit. The levels of Leu, Ile, and to a lesser extent Val increased in the transgenic lines (Fig. 5B). This observation is consistent with a catabolic function for SlBCAT1 and loss of function resulting in higher amino acid levels.
Figure 5.
Characterization of SlBCAT1 antisense lines. A, RT-PCR analyses of SlBCAT1 expression in transgenic plants. B, Content of BCAAs in red tomato fruits (40 dpa). Data represent means ± se from three independent biological replicates with two technical replicates for each. Asterisks show statistically significant changes according to Student's t test (P < 0.05). FW, Fresh weight; WT, wild type.
In order to determine if an increase in either a single synthetic or catabolic SlBCAT could alter fruit metabolism, constitutive overexpression (OE) constructs of two cDNAs, SlBCAT1 and SlBCAT3, were transformed into tomato plants. These two cDNAs were chosen due to their high expression in ripening fruits and because they represent a primarily catabolic and primarily anabolic enzyme, respectively. Ripe fruits from field-grown T1 plants were analyzed for amino acid content in comparison with M82 controls. Three independent lines from each construct were chosen for this analysis, and RNA from each line was analyzed by qRT-PCR. Although there were significant increases in expression of the transgenes (Supplemental Fig. S3), there were no consistent differences from the control in amino acid content in any of the SlBCAT1-OE lines (Supplemental Table S1), nor was there any visible phenotype. The only significant change was an increase in Ile in the SlBCAT3-OE lines, which is consistent with SlBCAT3 having the highest enzyme activity on KMV, the precursor to Ile. Increased expression of an individual SlBCAT does not necessarily change amino acid metabolism in tomato fruit, likely due to a tight enzymatic regulation of the BCAA pathways and/or redundancy of individual SlBCATs in fruit.
Genetic Analysis of BCATs in Tomato
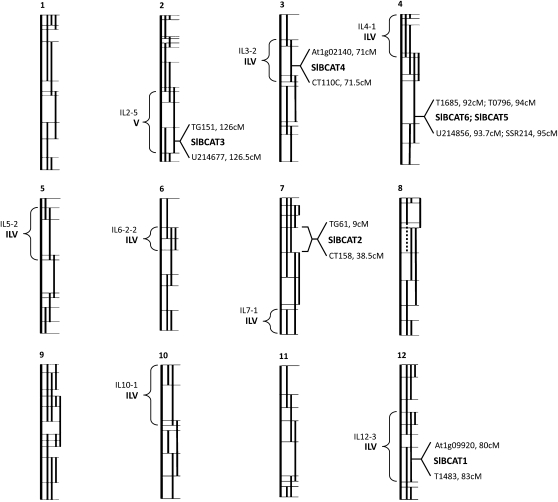
We previously identified quantitative trait loci (QTLs) for the BCAAs in tomato fruit pericarp of introgression lines (ILs) resulting from the interspecific cross of S. lycopersicum and its wild relative Solanum pennellii (Schauer et al., 2006, 2008). There are 13 Ile, 17 Leu, and 18 Val QTLs, with seven of these coordinately altering all three BCAAs (Fig. 6). Given that BCAT enzymes participate both in the biosynthesis and degradation of all three BCAAs, we determined whether any of the SlBCAT genes colocalized with the seven QTLs simultaneously affecting all three amino acids. A draft tomato genome is available (http://solgenomics.net/). We were able to precisely map five of the SlBCAT genes by their proximities to previously mapped markers within scaffolds (Fig. 6). The scaffold containing SlBCAT2 did not contain any mapped marker. SlBCAT2 was mapped to an IL bin on chromosome 7 using a polymorphic marker. Two of the major coordinate QTLs for BCAA content (i.e. those in which the change in amino acid content was consistent for Ile, Leu, and Val) were found to colocalize with SlBCAT genes (IL3-2:SlBCAT4 and IL12-3:SlBCAT1).
Figure 6.
Map positions of SlBCATs and BCAA QTLs. cM, Centimorgan.
Evaluation of Amino Acid and Transcript Levels of Sublines of IL3-2 Containing BCAT4
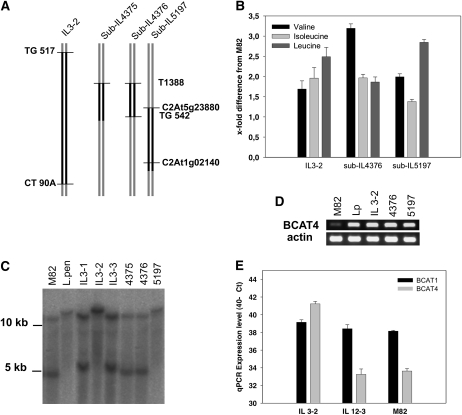
To gain better genetic resolution of the BCAA QTLs, we searched for recombinant lines containing smaller S. pennellii introgressions in the region of interest. Only for the QTL identified in IL3-2 was such material available, with three as yet undescribed sublines spanning this region (Fig. 7A). Only two of these lines were marker delimited, subIL5197 and subIL4376; therefore, we chose these for further characterization. While subIL5197 expressed the S. pennellii BCAT4 allele, subIL4376 did not. IL3-2, subIL4376-1, and subIL5197-5 carried the coordinate QTL for Ile, Leu, and Val, displaying more than 160% of the levels of these amino acids than the S. lycopersicum control (Fig. 7B). Semiquantitative comparisons of the expression levels of BCAT4 in IL3-2 and the two sublines to M82 revealed that all three displayed enhanced expression of BCAT4 (Fig. 7D). Further quantitative analysis by qRT-PCR revealed that the expression level of BCAT4 is 120.1-fold (±7.4 se) higher in IL3-2 than M82, as determined by the cycle thresholds of the samples normalized to the ubiquitin endogenous control gene (ΔCt; Fig. 7E). Fold difference was derived from the ΔCt values of BCAT4 in IL3-2 and M82 as described in “Materials and Methods.” Sequencing of the S. lycopersicum and S. pennellii BCAT4 revealed no polymorphism in translated regions of the gene (data not shown). This finding is highly suggestive of an expression QTL mediated by a polymorphism in the promoter or another regulatory element that directly controls BCAT4 expression. We were able to delimit this QTL to a 1-centimorgan region (distance between markers C2At5g23880 and TG542) immediately adjacent to the BCAT4 gene.
Figure 7.
Mapping of the gene encoding SlBCAT4 and characterization of respective ILs. A, Schematic presentation of the introgressed region. B, Analysis of BCAA content in fruits by GC-MS. Data represent means ± se from six independent biological replicates. C, DNA-blot analysis of ILs. Ten micrograms of genomic DNA was digested with BfrI restriction enzyme, blotted, and hybridized with radiolabeled gene-specific probe. D, Analysis of the level of expression by RT-PCR. E, qRT-PCR analyses of BCAT1 and BCAT4 transcripts in tomato fruits of S. lycopersicum cv M82 and different ILs. Data represent means ± se from three independent biological replicates with two technical replicates for each point.
Sequence Comparison, Expression Analysis, and Kinetic Characterization of SlBCAT1 in IL12-3 and S. lycopersicum
Comparative analysis of BCAT1 expression in S. lycopersicum and IL12-3 revealed that, unlike the IL3-2 QTL, expression of this gene was invariant (Fig. 7E). Therefore, we evaluated the nucleotide sequences of the translated regions of BCAT1 amplified from M82 and IL12-3-1 (Supplemental Fig. S3). This analysis revealed the presence of six nucleotide polymorphisms between the S. lycopersicum and S. pennellii alleles resulting in three amino acid changes; the S. lycopersicum allele harbors Ser-220, Asp-236, and Arg-237, whereas the S. pennellii allele harbors Ala-220, Glu-236, and Lys-237. Interestingly, all three residues lie on the fourth and fifth α-helices of the protein, very close to the conserved active site Lys residue (Tremblay and Blanchard, 2009).
In order to determine if differences in these three residues confer altered enzymatic function, both forms were expressed and purified from E. coli cells and assayed. Since SlBCAT1 is thought to function primarily in amino acid catabolism in fruit, assays were performed using BCAA substrates (Table III). The S. pennellii Km values were significantly (P < 0.05) higher than the S. lycopersicum enzyme. Consistent with the Km values, the S. pennellii catalytic efficiencies were lower with all three substrates. These small but significantly different values suggest that the S. lycopersicum enzyme is more catabolically efficient and could explain the higher BCAA levels in IL12-3-1.
Table III. Kinetic parameters of SlBCAT1 enzymes.
Activities of purified recombinant SlBCAT1 and SpBCAT1 proteins on BCAA substrates. Km is presented as average ± se. Kinetic data were obtained using GraphPad Prism5 software. Enzymes contained an N-terminal glutathione S-transferase tag. Km values of S. pennellii are significantly different from values of M82 for the same substrate. Asterisks show significance as determined by Student's t test (P < 0.05).
| Allele | Substrate | Km | Vmax | Kcat | Kcat/Km |
| mm | nkat mg−1 | s−1 | μm−1 s−1 | ||
| M82 | Leu | 0.56 ± 0.05 | 2.7 | 63.1 | 0.113 |
| Ile | 0.60 ± 0.09 | 2.8 | 66.4 | 0.111 | |
| Val | 0.92 ± 0.13 | 1.9 | 45.8 | 0.050 | |
| S. pennellii | Leu | 0.84 ± 0.07* | 3.4 | 80.2 | 0.095 |
| Ile | 0.84 ± 0.09* | 2.9 | 69.9 | 0.083 | |
| Val | 1.32 ± 0.25* | 2.3 | 55.0 | 0.042 |
DISCUSSION
Here, we describe six tomato BCAT cDNAs, distinguished by their unique patterns of expression and subcellular locations. SlBCAT1 and SlBCAT2 localize to mitochondria. Given that mitochondria are the primary location of BCAA catabolism in plant cells and that the activities of these two enzymes are primarily catabolic, only partially restoring growth in E. coli auxotrophs, these are likely to be the primary BCAA catabolic enzymes in tomato. Since SlBCAT1 has the highest expression in ripening fruit, it likely is the primary enzyme for recycling of BCAAs generated by protein degradation. This conclusion is further supported by the elevation of BCAAs in SlBCAT1 antisense transgenic tomato. Since SlBCAT2 is expressed in all green tissues examined, it is likely a more general contributor to BCAA catabolism. We expect a catabolic mitochondrial housekeeping gene to exist, since BCAAs are used as precursors to the tricarboxylic acid cycle intermediates succinyl-CoA and acetyl-CoA as well as direct electron donors of the mitochondrial electron transport chain (Ishizaki et al., 2005). This enzyme may also function in the crucial process of regulating steady-state levels of BCAAs in cells.
SlBCAT3 and SlBCAT4 are localized to chloroplasts and were able to effectively restore the growth of E. coli BCAA auxotrophs, consistent with a primary role in BCAA synthesis. The kinetic properties of these two enzymes, especially of SlBCAT3, further support this conclusion. It is possible, however, that under certain conditions these two enzymes may be active in BCAA catabolism, evident from their ability to use BCAAs as substrates. SlBCAT3 likely functions as the major enzyme for BCAA synthesis, since it is expressed nearly equally in all tissues, including all stages of fruit development. SlBCAT4 appears to be more specialized in function, given that its expression is by far highest in flowers and is relatively low in other tissues compared with the other SlBCATs. This expression pattern may reflect high demand for amino acid synthesis in reproductive tissue. Alternatively, it may have a specific role beyond primary metabolism, as has been suggested for Arabidopsis AtBCAT3 and AtBCAT4 (Schuster et al., 2006; Knill et al., 2008). The NbBCAT, recently found to have a role in hormonal regulation, is also expressed highly in flowers and localized to chloroplasts (Gao et al., 2009).
SlBCAT5 and SlBCAT6 expression was not detected in any of the tissues analyzed. These two unigenes are represented by only five and two ESTs, respectively. Both ESTs for SlBCAT6 were isolated from callus tissue. These genes may be expressed under specific growth, hormonal, or environmental conditions or may be linked to specific secondary metabolic pathways. Their distinct localization patterns and demonstrated functionality on branched-chain substrates makes these two genes interesting candidates for future research. The vacuolar localization of SlBCAT6 suggests that it may function in the recycling of proteolytically derived BCAAs. This function is further supported by its relatively high affinity for all branched-chain substrates and by the fact that BCAAs accumulate in this organelle (Farre et al., 2001). Of particular interest is this enzyme's efficiency using KIV, which is extremely high compared with all other SlBCATs and all other substrates. It is unclear why this enzyme is more active in Val synthesis.
Overexpression of the most highly expressed anabolic (SlBCAT3) or catabolic (SlBCAT1) cDNAs in fruits did not significantly alter the BCAA pool. As in bacteria (Massey et al., 1976), the enzymes in the pathway of BCAA synthesis are tightly regulated by substrate feedback. Increased BCAA catabolism may stimulate coincident increased BCAA synthesis. The opposite effect may occur with overexpression of SlBCAT3, a primarily anabolic enzyme, where increased BCAA synthesis might have an effect on feedback to earlier steps in the BCAA synthesis pathway, such as Thr deaminase or acetolactate synthase, inhibiting BCAA synthesis.
BCAT4 transcript is substantially increased in IL3-2 and the sublines relative to S. lycopersicum. These lines also have elevated levels of Leu, Ile, and Val (Fig. 7B). However, sequencing of the coding region of the gene amplified from IL3-2 and from S. lycopersicum revealed no polymorphisms. Furthermore, subline 4367 carries the S. lycopersicum BCAT4 allele. One possible explanation for the BCAT4 expression QTL is that this very large increase in expression is caused by a difference in the noncoding region of the gene. Although subline 4376 contains the S. lycopersicum allele, the point of recombination is in the vicinity of the structural gene. Analysis of the genome scaffolds in the region indicates that this is the only structural gene associated with BCAA metabolism mapping to this locus. We cannot, however, exclude the possibility that the region between markers C2At5g23880 and TG542 encodes a trans-acting factor that is responsible for elevated BCAT4 expression. These data, alongside the localization of the protein in the chloroplast, are consistent with BCAT4 operating predominantly in the synthetic direction in vivo. By contrast, BCAT1, which encodes a mitochondria-localized protein, is equivalently expressed in IL12-3 and S. lycopersicum. However, sequencing of the IL12-3 and S. lycopersicum BCAT1 coding regions revealed three variant amino acid residues. Intriguingly, when aligning these two protein sequences with that from Mycobacterium tuberculosis, for which a high-resolution crystal structure exists (Tremblay and Blanchard, 2009), the variant residues are very close to the active site of the protein. The S. pennellii allele exhibits higher Km values for Leu, Ile, and Val that may explain the increased levels of BCAAs in fruits. However, results from the SlBCAT1 antisense transgenic lines do indicate that SlBCAT1 activity is a major determinant of BCAA levels in mature tomato fruits.
CONCLUSION
The results of these experiments give important new information about BCAA metabolism and the BCAT gene family. They support anabolic functions for SlBCAT3/SlBCAT4, catabolic functions for SlBCAT1/SlBCAT2, and potentially novel functions for SlBCAT5/SlBCAT6. They also suggest possible roles of SlBCAT1 and SlBCAT4 in the regulation of BCAAs in tomato.
MATERIALS AND METHODS
All chemicals and reagents used were purchased from Sigma-Aldrich unless otherwise noted. Oligonucleotides were purchased from Integrated DNA Technologies and are listed in Supplemental Table S2.
Cloning of SlBCAT cDNAs
ESTs for each SlBCAT were found by searching the SOL Genomics Network tomato (Solanum lycopersicum) EST database (http://solgenomics.net/index.pl) for sequences that share homology with known plant BCATs. The full-length clones of each SlBCAT were obtained using RACE PCR with the SMART RACE cDNA synthesis kit (Clontech Laboratories). PCR with Advantage HF2 polymerase (Clontech Laboratories) was used to amplify the full-length open reading frames from cDNA. These were cloned into pGEMT (Promega) and sequenced. Alignments of protein sequences were produced using ClustalW (Larkin et al., 2007).
Constructs
Open reading frames for each construct were amplified from cDNA by PCR and cloned into pGEMT-easy vector (Promega). SlBCAT expression constructs were made by cloning into the NheI and SalI restriction sites of pET-28b (Invitrogen), which contains an N-terminal 6xHis tag. Protein expressionconstructs for assaying alleles of SlBCAT1 and Solanum pennellii BCAT1 were constructed by subcloning into pENTR/SD/D-TOPO (Invitrogen) and then into pDEST15 (Invitrogen), which contains an N-terminal glutathione S-transferase tag. Primers were designed to omit signal peptides, as predicted by SignalP software (Emanuelsson et al., 2007), and are listed in Supplemental Table S2.
Bacterial complementation constructs were made by excising the inserts from pET28b and inserting them into pBAD24 (Guzman et al., 1995) using SalI and NotI restriction sites, resulting in a pBAD24 construct containing a 6xHis tag.
For plant overexpression constructs, SlBCAT1 and SlBCAT3 cDNAs were cloned in the sense orientation into pENTR/D-TOPO and cloned using Gateway LR Recombinase (Invitrogen) into a vector containing the figwort mosaic virus promoter (Richins et al., 1987), a kanamycin resistance gene, and an Agrobacterium tumefaciens nopaline synthase 3′ terminator. The overexpression constructs were introduced into S. lycopersicum cv M82 by Agrobacterium-mediated transformation as described (McCormick et al., 1986). Primary transgenic tomato plants were grown in greenhouses under standard conditions and supplemented with slow-release fertilizer. Subsequent generations of transgenic and control tomato plants were grown at the North Florida Research and Education Center. The SlBCAT1 antisense plants as well as all IL and subIL materials were grown in a greenhouse at the Max Planck Institute of Molecular Plant Physiology under long-day conditions (16-/8-h day/night cycle), temperature of 22°C, and 50% humidity.
For the antisense SlBCAT1 construct, a 1,138-bp fragment of SlBCAT1 was amplified from tomato. It was cloned first in the pENTR/SD/D-TOPO vector (Invitrogen) and then subcloned into the binary Gateway vector pK2WG7 (Karimi et al., 2002) in the antisense orientation under the control of the 35S promoter, using the Gateway Technology system (Invitrogen). Transgenic cv Moneymaker plants were selected on kanamycin-containing medium (50 mg mL−1).
C-terminal GFP constructs were made by cloning full-length SlBCAT open reading frames into pDONR221 (Invitrogen) and then cloning into the pK7WGF2 Gateway binary destination vector (Karimi et al., 2002). GFP constructs were transformed into Agrobacterium pMP90RK (Koncz and Schell, 1986).
Protein Production and Purification
Protein expression constructs were transformed into BL21(DE3) competent cells (Invitrogen). Expression of protein was induced with isopropylthio-β-galactoside according to the BL21(DE3) manufacturer's instructions. Cells were pelleted and lysed by sonication in phosphate-buffered saline and then treated with Protease Inhibitor Cocktail (Sigma-Aldrich) according to the manufacturer's directions. Proteins were purified from cell lysates using gravity flow with TALON Affinity Purification Resin or Glutathione-Superflow Resin (Clontech) according to the manufacturer's instructions. Protein elutions were quantified using the Bradford method (Bradford, 1976). Protein purity was determined to be at least 95% by analysis with SDS-PAGE and staining with Coomassie Brilliant Blue Safestain (Invitrogen).
Enzyme Assays
Forward and reverse assays were performed as described previously (Prohl et al., 2000). One microgram of purified SlBCAT was used in each reaction, which was carried out at 25°C for 5 min. Samples were read in a SmartSpec (Bio-Rad) spectrophotometer. Forward assays were recorded at 340 nm in quartz cuvettes and reverse assays at 440 nm in plastic cuvettes. Samples with heat-denatured enzymes were used to obtain blank readings. For both assays, reactions lacking substrate or enzyme or containing boiled enzyme were used as controls. Kinetic data were calculated using GraphPad Prism5 software (Graphpad Software).
Microscopy and Subcellular Localization
Agrobacterium cultures transformed with SlBCAT-GFP constructs were grown overnight in Luria broth, then pelleted and resuspended in infiltration solution (10 mm MgCl2 and 10 mm MES) to an optical density at 600 nm (OD600) of 0.4. Agrobacterium solutions were injected into the underside of young fully expanded Nicotiana benthamiana leaves with a 2-mL syringe. Plants were grown for 4 d after infection. Protoplasts were released from N. benthamiana leaves using the protocol of Yoo et al. (2007). Protoplasts transformed with SlBCAT1-GFP and SlBCAT2-GFP constructs were stained with 500 nm MitoTracker Orange, as directed by the manufacturer (Invitrogen). Cells were visualized using a Zeiss Pascal LSM5 Confocal Laser Scanning Microscope (Carl Zeiss MicroImaging) with a 40× objective. GFP was visualized with an argon laser exciting at 488 nm and detected between 500 and 530 nm. A helium-neon laser, exciting at 543 nm, was used to visualize chlorophyll autofluorescence, detected at 633, and MitoTracker Orange, detected at 576 nm.
Gas Chromatography-Mass Spectrometry Analyses of Nonvolatile Plant Metabolites
Metabolite extraction, derivatization, gas chromatography-mass spectrometry (GC-MS) analysis, and data processing were performed as described previously (Lisec et al., 2006; Schauer et al., 2006), with the exception that, for low-abundance metabolites, a substantially higher extract concentration was injected onto the GC-MS system. The absolute concentration of metabolites was determined by comparison with standard concentration curves as defined by Schauer et al. (2005b). Metabolites were identified in comparison with database entries of authentic standards (Kopka et al., 2005; Schauer et al., 2005a). In addition, the metabolites KIC, KMV, and KIV, for which no mass spectral tag information was available, were identified by analysis of identically derivatized authentic standards.
Expression Analysis
RNA was isolated from tomato fruit tissue using the RNeasy Plant RNA Extraction Kit (Qiagen) followed by DNase treatment to rid samples of contaminating DNA. RNA was quantified using a Nanodrop spectrophotometer (Thermo Fisher Scientific). Omniscript reverse transcriptase (Qiagen) was used with 1 μg of each RNA sample to synthesize oligo(dT)-primed cDNA. 1× SYBR Green Master Mix (Applied Biosystems) was used with 1 μL of each cDNA sample and 500 nm gene-specific primers for qRT-PCR on the Applied Biosystems StepOnePlus real-time PCR machine. For RT-PCR of SlBCATs in different plant tissues and for overexpression transgenics, five-point standard curves were made for each SlBCAT to calculate the ratio of transcript. For comparison of BCATs in ILs and antisense transgenics, ΔCt values were determined by comparison with the ubiquitin control gene. For BCAT4 analysis in IL3-2 and M82, fold differences were derived by comparison with the ubiquitin calibrator and calculated using the ΔΔCt quantification algorithm (Livak and Schmittgen, 2001). Amplification conditions were as follows: 2 min at 50°C; 10 min at 95°C; 40 cycles each of 15 s at 95°C followed by 1 min at 60°C; 15 s at 95°C; 20 s at 60°C; 15 s at 95°C. Primer specificity was confirmed with melting curve analysis on the StepOnePlus real-time PCR machine.
Escherichia coli Complementation
E. coli strain BW25113 with knockouts in ilvE (JW5606-1) or tyrB (JW4014-2) were purchased from the Keio Collection (Baba et al., 2006). Double knockouts were constructed as described previously (Cherepanov and Wackernagel, 1995; Baba et al., 2006) and validated by PCR with primers flanking the sites of the two genes (Supplemental Table S2). Constructs of SlBCATs in pBAD24 were transformed into ΔilvE/ΔtyrB cells. Cells were first grown in liquid M9 minimal medium supplemented with 0.2% casamino acids and 1 mm thiamine hydrochloride and then were washed and transferred to M9 minimal medium lacking amino acids (Sambrook et al., 1989) and supplemented with 0.2% (w/v) Ara for induction, 0.4% (w/v) glycerol for carbon source, and 50 μg mL−1 carbenicillin. Cell culture density in minimal medium lacking amino acids was measured by OD600 every hour with shaking at 37°C. The experiment was repeated three times with similar results. Protein expression levels of SlBCATs were confirmed by protein gel blotting of cells normalized by OD600 probed with mouse anti-His antibody (Invitrogen).
Amino Acid Analysis of Tomato Fruit by GC-MS
Amino acid levels in M82 control and SlBCAT1-OE and SlBCAT3-OE transgenic ripe tomato fruit were determined by derivatization with methyl chloroformate and quantification by GC-MS according to the method of Chen et al. (W.P. Chen, X.Y. Yang, W. Gray, and J.D. Cohen, unpublished data), using an Agilent 6890N gas chromatograph and 5975 mass spectrometer. Three technical replicates of each of three biological replicates were analyzed for each transgenic line. Fruits were grown in fields at the North Florida Research and Education Center.
Extraction and Analysis of BCAAs by HPLC
Amino acids were measured using HPLC after labeling with o-phthalaldehyde according to the method of Kreft et al. (2003). Detection and quantification were based on the conversion of the primary amino group with o-phthalic acid dialdehyde to a fluorescing derivative. Peak areas were integrated using Chromeleon software 6.8 (Dionex) and subjected to quantification by comparison with calibration curves generated following serial runs of a dilution series of mixed standards.
IL Characterization
The map positions of SlBCAT2 as well as SlBCAT4 allele scoring were determined by genomic DNA-blot analysis. Genomic DNA was isolated from leaves using the cetyl-trimethyl-ammonium bromide method (Doyle and Doyle, 1990). Ten micrograms of DNA was digested with restriction endonuclease, separated on 0.7% Tris-acetate EDTA agarose gels, and alkali blotted onto Porablot NY Amp nylon membranes (Macherey-Nagel). Hybridization was performed with a 32P-labeled cDNA clone. Genes were mapped by RFLP after screening for polymorphism between the parental lines (M82 and S. pennellii) with more than 20 restriction enzymes.
Statistical Analysis
Statistical analyses were performed by algorithms in GraphPad Prism5 software (enzyme kinetics) or Microsoft Excel. Significant differences were determined by Student's t test (P < 0.05).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Evolutionary relationships of mature SlBCAT proteins.
Supplemental Figure S2. Evolutionary relationships of mature SlBCAT and AtBCAT proteins.
Supplemental Figure S3. Transcript analysis in fruits of transgenic plants overexpressing SlBCATs.
Supplemental Figure S4. Comparison of nucleotide sequences of BCAT1 from S. lycopersicum cv M82 and S. pennellii.
Supplemental Table S1. Levels of free amino acids in red ripe fruit of M82 and SlBCAT overexpression lines.
Supplemental Table S2. Primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank Valerie De Crecy-Lagard and Basma El Yacoubi for providing us with the pBAD24 plasmid and for help with making E. coli knockouts, members of the University of Florida Electron Microscopy and Bioimaging facility, Romain Fouquet for assistance with subcellular localization, Peter Bliss for assistance with field and greenhouse work, Bala Rathinasabapathi for help with enzyme assays, and Jim Giovannoni for providing EST clones of SlBCATs.
References
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio Collection. Mol Syst Biol 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck HC, Hansen AM, Lauritsen FR. (2004) Catabolism of leucine to branched-chain fatty acids in Staphylococcus xylosus. J Appl Microbiol 96: 1185–1193 [DOI] [PubMed] [Google Scholar]
- Binder S, Knill T, Schuster J. (2007) Branched-chain amino acid metabolism in higher plants. Physiol Plant 129: 68–78 [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, Urbanczyk-Wochniak E, Zanor MI, Nunes-Nesi A, Nikiforova V, Centeno DC, Ratzka A, Pauly M, et al. (2006) Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142: 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158: 9–14 [DOI] [PubMed] [Google Scholar]
- Daschner K, Thalheim C, Guha C, Brennicke A, Binder S. (1999) In plants a putative isovaleryl-CoA-dehydrogenase is located in mitochondria. Plant Mol Biol 39: 1275–1282 [DOI] [PubMed] [Google Scholar]
- Diebold R, Schuster J, Daschner K, Binder S. (2002) The branched-chain amino acid transaminase gene family in Arabidopsis encodes plastid and mitochondrial proteins. Plant Physiol 129: 540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15 [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Engqvist M, Drincovich MF, Flugge UI, Maurino VG. (2009) Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities participate in the last reactions of the methylglyoxal and beta-oxidation pathways. J Biol Chem 284: 25026–25037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre EM, Tiessen A, Roessner U, Geigenberger P, Trethewey RN, Willmitzer L. (2001) Analysis of the compartmentation of glycolytic intermediates, nucleotides, sugars, organic acids, amino acids and sugar alcohols in potato tubers using a non-aqueous fractionation method. Plant Physiol 127: 685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Wang C, Wei C, Li Y. (2009) A branched-chain aminotransferase may regulate hormone levels by affecting KNOX genes in plants. Planta 230: 611–623 [DOI] [PubMed] [Google Scholar]
- Gelfand DH, Steinberg RA. (1977) Escherichia coli mutants deficient in the aspartate and aromatic amino acid aminotransferases. J Bacteriol 130: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL. (2010) Broad connections in the Arabidopsis seed metabolic network revealed by metabolite profiling of an amino acid catabolism mutant. Plant J 61: 579–590 [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177: 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagelstein P, Sieve B, Klein M, Jans H, Schultz G. (1997) Leucine synthesis in chloroplasts: leucine/isoleucine aminotransferase and valine aminotransferase are different enzymes in spinach chloroplasts. J Plant Physiol 150: 23–30 [Google Scholar]
- Holmberg S, Petersen JG. (1988) Regulation of isoleucine-valine biosynthesis in Saccharomyces cerevisiae. Curr Genet 13: 207–217 [DOI] [PubMed] [Google Scholar]
- Ishizaki K, Larson TR, Schauer N, Fernie AR, Graham IA, Leaver CJ. (2005) The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17: 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandra G, Severson R, Wagner GJ. (1990) Modified branched-chain amino acid pathways give rise to acyl acids of sucrose esters exuded from tobacco leaf trichomes. Eur J Biochem 188: 385–391 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Knill T, Schuster J, Reichelt M, Gershenzon J, Binder S. (2008) Arabidopsis branched-chain aminotransferase 3 functions in both amino acid and glucosinolate biosynthesis. Plant Physiol 146: 1028–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhaw GB. (2003) Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol Mol Biol Rev 67: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. (1986) The promoter of Tl-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmuller E, Dormann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) Gmd@csb.Db: the Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kreft O, Hoefgen R, Hesse H. (2003) Functional analysis of cystathionine γ-synthase in genetically engineered potato plants. Plant Physiol 131: 1843–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroumova AB, Xie Z, Wagner GJ. (1994) A pathway for the biosynthesis of straight and branched, odd- and even-length, medium-chain fatty acids in plants. Proc Natl Acad Sci USA 91: 11437–11441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li L, Thipyapong P, Breeden DC, Steffens JC. (2003) Overexpression of a bacterial branched-chain alpha-keto acid dehydrogenase complex in Arabidopsis results in accumulation of branched-chain acyl-CoAs and alteration of free amino acid composition in seeds. Plant Sci 165: 1213–1219 [Google Scholar]
- Liepman AH, Olsen LI. (2004) Genomic analysis of aminotransferases in Arabidopsis thaliana. Crit Rev Plant Sci 23: 73–89 [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Massey LK, Sokatch JR, Conrad RS. (1976) Branched-chain amino-acid catabolism in bacteria. Bacteriol Rev 40: 42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S, Niedermeyer J, Fry J, Barnason A, Horsch R, Fraley R. (1986) Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep 5: 81–84 [DOI] [PubMed] [Google Scholar]
- Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin CW, Wright MH, Ahrens R, Wang Y, et al. (2005) The SOL Genomics Network: a comparative resource for Solanaceae biology and beyond. Plant Physiol 138: 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JT, Morrison JF. (1978) Role of the Escherichia coli aromatic amino acid aminotransferase in leucine biosynthesis. J Bacteriol 136: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prohl C, Kispal G, Lill R. (2000) Branched-chain-amino-acid transaminases of yeast Saccharomyces cerevisiae. Methods Enzymol 324: 365–375 [DOI] [PubMed] [Google Scholar]
- Richins RD, Scholthof HB, Shepherd RJ. (1987) Sequence of figwort mosaic virus DNA (caulimovirus group). Nucleic Acids Res 15: 8451–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schauer N, Semel Y, Balbo I, Steinfath M, Repsilber D, Selbig J, Pleban T, Zamir D, Fernie AR. (2008) Mode of inheritance of primary metabolic traits in tomato. Plant Cell 20: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al. (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al. (2005a) GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Schauer N, Zamir D, Fernie AR. (2005b) Metabolic profiling of leaves and fruit of wild species tomato: a survey of the Solanum lycopersicum complex. J Exp Bot 56: 297–307 [DOI] [PubMed] [Google Scholar]
- Schulze-Siebert D, Heineke D, Scharf H, Schultz G. (1984) Pyruvate-derived amino acids in spinach chloroplasts: synthesis and regulation during photosynthetic carbon metabolism. Plant Physiol 76: 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster J, Binder S. (2005) The mitochondrial branched chain amino transferase (AtBCAT-1) is capable to initiate degradation of leucine, isoleucine and valine in almost all tissues of Arabidopsis. Plant Mol Biol 57: 241–254 [DOI] [PubMed] [Google Scholar]
- Schuster J, Knill T, Reichelt M, Gershenzon J, Binder S. (2006) Branched-chain aminotransferase4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. Plant Cell 18: 2664–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Day DA, Millar AH. (2004) Lipoic acid-dependent oxidative catabolism of alpha-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol 134: 838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay LW, Blanchard JS. (2009) The 1.9 angstrom structure of the branched chain amino acid transaminase (IlvE) from Mycobacterium tuberculosis. Acta Crystallogr Sect F Struct Biol Cryst Commun 65: 1071–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartak NB, Liu L, Wang BM, Berg CM. (1991) A functional leuABCD operon is required for leucine synthesis by the tyrosine-repressible transaminase in Escherichia coli K-12. J Bacteriol 173: 3864–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters DS, Steffens JC. (1990) Branched chain amino acid metabolism in the biosynthesis of Lycopersicon pennellii glucose esters. Plant Physiol 93: 1544–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.