Abstract
Shotgun proteomics analysis allows hundreds of proteins to be identified and quantified from a single sample at relatively low cost. Extensive DNA sequence information is a prerequisite for shotgun proteomics, and it is ideal to have sequence for the organism being studied rather than from related species or accessions. While this requirement has limited the set of organisms that are candidates for this approach, next generation sequencing technologies make it feasible to obtain deep DNA sequence coverage from any organism. As part of our studies of specialized (secondary) metabolism in tomato (Solanum lycopersicum) trichomes, 454 sequencing of cDNA was combined with shotgun proteomics analyses to obtain in-depth profiles of genes and proteins expressed in leaf and stem glandular trichomes of 3-week-old plants. The expressed sequence tag and proteomics data sets combined with metabolite analysis led to the discovery and characterization of a sesquiterpene synthase that produces β-caryophyllene and α-humulene from E,E-farnesyl diphosphate in trichomes of leaf but not of stem. This analysis demonstrates the utility of combining high-throughput cDNA sequencing with proteomics experiments in a target tissue. These data can be used for dissection of other biochemical processes in these specialized epidermal cells.
Plants synthesize a wide variety of specialized (“secondary”) chemicals that influence their interactions with the environment. Commercial interest in these plant natural products stems from their importance as pharmaceuticals, in plant aromas and flavors, and as feedstocks for industrial chemicals. The synthesis of these metabolites typically occurs in specialized structures such as laticifers (Hagel et al., 2008), resin ducts (Trapp and Croteau, 2001), or trichomes (Werker, 2000). Trichomes are specialized cells present on the surfaces of many plants and are capable of synthesizing and either storing or secreting large amounts of specialized metabolites (Schilmiller et al., 2008). These include compounds of interest to humans such as food flavors from mint (Mentha spp.) and basil (Ocimum basilicum; Rios-Estepa et al., 2008; Xie et al., 2008), the antimalarial drug artemisinin from Artemisia annua (Zhang et al., 2008), and tetrahydrocannabinol in medical marijuana (Cannabis sativa; Flores-Sanchez and Verpoorte, 2008). Because trichomes are located on plant surfaces and produce biologically active metabolites, they can protect against a number of environmental stresses, including herbivores and pathogens (Werker, 2000; Wagner et al., 2004).
Unlike Arabidopsis, which only makes nonglandular trichomes (Wagner et al., 2004), cultivated tomato (Solanum lycopersicum) also contains glandular trichomes (Luckwill, 1943). There are two abundant types of glandular trichomes on tomato organs. Type I trichomes have a multicellular stalk with a single, small gland cell at the tip (Fig. 1A). Type VI trichomes have a unicellular stalk with a four-cell glandular head (Fig. 1, A and C). Together, these glands produce a variety of compounds, including terpenes, acyl sugars, and phenylpropanoid-derived metabolites (Slocombe et al., 2008; Besser et al., 2009; Schilmiller et al., 2009, 2010). In addition to documented chemical diversity, an increasing number of tomato genetic and genomic resources are becoming available, including an assembly of the tomato genome shotgun sequence (http://solgenomics.net/). These and other tools make tomato trichomes amenable to studies of glandular trichome function and the synthesis of specialized metabolites.
Figure 1.
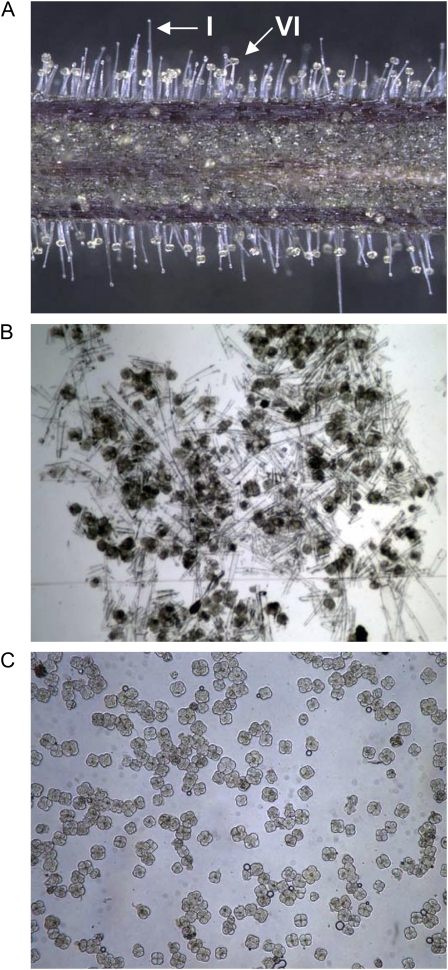
Tomato stem and petiole trichomes. A, Tomato stems and petioles are covered by multiple trichome types, including the glandular type I and type VI trichomes. B and C, Mixed-type trichome samples (B) were isolated by scraping frozen tissue in liquid nitrogen, and type VI gland samples (C) were isolated by glass bead abrasion and filtering with nylon mesh screens.
Their epidermal location simplifies the analysis of trichome RNAs, proteins, and metabolites (Schilmiller et al., 2008). Using isolated glands, many of the genes involved in essential oil production in mint were biochemically and molecularly characterized (Colby et al., 1993; Burke et al., 1999; Lange et al., 2000). EST sequencing played a critical role in identifying enzymes of trichome-specialized metabolism, including artemisinin (Teoh et al., 2006), germacrene A synthesis in A. annua (Bertea et al., 2006), geraniol synthesis in basil (Iijima et al., 2004b), eugenol synthesis in basil and isoeugenol synthesis in petunia (Petunia hybrida; Koeduka et al., 2006), methyl ketone production in Solanum habrochaites (Fridman et al., 2005), and xanthohumol synthesis in hops (Humulus lupulus; Nagel et al., 2008).
Proteomics techniques are another useful set of tools for the discovery of enzymes and pathways. These techniques are especially powerful in studies of enriched cell types. Two-dimensional gel electrophoresis and shotgun proteomics approaches were utilized for tomato pollen (Sheoran et al., 2007), elicitor-treated opium poppy (Papaver somniferum) cell culture (Zulak et al., 2009), guard cells of Arabidopsis (Arabidopsis thaliana) and Brassica napus (Zhao et al., 2008; Zhu et al., 2009), and soybean (Glycine max) root hairs (Brechenmacher et al., 2009). Laser capture microdissection combined with proteomics was used to analyze vascular bundles from Arabidopsis stems (Schad et al., 2005) and pericycle cells from maize (Zea mays) primary roots (Dembinsky et al., 2007). Proteomics analysis of isolated trichomes revealed insights into their metabolism and function. For example, several stress-related proteins were found in tobacco (Nicotiana tabacum) trichomes (Amme et al., 2005), and proteins involved in sulfur metabolism and detoxification were revealed in Arabidopsis trichomes (Wienkoop et al., 2004). An in-depth analysis of basil peltate trichomes resulted in the identification of 881 proteins across four different basil lines (Xie et al., 2008).
The lack of genome sequence information limits the ability to discover proteins representing new genes in nonmodel species (Carpentier et al., 2008). Although the number of plants with high-coverage genomic sequences is increasing at a fast rate, proteomics studies in many plants continue to rely on EST sequences (Faurobert et al., 2007; Alvarez et al., 2008; Xie et al., 2008; Zulak et al., 2009). Because ESTs are enriched in protein-coding sequences, they can be especially useful for analyzing proteomics data. However, most species have a relatively small amount of transcriptome sequence available. This problem is compounded by the fact that ESTs typically are derived from many different tissues, limiting the representation of sequences expressed in specific cell types. Technological advances now enable deep cDNA sequencing, permitting more complete representation of expressed genes compared with traditional Sanger sequencing of cDNA clones (Cheung et al., 2006; Weber et al., 2007; Lister et al., 2009; Rounsley and Last, 2010). The utility of combining proteomics and deep cDNA sequencing was recently demonstrated using isolated pea (Pisum sativum) chloroplast envelope protein and total leaf and hypocotyl cDNA sequencing using 454 technology (Bräutigam et al., 2008).
Our systematic studies on glandular trichome function (www.trichome.msu.edu; Schilmiller et al., 2009, 2010) led to a need for a comprehensive RNA and protein “parts list.” A large trichome-specific EST sequence collection was generated using 454 pyrosequencing (http://bioinfo.bch.msu.edu/trichome_est; Schilmiller et al., 2009) and used to analyze shotgun proteomics data from isolated trichomes. The combined transcriptome and proteome data set was mined to identify genes and proteins expressed in trichomes. The utility of this information was demonstrated by characterization of a new sesquiterpene synthase that produces β-caryophyllene and α-humulene in tomato trichomes.
RESULTS
Sequencing and Assembly of Tomato cv M82 Trichome cDNA
Glandular trichomes express genes encoding proteins unique to these specialized cell types (Wang et al., 2001; Shepherd et al., 2005). Because trichomes represent a small fraction of cells on vegetative and reproductive structures, the existing collections of tomato cDNA sequences do not have strong representation of trichome-specific transcripts. We previously generated tomato mixed-type stem and petiole trichome EST assemblies by deep sequencing of cDNA by 454 pyrosequencing (Schilmiller et al., 2009). cDNA from isolated leaf type VI glands was sequenced to complement the mixed-type stem trichome library and enable searching for trichome type-specific transcripts and proteins. A combined total of 467,677 reads (244,655 from the mixed trichome library and 223,022 from the type VI library) were generated and submitted to the GenBank Short Read Archive (SRP000732). Reads from the combined libraries were assembled into 23,585 contigs with an average length of approximately 300 bp. BLASTX searches were conducted against the National Center for Biotechnology Information (NCBI) nonredundant protein database and a subset of this database containing only green plant sequences (viridiplantae, taxid:33090). Each contig was annotated according to the top BLASTX match (Supplemental Table S1), and the top 25 contigs with the highest number of assembled sequence reads are shown in Table I.
Table I. Top 25 trichome EST contigs with the most reads and their closest BLASTN match in the TIGR tomato TA.
Percentage of total trichome or TA ESTs was calculated for the 454 contigs and the TAs, and their ratio (%454/%TA) is shown. Genes that have a high %454/%TA value represent genes whose expression is likely enriched in trichome tissue.
| Contig | Contig Length | Contig Annotationa | TIGR TA | %TAb | %454c | %454/%TA | TA Annotation |
| M01000000389 | 1,024 | Metallocarboxypeptidase inhibitor protein | TA29971_4081 | 0.0015 | 4.5818 | 3,058 | Metallocarboxypeptidase inhibitor |
| M01000017652 | 813 | Wound-induced protein Sn-1, vacuolar membrane: pepper | TA23682_4081 | 0.0045 | 2.2586 | 503 | Sn-1 protein [Capsicum annuum (bell pepper)] |
| M01000001081 | 3,596 | No hits found | TA24712_4081 | 0.0030 | 1.7728 | 592 | |
| M01000000512 | 3,014 | Lipoxygenase C | TA18073_4081 | 0.0135 | 1.5945 | 118 | Lipoxygenase [Lycopersicon esculentum (tomato)] |
| M01000000650 | 1,934 | Putative S-adenosylmethionine decarboxylase proenzyme [Lycopersicon chilense] | TA18118_4081 | 0.1938 | 1.3360 | 7 | S-Adenosylmethionine decarboxylase [Lycopersicon chilense (Solanum chilense)] |
| M01000012828 | 786 | Hypothetical 16.5 K protein: common tobacco | TA25836_4081 | 0.0035 | 1.3003 | 372 | Orf protein [Nicotiana tabacum (common tobacco)] |
| M01000000329 | 672 | Major latex-like protein [Prunus persica] | AW624755 | 0.0005 | 1.2902 | 2,584 | Major latex-like protein [Prunus persica (peach)] |
| M01000000347 | 1,989 | Proteinase inhibitor [Solanum tuberosum] | TA17990_4081 | 0.0594 | 1.0928 | 18 | Cathepsin D inhibitor precursor [Lycopersicon esculentum (tomato)] |
| M01000001067 | 651 | Wound-induced protein Sn-1, vacuolar membrane: pepper | TA23682_4081 | 0.0045 | 1.0569 | 235 | Sn-1 protein [Capsicum annuum (bell pepper)] |
| M01000022834 | 885 | Major latex-like protein [Prunus persica] | BI930179 | 0.0005 | 0.8775 | 1,757 | |
| M01000000118 | 1,194 | DS2 protein [Solanum tuberosum] | TA20485_4081 | 0.0110 | 0.8493 | 77 | ASR4 [Lycopersicon esculentum (tomato)] |
| M01000003167 | 765 | No hits found | TA24589_4081 | 0.0025 | 0.8290 | 332 | |
| M01000000394 | 1,145 | Neryl diphosphate synthase1 | TA23792_4081 | 0.0045 | 0.7095 | 158 | Dehydrodolichyl diphosphate synthase 5 [Arabidopsis thaliana (mouse-ear cress)] |
| M01000000349 | 948 | Early light-inducible protein [Lycopersicon esculentum] | TA18127_4081 | 0.0295 | 0.6879 | 23 | Early light-inducible protein [Lycopersicon esculentum (tomato)] |
| M01000000473 | 836 | Nonspecific lipid transfer protein [Lycopersicon esculentum] | TA17713_4081 | 0.0839 | 0.6648 | 8 | Nonspecific lipid transfer protein [Lycopersicon esculentum (tomato)] |
| M01000001122 | 2,034 | Epidermal germacrene C synthase [Lycopersicon esculentum] | TA25209_4081 | 0.0015 | 0.5968 | 398 | Epidermal germacrene C synthase [Lycopersicon esculentum (tomato)] |
| M01000001159 | 1,817 | PPO precursor [Lycopersicon esculentum] | TA17467_4081 | 0.0035 | 0.5923 | 169 | PPO E, chloroplast precursor [Lycopersicon esculentum (tomato)] |
| M01000013214 | 824 | No hits found | TA28678_4081 | 0.0010 | 0.5820 | 583 | |
| M01000000759 | 1,841 | UDP Rha-anthocyanidin-3-glucoside rhamnosyltransferase: garden petunia (fragment) | TA17542_4081 | 0.0065 | 0.5559 | 86 | Anthocyanidin 3-O-glucosyltransferase [petunia hybrida (petunia)] |
| M01000000483 | 1,359 | PPO precursor [Lycopersicon esculentum] | TA17484_4081 | 0.0584 | 0.4969 | 9 | PPO F, chloroplast precursor [Lycopersicon esculentum (tomato)] |
| M01000000500 | 1,577 | D12 oleate desaturase [Solanum commersonii] | TA18969_4081 | 0.0340 | 0.4858 | 14 | D12 oleate desaturase [Solanum commersonii (Commerson's wild potato)] |
| M01000000955 | 1,867 | β-Primeverosidase [Camellia sinensis] | TA19521_4081 | 0.0170 | 0.4762 | 28 | β-Glucosidase [Prunus avium (cherry)] |
| M01000000211 | 1,971 | Pectin methylesterase [Sesbania rostrata] | TA18816_4081 | 0.0390 | 0.4734 | 12 | Pectin methylesterase precursor [Sesbania rostrata] |
| M01000000185 | 1,453 | Stearoyl acyl carrier protein desaturase Lldd3A20 [Lupinus luteus] | TA22497_4081 | 0.0065 | 0.4616 | 71 | Stearoyl acyl carrier protein desaturase Lldd3A20 [Lupinus luteus (European yellow lupin)] |
| M01000000073 | 1,478 | Polyubiquitin [Elaeagnus umbellata] | TA17549_4081 | 0.1298 | 0.4507 | 3 | Ubiquitin C variant [Homo sapiens (human)] |
Contig annotation is based on best BLASTX match in GenBank or manually annotated.
Total number of TA ESTs is 200,250.
Total number of 454 trichome ESTs is 467,677.
We sought to identify trichome-specific transcripts by looking for contigs with an overrepresentation of reads in the trichome 454 assembly. The Institute for Genomic Research (TIGR) transcript assemblies (TAs; http://plantta.jcvi.org/index.shtml; Childs et al., 2007) were used for this comparison because they were generated from 200,250 ESTs derived from a broad collection of tissues. A BLASTN match to a TIGR TA was found for 87% of the trichome contigs using an E-value cutoff of less than 1 × 10−10 (Supplemental Table S1). Because trichomes constitute a relatively small part of the tissues used to generate the TAs, trichome-specific ESTs are underrepresented in those libraries. The monoterpene biosynthetic enzyme Neryl Diphosphate Synthase1 (TA23792_4081) is an example of a trichome-enriched transcript: it represents 0.71% of the trichome ESTs from the combined libraries and only 0.005% of TA ESTs, or a 160-fold enrichment in trichomes (Schilmiller et al., 2009).
A total of 3,052 trichome EST contigs did not have a BLASTN match to a TA, suggesting that our sequencing revealed other good candidates for trichome-enriched genes (Supplemental Table S2). Many of the contigs on the list of 3,052 are relatively short (2,668 have a length less than 300 bp) with few reads and are mathematically less likely to have a match with an E-value below 1 × 10−10. Out of the 3,052 contigs with no TA match, there were 651 contigs with a BLASTX match of E-value less than 1 × 10−10 in the nonredundant green plants (viridiplantae) protein database at GenBank (Supplemental Table S2). These contigs represent genes that are potentially trichome specific and have some related sequences with annotation that may help in determining their function.
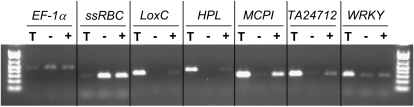
To validate the comparison of trichome reads against the TAs experimentally, five genes with a high enrichment of trichome ESTs (Table II) were chosen for reverse transcription (RT)-PCR analysis (Fig. 2). A putative metallocarboxypeptidase inhibitor and a gene of unannotated function (TA24712) represent the class of genes that are very highly expressed in the trichome based on the number of 454 reads. A second category is represented by lipoxygenase C and hydroperoxide lyase (HPL). These together catalyze the production of volatile aldehydes and are known to be expressed in various tissues, including leaf, flower, and fruit (Howe et al., 2000; Chen et al., 2004), consistent with the presence of ESTs in the TA assemblies. Interestingly, these were not previously reported to be active in trichomes, yet our data show that they have trichome-enriched mRNA accumulation. We also analyzed a putative WRKY transcription factor gene that appears to be modestly expressed yet is enriched in trichome ESTs. RT-PCR was used to compare expression in isolated stem and petiole trichomes with that in stems and petioles with their trichomes removed (Fig. 2). These results confirmed that all five genes have relatively high expression in trichomes compared with the shaved tissue, despite their broad range of expression levels in trichomes.
Table II. Trichome-enriched genes: EST counts from the tomato 454 assembly and TIGR tomato TA release 2.
| Sample | TA | Contig No. | 454 Reads (% of Total)a | ESTs in TIGR Tomato TA (% of Total) | % 454 Reads/% ESTs in TA |
| Putative metallocarboxypeptidase inhibitor (MCPI) | TA29971_4081 | M01000000389 | 27,547 (5.9%) | 3 (0.002%) | 2,950 |
| Unknown protein | TA24712_4081 | M01000001081 | 8,479 (1.8%) | 6 (0.003%) | 600 |
| Lipoxygenase C (LoxC) | TA18073_4081 | M01000000512 | 7,591 (1.6%) | 27 (0.01%) | 160 |
| Hydroperoxide lyase (HPL) | TA20291_4081 | M01000001141 | 1,625 (0.3%) | 24 (0.01%) | 30 |
| WRKY transcription factor | AY157061b | M01000001133 | 46 (0.01%) | 1 (0.0005%) | 20 |
Reads from all contigs matching appropriate TA were summed.
Singleton EST.
Figure 2.
RT-PCR analysis of gene expression in trichomes. M82 tomato plants were grown to 3 weeks old, and RNA was extracted from isolated stem and petiole trichomes (T), stem and petiole tissue after removal of trichomes (−), and stem and petiole tissue with intact trichomes (+). Expression was analyzed for genes with ESTs enriched in trichome tissue compared with the TIGR TA ESTs. LoxC, Lipoxygenase C; MCPI, putative metallocarboxypeptidase inhibitor. Translation elongation factor-1α (EF-1α) was used as a constitutive control, and small subunit of Rubisco (ssRBC; TA17646_4081) was included as a control not expected to be highly expressed in trichome tissue. All reactions were done for 20 cycles except for WRKY, which was 25 cycles.
Shotgun Proteomics Analysis of Mixed-Type Trichomes and Isolated Type VI Glands
The availability of a large number of tomato cDNA contigs from varied tissue types created a DNA sequence data set for use in shotgun proteomics analysis. Protein was purified from trichomes isolated from stem and petiole tissues of 3-week-old tomato plants, the same developmental stage used for cDNA sequencing (Fig. 1A). Mixed-type trichome samples, including stalk and gland cells free from detectable underlying epidermal pavement cells, were collected in bulk by gently abrading stem and petiole sections in liquid nitrogen (Fig. 1B). Type VI glands, which are four cell clusters that are easily removed from the stalk cell, were size selected using nylon mesh screens and could be collected at greater than 98% purity based on microscopy (Fig. 1C). Mixed-type trichome and type VI gland protein samples were analyzed using a gel-enhanced liquid chromatography-tandem mass spectrometry (LC-MS/MS) approach (Lee and Cooper, 2006).
A custom protein sequence database was created using translated sequences from the assembly of the M82 mixed-type and type VI trichome 454 reads to analyze the proteomics data. Mascot (www.matrixscience.com) searches were performed using the translated DNA sequence followed by validation using the Scaffold software package (www.proteomesoftware.com), which uses the PeptideProphet and ProteinProphet algorithms (Keller et al., 2002; Nesvizhskii et al., 2003). A stringent set of protein sequence validation criteria was employed to reduce the rate of false protein discovery: the protein identification threshold was set to 99% probability and a minimum of two peptides identified with at least 95% peptide probability.
A total of 1,973 proteins were identified from the combined total and type VI trichome data sets using the 454 sequence database (Supplemental Table S3). A significant number of these proteins are represented more than once due to disjointed contigs of the same gene. This is because the short reads from the 454 sequencer did not assemble as well as longer Sanger sequenced ESTs. A manual inspection was done for those contigs that had a BLASTN match to the same TA, and spectral counts were combined when multiple records represented the same protein. This resulted in condensing the list to 1,552 proteins identified using the 454 contigs (Supplemental Table S4). Of the 1,552 proteins identified, 1,360 were found in both samples, with 67 proteins only in the type VI trichome sample and 125 specific to the mixed-type preparation. The high degree of overlap between the two data sets is not surprising, as type VI glands make up a significant fraction of cells in the mixed-type trichome sample (Kang et al., 2010).
Correlation of Trichome Chemistry with mRNA and Protein Accumulation
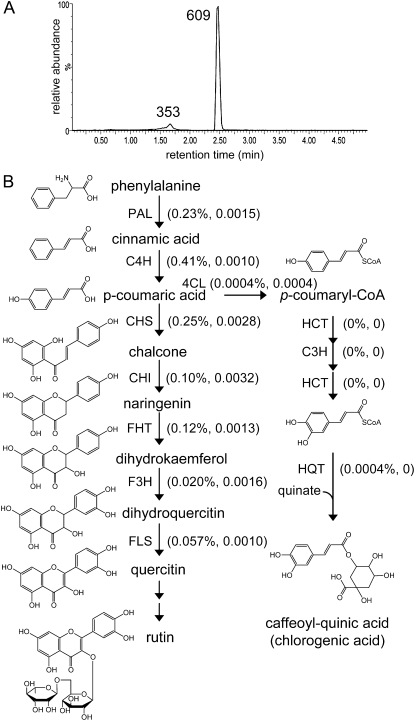
The phenylpropanoid pathway-derived compounds rutin and chlorogenic acid are detected in extracts from mixed-type trichomes, with higher MS signal intensity for rutin (mass-to-charge ratio [m/z] 609; Fig. 3A). Similar quantitative differences were recently reported for these compounds in tomato leaf trichomes (Kang et al., 2010). We tested the quality of the transcriptome and proteome data by quantifying the number of ESTs and spectral counts for the enzymes in these biosynthetic pathways. Spectra-matching peptides for the first seven enzymes leading from Phe to rutin were found (Fig. 3B). In contrast, no peptides for the known enzymes of chlorogenic synthesis were observed when searching the proteomics data using the M82 trichome EST assemblies. Thus, the relative abundance of these metabolites in these tomato trichomes predicts our ability to detect the mRNAs and peptides encoding their biosynthetic enzymes.
Figure 3.
Rutin and chlorogenic acid biosynthetic pathways. A, LC-MS-extracted ion chromatograms showing the presence of rutin (m/z 609) and chlorogenic acid (m/z 353) in isolated total stem and petiole trichome extracts. B, Biosynthetic pathways for the production of rutin and chlorogenic acid. For each pathway step, the percentage of ESTs and normalized spectral abundance factors from the mixed-type trichome data are shown in parentheses. PAL, Phe ammonia-lyase; C4H, cinnamate 4-hydroxylase; CHS, chalcone synthase; CHI, chalcone isomerase; FHT, flavanone 3-hydroxylase; F3H, flavonoid 3′-hydroxylase; FLS, flavonol synthase; 4CL, 4-coumaryl-CoA ligase; HCT, cinnamoyl-CoA shikimate/quinate transferase; C3H, p-coumaryl 3′-hydroxylase; HQT, hydroxycinnamoyl-CoA quinate transferase.
Tomato trichomes accumulate polyphenol oxidase (PPO) protein and activity in addition to the production of phenolic compounds that are PPO substrates (Yu et al., 1992). These compounds are thought to be polymerized by PPO after a trichome gland is ruptured by an insect (Tingey, 1991). Tomato PPO is encoded by a seven-member gene family (Thipyapong et al., 1997). Of these, transcripts (454 reads) and proteins (MS/MS spectra) matching PPO-E and PPO-F were detected in the mixed-type trichome and type VI gland data (Supplemental Tables S1 and S3). This is consistent with in situ hybridization demonstrating the presence of PPO-E and PPO-F transcripts in type I and VI trichomes (Thipyapong et al., 1997).
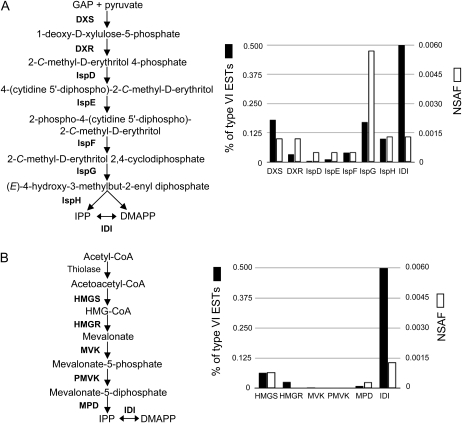
Previous work on trichomes demonstrated that isopentenyl diphosphate and dimethylallyl diphosphate are synthesized primarily through the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway in plastids rather than via the mevalonate pathway in the cytosol (Iijima et al., 2004a; Xie et al., 2008). In the plastids of M82 tomato trichomes, isopentenyl diphosphate and dimethylallyl diphosphate are used by Neryl Diphosphate Synthase1 enzyme, leading to the synthesis of monoterpenes, the major volatile isoprenoid class detected in glands (Schilmiller et al., 2009). Consistent with the plastidic synthesis of trichome monoterpenes, ESTs and spectra matching the proteins of the MEP pathway are significantly more abundant than those of the mevalonate pathway (Fig. 4).
Figure 4.
EST abundance and proteomics-normalized spectral abundance factors (NSAF) for steps of the MEP and mevalonate pathways in type VI trichomes. EST counts (percentage of the total type VI ESTs) and NSAF for genes and proteins of the MEP pathway (A) are more abundant than those of the mevalonate pathway (B) in type VI trichomes. DXS, 1-Deoxy-d-xylulose-5-phosphate synthase; DXR, 1-deoxy-d-xylulose-5-phosphate reductoisomerase; IspD, 4-diphosphocytidyl-2-C-methyl-d-erythritol synthase; IspE, 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol kinase; IspF, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase; IspG, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase; IspH, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; IDI, isopentenyl/dimethylallyl diphosphate isomerase; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; HMGR, HMG-CoA reductase; MVK, mevalonate kinase; PMVK, diphosphomevalonate kinase; MPD, diphosphomevalonate decarboxylase.
Using Combined Genomics, Proteomics, and Metabolite Data for Enzyme Discovery
The availability of EST, proteomics, and metabolite data allows for a detailed analysis of trichome biosynthetic pathways. As shown in Table III, a search of the trichome EST database revealed high-level expression of the gene for the previously characterized tomato sesquiterpene synthase SSTLE1 (AF279453; 0.6% of trichome ESTs). This gene encodes a protein that catalyzes the production of germacrene C in vitro (Colby et al., 1998; van der Hoeven et al., 2000). Mass spectra from both the mixed-type trichome and the type VI proteomics samples matched this contig (Table III). Another contig (M01000000371) that is 91% identical to SSTLE1 at the nucleotide level was also found in the trichome EST database. Similar to SSTLE1, M01000000371 mRNA accumulated to a much higher level in trichomes than in nontrichome tissues; this gene represents only 0.0005% of the TA ESTs; however, it represents 0.11% of the trichome ESTs (Table III).
Table III. EST counts and proteomics spectral counts for SSTLE1 and CAHS.
Percentage of the total ESTs or normalized spectral abundance factor is shown.
| Sample | Stem Mixed-Type Trichome ESTs | Leaf Type VI Trichome ESTs | Total Trichome ESTs | ESTs in TIGR Tomato TA | Stem Mixed-Type Trichome Spectra | Stem Type VI Spectra |
| M01000001122 (SSTLE1) | 749 (0.31%) | 2,042 (0.92%) | 2,791 (0.60%) | 3 (0.0015%) | 333 (0.0029) | 274 (0.0037) |
| M01000000371 (CAHS) | 43 (0.018%) | 465 (0.21%) | 508 (0.11%) | 1 (0.0005%) | 0 | 0 |
Previous studies showed that sesquiterpene production requires genes located on chromosome 6, and SSTLE1 was mapped to this region (van der Hoeven et al., 2000). A search of the tomato genome scaffold sequences (http://solgenomics.net/, version 1.0) using M01000000371 revealed a scaffold sequence containing this gene separated from the SSTLE1 sequence by approximately 38 kb. The presence of SSTLE1 along with other markers on the scaffold indicates that M0100000371 is also located on chromosome 6, within regions 6-2 and 6-2-2 in the tomato cv M82 × Solanum pennellii LA0716 introgression lines (Eshed and Zamir, 1994, 1995). This region is known to control sesquiterpene production in tomato (van der Hoeven et al., 2000; Schilmiller et al., 2010).
While M01000000371 was well represented in the trichome EST database, it was surprising that no mass spectra matched this contig in the proteomics data. However, as shown in Table III, the majority of the reads in contig M01000000371 were from type VI gland cDNA prepared from leaf trichomes rather than stem. It was of interest to ask whether the observed difference in expression between stem and leaf trichomes resulted in altered terpene accumulation. In fact, the sesquiterpenes β-caryophyllene and α-humulene were nearly undetectable in stem type VI glands, whereas leaf type VI glands produced significant levels of these compounds (Table IV). In contrast, the levels of δ-elemene (the compound that is detected due to thermal degradation of germacrene C in the gas chromatograph) were the same for stem and leaf.
Table IV. Terpene levels in type VI glands from leaf and stem tissue, which is the same developmental stage as used for EST sequencing and proteomics.
Amounts shown are averages and sd from four replicates.
| Terpene | Leaf | Stem |
| pg trichome−1 | ||
| β-Caryophyllene | 39.1 ± 9.3 | 1.1 ± 0.3 |
| α-Humulene | 29.8 ± 7.0 | 2.2 ± 0.2 |
| δ-Elemene | 82.1 ± 17.3 | 81.1 ± 12.0 |
| α-Pinene | 24.5 ± 11.0 | 105 ± 16.3 |
| 2-Carene | 44.2 ± 25.8 | 189 ± 45.0 |
| α-Phellandrene | 25.0 ± 13.2 | 107 ± 20.1 |
| α-Terpinene | 6.2 ± 3.2 | 19.3 ± 6.6 |
| Limonene + β-phellandrene | 358 ± 197 | 1,690 ± 378 |
These data strongly suggested that M01000000371 encodes a sesquiterpene synthase producing β-caryophyllene and α-humulene in leaf type VI glands. To test this hypothesis, a His-tagged version of the predicted open reading frame was expressed in Escherichia coli. The affinity-purified enzyme was incubated with E,E-farnesyl diphosphate (FPP), and the reaction products were analyzed by gas chromatography (GC)-MS. Two peaks were detected and identified as β-caryophyllene and α-humulene by comparison of retention times and mass spectra with authentic standards (Fig. 5; Supplemental Fig. S1). The in vitro reaction products were also identical to sesquiterpenes from analysis of tomato leaf-disc head space (Fig. 5; Supplemental Fig. S1). Together, these data show that this enzyme, which we designate as CAHS (for β-caryophyllene/α-humulene synthase; GenBank accession no. GU647162), is responsible for β-caryophyllene and α-humulene production in leaf type VI trichomes.
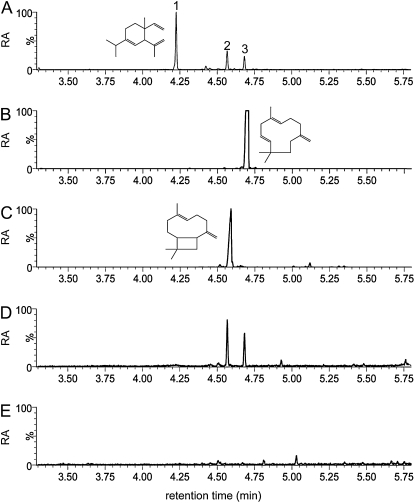
Figure 5.
In vitro assay with purified recombinant CAHS. A, Sesquiterpenes produced by M82 leaf trichomes were collected from a small leaflet in a sealed vial using a solid-phase microextraction fiber. Peak 1, δ-Elemene; peak 2, β-caryophyllene; peak 3, α-humulene. B, α-Humulene standard. C, β-Caryophyllene standard. D, Reaction products from incubation of purified CAHS with FPP. E, Solid-phase microextraction fiber negative control. All data shown are extracted ion chromatograms for m/z 93. RA, Relative abundance.
DISCUSSION
Plants make tens of thousands of structurally diverse specialized metabolites, with known biological roles ranging from plant stress tolerance to pharmaceutically important drugs. Production and storage of these compounds are often limited to specific subcellular compartments, specialized cells, or tissue types. This allows the metabolites to be sequestered (e.g. toxic latex in laticifers) and permits them to exert their biological functions where and when needed (e.g. hydrolysis of glucosinolates to toxic products by myrosinase upon tissue damage by chewing insects). However, these specialized cell types can be difficult to isolate and study. In contrast, secreting glandular trichomes are easy-to-isolate specialized epidermal appendages that make large amounts of specific classes of specialized metabolites. This makes them excellent tissues for systematic investigation of these biosynthetic pathways by analyzing metabolite accumulation and gene expression patterns. In this study, we took advantage of the ease of gland isolation to perform deep EST sequencing by 454 pyrosequencing and shotgun proteomics to comprehensively analyze gene expression in tomato type VI and mixed-type trichomes.
This deep analysis of the trichome transcriptome and proteome revealed details of several well-characterized pathways of specialized metabolism. For example, proteins were identified for all eight steps of the MEP pathway, which produces precursors for isoprenoid biosynthesis (Fig. 4). Similarly, cDNAs and proteins were found for nearly all known steps leading to the synthesis of rutin (Fig. 4). These examples validate the hypothesis that the EST and proteomics data should represent the majority of enzymes for pathways active in trichome cells. Thus, these data, coupled with metabolite profiling to reveal classes of compounds that accumulate in the glands, will be useful for discovering new enzymes and regulators of specialized metabolic pathways.
The EST and proteomics results revealed that the previously characterized enzymes of short-chain oxylipin biosynthesis are strongly expressed in tomato trichomes. These oxidized lipid products are known to be part of the “leafy green” smell associated with cut grass (Howe and Schilmiller, 2002). The mRNAs for the enzymes that produce aldehydes (lipoxygenase and HPL) are known to accumulate in several tomato tissues, including modest expression of HPL in whole stems (Heitz et al., 1997; Howe et al., 2000). Our results indicate that this low-level expression in whole stems is the result of strong trichome expression and weak or no expression in nontrichome stem tissue (Fig. 2). Synthesis of volatile aldehydes on the surface of the plant is consistent with the involvement of these oxylipins in interactions with insects and pathogens (Matsui, 2006).
The combination of trichome EST and proteomics data analysis with targeted metabolite measurements revealed the function of a previously uncharacterized tomato trichome sesquiterpene synthase. Several lines of evidence led to the hypothesis that CAHS controls β-caryophyllene and α-humulene synthesis. First, β-caryophyllene and α-humulene are produced at significantly higher levels in leaf trichomes compared with stem trichomes, which correlates with differences in EST representation in the glands of these organs. Second, analysis of tomato genomic DNA assemblies revealed that CAHS is located on chromosome 6 in a region previously shown to control sesquiterpene production in tomato (van der Hoeven et al., 2000; Schilmiller et al., 2010). In vitro assays of the recombinant protein showed that it converts FPP to the sesquiterpenes β-caryophyllene and α-humulene in ratios similar to those found in leaf trichomes (Fig. 5; Supplemental Fig. S1).
Recently published results showed that virus-induced gene silencing using a SSTLE1 sequence resulted in reduction of accumulation of all sesquiterpenes, including β-caryophyllene and α-humulene (Besser et al., 2009). Our results are consistent with the hypothesis that the sequence used for the virus-induced gene silencing experiment resulted in suppression of both SSTLE1 and CAHS, presumably due to the high identity of the silencing sequence used (94% at the nucleotide level). We propose that tomato trichome expression of SSTLE1 normally results in germacrene accumulation, whereas CAHS is responsible for β-caryophyllene and α-humulene.
An important insight from this work is that there are differences in metabolite accumulation and gene expression of type VI glands between leaf and stem (Tables III and IV) despite the observation that they are morphologically indistinguishable. These differences indicate that the developmental state of the surrounding tissue influences gene expression and metabolite accumulation in gland cells. Thus, development of methods for analyzing the chemistry and gene expression profiles of individual gland cells will advance the discovery of metabolic pathways and physiological roles of these morphologically indistinguishable cell types. These results reinforce the importance of developing comprehensive data sets of mRNA, protein, and metabolite accumulation across tissues and developmental stages.
Identification of approximately 1,500 proteins from two shotgun proteomics experiments using trichome 454 EST assemblies confirms the power of combining deep mRNA and protein sequencing for plants lacking substantial genomics data resources. Improvements in assembly and annotation of the tomato genome will increase the utility of the proteomics data, for example, by revealing proteins encoded by low-abundance mRNAs. Our results also reinforce the value of combining mRNA and protein sequencing with metabolite analysis for the discovery of new metabolic enzymes. Finally, these deep sequence resources will be useful in gene model validation and annotation of the emerging tomato genome sequence.
MATERIALS AND METHODS
Plant Growth and Trichome isolation
Tomato (Solanum lycopersicum ‘M82’) seed was germinated on moistened filter paper and then transferred to Jiffy peat pots (Hummert). Plants were grown in a growth chamber maintained under 16 h of light (250 μE m−2 s−1) at 28°C and 8 h of dark at 20°C. Mixed-type trichomes from petioles and stems of 21-d-old plants were isolated by gently scraping tissue with a spatula frozen in liquid nitrogen using a flat-end plastic spatula. Glandular heads from type VI trichomes were collected using a procedure modified from Gang et al. (2001). After removal of leaflets, petiole and stem tissue from 21-d-old plants was quickly collected into a 500-mL volume glass bottle containing ice-cold gland isolation buffer (50 mm Tris-Cl, pH 7.5, 200 mm sorbitol, 20 mm Suc, 10 mm KCl, 5 mm MgCl2, 0.5 mm K2HPO4, 5 mm succinic acid, 1 mm EGTA, and 14 mm β-mercaptoethanol) with 0.5-mm glass beads. Type VI glands were removed by shaking the bottle by hand two times for 30 s each with a 1-min period on ice between shakings. The material was filtered first through a 350-μm nylon mesh followed by a 100-μm mesh to remove stem and petiole tissue and glass beads. The type VI glands were collected onto a 40-μm mesh and washed with isolation buffer. To minimize collection time, the single-layer nylon filters were clamped in embroidery hoops and held one over the other using a ring stand to allow quick sequential filtering. After a brief spin in a microcentrifuge to concentrate gland cells, the buffer was removed using a pipette and the pellet was frozen in liquid nitrogen.
Metabolite Analysis
For analysis of total stem trichomes from 3-week-old M82 plants, stem sections (approximately 2 cm long) were dipped in 1 mL of acetonitrile:isopropanol:water (3:3:2, v/v) for 1 min with gentle rocking. LC-MS analysis of samples was performed as recently described (Gu et al., 2010). Terpenes were analyzed from type VI trichomes on stems and leaves of 3-week-old plants by picking glands using a pulled Pasteur pipette and dipping into tert-butyl methyl ether solvent containing 10 ng μL−1 tetradecane as internal standard. Each sample consisted of 200 glands picked into 100 μL of solvent. Samples were analyzed by GC-MS by injection onto a DB-5 column (Agilent; 10 m length, 0.33 μm film thickness, 100 μm internal diameter) on a 6890 gas chromatograph (Agilent) coupled to a 5975b mass spectrometer (Agilent). Injector temperature was 280°C working in splitless mode. The GC temperature program was as follows: 40°C for 1 min, 30°C min−1 to 90°C, 5°C min−1 to 110°C, 40°C min−1 to 165°C, 5°C min−1 to 180°C, 40°C min−1 to 320°C, hold for 2 min. The mass spectrometer was operated in selected ion monitoring mode for m/z 85 and 93. Amounts of compounds were normalized to a tetradecane internal standard and quantified using an external standard curve of γ-terpinene.
cDNA Library Construction and Sequencing
Frozen trichomes were ground to a powder in liquid nitrogen using a mortar and pestle. RNA was extracted using the RNeasy Plant mini kit (Qiagen) according to the manufacturer's instructions and including the on-column DNaseI treatment step. RNA quality was assessed using the Agilent 2100 Bioanalyzer RNA chip (Agilent Technologies). Preparation and sequencing of cDNA from the mixed-type trichome sample were described previously (Schilmiller et al., 2009). Preparation of cDNA from total RNA isolated from type VI glands was done using a slightly modified protocol from the SMART cDNA library construction kit (Clontech). First-strand cDNA was synthesized from 1 μg of total RNA as described in the kit protocol. For first-strand synthesis, a modified oligo(dT) primer (5′-TAGAGGCCGAGGCGGCCGACATGTTTTGTTTTTTTTTCTTTTTTTTTTVN-3′, where V = A, C, or G and N = A, T, C, or G) was used in a 10-μL reaction using SuperScript II reverse transcriptase (Invitrogen) and the SMART IV primer from the SMART cDNA library construction kit. Double-stranded cDNA was synthesized as described in the SMART cDNA kit protocol using the modified oligo(dT) primer and the 5′PCR primer from the kit. Amplified cDNA was purified using Qiagen QIAquick PCR purification spin columns and pooled to give 5 μg of cDNA. Preparation of type VI gland trichome cDNA for sequencing using the GS-FLX sequencer was done by the Michigan State University Research Technology Support Facility according to the manufacturer's protocol. Reads from both the mixed-type trichome library and the type VI gland library were processed and trimmed to remove low-quality and primer sequences using SeqClean (Pertea et al., 2003). The cleaned reads were assembled using CAP3 (Huang and Madan, 1999) followed by a second round of CAP3 to cluster the first-pass contigs and singletons. First-round CAP3 parameter settings for percentage match, overlap length, maximum overhang percentage, gap penalty, and base quality cutoff for clipping were -p 90, -o 50, -h 15, -g 2, and -c 17, respectively. For the second round CAP3 parameters, -o was changed to 100. A translated BLAST (BLASTX) search was then performed with the final contigs against the nonredundant and green plants databases at NCBI.
Protein Extraction and Proteomics Analysis
Protein was extracted from frozen ground trichomes with warm (80°C) buffer consisting of 100 mm Tris-Cl (pH 6.8), 150 mm NaCl, 10% glycerol, 4% SDS, and 200 mm dithiothreitol supplemented with Complete Mini EDTA-free proteinase inhibitors (one tablet per 10 mL; Roche). Lipophilic contaminants that would interfere with electrophoresis were removed by chloroform-methanol extraction of the solubilized protein. Protein pellets from the chloroform-methanol extraction were resuspended in extraction buffer, and protein concentrations were determined using the Coomassie Plus (Pierce) protein assay. Trichome proteins (mixed type, 120 μg; type VI, 190 μg) were separated on a 4% to 15% one-dimensional SDS-PAGE gradient gel (18-well; Criterion; Bio-Rad). The lane was excised and cut into 12 4-mm slices followed by in-gel trypsin digestion (Shevchenko et al., 1996). The extracted peptides were then injected by a Waters nanoAcquity Sample Manager and loaded for 5 min onto a 5-μm particle, 180-μm × 20-mm Waters Symmetry C18 peptide trap at 4 μL min−1 in 2% acetonitrile/0.1% formic acid. The bound peptides were then eluted onto a 1.7-μm particle, 100-μm × 100-mm Waters BEH C18 nanoAcquity column and eluted over 240 min with a gradient of 5% B to 35% B over a period of 205 min, ramping to 90% B from 205 to 215 min, and immediately back to 5% B using a Waters nanoAcquity ultra-HPLC system (buffer A = 99.9% water/0.1% formic acid, buffer B = 99.9% acetonitrile/0.1% formic acid) with a flow rate of 300 nL min−1. Column eluate was introduced into a ThermoScientific linear ion trap Fourier transform ion cyclotron resonance mass spectrometer outfitted with a Thermo Nanospray I spray source. Survey scans were taken in the Fourier transform ion cyclotron resonance cell at 25,000 resolution at m/z 400, and the top 10 ions in each survey scan were subjected to automatic low-energy collision-induced dissociation in the linear ion trap.
The resulting MS/MS spectra were converted to peak lists using BioWorks Browser version 3.2 (ThermoFisher) and searched against an in-house-generated tomato cv M82 trichome cDNA database (http://bioinfo.bch.msu.edu/trichome_est) using the Mascot searching algorithm (version 2.2; Matrix Science). The database employed for this analysis contained the assembled M82 mixed-type trichome and type VI trichome reads. Searches allowed for the identification of carbamidomethyl Cys as a fixed peptide modification and Met oxidation as a variable modification. Up to two missed tryptic cleavages were allowed. Peptide tolerance was set to 10 ppm, and MS/MS tolerance to 0.8 D. The Mascot output was then analyzed using Scaffold (Proteome Software), which calculates the probability of protein identification based on ProteinProphet and PeptideProphet algorithms (Keller et al., 2002; Nesvizhskii et al., 2003). The protein and peptide identification thresholds were set to 99% and 95% probability, respectively, with a minimum of two peptides per protein required. Proteomics data were deposited to the NCBI Peptidome database with the accession number PSE138. A normalized spectral abundance factor for each protein was calculated to adjust for protein length according to Paoletti et al. (2006).
RT-PCR Analysis
Total RNA was extracted as described above from mixed-type trichomes isolated from stem and petiole tissues, stem and petiole with intact trichomes, and stem and petiole tissue after removal of trichomes by rubbing frozen tissue by hand with a latex laboratory glove. RT of 1 μg of total RNA was performed using SuperScript II (Invitrogen) and an oligo(dT)12-18 primer. PCR amplification of select transcripts was performed using gene-specific primers (Supplemental Table S5) designed using GenBank or TIGR tomato TA (http://plantta.jcvi.org/) sequences. All reactions were performed for 20 cycles except for WRKY, where 25 cycles were used.
CAHS Expression and Enzyme Assay
The CAHS open reading frame (GenBank accession no. GU647162) was amplified using the forward primer 5′-ATGGCTAGCGCTTCTTCTTCTGCT-3′, which introduces an NheI site, and the reverse primer 5′-AAGCTTTCATATTTCGACAGACTCA-3′, which creates a HindIII site. The amplified fragment was subcloned into pGEM-Teasy (Promega) for sequence verification, followed by subcloning into the NheI and HindIII sites of the expression vector pET28b (Novagen), creating a fusion open reading frame with an N-terminal 6× His tag. The construct was mobilized into Escherichia coli Rosetta-gami 2 (Novagen) cells, and His-tagged recombinant CAHS was purified as follows. A 500-mL culture was grown in terrific broth medium supplemented with 12.5 μg mL−1 tetracycline, 50 μg mL−1 streptomycin, 34 μg mL−1 chloramphenicol, and 15 μg mL−1 kanamycin at 37°C with shaking at 200 rpm to an optical density at 600 nm of 0.4. Expression of CAHS was induced with 0.1 mm isopropylthio-β-galactoside, and the culture was grown for 20 h at 25°C with shaking at 120 rpm. Cells were harvested by centrifugation and resuspended in lysis buffer (50 mm HEPES, pH 8.0, 5% glycerol, 100 mm KCl, and 7.5 mm MgCl2) followed by sonication. His-tagged CAHS was purified from the cleared lysate by nickel affinity chromatography (Qiagen) according to the manufacturer's protocols. Protein measurements were performed using a bicinchoninic acid assay (Pierce) using bovine serum albumin as a standard.
For terpene product identification, purified CAHS (2 μg) was incubated in a sealed glass vial in lysis buffer containing 10 μm FPP in a final volume of 100 μL at 30°C for 30 min. The gas phase of the glass vials was then extracted with a solid-phase microextraction fiber (polydimethylsiloxane/divinylbenzene; 65 μm; Supelco) for 5 min at 42°C and analyzed by GC-MS. The GC analysis was as described above with modifications as follows: inlet set at 250°C; and oven at 50°C for 0.5 min, 40°C min−1 to 250°C, hold for 0.3 min. The mass spectrometer was set to scan m/z range 45 to 350. Blanks with buffer without enzyme, boiled enzyme controls, and commercial standards were also run to confirm the identity of peaks.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GU647162 (CAHS), SRP000732 (454 sequences), and PSE138 (proteomics data).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Mass spectra of sesquiterpene standards and reaction products.
Supplemental Table S1. List of assembled 454 contigs and associated annotation, closest TIGR TA BLASTN match, number of ESTs in TA, number of reads in 454 contig, percentage of total ESTs in each assembly, and ratio of percentage totals for each assembly.
Supplemental Table S2. List of the 3,052 contigs from 454 sequencing that did not have a significant BLASTN match to a TIGR TA (E-value < 1e−10).
Supplemental Table S3. List of 1,973 proteins identified from total and type VI proteins using the assembled 454 contigs.
Supplemental Table S4. List of 1,552 proteins after manually combining records where 454 contigs matched the same TA.
Supplemental Table S5. Primers used for expression analysis by RT-PCR.
Supplementary Material
Acknowledgments
EST sequencing and proteomics experiments were performed with help from the Michigan State University Research Technology Support Facility (RTSF). We thank RTSF member Doug Whitten for assistance with proteomics data analysis. Amanda Charbonneau, Eran Pichersky, Ines Schauvinhold, and Jeremy Kapteyn, members of the Solanum Trichome Project (www.trichome.msu.edu), provided helpful support during the project. We are grateful to the International Tomato Sequencing Consortium for providing genome sequence assemblies to the public prior to publication.
References
- Alvarez S, Marsh EL, Schroeder SG, Schachtman DP. (2008) Metabolomic and proteomic changes in the xylem sap of maize under drought. Plant Cell Environ 31: 325–340 [DOI] [PubMed] [Google Scholar]
- Amme S, Rutten T, Melzer M, Sonsmann G, Vissers JPC, Schlesier B, Mock HP. (2005) A proteome approach defines protective functions of tobacco leaf trichomes. Proteomics 5: 2508–2518 [DOI] [PubMed] [Google Scholar]
- Bertea CM, Voster A, Verstappen FWA, Maffei M, Beekwilder J, Bouwmeester HJ. (2006) Isoprenoid biosynthesis in Artemisia annua: cloning and heterologous expression of a germacrene A synthase from a glandular trichome cDNA library. Arch Biochem Biophys 448: 3–12 [DOI] [PubMed] [Google Scholar]
- Besser K, Harper A, Welsby N, Schauvinhold I, Slocombe S, Li Y, Dixon RA, Broun P. (2009) Divergent regulation of terpenoid metabolism in the trichomes of wild and cultivated tomato species. Plant Physiol 149: 499–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Shrestha RP, Whitten D, Wilkerson CG, Carr KM, Froehlich JE, Weber APM. (2008) Low-coverage massively parallel pyrosequencing of cDNAs enables proteomics in non-model species: comparison of a species-specific database generated by pyrosequencing with databases from related species for proteome analysis of pea chloroplast envelopes. J Biotechnol 136: 44–53 [DOI] [PubMed] [Google Scholar]
- Brechenmacher L, Lee J, Sachdev S, Song Z, Nguyen THN, Joshi T, Oehrle N, Libault M, Mooney B, Xu D, et al. (2009) Establishment of a protein reference map for soybean root hair cells. Plant Physiol 149: 670–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke CC, Wildung MR, Croteau R. (1999) Geranyl diphosphate synthase: cloning, expression, and characterization of this prenyltransferase as a heterodimer. Proc Natl Acad Sci USA 96: 13062–13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier SC, Panis B, Vertommen A, Swennen R, Sergeant K, Renaut J, Laukens K, Witters E, Samyn B, Devreese B. (2008) Proteome analysis of non-model plants: a challenging but powerful approach. Mass Spectrom Rev 27: 354–377 [DOI] [PubMed] [Google Scholar]
- Chen G, Hackett R, Walker D, Taylor A, Lin Z, Grierson D. (2004) Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol 136: 2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung F, Haas B, Goldberg S, May G, Xiao Y, Town C. (2006) Sequencing Medicago truncatula expressed sequenced tags using 454 Life Sciences technology. BMC Genomics 7: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs KL, Hamilton JP, Zhu W, Ly E, Cheung F, Wu H, Rabinowicz PD, Town CD, Buell CR, Chan AP. (2007) The TIGR Plant Transcript Assemblies database. Nucleic Acids Res 35: D846–D851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby SM, Alonso WR, Katahira EJ, McGarvey DJ, Croteau R. (1993) 4S-limonene synthase from the oil glands of spearmint (Mentha spicata): cDNA isolation, characterization, and bacterial expression of the catalytically active monoterpene cyclase. J Biol Chem 268: 23016–23024 [PubMed] [Google Scholar]
- Colby SM, Crock J, Dowdle-Rizzo B, Lemaux PG, Croteau R. (1998) Germacrene C synthase from Lycopersicon esculentum cv. VFNT Cherry tomato: cDNA isolation, characterization, and bacterial expression of the multiple product sesquiterpene cyclase. Proc Natl Acad Sci USA 95: 2216–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembinsky D, Woll K, Saleem M, Liu Y, Fu Y, Borsuk LA, Lamkemeyer T, Fladerer C, Madlung J, Barbazuk B, et al. (2007) Transcriptomic and proteomic analyses of pericycle cells of the maize primary root. Plant Physiol 145: 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Zamir D. (1994) A genomic library of Lycopersicon pennellii in L. esculentum: a tool for fine mapping of genes. Euphytica 79: 175–179 [Google Scholar]
- Eshed Y, Zamir D. (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faurobert M, Mihr C, Bertin N, Pawlowski T, Negroni L, Sommerer N, Causse M. (2007) Major proteome variations associated with cherry tomato pericarp development and ripening. Plant Physiol 143: 1327–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Sanchez I, Verpoorte R. (2008) Secondary metabolism in cannabis. Phytochem Rev 7: 615–639 [Google Scholar]
- Fridman E, Wang J, Iijima Y, Froehlich JE, Gang DR, Ohlrogge J, Pichersky E. (2005) Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant Cell 17: 1252–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang DR, Wang J, Dudareva N, Nam KH, Simon JE, Lewinsohn E, Pichersky E. (2001) An investigation of the storage and biosynthesis of phenylpropenes in sweet basil. Plant Physiol 125: 539–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL. (2010) Broad connections in the Arabidopsis seed metabolic network revealed by metabolite profiling of an amino acid catabolism mutant. Plant J 61: 579–590 [DOI] [PubMed] [Google Scholar]
- Hagel JM, Yeung EC, Facchini PJ. (2008) Got milk? The secret life of laticifers. Trends Plant Sci 13: 631–639 [DOI] [PubMed] [Google Scholar]
- Heitz T, Bergey DR, Ryan CA. (1997) A gene encoding a chloroplast-targeted lipoxygenase in tomato leaves is transiently induced by wounding, systemin, and methyl jasmonate. Plant Physiol 114: 1085–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Lee GI, Itoh A, Li L, DeRocher AE. (2000) Cytochrome P450-dependent metabolism of oxylipins in tomato: cloning and expression of Allene Oxide Synthase and Fatty Acid Hydroperoxide Lyase. Plant Physiol 123: 711–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Schilmiller AL. (2002) Oxylipin metabolism in response to stress. Curr Opin Plant Biol 5: 230–236 [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. (1999) CAP3: a DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y, Davidovich-Rikanati R, Fridman E, Gang DR, Bar E, Lewinsohn E, Pichersky E. (2004a) The biochemical and molecular basis for the divergent patterns in the biosynthesis of terpenes and phenylpropenes in the peltate glands of three cultivars of basil. Plant Physiol 136: 3724–3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y, Gang DR, Fridman E, Lewinsohn E, Pichersky E. (2004b) Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiol 134: 370–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Shi F, Jones AD, Marks MD, Howe GA. (2010) Distortion of trichome morphology by the hairless mutation of tomato affects leaf surface chemistry. J Exp Bot 61: 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Koeduka T, Fridman E, Gang DR, Vassao DG, Jackson BL, Kish CM, Orlova I, Spassova SM, Lewis NG, Noel JP, et al. (2006) Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc Natl Acad Sci USA 103: 10128–10133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Wildung MR, Stauber EJ, Sanchez C, Pouchnik D, Croteau R. (2000) Probing essential oil biosynthesis and secretion by functional evaluation of expressed sequence tags from mint glandular trichomes. Proc Natl Acad Sci USA 97: 2934–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Cooper B. (2006) Alternative workflows for plant proteomic analysis. Mol Biosyst 2: 621–626 [DOI] [PubMed] [Google Scholar]
- Lister R, Gregory BD, Ecker JR. (2009) Next is now: new technologies for sequencing of genomes, transcriptomes, and beyond. Curr Opin Plant Biol 12: 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckwill L. (1943) The Genus Lycopersicon: A Historical, Biological, and Taxonomic Survey of the Wild and Cultivated Tomatoes. Aberdeen University Press, Aberdeen, Scotland [Google Scholar]
- Matsui K. (2006) Green leaf volatiles: hydroperoxide lyase pathway of oxylipin metabolism. Curr Opin Plant Biol 9: 274–280 [DOI] [PubMed] [Google Scholar]
- Nagel J, Culley LK, Lu Y, Liu E, Matthews PD, Stevens JF, Page JE. (2008) EST analysis of hop glandular trichomes identifies an O-methyltransferase that catalyzes the biosynthesis of xanthohumol. Plant Cell 20: 186–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. (2006) Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci USA 103: 18928–18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B, et al. (2003) TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19: 651–652 [DOI] [PubMed] [Google Scholar]
- Rios-Estepa R, Turner GW, Lee JM, Croteau RB, Lange BM. (2008) A systems biology approach identifies the biochemical mechanisms regulating monoterpenoid essential oil composition in peppermint. Proc Natl Acad Sci USA 105: 2818–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley SD, Last RL. (2010) Shotguns and SNPs: how fast and cheap sequencing is revolutionizing plant biology. Plant J 61: 922–927 [DOI] [PubMed] [Google Scholar]
- Schad M, Lipton MS, Giavalisco P, Smith RD, Kehr J. (2005) Evaluation of two-dimensional electrophoresis and liquid chromatography-tandem mass spectrometry for tissue-specific protein profiling of laser-microdissected plant samples. Electrophoresis 26: 2729–2738 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Last RL, Pichersky E. (2008) Harnessing plant trichome biochemistry for the production of useful compounds. Plant J 54: 702–711 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Schauvinhold I, Larson M, Xu R, Charbonneau AL, Schmidt A, Wilkerson C, Last RL, Pichersky E. (2009) Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc Natl Acad Sci USA 106: 10865–10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Shi F, Kim J, Charbonneau AL, Holmes D, Jones AD, Last RL. (2010) Mass spectrometry reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J 62: 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheoran IS, Ross ARS, Olson DJH, Sawhney VK. (2007) Proteomic analysis of tomato (Lycopersicon esculentum) pollen. J Exp Bot 58: 3525–3535 [DOI] [PubMed] [Google Scholar]
- Shepherd RW, Bass WT, Houtz RL, Wagner GJ. (2005) Phylloplanins of tobacco are defensive proteins deployed on aerial surfaces by short glandular trichomes. Plant Cell 17: 1851–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. (1996) Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Slocombe SP, Schauvinhold I, McQuinn RP, Besser K, Welsby NA, Harper A, Aziz N, Li Y, Larson TR, Giovannoni J, et al. (2008) Transcriptomic and reverse genetic analyses of branched-chain fatty acid and acyl sugar production in Solanum pennellii and Nicotiana benthamiana. Plant Physiol 148: 1830–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh KH, Polichuk DR, Reed DW, Nowak G, Covello PS. (2006) Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett 580: 1411–1416 [DOI] [PubMed] [Google Scholar]
- Thipyapong P, Joel DM, Steffens JC. (1997) Differential expression and turnover of the tomato polyphenol oxidase gene family during vegetative and reproductive development. Plant Physiol 113: 707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingey WM. (1991) Potato glandular trichomes: defensive activity against insect attack. Hedin PA, , Naturally Occurring Pest Bioregulators. American Chemical Society, Washington, DC, pp 126–135 [Google Scholar]
- Trapp S, Croteau R. (2001) Defensive resin biosynthesis in conifers. Annu Rev Plant Physiol Plant Mol Biol 52: 689–724 [DOI] [PubMed] [Google Scholar]
- van der Hoeven RS, Monforte AJ, Breeden D, Tanksley SD, Steffens JC. (2000) Genetic control and evolution of sesquiterpene biosynthesis in Lycopersicon esculentum and L. hirsutum. Plant Cell 12: 2283–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GJ, Wang E, Shepherd RW. (2004) New approaches for studying and exploiting an old protuberance, the plant trichome. Ann Bot (Lond) 93: 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Wang R, DeParasis J, Loughrin JH, Gan S, Wagner GJ. (2001) Suppression of a P450 hydroxylase gene in plant trichome glands enhances natural-product-based aphid resistance. Nat Biotechnol 19: 371–374 [DOI] [PubMed] [Google Scholar]
- Weber APM, Weber KL, Carr K, Wilkerson C, Ohlrogge JB. (2007) Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol 144: 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werker E. (2000) Trichome diversity and development. Adv Bot Res 31: 1–35 [Google Scholar]
- Wienkoop S, Zoeller D, Ebert B, Simon-Rosin U, Fisahn J, Glinski M, Weckwerth W. (2004) Cell-specific protein profiling in Arabidopsis thaliana trichomes: identification of trichome-located proteins involved in sulfur metabolism and detoxification. Phytochemistry 65: 1641–1649 [DOI] [PubMed] [Google Scholar]
- Xie Z, Kapteyn J, Gang DR. (2008) A systems biology investigation of the MEP/terpenoid and shikimate/phenylpropanoid pathways points to multiple levels of metabolic control in sweet basil glandular trichomes. Plant J 54: 349–361 [DOI] [PubMed] [Google Scholar]
- Yu H, Kowalski SP, Steffens JC. (1992) Comparison of polyphenol oxidase expression in glandular trichomes of Solanum and Lycopersicon species. Plant Physiol 100: 1885–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Teoh KH, Reed DW, Maes L, Goossens A, Olson DJH, Ross ARS, Covello PS. (2008) The molecular cloning of artemisinic aldehyde Δ11(13) reductase and its role in glandular trichome-dependent biosynthesis of artemisinin in Artemisia annua. J Biol Chem 283: 21501–21508 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zhang W, Stanley BA, Assmann SM. (2008) Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 20: 3210–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Dai S, McClung S, Yan X, Chen S. (2009) Functional differentiation of Brassica napus guard cells and mesophyll cells revealed by comparative proteomics. Mol Cell Proteomics 8: 752–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulak KG, Khan MF, Alcantara J, Schriemer DC, Facchini PJ. (2009) Plant defense responses in opium poppy cell cultures revealed by liquid chromatography-tandem mass spectrometry proteomics. Mol Cell Proteomics 8: 86–98 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.