Abstract
Core histones are susceptible to a variety of post-translational modifications (PTMs), among which methylation and acetylation play critical roles in various chromatin-dependent processes. The nature and biological functions of these PTMs have been extensively studied in plants, animals and yeasts. In contrast, the histone modifications in Neurospora crassa, a convenient model organism for multicellular eukaryotes, remained largely undefined. In the present study, we used several mass spectrometric techniques, coupled with HPLC separation and multiple protease digestion, to identify the methylation and acetylation sites in core histones isolated from Neurospora. Electron transfer dissociation (ETD) was employed to fragment the heavily modified long N-terminal peptides. In addition, accurate mass measurement of fragment ions allowed for unambiguous differentiation of modification by acetylation or tri-methylation. Many modification sites conserved in other organisms were identified in Neurospora. In addition, some unique modification sites in histone H2B, including N-terminal α methylation, methylation at K3 and acetylation at K19, K28 and K29, were observed. Our analysis provides a potentially comprehensive picture of methylation and acetylation of core histones in Neurospora, which should serve as a foundation for future studies on the function of histone PTMs in this model organism.
The eukaryotic nucleosome, the fundamental unit of chromatin, plays an important role in packaging and organizing the genetic material (1). Each nucleosome consists of 146 bp of DNA wrapped around an octameric core histone complex consisting of two H2A-H2B dimers flanking a (H3-H4)2 tetramer (1, 2). All core histones have a basic N-terminal domain, a globular domain organized by the histone fold and a C-terminal tail (1, 2). The histone N-terminal tails, which extend out from the core particle, are involved in the establishment of chromatin structural states, whereas their histone fold domains mediate histone-histone and histone-DNA interactions (2).
Core histones are susceptible to a variety of post-translational modifications (PTMs), which include methylation, acetylation, phosphorylation, ubiquitination, SUMOylation and ADP-ribosylation. These modifications, which occur mainly on the N-terminal tails (3, 4), can affect interactions of nucleosomes with trans-acting factors, and are thought to play a role in the assembly and disassembly of chromatin states, and can control the accessibility of DNA for important cellular processes including transcription, replication, gene silencing and DNA repair (5-9). Distinct modifications of the histone tails can recruit specific chromatin-binding proteins, and modifications on the same or different histone tails may be interdependent and generatevarious combinations on any individual nucleosome (10-12).
Mass spectrometry has been widely used for assessing histone PTMs. It provides direct information about the sites and types of modifications, differentiates isobaric modifications (e.g., acetylation vs. tri-methylation) (13), and allows for quantitative analysis (14). PTM information obtained by mass spectrometric analysis facilitates genome-wide functional studies and provides the foundation for studies of modifications in specific regions of genomes using chromatin immunoprecipitation (ChIP) with antibodies recognizing specifically modified histones (15-17).
Neurospora crassa is a convenient model eukaryote, showing genomic features lacking in some other simple eukaryotes (e.g. yeast), including DNA methylation and certain histone modifications (e.g. H3K27 methylation). Studies using this organism have contributed significantly to the fundamental understanding of circadian rhythms, DNA methylation, genome defense systems, mitochondrial protein import, post-transcriptional gene silencing, DNA repair and other processes (18). Being a multicellular filamentous fungus, Neurospora also provides a system to study cellular differentiation and development (19). However, no comprehensive investigation of the PTMs of Neurospora core histones has been reported.
In the present study, we extracted core histones from N. crassa and systematically mapped histone methylation and acetylation, using a combination of digestion with various proteases and analyses with multiple types of mass spectrometers. Our results allowed for the identification of acetylation and methylation sites of lysine residues that were found previously in Arabidopsis thaliana, Saccharomyces cerevisiae and humans. More importantly, we identified several unique acetylation and methylation sites in core histones of Neurospora. Our analysis on core histone PTMs provides a foundation for examining the regulation of histone modifications and for genome-wide functional studies in this model organism.
Experimental Procedures
Extraction of core histones from Neurospora crassa
Neurospora crassa was cultured as described previously (20-22) and stored at -80°C. The core histones were obtained by using procedures reported for nuclei isolation by Emmett et al. (23) and histone extraction by Goff (24) with some modifications. Briefly, frozen Neurospora tissue (5 g) was ground to fine powder with a pestle in a cold mortar under liquid nitrogen. To the powder was subsequently added 20 mL nuclei isolation buffer containing 0.3 M sucrose, 40 mM HaHSO3, 25 mM Tris-HCl (pH 7.4), 10 mM MgSO4, 0.5 mM EDTA, 0.5% NP40, and the suspension was stirred vigorously. Cells were disrupted by intermittent exposure of the homogenate to sonication. The resulting mixture was filtered through two layers of silk in a Büchner funnel to remove whole cells and cell debris.
The above filtrate was centrifuged at 7,500g for 10 min and the supernatant was removed. To the precipitate was subsequently added 50 mL nuclei isolation buffer, and the resulting mixture was gently shaked for 30 min and centrifuged again. The precipitate, which contained the nuclei, was resuspended in 1-mL ice-cold NaHSO3 solution (25 mM, pH 7.2), followed by brief sonication (5 s). Hydrochloric acid was added immediately into the suspension to a final concentration of 0.3 M, and the resulting mixture was incubated at 4°C for 1 hr with continuous vortexing on an automatic vortexing machine. The histones in the supernatant were precipitated with cold acetone, centrifuged, dried and redissolved in water.
HPLC separation and protease digestion
Core histones were isolated by HPLC on an Agilent 1100 system (Agilent Technologies, Santa Clara, CA) as described previously (25). A 4.6×250 mm C4 column (Grace Vydac, Hesperia, CA) was used. The wavelength for the UV detector was 220 nm. The flow rate was 0.8 mL/min, and a 60-min linear gradient of 30-60% acetonitrile in 0.1% trifluoroacetic acid (TFA) was employed.
In order to obtain high sequence coverage of the proteins, purified histones were digested separately with several proteases, including trypsin, Arg-C, Glu-C, Asp-N, and chymotrypsin. A protein/enzyme ratio of 50:1 (w/w) was employed for trypsin and 20:1 for other proteases. The different buffers used for the digestions were 100 mM NH4HCO3 (pH 8.0) for trypsin, Arg-C or Glu-C; 50 mM sodium phosphate (pH 8.0) for Asp-N; and 100 mM Tris-HCl (pH 7.8) along with 10 mM CaCl2 for chymotrypsin. The digestion was carried out overnight at room temperature for chymotrypsin and at 37°C for other proteases. The peptide mixtures were subjected directly to LC-MS/MS analysis, or to a further peptide fractionation by HPLC and then analyzed by MALDI-MS/MS on a Q-STAR instrument or ESI-MS/MS on an LTQ-Orbitrap (See below).
Peptide fractionation was performed on the same HPLC system with a Zorbax SB-C18 capillary column (0.5×150 mm, 5 μm in particle size, Agilent Technologies), and a 60-min gradient of 2-60% acetonitrile in 0.6% acetic acid was used. The flow rate was 10 μL/min.
Mass spectrometry
MALDI-MS/MS measurements were performed on a QSTAR XL quadrupole/time-of-flight instrument equipped with an o-MALDI ion source (Applied Biosystems, Foster City, CA). The laboratory collision energy applied for MS/MS varied from 50 to 75 eV depending on peptide sequences and modification levels. The collision gas was nitrogen.
LC-MS/MS experiments were performed on three different instruments, including a 6510 QTOF LC/MS system with HPLC-Chip Cube MS interface (Agilent Technologies), an LTQ linear ion trap mass spectrometer and an LTQ-Orbitrap XL mass spectrometer with electron transfer dissociation (ETD) capability (Thermo Electron Co., San Jose, CA). The same 60-min linear gradient of 2-60% acetonitrile in 0.1% formic acid was applied for peptide separation.
In the 6510 QTOF LC/MS system, the sample enrichment, desalting, and HPLC separation were carried out automatically on the Agilent HPLC-Chip with an integrated trapping column (40 nL) and a separation column (Zorbax 300SB-C18, 75 μm×150 mm, 5 μm in particle size). The Chip spray voltage (VCap) was set at 1950 V and varied depending on chip conditions. MS/MS experiments were carried out in either the data-dependent scan mode or the pre-selected ion mode. The width for precursor ion selection was 4 m/z units. The temperature and flow rate for the drying gas were 325°C and 4 L/min, respectively. Nitrogen was used as collision gas, and collision energy followed a linear equation with a slope of 3 V per 100 m/z units and an offset of 2.5 V. The raw data obtained in the data-dependent scan mode were converted to Mascot generic format files, and submitted to the Mascot Database search engine (Matrix Science, Boston, MA) for protein and PTM identification.
For LC-MS/MS analysis on the LTQ, peptides were separated with a Zorbax SB-C18 capillary column (0.5×150 mm, 5 μm in particle size, Agilent Technologies), and the mobile phases were delivered by the Agilent 1100 capillary HPLC pump at a flow rate of 6 μL/min. Helium was employed as the collision gas, and the normalized collision energy was 30%. The spray voltage was 4.5 kV, and the temperature for the ion transport tube was 275°C.
ETD spectra were acquired on an LTQ-Orbitrap (Thermo Electron Co., San Jose, CA). Both online LC-MS/MS and offline direct infusion analyses were performed. In brief, during the online LC-MS/MS analysis, samples were redissolved in 15 μL of 0.1% formic acid and loaded to a Biobasic C18 Picofrit capillary column (75 μm×100 mm, 15 μm in particle size, New Objective, Woburn, MA) at a flow rate 0.3 μL/min. The ETD reaction time was 100 ms. In the offline analysis, the pre-fractionated H2B and H4 N-terminal peptides were redissolved in 20 μL 50:50 acetonitrile:H2O with 0.1% formic acid. Samples were directly infused. Different ETD reaction time was used to optimize peptide fragmentation.
Results
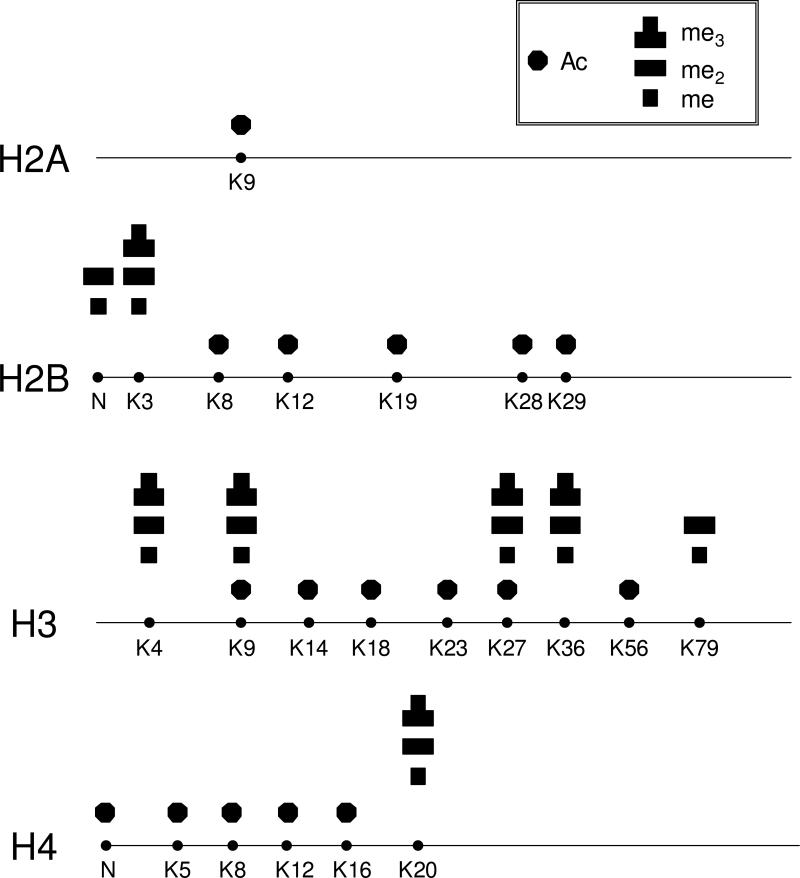
Neurospora crassa is a convenient model for multicellular eukaryotes, and it has been frequently used for studying the regulation of various cellular processes (19). To improve the foundation of information on chromatin structure and function in Neurospora, we initiated a systematic investigation of the PTMs of core histones in this organism. We first extracted core histones from Neurospora tissues and fractionated individual core histones by using reverse-phase HPLC. The core histones were eluted in the order of H2B, H4, H2A, and H3 (Figure S1). We then digested the core histones individually with different proteases and analyzed the peptide mixtures with LC-MS/MS on various instrument platforms to obtain high sequence coverage and achieve unambiguous PTM assignment. The detected PTM sites are summarized in Figure 1 and the sequence coverage is shown in Figure S2.
Figure 1.
Summaries of the detected PTMs of Neurospora core histones. The modified residues are labeled, and “N” represents N terminus. Acetylation is designated with solid octagon, and mono-, di-, and tri-methylation are represented by one-, two, and three square boxes, respectively.
Identification of PTMs in histones H2B and H2A
Isolated histone H2B was digested with trypsin, Asp-N and Glu-C separately to obtain peptides in appropriate lengths and good sequence coverage. The digestion mixtures were subjected subsequently to LC-MS/MS analysis, and the acquired mass spectra were searched with Mascot search engine and the results manually verified. A sequence coverage of 100% was reached and multiple modification sites were identified.
Acetylation of histone H2B was reported for several organisms, including human, budding yeast, and Arabidopsis (26-29). Here we identified many acetylation sites, including K7 and K12, which appear acetylated among different organisms, and K19, K28 and K29, which have not been reported.
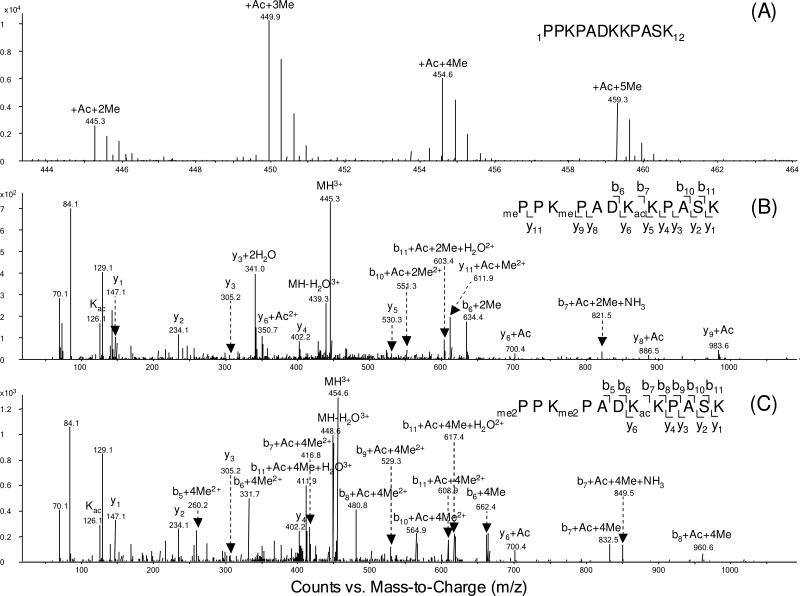
Aside from acetylation, we also observed methylation of K3 and of the N-terminus of Neurospora H2B. Example mass spectra for the H2B N-terminal peptide 1PPKPADKKPASK12 are depicted in Figure 2 and Figure S3. The positive-ion ESI-MS (Figure 2A) revealed the presence of one acetyl group and 2, 3, 4, or 5 methyl groups in this peptide segment. The MS/MS of this group of peptides were all obtained with the selected-ion monitoring (SIM) mode of analysis. The modification sites could be easily determined from fragment ions, with the consideration of mass shifts introduced by PTMs, e.g. 14.0157 Da for monomethylation and 42.0106 Da for acetylation. MS/MS results revealed that the N-terminus was mono- or dimethylated, K3 was mono-, di- or tri-methylated, and K7 was completely acetylated. In the MS/MS of the di-methylated and mono-acetylated peptide (Figure 2B), the existence of the b6+2Me, b10+Ac+2Me and y11+Ac+Me ions provides evidence of mono-methylation on the N-terminus and K3. We also observed y4, y5, y6+Ac ions, providing evidence for acetylation of K7. Similar methylation of an N-terminal proline has been observed in H2B from Drosophila melanogaster (30) and from gonads of the starfish Asterias rubens (31). Along this line, N-terminal alanine methylation was found for Tetrahymena histone H2B (32) and Arabidopsis histone H2B variants HTB-9 and HTB-11 (33). Moreover, N-terminal α-methylation of RCC1 protein is believed to promote stable association of RCC1 with chromatin through DNA binding in an α-methylation-dependent manner (34). This is required for correct spindle assembly and chromosome segregation during mitosis. The methylation at the N-terminus of H2B may have additional functions that are unknown.
Figure 2.
(A) ESI-MS of N-terminal tryptic peptide 1PPKPADKKPASK12 of histone H2B extracted from Neurospora. (B-C) The MS/MS of the di-methylated, mono-acetylated (B) and tetra-methylated, mono-acetylated (C) H2B peptide with residues 1-12 obtained by Q-TOF analysis.
Differentiation of tri-methylation from acetylation is essential in PTM studies of histones (13). A typical method to distinguish these two modifications is based on the immonium ion with m/z 126.1 from acetylated lysine and the neutral loss of a trimethylamine [N(CH3)3] from trimethyl lysine-containing precursor and fragment ions (13). However, the method does not work effectively with peptides containing more than one acetylation or tri-methylation sites, such as the H2B N-terminal peptide, which includes multiple modified lysines. In our study, we were able to differentiate these two modifications based on subtle difference in mass increase of the lysine residue induced by the acetylation and tri-methylation. For instance, the measured mass difference between the b6 and b7 ions observed in Figure 2C for the peptide segment including residues 1-12 was 832.5055-662.3982 = 170.1073 Da. This mass difference is closer to that expected for K7 acetylation (170.1056 Da, with a relative deviation of 9.9 ppm) than that expected for tri-methylation of K7 (170.1420 Da, with a relative deviation of 204 ppm). We therefore concluded that K7 is acetylated. All the acetylation and tri-methylation sites of Neurospora core histones were unambiguously established in this way, and more sample results are displayed in Table S1.
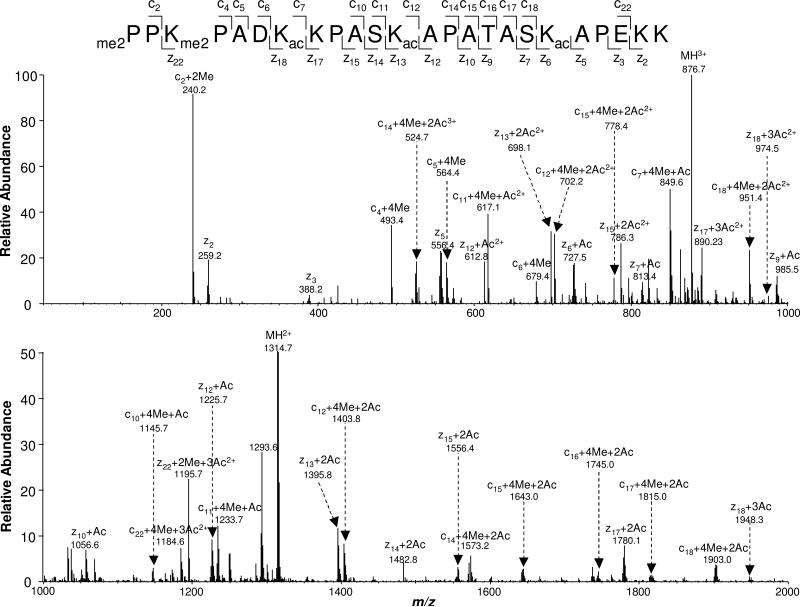
The presence of an acetylated lysine can block trypsin cleavage of its C-terminal side amide bond. With many lysine residues being acetylated, tryptic cleavage of H2B gives rise to a very long N-terminal peptide. Traditional collisionally induced dissociation (CID) cannot provide a complete series of fragment ions due to its poor ability to cleave the backbone of large peptides, rendering it difficult to identify modification sites. For instance, the exact numbers of methyl groups on the N-terminus and K3 cannot be delineated unambiguously based on the CID-produced MS/MS on the tetra-methylated peptide with residues 1-12 (Figure 2C). To overcome this limitation, we applied ETD, which can afford efficient cleavage along the backbone of long peptides or even intact proteins (35, 36), to analyze those large N-terminal peptides. Figure 3 shows the ETD MS/MS of the [M+5H]5+ ion of 1PPKPADKKPASKAPATASKAPEKK24 with the N-terminus and K3 being di-methylated, as well as with K7, K12 and K19 being acetylated. Nearly complete series of c and z ions were formed from ETD. The observation of c2+2Me and c4+4Me ions reveals the dimethylation of the N terminus and K3. The presence of the c6+4Me, c7+4Me+Ac, z17+2Ac, and z18+3Ac ions supported the K7 acetylation, while the existence of the c11+4Me+Ac, c12+4Me+2Ac, z12+Ac, z13+2Ac ions demonstrated the K12 acetylation. Along this line, the acetylation of K19 was identified based on the observation of the z5 and z6+Ac ions. K28 and K29 were also found to be acetylated heterogeneously in Neurospora H2B, as supported by the coexistence of y5, y5+Ac and b4, b4+Ac ions in the MS/MS of the Asp-N-produced peptide 25DAGKKTAASG34 (Figure S4A).
Figure 3.
The ETD MS/MS of the Neurospora H2B N-terminal peptide with residues 1-24 with the N-terminus and K3 being dimethylated, and with K7, K12 and K19 being acetylated.
Purified H2A was digested individually with trypsin, Arg-C or chymotrypsin under optimized conditions, and analyzed by LC-MS/MS. We found that only K9 was acetylated (MS/MS for the [M+2H]2+ ion of 6SGGKASGSKNAQSR19 is shown in Figure S4B, which reveals the formation of the b4+Ac, y10 and y11+Ac ions).
Identification of PTMs in histone H3
The HPLC-purified histone H3 was digested with Arg-C and Glu-C, and the digestion mixture was analyzed by LC-MS/MS directly, or further fractionated by HPLC and analyzed by MALDI-MS/MS. All the conserved methylation and acetylation sites reported in other organisms were identified in Neurospora, including methylation at K4, K9, K27, K36, K79 and acetylation at K9, K14, K18, K23, K27 and K56. K4 was found to be mono-, di- and tri-methylated, whereas K79 was found to be mono- and di-methylated in Neurospora (Figure S5A&B). In the MS/MS of the [M + 2H]2+ ion (m/z 366.7) of the tri-methylated peptide 3TKQTAR8 (Figure S5C), we observed the b3+3Me, b4+3Me, and y4 ions, supporting the observed tri-methylation of K4. MS/MS of the dimethylated peptide segment 73EIAQDFKSDLR83 displayed the presence of a complete series of y ions, revealing di-methylation of K79 (Figure S5D). Figure S5E illustrates the fragmentation of the precursor ion at m/z 646.9, which is consistent with the acetylation of K56 in peptide 54YQKSTELLIR63. In the MS/MS of the triply charged peptide 9KSTGGKAPRKQLASKAAR26, K9, K14, K18 and K23 were determined to be acetylated (Figure S5F), with the observation of b2+Ac ion for supporting K9 acetylation, the b5+Ac and b7+2Ac ions for K14 acetylation, as well as y1, y5+Ac, y11+2Ac ions for K18 and K23 acetylation.
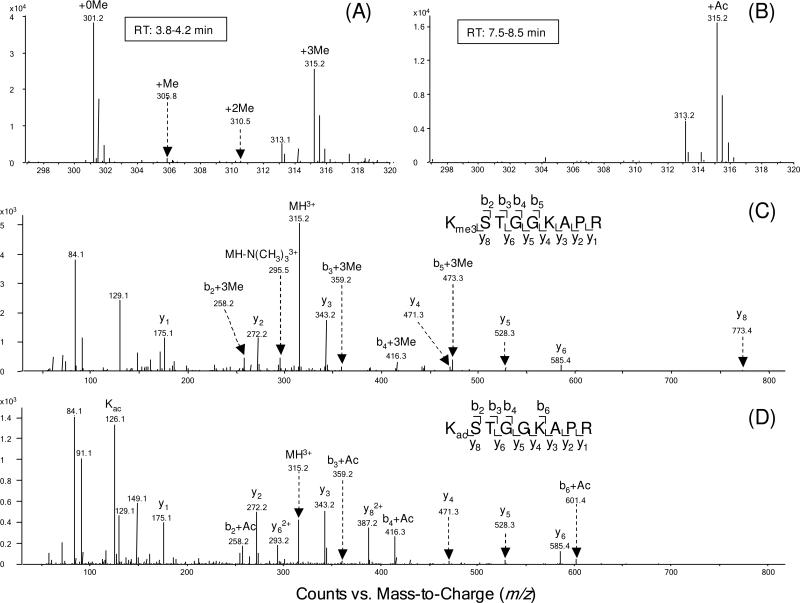
It is worth noting that K9 and K27 were found to be either methylated or acetylated. The methylated and acetylated peptides exhibited different retention time on the reverse-phase column used for LC-MS/MS analysis; the retention times for the methylated and acetylated peptides were approximately 4 and 8 min, respectively (Figure S6). Thus, methylated and acetylated peptides could be well isolated, and unambiguous MS/MS spectra could be obtained. Figure 4A shows the ESI-MS of the triply charged ions of the unmodified, mono-, di-, and trimethylated peptides with residues 9KSTGGKAPR17, while Figure 4B shows the MS for the acetylated peptide with same sequence. In the MS/MS of the [M+3H]3+ ions of the tri-methylated and acetylated peptides, a series of tri-methylated and acetylated b ions and unmodified y ions were observed, supporting the conclusion that K9 was tri-methylated and acetylated, respectively (Figure 4C&D). The presence of the immonium ion at m/z 126, the neutral loss of N(CH3)3 (59 Da) from the precursor ion, as well as the mass difference between tri-methylation and acetylation helped differentiate these two types of isobaric modifications.
Figure 4.
Positive-ion ESI-MS of the Arg-C-produced Neurospora H3 peptide 9KSTGGKAPR17 with K9 being methylated (A) or acetylated (B). Shown in (C) and (D) are the MS/MS, obtained on the Q-TOF mass spectrometer, of the tri-methylated and acetylated peptides with residues 9-17.
K27 was also found to be mono-, di- or tri-methylated or acetylated in Neurospora (Figure S7A, B). The MS/MS of the peptide 27KSAPSTGGVKKPHR42 with different modification levels were obtained by the selected-ion monitoring. The MS/MS of the [M+4H]4+ ion at m/z 384.2 eluted at 16 and 18 min revealed the tri-methylation on K36 as well as the trimethylation or acetylation on K27. In this context, the presence of the y7+3me, y8+3me, y9+3me, y10+3me and y11+3me, along with the observation of the y1, y2, and y4 ions, demonstrates the tri-methylation on K36 (Figure S7C, D). On the other hand, the acetylation and tri-methylation on K27 are manifested by the presence of b2 and b3 ions bearing an acetyl group and three methyl groups, respectively (Figure S7C, D).
We also attempted to examine the phosphorylation of core histones by enriching phosphorylated peptides using TiO2-coated magnetic beads [Phosphopeptide enrichment kit (PerkinElmer, Waltham, MA)] (37). We were able to detect very low level of phosphorylation of H3 S10 in a phosphatase-deficient (PP1-deficient) Neurospora strain (Figure S8) (20); H3 S10 phosphorylation in wild-type Neurospora was, however, below the detection limit of the instruments.
Identification of PTMs in histone H4
Purified H4 was digested by trypsin and Asp-N separately and subjected to LC- or MALDI-MS/MS analysis. All the modifications were located on the N-terminal segment, similar to observations for other organisms. Asp-N digestion produced a long N-terminal peptide with residues 1TGRGKGGKGLGKGGAKRHRKILR23, containing all the modification sites in H4, which include acetylation at the N-terminus and at K5, K8, K12 and K16, along with methylation at K20 (Figure S9A). A LTQ-Orbitrap with ETD provided high-quality tandem mass spectra of the [M+6H]6+ ions of the peptide with a nearly complete series of c and z ions. Figure S9B shows an ETD MS/MS of the di-acetylated and tri-methylated peptide with residues 1-23. The formation of the mono-acetylated small c ions and z20+3Me+Ac ions suggests N-terminal acetylation. Additionally, the presence of c15+Ac, c16+2Ac, z7+3Me, z8+3Me+Ac ions, and c19+2Ac, c20+2Ac+3Me, z3, z4+3Me ions supports the conclusions of K16 acetylation and K20 tri-methylation, respectively.
LC-MS/MS analysis, with the use of the QTOF mass spectrometer, of the tryptic digestion mixture led to the identification of relatively low levels of acetylation on K5, K8 and K12, which cannot be identified from the corresponding analyses of the Asp-N digestion mixture of histone H4. In the MS/MS of the doubly charged peptide 4GKGGKGLGK12, K5 and K8 were determined to be acetylated, as exemplified by the observations of the b2+Ac, b5+2Ac, y4, y5+Ac, and y7+Ac ions (Figure S10A). MS/MS of the doubly charged ion of the di-acetylated peptide 9GLGKGGAKR17 revealed the formation of b3, b4+Ac, y5+Ac and y6+2Ac ions supporting the K12 acetylation, and the presence of the y1 and y2+Ac ions demonstrating the acetylation of K16 (Figure S10B).
Discussion
We used a combination of various mass spectrometric methods including MALDI-TOF, LC-MS/MS with CID and ETD, coupled with different protease digestions and HPLC purification, to produce a thorough map of lysine methylation and acetylation sites in the core histones of the filamentous fungus Neurospora crassa. Our results show that the core histones in this organism are extensively acetylated and/or methylated on the lysine residues that have been found acetylated and/or methylated in mammals, budding yeast and plants. In addition, some novel modifications were detected. The methylation and acetylation sites found in Neurospora H2B include N-terminal and K3 methylation and K7, K12, K19, K28, K29 acetylation. Some of these sites, along with additional sites in the globular and C-terminal domain of the protein, were also found to be modified in the accompanying study, where the histone proteins were fractionated with two-dimensional AUT × AU polyacrylamide gels containing acetic acid/urea/Triton X-100 prior to mass spectrometric study (38). H2A was shown to have acetylation on K9. H3 was found to bear methylation on K4, K9, K27, K36 and K79, and to be subject to acetylation on K9, K14, K18, K23, K27 and K56. Finally, H4 was found to sport methylation of K20 and acetylation of the N-terminus, K5, K8, K12, and K16 (summarized in Figure 1). In this context, it is worth noting that some low levels of modification in Neurospora might not have been detected even with the combination of multiple protease digestions and various mass spectrometric techniques. A detailed comparison of known PTMs of core histones in different organisms is summarized in Table 1 (14, 26, 39, 40). The N-terminal and K3 methylation that we observed in H2B appears to be novel. H2A was found to be only acetylated on K9 in Neurospora, as in humans. As noted in Table 1, we found conserved acetylation of H2B at K7 and K12 (the corresponding residues in other organisms are K6 and K11, respectively), and unique acetylation of Neurospora H2B at K19, K28, and K29. It has been reported that acetylated lysine residues in yeast H2B activate the transcription of genes involved in NAD biosynthesis and vitamin metabolism (41). Thus, it will be interesting to assess the role of Neurospora H2B acetylation in transcription activation. Indeed, a detailed comparison of histone H2B from a histone deacetylase mutant and wild-type Neurospora revealed differences that might be responsible for defects in DNA methylation observed in this mutant (K. M. Smith, J. R. Dobosy, D. Do, J. E. Riefsnyder, D. C. Anderson, G. R. Green and E. U. Selker, in preparation; and the accompanying paper (38)).
Table 1.
Comparison of core histone methylation and acetylation among different organisms including Neurospora crassa, Saccharomyces cerevisiae, Arabidopsis thaliana and Homo sapiens (14, 26, 39, 40).
| Histones | Modifications | Modification sites | |||
|---|---|---|---|---|---|
| N. crassa | S. cerevisiae | A. thaliana | H. sapiens | ||
| H2B | Methylation | N terminus, K3 | |||
| Acetylation | K7, K12, K19, K28, K29 | K3, K6, K11, K16, K21, K22 | K6, K11, K27, K32 | K5, K11, K12, K15, K16, K23 | |
| H2A | Acetylation | K9 | K4, K7 | K5, K144 | K5, K9 |
| H3 | Methylation | K4, K9, K27, K36, K79 | K4, K36, K79 | K4, K9, K27, K36 | K4, K9, K27, K36, K79 |
| Acetylation | K9, K14, K18, K23, K27, K56 | K9, K14, K18, K23, K27, K56 | K9, K14, K18, K23 | K9, K14, K18, K23, K27 | |
| H4 | Methylation | K20 | K20 | K20 | |
| Acetylation | N terminus, K5, K8, K12, K16 | N terminus, K5, K8, K12, K16 | K5, K8, K12, K16 | N terminus, K5, K8, K12, K16 | |
In Neurospora H3, we found all the commonly conserved N-terminal modifications including methylation of K4, K9, K27 and K36, as well as acetylation of K9, K14, K18, K23 and K27. K4 was mostly unmodified, but approximately 10% of the K4 peptide was mono-, di- or tri-methylated. K9 was predominantly tri-methylated. K27 and K36 were mono-, di- and trimethylated. The methylated and acetylated peptides housing the same residues had different retention times during HPLC separation, allowing the modification types to be easily determined. Moreover, we observed acetylation on K56. The predominant form of H3 K9 methylation observed in our data, trimethylation, was previously shown to be enriched in regions of DNA methylation in the Neurospora genome (42, 43). The SET domain methyltransferase DIM-5 catalyzes this modification and is required for DNA methylation (44). Unlike plants and mammals, in Neurospora H3K36 mono-, di- and tri- methylation is catalyzed by a single enzyme, SET-2 (21). H3K36 methylation is enriched in coding regions of the genome and is required for normal vegetative and sexual development (21).
H3K79 is mono-, di- or tri-methylated in many mammalian and non-mammalian cell lines and in budding yeast. However, K79 was found only mono- and di-methylated in Neurospora. It would be interesting to explore the functional implications of the lack of K79 trimethylation, since the H3 K79 methyltransferase, DOT1, is involved in telomeric silencing in yeast (45). In addition, it is worth noting that K64 of H3 of mammalian cells was recently found to be tri-methylated; this methylation is associated with heterochromatin, and it is lost during developmental reprogramming (46). We monitored specifically the fragmentation of the peptide containing this putative modification by MS/MS but did not find this modification in Neurospora. It is possible that the level of this modification is below the detection limits of the method; conceivably, it could also have been lost during extraction process.
Histone H4 acetylation sites, including the N-terminus, and residues K5, K8, K12 and K16, are conserved in almost all organisms, including Neurospora. They play important roles in many processes including transcriptional activation, DNA double strand break repair and cellular lifespan regulation (47, 48). H4 K20 methylation, conserved in almost all multi-cellular organisms, was also found in Neurospora, in the form of mono-, di- and mainly tri-methylation. While its role in heterochromatin silencing and DNA damage response has been extensively studied in humans, Drosophila melanogaster and Schizosaccharomyces pombe (49-52), it is also important to study its function in Neurospora.
In summary, a systematic mapping of histone methylation and acetylation in Neurospora crassa was obtained by mass spectrometric analyses. The rigorous identification of modification sites provides a foundation for further studies on the regulation and functions of histone modifications in this model organism.
Supplementary Material
Acknowledgment
The authors wish to thank Dr. Lihua Jiang at Thermo for performing the ETD experiments and Dr. David C. Anderson for communicating his findings prior to publication.
Funding Support: This work was supported by the National Institutes of Health (grants GM068087, CA116522 and GM025690).
Abbreviations
- PTM
post-translational modification
- ETD
electron transfer dissociation
- ChIP
chromatin immunoprecipitation
- CID
collisionally induced dissociation
Footnotes
Supporting Information Available: HPLC trace for the fractionation of core histones, and LCMS/MS results. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.McGhee JD, Felsenfeld G. Nucleosome structure. Annu. Rev. Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 4.Berger SL. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 5.Geiman TM, Robertson KD. Chromatin remodeling, histone modifications, and DNA methylation-how does it all fit together? J. Cell. Biochem. 2002;87:117–125. doi: 10.1002/jcb.10286. [DOI] [PubMed] [Google Scholar]
- 6.Nacheva GA, Guschin DY, Preobrazhenskaya OV, Karpov VL, Ebralidse KK, Mirzabekov AD. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 1989;58:27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- 7.Irvine RA, Lin IG, Hsieh CL. DNA methylation has a local effect on transcription and histone acetylation. Mol. Cell. Biol. 2002;22:6689–6696. doi: 10.1128/MCB.22.19.6689-6696.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peterson CL, Laniel MA. Histones and histone modifications. Curr. Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 10.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 11.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell. Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 12.Fillingham J, Greenblatt JF. A histone code for chromatin assembly. Cell. 2008;134:206–208. doi: 10.1016/j.cell.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Zhang K, Yau PM, Chandrasekhar B, New R, Kondrat R, Imai BS, Bradbury ME. Differentiation between peptides containing acetylated or trimethylated lysines by mass spectrometry: an application for determining lysine 9 acetylation and methylation of histone H3. Proteomics. 2004;4:1–10. doi: 10.1002/pmic.200300503. [DOI] [PubMed] [Google Scholar]
- 14.Beck HC, Nielsen EC, Matthiesen R, Jensen LH, Sehested M, Finn P, Grauslund M, Hansen AM, Jensen ON. Quantitative proteomic analysis of post-translational modifications of human histones. Mol. Cell. Proteomics. 2006;5:1314–1325. doi: 10.1074/mcp.M600007-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 17.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 18.Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg RL, Perkins DD, Kroken S, Cogoni C, Macino G, Catcheside D, Li W, Pratt RJ, Osmani SA, DeSouza CP, Glass L, Orbach MJ, Berglund JA, Voelker R, Yarden O, Plamann M, Seiler S, Dunlap J, Radford A, Aramayo R, Natvig DO, Alex LA, Mannhaupt G, Ebbole DJ, Freitag M, Paulsen I, Sachs MS, Lander ES, Nusbaum C, Birren B. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 19.Davis RH, Perkins DD. Timeline: Neurospora: a model of model microbes. Nat. Rev. Genet. 2002;3:397–403. doi: 10.1038/nrg797. [DOI] [PubMed] [Google Scholar]
- 20.Adhvaryu KK, Selker EU. Protein phosphatase PP1 is required for normal DNA methylation in Neurospora. Genes Dev. 2008;22:3391–3396. doi: 10.1101/gad.1738008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adhvaryu KK, Morris SA, Strahl BD, Selker EU. Methylation of histone H3 lysine 36 is required for normal development in Neurospora crassa. Eukaryot. Cell. 2005;4:1455–1464. doi: 10.1128/EC.4.8.1455-1464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selker EU, Fritz DY, Singer MJ. Dense nonsymmetrical DNA methylation resulting from repeat-induced point mutation in Neurospora. Science. 1993;262:1724–1728. doi: 10.1126/science.8259516. [DOI] [PubMed] [Google Scholar]
- 23.Emmett N, Williams CM, Frederick L, Williams LS. Arginyl-transfer ribonucleic acid and synthetase of Neurospora crassa. Mycologia. 1972;64:499–509. [PubMed] [Google Scholar]
- 24.Goff CG. Histones of Neurospora crassa. J. Biol. Chem. 1976;251:4131–4138. [PubMed] [Google Scholar]
- 25.Xiong L, Ping L, Yuan B, Wang Y. Methyl group migration during the fragmentation of singly charged ions of trimethyllysine-containing peptides: precaution of using MS/MS of singly charged ions for interrogating peptide methylation. J. Am. Soc. Mass Spectrom. 2009;20:1172–1181. doi: 10.1016/j.jasms.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K, Sridhar VV, Zhu J, Kapoor A, Zhu JK. Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS One. 2007;2:e1210. doi: 10.1371/journal.pone.0001210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang KL. Characterization of acetylation of Saccharomyces cerevisiae H2B by mass spectrometry. Int. J. Mass Spectrom. 2008;278:89–94. [Google Scholar]
- 28.Clayton AL, Hebbes TR, Thorne AW, Cranerobinson C. Histone Acetylation and Gene Induction in Human-Cells. FEBS Lett. 1993;336:23–26. doi: 10.1016/0014-5793(93)81601-u. [DOI] [PubMed] [Google Scholar]
- 29.Grimes SR, Henderson N. Acetylation of rat testis histones H2B and Th2B. Dev. Biol. 1984;101:516–521. doi: 10.1016/0012-1606(84)90165-9. [DOI] [PubMed] [Google Scholar]
- 30.Desrosiers R, Tanguay RM. Methylation of Drosophila histones at proline, lysine, and arginine residues during heat shock. J. Biol. Chem. 1988;263:4686–4692. [PubMed] [Google Scholar]
- 31.Martinage A, Briand G, Van Dorsselaer A, Turner CH, Sautiere P. Primary structure of histone H2B from gonads of the starfish Asterias rubens. Identification of an N-dimethylproline residue at the amino-terminal. Eur. J. Biochem. 1985;147:351–359. doi: 10.1111/j.1432-1033.1985.tb08757.x. [DOI] [PubMed] [Google Scholar]
- 32.Nomoto M, Kyogoku Y, Iwai K. N-Trimethylalanine, a novel blocked N-terminal residue of Tetrahymena histone H2B. J. Biochem. 1982;92:1675–1678. doi: 10.1093/oxfordjournals.jbchem.a134096. [DOI] [PubMed] [Google Scholar]
- 33.Bergmuller E, Gehrig PM, Gruissem W. Characterization of post-translational modifications of histone H2B-variants isolated from Arabidopsis thaliana. J. Proteome Res. 2007;6:3655–3668. doi: 10.1021/pr0702159. [DOI] [PubMed] [Google Scholar]
- 34.Chen T, Muratore TL, Schaner-Tooley CE, Shabanowitz J, Hunt DF, Macara IG. N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat. Cell Biol. 2007;9:596–603. doi: 10.1038/ncb1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochim. Biophys. Acta. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Anderson DC, Green GR, Smith K, Selker EU. Neurospora crassa histone H2B is extensively methylated and some post-translational modifications are sensitive to histone deacetylase 1 inactivation. Biochemistry. 2010;49 doi: 10.1021/bi100391w. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, Shabanowitz J, Mishra N, Strahl BD, Allis CD, Hunt DF. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 40.Sinha I, Wiren M, Ekwall K. Genome-wide patterns of histone modifications in fission yeast. Chromosome Res. 2006;14:95–105. doi: 10.1007/s10577-005-1023-4. [DOI] [PubMed] [Google Scholar]
- 41.Parra MA, Kerr D, Fahy D, Pouchnik DJ, Wyrick JJ. Deciphering the roles of the histone H2B N-terminal domain in genome-wide transcription. Mol. Cell. Biol. 2006;26:3842–3852. doi: 10.1128/MCB.26.10.3842-3852.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- 43.Lewis ZA, Honda S, Khlafallah TK, Jeffress JK, Freitag M, Mohn F, Schubeler D, Selker EU. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 2009;19:427–437. doi: 10.1101/gr.086231.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 45.Ng HH, Feng Q, Wang HB, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daujat S, Weiss T, Mohn F, Lange UC, Ziegler-Birling C, Zeissler U, Lappe M, Schubeler D, Torres-Padilla ME, Schneider R. H3K64 trimethylation marks heterochromatin and is dynamically remodeled during developmental reprogramming. Nat. Struct. Mol. Biol. 2009;16:777–781. doi: 10.1038/nsmb.1629. [DOI] [PubMed] [Google Scholar]
- 47.Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 49.Fang J, Feng Q, Ketel CS, Wang HB, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 2002;12:1086–1099. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- 50.Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 2002;16:2225–2230. doi: 10.1101/gad.1014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 52.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.