Abstract
Here we describe a novel microarray platform that integrates all functions needed to perform any array-based experiment in a compact instrument on the researcher’s laboratory benchtop. Oligonucle otide probes are synthesized in situ via a light- activated process within the channels of a three-dimensional microfluidic reaction carrier. Arrays can be designed and produced within hours according to the user’s requirements. They are processed in a fully automatic workflow. We have characterized this new platform with regard to dynamic range, discrimination power, reproducibility and accuracy of biological results. The instrument detects sample RNAs present at a frequency of 1:100 000. Detection is quantitative over more than two orders of magnitude. Experiments on four identical arrays with 6398 features each revealed a mean coefficient of variation (CV) value of 0.09 for the 6398 unprocessed raw intensities indicating high reproducibility. In a more elaborate experiment targeting 1125 yeast genes from an unbiased selection, a mean CV of 0.11 on the fold change level was found. Analyzing the transcriptional response of yeast to osmotic shock, we found that biological data acquired on our platform are in good agreement with data from Affymetrix GeneChips, quantitative real-time PCR and—albeit somewhat less clearly—to data from spotted cDNA arrays obtained from the literature.
INTRODUCTION
Microarrays have become a standard tool in molecular biology that has revolutionized genomics research. Microarrays are used extensively for gene expression profiling (1,2) in many applications including the discovery of gene function (3,4), drug evaluation (4–6), pathway dissection (7), classification of clinical samples (8–10), exon mapping (11) and investigation of splicing events (12). Arrays may be produced either by deposition of presynthesized material (1,13–15) or by in situ oligonucleotide synthesis (16,17). DNA arrays manufactured by physical deposition of presynthesized material require labor-intensive preparation and record-keeping of DNA probes. In contrast, oligonucleotide arrays synthesized in situ using a photolithographic method (18) only require DNA sequence data. However, cost and time spent in generating the photolithographic masks render this approach as slow and inflexible as the deposition methods. Recently, more flexible microarray technologies have been developed. These employ either ink-jet printing (19) or micromirror devices (20,21) for in situ synthesis of customized oligonucleotide arrays. Although these techniques provide full flexibility with respect to the array design, the actual generation of the array and in some cases even the hybridization and detection steps are restricted to centralized manufacturer facilities. Again, the investigator’s flexibility remains limited. In addition, array synthesis and subsequent processing steps are not physically linked and require error-prone manual handling. The geniom platform described here is the first system to overcome these restrictions. The investigator gains full control of the complete workflow of any microarray experiment. The technology integrates microarray production, hybridization and detection in a compact benchtop unit. Automation of these processes and a powerful software interface allow the scientist to design and perform microarray-based experiments using sequence information derived from public databases. Microarrays are generated by in situ oligonucleotide synthesis via a light-activated process employing a digital micromirror device and highly efficient photochemistry (22,23). Instead of a conventional microscope slide, a truly micro-machined three-dimensional microstructure bearing four individual channel-like chambers (arrays) is used as a reaction carrier. This approach allows one to run several array experiments on a single carrier since up to four individual microarrays are generated and may be hybridized sequentially or in parallel. In contrast to the recently described maskless array synthesizer, which also uses a micromirror device for in situ oligonucleotide synthesis (20), geniom is highly automated and integrates all functions required to perform an array-based experiment within a single device on the investigator’s laboratory benchtop. A more detailed description of this technology is presented by Stähler et al. (24) and can also be found in Supplementary Material Figure 1.
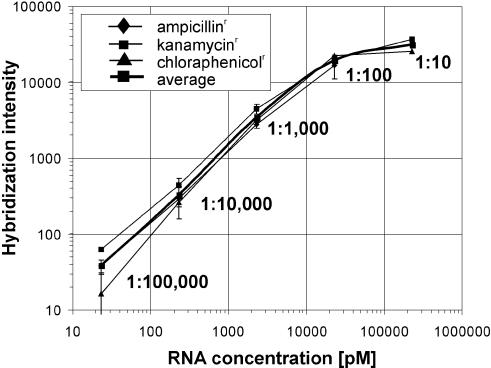
Figure 1.
Dynamic range of oligonucleotide arrays from the geniom platform. Log–log plot of the normalized hybridization intensity (average of the 20 PM–MM intensity differences for each gene) versus concentration for three different prokaryotic cRNA targets. The three cRNA targets (ampr, kanr, cmr) were spiked into labeled yeast cRNA at molar ratios of 1:100 000–1:100 and each dilution was measured six times. kanr and cmr cRNA was additionally spiked once at a molar ratio of 1:10. The error bars indicate the standard deviation calculated across the replicates after elimination of outliers.
In the study presented here, we characterized the geniom technology on a technical level with regard to dynamic range, discrimination power and reproducibility. In addition, we validated complex biological results acquired on the geniom platform by comparison with existing technologies and conventional standards. Analyzing the transcriptional response of Saccharomyces cerevisiae to osmotic shock, we found a good agreement of data obtained on geniom arrays, Affymetrix GeneChip data, and expression results obtained by quantitative real-time PCR. Our study also revealed a high concordance of geniom results and cDNA data from the literature (25). While the actual fold-change values are less consistent in this latter comparison, the vast majority of genes included in our study showed the same trend of regulation in both assay systems.
MATERIALS AND METHODS
Oligonucleotide arrays
Light-activated in situ oligonucleotide synthesis was performed essentially as described by Singh-Gasson et al. (20) using a digital micromirror device (Texas Instruments). The synthesis was performed within the geniom device on an activated three-dimensional reaction carrier consisting of a glass-silicon-glass sandwich (DNA processor; see Supplementary Material Fig. 1). Four individually accessible microchannels (referred to as arrays) etched into the silicon layer of the DNA processor are connected to the microfluidic system of the geniom device. Using standard DNA synthesis reagents (Proligo) and 3′-phosphoramidites carrying a 5′-photolabile protective group (22,23), oligonucleotides were synthesized in parallel in all four translucent arrays of one reaction carrier. Prior to synthesis, the glass surface was activated by coating with a spacer. The synthesized probe sets may be the same or different for all four arrays. Actually, the time needed for synthesis of standard arrays used in this study is independent of the number of different probe sets, the probe sequences and the number of probes synthesized within one probe set (current limit: 14 000 features per array; corresponding to 4 × 14 000 = 56 000 features per reaction carrier). However, the probe length substantially influences synthesis time. According to the conservative protocol used in this study, the synthesis of four typical 25mer arrays (with 12 880 features each) takes ∼15.5 h (including 1.5 h for the final deprotection step). The yeast probe set (ten 25mer probes per transcript) was calculated based on the full genome sequence (retrieved online from http://genome-www.stanford.edu/Saccharomyces/) using a combination of sequence uniqueness criteria and rules for selection of oligonucleotides likely to hybridize with high specificity and sensitivity. The selection criteria were essentially as described in Lockhart et al. (2) with modifications for the longer probes used here (25mers instead of 20mers).
Yeast strain and growth conditions
Saccharomyces cerevisiae, wild-type strain W303-1A, MATa, ura3-52, trp1Δ2, leu2-3_112, his3-11, ade2-1, can1-100 (accession no. 20000A; EUROSCARF, Frankfurt a.M., Germany) was grown in 240 ml batch cultures at 30°C in YPD (1% yeast extract, 2% peptone, 2% glucose) to an A600 of 1.0. At this point, cells were collected for determination of expression profiles under baseline conditions. Osmotic stress was applied by adding prewarmed (30°C) 5 M NaCl to a final concentration of 0.7 M NaCl. Cells were collected 45 min after the addition of NaCl. Ten milliliters of suspension culture were chilled on ice, cells were pelleted, washed once with ice-cold water, frozen in a dry ice/ethanol bath and stored at –20°C until use.
RNA extraction and preparation for hybridization
Total RNA was extracted from frozen cell pellets using a hot phenol method (26). Amplification and labeling was achieved using a modification of the procedure first described by Van Gelder et al. (27) and Eberwine et al. (28). In brief, 5 µg of total RNA were used as a starting material and converted into double-stranded cDNA using an oligo(dT) primer with a 5′ T7 RNA polymerase promoter sequence and the Superscript II system for cDNA synthesis (Invitrogen). Double-stranded cDNA was purified by phenol–chloroform extraction followed by ethanol precipitation. Using the purified double-stranded cDNA as a template, in vitro transcription was performed using T7 RNA Polymerase (T7 Megascript Kit, Ambion) in the presence of a mixture of unlabeled ATP, CTP, GTP and UTP and biotin-labeled CTP and UTP [biotin-11-CTP (PerkinElmer); biotin-16-UTP (Roche)]. Biotinylated cRNA was purified on an affinity resin (RNeasy, Qiagen). The cRNA yield was determined by measuring the light absorbance at 260 nm (1 OD at 260 nm corresponds to 40 µg/ml RNA). Prior to hybridization, 15 µg of cRNA were fragmented randomly to an average length of ∼100 nt by incubating at 94°C for 35 min in a 5 µl volume of 40 mM Tris-acetate pH 8.1, 100 mM potassium acetate and 30 mM magnesium acetate. A detailed description of the labeling protocol will be provided upon request. Transcripts of the ampicillinr (ampr), kanamycinr (kanr) and chloramphenicolr (cmr) resistance genes used for the determination of the dynamic range were prepared as follows. Each gene was PCR amplified from a plasmid vector (ampr from pBR322; cmr from pDNR-LIB; kanr from pLP-GBKT7) and the PCR product was cloned into pBluescript II SK (+) (downstream of the T3 polymerase promotor sequence; between the BamHI and the EcoRI restriction sites). In addition, an A(50) sequence was inserted between the EcoRI and HindIII sites of the same vector (immediately downstream of the resistance gene). Run-off transcripts [with a 3′ A(50) tail] were generated using the T3 Megascript Kit (Ambion) and 1 µg of the HindIII-digested construct as a template. One microgram of the in vitro transcript was used as a template for cRNA synthesis as described above. Different amounts of the biotinylated cRNA were then spiked into the yeast cRNA samples (prior to fragmentation).
Array hybridization, detection and data analysis
Microarrays were hybridized with 15 µg of fragmented cRNA in a final volume of 20 µl. Hybridization solutions contained 100 mM MES (pH 6.6), 0.9 M NaCl, 20 mM EDTA and 0.01% (v/v) Tween-20 (referred to as MES-hyb). In addition, the solutions contained 0.1 mg/ml sonicated herring sperm DNA (Promega) and 0.5 mg/ml BSA (Sigma). RNA samples were heated in the hybridization solution to 95°C for 3 min followed by 45°C for 3 min before being placed in an array which had been prehybridized for 15 min with 1% (w/v) BSA in MES-hyb at RT. Hybridizations were carried out at 45°C for 16 h without agitation. After removing the hybridization solutions, arrays were first washed with non-stringent buffer [0.005% (v/v) Triton X-100 in 6× SSPE] for 20 min at 25°C and subsequently with stringent buffer [0.005% (v/v) Triton X-100 in 0.5× SSPE] for 20 min at 45°C. After washing, the hybridized RNA was fluorescence-stained by incubating with 10 µg/ml streptavidin–phycoerythrin (Molecular Probes) and 2 µg/µl BSA in 6× SSPE at 25°C for 15 min. Unbound streptavidin–phycoerythrin was removed by washing with non-stringent buffer for 20 min at 25°C. Detection and feature readout were performed using the CCD-based detection system of the geniom device (Cy3 filter set). Processing of raw data including background correction, array to array normalization and determination of gene expression levels as well as calculation of fold-change values were essentially as described by Zhou and Abagyan (29). All steps were carried out using the PROP algorithm of the geniom application software which is based on the MOID algorithm described by Zhou and Abagyan (29).
Affymetrix GeneChip reference data
Aliquots of the same biotinylated cRNA samples analyzed on the geniom platform were sent to a service provider. The samples were hybridized to Affymetrix yeast GeneChips (YG-S98) according to the protocol in the Affymetrix GeneChip Expression Manual. Starting from the raw data files (.cel files), analysis was performed using both the Affymetrix MAS4 algorithm (at the service provider) and the PROP algorithm (at febit).
Quantitative PCR
In vitro transcripts [with a 3′ A(50) tail] of ampr (250 pg), kanr (25 pg) and cmr (2.5 pg) were spiked into 5 µg of total RNA from yeast (control and treated). cRNA was prepared as described above but omitting the biotin labeling. The cRNA was then converted into cDNA using random hexamer primers and the Superscript II Kit. Quantitative PCR was performed using the iCycler iQ™ (Bio-Rad). Reactions contained ∼250 pg non-purified cDNA, 300 nM forward and reverse primers (designed using the DNAMAN software; sequences will be provided upon request) and 25 µl of 2× QuantiTect SYBR Green PCR Master Mix (Qiagen) in a final volume of 50 µl. Samples were incubated for 13.5 min at 95°C followed by 50 cycles of denaturation (30 s at 95°C), annealing (30 s at 62°C) and extension (45 s at 72°C). The data obtained were normalized using all three spike-in controls. Fold-change values were calculated taking the PCR efficiencies into account (30,31).
RESULTS
Dynamic range and discrimination power
Spiking experiments were performed to determine the dynamic range of oligonucleotide arrays processed on the geniom platform. Biotinylated cRNAs from three prokaryotic genes (antibiotic resistance genes: ampr, kanr, cmr) were mixed and spiked into 0.75 µg/µl biotinylated cRNA background from yeast total RNA at molar ratios of 1:100– 1:100 000. In addition, kanr and cmr cRNAs were spiked at a molar ratio of 1:10. Using an estimate of 15 000 copies of mRNA per yeast cell (32–34) a frequency of 1:100 000 corresponds to that of an mRNA present at a density of one copy per six to seven cells. In 15 µg of cRNA background and a hybridization volume of 20 µl, a frequency of 1:100 000 corresponds to a concentration of ∼22.7 pM and an absolute amount of 0.45 fmol (approximately 2.7 × 108 molecules or ∼0.15 ng) of specific RNA. Each combination of dilution and background was hybridized six times with the exception of the 1:10 ratios which do not reflect situations encountered in normal cells and therefore were hybridized only once. In order to ensure optimal comparability of the data generated with the geniom instrument to those from other in situ synthesized short oligonucleotide arrays that mostly include mismatch (MM) controls, all samples were hybridized to arrays containing 16 perfect match (PM)/MM probe pairs (25mers) for each of 100 randomly chosen yeast genes, and 20 PM/MM probe pairs (25mers) for each of the three prokaryotic genes, although the geniom application software does not necessarily require MM probes for gene expression analysis. The arrays had been pretested for cross-hybridization. Yeast probes cross-hybridizing to the spiked-in transcripts as well as probes designed for these transcripts cross-hybridizing to the yeast background had been removed.
As indicated in Figure 1, the hybridization intensity is linearly correlated to the cRNA target concentration in the range of 1:100 000–1:1000. In the range of 1:1000–1:100, the signal increases by a factor of approximately six rather than 10 because the probes immobilized on the array are beginning to saturate. Between 1:100 and 1:10, saturation proceeds and the hybridization signal only increases by a factor of 1.5. At a molar ratio of approximately 1:100 000, the critical level for the discrimination power of the system is reached. While the presence of the prokaryotic transcripts was detected above the background in 14 out of 18 experiments at this level (six replicate hybridizations for each of the three genes), the remaining four experiments (three times kanr and once ampr) indicate that a ratio of 1:100 000 is the threshold level for at least some probe sets. In experiments lacking the complex cRNA background, the transcripts could be detected at concentrations corresponding to a ratio of 1:1 000 000 (data not shown). The dynamic range of two to three orders of magnitude and the discrimination power of 1:100 000 measured here for arrays of the geniom platform compare very well to data obtained with other commercially available in situ synthesized (35) or spotted (13) oligonucleotide arrays. For the Affymetrix GeneChips a dynamic range of three to four orders of magnitude was initially reported (2). However, these data were obtained using a customized array containing probe sets with more than 500 PM/MM probe pairs per transcript. In a more recent study on commercial GeneChips with 20 PM/MM probe pairs per gene, a linear relationship between transcript abundance and signal intensity was observed at ratios of 1:150 000–1:15 000. Linearity ceased above the 1:15 000 ratio and saturation emerged around the 1:150 level (36).
Reproducibility of raw data
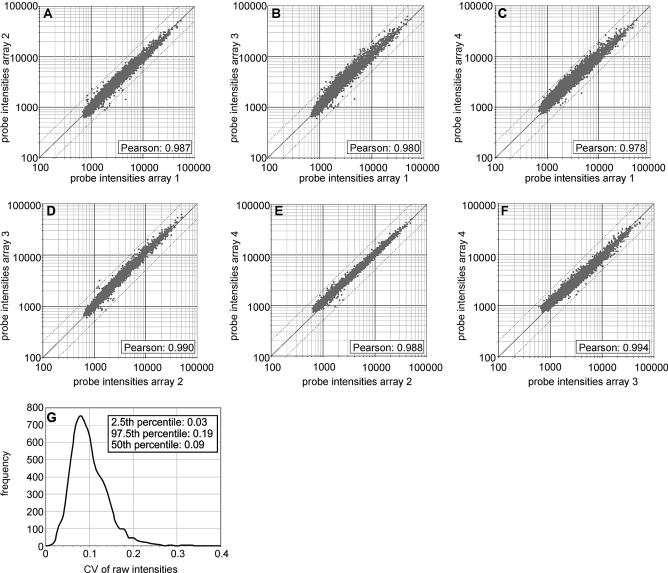
Replicate experiments were performed to determine the reproducibility of array synthesis, hybridization and technical readout. Aliquots of the same cRNA sample were hybridized to four identical arrays and the coefficient of variation (CV) for each individual feature was calculated based on the raw fluorescence intensities across the four replicates without applying any data preprocessing steps like background correction, array-to-array normalization, removal of outliers or removal of low-intensity spots. Since we expected the CV to be higher for features with a low intensity and lower for features with a high intensity we again designed the arrays with PM/MM probe pairs to obtain a balanced ratio between high intensity (PM probes) and low intensity features (MM probes). The four arrays each contained 6398 25mer probes (corresponding to 3199 PM/MM probe pairs). The probe sequences were derived from the Affymetrix HuGeneFL and the Test2 GeneChips. In addition, each array included 154 negative control features where a single ‘T’ mononucleotide was synthesized instead of a 25mer probe. The arrays were hybridized to aliquots of a cRNA sample from a pool of total RNAs (Homo sapiens, Arabidopsis thaliana, Drosophila melanogaster). This sample may be inappropriate for meaningful biological experiments focusing on the expression of specific genes but is very well suited for experiments with a technical scope. Due to its high complexity, this sample is likely to undergo specific hybridization, unspecific cross-hybridization (including cross-species hybridization) as well as extensive target–target interactions and thus will serve as a good indicator for the reproducibility of the array synthesis and the hybridization process in particular. For the analysis, we first performed a pairwise comparison of the four arrays (Fig. 2A–F). The average Pearson correlation coefficient calculated on the raw intensities for all possible combinations of two arrays was 0.986. To further investigate the reproducibility of the system on the raw data level, the CV for each of the 6398 features was calculated across the four replicates and CVs were plotted as a frequency distribution (Fig. 2G): 95% of all 6398 values were in the range of 0.03 (2.5th percentile) to 0.19 (97.5th percentile), the median CV being 0.09. A slightly higher median CV of 0.10 was found when the analysis was restricted to the 10% of features with the lowest intensities. These features do not represent the lowest features within a group consisting of only high-intensity features but indeed have very low intensities close to non-specific background. This is evident from the comparison of the average intensity of these features to the local background and to the negative control spots, where a single ‘T’ was synthesized instead of a 25mer probe. The average intensity of the 10% lowest features within the total of 6398 features (value: 911), the average of the local background of all spots on the array (value: 1198) and the average intensity for the negative controls (value: 1040) were all in the same range. Actually, the average of the 10% lowest features is even slightly lower than the average of the negative controls and the average of the local background. The latter phenomenon is due to the fact that the local background—at least for high intensity features—is increased by a ‘neighborhood’ effect caused by blooming of the hybridization signal. This is in agreement with a recent study published by Machl et al. that describes a similar ‘neighborhood’ effect for cDNA arrays spotted on nylon membranes and hybridized with radioactive labeled samples (37). Why the average intensity of the negative control features somewhat exceeds the average intensity found for the 10% lowest features is less obvious. A possible explanation could result from the higher negative charge of a 25mer probe as compared with a single ‘T’ nucleotide. In this case, the higher density of negative charges would lead to an increased repulsion of the equally negatively charged non-cognate targets that might reduce unspecific binding of non-cognate targets at the 25mer features. Another possible explanation is that steric hindrance for non-specific binding of the streptavidin–phycoerythrin complex to the glass surface might be higher for a feature with 25mers than for a feature carrying ‘T’ mononucleotides. This could result in a slightly higher blocking effect of 25mers as compared with ‘T’ mononucleotides. In summary, our technical experiments indicate a high reproducibility of geniom arrays on the raw data level and suggest that the good reproducibility is retained when applying geniom arrays to complex biological expression profiling experiments with the majority of features being in the low intensity range. However, in this case the average CV value might be slightly higher compared to our analysis with an unbiased distribution of raw intensity data across the entire intensity range.
Figure 2.
Reproducibility of the geniom platform on the raw data level. Aliquots of a single cRNA sample from a pool of total RNA (H.sapiens, D.melanogaster, A.thaliana) have been hybridized to four different arrays with 6398 features each. Raw intensity values (Supplementary Material Table 1) represent the median of approximately 30 CCD pixels for each feature. No data preprocessing (such as background correction, normalization, elimination of outliers or removal of low intensity features) was performed. (A–F) Pairwise comparison of raw intensities from the four arrays as scatter plots. (G) Frequency distribution of CVs. The CV for each of the 6398 features (probes) was calculated across the four replicates and CVs were plotted as a frequency distribution.
Reproducibility of fold-change and expression level values
Having demonstrated a high reproducibility for the raw intensity data, we evaluated the variability of fold-change values, the ultimate result of standard gene expression profiling experiments. We therefore measured the transcriptional response of 1125 randomly chosen yeast genes to osmotic shock in four identical experiments on eight arrays. In contrast to the technical experiments described in the previous sections, this experiment was designed as a real-world gene expression profiling. As a consequence, the array design, which included MM controls beforehand, was adapted to our standard for expression arrays and the MM controls were omitted. This approach was supported by the geniom application software which operates on a algorithm similar to the MOID principle (29) for gene expression profiling experiments and thus does not require MM controls for calculating expression levels and fold-change values. The eight arrays used in this study each contained 12 880 features (including all controls) with ten 25mer PM probes per transcript. Following hybridization with aliquots of either a control sample or a treated sample, we first calculated the CV of the 12 880 unprocessed raw intensities across the four arrays hybridized with the same sample. Mean CV values of 0.12 and 0.10 were found for the arrays hybridized with the control sample and the treated sample, respectively. A pairwise comparison of raw intensity data from the four control arrays in each possible combination revealed a mean Pearson correlation coefficient of 0.984 (min: 0.979; max: 0.993). In an identical analysis performed on the four arrays, hybridized with the cRNA sample from osmotically shocked yeast cells, we found a mean Pearson correlation coefficient of 0.986 (min: 0.977; max: 0.995). In conclusion, these values confirm the high reproducibility of the raw intensity data demonstrated in the last section and also suggest that the CV of the raw data is almost the same for arrays designed with PM/MM probe pairs (last section) and arrays with PM probes only (this section).
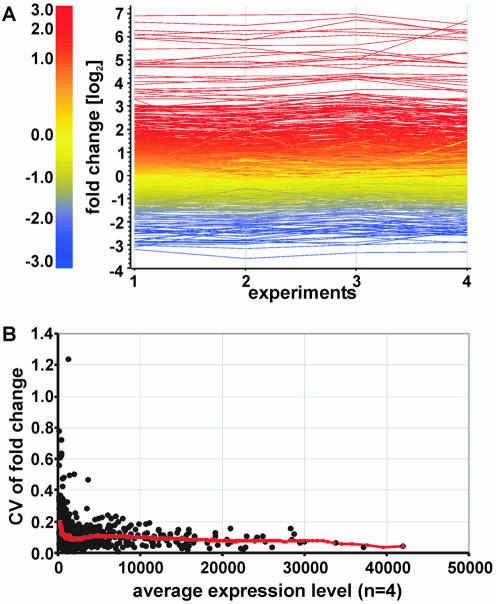
We next focused on the reproducibility of fold-change values obtained from geniom arrays. Fold-change values were calculated based on background-corrected and normalized intensities of one control and one treated array. They were subsequently compared between the four experiments (Fig. 3A). For this purpose, the CV of the fold-change value for each of the 1125 genes was calculated across the four replicates and graphed as a function of the gene’s expression level (Fig. 3B). As expected, the CV was highest for genes expressed at low levels (low Ek values) and decreased with rising expression levels (high Ek values). Table 1 shows the average CV for each of five different classes of 1125 genes classified according to their expression level. With the exception of genes expressed at very low levels (Ek < 400), the average CV value remains below 0.2 throughout all classes and even drops below 0.1 for highly expressed genes (Table 1). The probe sets for the three prokaryotic spike-in controls (ampr, kanr, cmr; see Dynamic range and discrimination power) produce Ek values of ∼350 in the absence of these transcripts. Ek values below 400 therefore indicate genes expressed at very low levels or not at all. As shown in Figure 3B and in Table 1, the distribution of CV values within a class is considerably wider for classes with genes expressed at low levels and narrower for classes including highly expressed genes. For genes with Ek values below 400, for instance, 95% of CVs fall into the range between 0.05 (2.5th percentile) and 0.63 (97.5th percentile), whereas for genes with an Ek level above 5000, the 95% range of the CVs is 0.03 and 0.22. The wider distribution together with the higher average CV render fold-change values for genes expressed at low levels less reliable than those of genes expressed at high levels. This limitation is shared by most if not all array platforms and is also documented for in situ synthesized 24mer arrays (38) and the Affymetrix GeneChip arrays (39). The average CV calculated for all 1125 genes irrespective of the expression levels is 0.11. It is worth noting, however, that this value is strongly influenced by the selection of genes. Adding more highly expressed genes would lower this value. On the contrary, a biased selection of genes expressed at low level would lead to a considerably higher CV. The selection of genes included in our study was unbiased and spans the entire expression range (Table 1). Thus, the average value of 0.11 presented in this study is likely to reflect the level of reproducibility encountered in typical gene expression profiling experiments on geniom arrays. In summary, our study revealed CV values that suggest a high reproducibility of fold-change values and compare favorably to data from spotted 35mer arrays where an average CV for the fold-change values of ∼0.3 was found (13). In addition, the CV values found on the geniom platform are significantly lower than those obtained with 24mer arrays synthesized on microscopic slides using a maskless photolithographic instrument. For these arrays, average CVs of the fold-change data typically are in the range between 0.45 (average for low expressed genes) and 0.29 (average for highly expressed genes) (38). Besides the fold-change data, the gene expression level is the most important result of a gene expression profiling experiment. This is particularly true for experiments which determine relative mRNA levels within a single sample rather than comparing two or more samples. In the experiment described above, four arrays were hybridized with aliquots of a yeast control sample and another four arrays were hybridized with aliquots of a sample from yeast cells treated with an osmotic shock. In order to investigate the reproducibility of expression levels (Ek values) obtained with our platform, we calculated CVs of the Ek values across the four replicates hybridized with the same sample for each of the 1125 genes included in the experiment. In agreement with results of a recent study performed on Affymetrix GeneChips (40), the CV of the expression levels was higher for genes expressed at low levels (low Ek values) and lower for genes expressed at high levels (high Ek values). As described above, we grouped the genes into five different classes according to their expression level. The average CV values calculated for these classes were in the 0.17–0.10 range. As shown in Table 2, a trend towards higher CVs for genes expressed at low levels and towards lower CVs for highly expressed genes is evident in the arrays hybridized with the control sample as well as in the arrays hybridized with the treated sample. This is a remarkable finding because the same gene may have different Ek levels on the ‘control’ and the ‘treated’ array: the genes that make up a certain expression class are not necessarily the same for the control and the treated sample. We therefore conclude that the high variability found for genes expressed at low levels is indeed due to technical parameters and is only slightly influenced by the individual genes analyzed.
Figure 3.
Reproducibility of the geniom platform on the fold-change level. The transcriptional response of 1125 yeast genes to osmotic shock was analyzed in four identical experiments and the fold-change values (Supplementary Material Table 2A) were compared. (A) Diagram of log2 fold-change values. Each transcript is represented as a line colored according to the log2 fold-change value. The color code is given on the left. (B) CVs of fold-change values. The CV for each of the 1125 genes was calculated across the four experiments and graphed as a function of the gene’s expression level (Ek value). The gene’s expression level represents the average of the Ek values from the four control arrays. A trend line representing the moving average of 100 genes is shown.
Table 1. CV of fold-change values as a function of the expression level (Ek value)a.
| Classes of Ek values | Number of genes | Average CV | 95% of CVs between |
|---|---|---|---|
| Up to 400 | 104 | 0.20 | 0.05–0.63 |
| 400–1000 | 391 | 0.12 | 0.03–0.29 |
| 1000–2000 | 299 | 0.10 | 0.02–0.20 |
| 2000–5000 | 185 | 0.10 | 0.03–0.21 |
| >5000 | 146 | 0.09 | 0.03–0.22 |
| All genes | 1125 | 0.11 | 0.03–0.29 |
aThe CVs were calculated across four replicates (Fig. 3). Genes were grouped into five different classes according to the average Ek value on the four control arrays. The range that includes 95% of the CV values of a certain class was determined by calculating the 2.5th and 97.5th percentiles on all CVs within this class.
Table 2. CVs of expression levels as a function of the expression levela.
| Classes of Ek values | Control | Treated | ||||
|---|---|---|---|---|---|---|
| Number of genes | Average CV | 95% of CVs between | Number of genes | Average CV | 95% of CVs between | |
| <400 | 104 | 0.17 | 0.04–0.38 | 128 | 0.15 | 0.05–0.34 |
| 400–1000 | 391 | 0.13 | 0.04–0.30 | 383 | 0.12 | 0.03–0.25 |
| 1000–2000 | 299 | 0.13 | 0.05–0.25 | 274 | 0.11 | 0.04–0.21 |
| 2000–5000 | 185 | 0.14 | 0.06–0.24 | 167 | 0.12 | 0.06–0.20 |
| >5000 | 146 | 0.12 | 0.05–0.20 | 173 | 0.10 | 0.04–0.18 |
| All genes | 1125 | 0.14 | 0.05–0.28 | 1125 | 0.12 | 0.04–0.26 |
aThe CVs were calculated across four arrays hybridized with aliquots of a control sample (control) and another four arrays hybridized with aliquots of a sample from yeast cells which were harvested after an osmotic shock (treated). The genes were grouped into five different classes according to their mean expression level on the four arrays. The range that includes 95% of the CV values of a certain class was determined by calculating the 2.5th and 97.5th percentiles on all CVs within this class.
Accuracy of biological results
In an attempt to validate the accuracy of results from the geniom platform we have analyzed the transcriptional response of yeast to osmotic shock. The data acquired with the geniom platform were compared with data from cDNA arrays published by Rep et al. (25) and to reference data from Affymetrix GeneChips which were generated as described in the experimental protocol. Our study comprised 4857 genes which were all analyzed twice on standard gene expression arrays containing 10 PM probes per gene (25mers; without MM controls). Using the same type of arrays we also measured an additional group of 203 genes in 10 replicates.
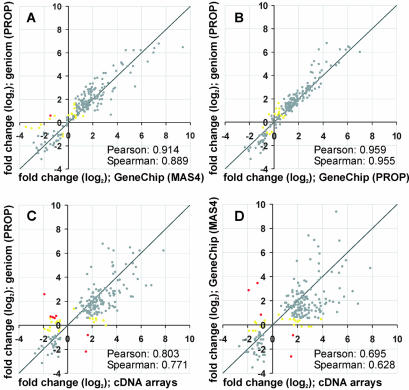
These 203 genes were found to be involved in the cellular response of yeast to osmotic shock in the experiments on spotted cDNA arrays published by Rep et al. (25). This selection of genes thus is biased with respect to the expected fold-change values and is also likely to be biased with respect to the expected expression level. However, since we were interested in the accuracy of biological results obtained from geniom arrays and the regulation of these 203 genes is known to be the major response of yeast cells to osmotic shock, we first focused the data analysis on these particular genes before extending it to the total of 4857 genes. Figure 4 shows fold-change values for these genes compared pair-wise between geniom arrays, Affymetrix GeneChips and the cDNA arrays used by Rep et al. (25). As indicated by a Pearson correlation coefficient of 0.914 and a Spearman rank correlation coefficient of 0.889, a high conformity was found between the geniom data and the GeneChip data despite comparing two completely independent array platforms (Fig. 4A). Note that the only parameter kept constant on both platforms was the biological sample. When reducing the complexity by applying the same analysis algorithm to both the raw intensity values from the geniom arrays and the raw data from the GeneChips (as found in the .cel file) an even higher similarity was found and the Pearson correlation coefficient increased to 0.959 (Fig. 4B). For further analysis, we again focused on the comparison of independent platforms (Fig. 4A, C and D) grouping the genes into three different categories. Genes with fold-change values ≥1.5 (log2 value: 0.58) were considered to be upregulated. Genes with fold-change values ≤–1.5 (log2 value: –0.58) were considered to be downregulated and genes with fold-change values between –1.5 and 1.5 (log2 value: –0.58 to 0.58) were considered to be unaffected. Based on this categorization, 184 out of 203 genes showed the same tendency on Affymetrix and geniom arrays (142 upregulated, 30 downregulated, 12 unchanged). From the remaining genes, nine were found to be regulated on the Affymetrix GeneChip but unaffected on the geniom arrays and nine genes behaved vice versa. Only one gene switched between the upregulated and the downregulated categories. As indicated by the correlation coefficients, the geniom data closely match the GeneChip data. In addition, they are very similar to the data obtained with cDNA arrays. A total of 174 out of 203 genes showed the same tendency in the geniom and the cDNA data set. A minority of 21 genes switched between unchanged on the geniom arrays and regulated on the cDNA arrays, one gene vice versa, and seven genes were found to be regulated in the opposite sense on both platforms. The conformity between geniom data and GeneChip data, however, is greater than the similarity found between the cDNA data and either of the oligonucleotide arrays (Fig. 4). In general, most genes showed the same tendency on the spotted cDNA arrays and on both oligonucleotide array formats. Thus, the major findings described by Rep et al. (25) could be reproduced on geniom arrays (Supplementary Material Table 2B). The actual fold-change values, however, differ significantly between the cDNA arrays and the oligonucleotide arrays. This is in good agreement with studies that revealed substantial differences in the overall performance of cDNA arrays and oligonucleotide arrays. Generally, spotted cDNA arrays show a higher sensitivity than short oligonucleotide arrays (19,41). Conversely, spotted cDNA arrays are known to exhibit lower specificity than short oligonucleotide arrays, primarily because of cross-hybridization of highly homologous transcripts and non-cognate cDNA probes and due to varying hybridization efficiencies of long cDNA probes (42–45). An additional factor that might contribute to the variance in the fold-change values observed in our study is the biological sample itself. The cDNA data were taken from the literature. Therefore, the total RNA source used for the experiments on the cDNA arrays was not identical to that used for the geniom and the Affymetrix oligonucleotide arrays. A recently published, extensive study designed as an interlaboratory comparison revealed that variations introduced by in vitro handling steps and variations between replicate cultures in particular can significantly influence the result of a gene expression experiment (46). In addition, the labeling procedures differ significantly: the oligonucleotide arrays used in this study were hybridized to an amplified biotinylated cRNA sample (synthesized starting from the total RNA, as described in Materials and Methods) while the cDNA arrays used by Rep et al. (25) were hybridized with a non amplified, [33P]CTP-labeled cDNA sample (synthesized from the total RNA via reverse transcription). Taken together, the first part of our study focusing on the 203 genes known to be regulated in the cellular response of yeast to osmotic shock suggests a high conformity of biological data obtained on geniom arrays and data aquired on Affymetrix GeneChips. We also found that the great majority of the 203 genes (86% when applying the categorization criteria described above) showed the same tendency on geniom arrays and spotted cDNA arrays. The significant variation of the actual fold-change values found in the latter comparison is likely to be caused by differences in the general performance of the two array formats and by differences in the samples used for the experiments on the respective platforms.
Figure 4.
Log–log plots comparing fold-change data from three different array formats. The transcriptional response of 203 yeast genes to osmotic shock was analyzed on the geniom platform in 10 replicates. The average fold-change values were compared with data from Affymetrix GeneChips and to cDNA array data from the literature (25) (Supplementary Material Table 2B). Genes that fall into the same category of regulation on the respective platforms are shown in gray (cut-offs for categorization: fold-change ≤–0.58, downregulated; fold-change >–0.58 but <0.58, unchanged; fold-change ≥0.58, upregulated). Genes that were found to be up- or downregulated on one platform but unchanged on the other are shown in yellow. Genes that behave the opposite way are shown in red. (A) Comparison of geniom data and MAS4 calculated fold-change values from the Affymetrix GeneChips. (B) Comparison of geniom data and PROP-calculated fold-change values from the Affymetrix GeneChips. (C) Comparison of geniom data and the cDNA array data from the literature (25). (D) Comparison of Affymetrix GeneChip data and cDNA array data from the literature (25).
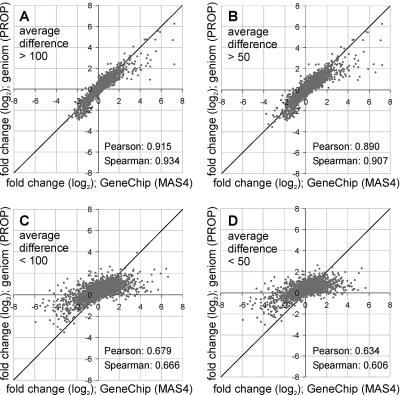
So far, we have restricted our analysis to the 203 genes known to be involved in the cellular response of yeast to osmotic shock. Most of these genes are highly regulated and tend to be expressed at higher levels. They are therefore much more likely to show the same trends on different platforms than randomly selected genes. In order to investigate if the high concordance of geniom and GeneChip data is confirmed in experiments with a completely unbiased selection of genes, we extended the analysis to all 4857 genes included in our study. We compared the average fold-change values calculated on the two replicate experiments performed on the geniom instrument to the fold-change values obtained from the Affymetrix GeneChips. 3276 (68%) out of 4857 genes fell into the same category; 1076 genes (22%) were unchanged on the febit arrays but downregulated or upregulated on the GeneChips; 436 genes (9%) were unchanged on the Affymetrix GeneChips but regulated on the geniom arrays and 69 genes (1%) were found to be regulated in the opposite sense on both platforms (Supplementary Material Table 2C). Overall, a mean Pearson correlation coefficient of 0.742 and an average Spearman rank correlation coefficient of 0.759 were calculated on the fold-change level. Taken together, these data indicate a considerably lower agreement of the fold-change values for the 4857 randomly selected genes than for the 203 genes from a biased selection. To address the question of whether the poor conformity applies to all 4857 genes analyzed or if it is restricted to a certain subgroup of genes, we refined our analysis taking the expression levels into account. Mills and Gordon (39) investigated false-positive rates using Affymetrix Mu11KsubA and Mu11KsubB GeneChips. All genes recognized as increased or decreased in same-to-same comparisons were defined as noise. Most of these genes were clustered at expression levels below 250 (measured by the average difference between PM and MM of all PM/MM probe pairs for one transcript). Grundschober et al. (40) used GeneChip U34 and estimated CVs of triplicate hybridizations to determine significant fold-changes thresholds. They found the fold-change value to be reliable above a cut-off expression level of 100. We applied this 100 cut-off as well as a less stringent cut-off at 50 to our analysis. We classified the 4857 genes according to their average difference (expression level) on the GeneChip array (base array) into ‘expressed at low level’ (below the respective cutoff) and ‘expressed at a higher level’ (above the respective cut-off). Then, we analyzed the agreement of the fold-change values obtained with the GeneChips and the fold-change values acquired from geniom arrays within these groups. As shown in Figure 5A and B, we found a substantial correlation between GeneChip data and geniom data for genes expressed at an elevated level: for genes with an expression level above 100, a Pearson correlation coefficient of 0.915 was calculated and, applying the same categorization criteria as used for the 203 genes, 83% of all genes (1401 out of 1688) showed the same tendency on both platforms (Fig. 5A). A slightly lower but still significant conformity was found for genes with an expression level above the less stringent cut-off at 50: for these genes, the Pearson correlation coefficient was 0.890, and 80% of all genes (2067 out of 2596) showed the same tendency of regulation (Fig. 5B). In contrast, we found only poor correlations of fold-change values for the genes with an expression level lower than the respective cut-offs: for genes with an expression level lower than 100 (3169 genes) or lower than 50 (2261 genes) we observed a Pearson correlation coefficient of 0.679 and 0.634, respectively (Fig. 5C and D).
Figure 5.
Log–log plots comparing fold-change data from geniom arrays and Affymetrix GeneChips. The transcriptional response of 4857 randomly chosen yeast genes to osmotic shock was analyzed on geniom arrays in two replicates and the average fold-change values were compared with data from Affymetrix GeneChips (Supplementary Material Table 2C). For this comparison, the genes were grouped into ‘expressed at low level’ and ‘expressed at a higher level’ according to their expression level (average difference) on the GeneChip base array. Cut-offs at either 100 or 50 were used for the categorization. (A) Log–log plot of the 1688 genes with an average difference above a cut-off at 100. (B) Log–log plot of the 2596 genes with an average difference above a cut-off at 50. (C) Log–log plot of the 3169 genes with an average difference below 100. (D) Log–log plot of the 2261 genes with an average difference below 50.
From these data we conclude that—at least for yeast— fold-change values obtained from geniom arrays are in good concordance with fold-change values acquired with Affymetrix GeneChips (with the exception of genes expressed at very low levels). This is a remarkable finding if the context of the experimental design is considered. The only parameter kept constant between the two platforms was the biological sample. All other parameters, including the probe design and the algorithm used for data analysis, were different for both platforms. Despite this high correlation found for genes expressed at elevated levels, our comparison also revealed substantial differences in the fold-change values obtained with both platforms with regard to genes expressed at low levels. This finding was not unexpected and is likely to be caused by a higher variation of fold-change values calculated on low signal intensities. The fact that calculations based on such low signal intensities are prone to increased variation is known for most if not all array formats, including spotted 35mer arrays (13), in situ synthesized 24mer arrays (38) and GeneChips (39,40)—and was also found for the geniom platform in this study.
We further demonstrated that geniom data not only match data acquired with other array formats but also reflect the true gene expression pattern of the biological system analyzed. We used a non-array reference system and compared the gene expression data from the geniom platform with those obtained by quantitative RT–PCR (SYBR Green assay). For this experiment, a subset of 56 genes from the 203 genes shown in Figure 4 was selected. The choice was based on the fold-change distribution in the array experiments, such that the validated data set spans the entire range of fold-change values observed. The selection was otherwise unbiased and random. The quantitative RT–PCR analysis was performed with the same RNA samples used for the array experiments. Seven out of the 56 genes were excluded from the analysis due to PCR efficiencies below 1.70. Table 3 compares the fold-change values of the evaluable genes to the average fold-change values from the 10 replicate experiments on the geniom platform described above (Fig. 4). As indicated by the Pearson correlation coefficient of 0.966 and the Spearman rank correlation coefficient of 0.972, a very high conformity was found between the two data sets. Due to the lower dynamic range of oligonucleotide arrays as compared with quantitative RT–PCR, the fold-change values for highly regulated genes are compressed on the geniom platform. This phenomenon has been described before for other spotted (13) or in situ synthesized (38) oligonucleotide arrays. Despite those differences in the fold-change values of highly regulated genes, our study provided evidence that geniom arrays generate accurate and reliable results and thus enable scientists to address complex biological questions.
Table 3. Comparison of fold-change data from geniom arrays and quantitative RT–PCRa.
| Gene | Geniom arrays | Quantitative RT–PCR | ||
|---|---|---|---|---|
| Average fold-change | Average fold-change (log2) | Average fold-change | Average fold-change (log2) | |
| YMR175W | 111.05 | 6.80 | 164.81 | 7.36 |
| YBR117C | 85.37 | 6.42 | 2112.88 | 11.04 |
| YER150W | 34.41 | 5.10 | 76.91 | 6.27 |
| YDL223C | 29.00 | 4.86 | 42.23 | 5.40 |
| YAL061W | 21.70 | 4.44 | 23.62 | 4.56 |
| YDL204W | 19.18 | 4.26 | 33.15 | 5.05 |
| YGR248W | 19.12 | 4.26 | 2.04 | 4.46 |
| YKL151C | 13.54 | 3.76 | 10.67 | 3.42 |
| YHR087W | 9.88 | 3.30 | 33.58 | 5.07 |
| YGR066C | 8.57 | 3.10 | 16.75 | 4.07 |
| YML054C | 7.25 | 2.86 | 4.39 | 2.14 |
| YHR094C | 6.44 | 2.69 | 8.31 | 3.05 |
| YML100W | 6.35 | 2.67 | 7.08 | 2.82 |
| YLR267W | 5.12 | 2.36 | 3.06 | 1.61 |
| YER103W | 4.91 | 2.30 | 4.32 | 2.11 |
| YKL150W | 4.21 | 2.07 | 5.48 | 2.45 |
| YHR022C | 4.02 | 2.01 | 5.63 | 2.49 |
| YLR031W | 3.98 | 1.99 | 4.36 | 2.12 |
| YEL039C | 3.71 | 1.89 | 2.96 | 1.56 |
| YMR031C | 3.06 | 1.61 | 1.50 | 0.59 |
| YCL040W | 2.91 | 1.54 | 4.25 | 2.09 |
| YER054C | 2.62 | 1.39 | 3.18 | 1.67 |
| YDR533C | 2.31 | 1.21 | 2.06 | 1.04 |
| YGR170W | 2.27 | 1.18 | 1.69 | 0.76 |
| YJL149W | 2.22 | 1.15 | 3.30 | 1.72 |
| YDR100W | 2.09 | 1.06 | 1.84 | 0.88 |
| YDR463W | 2.08 | 1.06 | 1.50 | 0.58 |
| YLR042C | 2.06 | 1.04 | 2.13 | 1.09 |
| YER041W | 1.72 | 0.78 | –1.05 | –0.07 |
| YGR146C | 1.54 | 0.63 | 1.46 | 0.55 |
| YMR030W | 1.45 | 0.54 | 2.11 | 1.08 |
| YHR086W | 1.30 | 0.38 | –1.09 | –0.12 |
| YDL135C | 1.15 | 0.21 | –1.93 | –0.95 |
| YKL160W | –1.17 | –0.22 | –2.56 | –1.35 |
| YBL002W | –1.18 | –0.24 | –1.53 | –0.61 |
| YGR138C | –1.24 | –0.31 | –2.64 | –1.40 |
| YDR324C | –1.39 | –0.48 | –10.36 | –3.37 |
| YKL109W | –1.84 | –0.88 | –4.99 | –2.32 |
| YER165W | –2.19 | –1.13 | –7.59 | –2.92 |
| YGR155W | –3.04 | –1.61 | –7.15 | –2.84 |
| YGL055W | –3.29 | –1.72 | –10.69 | –3.42 |
| YDL198C | –3.32 | –1.73 | –37.42 | –5.23 |
| YHR128W | –3.68 | –1.88 | –12.13 | –3.60 |
| YJL217W | –3.92 | –1.97 | –7.19 | –2.85 |
| YDL014W | –4.47 | –2.16 | –22.89 | –4.52 |
| YGR060W | –4.55 | –2.19 | –16.32 | –4.03 |
| YKR013W | –4.60 | –2.20 | –1.96 | –0.97 |
| YER052C | –6.24 | –2.64 | –21.92 | –4.45 |
| YGR234W | –8.91 | –3.16 | –33.48 | –5.07 |
aValues in the geniom columns represent averages from 10 identical experiments (Fig. 4). All fold-change values can be found in Supplementary Material Table 2B.
DISCUSSION
This study was designed to validate the geniom technology, a novel and fully integrated oligonucleotide array platform for gene expression profiling applications. We first focused on the technical aspects and evaluated the discrimination power, the dynamic range, and the reproducibility of the system. The system is able to detect RNAs present at a frequency of 1:100 000. In good agreement with data published for other oligonucleotide array platforms (13,35,36), detection is quantitative over more than two orders of magnitude. The geniom technology integrates array synthesis, hybridization and detection in a single benchtop device located in the investigator’s laboratory. As quality assurance is a more demanding issue for benchtop instruments compared with centralized facilities, special attention was paid to data reproducibility. Primary experiments on four identical arrays with 6398 features each revealed a mean CV value of 0.09 for the non-processed raw intensities with an unbiased distribution across the entire intensity range. In a more elaborate experiment targeting 1125 randomly chosen yeast genes, we found the CV for the fold-change values to be substantially influenced by the expression level. The average CV values range between 0.20 for genes expressed at very low levels and 0.09 for genes expressed at high levels. The CVs for the expression levels range between 0.19 (average for genes expressed at very low levels) and 0.10 (average for genes expressed at high levels). Taken together, the CV values indicate a good reproducibility of raw data, fold-change values and expression levels but also revealed that expression results for genes expressed at low level are considerably less consistent than those of genes expressed at higher levels. This phenomenon is common to most if not all array platforms and is known for the widely used GeneChip arrays (39,40), in situ synthesized 24mer arrays (38) and spotted 35mer arrays (13). By extending our study from inter-array to inter-instrument comparisons we demonstrated that different individual geniom instruments perform equally well. For all four instruments included in our study, the mean CVs for the fold-change values (mean value across the entire expression range) were in the range of 0.11–0.18 (data not shown). As a next step, the accuracy of biological data was demonstrated by comparing the geniom data from a real-world experiment to reference data obtained from Affymetrix GeneChips, data from quantitative RT–PCR and cDNA array data from the literature (25). In this experiment, we were able to reproduce the major findings of Rep et al., who investigated the transcriptional response of yeast to osmotic shock in great detail on cDNA arrays and generated a list of 203 genes which they identified as the main responders to the osmotic shock treatment (25). Despite substantial differences in the actual fold-change values, the great majority of the 203 genes showed the same tendency of regulation on the geniom oligonucleotide arrays. By comparing the geniom data for these genes to reference data acquired on Affymetrix GeneChips we found a high conformity of fold-change data. A larger experiment comprising 4857 yeast genes from a random selection, confirmed the high correlation of geniom data and Affymetrix data. Despite a high correlation of fold-change data for highly expressed genes, however, substantial differences in the fold-change values were evident for genes in the low expression level. This was not an unexpected finding and is in good agreement with a higher variation of fold-change data found for genes expressed at low levels on both the Affymetrix GeneChips (39) and the geniom arrays. In an attempt to demonstrate that geniom data not only match data obtained from other array formats but also reflect the gene expression pattern of the biological system analyzed, we used quantitative real-time PCR to measure the fold-change of 56 yeast genes that span the entire expression range. Due to the lower dynamic range of geniom arrays as compared with real-time PCR we observed some differences in the fold-change values of highly regulated genes, reflecting the compression of geniom data in the high-intensity range. Nevertheless, a Pearson correlation coefficient of 0.966 clearly indicated a high concordance between the geniom data and the data obtained by quantitative real-time PCR. In conclusion, our data suggest that the geniom technology produces reproducible and reliable results and complements other well established array platforms. Due to its design, however, it provides a number of new opportunities. The sequence of each individual probe may be varied on each array and all that is required to generate a new array is sequence information. Sequence updates or results from a previously performed array experiment can be incorporated into new array designs. The automation ensures convenient handling of the machine and thus may contribute to a more widespread use of the complex array technologies.
In this study, we have validated the geniom platform for gene expression profiling experiments. Supported by the small reaction volumes and the design of the arrays as three-dimensional microchannels, however, the system is also well suited for other applications involving enzymatic reactions such as primer extension, ligation or on-chip PCR.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
Part of the work presented here was supported by a grant from the German BMBF (Bundesministerium für Bildung und Forschung, Deutschland) to febit ag.
REFERENCES
- 1.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 2.Lockhart D.J., Dong,H., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittmann,M., Wang,C., Kobayashi,M., Horton,H. and Brown,E.L. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 3.Chu S., DeRisi,J., Eisen,M., Mulholland,J., Botstein,D., Brown,P.O. and Herskowitz,I. (1998) The transcriptional program of sporulation in budding yeast. [Erratum: Science, 282, 5393] Science, 282, 699–705. [DOI] [PubMed] [Google Scholar]
- 4.Hughes T.R., Marton,M.J., Jones,A.R., Roberts,C.J., Stoughton,R., Armour,C.D., Bennett,H.A., Coffey,E., Dai,H., He,Y.D., Kidd,M.J., King,A.M., Meyer,M.R., Slade,D., Lum,P.Y., Stepaniants,S.B., Shoemaker,D.D., Gachotte,D., Chakraburtty,K., Simon,J., Bard,M. and Friend,S.H. (2000) Functional discovery via a compendium of expression profiles. Cell, 102, 109–126. [DOI] [PubMed] [Google Scholar]
- 5.Gray N.S., Wodicka,L., Thunnissen,A.M., Norman,T.C., Kwon,S., Espinoza,F.H., Morgan,D.O., Barnes,G., LeClerc,S., Meijer,L., Kim,S.H., Lockhart,D.J. and Schultz,P.G. (1998) Exploiting chemical libraries, structure and genomics in the search for kinase inhibitors. Science, 218, 533–538. [DOI] [PubMed] [Google Scholar]
- 6.Marton M.J., DeRisi,J.L., Bennett,H.A., Iyer,V.R., Meyer,M.R., Roberts,C.J., Stoughton,R., Burchard,J., Slade,D., Dai,H., Bassett,D.E.,Jr, Hartwell,L.H., Brown,P.O. and Friend,S.H. (1998) Drug target validation and identification of secondary drug target effects using DNA microarrays. Nature Med., 4, 1293–1301. [DOI] [PubMed] [Google Scholar]
- 7.Roberts C.J., Nelson,B., Marton,M.J., Stoughton,R., Meyer,M.R., Bennett,H.A., He,Y.D., Dai,H., Walker,W.L., Hughes,T.R., Tyers,M., Boone,C. and Friend,S.H. (2000) Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science, 287, 873–880. [DOI] [PubMed] [Google Scholar]
- 8.Khan J., Simon,R., Bittner,M., Chen,Y., Leighton,S.B., Pohida,T., Smith,P.D., Jiang,Y., Gooden,G.C., Trent,J.M. and Meltzer,P.S. (1998) Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res., 58, 5009–5013. [PubMed] [Google Scholar]
- 9.Perou C.M., Jeffrey,S.S., van de Rijn,M., Rees,C.A., Eisen,M.B., Ross,D.T., Pergamenschikov,A., Williams,C.F., Zhu,S.X., Lee,J.C., Lashkari,D., Shalon,D., Brown,P.O. and Botstein,D. (1999) Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc. Natl Acad. Sci. USA, 96, 9212–9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golub T.R., Slonim,D.K., Tamayo,P., Huard,C., Gaasenbeek,M., Mesirov,J.P., Coller,H., Loh,M.L., Downing,J.R., Caligiuri,M.A., Bloomfield,C.D. and Lander,E.S. (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science, 286, 531–537. [DOI] [PubMed] [Google Scholar]
- 11.Shoemaker D.D., Schadt,E.E., Armour,C.D., He,Y.D., Garrett-Engele,P., McDonagh,P.D., Loerch,P.M., Leonardson,A., Lum,P.Y., Cavet,G., Wu,L.F., Altschuler,S.J., Edwards,S., King,J., Tsang,J.S., Schimmack,G., Schelter,J.M., Koch,J., Ziman,M., Marton,M.J., Li,B., Cundiff,P., Ward,T., Castle,J., Krolewski,M., Meyer,M.R., Mao,M., Burchard,J., Kidd,M.J., Dai,H., Phillips,J.W., Linsley,P.S., Stoughton,R., Scherer,S. and Boguski,M.S. (2001) Experimental annotation of the human genome using microarray technology. Nature, 409, 922–927. [DOI] [PubMed] [Google Scholar]
- 12.Hu G.K., Madore,S.J., Moldover,B., Jatkoe,T., Balaban,D., Thomas,J. and Wang,Y. (2001) Predicting splice variants from DNA chip expression data. Genome Res., 11, 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramakrishnan R., Dorris,D., Lublinsky,A., Nguyen,A., Domanus,M., Prokhorova,A., Gieser,L., Touma,E., Lockner,R., Tata,M., Zhu,X., Patterson,M., Shippy,R., Sendera,T.J. and Mazumder,A. (2002) An assessment of Motorola CodeLink™ microarray performance for gene expression profiling applications. Nucleic Acids Res., 30, e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yue H., Eastman,P.S., Wang,B.B., Minor,J., Doctolero,M.H., Nuttall,R.L., Stack,R., Becker,J.W., Montgomery,J.R., Vainer,M. and Johnston,R. (2001) An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res., 29, e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guckenberger M., Kurz,S., Aepinus,C., Theiss,S., Haller,S., Leimbach,T., Panzner,U., Weber,J., Paul,H., Unkmeir,A., Frosch,M. and Dietrich,G. (2002) Analysis of the heat shock response of Neisseria meningitidis with cDNA- and oligonucleotide-based DNA microarrays. J. Bacteriol., 184, 2546–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Southern E.M., Maskos,U. and Elder,J.K. (1992) Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: evaluation using experimental models. Genomics, 13, 1008–1017. [DOI] [PubMed] [Google Scholar]
- 17.Maskos U. and Southern,E.M. (1992) Oligonucleotide hybridizations on glass supports: a novel linker for oligonucleotide synthesis and hybridization properties of oligonucleotides synthesized in situ. Nucleic Acids Res., 20, 1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fodor S.P., Read,J.L., Pirrung,M.C., Stryer,L., Lu,A.T. and Solas,D. (1991) Light-directed, spatially addressable parallel chemical synthesis. Science, 251, 767–773. [DOI] [PubMed] [Google Scholar]
- 19.Hughes T.R., Mao,M., Jones,A.R., Burchard,J., Marton,M.J., Shannon,K.W., Lefkowitz,S.M., Ziman,M., Schelter,J.M., Meyer,M.R., Kobayashi,S., Davis,C., Dai,H., He,Y.D., Stephaniants,S.B., Cavet,G., Walker,W.L., West,A., Coffey,E., Shoemaker,D.D., Stoughton,R., Blanchard,A.P., Friend,S.H. and Linsley,P.S. (2001) Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol., 19, 342–347. [DOI] [PubMed] [Google Scholar]
- 20.Singh-Gasson S., Green,R.D., Yue,Y., Nelson,C., Blattner,F., Sussman,M.R. and Cerrina,F. (1999) Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol., 17, 974–978. [DOI] [PubMed] [Google Scholar]
- 21.Pellois J.P., Zhou,X., Srivannavit,O., Zhou,T., Gulari,E. and Gao,X. (2002) Individually addressable parallel peptide synthesis on microchips. Nat. Biotechnol., 20, 922–926. [DOI] [PubMed] [Google Scholar]
- 22.Beier M. and Hoheisel,J.D. (2000) Production by quantitative photolithographic synthesis of individually quality-checked DNA microarrays. Nucleic Acids Res., 28, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasan A. Stengele,K.-P., Giegrich,H., Cornwell,P., Isham,K.R., Sachleben,R.A., Pfleiderer,W. and Foote,S. (1997) Photolabile protecting groups for nucleotides: synthesis and photodeprotection rates. Tetrahedron, 53, 4247–4264. [Google Scholar]
- 24.Stähler C.F., Stähler,P.F., Müller,M., Stähler,F. and Lindner,H. (1999) Patent DE-19940750.9-52; PCT/WO/EP/99/0617; AU-749884B2.
- 25.Rep M., Krantz,M., Thevelein,J.M. and Hohmann,S. (2000) The transcriptional response of Saccharomyces cerevisiae to osmotic shock. J. Biol. Chem., 275, 8290–8300. [DOI] [PubMed] [Google Scholar]
- 26.Schmitt M.E., Brown,T.A. and Trumpower,B.L. (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res., 18, 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Gelder R.N., von Zastrow,M.E., Yool,A., Dement,W.C., Barchas,J.D., Eberwine,J.H. (1990) Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Science, 87, 1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eberwine J., Yeh,H., Miyashiro,K., Cao,Y., Nair,S., Finnell,R., Zettel,M. and Coleman,P. (1992) Analysis of gene expression in single live neurons. Proc. Natl Acad. Sci. USA, 89, 3010–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y. and Abagyan,R. (2002) Match-only integral distribution (MOID) algorithm for high-density oligonucleotide array analysis. BMC Bioinformatics, 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasmussen R. (2001) Quantification on the lightCycler. In Meuer,S., Wittwer,C. and Nakagawara,K. (eds), Rapid Cycle Real-time PCR, Methods and Applications. Springer Press, Heidelberg, pp. 21–34. [Google Scholar]
- 31.Muller P.Y., Janovjak,H., Miserez,A.R. and Dobbie,Z. (2002) Processing of gene expression data generated by quantitative real-time RT-PCR. [Erratum: Biotechniques, 33, 514] Biotechniques, 32, 1372–1374, 1376,, 1378–1379. [PubMed] [Google Scholar]
- 32.Iyer V. and Struhl,K. (1996) Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 93, 5208–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hereford L.M. and Rosbash,M. (1997) Number and distribution of polyadenylated RNA sequences in yeast. Cell, 10, 453–462. [DOI] [PubMed] [Google Scholar]
- 34.Lewin B. (1980) Gene Expression. Wiley-Interscience, New York, NY, Vol. 2. [Google Scholar]
- 35.Albert T.J., Norton,J., Ott,M., Richmond,T., Nuwaysir,K., Nuwaysir,E.F., Stengele,K.P. and Green,R.D. (2003) Light-directed 5′→3′ synthesis of complex oligonucleotide microarrays. Nucleic Acids Res., 31, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chudin E., Walker,R., Kosaka,A., Wu,S.X., Rabert,D., Chang,T.K. and Kreder,D.E. (2002) Assessment of the relationship between signal intensities and transcript concentration for Affymetrix GeneChip arrays. Genome Biol., 3, research0005.1–research0005.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Machl A.W., Schaab,C. and Ivanov,I. (2002) Improving DNA array data quality by minimising ‘neighbourhood’ effects. Nucleic Acids Res., 30, e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nuwaysir E.F., Huang,W., Albert,T.J., Singh,J., Nuwaysir,K., Pitas,A., Richmond,T., Gorski,T., Berg,J.P., Ballin,J., McCormick,M., Norton,J., Pollock,T., Sumwalt,T., Butcher,L., Porter,D., Molla,M., Hall,C., Blattner,F., Sussman,M.R., Wallace,R.L., Cerrina,F. and Green,R.D. (2002) Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res., 12, 1749–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mills J.C. and Gordon,J.I. (2001) A new approach for filtering noise from high-density oligonucleotide microarray datasets. Nucleic Acids Res., 29, e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grundschober C., Malosio,M.L., Astolfi,L., Giordano,T., Nef,P. and Meldolesi,J. (2002) Neurosecretion competence. A comprehensive gene expression program identified in PC12 cells. J. Biol. Chem., 277, 36715–36724. [DOI] [PubMed] [Google Scholar]
- 41.Schulze A. and Downward,J. (2001) Navigation gene expression using microarrays—a technology review. Nature Cell Biol., 3, E190–E195. [DOI] [PubMed] [Google Scholar]
- 42.Li J., Pankratz,M. and Johnson,J.A. (2002) Differential gene expression patterns revealed by oligonucleotide versus long cDNA arrays. Toxicol. Sci., 69, 383–390. [DOI] [PubMed] [Google Scholar]
- 43.Bartosiewicz M., Trounstine,M., Barker,D., Johnston,R. and Buckpitt,A. (2000) Development of a toxicological gene array and quantitative assessment of this technology. Arch. Biochem. Biophys., 376, 66–73. [DOI] [PubMed] [Google Scholar]
- 44.Heller R.A., Schena,M., Chai,A., Shalon,D., Bedilion,T., Gilmore,J., Woolley,D.E. and Davis,R.W. (1997) Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc. Natl Acad. Sci. USA, 94, 2150–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richmond C.S., Glasner,J.D., Mau,R., Jin,H. and Blattner,F.R. (1999) Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res., 27, 3821–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piper M.D.W., Daran-Lapujade,P., Bro,C., Regenberg,B., Knudsen,S., Nielsen,J. and Pronk,J.T. (2002) Reproducibility of oligonucleotide microarray transcriptome analyses. An interlaboratory comparison using chemostat cultures of Saccharomyces cerevisiae. J. Biol. Chem., 277, 37001–37008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.