Abstract
Because intracellular [Na+] is kept low by Na+/K+-ATPase, Na+ dependence is generally considered a property of extracellular enzymes. However, we found that p94/calpain 3, a skeletal-muscle-specific member of the Ca2+-activated intracellular “modulator proteases” that is responsible for a limb-girdle muscular dystrophy (“calpainopathy”), underwent Na+-dependent, but not Cs+-dependent, autolysis in the absence of Ca2+. Furthermore, Na+ and Ca2+ complementarily activated autolysis of p94 at physiological concentrations. By blocking Na+/K+-ATPase, we confirmed intracellular autolysis of p94 in cultured cells. This was further confirmed using inactive p94:C129S knock-in (p94CS-KI) mice as negative controls. Mutagenesis studies showed that much of the p94 molecule contributed to its Na+/Ca2+-dependent autolysis, which is consistent with the scattered location of calpainopathy-associated mutations, and that a conserved Ca2+-binding sequence in the protease acted as a Na+ sensor. Proteomic analyses using Cs+/Mg2+ and p94CS-KI mice as negative controls revealed that Na+ and Ca2+ direct p94 to proteolyze different substrates. We propose three roles for Na+ dependence of p94; 1) to increase sensitivity of p94 to changes in physiological [Ca2+], 2) to regulate substrate specificity of p94, and 3) to regulate contribution of p94 as a structural component in muscle cells. Finally, this is the first example of an intracellular Na+-dependent enzyme.
Keywords: Calcium, Calpain, Muscular Dystrophy, Proteolytic Enzymes, Skeletal Muscle, Sodium Ion
Introduction
Na+ is essential for all life. Its extracellular concentration is relatively high (∼145 mm), and a number of enzymes, such as blood coagulation proteases, are regulated by the [Na+] (1–3). The relative intra- and extracellular concentrations of Na+, K+, and Cl− constitute the basis of the membrane potential, which is especially important for excitatory cells such as muscle cells (4, 5). Depolarization of the muscle cell membrane caused by the opening of acetylcholine receptors results in the cell excitation-coupled contraction. Intracellular [Na+] ([Na+]i) is maintained at rather low levels (∼15 mm). To achieve this, cells express Na+/K+-ATPases on their cytoplasmic membrane and pump Na+ out of the cell in exchange for K+ (6). Therefore, it has been believed that all Na+-dependent enzymes are extracellular.
Ca2+, like Na+, is essential for life, and the calpains constitute a superfamily of Ca2+-activated intracellular modulator proteases that regulate various cellular functions, such as signal transduction, autophagy, and apoptosis (7–10). However, the most abundant and ubiquitous calpains (μ-calpain and m-calpain, the “conventional” calpains) require Ca2+ concentrations that are much higher (10 μm ∼ 1 mm) than the physiological intracellular [Ca2+] ([Ca2+]i, ∼100 nm) making them almost inactive in the cytosol (7, 10). On the other hand, skeletal muscle-specific calpain, p94/calpain 3 (11, 12), is unique in that when generated by in vitro translation, it undergoes very rapid (t½ < 10 min) autolytic degradation even in the absence of Ca2+ (13). When expressed in cultured cells, such as COS7 cells, p94, but not an active site-inactive mutant (p94:C129S), also autolyses extensively, without stimulation, almost disappearing from the cell (13). These findings indicate that the physiological intracellular ionic environment is sufficient for p94 to be activated, unlike conventional calpains. It should be noted, however, that rapid autolytic activity of p94 is suppressed in skeletal muscle cells in vivo, probably by its binding to connectin/titin, a gigantic elastic protein connecting the Z-band and the M-line (14, 15).
p94 and the conventional calpains are expressed in skeletal muscle. However, the fact that p94 is a product of the gene, CAPN3, responsible for limb-girdle muscular dystrophy type 2A (also called calpainopathy) (12, 16, 17) clearly indicates that p94 plays roles that are distinct from those of the conventional calpains but more critical for skeletal muscle function. One intriguing feature of calpainopathy is that pathogenic missense mutations have been located along the entire p94 molecule (18, 19) (see supplemental Fig. S1), which makes it difficult to explain what kind of defects in this protease lead to calpainopathy. We have shown that the impairment of proteolytic activity of p94 by gene mutations is an essential cause of calpainopathy (20). Thus, unique autolytic activity of p94 must be related to the pathogenic mechanisms of calpainopathy. However, the nature of this relationship remains elusive, especially at the molecular level.
One of the largest stumbling blocks for the analysis of p94 is that its rapid autolysis makes biochemical experiments almost impossible to conduct. To address the relevance of autolysis of p94 to the pathogenesis of calpainopathy, we used an activity-attenuated mutant, p94:N358D, in which one of the active-site residues, Asn-358, is replaced by Asp. In contrast to wild-type (WT)5 p94, which autolyses quickly in the absence of Ca2+, p94:N358D expressed in Sf-9 cells can be partially purified and, contrary to our expectation, shows a Ca2+-dependent autolysis, unlike p94:C129S (15). p94:N358D is very unstable in solutions with high salt concentrations. Therefore, we examined the relationship between autolysis and the salt concentration. We found that p94 is activated by Na+, making it the first example of an intracellular Na+-dependent enzyme. This finding helps to explain some of unique features of p94 and calpainopathy. This report characterizes Na+ dependence of p94, focusing on the possibility that the coordinated sensitivity of p94 to Na+ and Ca2+ enables its regulation to be quite sophisticated and that it governs a multiplicity of p94 functions.
EXPERIMENTAL PROCEDURES
Experimental Animals
All procedures using experimental animals were approved by the Experimental Animal Care and Use Committee of Rinshoken, and the animals were treated in accordance with the committee guidelines. C57BL/6 mice were purchased from Nihon CLEA Inc. Cre-recombinase-expressing Tg mice were provided by The Jackson Laboratory. p94CS-KI mice (Capn3C129S/C129S) were generated by introducing a missense mutation corresponding to C129S by gene targeting using 129S ES cells and the Cre-loxP system so that only mouse p94-C129S, instead of WT p94, was expressed in these mutant mice under the normal transcriptional regulation of Capn3, which will be described elsewhere.6
Biochemical Experiments
Pellet (Ppt) and supernatant (Sup) fractions from the gastrocnemius (GC) of WT and p94CS-KI mice were prepared as described previously (21) but without adding the calpastatin peptide. The p94-N358D and p94-C129S/N358D proteins were partially purified as described previously (15). Partial purification of rat wild-type p94 was performed as follows; 150 mg of rat GC was homogenized in homogenization buffer (10 mm Tris/Cl (pH 8.0), 1 mm EDTA/KOH (pH 7.5), 1 mm dithiothreitol) containing protease inhibitors, in this case (20 μg/ml soybean trypsin inhibitor (Sigma), 28 μm E64 (Peptide Institute Inc.), 0.5 mm 4-(2-aminoethyl)benzenesulfonyl fluoride (Sigma), 0.05 mg/ml calpastatin (Takara), and 2 mm phenylmethylsulfonyl fluoride (Sigma)). The Sup obtained by centrifugation (12,500 × g, 15 min, twice) was applied to a DEAE-Toyopearl (Tosoh) column (6.3 ml) and separated into 4-ml fractions eluted by 0–0.4 m CsCl in 10 mm Tris/Cl (pH 7.5), 1 mm EDTA/KOH (pH 7.5), and 1 mm dithiothreitol (see supplemental Fig. S2). The fractions were subjected to Western blot analysis, and the fraction containing the most abundant p94 (∼0.15 m CsCl elution) was concentrated 40-fold by ultrafiltration using a Millipore Microcon YM-10 (10-kDa cut) and used for the autolysis (incubation) assay.
The autolysis assay was performed by incubating partially purified p94-N358D or p94-C129S/N358D protein (∼10 ng/μl for the 94-kDa band) (Fig. 1), the pellet and supernatant fractions from WT and p94CS-KI mouse GC (prepared as above) (Fig. 2A), a partially purified rat wild-type p94 fraction prepared as above (∼1 ng/μl) (Fig. 2B), or the lysates of the cells expressing p94 mutants (Fig. 4; see below) in 20 mm Tris/Cl (pH 7.5) and 1 mm dithiothreitol for the indicated times at the indicated temperatures with the indicated concentrations of salts (0∼300 mm NaCl, KCl, or CaCl2, or 2 mm EDTA/KOH (pH 8.0)). For CaCl2, 2× Ca2+/EGTA buffer (40 mm PIPES/KOH (pH7.2), 4 mm MgCl2, 2 mm CaCl2, and 10.0, 5.00, 2.30, 2.01, 1.95, 1.80, 1.40, and 0.00 mm EGTA/KOH (pH 7.2) for pCa = ∞, 7, 6, 5, 4.5, 4, 3.5, and 3, respectively; see Ref. 22) was used. The intensities of the bands detected by Western analysis using the p94-specific anti-pIS2 antibody (21) were quantified by Photoshop CS2 (Adobe). The relative activities were calculated by normalization in which the background intensity and band intensity at [NaCl] = [CaCl2] = 0 (shown in Fig. 2B, lane 1) are set to 0 and 100%, respectively. The lysates of the cells expressing p94 mutants were prepared by homogenizing the cells in 20 mm Tris/Cl (pH 7.5), 1 mm EDTA/KOH (pH 8.0), and 1 mm dithiothreitol (Fig. 4).
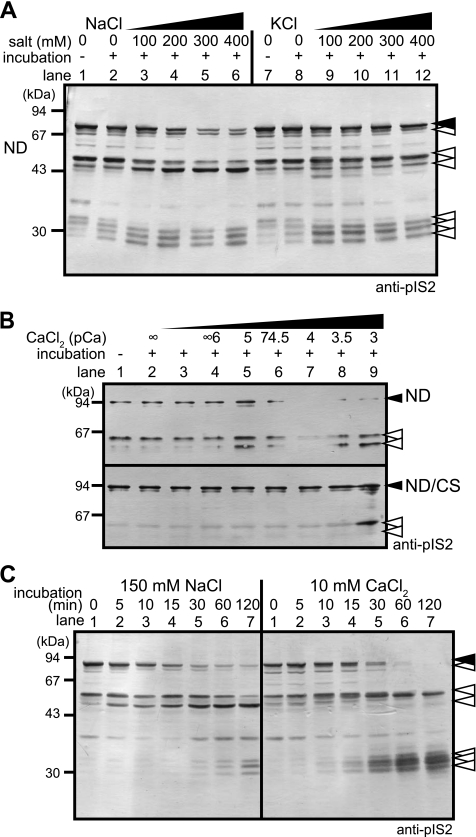
FIGURE 1.
Na+-dependent autolysis of p94. A, partially purified p94:N358D protein was incubated for 0 h (lanes 1 and 7) or 15 h (lanes 2–6 and 8–12) at 4 °C at the indicated [NaCl] or [KCl]. B, p94:N358D (ND, upper) or the inactive p94:C129S/N358D double mutant (ND/CS, lower; negative control) was incubated at the indicated [CaCl2] (pCa = −log[CaCl2]) at 37 °C for 30 min. C, p94:N358D was incubated at 37 °C for the indicated times with 150 mm NaCl or 10 mm CaCl2. Note that the autolysis induced by NaCl or CaCl2 at these concentrations proceeded at similar speeds but resulted in slightly different autolytic fragment patterns. Closed and open arrowheads indicate the full-length and autolytic fragments of p94, respectively, detected by Western blotting using an anti-pIS2 antibody.
FIGURE 2.
Na+- and Ca2+-dependent autolysis of intact p94. A, Ppt (lanes 1–16) and Sup (17–32) fractions of skeletal muscle extracts from WT and p94CS-KI (CS) mice were incubated with 2 mm EDTA (CaCl2, −), 4 mm CaCl2 (CaCl2, +), and/or 150 mm NaCl (NaCl, +) at 37 °C for 0 (incubation, −) or 40 min (incubation, +). Asterisks denote nonspecific signals (bands around 110 and 44 kDa were derived from SERCA and actin, respectively; the three other nonspecific signals in the Sup were from the secondary antibody). B, partially purified WT p94 protein from rat skeletal muscle extract (see supplemental Fig. S2) was incubated with various combinations of NaCl and CaCl2 as indicated at 37 °C for 10 min. The asterisk denotes a nonspecific signal from an antigen that was concentrated in this partially purified fraction. C, intensities of the p94 bands (B, closed arrowhead) were quantified, and the relative activities were plotted as a function of [NaCl] at a given [CaCl2]. The [NaCl] that gave half-maximal activity (Ka(Na+)) at [CaCl2] = 100 nm was ∼15 mm. D, skeletal muscle primary culture cells from WT (lanes 1–7) or p94CS-KI (8–10) mice were grown in differentiation medium for 6 days to form myotubes (24) and incubated without (lanes 1 and 8) or with 1 μm (lane 2), 5 μm (lane 3), or 10 μm (lanes 4 and 9) monensin or 10 μm (lane 5), 100 μm (lane 6), or 1 mm (lane 7 and 10) ouabain. Cells were harvested after 1 h. anti-pIS2 and CBB under the panels indicate detection was by Western blot analysis using anti-pIS2 and by Coomassie Brilliant Blue staining, respectively. Closed and open arrowheads indicate the full-length and autolytic fragments of p94, respectively, detected by an anti-pIS2 antibody.
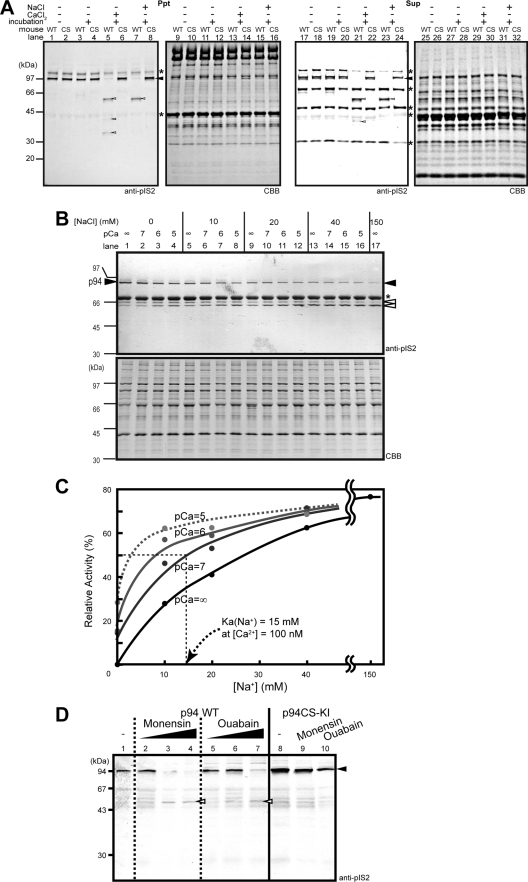
FIGURE 4.
Western blot analysis of p94 mutants expressed in COS7 cells. Where indicated, the lysates of the cells expressing p94 mutants were incubated with 2 mm EDTA (E), 10 mm CaCl2 (Ca), or 150 mm NaCl (Na) at 37 °C. Vc indicates the negative control using an empty vector. Closed and open arrowheads indicate the full-length and autolytic fragments of the p94 mutants, respectively, detected by an anti-pIS2 or anti-p94-dI antibody, as indicated. Note that plural autolytic sites exist in the IS1 and IS2 of p94, and the Mr of the autolytic fragments varies depending on mutations (15). A, IS deletion mutants, Δex6 (= ΔIS1), Δex15, 16 (= ΔIS2), and Δex6, 15, 16 (= Δ(IS1+IS2)), were incubated in the absence (Substrate (C129S), −) or presence (Substrate (C129S), +) of p94:C129S as an intermolecular autolytic substrate. This is because IS1 deletion almost completely suppressed the intramolecular autolysis (lanes 7 and 8). Note that Δ(IS1+IS2) compromised the Na+, but not Ca2+, dependence of the mutant (lanes 27 and 28). Gray arrowheads indicate the full-length of the substrate p94:C129S (94 kDa). B, the spontaneous autolytic and fodrinolytic (lower panel) activities of p94 C-terminal deletion mutants, Δ(IS2+dIV) and Δ(dIII+IS2+dIV), were compared with those of p94 WT (lane 2) and their C129S double-mutant versions (lanes 4 and 6). Qualitative evaluations of autolysis and fodrinolysis are indicated by + and −. Although Δ(dIII+IS2+dIV) showed neither spontaneous autolysis nor fodrinolysis, it showed Ca2+- but not Na+-dependent autolysis (lanes 8 and 9), which was also blocked by EDTA (lane 7) as in the case of Δ(IS1+IS2) (A, lanes 25–28)) (28). C, involvement of CBS-IIa in activation of p94 by Na+ and Ca2+ was examined using a series of p94 mutants as in B. None of the single mutants in CBS-IIa or -IIb (lanes 3–6) showed substantially compromised activities of spontaneous autolysis and fodrinolysis, strongly suggesting that they retained both their Na+ and Ca2+ dependence, which could not be examined further by incubation due to the low signal level as in the case of WT. In contrast, the double mutant in CBS-IIa, D120A/E199A (lane 7), lost both the activities, but that in CBS-IIb (E364A/D371A, lane 8) did not. D, shown is Ca2+ and Na+ dependence of the autolysis in D120A/E199A and E364A/D371A compared with that of p94:C129S and p94:N358D. Again, D120A/E199A completely lost both its Ca2+ and Na+ dependence (lanes 13 and 4), but E364A/D371A retained them though very weak (lanes 17 and 18).
For intramolecular autolysis assay, they were incubated in the presence of 2 mm EDTA/KOH (pH 8.0), 10 mm CaCl2, or 150 mm NaCl at 37 °C for 40 min (Figs. 4, B, right panel, and D). To evaluate “intermolecular autolytic” activity of IS1 deletion mutants, of the amount of the lysate prepared from cells expressing p94:C129S mutant was added before the incubation (Fig. 4A).
Spontaneous autolysis of p94 and fodrinolysis by p94 were detected by Western analysis of the cell lysates without incubation using an anti-pIS2 antibody (for all except for C-terminal deletion mutants, which do not contain the antigen region), an anti-p94-dI antibody (Abcam, ab38960; for C-terminal deletion mutants), and an anti-Gly-Met-Met-Pro-Arg (GMMPR) antiserum, which was raised for this study using the penta-peptide (NH2-GMMPR-COOH) corresponding to the N-terminal sequence of calpain-proteolyzed fodrin as previously described (23) (Figs. 4, B, left panels, and C).
Construction of p94 Mutant Expression Vectors (Figs. 3B and 4)
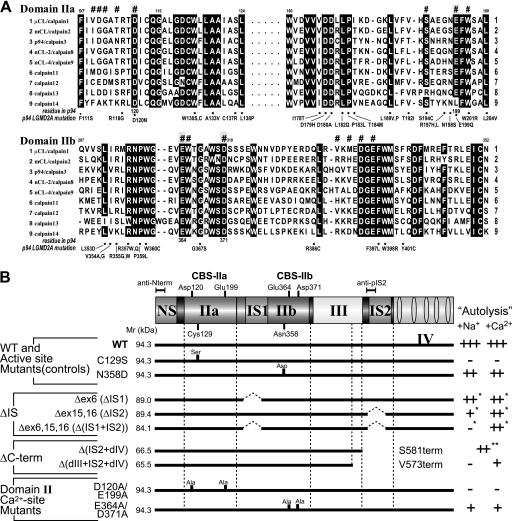
FIGURE 3.
Regions responsible for the Na+ dependence of p94. A, alignment of the amino acid sequences of nine typical human calpains is shown. Parts of the protease subdomains IIa and IIb, which include the residues responsible for Ca2+ binding (indicated by #; named CBS-IIa and CBS-IIb, respectively) are shown. The residues shown in gray on a black background were mutated in subsequent experiments. Residues conserved among more than seven sequences are shown on a black background. Residue numbers for μCL/calpain1 are given above the sequences. Residue numbers for p94/calpain3 and calpainopathy-related mutations (from Leiden Muscular Dystrophy pages) are shown below the alignment (see also supplemental Fig. S1). B, summary of mutants used in this study and the dependence of their autolysis on Na+ and Ca2+ (see Fig. 4). For ΔIS mutants corresponding to splicing isoforms, Δex6, Δex15,16, and Δex6,15,16, the dependence was evaluated by their proteolytic activity against p94:C129S (*, see Fig. 4A). The dependence of the Δ(IS2+dIV) mutant was determined by comparing its level of expression and fodrinolytic activity with those of the Δ(IS2+dIV)/C129S double mutant (**, see Fig. 4B). Note that ΔIS2, but not Δ(IS2+dIV), can be detected by anti-pIS2 as ΔIS2 retains a part of the antigen region for anti-pIS2.
IS1 and/or IS2 deletion mutants were produced using human WT p94 cDNA, as previously described (15). Missense mutants were produced as previously described (20). C-terminal deletion mutants were produced by introducing a termination codon into Ser-581 or Val-573 using the same mutation protocol as for the missense mutants. These cDNAs were inserted into a pSRD vector and expressed in COS7 cells as previously described (21).
Muscle Cell Cultures
The preparation of mouse skeletal muscle primary cell cultures, and their transfection were carried out as previously described (24). They were incubated with or without the indicated concentrations of monensin or ouabain. The cells were harvested after 1 h and subjected to Western blot analysis using the anti-pIS2 antibody (Fig. 2D). For real-time observation of the p94 autolysis (supplemental Videos S1 and S2), the full-length human WT p94 cDNA and its protease-inactive form, p94-C129S, were cloned into the pEGFP vector (Clontech). Four days after switching to differentiation medium, myocytes were subjected to analysis. To increase the [Na+]i, 1 mm ouabain was added to the culture medium. Images were scanned and recorded using an LSM510 confocal microscope (Zeiss) equipped with an incubation system (37 °C, 5% CO2; Tohkaihit Co.).
Proteomic Analysis (Figs. 5–7)
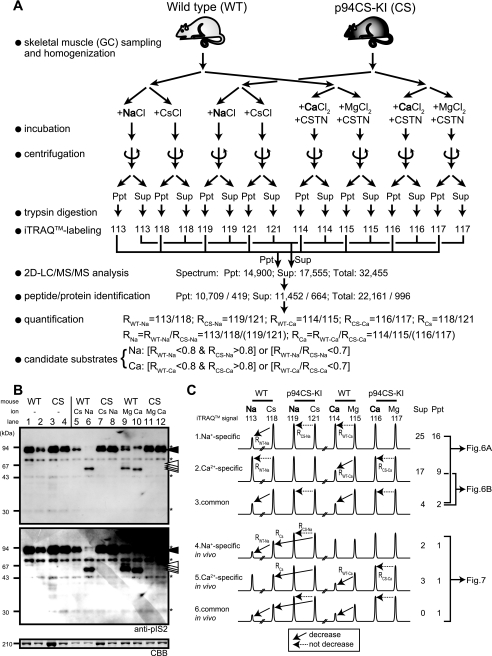
FIGURE 5.
Scheme for the differential proteome analysis to determine p94 substrates specific for the activating ions. A, total extracts from the GC muscles of WT and p94CS-KI mice were incubated in the presence of NaCl or CsCl and EDTA or of CaCl2 or MgCl2 and calpastatin. The samples were spun to obtain the Sup and Ppt fractions, digested by trypsin, and labeled with iTRAQTM 8-plex reagent. The labeled samples were mixed and analyzed by two-dimensional/tandem mass spectometry, and peptides/proteins were identified by ProteinPilotTM. Candidates for substrates were selected as those showing a significant decrease after incubation with NaCl or CaCl2 compared with the negative control (CsCl or MgCl2, respectively) only for WT but not for p94CS-KI, as indicated in the equations. B, shown is a Western blot analysis of the above samples using anti-pIS2. The above samples (total extracts before centrifugation) before (lanes 1–4) or after (lanes 5–12) incubation with CsCl (lanes 5 and 7), NaCl (lanes 6 and 8), MgCl2 (lanes 9 and 11), or CaCl2 (lanes 10 and 12) were examined by Western blot analysis using the anti-IS2 antibody. The upper and middle panels show 30-s and 3-min exposures. Asterisks denote nonspecific signals. The lowest panel, the 210-kDa myosin heavy chain stained by Coomassie Brilliant Blue (BBC), shows that each sample had a different protein concentration before the incubation (the second WT sample was especially skewed). The amounts of p94 in the WT and p94CS-KI mouse muscle were roughly equal (see Fig. 2A). C, theoretical signal patterns for p94 substrates that were recognized in a Na+-specific (1), Ca2+-specific (2), or Na+/Ca2+ (3) manner (for a list of substrates, see Fig. 6 as indicated). If substrates were constitutively proteolyzed in vivo, their amounts should be lower in WT than in p94CS-KI muscle without in vitro p94 activation. Therefore, they should be lower in WT-CsCl than in p94CS-KI-CsCl (4∼6). For candidate in vivo substrates, those identified by only a single kind of peptide were eliminated to avoid false positives (for a list, see Fig. 7). The numbers in the right-hand column indicate the number of proteins in each fraction (Sup or Ppt) that showed the corresponding patterns.
FIGURE 6.
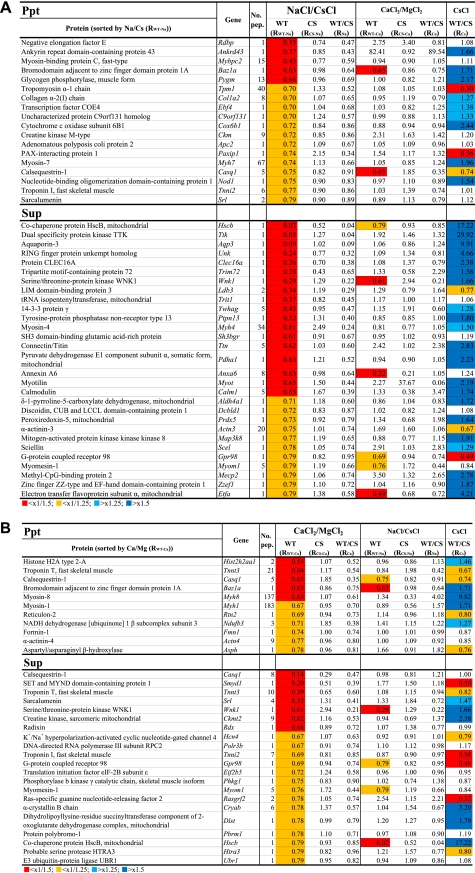
Summary of proteins that showed a Na+- and/or Ca2+-dependent decrease in WT, but not p94CS-KI, mouse skeletal-muscle fractions. A, Na+-dependent substrate candidates corresponding to those shown in Fig. 5C, 1 or 3. The lists were sorted by RWT-Na (the ratio of protein in NaCl to that in CsCl for WT mice corresponding to iTRAQTM signals at 113 and 118 for WT (see Fig. 5A). WT/CS is the ratio of the values obtained for WT and CS by each normalization (RWT-Na/RCS-Na and RWT-Ca/RCS-Ca, respectively). B, Ca2+-dependent substrate candidates corresponding to those shown in Fig. 5C, 2 or 3. Lists were sorted by RWT-Ca. Red, orange, sky blue, or dark blue backgrounds indicate that the values were less than 1/1.5 (0.67) or 1/1.25 (0.8) or more than 1.25 or 1.5, respectively. No. pep. indicates the number of kinds of peptides used to identify the corresponding protein.
FIGURE 7.
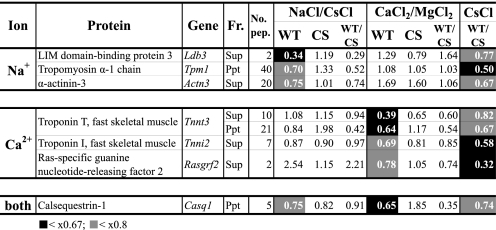
Potential in vivo substrates for p94 differentially activated by Na+ and Ca2+. Candidate in vivo substrates are selected from Fig. 6. The rationale for selecting candidates is shown schematically in Fig. 5C, 4–6. Black and gray backgrounds indicate values <1/1.5 (0.67) and <1/1.25 (0.8), respectively. No. pep. indicates the number of kinds of peptides used to identify the corresponding protein.
Skeletal muscle (GC) extracts from WT and p94CS-KI mice (two mice for each species) were prepared by homogenizing them in 20 mm triethylammonium bicarbonate, 1 mm EDTA/K+, 1 mm Tris-(2-carboxylethyl) phosphine, and 1 mm phenylmethylsulfonyl fluoride, as described previously (21). The protein concentrations were determined for each sample, and equal amounts of two WT samples were mixed and used for further analysis. The p94CS-KI samples were treated similarly, so that both the WT and p94CS-KI samples had the same protein concentration (final 7.5 mg/ml). They were then incubated at 37 °C for 10 min with 150 mm NaCl or CsCl with 1 mm EDTA/KOH (pH 8.0) or 2 mm CaCl2 or MgCl2 with 0.15 mg/ml calpastatin (to suppress the activities of the conventional calpains; Takara). After the incubation, the Sup and Ppt fractions were obtained by centrifugation (20,000 × g, 30 min, 4 °C). Each fraction was then completely digested by trypsin (37 °C, 15 h) followed by labeling with iTRAQTM 8-plex reagent (ABI) according to the manufacturer's instructions. The samples were then analyzed using two-dimensional-liquid chromatography tandem mass spectometry as previously described (25). Peptides and proteins were identified by ProteinPilotTM Version 3 using a confidence value of 0.7 (80%). Ratios were calculated from quantified iTRAQTM signals: RWT-Na = iTRAQTM113/iTRAQTM118, RCS-Na = 119/121, RWT-Ca = 114/115, RCS-Ca = 116/117, RNa = RWT-Na/RCS-Na = 113/118/(119/121), RCa = RWT-Ca/RCS-Ca = 114/115/(116/117), RCs = 118/121. Candidates for substrates were selected as those showing a significant decrease (RWT-Na < 0.8, RWT-Ca < 0.8) after incubation with NaCl or CaCl2 compared with the negative control (CsCl or MgCl2, respectively) only for WT but not for p94CS-KI (RCS-Na > 0.8, RCS-Ca > 0.8, or RWT-Na/RCS-Na < 0.7, RWT-Ca/RCS-Ca < 0.7). (see Fig. 5A). The activation and autolysis of p94 was confirmed by Western analysis of each fraction (before separation by centrifugation) using the anti-pIS2 antibody and Super Signal West Dura detection (Thermo) (Fig. 5B). The rationale for selecting candidates for in vitro and in vivo substrates is shown schematically in Fig. 5C.
RESULTS
An Activity-attenuated Mutant of Skeletal Muscle-specific Calpain, p94/Calpain 3, Showed Na+-dependent Autolysis
In contrast to conventional calpains, whose autolysis is suppressed by high salt concentrations (26), autolysis of p94:N358D was NaCl-dependent (Fig. 1A). Surprisingly, KCl did not cause significant autolysis (Fig. 1A), indicating that the p94 autolysis was specifically dependent on Na+ and was not a general response to high salt concentrations. This explains the previous in vitro translation results showing that WT p94 undergoes autolysis in the absence of Ca2+ and the presence of NaCl (13). As described above, p94:N358D also shows Ca2+-dependent autolysis (Fig. 1B) (15). Ten millimolar CaCl2 and 150 mm NaCl were similarly effective (Fig. 1C). Notably, the autolysis patterns of p94 were different when induced by Ca2+ or Na+, suggesting that p94:N358D assumed distinct conformations when activated by the two ions. The [Na+] and [Ca2+] required for autolytic activity were ∼100 mm and 100 μm, respectively, in the absence of other ions (Fig. 1, A and B).
Native WT p94 Also Showed Na+-dependent Autolysis
We next examined whether native WT p94 showed the same activity profile. As a control, we used p94:C129S “knock-in” (p94CS-KI) mice, which have a missense mutation corresponding to C129S in Capn3 exon 3 and express a protease activity-null p94 mutant p94:C129S in place of WT p94.6 Total lysates of the skeletal muscles from WT and p94CS-KI mice were prepared in the absence of Ca2+ and Na+ and separated to Sup and Ppt fractions by centrifugation. Comparable levels of the 94-kDa pre-autolytic band were detected in both preparations and both fractions (Fig. 2A, lanes 1, 2, 17, and 18). When incubated with Ca2+ or Na+, the p94 expressed in WT muscle, but not the p94:C129S expressed in p94CS-KI muscle, autolyzed (lanes 3–8 and 19–24). When the incubations were carried out with both ions at varying concentrations using WT p94 partially purified from rat GC (supplemental Fig. S2), the ion effects were additive; that is, the [Na+] required for half-maximal autolytic activity was 15 mm at [Ca2+] = 100 nm, indicating that physiological cytosolic concentrations of Na+ and Ca2+ can activate WT p94 (Fig. 2, B and C).
As a more physiological condition, primary cultures of skeletal muscle cells from WT and p94CS-KI mice were treated with monensin or ouabain. Monensin is an ionophore specific for monovalent metal ions (Na+ ≫ K+ > Rb+) (27), and ouabain is a specific inhibitor of the Na+/K+-ATPase (16). Thus, both drugs increase [Na+]i, but by different mechanisms. When myocytes were incubated with either drug, p94 was autolyzed in the WT cells but not in the p94CS-KI cells (Fig. 2D). This indicated that p94 is activated by a small increase in [Na+]i under nearly physiological conditions. The increase in [Na+]i also caused increased [Ca2+]i, and it is also possible that the combined increase of [Na+]i and [Ca2+]i caused the autolysis of WT p94.
To investigate this phenomenon in real time, green fluorescent protein-fused WT p94 or p94:C129S was expressed in differentiated myocytes (myotubes) from WT or p94CS-KI mice, respectively, and time-lapse imaging was performed. The exogenous green fluorescent protein-p94s (both WT and p94:C129S) were predominantly observed at the M-line (24). When ouabain was added to the medium, the green fluorescent protein signals of p94WT, but not those of p94:C129S, disappeared within 30 min, indicating the rapid autolysis of p94 in the cell was stimulated by the increase in [Na+]i (see the supplemental videos and Fig. S5, A and B).
One of the Ca2+-binding Sites in the p94 Protease Domain Is Required for Na+-binding
To determine the region of p94 responsible for its Na+-dependent activation, we constructed various mutants (Fig. 3) and examined their Na+-dependent activities. The domain structure of p94 is typical of the conventional calpains (domains I to IV; dI∼dIV, see Fig. 3B and supplemental Fig. S1), and the sequences of its Ca2+-binding sites are well conserved (Fig. 3A). In addition, alternatively spliced exon 6 and exons 15–16, respectively, encode two insertion sequences in p94, IS1 and IS2, that are responsible for rapid autolysis of p94 (11, 13, 28, 29).
We first examined the autolysis of a mutant p94 lacking the IS1 and/or IS2 regions. Because the deletion of IS1 eliminates the major autolytic sites, p94:C129S was used as an alternative intermolecular autolytic substrate for the assay. In cultured cells, not only p94 itself, but also co-expressed p94:C129S is “intermolecularly autolyzed” by WT p94 (15, 30). ΔIS2 but not ΔIS1 showed a moderate reduction in Na+ dependence, and the Na+-dependent autolysis of Δ(IS1+IS2) was almost completely suppressed (Fig. 4A, lanes 12, 20, and 28). On the other hand, the expression of a C-terminal deletion mutant, Δ(IS2+dIV), showed spontaneous autolysis and fodrinolysis, similar to WT (Fig. 4B, lanes 2 and 3), i.e. at physiological [Na+]i + [Ca2+]i. Fodrin is one of in vivo substrates of conventional calpains and is also a good substrate for p94, and thus, the endogenous fodrin is proteolyzed at a specific position when p94 is expressed in cultured cells (20, 23). However, Δ(dIII+IS2+dIV), which is 8 amino acids shorter than Δ(IS2+dIV), lost both its autolytic and fodrinolytic functions (Fig. 4B, lanes 5 and 6). Incubation of Δ(dIII+IS2+dIV) with Ca2+, but not Na+, caused autolysis (Fig. 4B, lanes 7–9). These results indicated that dIV is dispensable for the Na+ binding of p94, and IS1, IS2, and dIII are involved in Na+ binding, with IS2 playing the most essential role.
Next, we replaced several well conserved Ca2+ binding residues with Ala (Fig. 3). Surprisingly, none of the single mutations (D120A or E199A, which correspond to the Ca2+-binding site in dIIa (CBS-IIa) or E364A or D371A in dIIb (CBS-IIb)) substantially compromised the spontaneous autolysis or fodrinolysis of p94 (Fig. 4C and supplemental Fig. S3). However, the double mutation D120A/E199A (in CBS-IIa) was insensitive to both Na+ and Ca2+, and E364A/D371A (in CBS-IIb) showed weak Na+- and Ca2+-dependent activity (Fig. 4D and supplemental S3). These results (summarized in Fig. 3B) strongly suggest that CBS-IIa is important for the p94 activity and that Na+ and Ca2+ both act at CBS-IIa.
We also tested 37 missense p94 mutants (including the ones described above), 3 of which corresponded to calpainopathy causative mutations. None showed Na+-dependent, Ca2+-insensitive autolysis (supplemental Fig. S4). This strongly suggests that the effect of Na+ is transduced via one of the Ca2+-binding sites. Altogether, our data indicated that IS1, IS2, and dIII contribute to the CBS-IIa-mediated Na+-dependence of p94 (supplemental Fig. S3). The involvement of other Ca2+-binding sites, however, cannot be completely eliminated. In any case, our findings suggest that the Na+ dependence of p94 is achieved by the insertion of IS1 and IS2 in the p94 molecule, which may change the relative conformations of dII, dIII, and dIV.
Na+ and Ca2+ Give p94 Differential Substrate Specificities, at Least in Vitro
Finally, to shed light on the physiological meaning of Na+ dependence of p94, we screened for p94 substrates by differential proteomic analysis using iTRAQTM. Extracts from WT and p94CS-KI skeletal muscles were incubated with NaCl, CsCl, CaCl2, or MgCl2, and 996 proteins were identified and quantified (Fig. 5A). Under the conditions used, Na+ and Ca2+ caused complete p94 autolysis, whereas Cs+ and Mg2+ resulted in no and weak autolysis, respectively (Fig. 5B). Among the proteins identified, candidates for substrates were selected as those showing a significant decrease (<×0.8) after incubation with NaCl or CaCl2 compared with their negative control (CsCl or MgCl2, respectively) in WT extract but not in p94CS-KI extract (Fig. 5C, 1–3). Relatively few (11∼29) proteins were identified for each ion and fraction. Strikingly, there was almost no overlap between the candidate substrates identified by incubation with Na+ versus Ca2+ (Fig. 6, A and B). Although these proteins may not have in vivo relevance, our data show that p94 has different substrate specificities in vitro depending on whether it is activated by Na+ or Ca2+ and on how these ions affect the substrates themselves.
Possible in Vivo Substrates for p94 Activated by Na+ and/or Ca2+
It is reasonable to think that the normal in vivo substrates for p94, if any, might be less abundant in muscle cells expressing WT p94 than in those expressing p94:C129S. To look for candidate in vivo substrates, we compared the levels of proteins present in the negative controls (WT/CS in CsCl incubation) to find those that were significantly reduced in muscle from WT mice compared with p94CS-KI mice (WT/CS-CsCl < 0.8 and NaCl/CsCl-WT < 0.8 for candidates for Na+; WT/CS-CsCl < 0.8 and CaCl2/MgCl2-WT < 0.8 for candidates for Ca2+, Fig. 5C, 4–6). This analysis yielded three candidate in vivo substrates for Na+-activated p94, three for Ca2+-activated p94, and one for p94 activated by either ion (Fig. 7). Notably, these candidates included several myofibrillar proteins, such as tropomyosin, troponins T and I, and α-actinin-3, suggesting roles for p94 in modulating the amounts of these proteins and, hence, in myofibril quality control. It is also noteworthy that calsequestrin-1, a Ca2+-binding/storing protein in the SR, is a candidate substrate for p94 activated by either ion. That calsequestrin probably has different conformations in the absence and presence of Ca2+ suggests p94 recognizes different substrates depending on whether it is activated by Na+ or Ca2+. On the other hand, it is possible that tropomyosin and troponins, also Ca2+-binding proteins, show different proteolytic patterns because they change their conformation depending on the presence or absence of Ca2+ rather than on a difference in substrate recognition of p94.
DISCUSSION
Because conventional μ- and m-calpains require ∼50 μm and 1 mm Ca2+, respectively, for their activity (7, 10), they can rarely be fully active in the cytosol, which may be a safety mechanism to prevent their overactivation. Although it is unclear which is the cause and which the result, the physiological [Na+]i-dependent low Ca2+ requirement of p94 is a property that facilitates its responsiveness to physiological [Ca2+]i changes in skeletal muscle. The rapid autolytic degradation of p94 (31), on the other hand, probably serves as a blockade against its overactivation. It is also possible that, at other times, a suppressor(s) of p94, which includes the connectin/titin N2A region (21, 32), attenuates p94 activity, enabling it to function immediately by changing its conformation in response to alterations in [Na+]i and/or [Ca2+]i without autolysis or the proteolysis of substrates.
In this respect, it is noteworthy that p94 is present in the SR (33). Although not generally high in the cell, the [Na+]i increases transiently immediately beneath the sarcolemma, during the depolarizing end-plate potential caused by the neuromuscular transmitter acetylcholine. This depolarization leads to increased [Ca2+]i as Ca2+ is released from the SR into the cytosol, but the excitation-coupled contraction is not accompanied by the influx of Ca2+ from outside the cells in most skeletal muscles (34). Instead, the sarcolemmal depolarization causes conformational changes in the skeletal muscle-specific dihydropyridine receptor at the T-tubule membrane, which interacts directly with the ryanodine receptor (RyR) at the SR membrane to elicit Ca2+ efflux from the SR (35, 36). However, it is largely unknown how depolarization causes the dihydropyridine receptor to change its conformation, and it is tempting to speculate that p94 at the SR membrane could contribute by sensing fluctuations in the local [Na+]i and/or [Ca2+]i. Indeed, p94 is predominantly expressed in skeletal, and not heart, muscle (11), and p94-KO mice have an impaired Ca2+ efflux from intracellular stores, but the influx of extracellular Ca2+ is not impaired (33).
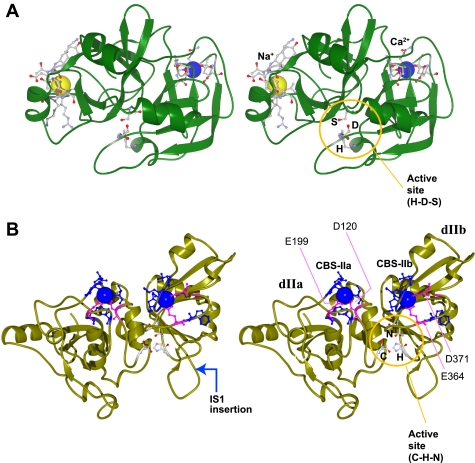
Several proteases involved in blood coagulation have Na+-dependent activity (1, 2). In particular, factor Xa (FXa) has one Ca2+- and one Na+-binding site (see Fig. 8). These sites are allosterically linked and have additive effects, although the concentrations required for activity of FXa are much higher than that of p94 because FXa is equipped to respond to extracellular conditions (apparent dissociation constant for [Na+] of FXa is 116 mm when [Ca2+] is 2.5 mm) (37). FXa is a serine protease whose primary structure has no similarity to that of the calpain protease domain (3, 38). Nevertheless, the loops responsible for the Ca2+/Na+ binding are similarly positioned relative to the active-site cleft in these two proteins (Fig. 8), which may be informative about their evolution.
FIGURE 8.
Three-dimensional structures of factor Xa and the μ-calpain/calpain 1 protease domain. Cross-eye stereo views were drawn by FiatLux MolFeat Version 4.5 using Protein Data Bank codes 2VVU for factor Xa (A) and 1KXR for Ca2+-bound μ-calpain (B). The active-site residues are indicated by the ball-and-stick drawings in the orange circles. The blue and yellow balls represent Ca2+ and Na+. The Ca2+ binding residues of the μ-calpain protease domain and those corresponding to the ones used for the mutagenesis study (see Fig. 3) are indicated by blue and pink ball-and-stick drawings, respectively. The position of the IS1 insertion in p94 is indicated by the blue arrow.
Because the metal ion radii of Ca2+ and Na+ are almost identical, a slight modification might enable a Ca2+-binding site to bind Na+. Indeed, Bonagura et al. (39, 40) reported that artificial mutagenesis of plural amino acid residues of cytochrome c peroxidase (CCP) makes CCP K+-, Na+-, or Ca2+-dependent. Our mutagenesis study (see Fig. 3) also strongly suggested that p94 CBS-IIa acquired its Na+-sensing ability by undergoing conformational changes induced not only by IS1 and residues in the vicinity of CBS-IIa but also by IS2 and dIII. This suggests that a large portion of the p94 molecule is involved in making p94 Na+-dependent. Therefore, various mutations along the entire p94 molecule can potentially destroy proper activity of p94, leading to calpainopathy. This may explain why calpainopathy patients have missense mutations scattered along the entire p94 molecule rather than clustered in the protease domain (18, 19) (see supplemental Fig. S1).
The evolutional process that led to acquisition of Na+ binding ability of p94 is unknown but interesting to contemplate. Obviously, the insertion of IS1 and IS2 by the addition of exons 6, 15, and 16 represents one or more important events. All vertebrates, but no other living organisms so far examined, have highly conserved CAPN3 genes (supplemental Fig. S1 and Table S1). The deduced amino acid sequence identity between the p94 paralogues of the most distant vertebrates, humans, and puffer fish, is 61.6% (supplemental Table S2), which is comparable with the interspecies identities of the representative intracellular proteases cathepsin L (56.4%), caspase 3 (64.4%), and proteasome subunit β5 (71.0%) (41). This level of sequence identity implies that p94 is indispensable in the skeletal muscles of all vertebrates. Strikingly, the N-terminal half of the IS2 region, one of the characteristic p94 protein domains, is highly conserved in all vertebrates (supplemental Fig. S1), and three human calpainopathy-causative mutations (S606L, R608K, and A609E) are found as a cluster in this region.
The protease domain (subdomains IIa+IIb) and domain III are also highly conserved across vertebrate species, but subdomain IIb is slightly less conserved than the two others. In particular, IIa and IIb differ most in their Ca2+-binding sites; i.e. five of eight residues constituting CBS-IIa are conserved in all sequences, whereas two of seven are conserved in CBS-IIb (supplemental Fig. S1). The importance of CBS-IIa is also supported by the fact that Thr-192 and Lys-193 adjacent to CBS-IIa are conserved in all p94s but not in any other vertebrate calpain (supplemental Fig. S1) (42). Accordingly, calpainopathy-associated mutations have been identified in four of the CBS-IIa residues and Thr-192 (D120N, S194C, E199Q, W201R, and T192I), but none has been found in the CBS-IIb residues (supplemental Fig. S1).
Altogether, these findings support the importance of IS2, CBS-IIa, and domain III for p94 function. On the other hand, the IS1 sequences are rather divergent. Considering our finding that p94:ΔIS1 had almost no effect on the Na+/Ca2+ dependence (see Figs. 3B and 4A) except for a loss of intramolecular autolysis, IS1 probably provides the intramolecular proteolytic sites that allow rapid autodegradation. In this sense, the amino acid sequence itself may not be so important, and any unstructured oligopeptide may function as the IS1. Then, why did vertebrates need an additional unique calpain, p94? Generally, vertebrates are terrestrial and larger than invertebrates, and thus, their skeletal muscles carry much higher loads/stress. It is possible that p94 is involved in the stress response of vertebrate skeletal muscles, e.g. by regulating stress-responsive proteins such as heat-shock proteins.
In light of our results, we propose three functional roles for Na+ dependence of p94; 1) the Na+ dependence could make p94 more responsive than conventional calpains to physiological changes in [Ca2+], 2) substrate specificity of p94 could be finely regulated by the balance between [Na+] and [Ca2+], and 3) a conformational change in p94 upon its binding to Na+/Ca2+ could be propagated through its interaction with structural components. Our findings are important for understanding how pathogenic/dystrophic situations are normally circumvented in skeletal muscle, and they may contribute to the development of novel therapeutic and diagnostic strategies for muscular dystrophies. At a minimum, our finding also provides the useful information that patient muscle samples should be analyzed in Na+-free buffers when a diagnosis of calpainopathy is suspected. Otherwise, p94 is rapidly degraded, which can result in a misdiagnosis. On the other hand, comparison of incubations of skeletal muscle extracts from patients in buffers with and without NaCl will provide us with simple and accurate detection of p94 activity, i.e. diagnosis of calpainopathy, which has been already suggested previously (43). Our results also reveal a new essential function of Na+ in the cytosol and raise the possibility that Na+ is involved in regulating other cytosolic Ca2+-dependent enzymes.
Supplementary Material
Acknowledgments
We thank all the members of the Calpain Project laboratory (Rinshoken) and Laboratory of Biological Function (University of Tokyo) for valuable support and discussions.
This work was supported in part by MEXT.KAKENHI Grants 18076007 (to H. S.) and 22770139 (to Y. O.) and JSPS.KAKENHI Grants 18700392 and 20500369 (to K. O.) and 19658057 and 20370055 (to H. S.), by Research Grant 20B-13 for Nervous and Mental Disorders from the Ministry of Health, Labour, and Welfare (to H. S.), and by a Takeda Science Foundation research grant (to H. S.).
We are most grateful to Dr. Koichi Suzuki (Professor Emeritus of The University of Tokyo), who established the basis of our calpain studies including this study and passed away on April 20, 2010. We dedicate this paper to the memory of him and will never forget his mentorship and generosity.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2, Figs. S1–S5, and Videos S1 and S2.
Ojima, K., Kawabata, Y., Nakao, H., Nakao, K., Doi, N., Kitamura, F., Ono, Y., Hata, S., Suzuki, H., Kawahara, H., Bogomolovas, J., Witt, C., Ottenheijm, C., Labeit, S., Granzier, H., Toyama-Sorimachi, N., Sorimachi, M., Suzuki, K., Maeda, T., Abe, K., Aiba, A., and Sorimachi, H. (2010) J. Clin. Invest. 120, in press..
- WT
- wild type
- [Ca2+]i
- intracellular [Ca2+]
- CBS-IIa
- Ca2+-binding site in dIIa
- CBS-IIb
- Ca2+-binding site in dIIb
- dI–VI
- domain I–VI
- FXa
- factor Xa
- GC
- gastrocnemius
- IS1/2
- insertion sequence 1/2
- [Na+]i
- intracellular [Na+]
- p94CS-KI mouse
- p94:C129S knock-in mouse
- Ppt
- pellet
- SR
- sarcoplasmic reticulum
- Sup
- supernatant
- PIPES
- 1,4-piperazinediethanesulfonic acid.
REFERENCES
- 1.Dang Q. D., Di Cera E. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 10653–10656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silva F. P., Jr., Antunes O. A., de Alencastro R. B., De Simone S. G. (2006) Biophys. Chem. 119, 282–294 [DOI] [PubMed] [Google Scholar]
- 3.Kamata K., Kawamoto H., Honma T., Iwama T., Kim S. H. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6630–6635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldin A. L. (2001) Annu. Rev. Physiol. 63, 871–894 [DOI] [PubMed] [Google Scholar]
- 5.Krishnamurthy H., Piscitelli C. L., Gouaux E. (2009) Nature 459, 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogawa H., Shinoda T., Cornelius F., Toyoshima C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13742–13747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goll D. E., Thompson V. F., Li H., Wei W., Cong J. (2003) Physiol. Rev. 83, 731–801 [DOI] [PubMed] [Google Scholar]
- 8.Beckmann J. S., Spencer M. (2008) Neuromuscul. Disord. 18, 913–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yousefi S., Perozzo R., Schmid I., Ziemiecki A., Schaffner T., Scapozza L., Brunner T., Simon H. U. (2006) Nat. Cell Biol. 8, 1124–1132 [DOI] [PubMed] [Google Scholar]
- 10.Suzuki K., Hata S., Kawabata Y., Sorimachi H. (2004) Diabetes 53, S12–S18 [DOI] [PubMed] [Google Scholar]
- 11.Sorimachi H., Imajoh-Ohmi S., Emori Y., Kawasaki H., Ohno S., Minami Y., Suzuki K. (1989) J. Biol. Chem. 264, 20106–20111 [PubMed] [Google Scholar]
- 12.Richard I., Broux O., Allamand V., Fougerousse F., Chiannilkulchai N., Bourg N., Brenguier L., Devaud C., Pasturaud P., Roudaut C., Hillaire D., Passos-Bueno M. R., Zatz M., Tischfield J. A., Fardeau M., E. J. C., Cohen D., Beckmann J. S. (1995) Cell 81, 27–40 [DOI] [PubMed] [Google Scholar]
- 13.Sorimachi H., Toyama-Sorimachi N., Saido T. C., Kawasaki H., Sugita H., Miyasaka M., Arahata K., Ishiura S., Suzuki K. (1993) J. Biol. Chem. 268, 10593–10605 [PubMed] [Google Scholar]
- 14.Labeit S., Kolmerer B. (1995) Science 270, 293–296 [DOI] [PubMed] [Google Scholar]
- 15.Ono Y., Torii F., Ojima K., Doi N., Yoshioka K., Kawabata Y., Labeit D., Labeit S., Suzuki K., Abe K., Maeda T., Sorimachi H. (2006) J. Biol. Chem. 281, 18519–18531 [DOI] [PubMed] [Google Scholar]
- 16.Richard I., Roudaut C., Marchand S., Baghdiguian S., Herasse M., Stockholm D., Ono Y., Suel L., Bourg N., Sorimachi H., Lefranc G., Fardeau M., Sébille A., Beckmann J. S. (2000) J. Cell Biol. 151, 1583–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kramerova I., Kudryashova E., Tidball J. G., Spencer M. J. (2004) Hum. Mol. Genet. 13, 1373–1388 [DOI] [PubMed] [Google Scholar]
- 18.Richard I., Roudaut C., Saenz A., Pogue R., Grimbergen J. E., Anderson L. V., Beley C., Cobo A. M., de Diego C., Eymard B., Gallano P., Ginjaar H. B., Lasa A., Pollitt C., Topaloglu H., Urtizberea J. A., de Visser M., van der Kooi A., Bushby K., Bakker E., Lopez de Munain A., Fardeau M., Beckmann J. S. (1999) Am. J. Hum. Genet. 64, 1524–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sáenz A., Leturcq F., Cobo A. M., Poza J. J., Ferrer X., Otaegui D., Camaño P., Urtasun M., Vílchez J., Gutiérrez-Rivas E., Emparanza J., Merlini L., Paisán C., Goicoechea M., Blázquez L., Eymard B., Lochmuller H., Walter M., Bonnemann C., Figarella-Branger D., Kaplan J. C., Urtizberea J. A., Martí-Massó J. F., López de Munain A. (2005) Brain 128, 732–742 [DOI] [PubMed] [Google Scholar]
- 20.Ono Y., Shimada H., Sorimachi H., Richard I., Saido T. C., Beckmann J. S., Ishiura S., Suzuki K. (1998) J. Biol. Chem. 273, 17073–17078 [DOI] [PubMed] [Google Scholar]
- 21.Hayashi C., Ono Y., Doi N., Kitamura F., Tagami M., Mineki R., Arai T., Taguchi H., Yanagida M., Hirner S., Labeit D., Labeit S., Sorimachi H. (2008) J. Biol. Chem. 283, 14801–14814 [DOI] [PubMed] [Google Scholar]
- 22.Harafuji H., Ogawa Y. (1980) J. Biochem. 87, 1305–1312 [DOI] [PubMed] [Google Scholar]
- 23.Saido T. C., Yokota M., Nagao S., Yamaura I., Tani E., Tsuchiya T., Suzuki K., Kawashima S. (1993) J. Biol. Chem. 268, 25239–25243 [PubMed] [Google Scholar]
- 24.Ojima K., Ono Y., Doi N., Yoshioka K., Kawabata Y., Labeit S., Sorimachi H. (2007) J. Biol. Chem. 282, 14493–14504 [DOI] [PubMed] [Google Scholar]
- 25.Ono Y., Hayashi C., Doi N., Kitamura F., Shindo M., Kudo K., Tsubata T., Yanagida M., Sorimachi H. (2007) Biotechnol. J. 2, 565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Thompson V. F., Goll D. E. (2004) Biochim. Biophys. Acta 1691, 91–103 [DOI] [PubMed] [Google Scholar]
- 27.Yu X. M., Salter M. W. (1998) Nature 396, 469–474 [DOI] [PubMed] [Google Scholar]
- 28.Ono Y., Kakinuma K., Torii F., Irie A., Nakagawa K., Labeit S., Abe K., Suzuki K., Sorimachi H. (2004) J. Biol. Chem. 279, 2761–2771 [DOI] [PubMed] [Google Scholar]
- 29.Herasse M., Ono Y., Fougerousse F., Kimura E., Stockholm D., Beley C., Montarras D., Pinset C., Sorimachi H., Suzuki K., Beckmann J. S., Richard I. (1999) Mol. Cell Biol. 19, 4047–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorimachi H., Kinbara K., Kimura S., Takahashi M., Ishiura S., Sasagawa N., Sorimachi N., Shimada H., Tagawa K., Maruyama K., Suzuki K. (1995) J. Biol. Chem. 270, 31158–31162 [DOI] [PubMed] [Google Scholar]
- 31.Spencer M. J., Guyon J. R., Sorimachi H., Potts A., Richard I., Herasse M., Chamberlain J., Dalkilic I., Kunkel L. M., Beckmann J. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8874–8879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huebsch K. A., Kudryashova E., Wooley C. M., Sher R. B., Seburn K. L., Spencer M. J., Cox G. A. (2005) Hum. Mol. Genet. 14, 2801–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kramerova I., Kudryashova E., Wu B., Ottenheijm C., Granzier H., Spencer M. J. (2008) Hum. Mol. Genet. 17, 3271–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flucher B. E., Franzini-Armstrong C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8101–8106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanabe T., Beam K. G., Adams B. A., Niidome T., Numa S. (1990) Nature 346, 567–569 [DOI] [PubMed] [Google Scholar]
- 36.Cheng W., Altafaj X., Ronjat M., Coronado R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 19225–19230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rezaie A. R., He X. (2000) Biochemistry 39, 1817–1825 [DOI] [PubMed] [Google Scholar]
- 38.Hanna R. A., Campbell R. L., Davies P. L. (2008) Nature 456, 409–412 [DOI] [PubMed] [Google Scholar]
- 39.Bonagura C. A., Bhaskar B., Sundaramoorthy M., Poulos T. L. (1999) J. Biol. Chem. 274, 37827–37833 [DOI] [PubMed] [Google Scholar]
- 40.Bonagura C. A., Sundaramoorthy M., Pappa H. S., Patterson W. R., Poulos T. L. (1996) Biochemistry 35, 6107–6115 [DOI] [PubMed] [Google Scholar]
- 41.Mizushima N. (2007) Autophagy 3, 179–180 [DOI] [PubMed] [Google Scholar]
- 42.Sorimachi H., Suzuki K. (2001) J. Biochem. 129, 653–664 [DOI] [PubMed] [Google Scholar]
- 43.Fanin M., Nascimbeni A. C., Angelini C. (2007) J. Med. Genet. 44, 38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.