Abstract
Calcium signaling in neurons as in other cell types mediates changes in gene expression, cell growth, development, survival, and cell death. However, neuronal Ca2+ signaling processes have become adapted to modulate the function of other important pathways including axon outgrowth and changes in synaptic strength. Ca2+ plays a key role as the trigger for fast neurotransmitter release. The ubiquitous Ca2+ sensor calmodulin is involved in various aspects of neuronal regulation. The mechanisms by which changes in intracellular Ca2+ concentration in neurons can bring about such diverse responses has, however, become a topic of widespread interest that has recently focused on the roles of specialized neuronal Ca2+ sensors. In this article, we summarize synaptotagmins in neurotransmitter release, the neuronal roles of calmodulin, and the functional significance of the NCS and the CaBP/calneuron protein families of neuronal Ca2+ sensors.
Neuron-specific calcium sensors such as synaptotagmin and NCS proteins promote rapid changes in neuronal activity. Calmodulin and other ubiquitously expressed sensors also have neuronal functions, including ion channel regulation.
Calcium signaling in many cell types can mediate changes in gene expression, cell growth, development, survival, and cell death. However, neuronal calcium signaling processes have become adapted to modulate the function of important pathways in the brain, including neuronal survival, axon outgrowth, and changes in synaptic strength. Changes in the concentration of intracellular free Ca2+ ([Ca2+]i) are essential for the transmission of information through the nervous system as the trigger for neurotransmitter release at synapses. In addition, alterations in [Ca2+]i can lead to a wide range of different physiological changes that can modify neuronal functions over time scales of milliseconds through tens of minutes to days or longer (Berridge 1998). Many of these processes have been shown to be dependent upon the particular route of Ca2+ entry into the cell. It has long been known that the physiological outcome from a change in [Ca2+]i depends on its location, amplitude, and duration. The importance of location becomes even more pronounced in neurons because of their complex and extended morphologies. [Ca2+]i also regulates neuronal development and neuronal survival (Spitzer 2006). In addition, modifications to Ca2+ signaling pathways have been suggested to underlie various neuropathological disorders (Braunewell 2005; Berridge 2010).
Highly localized Ca2+ elevations (Augustine et al. 2003) formed following Ca2+ entry though voltage-gated Ca2+ channels (VGCCs) lead to synaptic vesicle fusion with the presynaptic membrane and thereby allow neurotransmitter release within less than a millisecond. Differently localized and timed Ca2+ signals can, for example, result in changes to the properties of the VGCCs (Catterall and Few 2008) or lead to changes in gene expression (Bito et al. 1997). Postsynaptic Ca2+ signals arising from activation of NMDA receptors give rise to two important processes in synaptic plasticity, long term potentiation (LTP) and long term depression (LTD). LTP and LTD are examples of the way synaptic transmission can change synaptic efficacy and are thought to be important in modulating learning and memory. Importantly, the Ca2+ signals that bring about either LTP or LTD differ only in their timing and duration. LTP is triggered by Ca2+ signals on the micromolar scale for shorter durations, whereas LTD is triggered by changes in [Ca2+]i on the nanomolar scale for longer durations (Yang et al. 1999). Specific Ca2+ signals are likely to be decoded by different Ca2+ sensor proteins. These are proteins that undergo a conformational change on Ca2+ binding and then interact with and regulate various target proteins. Among those Ca2+ sensors that are important for neuronal function are the synaptotagmins that control neurotransmitter release (Chapman 2008), the ubiquitous EF-hand containing sensor calmodulin that has many neuronal roles, and the more recently discovered neuronal EF-hand containing proteins, including the neuronal calcium sensor (NCS) protein (Burgoyne 2007) and the calcium-binding protein (CaBP)/calneuron (Haeseleer et al. 2002) families. We will briefly review synaptotagmins and the neuronal functions of calmodulin but concentrate on the NCS and CaBP families of Ca2+ sensors.
SYNAPTOTAGMINS AND NEUROTRANSMITTER RELEASE
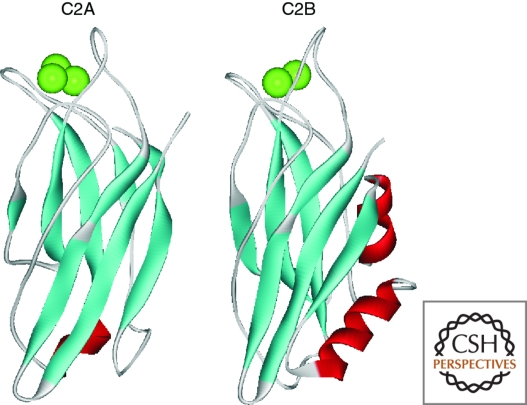
Synaptotagmins are transmembrane proteins mostly found associated with synaptic and secretory vesicles. There are multiple known isoforms of synaptotagmin (Craxton 2004) of which synaptotagmin I is the best studied. The role of synaptotagmins in neurotransmitter release has been the subject of intense investigations, which have been extensively reviewed (Chapman 2008; Rizo and Rosenmund 2008; Sudhof and Rothman 2009) and so only a brief outline is given here. Synaptotagmins bind Ca2+ with relatively low affinity (Kd > 10 µM) through their two C2 domains (C2A and C2B) (Shao et al. 1998; Fernandez et al. 2001), which are functional in many but not all synaptotagmin isoforms. Ca2+ binding by C2 domains requires coordination of Ca2+ by both the protein and membrane lipids and this lipid interaction is a key aspect for its function. In synaptotagmin I, the C2A and C2B domains (Fig. 1) bind three and two Ca2+ ions, respectively (Shao et al. 1998; Fernandez et al. 2001). It is now well established that synaptotagmin I is a key sensor for evoked, synchronous neurotransmitter release in many classes of neurons (Fernandez-Chacon et al. 2001). Structure–function studies based on expression of specific mutants have been carried out in mice, worms, and flies. For example, disruption of Ca2+ binding to the C2B domain of synaptotagmin I has been shown to have a more deleterious effect than disruption of Ca2+ binding to its C2A domain (Mackler et al. 2002; Robinson et al. 2002). The details of exactly how it triggers exocytosis and the function of other syntaptotagmin isoforms remain controversial. Membrane fusion requires the pairing and interaction of so-called SNARE proteins on vesicle and target membranes (Sollner et al. 1993). These can assemble into a SNARE complex that may form the minimal fusion machinery. For synaptic vesicle and neuroendocrine exocytosis, the SNARE proteins are SNAP-25, syntaxin 1, and synaptobrevin. In the case of regulated exocytosis, such as in neurotransmitter release, vesicle fusion is tightly regulated and requires a Ca2+ signal for activation. Ca2+ entry through VGCCs leading to Ca2+ elevation in local microdomains close to the mouth of the Ca2+ channel is able to trigger very rapid (within less than 1 ms) fusion of synaptic vesicles. Synaptotagmin can bind to both syntaxin and SNAP-25, and fast neurotransmitter release requires synaptotagmin (Geppert et al. 1994) probably prebound to assembled or partially assembled SNARE complexes (Schiavo et al. 1997; Rickman et al. 2006) so that Ca2+-induced interaction with phospholipids can occur rapidly (Xue et al. 2008). It is still under debate how important synaptotagmin is in vesicle docking (de Wit et al. 2009) and how it acts at the plasma membrane in fusion itself (Tang et al. 2006; Hui et al. 2009). Synaptotagmin could act as a brake on fusion that is relieved on Ca2+ binding or have a positive role in membrane fusion (Chicka et al. 2008). A recent focus has been on the combined role of synaptotagmin and another SNARE interacting protein complexin in timing synaptic vesicle fusion (Sudhof and Rothman 2009), but much still remains to be learnt about the molecular basis of its function. While synaptotagmin is a key sensor for evoked neurotransmitter release, an alternative C2-domain containing protein, Doc2b, has been identified as a Ca2+ sensor for spontaneous neurotransmitter release (Groffen et al. 2010). Like synaptotagmin, Doc2b appears to function via interaction with SNARE complexes.
Figure 1.
Structures of the C2A and C2B domains of synaptotagmin I. The structures show the isolated C2 domains in their Ca2+-loaded state with the bound Ca2+ ions shown in green. The coordinates for the structures for the C2A and C2B domains come from the PDB files 1BYN and 1K5W, respectively.
NEURONAL FUNCTIONS OF CALMODULIN
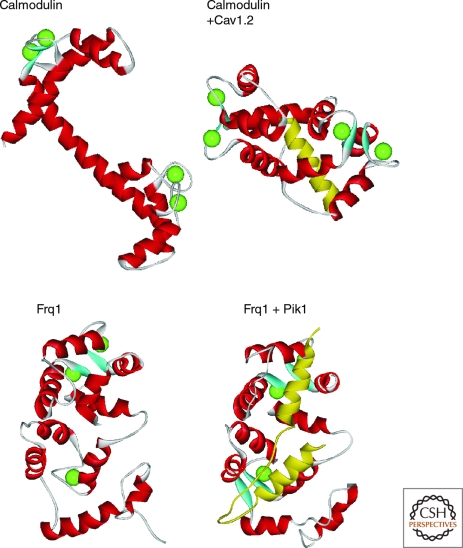
Calmodulin is a ubiquitously expressed Ca2+ -binding protein that can bind four Ca2+ ions through its four EF-hand domains (Chattopadhyaya et al. 1992). This protein has been highly conserved throughout evolution, is found in all eukaryotes, and is 100% identical across all vertebrates at the amino acid level. It is involved in the regulation of many essential physiological processes including cell motility, exocytosis, cytoskeletal assembly, and modulation of intracellular Ca2+ concentrations. The first two EF-hands of calmodulin form an amino-terminal globular domain that is joined by a flexible linker to a highly homologous carboxy-terminal region encompassing the third and fourth EF-hands. The carboxy-terminal pair of EF-hands has a much higher affinity for Ca2+ than the amino-terminal pair, which allows the two domains to behave independently at varying Ca2+ concentrations (Tadross et al. 2008). The highly flexible linker between the two domains can be bent dramatically upon binding to target proteins (Fig. 2) and is an essential property of calmodulin, which permits this protein to interact with a large and diverse array of interacting partners. The significant conformational changes on binding to its targets (Fallon et al. 2005) can increase its affinity for Ca2+.
Figure 2.
Comparison of the structures of Ca2+-loaded calmodulin and yeast frequenin with and without bound target peptides. The structures at the top are of Ca2+-bound calmodulin alone (PDB 1CLL) or in a complex with the IQ-like domain of the Cav1.2 Ca2+-channel α-subunit (PDB 2F3Z). The structures at the bottom are of the Ca2+-bound yeast frequenin (Frq1) alone (PDB 1FPW) or in a complex with the binding domain from Pik1 (PDB 2JU0). In each of the complexes, the target peptide is shown in yellow.
Calmodulin is present in brain at high concentrations (up to ∼100 µM). In addition to its more general functions, calmodulin also has a series of specific roles in transducing Ca2+ signals in neurons, including, for example, in the regulation of glutamate receptors (O'Connor 1999), ion channels (Saimi and Kung 2002), and proteins in signaling pathways such as neuronal nitric oxide synthase, and it can affect synaptic plasticity (Lisman et al. 2002; Xia and Storm 2005). One key direct function of calmodulin is in regulating the activity of VGCCs by binding to channel subunits (Catterall and Few 2008). Ca2+-binding to VGCC-associated calmodulin can have a range of effects on channel function, including mediating Ca2+-dependent facilitation or Ca2+-inactivation (Lee et al. 2000; DeMaria et al. 2001; Catterall and Few 2008; Liu et al. 2010). Calmodulin is also constitutively associated with and regulates opening of Ca2+-activated potassium channels (Xia 1998; Schumacher et al. 2001) and other types of potassium channels (Wen and Levitan 2002). Two other major modes of action of calmodulin are exerted indirectly through its target proteins Ca2+/calmodulin-dependent kinases (CaMKs) and calcineurin. CaMKs contribute to a number of regulatory pathways involving, for example, phosphorylation of AMPA receptors (Barria et al. 1997) and the nuclear transcription factor CREB (Deisseroth et al. 1998). Calmodulin also positively regulates presynaptic release probability and this is mediated via activation of CaMKII (Pang et al. 2010). The Ca2+-activated phosphatase calcineurin can dephosphorylate a wide range of neuronal proteins, leading to direct effects and effects through changes in gene transcription following activation of the transcription factor NFAT and its translocation into the nucleus. Calcineurin has also been implicated, for example, in synaptic plasticity (Malleret et al. 2001; Xia and Storm 2005). Although many aspects of neuronal function are known to be regulated by calmodulin, proteins related to calmodulin have been discovered in recent years, which are enriched or expressed exclusively in neurons. Duplication and diversification of the calmodulin gene may have given rise to these neuronal calcium sensing proteins so that they can carry out specific neuronal functions in higher organisms.
NCS PROTEIN FAMILY
Whereas calmodulin is ubiquitously expressed, the expression of other calcium sensing proteins can be restricted to particular tissues and cell types. A good example of this is the neuronal calcium sensor (NCS) family of proteins, which are primarily expressed in neurons or retinal photoreceptors. The NCS family of proteins is related to calmodulin but have distinct properties that allow them to carry out nonredundant roles that do not overlap with the functions of calmodulin. Members of the NCS protein family have been implicated in the regulation of neurotransmitter release, regulation of cell-surface receptors and ion channels, control of gene transcription, cell growth, and survival (Burgoyne 2007). The NCS proteins are encoded by a family of 14 genes in mammals with greater diversity stemming from alternative splicing of transcripts from a number of the genes. All NCS gene products harbor four EF-hand motifs and display limited similarity (<20%) to calmodulin (Burgoyne 2004).
NCS-1 is the most widely expressed of the NCS proteins and is thought to be the primordial NCS protein. The protein was first discovered (as frequenin) in Drosophila melanogaster (Pongs et al. 1993), where there are two very closely related genes (Sanchez-Gracia et al. 2010). Although initially thought to be neuronal specific (Nef et al. 1995), NCS-1 has also been identified in Saccharomyces cerevisiae (Hendricks et al. 1999). After this first appearance of NCS-1 in yeast, there has been a steady increase in the diversity of this family throughout evolution, which roughly correlates with increasing organism complexity. Five classes of NCS proteins have now been identified in higher organisms termed classes A-E (Burgoyne 2007). Class A contains NCS-1, which is present in yeast and all higher organisms. Class B consists of the visinin-like proteins (VSNLs), which appear first in Caenorhabditis elegans. Classes C and D evolved with the appearance of fish and comprise recoverin and the guanylyl-cyclase-activating proteins (GCAPs), respectively. Finally, class E contains the K+ channel-interacting proteins (KChIPs), which are found in insects and evolutionary subsequent species (Burgoyne 2004). Mammals have a single NCS-1, five VSNL proteins (hippocalcin, neurocalcin δ, and VILIPs1-3), a single recoverin, three GCAPs, and four KChIPs. Expression of the recoverins and GCAPs is restricted to the retina, whereas the rest of the NCS family is found in varied neuronal populations (Burgoyne 2007). Although localization and expression studies have proven difficult because of cross-reactivity of antibodies, it has been established that certain neurons express several or all of the NCS proteins, but in general, the expression profile for each of the NCS proteins is unique (Paterlini et al. 2000; Rhodes et al. 2004). This suggests that despite the high sequence homology between the proteins, each is likely to perform distinct functions in specific cell types (Burgoyne and Weiss 2001).
Unlike calmodulin, not all EF-hands are functional in the NCS proteins and the most amino-terminal EF-hand is unable to bind Ca2+ in all family members. In the case of recoverin and KChIP1, only two of its four EF-hand motifs are functional in Ca2+ binding (Burgoyne et al. 2004; Burgoyne 2007). Unlike the dumbbell structure of calmodulin, the NCS proteins are compact and globular when Ca2+-bound and they undergo limited conformational change on binding to their target proteins (Ames et al. 2006; Pioletti et al. 2006; Strahl et al. 2007; Wang et al. 2007) (Fig. 2). NCS proteins also differ from calmodulin in that many have motifs that allow membrane association. KChIP1 and all the members of classes A–D are N-myristoylated, whereas certain KChIP2, KChIP3, and KChIP4 isoforms harbor palmitoylation motifs. In some cases, the membrane association conferred by these moieties is dynamically regulated by Ca2+ binding when a sequestered mysristoyl chain becomes solvent-exposed following a Ca2+-driven shift in conformation as originally described for recoverin (Ames et al. 1997). VSNL proteins are also cytosolic at resting [Ca2+]i but localize to the plasma membrane or Golgi complex upon Ca2+ elevation (O'Callaghan et al. 2002; Spilker et al. 2002; O'Callaghan et al. 2003b). Each of the NCS proteins displays distinct subcellular localizations, which are in part determined by additional interactions with specific phosphoinositides mediated by basic amino-terminal residues immediately proximal to the site of acylation (O'Callaghan et al. 2003a; O'Callaghan et al. 2005).
NCS proteins are multifunctional regulators of various proteins involved in processes ranging from trafficking and ion channel modulation to gene transcription (Burgoyne 2004), and the function of NCS-1 in particular has been intensively studied. NCS-1, the primordial NCS protein, is highly evolutionarily conserved, retaining 59% identity with its yeast ortholog, frequenin. It displays a high Ca2+-binding affinity and is able to respond to small fluctuations in [Ca2+]i. NCS-1 is amino-terminally myristoylated and is constitutively associated with membranes including plasma and Golgi membranes (O'Callaghan et al. 2002), although in some cell lines, NCS-1 has been found to be partially cytosolic (de Barry et al. 2006) and it is able to rapidly exchange between membrane and cytosolic pools (Handley et al. 2010). In contrast to all other NCS family members, NCS-1 is not neuron specific and is expressed in neuroendocrine cells (McFerran et al. 1998) and at low levels in several nonneuronal cell types. NCS-1 has three functional EF-hand motifs, which have differing cation specificities. It has been suggested that under resting conditions when [Ca2+]i is low (≤0.1 µM), EF2 and EF3 are Mg++ bound, whereas EF4 is a Ca2+ specific binding site and remains vacant. In the Mg++ bound state, NCS-1 adopts a conformation, which reduces exposure of hydrophobic regions. This may be important in the prevention of nonspecific interactions in the absence of a specific Ca2+-signal. In the presence of elevated [Ca2+]i, EF2 and EF3 become Ca2+-occupied, simultaneously followed by Ca2+ binding to EF4 (Aravind et al. 2008). The Mg++ bound form of NCS-1 has a fivefold lower affinity for Ca2+ than the Mg++-free/Ca2+-free apo-form. This implies that Mg++ binding permits significant modulation of NCS-1 and is important in fine tuning its Ca2+-sensing properties (Aravind et al. 2008; Mikhaylova et al. 2009).
Much current understanding concerning the function of NCS-1 derives from overexpression or knockout studies. Overexpression in Drosophila caused a frequency-dependent facilitation of neurotransmitter release (Pongs et al. 1993) and its importance for neurotransmissions has been confirmed by knockout of the two Drosophila frequenin genes (Dason et al. 2009). In Xenopus, overexpression caused enhanced spontaneous and evoked transmission at neuromuscular junctions (Olafsson et al. 1995) and over-expression was also found to increase Ca2+-dependent exocytosis of dense core granules in PC12 cells (McFerran et al. 1998) and to enhance associative learning and memory in Caenorhabditis elegans (Gomez et al. 2001; Hilfiker 2003).
Knockout of NCS-1 (Frq1) in the yeast Saccharomyces cerevisiae is lethal because of its requirement for the activation of Pik1, one of the two yeast phosphatidylinositol-4 kinases (PI4Ks) (Hendricks et al. 1999). NCS-1 can also interact with the mammalian Golgi enzyme PI4KIIIβ and enhances its activity three- to 10-fold (Taverna et al. 2002; Haynes et al. 2005; de Barry et al. 2006). The interaction with Golgi-associated PI4KIIIβ suggests that it may regulate secretion through the modulation of phosphatidylinositol-dependent trafficking steps (Hendricks et al. 1999; Zhao et al. 2001; Haynes et al. 2005). In support of this, NCS-1 has also been demonstrated to associate with another PI4KIIIβ regulator ARF1, a small GTPase critical to multiple trafficking steps in mammalian cells (Haynes et al. 2005; Haynes et al. 2007).
Knockout of NCS-1 in other organisms is not lethal but does generate specific phenotypes. In Dictyostelium discoideum, loss of NCS-1 function alters developmental rate (Coukell et al. 2004) and in C. elegans results in impaired learning and memory (Gomez et al. 2001). Knockdown of one of the two NCS-1 genes in zebrafish, ncs-1, prevents formation of the semicircular canals of the inner ear (Blasiole et al. 2005). The signaling pathway involving NCS-1, ARF1, and PI4KIIIβ (Haynes et al. 2005) modulates the secretion of components important for the development of the vestibular apparatus of the inner ear (Petko et al. 2009). Knockdown of NCS-1 or expression of a dominant–negative inhibitor based on an EF-hand mutation (Weiss et al. 2000) disrupted the induction of long-term depression in rat cortical neurons (Jo et al. 2008).
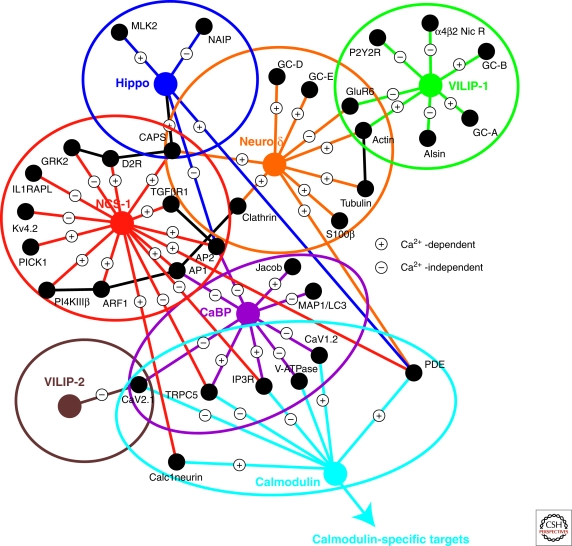
Many different binding partners have been identified for NCS-1 (Haynes et al. 2006; Haynes et al. 2007) (Fig. 3) and in some cases these interactions overlap with those of calmodulin (Schaad et al. 1996). This overlap using in vitro binding assays may not be physiologically meaningful because of substantial lower affinity of NCS-1 for known calmodulin targets (Schaad et al. 1996; Fitzgerald et al. 2008). NCS-1 has a higher affinity for calcium than calmodulin and therefore may preferentially interact with certain binding partners when the amplitude of a Ca2+-signal falls below the threshold for activation of calmodulin. For example, both calmodulin and NCS-1 have been shown to interact with and desensitize dopamine D2 receptors but are likely to mediate their effects at different [Ca2+]i (Kabbani et al. 2002; Woods et al. 2008). Functional analyses have confirmed that NCS-1 is a regulator of D2 receptors and that this function modulates learning in mice (Saab et al. 2009). Other NCS-1 target proteins (Fig. 3) appear to be specific for NCS-1 (Haynes et al. 2006). Various studies have implicated NCS-1 in the regulation of VGCCs (Weiss et al. 2000; Tsujimoto et al. 2002; Dason et al. 2009) but there is as yet no evidence for a direct interaction. Interestingly, in Drosophila, the effects of NCS-1 on both neurotransmission and nerve-terminal growth can be explained by a functional interaction with the VGCC cacophony, which is related to the mammalian P/Q-type VGCCs (Dason et al. 2009). In contrast, in this study, there was no evidence for an essential functional interaction with the fly PI4KIIIβ ortholog four wheel drive.
Figure 3.
An interaction map showing protein–protein interactions made by some NCS proteins and CaBP1 compared to calmodulin. Known protein interactions for CABP1, hippocalcin, NCS-1, neurocalcin δ,VILIP1, and VILIP2. Links indicate where these target proteins have also been found to interact with calmodulin. It is also indicated whether these interactions require the Ca2+-bound form of the protein or not.
The example of NCS-1 illustrates how evolutionary pressures have fine-tuned Ca2+ sensors to carry out specialized neuronal functions. The individual properties of NCS-1 allow this protein to localize to discrete domains within the cell and interact with distinct target proteins under conditions of Ca2+ stimulation, which would not activate the archetypal Ca2+ sensor, calmodulin. Although NCS-1 has been found not to be neuronal specific and may carry out more generalized functions conserved through evolution in organisms from yeast onwards, further adaptive mutations have given rise to many more members of the NCS protein family, each tasked with dedicated functions.
Much less is known about the VSNL or class B proteins, although these appear to modulate various signal transduction pathways such as cyclic nucleotide and MAPK signaling (Braunewell and Klein-Szanto 2009). VILIP-1 has been found to regulate a class of purinergic receptors (Chaumont et al. 2008). They have been shown to have effects on gene expression and are also involved in traffic of proteins to the plasma membrane (Lin et al. 2002; Brackmann et al. 2005). Hippocalcin has been suggested to be involved as a Ca2+sensor in long term depression in hippocampal neurons (Palmer et al. 2005) and shows a Ca2+/myristoyl switch for translocation within such neurons (Markova et al. 2008). It has also been implicated in protection from neuronal apoptosis (Mercer et al. 2000; Korhonen et al. 2004).
Recoverin is expressed exclusively in the retina and is believed to have a role in light adaptation and can enhance visual sensitivity (Polans et al. 1996; Sampath et al. 2005). Recoverin is found primarily in rod and cone cells of the retina (Yamagata et al. 1990; Dizhoor et al. 1991). Recoverin was predicted to prolong the lifetime of photolyzed rhodopsin by inhibiting its phosphorylation by rhodopsin kinase to extend the light response (Chen et al. 1995; Klenshin et al. 1995). The function of recoverin has been controversial and this hypothesis may be oversimplified. Discrepancies have been noted regarding the [Ca2+]i required for rhodopsin kinase interaction, which may lie outside normal physiological limits but analysis of recoverin knockout mice have shown changes in photoresponses consistent with a physiological role in inhibition of rhodopsin kinase (Makino et al. 2004).
The structure of recoverin has been extensively studied by X-ray crystallography and NMR studies to interrogate its structure in its Ca2+-bound and -free forms (Flaherty et al. 1993; Ames et al. 1995; Tanaka et al. 1995; Ames et al. 1997; Ames et al. 2002; Weiergraber et al. 2003). Recoverin is composed of two distinct domains connected through a bent linker and forms a compact structure in the absence of Ca2+. Unlike other NCS proteins, recoverin has only two functional EF-hand motifs. Upon binding of Ca2+, the amino-terminal domain comprising EF-1 and EF-2 rotates through 45° relative to the carboxy-terminal domain driving extrusion of its buried myristoyl group to permit association with membranes and revealing a hydrophobic surface, which can mediate interaction with the target protein rhodopsin kinase (Ames et al. 2006). The residues involved in the interaction of the myristoyl group with the hydrophobic pocket are also conserved in the other members of the NCS family, however not all of the other family members display this Ca2+/myristoyl switch (O'Callaghan et al. 2002; Stephen et al. 2007). NCS-1 and KChIP1 expose a similar hydrophobic surface upon Ca2+-binding, which could be similarly important for target interactions (Bourne et al. 2001; Scannevin et al. 2004; Zhou et al. 2004b; Pioletti et al. 2006). In contrast, other NCS proteins are able to interact with certain binding proteins in the absence of Ca2+ and therefore Ca2+-driven exposure of a hydrophobic surface cannot be the sole mechanism by which these proteins bind to effectors. Although extensive structural characterization of recoverin may go some way to inform an understanding of the general conserved structures of members of the NCS family, subtle differences in “active” surface residues of the individual proteins gives rise to their ability to interact specifically with a wide range of binding partners.
GCAPs are the only known activators of retinal guanylyl cyclases (GCs) (Palczewski et al. 2004) and are known to be physiological regulators of light adaptation (Mendez et al. 2001; Burns et al. 2002; Howes et al. 2002; Pennesi et al. 2003). They are unusual in that they activate GCs when in their Ca2+-free form but become inhibitors of GCs at higher Ca2+ concentrations (Dizhoor and Hurley 1996). GCAP3 is expressed in cone cells, whereas GCAP1 and GCAP2 are expressed in rod cells, and despite GCAP1 and GCAP2 having the same function in the same cell type, the two proteins have different Ca2+ binding affinities for GC activation. This means that both proteins are required for GC activation over the full physiological Ca2+ concentration range, thus maximizing the dynamic range of GC activity (Koch 2006). The GCAPs are an example of how calcium sensors have become adapted to increase the dynamic Ca2+ sensitivity of important regulatory mechanisms in specialized cell types (Palczewski et al. 2004).
KChIPs have been found to associate with transient voltage-gated potassium channels of the Kv4 family (An et al. 2000) and can stimulate their traffic to the plasma membrane (Hasdemir et al. 2005). Four KChIP genes and a large number of expressed splice variants are present in mammals (Pruunsild and Timmusk 2005). Knockout of KChIP1 has revealed a potential role in the GABAergic inhibitory system (Xiong et al. 2009). The KChIPs are expressed predominantly in the brain but KChIP2 is also expressed in the heart, and knockout of KChIP2 causes a complete loss of calcium-dependent transient outward potassium currents and susceptibility to ventricular tachycardia (Kuo et al. 2001). KChIP3 is also known as DREAM or calsenilin, and has documented roles in transcriptional regulation (Carrion et al. 1999; Mellstrom and Naranjo 2001) and in the processing of presenilins and amyloid precursor protein, which are important in the pathogenesis of Alzheimer's disease (Buxbaum et al. 1998; Jo et al. 2004). Despite KChIP3 being implicating in three quite different functions, it is likely that these are all physiologically relevant. KChIP3 knockout mice show reduced responses in acute pain models because of changes in prodynorphin synthesis (Cheng et al. 2002), decreased β-amyloid production, and physiological defects consistent with changes to the Kv4 channels (Lilliehook et al. 2003). Although many of the KChIPs and their isoforms may have overlapping functions, some differences between them are beginning to emerge (Holmqvist et al. 2002; Venn et al. 2008).
In support of key roles for the NCS family in higher organisms, a number of recent studies have implicated these proteins in the pathological progression of human neurological diseases in addition to the potential link with Alzheimer's disease via the interaction of KChIP3 with presenilins. VILIP1 may have a role in Alzheimer's disease because of an association with amyloid plaques in diseased brains (Schnurra et al. 2001). NCS-1 has been found to be up-regulated in patients with schizophrenia and bipolar disorders (Koh et al. 2003) and also interacts with interleukin-1 receptor accessory protein-like (IL1RAPL), a protein, which when mutated results in X-linked mental retardation (Bahi et al. 2003). The effects conferred by NCS proteins in all of these diseases would appear to be dependent on the up- or down-regulation of their expression. As yet few genetic links have been established between NCS proteins and the aforementioned diseases, suggesting epigenetic effects may be responsible. One idea is that epigenetic mediated alterations in NCS protein function may contribute to cognitive impairments observed in neurodegenerative states. Targeting of NCS protein function through this novel route may offer a future therapeutic approach for such diseases (Braunewell 2005).
The NCS protein family has evolved to carry out specialized neuronal functions that are separate to those of calmodulin. When attempting to decipher precisely why these proteins are particularly well adapted for carrying out functions in neurons, it is therefore relevant to compare their properties to those of calmodulin. Of note is their approximately 10-fold higher affinity for Ca2+ when compared to calmodulin. This higher affinity would allow the NCS proteins to be activated at much lower Ca2+ concentrations and, in combination with calmodulin, extends the dynamic range over which Ca2+ can regulate neuronal processes. In this way, responses to very slight or more dramatic changes in [Ca2+]i would depend on which populations of calcium binding proteins are activated under particular conditions (Burgoyne and Weiss 2001). As mentioned previously, the individual expression patterns and subcellular localization of each of the NCS proteins is also likely to represent a key factor in their specific roles in neuronal cell signaling. The characteristic amino-terminal myristoylation or palmitoylation modifications, which allow these proteins to associate with membranes, may spatially partition them to relevant subcellular sites within the cell, leading to a faster and more efficient response to particular Ca2+ signals. Specific physiological outcomes will be determined by their distinct target proteins. The various members of the NCS family arose at points in evolution corresponding to increasing neuronal sophistication in higher animals. As such, these proteins represent an example of how the properties of calcium binding proteins have been fine-tuned to act in specific neuronal signaling pathways.
CaBP Family
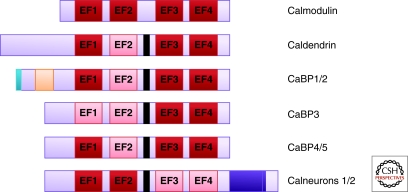
The CaBPs are a relatively recently discovered family of EF-hand containing Ca2+-binding proteins, which are only found in vertebrates (Haeseleer et al. 2000). They represent another example of a diverse family of Ca2+-sensors capable of regulating discrete processes in the nervous systems of higher organisms. The CaBPs share sequence homology with calmodulin and also display a similar structural arrangement of EF-hand motifs. Each of the CaBPs has four EF hands, although, like the NCS proteins, they display different patterns of EF-hand inactivation (Fig. 4). In CaBPs 1–5, the second EF-hand motif is inactive with the exception of CaBP3, which also has an inactive EF-1 motif. CaBP3 is believed to represent a pseudogene as only the mRNA has been detected in cells and as yet no protein product has been found (Haeseleer et al. 2000). Two proteins were named CaBP7 and CaBP8 (Haeseleer et al. 2002), but bioinformatic analysis is more consistent with them being a conserved and distinct subfamily of CaBPs (McCue et al. 2010). We will therefore refer to them by their alternative names, calneuron 2 and calneuron 1, respectively (Wu et al. 2001; Mikhaylova et al. 2006). The calneurons, by contrast to the CaBPs, have a different pattern of EF-hand inactivation with active EF-hands 1 and 2 and inactive EF-hands 3 and 4 (Mikhaylova et al. 2006). The CaBPs also differ from calmodulin in that their central α helical linker domain connecting the carboxy- and amino-terminal EF-hand pairs is extended by four amino acid residues. This has been suggested to allow these proteins to interact with unique targets (Haeseleer et al. 2000).
Figure 4.
Schematic diagram showing the domain structure of calmodulin and members of the CaBP/calneuron protein family. Active EF-hand motifs are shown in red and inactive EF-hand motifs are shown in pink. Compared to calmodulin, the CaBPs have an extended linker region between their first EF-hand pair and their second EF-hand pair (shown in black). CaBP1 and CaBP2 have an N-myristoylation site (shown in blue). CaBP1 and CaBP2 have alternative splice sites at their N-terminus, which give rise to long and short isoforms (shown in orange). Calneurons 1 and 2 possess a 38 amino acid extension at their C-terminus (shown in purple).
A major difference compared with calmodulin is the ability of CaBP 1 and 2 and calneurons 1 and 2 to target to specific cellular membranes (McCue et al. 2009). CaBP 1 and 2 are amino-terminally myristoylated, which permits localization to the plasma membrane and Golgi apparatus (Haeseleer et al. 2000; Haynes et al. 2004). The precise amino-terminal sequence to which the myristoyl group is attached is also important in the targeting of these two proteins, as exemplified by the long and short splice isoforms generated from their genes, which show subtle differences in their localization. CaBP1-Long localizes predominantly to the Golgi and also displays some cytosolic localization, whereas CaBP1-Short localizes most prominently to the plasma membrane and to Golgi structures (Haeseleer et al. 2000; McCue et al. 2009). Alternative splicing of the CaBP1 gene generates a third protein product, caldendrin (Seidenbecher et al. 1998). This splice isoform is significantly larger than either CaBP1-Long or CaBP1-Short because of an amino-terminal extension, but caldendrin mRNA lacks the exon required for N-myristoylation and as a result the protein displays a markedly different subcellular localization to its shorter relatives.
N-terminal acylation is important in the localization of some CaBPs, but the calneurons appear to be targeted via a different mechanism. Like CaBP1 and CaBP2, calneurons 1 and 2 localize to internal membranes that colabel with Golgi-specific markers and also to vesicular structures (McCue et al. 2009; Mikhaylova et al. 2009). Calneurons 1 and 2 do not possess an amino-terminal myristoylation motif and differ from the rest of the CaBP family because of a 38-amino acid extension at their carboxyl terminus. Analysis of this sequence revealed a predicted C-terminal transmembrane domain with a cytosolic amino terminus. The carboxy-terminal domain resembles tail-anchor motifs and directly localizes calneurons 1 and 2 to membranes particularly of the trans-Golgi network (McCue et al. 2009).
To date, only the structure of CaBP1-Short has been solved (Wingard et al. 2005; Li et al. 2009). This structural information may provide insight into the structures of the rest of the CaBPs. Analogy to calmodulin would suggest that the CaBPs should adopt a dumbbell like tertiary conformation consisting of an amino-terminal domain containing EF-1 and EF-2 and a carboxy-terminal domain containing EF-3 and EF-4, connected by a central linker. NMR analysis revealed that CaBP1 does indeed have two independent, noninteracting domains, joined by a flexible linker (Wingard et al. 2005). Investigation into the effects of Mg++ and Ca2+ binding has shown that as predicted the second EF hand of CaBP1 is incapable of binding divalent cations. EF-3 and EF-4 bind to both Mg++ and to Ca2+, whereas EF-1 is thought to be constitutively Mg++ bound. Binding to either Mg++ or Ca2+ induces distinct conformations of this protein. Mg++ binding results in a global conformational change, whereas Ca2+ binding results in only a localized change in EF-3 and EF-4. These two conformational states may allow CaBP1 to interact with different target molecules driven by the ratio of Mg++ to Ca2+ (Wingard et al. 2005; Li et al. 2009). The Mg++ bound form of CaBP1 is similar to that of apo-calmodulin, but the Ca2+ bound form appears markedly different. This is perhaps unsurprising as neither of the amino-terminal EF-hands of CaBP1 bind to Ca2+ under saturating conditions and only EF-1 binds to Mg++. This results in a constitutively closed conformation of the amino-terminal domain, whereas the carboxy-terminal domain can switch to an open conformation upon Ca2+ binding to EF-3 and EF-4. Comparison of the carboxy-terminal domain with that of calmodulin, however, still reveals differences in exposed hydrophobic residues thought to mediate target interactions (Wingard et al. 2005).
The structural differences between calmodulin and CaBP1 may go some way to explaining how they impose differing effects on the same target molecules. For instance, both CaBP1 and calmodulin bind to L-type Ca2+ channels with calmodulin causing Ca2+-induced channel closure but CaBP1 promoting channel opening (Zhou et al. 2004a; Zhou et al. 2005). Both calmodulin and CaBP1 also regulate inositol 1,4,5-trisphosphate receptors (IP3Rs) (Yang et al. 2002; Haynes et al. 2004; Kasri et al. 2004) with CaBP1 binding the type I IP3R with 100-fold higher affinity than calmodulin. This high affinity binding may result from the exposure of a distinct hydrophobic patch revealed in the carboxyl terminus of CaBP1 upon Ca2+-binding (Haynes et al. 2004; Li et al. 2009). This unique surface hydrophobicity profile is likely to be important for the specialization of CaBP1 function in the brain and retina, and the existence of splice isoforms is also likely to further fine-tune the actions of this Ca2+ sensor. The differing expression patterns, subcellular targeting mechanisms, and Ca2+ binding properties of the various members of the CaBP protein family would allow them to carry out highly specialized regulatory roles modulating important Ca2+-channels in the central nervous system.
The majority of studies to date on CaBP1 have examined the functions of the longest splice isoform caldendrin and it is not yet clear whether the other splice isoforms of CaBP1 can carry out the same functions. CaBP1-Long and -Short have been found to have roles in the regulation of various Ca2+-channels including P/Q-type (CaV2.1) channels (Lee et al. 2002), L-type (CaV1.2) channels (Zhou et al. 2005; Cui et al. 2007), TRPC5 channels (Kinoshita-Kawada et al. 2005), and IP3Rs (Yang et al. 2002), which they apparently inhibit (Haynes et al. 2004; Kasri et al. 2004). The interaction of CaV2.1 with CaBP1 appears to rely acutely upon amino-terminal myristoylation. Wild type, myristoylated, CaBP1-Long enhances channel inactivation and shifts the activation range to more depolarizing voltages (Lee et al. 2002). An N-myristoylation mutant, however, was unable to mediate these effects and instead modulated channels in a similar fashion to calmodulin (Few et al. 2005). Differential modulation of L-type channels depending on the splice isoform of CaBP1 has also been observed. CaBP1-Short has been shown to completely inhibit inactivation of CaV1.2 channels (Zhou et al. 2005), but caldendrin causes a more modest suppression and signals through a different set of molecular determinants (Tippens and Lee 2007). This suggests that the subcellular localization of CaBP1 splice variants is important for their function and there are likely to be individual roles for each protein. Interactions of caldendrin with other types of proteins have also been reported, such as its interaction with light chain 3 of MAP1A/B, a microtubule cytoskeletal protein (Seidenbecher et al. 2004), and an interaction with myo1c, a member of the myosin-1 family of motor proteins (Tang et al. 2007). Finally, a role for caldendrin in NMDA receptor (NMDAR) signaling has been reported via an interaction with a novel neuronal protein, Jacob. Upon extrasynaptic NMDAR activation, Jacob translocates to the nucleus to influence CREB activity, resulting in the stripping of synaptic contacts and an associated simplification of dendritic architecture. Synaptic NMDAR mediated synpatodendritic [Ca2+]i elevation induces caldendrin binding to Jacob, inhibiting nuclear trafficking and maintaining dendritic organization. This interaction represents a novel mechanism of synapse to nucleus communication and highlights the important roles of CaBP family members in the mammalian central nervous system (Dieterich et al. 2008).
Little information is available concerning the function of CaBP2 apart from in vitro studies suggesting that it might stimulate CaMK activity (Cui et al. 2007). Initially, CaBP2 was detected exclusively in the retina, although it has also been identified in auditory inner hair cells (Cui et al. 2007). CaBP5 was also detected in inner hair cells as well as in the retina, but in contrast to CaBP2, was found to have a modest inhibitory effect on the inactivation of CaV1.3 channels in transfected cells (Cui et al. 2007). Little is known about the functions of CaBP5, but knockout mice displayed reduced sensitivity of retinal ganglion cells to light responses, implicating CaBP5 in phototransduction pathways. CaBP5 was also found to interact with and suppress calcium-dependent inactivation of CaV1.2 channels (Rieke et al. 2008).
CaBP4 is the most extensively characterized of the CaBP family. It is expressed in the retina, where it localizes to synaptic terminals and has also been detected in auditory inner hair cells. CaBP4 modulates voltage gated Ca2+-channels and directly associates with the carboxyl terminus of the CaV1.4 α1 pore-forming subunit, shifting the activation range of the channel to more hyperpolarized voltages in transfected cells (Haeseleer et al. 2004). CaBP4 has also been shown to eliminate Ca2+-dependent inactivation of CaV1.3 channels, which is likely to be important in the modulation of these channels in inner hair cells, where Ca2+-dependent inactivation is weak or absent, probably allowing the audition of sustained sounds (Yang et al. 2006). A stronger inhibitory effect has been noted for CaBP1, however, suggesting that CaBP4 may not be the key Ca2+ sensor involved in this process (Cui et al. 2007). The function of CaBP4 is modulated by protein kinase C ζ in the retina, with increased CaBP4 phosphorylation in light-adapted tissue. Phosphorylation prolongs Ca2+ currents through CaV1.3 channels and suggests that light-stimulated phosphorylation of CaBP4 might help to regulate presynaptic Ca2+ signals in photoreceptors (Lee et al. 2007). CaBP4 has also been implicated in neurotransmitter release at synaptic terminals because of its interaction with unc119, a synaptic photoreceptor protein important for neurotransmitter release and maintenance of the nervous system (Haeseleer 2008). Knockout of CaBP4 results in mice with abnormalities in retinal function, where rod bipolar responses are approximately 100 times lower than those observed in wild-type animals (Haeseleer et al. 2004).
The functions of calneurons 1 and 2 have only recently begun to be investigated in detail. Both have been found to inhibit the activity of PI4KIIIβ at low or resting [Ca2+]i. Overexpression of the proteins was also found to inhibit Golgi-to-plasma membrane trafficking, caused enlargement of the trans-Golgi network (TGN), and reduced the number of Piccolo-Bassoon positive transport vesicles. A molecular switch for the production of phosphoinositides at the TGN is thought to be created by the opposing roles of NCS-1 and calneurons 1 or 2. At elevated Ca2+ levels, NCS-1 preferentially binds to PI4KIIIβ over the calneurons, activating the enzyme to drive enhanced TGN-to-plasma membrane trafficking (Mikhaylova et al. 2009). Patch clamping experiments have shown that over-expressed calneuron 1 can inhibit N-type Ca2+-channel currents in 293T cells and this inhibition was not observed with a truncated calneuron 1 lacking its hydrophobic C-terminus, suggesting normal localisation is important in carrying out this function (Shih et al. 2009).
CaBPs have been directly or indirectly implicated in multiple neuronal diseases. Post-mortem brains of chronic schizophrenics have lower numbers of caldendrin-immunoreactive neurons, which express the protein at a much higher level. This loss of caldendrin in some neurons and up-regulation in others is likely to profoundly change synapto-dendritic signalling in schizophrenic patients (Bernstein et al. 2007). Changes in the distribution of caldendrin have also been observed in kainate-induced epileptic seizures in rats. Caldendrin translocates to the postsynaptic density only in rats that suffered epileptic seizures and may implicate the protein in the pathophysiology of the disease (Smalla et al. 2003). CaBP4 function has been convincingly linked to disease and mutations in this gene generate defects in retinal function. Knockout of CaBP4 was shown to cause a phenotype similar to that of incomplete congenital stationary night blindness patients (Haeseleer et al. 2004) and mutations in CaBP4 can cause autosomal recessive night blindness (Zeitz et al. 2006). Patients with mutations in the CaBP4 gene have been identified, which display congenital stationary night blindness. However, some patients with mutations display different phenotypes (Zeitz et al. 2006). In particular, a novel homozygous nonsense mutation has been reported in two siblings that resulted in severely reduced cone function but only negligible effects on rod function (Littink et al. 2009). It appears, therefore, that genetic mutations in CaBP4 underlie cone-rod synaptic disorders (Littink et al. 2009).
CONCLUDING REMARKS
It has become increasingly clear that a full understanding of how specific aspects of neuronal function are regulated in response to spatially and temporally distinct Ca2+ signals will require a detailed knowledge of the Ca2+ sensors involved. Some of these, like synaptotagmin, are specialized for particular neuronal functions, whereas others such as calmodulin may be involved in multiple cellular processes. In recent years, much has been learnt about the functions of the NCS family of Ca2+ sensors, although the functions of some of the family members are still unknown. Nor is it clear what the significance is of the multiple genes and splice variants of these proteins. The CaBPs have so far been much less studied and much remains to be learnt about the functions of each of these sensors. Further advances will require new insights into the molecular targets of each of the Ca2+ sensors, the molecular basis for their regulation of these targets, and more detailed dissection of the functional roles of each protein in identified neurons.
ACKNOWLEDGMENTS
HVM was supported by a Wellcome Trust Prize PhD Studentship.
Footnotes
Editors: Martin Bootman, Michael J. Berridge, James W. Putney, and H. Llewelyn Roderick
Additional Perspectives on Calcium Signaling available at www.cshperspectives.org
REFERENCES
- Ames JB, Hamashima N, Molchanova T 2002. Structure and calcium-binding studies of a recoverin mutant (E85Q) in an allosteric intermediate state. Biochemistry 41: 5776–5787 [DOI] [PubMed] [Google Scholar]
- Ames JB, Ishima R, Tanaka T, Gordon JI, Stryer L, Ikura M 1997. Molecular mechanics of calcium-myristoyl switches. Nature 389: 198–202 [DOI] [PubMed] [Google Scholar]
- Ames JB, Levay K, Wingard JN, Lusin JD, Slepak VZ 2006. Structural basis for calcium-induced inhibition of rhodopsin kinase by recoverin. J Biol Chem 281: 37237–37245 [DOI] [PubMed] [Google Scholar]
- Ames JB, Tanaka T, Ikura M, Stryer L 1995. Nuclear magnetic resonance evidence for Ca2+-induced extrusion of the myristoyl group of recoverin. J Biol Chem 270: 30909–30913 [DOI] [PubMed] [Google Scholar]
- An WF, Bowlby MR, Bett M, Cao J, Ling HP, Mendoza G, Hinson JW, Mattsson KI, Strassle BW, Trimmer JS, et al. 2000. Modulation of A-type potassium channels by a family of calcium sensors. Nature 403: 553–556 [DOI] [PubMed] [Google Scholar]
- Aravind P, Chandra K, Reddy PP, Jeromin A, Chary KV, Sharma Y 2008. Regulatory and structural EF-hand motifs of neuronal calcium sensor-1: Mg2+ modulates Ca2+ binding, Ca2+-induced conformational changes, and equilibrium unfolding transitions. J Mol Biol 376: 1100–1115 [DOI] [PubMed] [Google Scholar]
- Augustine GJ, Santamaria F, Tanaka K 2003. Local calcium signaling in neurons. Neuron 40: 331–346 [DOI] [PubMed] [Google Scholar]
- Bahi N, Friocourt G, Carrié A, Graham ME, Weiss JL, Chafey P, Fauchereau F, Burgoyne RD, Chelly J 2003. IL1 receptor accessory protein like, a protein involved in X-linked mental retardation, interacts with Neuronal Calcium Sensor-1 and regulates exocytosis. Human Mol Genetics 12: 1415–1425 [DOI] [PubMed] [Google Scholar]
- Barria A, Muller D, Derkach V, Griffith LC, Soderling TR 1997. Regulatory phosphorylation of AMPA-type glutamate receptors by CaM-KII during long-term potentiation. Science 276: 2042–2045 [DOI] [PubMed] [Google Scholar]
- Bernstein HG, Sahin J, Smalla KH, Gundelfinger ED, Bogerts B, Kreutz MR 2007. A reduced number of cortical neurons show increased Caldendrin protein levels in chronic schizophrenia. Schizophr Res 96: 246–256 [DOI] [PubMed] [Google Scholar]
- Berridge MJ 1998. Neuronal calcium signalling. Neuron 21: 13–26 [DOI] [PubMed] [Google Scholar]
- Berridge MJ 2010. Calcium hypothesis of Alzheimer's disease. Pflugers Arch 459: 441–449 [DOI] [PubMed] [Google Scholar]
- Bito H, Deisseroth K, Tsien RW 1997. Ca2+-dependent regulation in neuronal gene expression. Current Opinion in Neurobiol 7: 419–429 [DOI] [PubMed] [Google Scholar]
- Blasiole B, Kabbani N, Boehmler W, Thisse B, Thisse C, Canfield V, Levenson R 2005. Neuronal calcium sensor-1 gene ncs-1 is essential for semicircular canal formation in zebrafish inner ear. J Neurobiol 64: 285–297 [DOI] [PubMed] [Google Scholar]
- Bourne Y, Dannenberg J, Pollmann V, Marchot P, Pongs O 2001. Immunocytochemical localisation and crystal structure of human frequenin (neuronal calcium sensor 1). J Biol Chem 276: 11949–11955 [DOI] [PubMed] [Google Scholar]
- Brackmann M, Schuchmann S, Anand R, Braunewell KH 2005. Neuronal Ca2+ sensor protein VILIP-1 affects cGMP signalling of guanylyl cyclase B by regulating clathrin-dependent receptor recycling in hippocampal neurons. J Cell Sci 118: 2495–2505 [DOI] [PubMed] [Google Scholar]
- Braunewell K-H 2005. The darker side of Ca2+ signaling by neuronal Ca2+-sensor proteins: From Alzheimer's disease to cancer. Trends Pharmacol Sci 26: 345–351 [DOI] [PubMed] [Google Scholar]
- Braunewell KH, Klein-Szanto AJ 2009. Visinin-like proteins (VSNLs): Interaction partners and emerging functions in signal transduction of a subfamily of neuronal Ca2+ -sensor proteins. Cell Tissue Res 335: 301–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne RD 2004. The neuronal calcium-sensor proteins. Biochim Biophys Acta 1742: 59–68 [DOI] [PubMed] [Google Scholar]
- Burgoyne RD 2007. Neuronal calcium sensor proteins: Generating diversity in neuronal Ca2+ signalling. Nat Rev Neurosci 8: 182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne RD, Weiss JL 2001. The neuronal calcium sensor family of Ca2+-binding proteins. Biochem J 353: 1–12 [PMC free article] [PubMed] [Google Scholar]
- Burgoyne RD, O'Callaghan DW, Hasdemir B, Haynes LP, Tepikin AV 2004. Neuronal calcium sensor proteins: Multitalented regulators of neuronal function. Trends Neurosci 27: 203–209 [DOI] [PubMed] [Google Scholar]
- Burns ME, Mendez A, Chen J, Baylor DA 2002. Dynamics of cyclic AMP synthesis in retinal rods. Neuron 36: 81–91 [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Choi EK, Luo YX, Lilliehook C, Crowley AC, Merriam DE, Wasco W 1998. Calsenilin: A calcium-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nature Medicine 4: 1177–1181 [DOI] [PubMed] [Google Scholar]
- Carrion AM, Link WA, Ledo F, Mellstrom B, Naranjo JR 1999. DREAM is a Ca2+-regulated transcriptional repressor. Nature 398: 80–84 [DOI] [PubMed] [Google Scholar]
- Catterall WA, Few AP 2008. Calcium channel regulation and presynaptic plasticity. Neuron 59: 882–901 [DOI] [PubMed] [Google Scholar]
- Chapman ER 2008. How does synaptotagmin trigger neurotransmitter release? Annu Rev Biochem 77: 615–641 [DOI] [PubMed] [Google Scholar]
- Chattopadhyaya R, Meador WE, Means AR, Quiocho FA 1992. Calmodulin structure refined at 1.7 A resolution. J Mol Biol 228: 1177–1192 [DOI] [PubMed] [Google Scholar]
- Chaumont S, Compan V, Toulme E, Richler E, Housley GD, Rassendren F, Khakh BS 2008. Regulation of P2X2 receptors by the neuronal calcium sensor VILIP1. Sci Signal 1: ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Inglese J, Lefkowitz RJ, Hurley JB 1995. Ca2+-dependent interaction of recoverin with rhodopsin kinase. J Biol Chem 270: 18060–18066 [DOI] [PubMed] [Google Scholar]
- Cheng H-YM, Pitcher GM, Laviolette SR, Whishaw IQ, Tong KI, Kockeritz LK, Wada T, Joza NA, Crackower M, Goncalves J, et al. 2002. DREAM is a critical transcriptional repressor for pain modulation. Cell 108: 31–43 [DOI] [PubMed] [Google Scholar]
- Chicka MC, Hui E, Liu H, Chapman ER 2008. Synaptotagmin arrests the SNARE complex before triggering fast, efficient membrane fusion in response to Ca2+. Nat Struct Mol Biol 15: 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coukell B, Cameron A, Perusini S, Shim K 2004. Disruption of the NCS-1/frequenin-related ncsA gene in Dictyostelium discoideum accelerates development. Develop Growth Differ 46: 449–458 [DOI] [PubMed] [Google Scholar]
- Craxton M 2004. Synaptotagmin gene content of the sequenced genomes. BMC Genomics 5: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Meyer AC, Calin-Jageman I, Neef J, Haeseleer F, Moser T, Lee A 2007. Ca2+-binding proteins tune Ca2+-feedback to Cav1.3 channels in mouse auditory hair cells. J Physiol 585: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dason JS, Romero-Pozuelo J, Marin L, Iyengar BG, Klose MK, Ferrus A, Atwood HL 2009. Frequenin/NCS-1 and the Ca2+-channel {α} 1-subunit co-regulate synaptic transmission and nerve-terminal growth. J Cell Sci 122: 4109–4121 [DOI] [PubMed] [Google Scholar]
- de Barry J, Janoshazi A, Dupont JL, Procksch O, Chasserot-Golaz S, Jeromin A, Vitale N 2006. Functional implication of neuronal calcium sensor-1 and PI4 kinase-β interaction in regulated exocytosis of PC12 cells. J Biol Chem 281: 18098–18111 [DOI] [PubMed] [Google Scholar]
- de Wit H, Walter AM, Milosevic I, Gulyas-Kovacs A, Riedel D, Sorensen JB, Verhage M 2009. Synaptotagmin-1 docks secretory vesicles to syntaxin-1/SNAP-25 acceptor complexes. Cell 138: 935–946 [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Heist EK, Tsien RW 1998. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature 392: 198–202 [DOI] [PubMed] [Google Scholar]
- DeMaria CD, Soong TW, Alselkhan BA, Alvania RS, Yue DT 2001. Calmodulin bifurcates the local Ca2+ signal that modulates P/Q-type Ca2+ channels. Nature 411: 484–489 [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Karpova A, Mikhaylova M, Zdobnova I, Konig I, Landwehr M, Kreutz M, Smalla KH, Richter K, Landgraf P, et al. 2008. Caldendrin-Jacob: A protein liaison that couples NMDA receptor signalling to the nucleus. PLoS Biol 6: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizhoor AM, Hurley JB 1996. Inactivation of EF-hands makes GCAP-2 (p24) a constitutive activator of photoreceptor guanulyl cyclase by preventing a Ca2+-induced “activator-to-inhibitor” transition. J Biol Chem 271: 19346–19350 [DOI] [PubMed] [Google Scholar]
- Dizhoor AM, Ray S, Kumar S, Niemi G, Spencer M, Brolley D, Walsh KA, Philipov PP, Hurley JB, Stryer L 1991. Recoverin: A calcium sensitive activator of retinal rod guanylate cyclase. Science 251: 915–918 [DOI] [PubMed] [Google Scholar]
- Fallon JL, Halling DB, Hamilton SL, Quiocho FA 2005. Structure of calmodulin bound to the hydrophobic IQ domain of the cardiac Ca(v)1.2 calcium channel. Structure 13: 1881–1886 [DOI] [PubMed] [Google Scholar]
- Fernandez-Chacon R, Konigstorfer A, Gerber SH, Garcia J, Matos MF, Stevens CF, Brose N, Rizo J, Rosenmund C, Sudhof TC 2001. Synaptotagmin I functions as a calcium regulator of release probability. Nature 410: 41–49 [DOI] [PubMed] [Google Scholar]
- Fernandez I, Arac D, Ubach J, Gerber SH, Shin O-h, Gao Y, Anderson RGW, Sudhof TC, Rizo J 2001. Three-dimensional structure of the synaptotagmin 1 C2B-domain: Synaptotagmin 1 as a phospholipid binding machine. Neuron 32: 1057–1069 [DOI] [PubMed] [Google Scholar]
- Few AP, Lautermilch NJ, Westenbroek RE, Scheuer T, Catterall WA 2005. Differential regulation of Cav2.1 channels by calcium binding protein 1 and visinin-like protein-2 requires N-terminal myristoylation. J Neurosci 25: 7071–7080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald DJ, Burgoyne RD, Haynes LP 2008. Neuronal calcium sensor proteins are unable to modulate NFAT activation in mammalian cells. Biochim Biophys Acta 1780: 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KM, Zoulya S, Stryer L, McKay DB 1993. 3-Dimensional structure of recoverin, a calcium sensor in vision. Cell 75: 709–716 [DOI] [PubMed] [Google Scholar]
- Geppert M, Goda Y, Hammer RE, Li C, Rosahl TW, Stevens CF, Sudhof TC 1994. Synaptotagmin I: A major Ca2+ sensor for transmitter release at a central synapse. Cell 79: 717–727 [DOI] [PubMed] [Google Scholar]
- Gomez M, De Castro E, Guarin E, Sasakura H, Kuhara A, Mori I, Bartfai T, Bargmann CI, Nef P 2001. Ca2+ signalling via the neuronal calcium sensor-1 regulates associative learning and memory in C.elegans. Neuron 30: 241–248 [DOI] [PubMed] [Google Scholar]
- Groffen AJ, Martens S, Arazola RD, Cornelisse LN, Lozovaya N, de Jong AP, Goriounova NA, Habets RL, Takai Y, Borst JG, et al. 2010. Doc2b is a high-affinity Ca2+ sensor for spontaneous neurotransmitter release. Science 327: 1614–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeseleer F 2008. Interaction and colocalization of CaBP4 and Unc119 (MRG4) in photoreceptors. Investigative Ophthalmol Visual Sci 49: 2366–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeseleer F, Imanishi Y, Maeda T, Possin DE, Maeda A, Lee A, Rieke F, Palczewski K 2004. Essential role of Ca2+-binding protein 4, a Cav1.4 channel regulator in photoreceptor synaptic function. Nature Neurosci 7: 1079–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeseleer F, Imanishi Y, Sokal I, Filipek S, Palczewski K 2002. Calcium-binding proteins: Intracellular sensors from the calmodulin superfamily. Biochem Biophys Res Comm 290: 615–623 [DOI] [PubMed] [Google Scholar]
- Haeseleer F, Sokal I, Verlinde CLMJ, Erdjument-Bromage H, Tempst P, Pronin AN, Benovic JL, Fariss RN, Palczewski K 2000. Five members of a novel Ca2+ binding protein (CABP) subfamily with similarity to calmodulin. J Biol Chem 275: 1247–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley MT, Lian LY, Haynes LP, Burgoyne RD 2010. Structural and functional deficits in a Neuronal Calcium Sensor-1 mutant identified in a case of Autistic Spectrum Disorder. PLoS ONE 5: e10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasdemir B, Fitzgerald DJ, Prior IA, Tepikin AV, Burgoyne RD 2005. Traffic of Kv4 K+ channels mediated by KChIP1 is via a novel post-ER vesicular pathway. J Cell Biol 171: 459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes LP, Tepikin AV, Burgoyne RD 2004. Calcium Binding Protein 1 is an inhibitor of agonist-evoked, inositol 1,4,5-trisphophate-mediated calcium signalling. J BiolChem 279: 547–555 [DOI] [PubMed] [Google Scholar]
- Haynes LP, Thomas GMH, Burgoyne RD 2005. Interaction of neuronal calcium sensor-1 and ARF1 allows bidirectional control of PI(4) kinase and TGN-plasma membrane traffic. J Biol Chem 280: 6047–6054 [DOI] [PubMed] [Google Scholar]
- Haynes LP, Fitzgerald DJ, Wareing B, O'Callaghan DW, Morgan A, Burgoyne RD 2006. Analysis of the interacting partners of the neuronal calcium-binding proteins L-CaBP1, hippocalcin, NCS-1 and neurocalcin. Proteomics 6: 1822–1832 [DOI] [PubMed] [Google Scholar]
- Haynes LP, Sherwood MW, Dolman NJ, Burgoyne RD 2007. Specificity, promiscuity and localization of ARF protein interactions with NCS-1 and phosphatidylinositol-4 kinase-IIIβ. Traffic 8: 1080–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks KB, Wang BQ, Schnieders EA, Thorner J 1999. Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nature Cell Biology 1: 234–241 [DOI] [PubMed] [Google Scholar]
- Hilfiker S 2003. Neuronal calcium sensor-1: A multifunctional regulator of secretion. Biochem Soc Trans 31: 828–832 [DOI] [PubMed] [Google Scholar]
- Holmqvist MH, Cao J, Hernandez-Pineda R, Jacobson MD, Carroll KI, Sung MA, Betty M, Ge P, Gilbride KJ, Brown ME, et al. 2002. Elimination of fast inactivation in Kv4 A-type potassium channels by an auxiliary subunit domain. Proc Natl Acad Sci USA 99: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes KA, Pennesi ME, Sokal I, Church-Kopish J, Schmidt B, Margolis D, Frederick JM, Rieke F, Palczewski K, Wu SM, et al. 2002. GCAP1 rescues rod photoreceptor response in GCAP1/GCAP2 knockout mice. EMBO Journal 21: 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui E, Johnson CP, Yao J, Dunning FM, Chapman ER 2009. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca(2+)-regulated fusion. Cell 138: 709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo J, Heon S, Kim MJ, Son GH, Park Y, Henley JM, Weiss JL, Sheng M, Collingridge GL, Cho K 2008. Metabotropic glutamate receptor-mediated LTD involves two interacting Ca2+ sensors, NCS-1 and PICK1. Neuron 60: 1095–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo D-G, Jang J, Kim B-J, Lundkvist J, Jung Y-K 2004. Overexpression of calsenilin enhances g-secretase activity. Neuroscience letters 378: 59–64 [DOI] [PubMed] [Google Scholar]
- Kabbani N, Negyessy L, Lin R, Goldman-Rakic P, Levenson R 2002. Interaction with the neuronal calcium sensor NCS-1 mediates desensitization of the D2 dopamine receptor. J Neurosci 22: 8476–8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasri NN, Holmes AM, Bultynck G, Parys JB, Bootman MD, Rietdorf K, Missiaen L, McDonald F, Smedt HD, Conway SJ, et al. 2004. Regulation of InsP3 receptor activity by neuronal Ca2+-binding proteins. EMBO J 23: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita-Kawada M, Tang J, Xiao R, Kaneko S, Foskett JK, Zhu MX 2005. Inhibition of TRPC5 channels by Ca2+ binding protein 1 in Xenopus oocytes. Pflugers Arch 450: 345–354 [DOI] [PubMed] [Google Scholar]
- Klenshin VA, Calvert PD, Bownds MD 1995. Inhibition of rhodopsin kinase by recoverin. J Biol Chem 270: 16147–16152 [DOI] [PubMed] [Google Scholar]
- Koch K-W 2006. GCAPs, the classical neuronal calcium sensors in the retina. A Ca2+-relay model of guanylate cyclase activation. Calcium Binding Proteins 1: 3–6 [Google Scholar]
- Koh PO, Undie AS, Kabbani N, Levenson R, Goldman-Rakic PS, Lidow MS 2003. Up-regulation of neuronal calcium sensor-1 (NCS-1) in the prefrontal cortex of schizophrenic and bipolar patients. Proc Natl Acad Sci 100: 313–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen L, Hansson I, Kukkonen JP, Brannvall K, Kobayashi M, Takamatsu K, Lindholm D 2004. Hippocalcin protects against caspase-12-induced and age-dependent neuronal degeneration. Mol Cell Neurosci 28: 85–95 [DOI] [PubMed] [Google Scholar]
- Kuo H-C, Cheng C-F, Clark RB, Lin JJ-C, Lin JL-C, Hoshijima M, Nguyen-Tran VTB, Gu Y, Ikeda Y, Chu P-H, et al. 2001. A defect in the Kv channel-interacting protein 2 (KChIP2) gene leads to a complete loss of Ito and confers susceptibility to ventricular tachycardia. Cell 107: 801–813 [DOI] [PubMed] [Google Scholar]
- Lee A, Scheuer T, Catterall WA 2000. Ca2+/calmodulin-dependent facilitation and inactivation of P/Q-type Ca2+ channels. J Neurosci 20: 6830–6838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Jimenez A, Cui G, Haeseleer F 2007. Phosphorylation of the Ca2+-binding protein CaBP4 by protein kinase C zeta in photoreceptors. J Neurosci 27: 12743–12754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Westenbroek RE, Haeseleer F, Palczewski K, Scheuer T, Catterall WA 2002. Differential modulation of Cav2.1 channels by calmodulin and Ca2+-binding protein 1. Nature Neurosci 5: 210–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Chan J, Haeseleer F, Mikoshiba K, Palczewski K, Ikura M, Ames JB 2009. Structural insights into Ca2+-dependent regulation of inositol 1,4,5-trisphosphate receptors by CaBP1. J Biol Chem 284: 2472–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilliehook C, Bozdagi O, Yao J, Gomez-Ramirez M, Zaidi NF, Wasco W, Gandy S, Santucci AC, Haroutunian V, Huntley GW, et al. 2003. Altered Ab formation and long-term potentiation in a calsenilin knock-out. J Neurosci 23: 9097–9106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Jeanclos EM, Treuil M, Braunewell K-H, Gundelfinger ED, Anand R 2002. The calcium sensor protein visinin-like protein-1 modulates the surface expression and agonist sensitivity of the a4b2 nicotinic acetylcholine receptor. J Biol Chem 277: 41872–41878 [DOI] [PubMed] [Google Scholar]
- Lisman J, Schulman H, Cline H 2002. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat Rev Neurosci 3: 175–190 [DOI] [PubMed] [Google Scholar]
- Littink KW, van Genderen MM, Collin RW, Roosing S, de Brouwer AP, Riemslag FC, Venselaar H, Thiadens AA, Hoyng CB, Rohrschneider K, et al. 2009. A novel homozygous nonsense mutation in CABP4 causes congenital cone-rod synaptic disorder. Investigative Ophthalmol Visual Sci 50: 2344–2350 [DOI] [PubMed] [Google Scholar]
- Liu X, Yang PS, Yang W, Yue DT 2010. Enzyme-inhibitor-like tuning of Ca2+ channel connectivity with calmodulin. Nature 463: 968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackler JM, Drummond JA, Loewen CA, Robinson IM, Reist NE 2002. The C2B Ca2+-binding motif of synaptotagmin is required for synaptic transmission in vivo. Nature 418: 340–344 [DOI] [PubMed] [Google Scholar]
- Makino CL, Dodd RL, Chen J, Burns ME, Roca A, Simon MI, Baylor DA 2004. Recoverin regulates light-dependent phosphodiesterase activity in retinal rods. J Gen Physiol 123: 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malleret G, Haditsch U, Genoux D, Jones MW, Bliss TV, Vanhoose AM, Weitlauf C, Kandel ER, Winder DG, Mansuy IM 2001. Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell 104: 675–686 [DOI] [PubMed] [Google Scholar]
- Markova O, Fitzgerald D, Stepanyuk A, Dovgan A, Cherkas V, Tepikin A, Burgoyne RD, Belan P 2008. Hippocalcin signaling via site-specific translocation in hippocampal neurons. Neuroscience letters 442: 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue HV, Burgoyne RD, Haynes LP 2009. Membrane targeting of the EF-hand containing calcium-sensing proteins CaBP7 and CaBP8. Biochem Biophys Res Commun 380: 825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue HV, Haynes LP, Burgoyne RD 2010. Bioinformatic analysis of CaBP/calneuron proteins reveals a family of highly conserved vertebrate Ca2+-binding proteins. BMC Res Notes 3: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFerran BW, Graham ME, Burgoyne RD 1998. NCS-1, the mammalian homologue of frequenin is expressed in chromaffin and PC12 cells and regulates neurosecretion from dense-core granules. J Biol Chem 273: 22768–22772 [DOI] [PubMed] [Google Scholar]
- Mellstrom B, Naranjo JR 2001. Ca2+-dependent transcriptional repression and derepression: DREAM, a direct effector. Seminars Cell Develop Biol 12: 59–63 [DOI] [PubMed] [Google Scholar]
- Mendez A, Burns ME, Sokal I, Dizhoor AM, Baehr W, Palczewski K, Baylor DA, Chen J 2001. Role of guanylate cyclase-activating proteins (GCAPs) in setting the flash sensitivity of rod photoreceptors. Proc Natl Acad Sci 98: 9948–9953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer WA, Korhonen L, Skoglosa Y, Olssen P-A, Kukknen JP, Lindholm D 2000. NAIP interacts with hippocalcin and protects neurons against calcium-induced cell death through caspase-3-dependent and -independent pathways. EMBO J 19: 3597–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova M, Reddy PP, Munsch T, Landgraf P, Suman SK, Smalla KH, Gundelfinger ED, Sharma Y, Kreutz MR 2009. Calneurons provide a calcium threshold for trans-Golgi network to plasma membrane trafficking. Proc Natl Acad Sci 106: 9093–9098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova M, Sharma Y, Reissner C, Nagel F, Aravind P, Rajini B, Smalla KH, Gundelfinger ED, Kreutz MR 2006. Neuronal Ca2+ signaling via caldendrin and calneurons. Biochim Biophys Acta 1763: 1229–1237 [DOI] [PubMed] [Google Scholar]
- Nef S, Fiumelli H, de Castro E, Raes M-B, Nef P 1995. Identification of a neuronal calcium sensor (NCS-1) possibly involved in the regulation of receptor phosphorylation. J Receptor Signal Trans 15: 365–378 [DOI] [PubMed] [Google Scholar]
- O'Callaghan DW, Tepikin AV, Burgoyne RD 2003b. Dynamics and calcium-sensitivity of the Ca2+-myristoyl switch protein hippocalcin in living cells. J Cell Biol 163: 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan DW, Haynes LP, Burgoyne RD 2005. High-affinity interaction of the N-terminal myristoylation motif of the neuronal calcium sensor protein hippocalcin with phosphatidylinositol 4,5-bisphosphate. Biochem J 391: 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan DW, Hasdemir B, Leighton M, Burgoyne RD 2003a. Residues within the myristoylation motif determine intracellular targeting of the neuronal Ca2+ sensor protein KChIP1 to post-ER transport vesicles and traffic of Kv4 K+ channels. J Cell Sci 116: 4833–4845 [DOI] [PubMed] [Google Scholar]
- O'Callaghan DW, Ivings L, Weiss JL, Ashby MC, Tepikin AV, Burgoyne RD 2002. Differential use of myristoyl groups on neuronal calcium sensor proteins as a determinant of spatio-temporal aspects of Ca 2+-signal transduction. J BiolChem 277: 14227–14237 [DOI] [PubMed] [Google Scholar]
- O'Connor V, El Far O, Bofill-Cardona E, Nanoff C, Freissmuth M, Karschin A, Airas JM, Betz H, Boehm S 1999. Calmodulin dependence of presynaptic metabotropic glutamate receptor signalling. Science 286: 1180–1184 [DOI] [PubMed] [Google Scholar]
- Olafsson P, Wang T, Lu B 1995. Molecular cloning and functional characterisation of the Xenopus Ca2+ binding protein frequenin. Proc Natl Acad Sci 92: 8001–8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Sokal I, Baehr W 2004. Guanylate cyclase-activating proteins: Structure, function and diversity. Biochem Biophys Res Comm 322: 1123–1130 [DOI] [PubMed] [Google Scholar]
- Palmer CL, Lim W, Hastie PG, Toward M, Korolchuk VI, Burbidge SA, Banting G, Collingridge GL, Isaac JT, Henley JM 2005. Hippocalcin functions as a calcium sensor in hippocampal LTD. Neuron 47: 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Cao P, Xu W, Sudhof TC 2010. Calmodulin controls synaptic strength via presynaptic activation of calmodulin kinase II. J Neurosci 30: 4132–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterlini M, Revilla V, Grant AL, Wisden W 2000. Expression of the neuronal calcium sensor protein family in the rat brain. Neurosci 99: 205–216 [DOI] [PubMed] [Google Scholar]
- Pennesi ME, Howes KA, Baehr W, Wu SM 2003. Guanylate cyclase-activating protein (GCAP) 1 rescues cone recovery kinetics in GCAP1/GCAP2 knockout mice. Proc Natl Acad Sci 100: 6783–6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petko JA, Kabbani N, Frey C, Woll M, Hickey K, Craig M, Canfield VA, Levenson R 2009. Proteomic and functional analysis of NCS-1 binding proteins reveals novel signaling pathways required for inner ear development in zebrafish. BMC Neurosci 10: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioletti M, Findeisen F, Hura GL, Minor DL 2006. Three-dimensional structure of the KChIP1–Kv4.3 T1 complex reveals a cross-shaped octamer. Nature Struct Mol Biol 13: 987–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polans A, Baehr W, Palczewski K 1996. Turned on by Ca2+! The physiology and pathology of Ca2+-binding proteins in the retina. Trends Neurosci 19: 547–554 [DOI] [PubMed] [Google Scholar]
- Pongs O, Lindemeier J, Zhu XR, Theil T, Endelkamp D, Krah-Jentgens I, Lambrecht H-G, Koch KW, Schwemer J, Rivosecchi R, et al. 1993. Frequenin - A novel calcium-binding protein that modulates synaptic efficacy in the Drosophila nervous system. Neuron 11: 15–28 [DOI] [PubMed] [Google Scholar]
- Pruunsild P, Timmusk T 2005. Structure, alternative splicing, and expression of the human and mouse KCNIP gene family. Genomics 86: 581–593 [DOI] [PubMed] [Google Scholar]
- Rhodes KJ, Carroll KI, Sung MA, Doliveira LC, Monaghan MM, Burke SL, Strassle BW, Buchwalder L, Menegola M, Cao J, et al. 2004. KChIPs and Kv4asubunitsSA as integral components of A-type potassium channels in mammalian brain. J Neurosci 24: 7903–7915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman C, Jiménez JL, Graham ME, Archer DA, Soloviev M, Burgoyne RD, Davletov B 2006. Conserved pre-fusion protein assembly in regulated exocytosis. Mol Biol Cell 17: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieke F, Lee A, Haeseleer F 2008. Characterization of Ca2+-binding protein 5 knockout mouse retina. Investigative Ophthalmol Visual Sci 49: 5126–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Rosenmund C 2008. Synaptic vesicle fusion. Nat Struct Mol Biol 15: 665–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson IM, Ranjan R, Schwarz TL 2002. Synaptotagmins I and IV promote transmitter release independently of Ca2+ binding in the C2A domain. Nature 418: 336–340 [DOI] [PubMed] [Google Scholar]
- Saab BJ, Georgiou J, Nath A, Lee FJ, Wang M, Michalon A, Liu F, Mansuy IM, Roder JC 2009. NCS-1 in the dentate gyrus promotes exploration, synaptic plasticity, and rapid acquisition of spatial memory. Neuron 63: 643–656 [DOI] [PubMed] [Google Scholar]
- Saimi Y, Kung C 2002. Calmodulin as an ion channel subunit. Ann Rev Physiol 64: 289–311 [DOI] [PubMed] [Google Scholar]
- Sampath A, Strissel KJ, Elias R, Arshavsky VY, McGinnis JF, Rieke F, Hurley JB 2005. Recoverin improves rod-mediated vision by enhancing signal transmission in the mouse retina. Neuron 46: 413–420 [DOI] [PubMed] [Google Scholar]
- Sanchez-Gracia A, Romero-Pozuelo J, Ferrus A 2010. Two frequenins in Drosophila: unveiling the evolutionary history of an unusual neuronal calcium sensor (NCS) duplication. BMC Evolutionary Biol 10: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannevin RH, Wang K-W, Jow F, Megules J, Kospco DC, Edris W, Carroll KC, Lu Q, Xu W, Xu Z, et al. 2004. Two N-terminal domains of Kv4 K+ channels regulate binding to and modulation by KChIP1. Neuron 41: 587–598 [DOI] [PubMed] [Google Scholar]
- Schaad NC, De Castro E, Nef S, Hegi S, Hinrichsen R, Martone ME, Ellisman MH, Sikkink R, Sygush J, Nef P 1996. Direct modulation of calmodulin targets by the neuronal calcium sensor NCS-1. Proc Natl Acad Sci 93: 9253–9258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavo G, Stenbeck G, Rothman JE, Sollner TH 1997. Binding of the synaptic vesicle v-SNARE, synaptotagmin, to the plasma membrane t-SNARE, SNAP-25 can explain docked vesicles at neurotoxin-treated synapses. Proc Natl Acad Sci 94: 997–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurra I, Bernstein H-G, Riederer P, Braunewell K-H 2001. The neuronal calcium sensor protein VILIP-1 is associated with amyloid plaques and promotes cell death and tau phosphorylation in vitro: A link between calcium sensors and alzheimers disease? Neurobiology Disease 8: 900–909 [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Rivard AF, Bachinger HP, Adelman JP 2001. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature 410: 1120–1124 [DOI] [PubMed] [Google Scholar]
- Seidenbecher CI, Landwehr M, Smalla K-H, Kreutz M, Dieterich DC, Zuschratter W, Reissner C, Hammarback JA, Bockers TM, Gundelfinger ED, et al. 2004. Caldendrin but not calmodulin binds to light chain 3 of MAP1A/B: An association with the microtubule cytoskeleton highlighting exclusive binding partners for neuronal Ca2+-sensor proteins. J Mol Biol 336: 957–970 [DOI] [PubMed] [Google Scholar]
- Seidenbecher CI, Langnaese K, Sanmarti-Vila L, Boeckers TM, Smalla K-H, Sabel BA, Garner CC, Gundelfinger ED, Kreutz MR 1998. Caldendrin, a novel neuronal calcium-binding protein confined to the somato-dendritic compartment. J Biol Chem 273: 21324–21331 [DOI] [PubMed] [Google Scholar]
- Shao X, Fernandez I, Sudhof TC, Rizo J 1998. Solution structures of the Ca2+-free and Ca2+-bound C2A domain of synaptotagmin I: Does Ca2+ induce a conformational change? Biochem 37: 16106–16115 [DOI] [PubMed] [Google Scholar]
- Shih PY, Lin CL, Cheng PW, Liao JH, Pan CY 2009. Calneuron I inhibits Ca2+ channel activity in bovine chromaffin cells. Biochem Biophys Res Commun 388: 549–553 [DOI] [PubMed] [Google Scholar]
- Smalla K-H, Seidenbecher CI, Tischmeyer W, Schicknick H, Wyneken U, Bockers TM, Gundelfinger ED, Kreutz MR 2003. Kainate-induced epileptic seizures induce a recruitment of caldendrin to the postsynaptic density in rat brain. Mol Brain Res 116: 159–162 [DOI] [PubMed] [Google Scholar]
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE 1993. SNAP receptors implicated in vesicle targeting and fusion. Nature 362: 318–324 [DOI] [PubMed] [Google Scholar]
- Spilker C, Dresbach T, Braunewell K-H 2002. Reversible translocation and activity-dependent localisation of the calcium-myristoyl switch protein VILIP-1 to different membrane compartments in living hippocampal neurons. J Neurosci 22: 7331–7339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer NC 2006. Electrical activity in early neuronal development. Nature 444: 707–712 [DOI] [PubMed] [Google Scholar]
- Stephen R, Bereta G, Golczak M, Palczewski K, Sousa MC 2007. Stabilizing function for myristoyl group revealed by the crystal structure of a neuronal calcium sensor, guanylate cyclase-activating protein 1. Structure 15: 1392–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl T, Huttner IG, Lusin JD, Osawa M, King D, Thorner J, Ames JB 2007. Structural insights into activation of phosphatidylinositol 4-kinase (pik1) by yeast frequenin (Frq1). J Biol Chem 282: 30949–30959 [DOI] [PubMed] [Google Scholar]
- Sudhof TC, Rothman JE 2009. Membrane fusion: Grappling with SNARE and SM proteins. Science 323: 474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadross MR, Dick IE, Yue DT 2008. Mechanism of local and global Ca2+ sensing by calmodulin in complex with a Ca2+ channel. Cell 133: 1228–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Ames JB, Harvey TS, Stryer L, Ikura M 1995. Sequestration of the membrane targeting myristoyl group of recoverin in the calcium-free state. Nature 376: 444–447 [DOI] [PubMed] [Google Scholar]
- Tang N, Lin T, Yang J, Foskett JK, Ostap EM 2007. CIB1 and CaBP1 bind to the myo1c regulatory domain. J Muscle Res Cell Motility 28: 285–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Maximov A, Shin OH, Dai H, Rizo J, Sudhof TC 2006. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell 126: 1175–1187 [DOI] [PubMed] [Google Scholar]
- Taverna E, Francolini M, Jeromin A, Hilfiker S, Roder J, Rosa P 2002. Neuronal calcium sensor 1 and phosphatidylinositol 4-OH kinase β interact in neuronal cells and are translocated to membranes during nucleotide-evoked exocytosis. J Cell Sci 115: 3909–3922 [DOI] [PubMed] [Google Scholar]
- Tippens AL, Lee A 2007. Caldendrin, a neuron-specific modulator of Cav/1.2 (L-type) Ca2+ channels. J Biol Chem 282: 8464–8473 [DOI] [PubMed] [Google Scholar]
- Tsujimoto T, Jeromin A, Satoh N, Roder JC, Takahashi T 2002. Neuronal calcium sensor 1 and activity-dependent facilitation of P/Q-type calcium channel currents at presynaptic nerve terminals. Science 295: 2276–2279 [DOI] [PubMed] [Google Scholar]
- Venn N, Haynes LP, Burgoyne RD 2008. Specific effects of KChIP3/calsenilin/DREAM but not KChIPs1, 2 and 4 on calcium signalling and regulated secretion in PC12 cells. Biochem J 413: 71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yan Y, Liu Q, Huang Y, Shen Y, Chen L, Chen Y, Yang Q, Hao Q, Wang K, et al. 2007. Structural basis for modulation of Kv4 K+ channels by auxiliary KChIP subunits. Nat Neurosci 10: 32–39 [DOI] [PubMed] [Google Scholar]
- Weiergraber OH, Senin II, Philippov PP, Granzin J, Koch K-W 2003. Impact of N-terminal myristoylation on the Ca2+-dependent conformational transition in recoverin. J Biol Chem 278: 22972–22979 [DOI] [PubMed] [Google Scholar]
- Weiss JL, Archer DA, Burgoyne RD 2000. NCS-1/frequenin functions in an autocrine pathway regulating Ca2+ channels in bovine adrenal chromaffin cells. J Biol Chem 275: 40082–40087 [DOI] [PubMed] [Google Scholar]
- Wen H, Levitan IB 2002. Calmodulin is an auxiliary subunit of KCNQ2/3 potassium channels. J Neurosci 22: 7991–8001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingard JN, Chan J, Bosanac I, Haeseleer F, Palczewski K, Ikura M, Ames JB 2005. Structural analysis of Mg2+ and Ca2+ binding to CaBP1, a neuron-specific regulator of calcium channels. J Biol Chem 280: 37461–37470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods AS, Marcellino D, Jackson SN, Franco R, Ferre S, Agnati LF, Fuxe K 2008. How calmodulin interacts with the adenosine A(2A) and the dopamine D(2) receptors. J Proteome Res 7: 3428–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YQ, Lin X, Liu CM, Jamrich M, Shaffer LG 2001. Identification of a human brain-specific gene, calneuron 1, a new member of the calmodulin superfamily. Mol Genet Metab 72: 343–350 [DOI] [PubMed] [Google Scholar]
- Xia Z, Storm DR 2005. The role of calmodulin as a signal integrator for synaptic plasticity. Nat Rev Neurosci 6: 267–276 [DOI] [PubMed] [Google Scholar]
- Xia X-M, Fakler B, Rivard A, Wayman G, Johnson-Pais T, Keen JE, Ishii T, Hirschberg B, Bond CT, Lutsenko S, Maylie J, Adelman JP 1998. Mechanisms of calcium gating in small conductance calcium activated potassium channels. Nature 395: 503–507 [DOI] [PubMed] [Google Scholar]
- Xiong H, Xia K, Li B, Zhao G, Zhang Z 2009. KChIP1: A potential modulator to GABAergic system. Acta Biochim Biophys Sin (Shanghai) 41: 295–300 [DOI] [PubMed] [Google Scholar]
- Xue M, Ma C, Craig TK, Rosenmund C, Rizo J 2008. The Janus-faced nature of the C(2)B domain is fundamental for synaptotagmin-1 function. Nat Struct Mol Biol 15: 1160–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Goto K, Kuo CH, Kondo H, Miki N 1990. Visinin: A novel calcium binding protein expressed in retianl cone cells. Neuron 4: 469–476 [DOI] [PubMed] [Google Scholar]
- Yang S-N, Tang Y-G, Zucker RS 1999. Selective induction of LTP and LTD by postsynaptic [Ca2+]i elevation. J Neurophysiol 81: 781–787 [DOI] [PubMed] [Google Scholar]
- Yang PS, Alseikhan BA, Hiel H, Grant L, Mori MX, Yang W, Fuchs PA, Yue DT 2006. Switching of Ca2+-dependent inactivation of Ca(V)1.3 channels by calcium binding proteins of auditory hair cells. J Neurosci 26: 10677–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, McBride S, Mak D-OD, Vardi N, Palczewski K, Haeseleer F, Foskett JK 2002. Identification of a family of calcium sensors as protein ligands of inositol trisphosphate receptor Ca2+ release channels. Proc Natl Acad Sci 99: 7711–7716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitz C, Kloeckener-Gruissem B, Forster U, Kohl S, Magyar I, Wissinger B, Matyas G, Borruat FX, Schorderet DF, Zrenner E, et al. 2006. Mutations in CABP4, the gene encoding the Ca2+-binding protein 4, cause autosomal recessive night blindness. Am J Hum Genet 79: 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Varnai P, Tuymetovna G, Balla A, Toth ZE, Oker-Blom C, Roder J, Jeromin A, Balla T 2001. Interaction of neuronal calcium sensor-1 (NCS-1) with phosphatidylinositol 4-kinase β stimulates lipid kinase activity and affects membrane trafficking in COS-7 cells. J Biol Chem 276: 40183–40189 [DOI] [PubMed] [Google Scholar]
- Zhou H, Kim S-A, Kirk EA, Tippens AL, Sun H, Haeseleer F, Lee A 2004a. Ca2+-binding protein-1 facilitates and forms a postsynaptic complex with Cav1.2 (L-type) Ca2+ channels. J Neurosci 24: 4698–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Yu K, McCoy KL, Lee A 2005. Molecular mechanism for divergent regulation of Cav1.2 Ca2+ channels by calmodulin and Ca2+-binding protein-1. J BiolChem 280: 29612–29619 [DOI] [PubMed] [Google Scholar]
- Zhou W, Qian Y, Kunjilwar K, Pfaffinger PJ, Choe S 2004b. Structural insights into the functional interaction of KChIP1 with shal-type K+ channels. Neuron 41: 573–586 [DOI] [PubMed] [Google Scholar]