Abstract
Inactivation of p53 is critical for the formation of most tumors. Illumination of the key function(s) of p53 protein in protecting cells from becoming cancerous is therefore a worthy goal. Arguably p53’s most important function is to act as a transcription factor that directly regulates perhaps several hundred of the cell’s RNA polymerase II (RNAP II)-transcribed genes, and indirectly regulates thousands of others. Indeed p53 is the most well studied mammalian transcription factor. The p53 tetramer binds to its response element where it can recruit diverse transcriptional coregulators such as histone modifying enzymes, chromatin remodeling factors, subunits of the mediator complex, and components of general transcription machinery and preinitiation complex (PIC) to modulate RNAPII activity at target loci (Laptenko and Prives 2006). The p53 transcriptional program is regulated in a stimulus-specific fashion (Murray-Zmijewski et al. 2008; Vousden and Prives 2009), whereby distinct subsets of p53 target genes are induced in response to different p53-activating agents, likely allowing cells to tailor their response to different types of stress. How p53 is able to discriminate between these different loci is the subject of intense research. Here, we describe key aspects of the fundamentals of p53-mediated transcriptional regulation and target gene promoter selectivity.
The p53 transcription factor regulates several hundred genes. It tailors responses to stimuli by employing various different cofactors, posttranslational modifications, and DNA binding sites.
That p53 protein is a critical tumor suppressor in cancer biology is evidenced by its high frequency of mutation in human cancers, presence as a germ-line mutation in Li-Fraumeni cancer prone families, and highly penetrant tumorigenic phenotype in p53 null mice. Centred in the core of a complex wiring of signalling pathways, p53 has been proposed as the master regulator of cell fate In unstressed cells, the activity of p53 is normally held in check by its negative regulator, Mdm2, an E3 ubiquitin ligase, which binds to p53 and targets it for proteasomal degradation (Toledo and Wahl 2006). In response to a plethora of stimuli, however, this inhibition is relieved and p53 target genes are transactivated to cause multiple outcomes such as cell cycle arrest (e.g., p21, 14-3-3), apoptosis (e.g., pig, bax, puma, noxa), senescence (e.g., pai-1), autophagy (e.g., dram), and others, or, they can regulate the p53 pathway itself (e.g., mdm2) (Murray-Zmijewski et al. 2008; Vousden and Prives 2009).
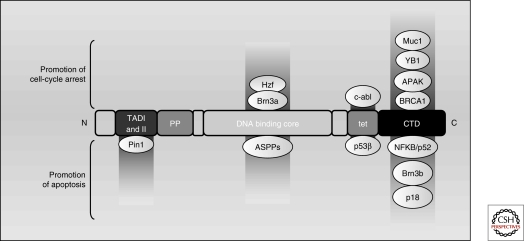
The architecture of the p53 protein itself (see Fig. 5) has features commonly associated with transcriptional regulators: a loosely structured amino-terminal transactivation domain (NTD; comprising two transactivation subdomain, TAD-I, residues 20–40; TAD-II, residues 40–60) a proline-rich region (residues 63–97), an evolutionarily conserved core DNA-binding domain (DBD) (residues 100–300), a linker region (residues 301–323), a tetramerization domain (residues 324–355), and finally, an unstructured basic domain located in the extreme carboxy-terminus, the CTD (residues 360–393).
Figure 5.
p53 target genes and cellular outcomes can be influenced by p53 binding partners. P53 activity can be modulated by different binding partners to induce differential transactivation of target genes and outcome. Proteins that interact with the TADs, the DNA binding core, the tetramerization domain, and the CTD are shown. Hzf, Brn3a, c-abl, Muc1 YB1, APAK, and BRCA1 induce a transcriptional program that facilitates cell cycle arrest, whereas Strap, JMY, Pin1, ASPP1 and ASPP2, p53β, NFκB/p52, and Brn3b induce a program that promotes apoptosis.
Over 80% of cancer-derived p53 mutations are found within the protein’s DNA binding domain (Olivier et al. 2002), underscoring p53’s main role as a sequence-specific DNA binding protein, which was uncovered almost 20 years ago (Vogelstein and Kinzler 1992). After this discovery, the field of p53-mediated transcriptional regulation quickly exploded. To date over 125 protein-coding genes and noncoding RNAs have been shown to be direct transcriptional targets of p53, i.e., ones defined as genes that contain specific sequences to which p53 binds leading to activation of their transcription on induction of p53 (Riley et al. 2008). Although originally characterized solely as a transcriptional coactivator, today p53’s functions have been expanded to include transcriptional repression (Ho and Benchimol 2003), regulation of translation (Ewen and Miller 1996) and homologous recombination (Bertrand et al. 2004), and even the induction of a transcription-independent apoptotic response (Vaseva and Moll 2009). Many mechanisms exist within the cell to fine-tune the p53 transcriptional program. In addition to locus-specific cis-regulatory elements, these include a dizzying number of posttranslational modifications of p53, covalent and noncovalent p53 binding partners, and p53 response elements of variable binding affinity. Each of these features dynamically contributes to the combinatorial regulation of the p53 response, and it is this sheer diversity of variables that presents such a daunting challenge to investigators wishing to study p53-mediated transcription (see diagram; Fig. 1). Here, we provide a basic overview of the mechanistic role of p53 in transcription, and address some of the ways in which promoter selectivity is accomplished. Although the activities of the p53 family members (p63, p73, and their various isoforms), as well as mutant p53, are also highly relevant to the regulation of p53-mediated transcription, they are outside the scope of this article and are discussed elsewhere.
Figure 1.
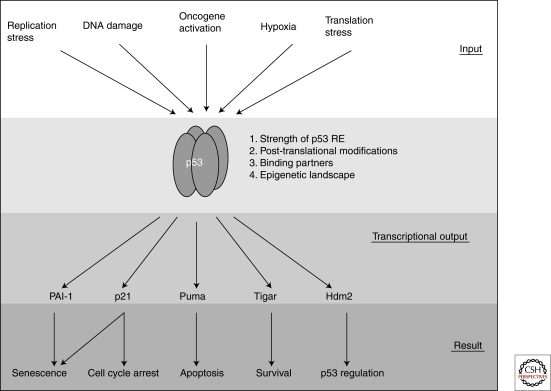
p53 lies at the center of a complex signalling network. In response to various inputs (top of figure), the p53 protein becomes stabilized. On stabilization of p53, various transcriptional outputs can be realized which may be determined by the strength of the p53 RE, the posttranslational modification status of p53, specific p53 binding partners, and the epigenetic landscape of the target gene promoter, among others. The transcriptional output of p53 is responsible for determining which cellular process(es) occur in response distinct genotoxic insults.
THE MECHANICS OF p53-MEDIATED TRANSCRIPTION
DNA Binding
The first step in p53-mediated transcription is the binding of the protein to its recognition site in DNA. After various types of genotoxic insults, p53 is stabilized, translocates to the nucleus, and binds as a dimer of dimers to its response element (RE) (McLure and Lee 1998; Kitayner et al. 2006). The p53 RE was originally defined as RRRCWWGYYY (n = 0–13) RRRCWWGYYY (where R is adenine or guanine, W is a purine base, and Y is a pyrimidine base) (el-Deiry et al. 1992; Funk et al. 1992), although the range of functional p53 binding sites includes many elements with one or more base pairs that do not match the consensus (Gohler et al. 2002). Noncanonical sites, such as the pig3 (TGYCC)n microsatellite response element (Contente et al. 2002), the triplet pairs of the pentameric element at the aqp3 locus (Zheng and Chen 2001), or the “head-to-tail” configuration of the mdr1 p53 site (Johnson et al. 2001), have also been described.
Although p53 REs tend to cluster within noncoding regions of the gene (Riley et al. 2008), they can be located practically anywhere within the target gene locus. P53 REs are most commonly found in the promoter at varying distances upstream (e.g., p21, noxa) from the transcription start site (TSS), although sometimes they are located very close (within ∼300 bp) to the TSS (e.g., hdm2, pcna), or within early intronic sequences (e.g., puma, pig3 microsatellite RE), but can even be found within exons (e.g., miR-34a). As a general—but not universal—rule, REs decrease in transactivation potential as they increase in distance from the TSS (Riley et al. 2008).
In addition to the primary sequence and the genomic location of the p53 RE, several other features of both p53 and DNA topology play a role in the affinity of p53 for its target site. It has been shown that p53 binding to DNA is dependent on the DBD’s ability to coordinate a single Zn++ ion via C176, C238, and C242, and, expectedly, p53 that is mutant in these residues is impaired in DNA binding (Hainaut and Milner 1993). Lending further support to the central role of p53’s DBD cysteine residues in transactivation, redox proteins such as Ref-1 can stimulate p53 binding in vitro by altering the redox state of p53 (Jayaraman et al. 1997; Seo et al. 2002). Notably also, several groups have shown that on binding to DNA, the DBD of p53 can induce a significant structural bend in the DNA, thus allowing the p53 tetramer to bind in a more sterically favorable fashion. It has therefore been postulated that REs with a flexible DNA conformation are favorable for p53 binding (Balagurumoorthy et al. 1995; Nagaich et al. 1997a; Nagaich et al. 1997b; Nagaich et al. 1999).
Although p53 binding to its target genes is clearly increased when the protein becomes stabilized after various forms of stress, surprisingly, a significant amount of p53 is also found bound to select sites under basal conditions despite its low level in the cell (Espinosa et al. 2003; Shaked et al. 2008). How does p53 locate its target sequence within this convoluted context of chromatin? One clue may be the fact that p53 contains two distinct DNA binding domains: the central core DBD with the ability to specifically recognize the p53 RE, as well as the highly basic CTD located in the last 30 residues of the protein that recognizes DNA (and RNA) nonspecifically. It has been shown that this nonspecific CTD binding region plays a role in the ability of p53 to linearly diffuse on naked DNA (Palecek et al. 1997; McKinney et al. 2004; Liu and Kulesz-Martin 2006; Tafvizi et al. 2008). Whether such sliding contributes to p53’s binding site localization remains to be determined. Nevertheless, the roles of the p53 CTD have been a subject of fascination for many years. Initial in vitro studies reported that this lysine rich region facilitates allosteric changes in p53 allowing increased binding by the core domain (Halazonetis and Kandil 1993; Hupp and Lane 1994). It was then shown that modification of the CTD negatively regulates the core domain’s ability to bind to short oligonucleotides in vitro and that acetylation of the CTD could enhance the transactivation of p53 targets (Avantaggiati et al. 1997; Gu and Roeder 1997; Sakaguchi et al. 1998; Liu et al. 1999); as a result, it was postulated that CTD acetylation enhances p53’s ability to bind to DNA. This hypothesis was challenged by the observation that carboxy-terminal modification affects p53 binding to short oligonucleotides but not to long segments of DNA such that are found in the cell (Espinosa and Emerson 2001). Although in vivo, acetylated p53 has been found enriched at promoters (Luo et al. 2004), it is not yet clear whether p53 needs to be acetylated to bind its RE, or whether DNA-bound p53 is more apt to be acetylated by HATs such as p300 (Dornan et al. 2003). Further, the CTD may also facilitate the enhanced recruitment of the TRRAP histone acetyltransferase containing complex as an alternate mechanism behind acetylated p53’s increased activity (Barlev et al. 2001). The CTD of p53 also positively regulates p53 binding to unique DNA structures such that are likely to be found in the cell, including single-stranded DNA overhangs, hemicatenated DNA, minicircular DNA, and supercoiled DNA (Mazur et al. 1999; Zotchev et al. 2000; Gohler et al. 2002; McKinney and Prives 2002; Stros et al. 2004). Along these lines, a carboxy-terminally truncated p53 (p53ΔC30) is markedly impaired in binding to chromatinized DNA templates in vitro (Espinosa and Emerson 2001) and to p53 target promoters in vivo when expressed at physiological levels (McKinney et al. 2004), indicating that the p53 CTD is in fact explicitly required for promoter binding in the context of chromatin.
Transcription Initiation
The ability of p53 to stimulate transcription on RNA polymerase II (RNAPII)-transcribed genes is certainly the most well-studied function of the tumor suppressor; a depiction of p53-dependent transactivation is shown in Figure 2. The modification of histones is necessary to open up chromatin so that the general transcription machinery can bind. In response to DNA damage, p53 is involved in the recruitment of the histone variant H2A.Z, an event which is required for full activation of p21 (Gevry et al. 2007). An elegant study has further elucidated a role for PRMT1 and CARM1, histone methyltransferases (HMTs) that cooperate with p300/CBP in a p53-dependent fashion, to facilitate transcription on the gadd45 locus after UV irradiation (An et al. 2004). It is possible that the action of these HMTs is required for full transcriptional activation of other p53-responsive genes as well. However, the most well-documented p53-dependent histone modification is acetylation: after p53 has bound to its recognition site, histone acetyltransferases (HATs) such as p300/CBP (Avantaggiati et al. 1997; Gu and Roeder 1997; Lill et al. 1997; Scolnick et al. 1997), pCAF (Scolnick et al. 1997; Barlev et al. 2001), GCN5 (Candau et al. 1997), or TIP60 (Gevry et al. 2007), are recruited in a p53-dependent fashion to acetylate the histones within the vicinity of p53 REs.
Figure 2.
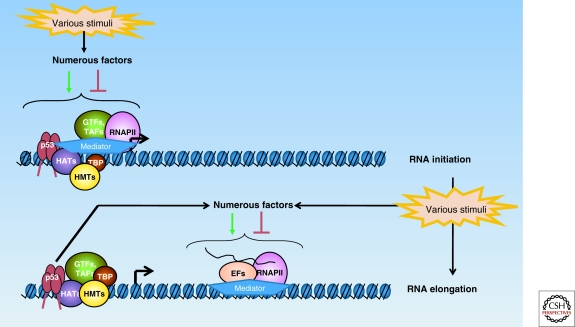
p53 can modulate transcription intiation and elongation at RNAPII-transcribed loci. p53 can direct preinitiation complex (PIC) assembly at certain target gene promoters under basal conditions, and at others only in response to stress. This process involves the ordered recruitment of histone methyltransferases (HMTs), histone acetyltransferase (HATs), and other coregulators in the vicinity of the p53 response element (RE) to open up chromatin so that RNAPII and its associated general transcription factors (GTFs) can bind to the transcription start site of the locus. In response to specific stimuli, p53 can also modulate transcription elongation via functional and physical interactions with various elongation factors.
The relationship of p53 with its most well-studied HATs, p300 and CBP, is fairly complex (Grossman 2001). As mentioned above, not only do p300/CBP acetylate histones, they also acetylate p53 itself, correlating with an increase in target transactivation (Avantaggiati et al. 1997; Gu and Roeder 1997; Scolnick et al. 1997). On DNA damage, p53 becomes phosphorylated in its amino-terminal region, and it has been shown that these damage-inducible modifications enhance p300/CBP-mediated acetylation at the CTD (Lambert et al. 1998; Sakaguchi et al. 1998). Furthermore, Mdm2, the main negative regulator of p53, can disrupt the interaction of p300 and p53 by competing with p300 for binding to the NTD of p53 (Grossman et al. 1998; Wadgaonkar and Collins 1999). Finally, to convolute matters even further, when complexed with Mdm2, p300/CBP can also serve as an E4 ubiquitin ligase for p53 (Grossman et al. 2003), thus targeting it for degradation. Taken together, these results demonstrate that p300 can paradoxically exert both positive and negative control over p53 function, perhaps implying that p300/CBP can switch the balance from p53 degradation to stabilization after DNA damage.
Once chromatin has been modified and remodeled (Li et al. 2007b), components of the preinititation complex (PIC) can be recruited or somehow altered to allow transcription initiation. TFIID is recruited to the promoter’s TATA region to nucleate the formation of the PIC, followed by TFIIB, and finally by the assembly of the other transcription initiation factors (TFIIF, TFIIE, TFIIH) complexed with unphosphorylated RNAPII (Orphanides et al. 1996; Woychik and Hampsey 2002). Adaptor complexes such as SAGA and Mediator aid in the recruitment of these general initation factors to the locus (Thomas and Chiang 2006). p53 itself is able to assist in the recruitment of various PIC components to the promoter, including TBP and its associated factors (Seto et al. 1992; Chen et al. 1993; Liu et al. 1993; Thut et al. 1995; Farmer et al. 1996), as well as TFIIA and TFIIH (Ko and Prives 1996; Xing et al. 2001). At least one TFIID subunit, TAF1, is recruited in an acetylated-p53-dependent fashion to the p53 RE on p21 before looping to the core promoter (Li et al. 2007a).
It is well-established that the kinetics of p53 binding after stress vary by promoter, cell type, and the nature of the stimulus. However, for many genes, such as p21, it is clear that the levels of p53 bound are not the sole determinant of the ensuing transcriptional response (Espinosa et al. 2003; Donner et al. 2007; Mattia et al. 2007; Hill et al. 2008b). In fact, it appears that similar levels of bound p53 can direct a differential PIC assembly that is both locus- and stimulus-specific (Espinosa 2008), as depicted in Figure 3. How is this accomplished? For one thing, low levels of RNAPII are poised at certain p53 targets and not others, lowering the barrier of activation for these genes (Espinosa et al. 2003). Furthermore, select basal components of the transcription initiation machinery are recruited to certain p53 target genes such as p21 regardless of the nature of the stimulus, whereas the assembly of other factors is strongly dependent on the nature of the stress. For example, both TFIIB and cdk8 are recruited to p21 after DNA damage or nutlin treatment, but they are absent after treatment with more mild forms of stress such as hydroxyurea or UVC (Espinosa et al. 2003; Donner et al. 2007; Beckerman et al. 2009); the recruitment of these two factors correlates well with transactivation of p21 RNA. In contrast, other cofactors such as TBP or MED1 show no such differential behavior under similar conditions (Donner et al. 2007; Mattia et al. 2007). These different stresses likely lead to the activation of different signaling cascades, thus altering the availability of key stimulus-specific coregulators—such as components of Mediator, GTFs, etc.—that are necessary for the transactivation of a certain locus. It will be of great interest to subject a large number of p53 target loci to this type of analysis, to determine whether any unifying pattern emerges between the nature of the p53 target and the stimulus-specific regulators it requires for transactivation.
Figure 3.
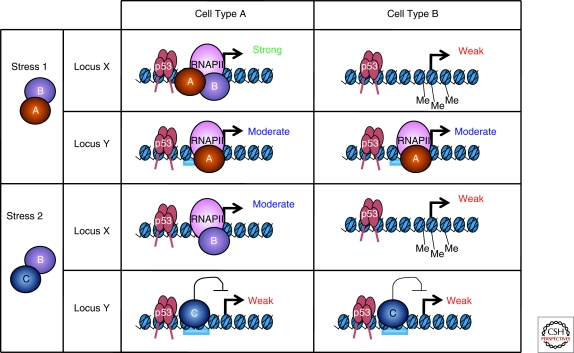
p53 target genes are regulated in a stimulus-, locus-, and context-specific manner. In cell types A and B, both locus X and Y exhibit similar amounts of bound p53, however, their transcriptional output differs. Stress 1 is able to induce a cellular environment where coactivators A and B are available. At locus X in cell type A, this produces a strong transcriptional response because both coactivators can bind; however, in cell type B, locus X is methylated and thus is not transcribed. Locus Y in cell types A and B cannot recruit recruit coactivator B, resulting in only a moderate transcriptional output. Stress 2 induces a transcriptional environment where coactivator B and corepressor C are available. Locus X does not contain a binding site for corepressor C, and therefore the transcriptional ouput is moderate. Locus Y, on the other hand, does contain a binding site for corepressor C, whose binding prevents the strong transactivation of gene Y in both cell types, despite high levels of bound p53.
It should be noted that other cis-regulatory elements within the locus can influence the transactivation potential of the target gene as well. For example, Myc is able to bind and repress the p21 locus through concerted action with its cofactor Miz. Importantly, the recruitment of Myc does not affect p53 binding to p21 (Seoane et al. 2002), nor does it affect the transactivation of a number of other p53 targets. Moreover, distinct epigenetic backgrounds can further define the response to p53 activation. For example, after treatment with Nutlin-3, a small molecule that antagonizes the p53:Mdm2 interaction, and greatly stabilizes p53 despite the absence of amino-terminal phosphorylation (Vassilev et al. 2004), comparable amounts of p53 are bound to the 14-3-3 p53 RE in both HCT116 and BV173 cells. However, RNAPII is only recruited to the 14-3-3 transcription start site in HCT116 cells. This is because of the fact that the 14-3-3 promoter is highly methylated in BV173 cells (Paris et al. 2008). Similarly, hCAS/Cse1L binds independently of p53 to various promoters, such as pig3, p53R2, and p53AIP1, and cooperates with p53 at these loci to reduce histone methylation of histone H3 at K27 (Tanaka et al. 2007). Thus, the very architecture of chromatin encompassing p53 target genes plays an important role in defining the transactivation potential of a locus.
Once bound to its cognate element, various regions of the p53 protein contribute in different ways to its transactivation ability. A p53 protein mutated at L22/W23 within TAD-I is largely inert, but surprisingly can still transactivate select proapoptotic genes (Johnson et al. 2005; Baptiste-Okoh et al. 2008), indicating that other regions such TAD-II and/or the proline-rich domain can independently function in transcriptional activation. On the other hand, a specific double mutation within TAD-II at W53/F54 selectively alters the transactivation of certain proapoptotic targets in yeast (Candau et al. 1997) and human cells (Zhu et al. 2000). TAD-I, TAD-II, and the CTD of p53 act in concert to recruit the STAGA complex to transactivate various target promoters (Gamper and Roeder 2008). In turn, the proline-rich domain of p53 is responsible for interactions with the corepressor mSin3A (Zilfou et al. 2001), as well as the p300/CBP histone acetyltransferase (Dornan et al. 2003) and the prolyl isomerase Pin1 (Zacchi et al. 2002; Zheng et al. 2002).
Transcription Elongation
Although the initiation phase of transcription was originally thought to be the stage at which most regulatory events occur, more recent findings show transcription elongation can also be an important point of control (Sims et al. 2004). Similarly, it is now becoming clear that p53-mediated transcriptional regulation is not confined to the initiation phase. After RNAPII clears the promoter, its processivity is facilitated by several elongation factors that function to repress the stalling and pausing of the polymerase. P53 interacts physically or functionally with several of these factors, including cdk9 (Claudio et al. 2006; Radhakrishnan and Gartel 2006), FACT (Keller et al. 2001), various components of the mediator complex (Gu et al. 1999; Zhang et al. 2005), and ELL (Shinobu et al. 1999). One study in budding yeast even establishes p53 as a bona fide elongation factor that tracks with RNAPII through gene coding regions (Balakrishnan and Gross 2008), although this function of p53 has not yet been shown in a mammalian system.
It is not surprising, then, that several recent studies have documented a role for p53 and its upstream signaling pathways in the stimulus- and locus-specific control of transcription elongation (Gomes et al. 2006; Donner et al. 2007; Mattia et al. 2007; Hill et al. 2008a; Beckerman et al. 2009). In the case of chromium exposure, the elongation of p21 but not puma RNA is inhibited, in a process that implicates DNA-PK (Hill et al. 2008b), whereas after replication stress, p21 elongation is inhibited in a Chk1-dependent fashion (Beckerman et al. 2009). Further work is required to extend these observations to other p53 targets, as the regulation of transcription elongation is emerging as a key layer of control in the fine-tuning of the p53 response.
p53-Mediated Repression
In addition to its well-documented role in transcription activation, p53 has also been shown to associate with and repress a wide range of targets. Although p53-mediated repression is still poorly understood, several mechanisms for the process have emerged.
(1) p53 can bind directly to its response element and recruit corepressors. One such example involves the recruitment of HDAC1 to specific promoters (such as map4, p21, stathmin, HSP90-beta, myc, or nanog) via a p53-dependent interaction with mSIN3A (Murphy et al. 1999; Zhang et al. 2004; Ho et al. 2005; Lin et al. 2005) and, in the case of p21 repression, also Zbtb4 (Weber et al. 2008). In this scenario, what prompts the bound p53 to promote repression of the target, rather than its activation? In some cases the answer may lie in the orientation of the p53 binding site (in which a “head to tail,” rather than the more common “head to head” orientation correlates with the repression of the target) (Johnson et al. 2001; Godar et al. 2008). The length of the spacer between the two half-sites may also contribute to the decision to repress a gene rather than activate it (Hoffman et al. 2002).
(2) p53 can inhibit expression of some genes via the activation of a repressor protein. The most well-studied example of this is the p53-mediated transactivation of p21 (Lohr et al. 2003), which inhibits CDK-dependent phosphorylation of the retinoblastoma protein (Xiong et al. 1993; Niculescu et al. 1998) to keep E2F-regulated genes in an inactive state (Delavaine and La Thangue 1999; Harbour and Dean 2000). Another example of this type of repression is the p53-dependent transactivation of the Slug protein. After high doses of γ-irradiation Slug binds and represses the puma promoter, preventing apoptosis (Wu et al. 2005).
(3) p53 can bind to its response element and squelch the activities of another transcriptional activator; in other words, p53 binding can occlude the binding of this other, stronger, transcription factor. For example, in response to hypoxic stress, p53 binds the alpha-fetoprotein promoter (AFP) and displaces the transcriptional activator HNF3 to repress AFP expression (Lee et al. 1999). This type of mechanism has been documented on a number of other genes, including cdc25C (St Clair et al. 2004), HBV (Ori et al. 1998), and pold1 (Li and Lee 2001).
(4) p53 can repress genes that do not contain a p53 response element via protein-protein interactions. The cyclin B2 promoter, for example, contains NF-Y recognition sites but no p53 RE. Promoter-bound NF-Y interacts with p53, and in this case p53 recruits HDAC1 to repress the cyclin B2 promoter (Imbriano et al. 2005).
Interestingly, in addition to acting on RNAPII-directed genes, p53 can also direct the repression of RNAPI and RNAPIII promoters. In the case of RNAPI, p53 is able to bind to TAF(I)110, preventing its association with SL1 and UBF, therefore repressing transcription on rRNA promoters by decreasing the rate of PIC assembly (Zhai and Comai 2000). In the case of RNAPIII-based transcription, the NTD of p53 is able to bind and sequester TFIIIB, preventing the transcription of 5S RNA and tRNA (Chesnokov et al. 1996; Budde and Grummt 1999; Gridasova and Henry 2005; White 2005).
PROMOTER SELECTIVITY WITHIN THE p53 TRANSCRIPTIONAL PROGRAM
One of the most intriguing questions in the field of p53-mediated transcription is how the protein is able to discriminate between its myriad promoters in response to differing stimuli. Here we focus on three established mechanisms that can contribute to the diversity of the transcriptional response: the p53 REs themselves, p53 posttranslational modifications, and p53 binding partners.
p53 Response Elements
With such a wide variety of REs in the genome, it is not surprising that p53 has different affinity for different consensus sequences. In fact, recent work has shown that tetrameric p53 is capable of binding to half- and even three quarter-consensus sites, such as those comprising the PIDD and Apaf-1 response elements (Jordan et al. 2008). Using a Saccharomyces cerevisiae model system, one study found as much as a 1000-fold difference in transactivation by p53 for its weaker versus its stronger sites, with this difference being largely dependent on the central sequence element in the p53 RE (Inga et al. 2002). On the whole, cell cycle target genes generally contain more robust binding elements than do proapoptotic targets (Weinberg et al. 2005), perhaps because they are more evolutionarily conserved (Horvath et al. 2007). This differential affinity of p53 for its REs allows the nuclear concentration of the protein to be one determining factor in how target genes are activated, and hence also for which cellular outcome occurs after genotoxic stress (Szak et al. 2001; Lokshin et al. 2005). The proclivity of a p53 RE to bend may also play a large role in determining the affinity and stability of p53 with its cognate element (Batta and Kundu 2007; Pan and Nussinov 2008). Intriguingly, a genome-wide study has shown that not all sites bound by p53 are in fact transcriptionally activated (Wei et al. 2006a), raising the possibility that other coactivators and/or a specific modification status of p53 are needed to induce robust activation of select p53 target genes (discussed later).
Modifications
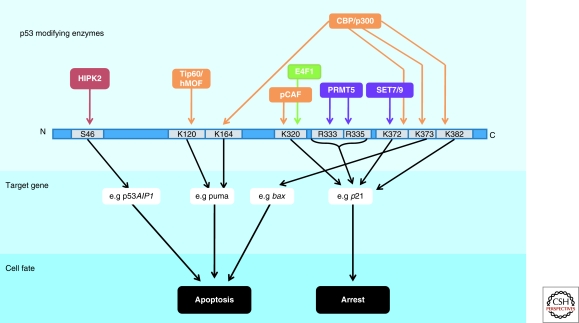
p53 is regulated spatially and temporally by many posttranslational modifications, primarily at the amino- and carboxy-termini, but also within the DBD. These modifications integrate the various cell signaling pathways that converge on the p53 protein (Sakaguchi et al. 1998; Bode and Dong 2004; Kruse and Gu 2009). Many modifications have been shown to predispose p53 to selectively activate or repress certain targets; however, the precise combinatorial impact of the numerous p53 modifications on transactivation is only weakly characterized. Here, only modifications with a direct, established impact on transcription are discussed (summarized in Fig. 4).
Figure 4.
Selective impact of p53 modifications on transactivation Several examples are depicted of residues within p53 that can be modified by acetylation (orange), phosphorylation (red), ubiquitylation (green), or methylation (purple). The preferential activation of the indicated target genes (and others, not depicted) can result in specific cell fates, such as apoptosis or cell cycle arrest.
One modification that has generated much attention is the phosphorylation of S46. After treating cells with UV, p53 can be phosphorylated at this residue to specifically induce its ability to transactivate a pro-apoptotic gene AIP-1 (Oda et al. 2000). HIPK2 is the main kinase implicated in the modification of this residue (D’Orazi et al. 2002), although AMPK (Okoshi et al. 2008), PKC-δ (Yoshida et al. 2006), p38 (Perfettini et al. 2005), and DYRK-2 (Taira et al. 2007) can also phosphorylate this site. Phosphorylation of S46 is the earliest and perhaps the most clear-cut example of a modification promoting promoter selectivity by p53.
Furthermore, two groups have shown that p53 can be acetylated in the DNA-binding domain in response to genotoxic stress at either K120 or K164 (Sykes et al. 2006; Tang et al. 2006; Tang et al. 2008). The K120 modification is catalyzed by hMOF or TIP60, and this acetylation is required for the p53-mediated transactivation of the proapoptotic targets puma and bax, at a post-binding step. The K164 residue is modified by p300/CBP and, in concert with the acetylation of the extreme CTD, is indispensible for the activation of most p53 target genes (including p21, pig3, puma, and bax) with the remarkable exception of mdm2. It is possible that the unique, nucleosome-free architecture of the mdm2 p53 RE contributes to this distinctive phenomenon. To that end, it would be informative to assess the transactivation capability of the p53-8KR mutant (which abrogates acetylation on K164, K120, and six lysines in the extreme CTD) on other p53 target genes with a similar nucleosome-free status, such as pcna.
Various investigators have attempted to determine the discrete effects of the acetylation of individual CTD lysine residues on transcription. Within the CTD, lysine residues 305, 370, 372, 373, 381, and 382 can each be acetylated by p300/CBP whereas K320 is acetylated by pCAF. The Avantagiatti group describes a model in which distinct p53 CTD acetylation cassettes can differentially influence gene-expression patterns and cell fate (Knights et al. 2006). By interacting with different coactivators and repressors, the acetylation-mimicking mutant p53-K320Q promotes cell cycle arrest by predominantly transactivating high-affinity p53 binding sites such as p21; in contrast, p53-K373Q promotes apoptosis by favoring interaction with lower-affinity binding sites that included such promoters as bax and p53AIP1. Similarly, others report an increased association of K320-acetylated p53 with p21 after etoposide treatment compared with K382-acetylated p53 (Xenaki et al. 2008). These data agree with the K317R knock-in mice (the equivalent of human K320) that show reduced transactivation of various proapoptotic, but not cell cycle, p53 targets (Chao et al. 2006). In line with this work, treatment of LNCaP cells, which contain constitutively K320-acetylated p53 (Roy et al. 2005), with two different HDAC inhibitors, CG-1521 and TSA, produces differentially acetylated p53 at K373 and K382 respectively. These distinctly modified versions of p53 promote differential association with various transcriptional cofactors leading to strong transcription from p21 and other targets in the case of p53-K373Ac but not in the case of p53-K382Ac (Roy and Tenniswood 2007). As a result, cells containing p53 modified at K373 are able to promote apoptosis, but cells containing p53 modified at K382 can only cause arrest (Roy et al. 2005).
To further complicate matters, the CTD lysines of p53 can also be methylated. When p53 is monomethylated at K382 by Set8, strong targets such as p21 and puma are inhibited, whereas the activation of other weaker targets remains unaffected (Shi et al. 2007). On the other hand, Set7- and Set9-mediated methylation of Lys372 increases transcription from the p21 locus (Chuikov et al. 2004), and is also an important step in the acetylation of p53 CTD lysine residues and subsequent stabilization of the protein (Ivanov et al. 2007). Adding another layer of complexity, p53 methylation at K372 is refractory to Smyd2-mediated monomethylation at K370, which itself causes inhibition of p53 activity (Huang et al. 2006). Dimethylation at K370, however, increases p53 activity by promoting interaction with its coactivator 53BP1. LSD1, a lysine demethylase, can reverse K370 dimethylation, inhibiting this interaction (Huang et al. 2007). Furthermore, arginine methylation of the p53 tetramerization domain at R333, R335 and R337 has been implicated in transcriptional control: depletion of the PRMT5 methyltransferase results in reduced transcription of p21, but not puma or noxa, resulting in increased apoptosis (Jansson et al. 2008).
Even ubiquitylation can affect p53’s ability to regulate transcription. Unlike Mdm2-mediated monoubiquitylation of p53, which promotes export of p53 from the nucleus, the effect of E4F1-mediated monoubiquitylation at K320 of p53 is transcriptional. K320 ubiquitylation predisposes the p53 protein to transactivate cell cycle targets such as p21 and cyclinG1 while having no impact on noxa transcription, prompting the cell to undergo arrest rather than apoptosis (Le Cam et al. 2006).
Clearly, great complexity exists in the world of p53 posttranslational modifications, convoluted by the fact that many of the modifications appear to be stress- and cell-type specific. However, these studies (and others) put forth the exciting idea of a p53 modification code, analogous to the histone modification code, whereby diverse modifications of p53 at specific residues can direct it toward activating or repressing select target genes.
p53 Binding Partners
Many cofactors have been described that selectively alter p53’s transcriptional program (summarized in Fig. 5). These binding partners can function either by altering the ability of p53 to recognize a specific subset of REs, or by influencing the ability of p53 to recruit transcriptional coactivators at certain loci. In particular cases, an interplay exists between the modification status of p53 modification and its ability to selectively interact with a binding partner.
One of the more clear-cut examples of cofactor-induced promoter selectivity involves the three members of the ASPP family of proteins, ASPP1, ASPP2 and iASPP. ASPP1 and ASPP2 bind the p53 core domain and predispose the cell toward an apoptotic phenotype, by causing transactivation of bax but not p21 (Samuels-Lev et al. 2001). On the other hand, iASPP binds to p53 and inhibits the transactivation of proapoptotic genes (Bergamaschi et al. 2006). Highlighting the interplay between p53 posttranslational modifications and selective cofactor binding, p53 phosphorylated at S46 is recognized by the prolyl-isomerase Pin1, and binding of Pin1 to thus modified p53 leads to its dissociation from iASPP and the consequent induction of apoptosis (Mantovani et al. 2007). Moreover, a well-studied p53 polymorphism, P72, shows increased association with the iASPP protein when compared with the R72 version, perhaps providing insight as to why R72 is more pro-apoptotic than P72 (Bergamaschi et al. 2006).
Such symmetrical phenotypes are not unique to the ASPP family of proteins. An isoform of p53, p53-β, which can form heterotetramers with wild-type p53, promotes binding to the bax but not the p21 RE (Bourdon et al. 2005); conversely, phosphorylation of p53 by c-abl stabilizes the homotetramer, and promotes binding to p21 rather than bax (Wei et al. 2005). Brn3a and Brn3b have similarly opposing phenotypes: Brn3a influences p53 to transactivate bax and not p21, whereas Brn3b does the reverse (Budhram-Mahadeo et al. 2006).
Certain proteins are able to bind to and predispose p53 exclusively toward an apoptotic program. For instance, YB1, a Y-box binding protein, interacts with p53 to block the transactivation of bax, but not p21 (Homer et al. 2005). Similarly, the p52 subunit of NF-κB has been shown to inhibit p21 activation, but can also act in concert with p53 to increase gadd45, puma, and DR5/Killer expression (Schumm et al. 2006). Furthermore, the p38-regulated p18/hamlet protein binds to p53 and shows increased transactivation of target promoters such as noxa, but not bax, p21, or puma, resulting in an increased apoptotic response (Cuadrado et al. 2007; Lafarga et al. 2007).
On the flip side, other p53 binding partners selectively promote cell cycle arrest. BRCA1 is one such case. By interacting with the p53 CTD (Zhang et al. 1998), BRCA1 selectively redirects p53 to activate its cell cycle arrest and DNA repair genes (MacLachlan et al. 2002), perhaps by recruiting acetyltransferases such as CBP/p300 to the locus (Pao et al. 2000). Notably, BRCA1 has also been implicated in stabilizing S15-phosphorylated p53 and the subsequent maintenance of G1 arrest (Fabbro et al. 2004). Similarly, the membrane glycoprotein Muc1 is able to bind to and activate p21 in a p53-dependent fashion while at the same time repressing Bax in a p53-independent manner (Wei et al. 2006b). An analogous function is ascribed to Hzf, a zinc-finger protein itself induced by p53, which promotes p53 binding and transactivation of p21 and 14-3-3, but not bax (Das et al. 2007). APAK, a KRAB-type zinc finger protein, binds p53 under basal conditions and prevents the transactivation of proapoptotic targets (Tian et al. 2009).
Several p53 cofactors, although indeed displaying the ability to discriminate between promoters, nonetheless show no clear predisposition toward either pro- or antiapoptotic targets. For instance, it should be noted that endogenously expressed Mdm2 can be found at promoters such as p21 (Minsky and Oren 2004; Arva et al. 2005; Ohkubo et al. 2006; White et al. 2006; Tang et al. 2008), negatively influencing p53’s transcriptional output. The mechanism by which this occurs is unclear, but perhaps Mdm2 functions through recruitment of the histone deacetylases KAP1 (Wang et al. 2005) and HDAC1 (Ito et al. 2002), the disruption of HAT interactions with p53 (Ito et al. 2002; Jin et al. 2004), or the monoubiquitylation of histone H2B within the vicinity of the p53 RE (Minsky and Oren 2004). Notably, a role for a Mdm2/Rb/p53 trimeric complex has been described in transrepressing pro-apoptotic targets (Hsieh et al. 1999). Interestingly, when both Mdm2 and MdmX are overexpressed, they can bind many p53 target promoters with the notable exception of Mdm2 gene itself. Although the precise influence of MdmX promoter binding on p53 transcriptional output is unclear (Tang et al. 2008), it remains to be seen whether the disruption of the p53/Mdm2 interaction at the promoter is required for the antirepression of certain target genes.
Although it is tempting to invoke cofactors to explain p53’s decision to effectuate cell cycle arrest or apoptosis, we must keep in mind that for most binding partners, simply too few targets have been looked at to draw any firm conclusions to this end. It is likely that as the influence of these p53-interacting proteins is assessed on more targets, their ability to strictly discriminate between pro- and antiapoptotic targets genes may have to be refined.
CONCLUDING REMARKS
Even more than 20 years on, our understanding of p53-mediated transcription continues to grow by leaps and bounds. It is now clear the p53 can both activate and repress its target genes; that p53’s regulatory influence extends not just to transcription initiation, but also to elongation; and that several features—including but not limited to the epigenetic landscape of the locus, p53 posttranslational modifications, the nature of the p53 RE, and p53-interacting partners – function in concert to determine the specificity of the p53 transcriptional response. Although it will take a massive effort to integrate the individual effects of each of these factors into a coherent model for transcriptional regulation by p53, great insight into cancer therapy will undoubtedly be gained from doing so.
ACKNOWLEDGMENTS
This review was written under the auspices of CA77742 and CA87497; RB is supported by a Canadian postgraduate scholarship from NSERC.
Footnotes
Editors: Arnold J. Levine and David P. Lane
Additional Perspectives on The p53 Family available at www.cshperspectives.org
REFERENCES
- An W, Kim J, Roeder RG 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117: 735–748 [DOI] [PubMed] [Google Scholar]
- Arva NC, Gopen TR, Talbott KE, Campbell LE, Chicas A, White DE, Bond GL, Levine AJ, Bargonetti J 2005. A chromatin-associated and transcriptionally inactive p53-Mdm2 complex occurs in mdm2 SNP309 homozygous cells. J Biol Chem 280: 26776–26787 [DOI] [PubMed] [Google Scholar]
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89: 1175–1184 [DOI] [PubMed] [Google Scholar]
- Balagurumoorthy P, Sakamoto H, Lewis MS, Zambrano N, Clore GM, Gronenborn AM, Appella E, Harrington RE 1995. Four p53 DNA-binding domain peptides bind natural p53-response elements and bend the DNA. Proc Natl Acad Sci 92: 8591–8595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan SK, Gross DS 2008. The tumor suppressor p53 associates with gene coding regions and co-traverses with elongating RNA polymerase II in an in vivo model. Oncogene 27: 2661–2672 [DOI] [PubMed] [Google Scholar]
- Baptiste-Okoh N, Barsotti AM, Prives C 2008. A role for caspase 2 and PIDD in the process of p53-mediated apoptosis. Proc Natl Acad Sci 105: 1937–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell 8: 1243–1254 [DOI] [PubMed] [Google Scholar]
- Batta K, Kundu TK 2007. Activation of p53 function by human transcriptional coactivator PC4: Role of protein-protein interaction, DNA bending, and posttranslational modifications. Mol Cell Biol 27: 7603–7614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerman R, Donner AJ, Mattia M, Peart MJ, Manley JL, Espinosa JM, Prives C 2009. A role for Chk1 in blocking transcriptional elongation of p21 RNA during the S-phase checkpoint. Genes Develop 23: 1364–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi D, Samuels Y, Sullivan A, Zvelebil M, Breyssens H, Bisso A, Del Sal G, Syed N, Smith P, Gasco M, et al. 2006. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat Gen 38: 1133–1141 [DOI] [PubMed] [Google Scholar]
- Bertrand P, Saintigny Y, Lopez BS 2004. p53’s double life: Transactivation-independent repression of homologous recombination. Trends Genet 20: 235–243 [DOI] [PubMed] [Google Scholar]
- Bode AM, Dong Z 2004. Posttranslational modification of p53 in tumorigenesis. Nature reviews 4: 793–805 [DOI] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP 2005. p53 isoforms can regulate p53 transcriptional activity. Genes Develop 19: 2122–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde A, Grummt I 1999. p53 represses ribosomal gene transcription. Oncogene 18: 1119–1124 [DOI] [PubMed] [Google Scholar]
- Budhram-Mahadeo VS, Bowen S, Lee S, Perez-Sanchez C, Ensor E, Morris PJ, Latchman DS 2006. Brn-3b enhances the pro-apoptotic effects of p53 but not its induction of cell cycle arrest by cooperating in trans-activation of bax expression. Nucleic Acids Res 34: 6640–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candau R, Scolnick DM, Darpino P, Ying CY, Halazonetis TD, Berger SL 1997. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene 15: 807–816 [DOI] [PubMed] [Google Scholar]
- Chao C, Wu Z, Mazur SJ, Borges H, Rossi M, Lin T, Wang JY, Anderson CW, Appella E, Xu Y 2006. Acetylation of mouse p53 at lysine 317 negatively regulates p53 apoptotic activities after DNA damage. Mol Cell Biol 26: 6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Farmer G, Zhu H, Prywes R, Prives C 1993. Cooperative DNA binding of p53 with TFIID (TBP): A possible mechanism for transcriptional activation. Genes Develop 7: 1837–1849 [DOI] [PubMed] [Google Scholar]
- Chesnokov I, Chu WM, Botchan MR, Schmid CW 1996. p53 inhibits RNA polymerase III-directed transcription in a promoter-dependent manner. Mol Cell Biol 16: 7084–7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, et al. 2004. Regulation of p53 activity through lysine methylation. Nature 432: 353–360 [DOI] [PubMed] [Google Scholar]
- Claudio PP, Cui J, Ghafouri M, Mariano C, White MK, Safak M, Sheffield JB, Giordano A, Khalili K, Amini S, et al. 2006. Cdk9 phosphorylates p53 on serine 392 independently of CKII. J Cell Physiol 208: 602–612 [DOI] [PubMed] [Google Scholar]
- Contente A, Dittmer A, Koch MC, Roth J, Dobbelstein M 2002. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat Gen 30: 315–320 [DOI] [PubMed] [Google Scholar]
- Cuadrado A, Lafarga V, Cheung PC, Dolado I, Llanos S, Cohen P, Nebreda AR 2007. A new p38 MAP kinase-regulated transcriptional coactivator that stimulates p53-dependent apoptosis. EMBO J 26: 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G, et al. 2002. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol 4: 11–19 [DOI] [PubMed] [Google Scholar]
- Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, Lee SW 2007. Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell 130: 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delavaine L, La Thangue NB 1999. Control of E2F activity by p21Waf1/Cip1. Oncogene 18: 5381–5392 [DOI] [PubMed] [Google Scholar]
- Donner AJ, Szostek S, Hoover JM, Espinosa JM 2007. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell 27: 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornan D, Shimizu H, Burch L, Smith AJ, Hupp TR 2003. The proline repeat domain of p53 binds directly to the transcriptional coactivator p300 and allosterically controls DNA-dependent acetylation of p53. Mol Cell Biol 23: 8846–8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B 1992. Definition of a consensus binding site for p53. Nat Gen 1: 45–49 [DOI] [PubMed] [Google Scholar]
- Espinosa JM 2008. Mechanisms of regulatory diversity within the p53 transcriptional network. Oncogene 27: 4013–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JM, Emerson BM 2001. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol Cell 8: 57–69 [DOI] [PubMed] [Google Scholar]
- Espinosa JM, Verdun RE, Emerson BM 2003. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell 12: 1015–1027 [DOI] [PubMed] [Google Scholar]
- Ewen ME, Miller SJ 1996. p53 and translational control. Biochim Biophys Acta 1242: 181–184 [DOI] [PubMed] [Google Scholar]
- Fabbro M, Savage K, Hobson K, Deans AJ, Powell SN, McArthur GA, Khanna KK 2004. BRCA1-BARD1 complexes are required for p53Ser-15 phosphorylation and a G1/S arrest following ionizing radiation-induced DNA damage. J Biol Chem 279: 31251–31258 [DOI] [PubMed] [Google Scholar]
- Farmer G, Colgan J, Nakatani Y, Manley JL, Prives C 1996. Functional interaction between p53, the TATA-binding protein (TBP), and TBP-associated factors in vivo. Mol Cell Biol 16: 4295–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk WD, Pak DT, Karas RH, Wright WE, Shay JW 1992. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol 12: 2866–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper AM, Roeder RG 2008. Multivalent binding of p53 to the STAGA complex mediates coactivator recruitment after UV damage. Mol Cell Biol 28: 2517–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L 2007. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Develop 21: 1869–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godar S, Ince TA, Bell GW, Feldser D, Donaher JL, Bergh J, Liu A, Miu K, Watnick RS, Reinhardt F, et al. 2008. Growth-inhibitory and tumor- suppressive functions of p53 depend on its repression of CD44 expression. Cell 134: 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohler T, Reimann M, Cherny D, Walter K, Warnecke G, Kim E, Deppert W 2002. Specific interaction of p53 with target binding sites is determined by DNA conformation and is regulated by the C-terminal domain. J Biol Chem 277: 41192–41203 [DOI] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Develop 20: 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gridasova AA, Henry RW 2005. The p53 tumor suppressor protein represses human snRNA gene transcription by RNA polymerases II and III independently of sequence-specific DNA binding. Mol Cell Biol 25: 3247–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR 2001. p300/CBP/p53 interaction and regulation of the p53 response. Eur J Biochem FEBS 268: 2773–2778 [DOI] [PubMed] [Google Scholar]
- Grossman SR, Deato ME, Brignone C, Chan HM, Kung AL, Tagami H, Nakatani Y, Livingston DM 2003. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 300: 342–344 [DOI] [PubMed] [Google Scholar]
- Grossman SR, Perez M, Kung AL, Joseph M, Mansur C, Xiao ZX, Kumar S, Howley PM, Livingston DM 1998. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell 2: 405–415 [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90: 595–606 [DOI] [PubMed] [Google Scholar]
- Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG 1999. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell 3: 97–108 [DOI] [PubMed] [Google Scholar]
- Hainaut P, Milner J 1993. A structural role for metal ions in the “wild-type” conformation of the tumor suppressor protein p53. Cancer Res 53: 1739–1742 [PubMed] [Google Scholar]
- Halazonetis TD, Kandil AN 1993. Conformational shifts propagate from the oligomerization domain of p53 to its tetrameric DNA binding domain and restore DNA binding to select p53 mutants. EMBO J 12: 5057–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Dean DC 2000. The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Develop 14: 2393–2409 [DOI] [PubMed] [Google Scholar]
- Hill R, Leidal AM, Madureira PA, Gillis LD, Cochrane HK, Waisman DM, Chiu A, Lee PW 2008a. Hypersensitivity to chromium-induced DNA damage correlates with constitutive deregulation of upstream p53 kinases in p21-/-HCT116 colon cancer cells. DNA Repair 7: 239–252 [DOI] [PubMed] [Google Scholar]
- Hill R, Leidal AM, Madureira PA, Gillis LD, Waisman DM, Chiu A, Lee PW 2008b. Chromium-mediated apoptosis: Involvement of DNA-dependent protein kinase (DNA-PK) and differential induction of p53 target genes. DNA Repair 7: 1484–1499 [DOI] [PubMed] [Google Scholar]
- Ho J, Benchimol S 2003. Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differentiation 10: 404–408 [DOI] [PubMed] [Google Scholar]
- Ho JS, Ma W, Mao DY, Benchimol S 2005. p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol Cell Biol 25: 7423–7431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M 2002. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem 277: 3247–3257 [DOI] [PubMed] [Google Scholar]
- Homer C, Knight DA, Hananeia L, Sheard P, Risk J, Lasham A, Royds JA, Braithwaite AW 2005. Y-box factor YB1 controls p53 apoptotic function. Oncogene 24: 8314–8325 [DOI] [PubMed] [Google Scholar]
- Horvath MM, Wang X, Resnick MA, Bell DA 2007. Divergent evolution of human p53 binding sites: Cell cycle versus apoptosis. PLoS Genet 3: e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JK, Chan FS, O’Connor DJ, Mittnacht S, Zhong S, Lu X 1999. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol Cell 3: 181–193 [DOI] [PubMed] [Google Scholar]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL 2006. Repression of p53 activity by Smyd2-mediated methylation. Nature 444: 629–632 [DOI] [PubMed] [Google Scholar]
- Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, et al. 2007. p53 is regulated by the lysine demethylase LSD1. Nature 449: 105–108 [DOI] [PubMed] [Google Scholar]
- Hublitz P, Kunowska N, Mayer UP, Muller JM, Heyne K, Yin N, Fritzsche C, Poli C, Miguet L, Schupp IW, et al. 2005. NIR is a novel INHAT repressor that modulates the transcriptional activity of p53. Genes Develop 19: 2912–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupp TR, Lane DP 1994. Allosteric activation of latent p53 tetramers. Curr Biol 4: 865–875 [DOI] [PubMed] [Google Scholar]
- Imbriano C, Gurtner A, Cocchiarella F, Di Agostino S, Basile V, Gostissa M, Dobbelstein M, Del Sal G, Piaggio G, Mantovani R 2005. Direct p53 transcriptional repression: In vivo analysis of CCAAT-containing G2/M promoters. Mol Cell Biol 25: 3737–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inga A, Storici F, Darden TA, Resnick MA 2002. Differential transactivation by the p53 transcription factor is highly dependent on p53 level and promoter target sequence. Mol Cell Biol 22: 8612–8625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP 2002. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J 21: 6236–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov GS, Ivanova T, Kurash J, Ivanov A, Chuikov S, Gizatullin F, Herrera-Medina EM, Rauscher F 3rd, Reinberg D, Barlev NA 2007. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol 27: 6756–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, La Thangue NB 2008. Arginine methylation regulates the p53 response. Nat Cell Biol 10: 1431–1439 [DOI] [PubMed] [Google Scholar]
- Jayaraman L, Murthy KG, Zhu C, Curran T, Xanthoudakis S, Prives C 1997. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Develop 11: 558–570 [DOI] [PubMed] [Google Scholar]
- Jin Y, Zeng SX, Lee H, Lu H 2004. MDM2 mediates p300/CREB-binding protein-associated factor ubiquitination and degradation. J Biol Chem 279: 20035–20043 [DOI] [PubMed] [Google Scholar]
- Johnson RA, Ince TA, Scotto KW 2001. Transcriptional repression by p53 through direct binding to a novel DNA element. J Biol Chem 276: 27716–27720 [DOI] [PubMed] [Google Scholar]
- Johnson TM, Hammond EM, Giaccia A, Attardi LD 2005. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat Gen 37: 145–152 [DOI] [PubMed] [Google Scholar]
- Jordan JJ, Menendez D, Inga A, Noureddine M, Bell DA, Resnick MA 2008. Noncanonical DNA motifs as transactivation targets by wild type and mutant p53. PLoS Genet 4: e1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller DM, Zeng X, Wang Y, Zhang QH, Kapoor M, Shu H, Goodman R, Lozano G, Zhao Y, Lu H 2001. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol Cell 7: 283–292 [DOI] [PubMed] [Google Scholar]
- Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, Shakked Z 2006. Structural basis of DNA recognition by p53 tetramers. Mol Cell 22: 741–753 [DOI] [PubMed] [Google Scholar]
- Knights CD, Catania J, Giovanni SD, Muratoglu S, Perez R, Swartzbeck A, Quong AA, Zhang X, Beerman T, Pestell RG, et al. 2006. Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J Cell Biol 173: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko LJ, Prives C 1996. p53: Puzzle and paradigm. Genes Develop 10: 1054–1072 [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W 2009. Modes of p53 regulation. Cell 137: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarga V, Cuadrado A, Nebreda AR 2007. p18(Hamlet) mediates different p53-dependent responses to DNA-damage inducing agents. Cell Cycle 6: 2319–2322 [DOI] [PubMed] [Google Scholar]
- Lambert PF, Kashanchi F, Radonovich MF, Shiekhattar R, Brady JN 1998. Phosphorylation of p53 serine 15 increases interaction with CBP. J Biol Chem 273: 33048–33053 [DOI] [PubMed] [Google Scholar]
- Laptenko O, Prives C 2006. Transcriptional regulation by p53: One protein, many possibilities. Cell Death Differ 13: 951–961 [DOI] [PubMed] [Google Scholar]
- Le Cam L, Linares LK, Paul C, Julien E, Lacroix M, Hatchi E, Triboulet R, Bossis G, Shmueli A, Rodriguez MS, et al. 2006. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell 127: 775–788 [DOI] [PubMed] [Google Scholar]
- Lee KC, Crowe AJ, Barton MC 1999. p53-mediated repression of alpha-fetoprotein gene expression by specific DNA binding. Mol Cell Biol 19: 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Lee MY 2001. Transcriptional regulation of the human DNA polymerase delta catalytic subunit gene POLD1 by p53 tumor suppressor and Sp1. J Biol Chem 276: 29729–29739 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007b. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li AG, Piluso LG, Cai X, Gadd BJ, Ladurner AG, Liu X 2007a. An acetylation switch in p53 mediates holo-TFIID recruitment. Mol Cell 28: 408–421 [DOI] [PubMed] [Google Scholar]
- Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM 1997. Binding and modulation of p53 by p300/CBP coactivators. Nature 387: 823–827 [DOI] [PubMed] [Google Scholar]
- Liu Y, Kulesz-Martin MF 2006. Sliding into home: Facilitated p53 search for targets by the basic DNA binding domain. Cell Death and Differentiation 13: 881–884 [DOI] [PubMed] [Google Scholar]
- Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y 2005. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol 7: 165–171 [DOI] [PubMed] [Google Scholar]
- Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol 19: 1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Miller CW, Koeffler PH, Berk AJ 1993. The p53 activation domain binds the TATA box-binding polypeptide in Holo-TFIID, and a neighboring p53 domain inhibits transcription. Mol Cell Biol 13: 3291–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr K, Moritz C, Contente A, Dobbelstein M 2003. p21/CDKN1A mediates negative regulation of transcription by p53. J Biol Chem 278: 32507–32516 [DOI] [PubMed] [Google Scholar]
- Lokshin M, Tanaka T, Prives C 2005. Transcriptional regulation by p53 and p73. Cold Spring Harb Symp Quant Biol 70: 121–128 [DOI] [PubMed] [Google Scholar]
- Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W 2004. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci 101: 2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLachlan TK, Takimoto R, El-Deiry WS 2002. BRCA1 directs a selective p53-dependent transcriptional response towards growth arrest and DNA repair targets. Mol Cell Biol 22: 4280–4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani F, Tocco F, Girardini J, Smith P, Gasco M, Lu X, Crook T, Del Sal G 2007. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat Struct Mol Biol 14: 912–920 [DOI] [PubMed] [Google Scholar]
- Mattia M, Gottifredi V, McKinney K, Prives C 2007. p53-Dependent p21 mRNA elongation is impaired when DNA replication is stalled. Mol Cell Biol 27: 1309–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur SJ, Sakaguchi K, Appella E, Wang XW, Harris CC, Bohr VA 1999. Preferential binding of tumor suppressor p53 to positively or negatively supercoiled DNA involves the C-terminal domain. J Mol Biol 292: 241–249 [DOI] [PubMed] [Google Scholar]
- McKinney K, Prives C 2002. Efficient specific DNA binding by p53 requires both its central and C-terminal domains as revealed by studies with high-mobility group 1 protein. Mol Cell Biol 22: 6797–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney K, Mattia M, Gottifredi V, Prives C 2004. p53 linear diffusion along DNA requires its C terminus. Mol Cell 16: 413–424 [DOI] [PubMed] [Google Scholar]
- McLure KG, Lee PW 1998. How p53 binds DNA as a tetramer. EMBO J 17: 3342–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minsky N, Oren M 2004. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell 16: 631–639 [DOI] [PubMed] [Google Scholar]
- Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, Wen SF, Wang L, Kirschmeier P, Bishop WR, et al. 2002. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene 21: 2613–2622 [DOI] [PubMed] [Google Scholar]
- Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ, George DL 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Develop 13: 2490–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Slee EA, Lu X 2008. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev 9: 702–712 [DOI] [PubMed] [Google Scholar]
- Nagaich AK, Appella E, Harrington RE 1997a. DNA bending is essential for the site-specific recognition of DNA response elements by the DNA binding domain of the tumor suppressor protein p53. J Biol Chem 272: 14842–14849 [DOI] [PubMed] [Google Scholar]
- Nagaich AK, Zhurkin VB, Durell SR, Jernigan RL, Appella E, Harrington RE 1999. p53-induced DNA bending and twisting: p53 tetramer binds on the outer side of a DNA loop and increases DNA twisting. Proc Natl Acad Sci 96: 1875–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaich AK, Zhurkin VB, Sakamoto H, Gorin AA, Clore GM, Gronenborn AM, Appella E, Harrington RE 1997b. Architectural accommodation in the complex of four p53 DNA binding domain peptides with the p21/waf1/cip1 DNA response element. J Biol Chem 272: 14830–14841 [DOI] [PubMed] [Google Scholar]
- Niculescu AB 3rd, Chen X, Smeets M, Hengst L, Prives C, Reed SI 1998. Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol Cell Biol 18: 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, et al. 2000. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102: 849–862 [DOI] [PubMed] [Google Scholar]
- Ohkubo S, Tanaka T, Taya Y, Kitazato K, Prives C 2006. Excess HDM2 impacts cell cycle and apoptosis and has a selective effect on p53-dependent transcription. J Biol Chem 281: 16943–16950 [DOI] [PubMed] [Google Scholar]
- Okoshi R, Ozaki T, Yamamoto H, Ando K, Koida N, Ono S, Koda T, Kamijo T, Nakagawara A, Kizaki H 2008. Activation of AMP-activated protein kinase induces p53-dependent apoptotic cell death in response to energetic stress. J Biol Chem 283: 3979–3987 [DOI] [PubMed] [Google Scholar]
- Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P 2002. The IARC TP53 database: New online mutation analysis and recommendations to users. Hum Mutat 19: 607–614 [DOI] [PubMed] [Google Scholar]
- Ori A, Zauberman A, Doitsh G, Paran N, Oren M, Shaul Y 1998. p53 binds and represses the HBV enhancer: An adjacent enhancer element can reverse the transcription effect of p53. EMBO J 17: 544–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D 1996. The general transcription factors of RNA polymerase II. Genes Develop 10: 2657–2683 [DOI] [PubMed] [Google Scholar]
- Palecek E, Vlk D, Stankova V, Brazda V, Vojtesek B, Hupp TR, Schaper A, Jovin TM 1997. Tumor suppressor protein p53 binds preferentially to supercoiled DNA. Oncogene 15: 2201–2209 [DOI] [PubMed] [Google Scholar]
- Pan Y, Nussinov R 2008. p53-Induced DNA bending: The interplay between p53-DNA and p53-p53 interactions. J Phys Chem B 112: 6716–6724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao GM, Janknecht R, Ruffner H, Hunter T, Verma IM 2000. CBP/p300 interact with and function as transcriptional coactivators of BRCA1. Proc Natl Acad Sci 97: 1020–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris R, Henry RE, Stephens SJ, McBryde M, Espinosa JM 2008. Multiple p53-independent gene silencing mechanisms define the cellular response to p53 activation. Cell Cycle 7: 2427–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfettini JL, Castedo M, Nardacci R, Ciccosanti F, Boya P, Roumier T, Larochette N, Piacentini M, Kroemer G 2005. Essential role of p53 phosphorylation by p38 MAPK in apoptosis induction by the HIV-1 envelope. J Exp Med 201: 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan SK, Gartel AL 2006. CDK9 phosphorylates p53 on serine residues 33, 315 and 392. Cell Cycle 5: 519–521 [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A 2008. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9: 402–412 [DOI] [PubMed] [Google Scholar]
- Roy S, Tenniswood M 2007. Site-specific acetylation of p53 directs selective transcription complex assembly. J Biol Chem 282: 4765–4771 [DOI] [PubMed] [Google Scholar]
- Roy S, Packman K, Jeffrey R, Tenniswood M 2005. Histone deacetylase inhibitors differentially stabilize acetylated p53 and induce cell cycle arrest or apoptosis in prostate cancer cells. Cell Death Differ 12: 482–491 [DOI] [PubMed] [Google Scholar]
- Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Develop 12: 2831–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels-Lev Y, O’Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X 2001. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell 8: 781–794 [DOI] [PubMed] [Google Scholar]
- Schumm K, Rocha S, Caamano J, Perkins ND 2006. Regulation of p53 tumour suppressor target gene expression by the p52 NF-kappaB subunit. EMBO J 25: 4820–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnick DM, Chehab NH, Stavridi ES, Lien MC, Caruso L, Moran E, Berger SL, Halazonetis TD 1997. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res 57: 3693–3696 [PubMed] [Google Scholar]
- Seo YR, Kelley MR, Smith ML 2002. Selenomethionine regulation of p53 by a ref1-dependent redox mechanism. Proc Natl Acad Sci 99: 14548–14553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Le HV, Massague J 2002. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419: 729–734 [DOI] [PubMed] [Google Scholar]
- Seto E, Usheva A, Zambetti GP, Momand J, Horikoshi N, Weinmann R, Levine AJ, Shenk T 1992. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc Natl Acad Sci 89: 12028–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H, Shiff I, Kott-Gutkowski M, Siegfried Z, Haupt Y, Simon I 2008. Chromatin immunoprecipitation-on-chip reveals stress-dependent p53 occupancy in primary normal cells but not in established cell lines. Cancer Res 68: 9671–9677 [DOI] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O 2007. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell 27: 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinobu N, Maeda T, Aso T, Ito T, Kondo T, Koike K, Hatakeyama M 1999. Physical interaction and functional antagonism between the RNA polymerase II elongation factor ELL and p53. J Biol Chem 274: 17003–17010 [DOI] [PubMed] [Google Scholar]
- Sims RJ 3rd, Belotserkovskaya R, Reinberg D 2004. Elongation by RNA polymerase II: The short and long of it. Genes Develop 18: 2437–2468 [DOI] [PubMed] [Google Scholar]
- St Clair S, Giono L, Varmeh-Ziaie S, Resnick-Silverman L, Liu WJ, Padi A, Dastidar J, DaCosta A, Mattia M, Manfredi JJ 2004. DNA damage-induced downregulation of Cdc25C is mediated by p53 via two independent mechanisms: One involves direct binding to the cdc25C promoter. Mol Cell 16: 725–736 [DOI] [PubMed] [Google Scholar]
- Stros M, Muselikova-Polanska E, Pospisilova S, Strauss F 2004. High-affinity binding of tumor-suppressor protein p53 and HMGB1 to hemicatenated DNA loops. Biochemistry 43: 7215–7225 [DOI] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB 2006. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 24: 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szak ST, Mays D, Pietenpol JA 2001. Kinetics of p53 binding to promoter sites in vivo. Mol Cell Biol 21: 3375–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafvizi A, Huang F, Leith JS, Fersht AR, Mirny LA, van Oijen AM 2008. Tumor suppressor p53 slides on DNA with low friction and high stability. Biophys J 95: L01–03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira N, Nihira K, Yamaguchi T, Miki Y, Yoshida K 2007. DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol Cell 25: 725–738 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Ohkubo S, Tatsuno I, Prives C 2007. hCAS/CSE1 L associates with chromatin and regulates expression of select p53 target genes. Cell 130: 638–650 [DOI] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W 2006. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 24: 827–839 [DOI] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W 2008. Acetylation is indispensable for p53 activation. Cell 133: 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM 2006. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41: 105–178 [DOI] [PubMed] [Google Scholar]
- Thut CJ, Chen JL, Klemm R, Tjian R 1995. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science 267: 100–104 [DOI] [PubMed] [Google Scholar]
- Tian C, Xing G, Xie P, Lu K, Nie J, Wang J, Li L, Gao M, Zhang L, He F 2009. KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat Cell Biol 11: 580–591 [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM 2006. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat Rev 6: 909–923 [DOI] [PubMed] [Google Scholar]
- Vaseva AV, Moll UM 2009. The mitochondrial p53 pathway. Biochim Biophys Acta 1787: 414–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303: 844–848 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW 1992. p53 function and dysfunction. Cell 70: 523–526 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C 2009. Blinded by the Light: The Growing Complexity of p53. Cell 137: 413–431 [DOI] [PubMed] [Google Scholar]
- Wadgaonkar R, Collins T 1999. Murine double minute (MDM2) blocks p53-coactivator interaction, a new mechanism for inhibition of p53-dependent gene expression. J Biol Chem 274: 13760–13767 [DOI] [PubMed] [Google Scholar]
- Wang C, Ivanov A, Chen L, Fredericks WJ, Seto E, Rauscher FJ 3rd, Chen J 2005. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. EMBO J 24: 3279–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Marquardt J, Elzi D, Forster N, Starke S, Glaum A, Yamada D, Defossez PA, Delrow J, Eisenman RN, et al. 2008. Zbtb4 represses transcription of P21CIP1 and controls the cellular response to p53 activation. EMBO J 27: 1563–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Li AG, Liu X 2005. Insights into selective activation of p53 DNA binding by c-Abl. J Biol Chem 280: 12271–12278 [DOI] [PubMed] [Google Scholar]
- Wei X, Xu H, Kufe D 2006b. MUC1 oncoprotein stabilizes and activates estrogen receptor alpha. Mol Cell 21: 295–305 [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. 2006a. A global map of p53 transcription-factor binding sites in the human genome. Cell 124: 207–219 [DOI] [PubMed] [Google Scholar]
- Weinberg RL, Veprintsev DB, Bycroft M, Fersht AR 2005. Comparative binding of p53 to its promoter and DNA recognition elements. J Mol Biol 348: 589–596 [DOI] [PubMed] [Google Scholar]
- White RJ 2005. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol 6: 69–78 [DOI] [PubMed] [Google Scholar]
- White DE, Talbott KE, Arva NC, Bargonetti J 2006. Mouse double minute 2 associates with chromatin in the presence of p53 and is released to facilitate activation of transcription. Cancer Res 66: 3463–3470 [DOI] [PubMed] [Google Scholar]
- Woychik NA, Hampsey M 2002. The RNA polymerase II machinery: Structure illuminates function. Cell 108: 453–463 [DOI] [PubMed] [Google Scholar]
- Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, Look AT 2005. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell 123: 641–653 [DOI] [PubMed] [Google Scholar]
- Xenaki G, Ontikatze T, Rajendran R, Stratford IJ, Dive C, Krstic-Demonacos M, Demonacos C 2008. PCAF is an HIF-1alpha cofactor that regulates p53 transcriptional activity in hypoxia. Oncogene 27: 5785–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Sheppard HM, Corneillie SI, Liu X 2001. p53 Stimulates TFIID-TFIIA-promoter complex assembly, and p53-T antigen complex inhibits TATA binding protein-TATA interaction. Mol Cell Biol 21: 3652–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D 1993. p21 is a universal inhibitor of cyclin kinases. Nature 366: 701–704 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Liu H, Miki Y 2006. Protein kinase C delta regulates Ser46 phosphorylation of p53 tumor suppressor in the apoptotic response to DNA damage. J Biol Chem 281: 5734–5740 [DOI] [PubMed] [Google Scholar]
- Zacchi P, Gostissa M, Uchida T, Salvagno C, Avolio F, Volinia S, Ronai Z, Blandino G, Schneider C, Del Sal G 2002. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature 419: 853–857 [DOI] [PubMed] [Google Scholar]
- Zhai W, Comai L 2000. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol 20: 5930–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Krutchinsky A, Fukuda A, Chen W, Yamamura S, Chait BT, Roeder RG 2005. MED1/TRAP220 exists predominantly in a TRAP/ Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol Cell 19: 89–100 [DOI] [PubMed] [Google Scholar]
- Zhang H, Somasundaram K, Peng Y, Tian H, Bi D, Weber BL, El-Deiry WS 1998. BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene 16: 1713–1721 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang JS, Chen LL, Cheng XK, Heng FY, Wu NH, Shen YF 2004. Repression of hsp90beta gene by p53 in UV irradiation-induced apoptosis of Jurkat cells. J Biol Chem 279: 42545–42551 [DOI] [PubMed] [Google Scholar]
- Zheng X, Chen X 2001. Aquaporin 3, a glycerol and water transporter, is regulated by p73 of the p53 family. FEBS letters 489: 4–7 [DOI] [PubMed] [Google Scholar]
- Zheng H, You H, Zhou XZ, Murray SA, Uchida T, Wulf G, Gu L, Tang X, Lu KP, Xiao ZX 2002. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature 419: 849–853 [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang S, Jiang J, Chen X 2000. Definition of the p53 functional domains necessary for inducing apoptosis. J Biol Chem 275: 39927–39934 [DOI] [PubMed] [Google Scholar]
- Zilfou JT, Hoffman WH, Sank M, George DL, Murphy M 2001. The corepressor mSin3a interacts with the proline-rich domain of p53 and protects p53 from proteasome-mediated degradation. Mol Cell Biol 21: 3974–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotchev SB, Protopopova M, Selivanova G 2000. p53 C-terminal interaction with DNA ends and gaps has opposing effect on specific DNA binding by the core. Nucleic acids Res 28: 4005–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]