Abstract
The true understanding of what we currently define as epigenetics evolved over time as our knowledge on DNA methylation and chromatin modifications and their effects on gene expression increased. The current explosion of research on epigenetics and the increasing documentation of the effects of various environmental factors on DNA methylation, chromatin modification, as well as on the expression of small non-coding RNAs (ncRNAs) have expanded the scope of research on the etiology of various diseases including cancer. The current review briefly discusses the molecular mechanisms of epigenetic regulation, and expands the discussion with examples on the role of environment, such as the immediate environment during development, in inducing epigenetic changes and modulating gene expression.
Keywords: DNA, drug metabolizing enzymes, histone modifications, methylation, transporters ncRNA, miRNA
INTRODUCTION
The term epigenetics was coined by Conrad Waddington in 1942 as a blend of two words, genetics and epigenesis, to indicate developmental events leading from fertilization to mature organism. The true understanding of what we currently define as epigenetics evolved over time as our knowledge on DNA methylation and chromatin modifications and their effects on gene expression increased (Felsenfeld, 2007; Reinberg, 2007). In the context of genetics and molecular biology, epigenetics can be defined as the “study of mitotically or meiotically heritable changes in gene function that cannot be explained by changes in the DNA sequence” (Riggs et al., 1996). Epigenetic inheritance involves the transmission of information (epigenetic mark) not encoded in the DNA sequence, from parent cell to daughter cells and from generation to generation. Epigenetic mark is like a bookmark that flags the chromatin state, “on” or “off”, “open” or “closed”, so it may be identified and maintained in the daughter cells (Choudhuri, 2009a). In the spirit of the term genomics, the term epigenomics has come into existence and is often used synonymously with the term epigenetics. However, epigenomics is a new frontier that studies epigenetic changes at the level of the entire genome (Callinan and Feinberg, 2006). At present, any discussions on epigenomics and epigenetics are thus invariably intertwined.
The current explosion of research on epigenetics and increasing documentation of effects of various environmental factors on DNA methylation and chromatin modification, as well as on the expression of small non-coding RNAs (ncRNAs), have allowed the field to forge ahead with rapid speed. Although much needs to be understood in terms of correlative effects versus causal effects between exposure to various environmental factors and epigenetic changes, available evidence nevertheless suggests that the influence of the environment on an organism at the molecular level can extend well beyond interactions with the DNA sequence (Reamon-Buettner et al., 2008). The present review summarizes the molecular basis of epigenetic regulation of gene expression and discusses some examples of epigenetic changes associated with alterated gene expression profile that underscore how studies on the environmentally-induced epigenetic changes may become increasingly relevant to development, health and disease.
MOLECULAR TARGETS AND MECHANISMS OF EPIGENETIC REGULATION
Molecular targets of epigenetic changes include DNA, histones, and non-coding RNAs (ncRNAs). Therefore, it is logical to first review the chromatin structure before discussing the molecular mechanisms of epigenetic regulation.
Structure of Chromatin
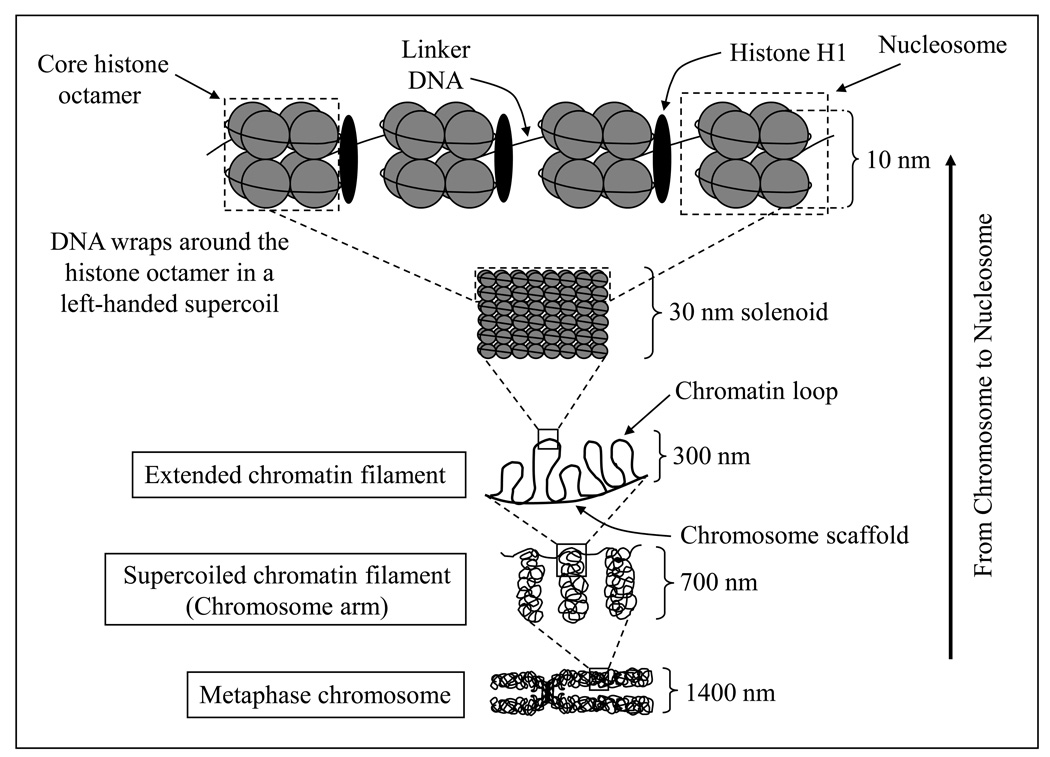
Chromatin (the DNA–histone complex in the nucleus) can be envisioned as a repeat of structural units called “nucleosomes”. The nucleosome core particle is composed of a histone octamer and the DNA that wraps around it. Histones are globular basic proteins with a flexible N-terminus (the so-called ‘tail’) that protrudes from the nucleosome. Histones are subject to various covalent modifications, and histone modifications occur primarily on the tail. The histone octamer contains two molecules each of histones H2A, H2B, H3, and H4. DNA wraps around the octamer in a left-handed supercoil of about 1.75 turns that contains approximately 150 bp. Histone H1 is the “linker histone” that, along with “linker DNA” (the DNA in between two nucleosome core particles), physically connects the adjacent nucleosome core particles. The length of linker DNA varies with species and cell types. Although the DNA associated with the histone octamer is approximately 150 bp, the entire nucleosome also includes part of the DNA on both sides of the core particle; hence the full nucleosome encompasses approximately 180- and 200-bp of DNA (Fig. 1).
Figure 1.
The hierarchy of organization from chromosome to nucleosome. The histone octamer contains two molecules each of histones H2A, H2B, H3 and H4. The DNA wraps around the octamer in a left-handed supercoil in about 1.75 turns that encloses about 150 bp. Histone H1 is the linker histone. Linker histone and linker DNA physically connect adjacent nucleosome core particles. The nucleosomes (10 nm each) are condensed into 30 nm solenoid fiber structure, which are condensed into 300-nm filament; the 300-nm filaments are further condensed into the 700-nm chromosome. During cell division, when the chromosomes duplicate, a 1,400-nm metaphase chromosome is produced containing two chromatids, each chromatid being 700 nm.
Chromatin can undergo changes in its conformation in response to various cellular metabolic demands. Altered chromatin conformation, in turn, can limit or enhance the accessibility and binding of the transcription machinery, thereby precipitating an epigenetic effect on transcription.
Molecular mechanisms of epigenetic regulation
The principal mechanisms that provide the molecular basis of epigenetic regulation of genome expression are (1) DNA methylation, (2) histone modification, and (3) regulation of gene expression by ncRNAs.
In addition, there are also other mechanisms that can affect gene expression epigenetically, such as the phenomenon of transvection in which gene expression is modulated through homologous chromosome pairing-mediated changes in promoter-enhancer interaction in trans. If both alleles of a gene are inactivated in such a way that the enhancer of one allele is non-functional but the promoter is intact, whereas in the other allele (in the homologous chromosome) the enhancer is intact but the promoter is non-functional, then the allele with the non-functional promoter will show no expression, whereas the allele with non-functional enhancer may show no expression or low-level expression. If the two homologous chromosomes undergo intimate synapsis (tight physical pairing), as happens in Dipteran insects, then these two alleles are brought in close physical proximity. Under these circumstances, the intact enhancer of one allele can interact in trans with the intact promoter of the other allele. Such interactions will result in enhanced gene expression from the allele that has the intact promoter. This phenomenon is known as transvection, and has been well studied in Drosophila (see reviews by Henikoff and Comai, 1998; Pirotta, 1999). The defining feature of transvection is its dependence on homologous chromosomal pairing. Therefore, chromosomal rearrangements that interfere with chromosome pairing also interfere with transvection. There is also a cis insulator bypass model of transvection in Drosophila (Morris et al., 1998; see also Choudhuri, 2009a). Transvection can also occur by the action of silencers in trans.
Transvection in Drosophila (and Dipteran insects in general) is possible because the chromosomes normally remain in intimate synapsis in somatic cells. In mammals, sustained somatic pairing (as seen in Drosophila) is generally absent except for some reported examples of tissue- and stage-specific pairing of particular loci, and homologous pairing of oppositely imprinted loci (Duncan, 2002).
The three principal mechanisms of epigenetic regulation of genome expression are discussed below.
1. DNA methylation and transcriptional silencing
DNA methylation machinery
In multicellular eukaryotes, DNA methylation involves covalent modification of cytosine (C) bases at the carbon-5 position of CG dinucleotides (5-MeC), referred to as CpG dinucleotides. The enzyme involved is DNA methyltransferase, known as DNMT, and the methyl donor is S-adenosylmethionine (SAM). The C of CpG is methylated in both strands of DNA. During replication, the parent strand remains methylated, but the newly synthesized daughter strand is not methylated. These hemimethylated double-stranded segments are recognized by maintenance methyltransferase, which methylates the (non-methylated) C on the daughter strand and restores the parental methylation pattern.
There are two types of DNMT enzymes: one is responsible for the de novo methylation that establishes the methylation pattern, and the other is responsible for maintenance methylation once the methylation pattern is established. Mammals have four different DNMTs: DNMT1, 2, 3a and 3b. Whereas DNMT3a and 3b are de novo DNA methyltransferases, DNMT1 is a maintenance methyltransferase. The true function of DNMT2 is not clear because it has weak methyltransferase activity, and its targeted deletion does not have any impact on the global DNA demethylation in the cell (Okano et al., 1998).
The original model of discrete and independent de novo and maintenance methylation functions has been questioned based on a number of recent observations. For example, up to 30% CpG sites remain hemimethylated following inactivation of the de novo methyltransferases DNMT3A and 3B (hence, inactivating de novo methylation function interferes with maintenance methylation function); DNMT 3A and 3B remain associated with the methylated DNA even after the enzymatic reaction has occurred; and DNMT3A and 3B, but not DNMT1, remain strongly anchored to the nucleosomes (hence, de novo methylation does not seem to end the job of de novo methyltransferases) (Jones and Liang, 2009). These observations suggest a cooperation between the de novo methyltransferase and maintenance methytransferase enzymes in maintaining genomic methylation pattern and status. The revised model of de novo and maintenance methylation functions proposes that the bulk of DNA methylation is indeed maintained by DNMT1– the most abundant DNMT in the cell. However, after the replication fork has advanced past a site that has been methylated by DNMT1, DNMT3A and DNMT3B are recruited by specific proteins. The recruited DNMT3 enzymes methylate sites that have been missed by DNMT1 (Jones and Liang, 2009).
Mechanism of transcriptional silencing by DNA methylation
In the genome, CpGs may or may not occur in clusters. CpG clusters, i.e., CpG-rich sequences of the genome are known as CpG islands. By definition, CpG islands are genomic regions that are at least 200-bp long with 50% or higher G+C content and 60% or higher observed/expected CpG ratio (Fazzari and Greally, 2004). In mammalian cells, the majority of CpG sites that do not exist as CpG clusters are methylated, such as satellite DNA, repetitive elements (e.g., transposons), non-repetitive intergenic DNA and exons of genes. Exceptions to this general CpG methylation paradigm are the CpG islands, which are unmethylated CpG clusters (Illingworth and Bird, 2009). In other words, isolated CpG sites are methylated but CpG clusters (CpG islands) are not methylated. Although CpG islands generally remain methylation-free, undermethylated state of CpG islands has also been reported (Wise and Pravtcheva, 1999; Straussman et al., 2009). A number of factors may dictate the undermethylated state of CpG islands, such as local sequence features, Sp1 binding sites, specific cis-acting enhancer elements, as well as specific histone methylation mark H3K4me3 (discussed later), which prevents the binding of de novo methylation complex (Straussman et al., 2009).
Methylation of the C of CpG is associated with transcriptional silencing, and the absence of methylation is associated with active transcription. Thus, unmethylated CpG islands are associated with the promoters of transcriptionally active genes, such as housekeeping genes and many regulated genes, such as genes showing tissue-specific expression. How CpG islands remain unmethylated remains unclear (Li and Bird, 2007).
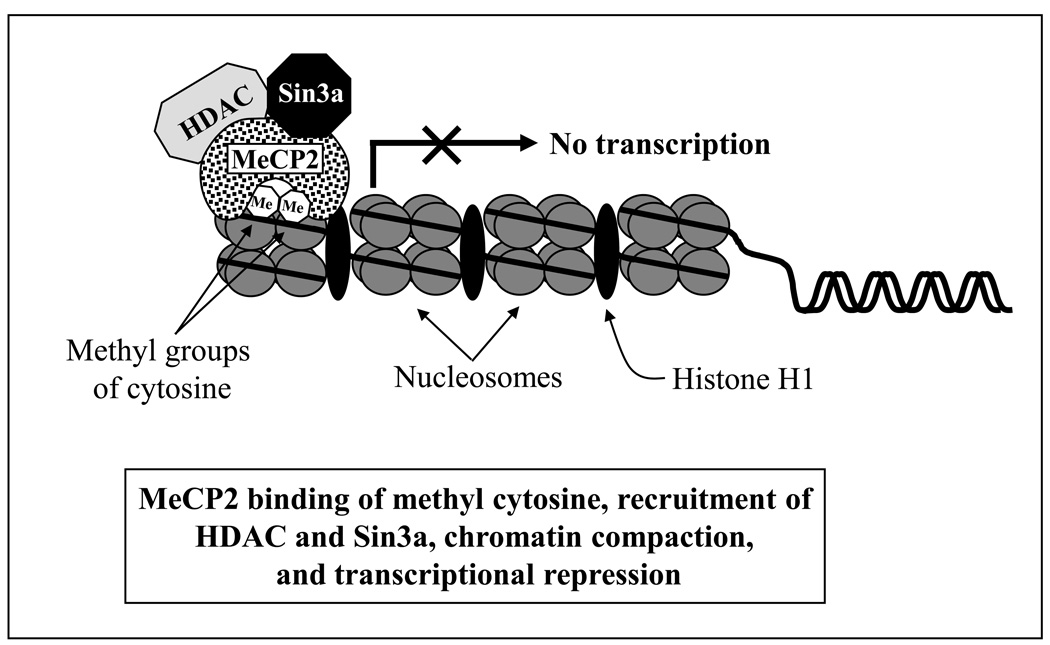
Transcriptional silencing is the result of a condensed state of chromatin brought about by DNA methylation. It is thought to be achieved by two mechanisms; both are supported by experimental evidence: (i) recruitment of methyl CpG-binding transcriptional repressors, and (ii) interference with the DNA binding of transcriptional activators. The first methyl CpG-binding protein purified and cloned was MeCP2, which was studied in detail in order to gain insight into the mechanism of methylation-mediated transcriptional repression (Nan et al., 1997, ’98; Wakefield et al., 1999). MeCP2 selectively binds 5-methyl cytosine in symmetrically positioned CpG dinucleotides in the mammalian genome; it is also able to bind to a single methylated CpG pair. It contains two functional domains: an 85-amino acid methyl-CpG-binding domain (MBD) essential for binding to 5-methyl cytosine, and a 104-amino acid transcriptional repression domain (TRD) that interacts with a corepressor complex containing histone deacetylases (HDACs) and the transcriptional repressor Sin3a. Recruitment of HDAC by MeCP2 causes deacetylation of histones, resulting in a more condensed chromatin conformation and transcriptional silencing (Fig. 2) (Nan et al., 1998). Thus, MeCP2 provides a mechanistic link between DNA methylation, histone deacetylation and consequent transcriptional repression (Nan et al., 1998; Galvão and Thomas, 2005).
Figure 2.
Mechanism of transcriptional repression by MeCP2. MeCP2 selectively binds 5-methyl cytosine in symmetrically positioned CpG dinucleotides in mammalian genome, and methyl CpG-binding protein (MeCP2) is able to bind to a single methylated CpG pair. The methyl-CpG-binding domain (MBD) binds to 5-methyl cytosine, and the transcriptional repression domain (TRD) interacts with a corepressor complex containing histone deacetylases (HDACs) and the transcriptional repressor Sin3a. Recruitment of HDAC by MeCP2 causes deacetylation of histones, resulting in a more condensed chromatin conformation and transcriptional silencing.
However, HDAC inhibitors (e.g., trichostatin A) do not fully relieve the repressive effects of HDAC recruited by MeCP2, thereby suggesting that mechanisms other than deacetylation also add to the transcription repressive ability of MeCP2 (Fuks et al., 2003). One such mechanism may involve preventing the access of transcriptional activators to the promoter and other regulatory sequences. Another mechanism was described by Fuks et al. (2003). Using L929 mouse fibroblast cells, they studied the role of MeCP2 in murine H19 gene repression. They observed that in addition to recruiting HDAC, MeCP2 also associates with histone methyltransferase activity and methylate Lys9 of histone H3, which is a transcription repressing chromatin modification (discussed later; see Table 1). These data demonstrate that MeCP2 reinforces a repressive chromatin state by acting as a bridge between two global epigenetic modifications, DNA methylation and repressive histone methylation.
Table 1.
Some transcriptional activating and repressing histone modifications
| Activating modifications | Repressing modifications |
|---|---|
| Acetylation: | |
| H2A: K5, K9, K13 | |
| H2B: K5, K12, K15, K20 | |
| H3: K9, K14, K18, K23, K56 | |
| H4: K5, K8, K13, K16 | |
| Methylation: | Methylation: |
| H3: K4, K36, K79 | H3: K9, K27 |
| H3: R17, R23 | H4: K20 |
| H4: R3 | |
| Phosphorylation: | |
| H3: T3 | |
| H3: S10, S28 | |
| H3: Y41 | |
| H2AX: S139 (for DNA repair) | |
| Ubiquitination: | Ubiquitination: |
| H2B: K120, | H2A: K119 |
| H2B: K123 (yeast) | |
| Sumoylation: | |
| H2A: K126 (yeast) | |
| H2B: K6, K7 (yeast) | |
| H4: K5, K8, K12, K16, K20 | |
MeCP2 can also bind to unmethylated DNA. Mutations in the MBD (frequently found in human Rett syndrome) that abolish the binding of MeCP2 to methylated DNA do not abolish its binding ability to unmethylated DNA (Galvão and Thomas, 2005; Nikitina et al., 2007).
In addition to MeCP2, there are other MBD proteins, such as MeCP1 and MBD1–4. MBD1–MBD4 were all discovered as EST clones with sequence similarity to the MBD motif of MeCP2 (Cross et al., 1997; Hendrich and Bird, 1998; Hendrich et al., 1999a). MBD2 is the DNA binding component of MeCP1 (Ng et al., 1999). MBD4 can participate in DNA repair as a thymine glycosylase to remove T/G mismatches generated after the deamination of the methyl C (Hendrich et al., 1999b). Several different experimental approaches, such as in vitro transcription assays on methylated and unmethylated DNA templates, transient transfection using methylated and unmethylated reporter constructs, functional analysis of mutants, HDAC inhibition, and transcriptional repression studies in different cell types have strongly suggested a role of these proteins in methylation-dependent transcriptional repression (Wade, 2001).
Methylation and metastable epialleles
Some alleles in mammals show variable expression between cells as well as between individuals despite the absence of genetic heterogeneity. The variable expression is driven by the epigenetic state of the allele. Such epigenetic mosaicism may result in a variegated (patchy) expression pattern of the allele in a tissue, and variable expressivity of the allele between individuals. These alleles were termed “metastable epialleles” by Rakyan et al. (2002). The term “metastable” refers to the labile nature of the epigenetic state, which is reflected in the variegated expression pattern of the allele; the term “epiallele” refers to the potential of these alleles in maintaining the epigenetic marks through generations. DNA methylation can result in the creation of metastable epialleles. An example is the agouti locus in mice.
The agouti (A) gene in mice is dominant over the allele (a) that determines the normal black coat color (due to the production of black eumelanin pigments in hair follicular melanocytes). The agouti gene is normally transcribed from a promoter in exon 2, and it causes a back and forth switch of developmental expression promoting the production of both black eumelanin and yellow/orange phaeomelanin, resulting in the characteristic agouti phenotype of black dorsal hairs (due to eumelanin) with a subapical yellow band (due to pheomelanin) (Duhl et al., 1994; Morgan et al., 1999).
Murine IAP (intracisternal A particle) retrotransposon contains nine CpG sites that can be methylated. Insertion of IAP retrotransposon upstream of the transcription start site of the agouti gene in opposite orientation results in the creation of a metastable epiallele Avy (the vy superscript indicates viable yellow progeny) (Duhl et al., 1994; Morgan et al., 1999). This metastable epiallele shows different degrees of methylation among the carriers, which is inversely correlated with its constitutive ectopic expression and the production of pheomelanin by melanocytes. Thus, the Avy/a mice show a range of coat color phenotypes from yellow (no CpG methylation) to full agouti (called pseudoagouti with hypermethylation of CpG sites 4–9), and different degrees of mottled (yellow/agouti) phenotypes in between. The constitutive ectopic expression of the Avy allele is driven by a cryptic promoter contained at the 5′-end of the IAP retrotransposon (i.e., proximal to the native promoter in exon 2). Differential methylation of the Avy allele is also associated with a range of increased body weight; the yellow coat color being associated with the highest obesity (Morgan et al., 1999; Rakyan et al., 2002).
2. Histone modifications, histone code, and transcriptional regulation
Histone code hypothesis
Covalent histone modification is a precisely regulated phenomenon that is also reversible, and it involves the participation of various proteins. In emphasizing the idea that specific histone modifications appear to act sequentially or in combination to form a recognizable “code” that is identified by specific proteins to bring about distinct downstream events like transcriptional activation or repression, Strahl and Allis (2000) coined the term “histone code”. The concept of such a code is analogous to the concept of a combination code of numbers in a lock. Only a specific combination of numbers will open a specific lock; changing just one number in that combination will fail to do the job.
Different types of known histone modifications
Histones are subject to many different types of reversible covalent post-translational modifications, such as acetylation, methylation, phosphorylation, ADP-ribosylation, ubiquitination, and sumoylation. Of these modifications, histone acetylation and its stimulatory effects on transcription has been known for the longest time. These modifications impact transcription through chromatin conformation. Some of these modifications are transcription activating but some are transcription repressing, yet others may have transcription activating or repressing effects depending on which amino acid residue of histone is modified.
The following summary of different types of histone modifications in chromatin is mainly based on a few recent reviews on this subject (Berger, 2007; Bhaumik et al., 2007; Keppler and Archer, 2008a, b). Other relevant references have been cited as necessary.
Histone acetylation (ac) is a transcription-activating modification that is achieved by the addition of acetyl group (−CH3CO) from acetyl Coenzyme A, to one or more lysine residues at the ε-amino group by histone acetyltransferases (HATs). Acetylation reduces the overall positive charge of histones by neutralizing the positive charge of the target lysine; therefore, it decreases the affinity of histone for the negatively charged DNA. A reduced DNA–histone interaction results in a decondensed, relaxed (i.e., open) chromatin conformation, which allows the transcriptional activators to gain access to their cognate recognition elements and initiate/enhance transcription. Many transcriptional coactivators have HAT activity. Table 1 shows the known histone acetylations at different lysine (K) residues. Acetylations are removed by histone deacetylases (HDACs).
Histone methylation (me) is catalyzed by histone methyltransferases (HMTs) at lysine and arginine residues primarily of histone H3 and H4 (Table 1). The methyl group donor is SAM. Methylation increases the bulk but does not interfere with the charge. Methylation can be mono- (me), di- (me2), or trimethylation (me3). Whereas arginine (R) methylation is presumably transcription activating, lysine methylation can cause either transcriptional activation or repression, depending on which lysine residue is methylated. H3K9 is an important amino acid because it can be acetylated as well as methylated (me, me2, and me3) but they have opposite transcriptional consequences; H3K9ac is transcription-activating whereas H3K9me, me2, and me3 are all transcription-repressing. However, H3K9me3 can also be associated with transcriptional activation, first shown by Vakoc et al. (2005) and subsequently corroborated by others. Therefore, a balance between H3K9 acetylation and methylation may be important in establishing specific chromatin domains. Trimethylation of H3K4 (H3K4me3) provides a binding site for the general transcription factor TFIID and enhances the recruitment and stability of the transcription preinitiation complex, thereby providing a functional basis to explain how histone code H3K4me3 activates transcription (Vermeulen et al., 2007). This finding was independently corroborated by Bing Ren and colleagues (Heintzman et al., 2007) who reported that active promoters are marked by H3K4me3, whereas enhancers are marked by H3K4me, but not trimethylation. They performed ChIP-chip analysis to determine the chromatin architecture along 44 human loci spanning 30 Mb, which was selected by the ENCODE consortium as common targets for genomic analysis. In a subsequent work using multiple human cell lines, the same group (Heintzman et al., 2009) found that the chromatin signatures at promoters are remarkably similar across all cell types. In contrast, enhancers are marked with cell-type-specific histone modification patterns. Histone methylation on lysine is removed by demethylases. Methylated arginine is not directly demethylated, but rather converted to citrulline by peptidylarginine deiminase 4 (PAD4/PADI4).
Histone phosphorylation (ph) is a transcription-activating modification. It is achieved by kinase-catalyzed addition of the negatively charged γ-phosphate, usually from ATP or GTP, to one or more serine and/or threonine residues of histone H3 (Table 1); serine 10 being a frequent target (H3S10ph). The addition of negatively charged phosphate to the N-terminal histone tails presumably disrupts the electrostatic interactions between histones and DNA, thereby destabilizing local chromatin conformation, and triggering transcriptional activation. Methylation at H3K9 can inhibit phosphorylation at H3S10; therefore, an interplay between these two modifications may be important in determining local chromatin modification and regulating transcription. Phosphorylation of histone H2AX at serine 139 forms γ-H2AX, which forms part of the histone code for DNA repair and accumulates at the site of DNA double-strand breaks to recruits various DNA repair proteins. Phosphorylation is removed by specific phosphatases. Recently, it has been reported that Janus kinase 2 (JAK2), which is a non-receptor tyrosine kinase regulating several cellular processes by inducing cytoplasmic signaling cascades, translocates to the nucleus and directly phosphorylates tyrosine 41 on histone H3 (H3Y41ph) (Dawson et al., 2009). Like all other known histone phosphorylations, H3Y41ph is also a transcription-activating modification and it prevents the binding of the transcription-repressive heterochromatin protein 1-alpha (HP1-alpha) to this region of H3. Because JAK2 phosphorylates H3Y41 and creates a transcription-activating modification, inhibition of JAK2 should have transcription repressive effect. This is what is observed in human leukemic cells where inhibition of JAK2 decreases H3Y41ph (as well as H3K4me3) at the promoter of the hematopoietic oncogene lmo2 and downregulates its expression. Decreased H3Y41ph and downregulation of lmo2 is coupled with a reciprocal increase in the binding of HP1-alpha around the lmo2 transcription start site (Dawson et al., 2009).
Histone ubiquitination (ub) is less well studied. In vivo, histone H1, H2A, H2B and H3 can all be ubiquitinated at lysine residues, but H2A and H2B ubiquitinations are the most common (Table 1). H2BK123ub in yeast is required for histone H3K4 and H3K79 methylation, which in turn activate transcription. Ubiquitination of histone H1 results in its release from the DNA; this helps reduce chromatin condensation and facilitates transcriptional activation. Histone deubiquitination is carried out by ubiquitin proteases.
Histone ADP-ribosylation, like histone ubiquitination, is also less well studied. It is catalyzed by ADP-ribosyltransferase and involves the transfer of ADP-ribose moiety of NAD+ to a specific amino acid of the acceptor protein with the simultaneous release of nicotinamide. Arginine, glutamate and lysine residues in the histone are frequently subject to ADP-ribosylation. Histones are ADP-ribosylated in response to DNA damage. DNA strand breaks are recognized by poly (ADP-ribose) polymerase-1 (PARP-1), which catalyzes histone ADP-ribosylation at the lysine residues. The resulting negative charge to the histone causes electrostatic repulsion between the histone and the negatively charged DNA. This leads to the pulling of the DNA away from the histones, hence loosening the chromatin structure and making it more accessible to repair enzymes (Edwards and Myers, 2007).
Histone sumoylation occurs at the lysine residues, and was first reported in 2003 in human cell lines (Shiio and Eisenman, 2003). Small ubiquitin-related modifier (SUMO) is a member of a growing family of ubiquitin-like proteins involved in posttranslational modifications of proteins implicated in crucial cellular processes like cell-cycle regulation, transcription, nucleocytoplasmic transport, DNA replication and repair, chromosome dynamics, apoptosis, and ribosome biogenesis (Vertegaal et al., 2006). Mammalian cells express three major SUMO proteins; SUMO-1, SUMO-2 and SUMO-3. Some SUMO targets are conjugated only to SUMO-1, others only to SUMO-2/3, and still others to all SUMO paralogues. Histone H4 can be conjugated to SUMO-1 and SUMO-3 (Shiio and Eisenman, 2003). As in ubiquitination, sumoylation also requires E1-activating (SAE1/SAE2) and E2-conjugating (UBC9) enzymes. The UBC9 interacts with the substrate to catalyze the formation of an isopeptide bond (peptide bond involving the ε-amino group of lysine instead of the usual α-amino group) between the C-terminal end of SUMO and the amino group of the target lysine. Of the four major core histones, histone H4 was found to be most efficiently sumoylated both in vivo and in vitro, whereas a relatively lower degree of sumoylation was observed for H2A, H2B, and H3 (Shiio and Eisenman, 2003). Histone sumoylation is a transcription-repressive modification (Table 1), and it apparently mediates transcriptional silencing through the recruitment of HDAC and HP1 (Garcia-Dominiguez and Reyes, 2009). It has been suggested that sumoylation-mediated transcriptional repression signal is provided by acetylation itself, because histone H4 sumoylation increases with increasing H4 acetylation. The extent of histone sumoylation in transcriptional regulation, however, is yet to be determined with certainty (Garcia-Dominiguez and Reyes, 2009). Because SUMO moiety is added through an isopeptide bond, removal of sumoylation requires isopeptidases, such as proteases of the Ubiquitin like protein protease (Ulp)/Sentrin specific proteases (SENPs) family, which catalyze de-sumoylation (Dasso, 2008).
Cross-talk between DNA methylation and histone modifications
Cross-talk between chromatin remodeling and histone modification is vital in transcriptional regulation. A number of histone methyltransferases, such as G9a, SUV39H1 [suppressor of variegation 3–9 homolog 1 (Drosophila)]1, PRMT5 (protein arginine methyltransferase 5), and also demethylases can regulate DNA methylation by either recruiting or regulating the stability of DNMTs. DNMTs, in turn, can recruit HDACs and MBPs to achieve chromatin condensation and gene silencing (Sharma et al., 2010). Such cross-talk between different arms of the epigenetic machinery makes the epigenetic orchestration of genome expression a tightly regulated process.
3. Non-coding RNA (ncRNA) and epigenetic regulation
Non-coding RNA (ncRNA)-mediated epigenetic regulation can be driven by long ncRNAs and small ncRNAs. Some of the long ncRNAs playing a role in epigenetic regulation have been known for some time, such as Xist RNA (17 kb) (Memili et al., 2001) and Tsix RNA (40 kb) (Lee et al., 1999) involved in X-chromosome inactivation in mammals. Several other long ncRNAs involved in genomic imprinting have also been described, such as Air (108 kb) (Sleutels et al., 2002), Kcnq1ot1 (>60 kb; the 3′-end is not fully known) (Mancini-Dinardo et al., 2006) and H19 (2.3 kb) (Cai and Cullen, 2007), and are discussed below. Both X-chromosome inactivation and genomic imprinting results in monoallelic gene expression.
The complexity of ncRNA-mediated epigenetic regulation has further expanded with the discovery of various small ncRNAs in animals and plants, such as microRNA (miRNA), small interfering RNA (siRNA), Piwi-interacting RNA (piRNA), repeat-associated siRNA (ra-siRNA), trans-acting siRNA (ta-siRNA), natural antisense transcript siRNA (nat-siRNA), heterochromatic siRNA (hc-siRNA), small scan RNA (scnRNA), and qiRNAs (QDE2-interacting small RNAs) (Choudhuri, 2009b).
Long ncRNAs and epigenetic regulation
The mechanism of epigenetic regulation by long ncRNAs like Air, Kcnq1ot1 and H19 involves genomic imprinting, which involves DNA methylation. Imprinted genes are expressed in a parent-of-origin-specific manner, that is, for maternally imprinted genes (i.e., maternal allele repressed), the allele inherited from the father is expressed; and for paternally imprinted genes, the allele inherited from the mother is expressed. Most imprinted genes are found in clusters that contain between 3 and 11 imprinted genes. In any imprinted cluster, the majority of the genes code for protein-coding mRNA, but there is at least one ncRNA-coding gene in the cluster. The ncRNAs in the cluster show reciprocal expression pattern with respect to the protein-coding genes in the cluster that are subject to silencing (Pauler and Barlow, 2006). Each imprinted cluster is regulated by an imprint control element (ICE), also called imprint control region (ICR). DNA methylation represses the activity of ICE in one parental chromosome (Pauler and Barlow, 2006).
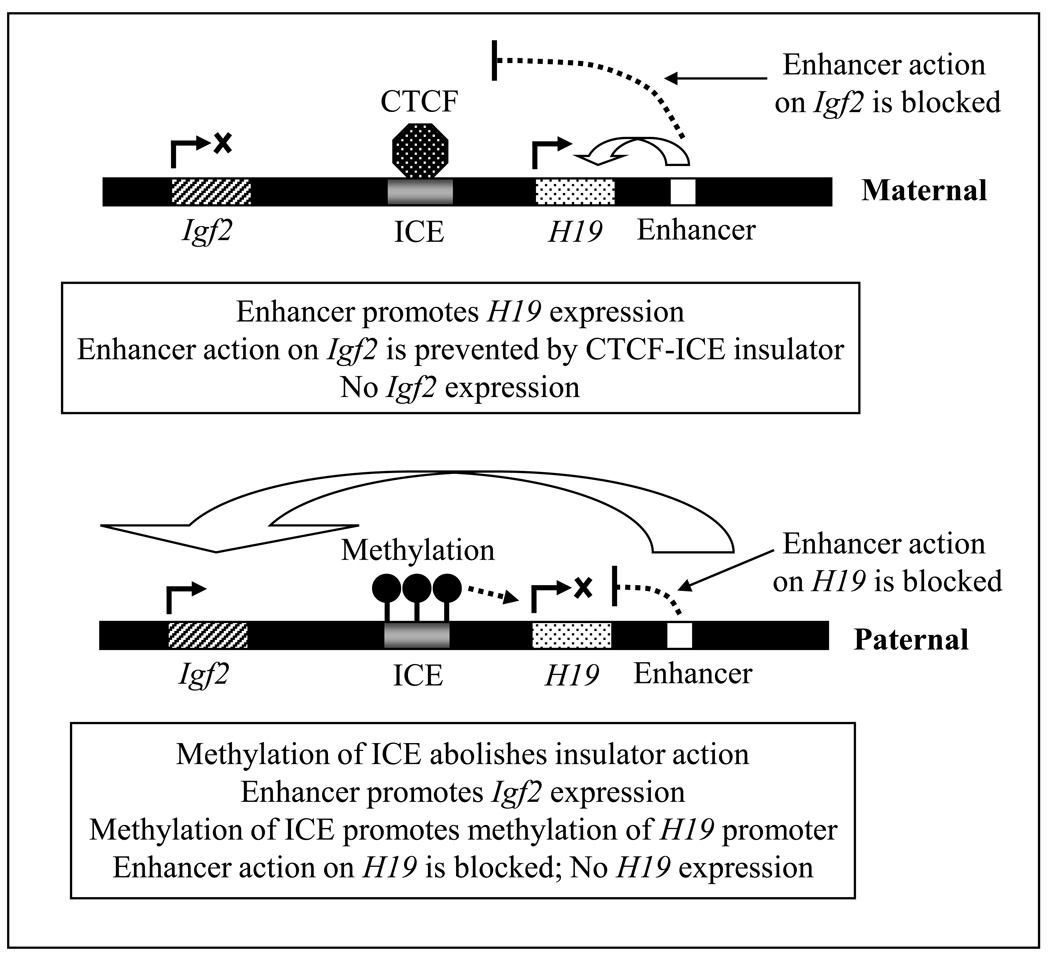
One of the best known examples of genomic imprinting is that of Igf2-H19, in which H19 is a fetal liver ncRNA, and Igf2 is the gene encoding insulin-like growth factor 2; hence, they show reciprocal expression patterns. In mice, H19 is paternally imprinted (repressed) but maternally expressed; thus Igf2 is paternally expressed but maternally imprinted (repressed) (Fig. 3). Igf2 is located ~80 kb upstream of H19 on the same chromosome, and the ICE is located in between them. The ICE is a ~2.4 kb long region located ~2 kb upstream of H19, and contains an insulator sequence that binds insulator protein. Insulators are specific DNA sequences that, when bound to specific insulator-binding protein(s), act as gene boundaries and shield a promoter from the effects of neighboring regulatory elements. There is also an enhancer region downstream from H19 that can enhance the transcription from both Igf2 and H19 loci. Monoallelic expression of Igf2 or H19 is regulated by the methylation status of ICE. On the maternal chromosome, the unmethylated ICE insulator binds the vertebrate insulator protein CTCF (CCCTC-binding factor). This mediates the silencing of Igf2 in cis by insulating it from the transcription activating effects of the downstream enhancer. On the paternal chromosome, the ICE is methylated, which inhibits CTCF binding (Schoenherr et al., 2003). Methylation of ICE also leads to secondary methylation of the H19 promoter. As a result, H19 becomes silenced in the paternal chromosome. Also, because the methylated paternal ICE lacks insulator activity, the paternal Igf2 promoter can interact with the enhancer to express Igf2 (Fig. 3).
Figure 3.
In mouse Igf2 and H19 are on the same chromosome such that Igf2 is located ~80-kb upstream of H19. The imprint control region (ICE), located in between Igf2 and H19, contains an insulator. There are also a set of enhancers located downstream from H19, which are utilized by both Igf2 and H19 for their expression. On the maternal chromosome, the unmethylated ICE binds the vertebrate insulator protein CCCTC-binding factor (CTCF). ICE-CTCF insulator prevents the enhancer from acting upon Igf2, essentially silencing its expression. On the paternal chromosome, the ICE is methylated preventing CTCF binding. Methylation of ICE also leads to secondary methylation of the H19 promoter, and silencing of H19. Because the methylated paternal ICE lacks insulator activity, the enhancer can interact with paternal Igf2 promoter and enhance Igf2 expression.
Similarly, Air ncRNA (paternally expressed) is associated with the silencing of the Igf2r cluster in paternal allele; hence the Igf2r cluster is expressed in the maternal allele. The Igf2r cluster has three protein-coding genes (Igf2r/Slc22a2/Slc22a3, where Slc22a2 and Slc22a3 are Oct2 and Oct3, respectively; Oct stands for organic cation transporter) (Sleutels et al., 2002; Alnouti et al., 2006). Another example is Kcnq1ot1 (potassium channel Q1 overlapping transcript 1) ncRNA, which is also paternally expressed and is associated with the silencing of the 800-kb Kcnq1 cluster, one of the largest imprinted clusters containing nine protein-coding genes (Mancini-Dinardo et al., 2006). The ICE of each cluster contains the promoter of the respective ncRNAs, such that when the ICE is methylated, the ncRNA is silenced and the protein-coding genes are expressed, but when the ICE is not methylated, the ncRNA is expressed and the protein-coding genes are silenced. The expression of these ncRNAs is correlated with the silencing of all the genes in the cluster. Premature termination of Air and Kcnq1ot1 transcription results in complete loss of silencing of the respective clusters and expression of the protein-coding genes (Sleutels et al., 2002; Mancini-Dinardo et al., 2006). At this time, the exact mechanism how Air and Kcnq1ot1 ncRNAs silence the expression of their respective clusters is not clear.
X chromosome inactivation in mammals by Xist (X-inactive specific transcript) RNA is another example of epigenetic regulation by long ncRNA. Before X inactivation, the Xist RNA is expressed from both X chromosomes. Eventually its expression is silenced on the X that remains active (Xa), but is maintained on the X that becomes inactivated (Xi). The Xist coats the entire 180-Mbp-long Xi in cis, which induces repressive chromatin modifications and DNA methylation in Xi. As a result, gene expression from Xi is silenced (Pauler and Barlow, 2006). Deletion analysis showed that a 0.9-kb region at the 5′ end of Xist RNA is essential for its transcriptional silencing activity; this region contains sequence motifs that can fold into two stem-loop structures that are repeated about 8 times. The stem-loop structures may represent binding sites for other factors aiding in the transcriptional silencing activity of Xist (Wutz et al., 2002).
Small ncRNAs and epigenetic regulation
Small ncRNAs play important roles in epigenetic regulation of gene and genome expression through different mechanisms, such as translational repression of mRNA, mRNA degradation, DNA methylation and chromatin modification. The following examples focus on the role of miRNAs (microRNAs) in epigenetic regulation.
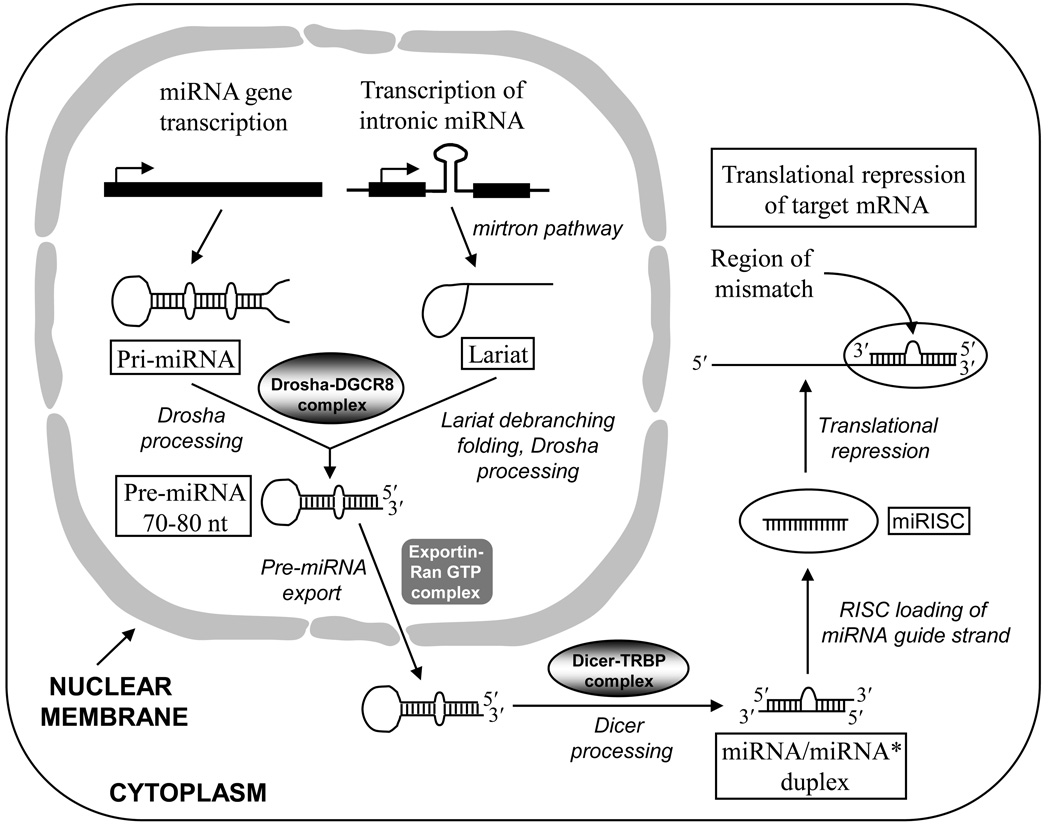
microRNAs (miRNAs, abbreviated as miR) are short (~22 nt) ncRNAs that regulate gene expression by binding to their cognate binding sites at the 3′-end of target mRNAs, and inhibiting their translation. microRNAs are transcribed from miRNA genes mostly by RNA polymerase II into long primary miRNA (pri-miRNA) transcripts, often containing thousands of nucleotides and forming hairpins (stem-loops). The pri-miRNA transcript is processed in the nucleus by Drosha–DGCR8 complex to produce 70–80 nt-long precursor miRNA (pre-miRNA) (Fig. 4). Drosha is an RNAse III-type enzyme, whereas DGCR8 helps in accurate Drosha processing by acting as a molecular ruler to determine the Drosha cleavage site, which is at the 11-nt position from the base of the stem structure. The 5′-end of the pre-miRNA has a bias for uridine, and the 3′-end has a 2-nt overhang. Pre-miRNAs are transported from the nucleus to the cytoplasm by Exportin-5 and Ran-GTP complex. In the cytoplasm, the pre-miRNAs are further processed into ~22 nt-long miRNA/miRNA* duplex by Dicer, which is another RNAse III-type enzyme. Dicer processing also produces a 2-nt overhang at the 3′-end of each strand of the miRNA/miRNA* duplex. The duplex undergoes unwinding. One of the two strands, called the guide strand (miRNA), is then loaded onto a protein complex called RNA-induced silencing complex (RISC) forming miRISC (Fig. 4). The other strand (miRNA*) that is not part of the RISC is called the passenger strand and is degraded. Guide strand selection is driven by the thermodynamic stability of the miRNA/miRNA* duplex. The unwinding of the duplex by Dicer begins at the 5′-end of the strand that has the lowest thermodynamic stability, and this strand acts as the guide strand. The guide strand binds to its cognate binding site of the target mRNA with a ~2 nt mismatch. The sequence specificity of miRNA guide strand for target recognition is determined by nt 2–8 of its 5′ region, and it is called the “seed sequence”. Binding of multiple miRISCs at the 3′-UTR of target mRNA represses its translation, resulting in the silencing of gene expression. Some pri-miRNAs that are encoded by introns are not processed by Drosha, but instead are processed by spliceosome in the nucleus (the mirtron pathway). Following spliceosome processing, the miRNA is released as lariat structure that first undergoes debranching followed by folding to form the pre-miRNA (see Choudhuri, 2010).
Figure 4.
Biogenesis and function of miRNA. The pri-miRNA transcript is processed in the nucleus by Drosha–DGCR8 complex to produce a 70–80 nt-long precursor miRNA (pre-miRNA). Pre-miRNA is transported to the cytoplasm by Exportin-5 and Ran-GTP. Some pri-miRNAs that are encoded by introns are processed by spliceosome (the mirtron pathway). Following spliceosome processing, the miRNA is released as lariat structure that first undergoes debranching followed by folding to form the pre-miRNA. In the cytoplasm, the 70–80 nt-long pre-miRNA is further processed into 22 nt-long duplex mature miRNA by Dicer, forming the miRNA/miRNA* duplex. From this duplex, only the guide strand is loaded onto the miRISC. The miRISC is targeted to the mRNA; the miRNA binds to the 3′-UTR of the mRNA and suppresses its translation.
4. Some examples of epigenetic phenomena that regulate gene and genome expression
Some of the relatively well-studied examples of epigenetic phenomena are transvection (discussed above), genomic imprinting (discussed above), X-chromosome inactivation (briefly discussed above), paramutation (not discussed in this review), and heterochromatin spread and position effect variegation (discussed below).
Transcriptionally inert heterochromatin sometimes spreads into transcriptionally active neighboring euchromatin, thereby silencing the genes that are adjacent to the heterochromatin. Such heterochromatin spread in a tissue is not complete because some cells escape it. As a result, genetically identical cells in a tissue display different cellular phenotypes; cells subjected to heterochromatin spread may display the silencing of certain genes adjacent to heterochromatin, whereas cells that escape it maintain the expression of those genes. This creates a mosaic gene expression pattern in the tissue, the so-called “variegated (patchy)” expression pattern. Because such variegated gene expression pattern is brought about by the proximity of the genes to the heterochromatin, it is called position effect variegation (PEV). Thus, gene silencing through heterochromatin spread is an epigenetic phenomenon. Various Suv or Su(var) (for “suppression of variegation”, sometimes written in all cap) proteins are modifiers of heterochromatinization and PEV through their interaction with the chromatin. Much of the current understanding of the function of these proteins came from studies in Drosophila. For example, in Drosophila Su(var)3–9 (also called Suv39) is a histone methyltransferase; it catalyzes the formation of H3K9me2 that promotes the binding of Su(var)2–5 to histone H3. Human homologue of Suv39 is called SUV39H1. Su(var)2–5 is also called Suv205 or HP1 (heterochromatin protein1), which is a chromodomain protein. Human HP1 is also called HP1hsa (hsa for Homo sapiens). Studies in Drosophila showed that H3K9me2 formation and its interaction with HP1 spread in tandem to ectopic locations on the chromosome arms, inducing heterochromatin formation and spread. Such H3K9me2–HP1 system-mediated heterochromatin spread can be antagonized by a mutant of the euchromatin-associated Su(var)3-1 (also called JIL-1), which is an euchromatic H3S10 kinase (Lerach et al., 2006; also see Choudhuri, 2009a). Some other examples of Su(var) proteins include Su(var)3–7, which is a Zn-finger protein, and histone deacetylase HDAC1, also called Su(var)3–26 (or Suv326).
Lessons from large-scale human genome-wide methylation studies
In a recent study, using Illumina’s GoldenGate methylation platform to investigate cytosine methylation, Christensen et al. (2009) analyzed 217 non-pathologic human tissues from 10 anatomic sites. A total of 1,413 autosomal CpG loci associated with 773 genes were analyzed. The goal was to document normal intra- and inter-individual differences in DNA methylation, and to discern how aging and exposures contribute to normal variations in methylation. The authors found highly significant CpG island–dependent correlations between age and methylation in solid tissues; loci in CpG islands gained methylation with age, whereas loci not in CpG islands lost methylation with age, and this pattern was consistent across tissues. The data clearly demonstrate age- and general environmental exposure-related differences in tissue-specific methylation and significant age-associated methylation patterns which are CpG island context-dependent. From their work, the authors concluded that large epigenetic changes occur in normal appearing tissues, and the relationship of these changes to companion genetic changes is of interest to study in the future.
In another study, De Bustos et al. (2009) reported the differences in methylation outside of CpG islands in nine different normal tissues from two human donors. They used HpaII-treated DNA; hence they focused on methylation differences in CCGG target sites. For this study, they focused on the euchromatic region of chromosome 1 representing ~8% of the genome, and compared the methylation profiles of different organs with that of spleen. The authors observed gross regional differences in methylation between tissues from the same individual, with the most striking differences between cerebellum and spleen. Profiles of the same tissue from different donors were found to be strikingly similar, as well as the profiles of different lobes of the brain.
Lister et al. (2009) provided the first complete DNA-methylation map of the human genome at a single-base-pair resolution. Using bisulfite treatment of the genomic DNA followed by high-throughput sequencing, the authors compared the methylation pattern in stem cells (pluripotent) and fibroblasts (differentiated). They found that despite comparable methylation pattern, significant differences exist between the two cell types. In fibroblasts, cytosine methylation occurs almost exclusively on CpG dinucleotides, which is associated with reduced gene activity. However, in stem cells ~25% of the methylation sites are found on C that neighbor bases other than G, and is mostly an A (NpCpN, where C is the methylated, and N can be A, C or T, but is mostly A). The authors showed that although the frequency of such non-CpG cytosine methylation is relatively low and varies between cells, it is enriched in transcriptionally active genes, particularly on the template DNA strand. Importantly, non-CpG methylation disappeared upon induced differentiation of the stem cells, and was restored in induced pluripotent stem cells. The existence of non-CpG methylation had been reported before in mouse stem cells (Ramsahoye et al., 2000), but its prevalence and location in the genome was not widely appreciated. The work by Lister et al. opens up the avenues for further functional studies of the human DNA methylome in the genome.
ENVIRONMENT, EPIGENETIC REGULATION AND ITS CONSEQUENCES
Epigenetic changes can be acquired throughout the life of an individual. Recent studies have demonstrated that epigenetic changes acquired in the intra-uterine environment and after birth, can persist throughout life and influence the physical and mental state as well as the ability to deal with the immediate environment, including the ability for drug and xenobiotic metabolism. This may determine clinical outcomes in health and disease.
Maternal nutritional status as a prenatal environment factor and its epigenetic effects
Multiple lines of evidence from in vitro and in vivo models have established that epigenetic modifications caused by in utero exposure to environmental factors, such as nutritional factors, environmental toxicants, or factors related to lifestyle (such as, tobacco smoke, alcohol, chemical carcinogens, infectious agents, UV radiation) can induce alterations in gene expression that may persist throughout life, as well as across multiple generations (Herceg, 2007).
Lillycrop et al. (2005) showed that feeding rats protein-restricted (hence unbalanced) diet during pregnancy resulted in persistent changes in DNA methylation and expression of the glucocorticoid receptor (GR) and peroxisomal proliferator-activated receptor-α (PPAR-α). Consumption of the protein-restricted diet during pregnancy resulted in ~21–22% lower CpG methylation in the hepatic PPAR-α and GR gene promoters in the offspring, compared to controls. In contrast, there was no difference in the methylation status of the PPAR-γ promoter, which is the major PPAR isoform expressed in liver. Gene methylation was determined by methylation-sensitive PCR and mRNA expression by semi-quantitative RT-PCR. Consistent with the fact that decreased promoter methylation is associated with an increase in gene expression, the expression of both PPAR-α and GR were upregulated in protein-restricted diet group, compared to controls. Acyl coA-oxidase (AOX) is directly regulated by PPAR-α; the authors also found that AOX mRNA expression was expectedly upregulated in the liver of offspring of the protein-restricted diet group, compared to controls. Because GR is involved in stress regulation, epigenetic changes affecting GR expression may interfere with an individual’s ability to cope with stress. In contrast, there was no change in the expression of PPAR-γ in which the promoter methylation was not different.
The same group (Burdge et al. 2007) followed up on the previous findings of Lillycrop et al. (2005), and investigated whether the altered methylation state of PPAR-α and GR promoters (caused by feeding the F0 females protein-restricted diet during pregnancy) is passed to the F2 offspring as well. They found that indeed the hypomethylation state of the target genes in F1 is also passed to the F2 offspring, even though the F1 offspring were fed a normal diet. This demonstrates that epigenetic marks can persist for at least two generations.
Nutritional factors can also act as environmental factors in adults to precipitate specific epigenetic changes. Aagaard-Tillery et al (2008) studied the effects of maternal diet on the epigenome of offspring in monkeys from the perspective of obesity. Chronic consumption of a maternal high-fat diet (35% fat, as opposed to 13% fat in controls) resulted in significant hyperacetylation of H3K14 in fetal hepatic tissues, and a trend towards increased acetylation of H3K9 and H3K18 as well, as revealed by chromatin immunoprecipitation (ChIP) with differential display PCR. The offspring of the obese monkeys were also obese. When the fat content of the diets were altered during pregnancy, maternal obesity still maintained but the epigenetic changes in offspring were no longer present. This study showed that in utero exposure to specific environmental factors (in this case, calorie-dense, high-fat maternal diet) could induce epigenetic changes, which in turn determines specific phenotypic/physiological outcome in the adults (in this case obesity). Microarray analysis showed that the expression of genes (such as GPT2, Rdh12, Npas2, Hsp and DNAJ2) involved in metabolism and associated response showed an appreciable increase. These results provide an epigenetic/molecular basis to the “fetal origins of adult disease” hypothesis.
Studies in humans have shown that manipulation of dietary folate causes detectable changes in global genomic DNA methylation status. Methylenetetrahydrofolate reductase (MTHFR) catalyzes the synthesis of 5-methyltetrahydrofolate (5-methyl-THF), the methyl donor for synthesis of methionine from homocysteine and precursor of S-adenosyl-L-methionine. Friso et al. (2002) sought to determine the effect of folate status on genomic DNA methylation with an emphasis on the interaction with the common C677T mutation in the MTHFR gene. By assessing genomic DNA methylation from 105 subjects homozygous for this mutation (T/T) and 187 homozygous for the wild-type (C/C) MTHFR genotype, they found that genomic DNA methylation directly correlated with plasma folate status and inversely with plasma homocysteine levels. T/T genotypes had a diminished level of DNA methylation (~32 ng 5-MeC/µg DNA) compared with those with the C/C wild-type (~62 ng 5-MeC/µg DNA). However, when analyzed according to folate status, only the T/T subjects with low plasma folate levels accounted for the diminished DNA methylation; this difference was not seen in T/T subjects with high plasma folate levels. Thus, low folate status in conjunction with the existing genetic polymorphism (the T/T genotype) precipitated the epigenetic effect. In other words, low plasma folate status acted as the selection pressure to reveal the epigenetic effect of the T/T polymorphism of the MTHFR gene.
Parental care as a postnatal environmental factor and its epigenetic effects
Recent research has shown the importance of parental care as an environmental factor that may determine the postnatal developmental outcome through epigenetic mechanisms. Subtle differences in the degree of parental care could lead to potentially profound epigenetic changes in animals that were raised in an otherwise identical environment. Young rats subjected to different degrees of licking and grooming (LG), and arched-back nursing (ABN) by the mother (equivalent to different degrees of motherly love and care) had differences in DNA methylation in the regulatory region of the glucocorticoid receptor (GR) gene in the hippocampus; higher methylation was found in pups that received less maternal care. The reason for studying the GR gene in hippocampus was that the GR is involved in stress regulation, and hippocampus, being rich in GR, is an important part of the brain involved in stress regulation. The observed epigenetic changes affected the regulation of stress hormone levels into adulthood, such that the pups that received greater maternal care (high LG–ABN) grew into calmer adults than their less-groomed counterparts (low LG–ABN). The causal relationship among the epigenetic state, GR expression and the maternal effect on stress responses was strongly suggested by the fact that these differences emerged over the first week of life, could be reversed with cross-fostering (where one mother showed higher LG–ABN behavior than the other), persisted into adulthood, and were associated with altered histone acetylation and transcription factor (nerve growth factor-inducible-A or NGFI-A) binding to the GR gene promoter. Significantly higher histone H3K9 acetylation and 3-fold higher NGFI-A binding to the hippocampal GR gene promoter was found in the adult offspring that received greater maternal care as pups (Weaver et al., 2004).
When the same group extended their work in humans to suicide victims with known history of childhood abuse, they observed similar results. Two genes were studied, ribosomal RNA (rRNA) genes and the human neuron-specific GR gene (nuclear receptor subfamily 3, group C, member 1 or NR3C1). The reason for choosing the rRNA gene was the fact that rRNA is part of the protein synthesis machinery and a number of studies suggested that the fraction of rRNA promoters that is active in a given cell is regulated through epigenetic markings (Brown and Szyf, 2008); the reason for choosing the GR gene has been mentioned already. It was found that in the brain of suicide victims, the rRNA gene in the hippocampus (but not in the cerebellum, which is involved in motor control) had highly methylated promoter and 5′ regulatory region (hence reduced rRNA expression) compared to healthy individuals, thereby demonstrating aberrant regulation of the protein synthesis machinery (McGowan et al., 2008). Similar results were found for the GR gene (NR3C1) expression level in the hippocampus. The authors found increased methylation of the NR3C1 gene promoter, and concomitant decrease in gene expression resulting in decreased levels of NR3C1 mRNA. Patch-methylated promoter constructs that mimicked the methylation state in samples from abused suicide victims showed decreased binding of NGFI-A transcription factor and decreased transcription of NGFI-A-inducible genes (McGowan et al., 2009). Sequencing of the NR3C1 gene promoter revealed identical sequence in all subjects, thereby eliminating the possible contribution of nucleotide sequence variations with differential gene expression outcomes.
Therefore, the findings in humans were consistent with that in rats, suggesting a common effect of parental care in childhood in determining behavior pattern in adulthood, which is apparently mediated by epigenetic regulation of the GR gene expression in hippocampus. This is reflected in restlessness and irritated behavior in adult rats that received less maternal care as pups, and suicidal behavior in adult humans who experienced childhood abuse. Although more data are needed in this regard investigating various other targets and behavioral endpoints, these findings nevertheless have important social, scientific and clinical implications. This work seems to provide clues to how epigenetic mechanisms may affect “genetically identical background” resulting in different phenotypic outcomes when exposed to different environments.
In the context of epigenetic alterations and their probable associations with behavioral changes, it may be pertinent to discuss here the effects of vinclozolin, a fungicide, as reported by Crews et al. (2007) and Skinner et al. (2008). In both studies, the authors administered pregnant rats with 100 mg/kg/day of vinclozolin intraperitonially from day 8–14 of gestation, corresponding to the period of gonadal sex determination. In a study investigating the transgenerational effects of vinclozolin on mate preference, Crews et al. (2007) observed that F3 generation females that were descendants of vinclozolin-treated F0 females preferred males who did not have a history of exposure to vinclozolin. In other words, females that were three generations removed from the exposure still discriminated and preferred males who did not have a history of exposure to vinclozolin. In contrast, males that were descendants of vinclozolin-treated F0 females did not show such mate preference. The differential mate preference behavior was ascribed to possible epigenetic changes caused by vinclozolin. The authors concluded that the consequences of exposure to endocrine disruptors are not just transgenerational but can be transpopulational as well. In another study, Skinner et al. (2008) reported that exposure to vinclozolin caused changes in brain transcriptome in both males and females through F3 generation. This was associated with increased anxiety behavior in rats as evidenced by decreased latency period in entering a dark box and spending more time in the dark side of the dark/light box. The authors hypothesized that such increased anxiety behavior in vinclozolin-treated rats, particularly in males, may be due to epigenetic changes induced in the germ line transgenerationally. In both these studies the authors did not actually investigate epigenetic changes. Instead, their speculation of epigenetic changes playing a role in mate preference and increased anxiety behavior was based on three indirect pieces of evidence: first, their previous study showed that vinclozolin treatment could alter DNA methylation pattern in the germ line transgenerationlly, which was associated with adverse reproductive function in males, including decreased spermatogenesis and increased infertility (Anway et al., 2005); secondly, the gene expression analysis in the study by Skinner and coworkers demonstrated transgenerational alterations in the gene expression profile, as evidenced by changes detected in the F3 generation; and thirdly, the behavioral changes were also found transgenerationally in the F3 generation offspring that were not exposed to vinclozolin themselves.
The initial observations of vinclozolin’s adverse transgenerational effects, major adverse effects in the F1 generation, as well as epigenetic effects could not be reproduced by two recent studies (Schneider et al., 2008; Inawaka et al., 2009). Schneider and coworkers demonstrated that when they used the same dose used by Anway and coworkers (100 mg/kg/day) but administered it orally, they could not reproduce any of the adverse reproductive effects (mating, fertility, sperm parameters, histopathology etc.) in F1, F2 or F3 males that were demonstrated by Anway and coworkers using intraperitoneal dosing. However, Schneider and coworkers did not determine the epigenetic effects but focused on the adverse male reproductive effects. In contrast, Inawaka et al. (2009) used the same dose and same route used by Anway and coworkers trying to reproduce vinclozolin’s reported dramatic transgenerational adverse reproductive effects as well as the corresponding epigenetic effects (altered DNA methylation) in male rats, but those earlier observations could not be reproduced either. In a recent review, Skinner et al. (2010) hypothesized that such differences in finding may be ascribed to the inbred versus outbred nature of the rat strain used in these studies. Interestingly, the current ADI (acceptable daily intake) of vinclozolin established by the World Health Organization is 10 µg/kg/day (Schneider et al., 2008). Therefore, the dose of vinclozolin used to demonstrate epigenetic changes and adverse reproductive effects in the original study was 10,000 times higher than the ADI. Evidently, more work is needed on this chemical using various doses to find out the extent and relevance of its epigenetic and genetic effects.
Epigenetic changes associated with drug- and xenobiotic-metabolizing enzymes and transporters and their consequences
Although the genetic regulation of various drug-metabolizing enzymes and transporters has been studied for many years now, studies on their epigenetic regulation is relatively recent. A number of publications have demonstrated various aspects of the epigenetic regulation of drug and xenobiotic enzymes and transporters. Because the expression of various cytochrome P450 (CYP) enzymes shows inter-individual variations, and can be influenced by environmental factors that include diet as well as environmental pollutants, an epigenetic component underlying their regulation is conceivable. Various CYP isoforms in humans that have been shown to have an epigenetic component in their regulation include CYP1A1, 1A2, 1B1, 2E1, 2W1, 2A13 (reviewed by Rodriguez-Antona et al., 2010). The epigenetic aspect of regulation of CYP enzymes may explain, at least in part, the observed inter-individual variability in their expression, and further modulate an individual’s genetic ability to cope with environmental chemicals, including drugs.
For example, DNA methylation is an important epigenetic mechanism regulating CYP1A1 expression. In prostate cancer cell line LNCaP, promoter methylation of the CYP1A1 gene prevents the binding of the Ah receptor (AhR) complex to the dioxin response element (DRE), resulting in repression of CYP1A1 expression (Okino et al. 2006). In contrast, hypomethylation of the promoter in noncancerous cell lines PWR-1E and RWPE-1 facilitates binding of the nuclear receptors to the DRE. For CYP1A2 gene, a single CpG in the sequence CCGG at position −2759 and next to the AP-1 binding site in the 5′-flanking region can reduce CYP1A2 gene expression (Hammons et al., 2001). The epigenetic regulation of CYP1A2 expression may explain the great degree of inter-individual variability observed in CYP1A2 expression, and the ability to metabolize CYP1A2 substrates. Similar promoter/enhancer methylation-driven alteration in expression has been reported for CYP1B1, CYP2E1, CYP2W1, CYP2A13 genes, in which hypermethylation is associated with decreased expression and hypomethylation is associated with increased expression (reviewed by Rodriguez-Antona et al., 2010). In addition to DNA methylation, it has been shown in the mouse Hepa-1 cell line that chromatin structure also plays an essential role in Cyp1a1 gene transcription. Specifically, induction of Cyp1a1 gene transcription is strongly associated with hyperacetylation of histone H3K14 and H4K16, as well as other modifications, such as H3K4me3 and H3S10ph (Schnekenburger et al., 2007). Using MCF-7 breast cancer cell line, it was demonstrated that the expression of CYP1B1 gene is also post-transcriptionally regulated by the microRNA miR-27b, which is a negative regulator of CYP1B1 gene expression. In breast cancer, miR-27b is downregulated; this allows increased translation of CYP1B1 mRNA and increased CYP1B1 protein expression (Tsuchiya et al., 2006). Using Human embryonic kidney (HEK)-293 cells co-transfected with CYP3A4 3′-UTR–luciferase reporter plasmid along with either miR-27b or miR-298 plasmid, Pan et al. (2009) demonstrated that the expression of CYP3A4 gene is also negatively regulated by miR-27b and miR-298.
In an effort to understand whether the developmental switch between Cyp3a16 (neonatal isoform) and Cyp3a11 (adult isoform) expression in mouse has an epigenetic basis, Li et al (2009) studied DNA methylation, and histone modifications (H3K4me2, H3K27me3) around the Cyp3a locus at various developmental stages from prenatal through neonatal to young adults. No DNA hypermethylation was observed at Cyp3a locus at any age. However, the expression of Cyp3a16 in neonatal livers and Cyp3a11 in adult livers was strongly correlated with increases in H3K4me2, which is a gene expression-promoting histone modification. Likewise, the suppression of Cyp3a16 expression in adult livers was correlated with decreases in H3K4me2 and increases in H3K27me3, the latter being a gene expression-repressing histone modification. Thus, the developmental switch between Cyp3a11 and Cyp3a16 gene expression is controlled by dynamic epigenetic regulation of these loci through histone modifications. An earlier study (Jin et al., 2004) showed that mouse Cyp1a2 gene expression coincides well with the methylation status of DNA during liver development.
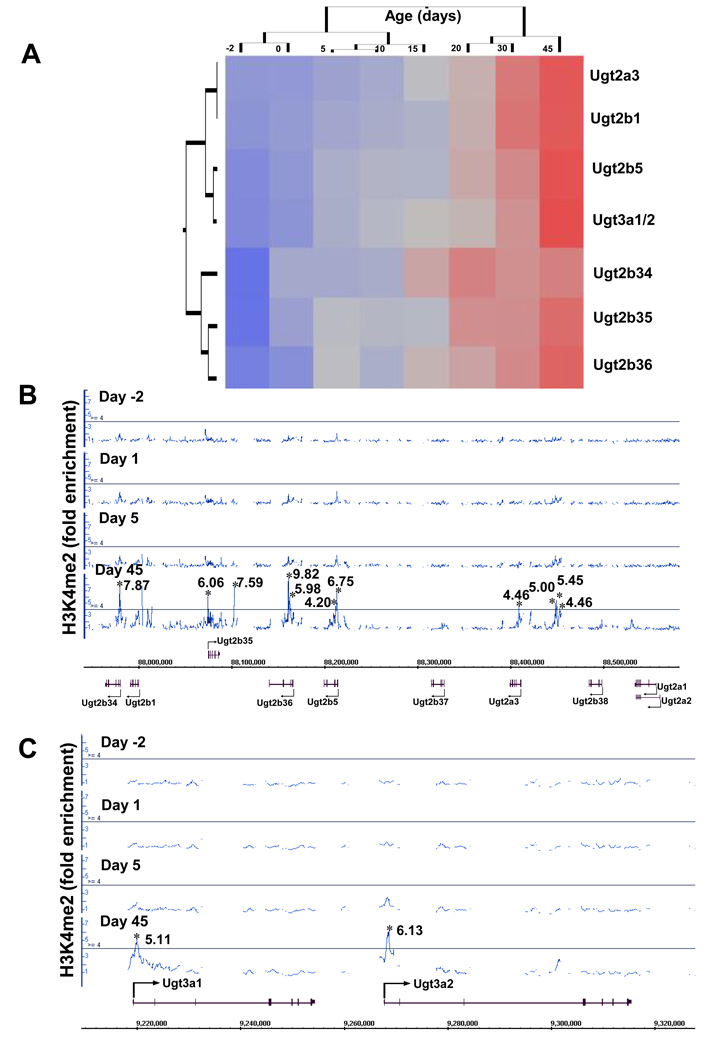
For phase-II drug and xenobiotic metabolizing enzymes, unpublished data from this laboratory showed that the adult-enriched permissive signal H3K4me2, in the absence of suppressive signals like DNA methylation or H3K27me3 at any age, marks the adult-specific expression of Ugt2 and Ugt3 (UDP-glucuronosyltransferase 2 and 3) gene polycistron clusters in mouse liver (Fig. 5A–C; supplemental Fig. 1). Taken together, the “time-clock” for the ontogeny of drug-metabolizing enzymes appears to be at least in part determined by distinct epigenetic signatures.
Figure 5.
Hepatic mRNA expression and enrichment of H3K4me2 around the Ugt2 and Ugt3 gene loci during development. (A) The ontogeny of Ugt2 and Ugt3 mRNAs in liver (from a sample size of n=5). The mRNA expression was determined by bDNA (branched DNA) assay. The average values were analyzed by a two-way hierachical clustering method (JMP v. 7.0) using Ward’s minimum variance and visualized by a dendrogram, which revealed adult-enriched expression patterns of these Ugts. Distances between genes reflect significance of associations. Blue color: low expression; red color: high expression. (B) and (C) H3K4me2 at the Ugt2 (B) and Ugt3 (C) gene loci during mouse liver development. ChIP-on-chip data were visualized by the Affymetrix Integrated Genome Browser (IGB) for H3K4me2 fold changes at day −2, 1, 5, and 45 of age. Solid lines through the signal enrichment peaks indicate the threshold value (4.0-fold compared to input background) for enriched intervals. Asterisks (*) indicate the peak center.
Just like the drug- and xenobiotic-metabolizing enzymes, the regulation of various drug and xenobiotic transporters also show an epigenetic component. Hypermethylation and silencing of Cd uptake transporter gene can induce Cd-resistant phenotype in cells that lack the Cd-binding protein metallothionein (MT), as was demonstrated by the MT-null yet Cd-resistant mouse A7 cells. In these cells, the Zn/Cd uptake transporter ZIP8 is not expressed because the slc39A8 gene that encodes ZIP8 is hypermethylated and silenced. Treatment of these cells with 5-aza-2′-deoxycytidine (DNMT inhibitor and a demethylating agent) reversed the silencing and resulted in an enhanced ZIP8 mRNA and protein expression, increased Cd uptake and accumulation, and increased sensitivity of these cells to Cd-induced toxicity (Fujishiro et al., 2009). This study demonstrates that an epigenetic mechanism can solely determine a specific cellular phenotype.
Recently, Imai et al. (2009) has reported the analysis of DNA methylation and histone modification profiles of various mouse liver-specific transporter genes, such as Oatp1b2 (Slco1b2), Ntcp (Slc10a1), Bsep (Abcb11), Pept2 (Slc15a2), and Abcg5 and Abcg8. Methylation analysis around the transcription start site (TSS) of these genes in liver, kidney and cerebrum showed that the CpG dinucleotides around the TSS of Oatp1b2, Ntcp, Bsep, and Abcg5/8 are hypomethylated in the liver but hypermethylated in the kidney and cerebrum. The opposite pattern was observed for Pept2, which is expressed in the kidney and cerebrum but not in the liver. Thus, the CpG methylation pattern directly correlates with the expression pattern of these transporters. Promoter histone modification status also correlates well with the expression of these transporters. Chromatin immunoprecipitation demonstrated histone H3 hyperacetylation in the promoters of hepatic Oatp1b2, Ntcp, Bsep and Abcg5/8, but little acetylation in the kidney and cerebrum. In contrast, the upstream region of Pept2 is hyperacetylated only in the kidney and cerebrum where it is expressed.
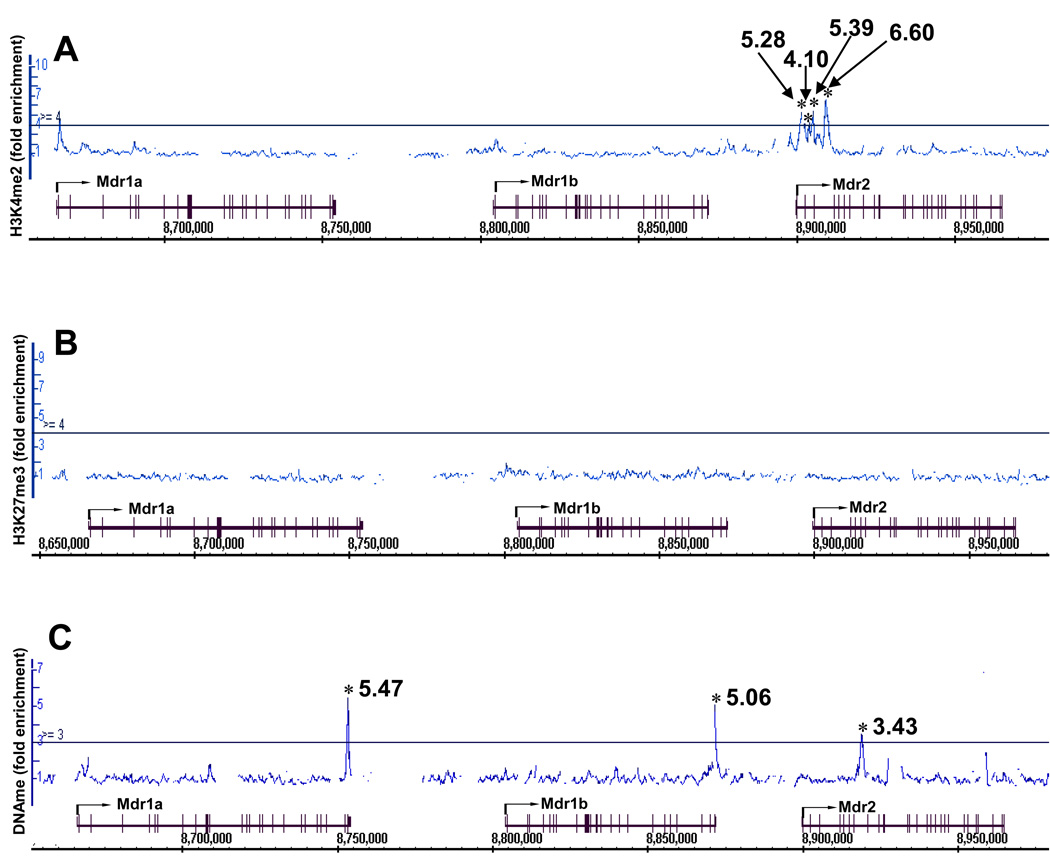
Data from our laboratory show that in mouse liver, the efflux transporters Mdr1a and 1b mRNAs are expressed at a low level but Mdr2 mRNA is expressed at a high level and is induced right after birth (Cui et al., 2009). Distinct epigenetic signatures were also identified around the Mdr gene cluster. The raw data (CEL files) on the epigenetic signatures on chromosome 5, where Cyp3a and Mdr genes are localized, were generated in our laboratory by ChIP-on-chip and published online in the Gene Expression Ominibus (GEO) database. The methods for expression and ChIP-on-chip analysis were described in two of our recent publications (Cui et al., 2009; Li et al., 2009). For this review, the CEL files were retrieved from the GEO database (accession number: GSE14620), and the epigenetic signatures around the Mdr gene loci were analyzed by the Integrated Genome Browser (IGB) and shown in Fig. 6. The permissive signal H3K4me2 was only observed in the Mdr2 gene locus, which is highly expressed (Fig. 6A); the non-permissive signal H3K27me3 was not observed at any regions within the gene cluster (Fig. 6B). In addition, DNA hypermethylation was observed within the 3′-UTR regions of Mdr1a and 1b, which are expressed at low mRNA levels. In contrast, DNA methylation was observed in the intragenic region of Mdr2 with relatively less enrichment (Fig. 6C). Thus, differential epigenetic signatures appear to correlate, at least in part, with the expressions of the Mdr genes in liver. Therefore, the data from the above studies demonstrate the existence of distinct epigenetic components in the regulation of tissue-specific expression of many transporters. Conceivably, the same principle may also apply for transporters of other species including humans.
Figure 6.
Distinct epigenetic signatures around the Mdr gene cluster in adult mouse liver (day 45 of age) (Mdr1a, 1b and 2). (A) H3K4me2, (B) H3K27Me3 and (C) DNA methylation (DNAme). ChIP-on-chip data were visualized by the Affymetrix Integrated Genome Browser (IGB) for H3K4me2, H3K27me3, and DNAme enrichment. Solid lines through the signal enrichment peaks indicate the threshold value (4.0-fold compared to input background for histone methylations, and 3.0-fold for DNAMe) for enriched intervals. Asterisks (*) indicate the peak center.
A similar promoter methylation-driven regulation of expression was reported earlier for mouse Abcc6 (Mrp6) gene (Douet et al., 2007), and human OAT3 gene (Kikuchi et al., 2006). In mouse, high and moderate levels of methylation of the Abcc6 promoter correlate with low levels of Abcc6 expression. Abcc6 expression in kidney, tail extremity, and skin was determined to be ~5%, 1%, and 0.1% of that in liver where it is expressed at the highest level. The mechanism of repression of Abcc6 gene expression was found to be CpG methylation-driven interference of the binding of the transcription factor Sp1, thereby inhibiting Sp1-dependent transcription. The epigenetic aspect of regulation of various drug and xenobiotic metabolizing enzymes and transporters may provide an explanation of the observed inter-individual variability in their expression, and further modulate an individual’s genetic ability to cope with environmental stress.
Epigenetic changes associated with diseased states
Epigenetic changes have been implicated in various disease etiologies, such as cancer, several developmental syndromes, cardiovascular diseases, type-2 diabetes, and obesity. Whereas the association between epigenetic changes and diseases other than cancer are increasingly populating the literature, studies on the epigenetics of cancer have the longest history. Therefore, the following discussion focuses on cancer-associated epigenetic changes. Because epigenetic changes can be reversed, altering disease-related epigenetic changes may influence the clinical outcome, thereby raising the possibility of epigenetic therapies for different diseases.
In cancer, the normal epigenetic landscape, that is, the pattern of DNA methylation, histone modifications, miRNA expression, as well as the expression of DNA methylation and histone modification enzymes are significantly altered. These epigenetic changes or epimutations, together with genetic alterations play an important role in the initiation and progression of cancer (Jones and Baylin, 2002). Most epigenetic changes translate into either up-regulation or silencing of gene expression which, in turn, may predispose the organism to mutational events, reduced ability to repair DNA damage, increased genomic instability, altered cellular response, such as aberrant cellular signaling, reduced apoptosis; all of these changes can contribute to tumorigenesis. DNA hypomethylation at repeat sequences can cause chromosomal rearrangements, thereby promoting genomic instability (Eden et al., 2003). Promoter hypermethylation and consequent silencing of the DNA mismatch repair gene (a tumor suppressor gene) MLH1 (mutL homolog 1) is also associated with microsatellite instability observed in colorectal cancer (Herman et al., 1998). An example of epigenetic changes resulting in mutation in DNA comes from the MGMT (O6-methylguanine-DNA methyltransferase) gene, which encodes the MGMT protein. MGMT protein removes the carcinogen-induced O6-methylguanine adducts from DNA. Failure to repair this DNA lesion results in G→A transition. Hypermethylation and silencing of MGMT gene increases the mutation rate in critical cellular regulators, including tumor suppressor genes and oncogenes (Estellar et al., 2001). Epigenetic changes may also precipitate a genetic (mutation) event by creating a platform, such as hypermethylated DNA, for the effector mediating the genetic change. For example, a metabolite of benzo[a]pyrene (B[a]P) is B[a]P diol epoxide (BPDE). BDPE is found in tobacco smoke and also in some overcooked food sources and it exhibits binding preference for methylated CpG sites. BPDE binding to methylated CpG sites results in the formation of increased DNA adducts and G→T transversions, which is often found in cancers of the aero-digestive tract in tobacco smokers (Yoon et al., 2001).
DNA hypermethylation and hypomethylation, histone modifications, and altered expression pattern of ncRNAs are all observed in cancers. A cancer epigenome is marked by genome-wide hypomethylation and site-specific promoter hypermethylation. Whereas DNA hypomethylation can lead to the activation of proto-oncogenes and growth promotion, DNA hypermethylation can down-regulate the expression of tumor suppressor genes. Examples of proto-oncogene activation in cancers include R-Ras in gastric cancer (Nishigaki et al., 2005), c-Neu in transgenic mouse models (Zhou et al., 2001), Hox11 in leukemia (Watt et al., 2000). A recent report by Smith et al. (2009) showed that aberrant expression of the transcription factor BORIS (Brother Of the Regulator of Imprinted Sites, which is a paralogue of the transcription factor CTCF, and is a novel member of the cancer-testis antigen family) results in coordinated promoter demethylation and simultaneous transcriptional up-regulation of a number of candidate proto-oncogenes and cancer testis antigens, including TKTL1 (transketolase-like enzyme 1), H19, different MAGE-A (melanoma associated antigen-A) family members (MAGE-A2, MAGE-A3/6, MAGE-A4, MAGE-A11). The upregulation was observed in multiple human malignancies including primary non-small cell lung cancers and head and neck squamous cell carcinoma. The authors performed functional validation of these genes using transient transfections to evaluate and/or confirm their growth promoting effects. Various tumor suppressor genes, such as RB (retinoblastoma), p16, MLH1 (mutL homolog1) and BRCA1 (breast cancer 1, early onset) undergo tumor-specific silencing by hypermethylation in human cancers (Sharma et al., 2010). Epigenetic silencing of tumor suppressor genes may provide the second hit in Knudson’s “two-hit” model of carcinogenesis (Knudson, 1971).
An example of epigenetic regulation by miRNA is provided by the regulation of Bcl2 gene and altered apoptosis of cancer cells by miR-15a and miR-16-1 (Calin et al., 2002; Cimmino et al., 2005; Iorio et al., 2005). miR-15a and miR-16-1 are tumor suppressor microRNAs (miRNAs) that can bind to the 3′-UTR of Bcl2 mRNA and down-regulate the expression of Bcl2 protein. About 65% of the patients diagnosed with B-cell chronic lymphocytic leukemia (B-CLL) have deletion or down-regulation of miR-15a and miR-16-1 genes located in a cluster in chromosome 13q14.3. The CLL cells also have a higher level of Bcl2 protein expression (Calin et al., 2002; Cimmino et al., 2005; Iorio et al., 2005). In a study using the leukemia-derived cell line MEG-01, which has no constitutive expression of miR-15a and miR-16-1 genes but has high level expression of Bcl2 gene and protein, Cimmino et al. (2005) demonstrated that transfecting this cell line with both miR-15a and miR-16-1 genes resulted in the expression of miR-15a and miR-16-1 with a concomitant decrease in Bcl2 protein expression, but no change in Bcl2 mRNA expression. Antisense knockdown of miR-15a and miR-16-1 expression resulted in an increase in Bcl2 protein, but no change in Bcl2 mRNA level. These results demonstrated that miR-15a and miR-16-1 are negative regulators of the anti-apoptotic protein Bcl2. The authors found a 9-bp Bcl2–complementarity sequence (the seed sequence) in these miRNAs. Because Bcl2 is an anti-apoptotic protein, repression of Bcl2 protein expression by miR-15a and miR-16-1 results in the apoptosis of cancerous cells. In addition to post-transcriptional gene silencing, miRNAs can also induce transcriptional gene silencing, likely through the binding of miRNA to the promoter of the target gene.
DISCUSSION
Epigenetic changes can be acquired over a period of time during the life of an individual and then passed on to the offspring. Thus, despite the triumph of Darwinism, epigenetics has ironically brought back Lamarckian flavor to biology.
Alterations in epigenetic pathways have been implicated in human diseases, especially in various types of cancers. In vitro, in vivo and epidemiological studies demonstrate that exposure to xenobiotics is frequently associated with various epigenetic changes. Once established, epigenetic marks can be transmitted to the offspring, and it is this transgenerational persistence of epigenetic changes and its potential impact that make the study of environmental effects on epigenetic regulation an important aspect of environmental and molecular toxicological studies.
Although the functional aspects of DNA methylation and various histone modifications are well documented, in the context of xenobiotic exposure a large number of the observed epigenetic changes in various studies have not been functionally linked to observed downstream events. Establishing a cause-and-effect relationship between xenobiotic exposure, epigenetic changes and physiological/pathological consequences becomes further confounded by the fact that epigenetic changes can be acquired over a period of time during the life of an organism. Documented associations between exposure to carcinogens, DNA methylation, and the onset of cancers have not always led to any mechanistic insight (Issa, 2004). Also, the differences in global CpG island methylation patterns between normal and cancer cells remain poorly understood (Toyota and Issa, 1999). Thus, even if specific epigenetic changes can be linked to downstream pathological events, it still remains unclear whether there is a temporal switch, or a threshold of epigenetic changes or specific combinations of epigenetic changes (epigenetic code) needed to trigger downstream events leading to specific pathological states. In many instances, an environmental chemical may have paradoxical epigenetic effects. For example, arsenic and nickel deplete cellular SAM levels, reduce DNMT activity, and causes global hypomethylation, which can all be mechanistically linked. However, at the same time, they also cause hypermethylation of specific gene promoters. Similarly, acute cadmium exposure results in DNA hypomethylation and decreased DNMT activity, whereas chronic cadmium exposure results in DNA hypermethylation and increased DNMT activity. The molecular regulation of such paradoxical effects remains to be explained.
The prevailing concept of DNA methylation as a static epigenetic mark on the chromatin may also be an oversimplification. Demonstration of periodic methylation and demethylation of transcriptionally active promoters with a periodicity of tens of minutes, brought to light some unanticipated dynamic role of DNA methylation in gene regulation in human cells (Kangaspeska et al., 2008; Métivier et al., 2008). Using human and mouse cell lines, Kim et al. (2009) recently described the mechanism of action of a methylation-demethylation switch for the cytochrome P450 gene CYP27B1. CYP27B1 [25(OH)D3 1α-hydroxylase] biotransforms vitamin D3 into its active form, 1α,25(OH)2D3 (Sawada et al., 2001). CYP27B1 expression is repressed by vitamin D and stimulated by parathyroid hormone (PTH). Vitamin D-mediated repression occurs via vitamin D receptor-interacting repressor (VDIR) complex that binds to the cis-acting negative vitamin D response element (nVDRE) of CYP27B1 gene promoter, and recruits DNMTs 1 and 3a, which methylate the DNA and cause repression of gene expression. PTH treatment stimulates CYP27B1 expression by causing dissociation of DNMTs from the VDIR complex, and triggering the phosphorylation of MBD4, which remains associated with the VDIR complex. Phosphorylation induces the glycosylase acivity of MBD4, which then actively demethylates DNA by a base excision repair mechanism.
Studies should also be designed to address whether the same disease outcome in subjects exposed to different environments (including different geographic locations) shows the same or similar epigenetic changes. In other words, there is a need to coordinate epidemiological epigenetic studies. For example, examining 85 tumor samples of hepatocellular carcinoma patients from China, Egypt, U.K. plus Europe, and the U.S., Shen et al. (2002) found a significant difference in p16 methylation in tumors from China and Egypt (34.4%) compared to tumors from Europe and the U.S. (12.2%).
Most of the studies on environmental epigenetics concentrated on documenting changes in DNA methylation and histone modifications. However, some recent reports also documented the epigenetic effects of chemicals mediated by ncRNA expression; for example, the expression of specific miRNAs (in response to RDX exposure) that can potentially mediate RDX-induced neurotoxicity through the modulation of BDNF mRNA (Zhang and Pan, 2009); Wy-14,643 (PPAR-alpha agonist)-mediated expression of the proto-oncogenic miR-17–92 cluster, which results in hepatocyte proliferation and liver carcinomas (Shah et al., 2007). The emerging significance of small ncRNA expression in toxicology were recently discussed in two reviews (Hudder and Novak, 2008; Choudhuri, 2010).
The study of epigenetic regulation of genome expression is currently undergoing major progress thanks to the development of new techniques. The study by Christensen et al. (2009) underscores the need for more studies to define the “normal” epigenetic state, if anything of that nature can be consistently defined. With our increasing ability to study epigenetic changes in response to xenobiotic exposure, and our increasing understanding of the effects of such changes in regulating genome expression, it is time that such changes are thoroughly documented. Such documentation, combined with an understanding of “normal” epigenetic state is probably the first step towards understanding and systematically analyzing the impact of environment in epigenetic regulation, and its potential physiological/pathological consequences.
Nevertheless, advances in epigenetics have provided a molecular basis for explaining the “nurture” element of the “Nature (genetics) versus Nurture (environment)” debate, and helped transform this debate from a philosophical one to a scientific one. What remains to be unveiled is the regulation of the regulator, that is, which and how signals induce epigenetic changes that, in turn, edit and modify the language of DNA.
Supplementary Material
Supplemental Figure 1: Absence of H3K27me3 and DNAme around the Ugt2 and Ugt3 gene loci during liver development. (A) H3K27me3 around the Ugt2 gene locus from day -2 to day 45 of age. (B) H3K27me3 around the Ugt3 gene locus from day -2 to day 45 of age. (C) DNAme around the Ugt2 gene locus from day -2 to day 45 of age. (D) DNAme around the Ugt3 gene locus from day -2 to day 45 of age. ChIP-on-chip data were visualized by the Affymetrix Integrated Genome Browser (IGB). Solid lines through the signal enrichment peaks indicate the threshold value (4.0-fold compared to input background) for enriched intervals. Asterisks (*) indicate the peak center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The opinions expressed in this article are the authors’ personal opinions and do not reflect those of FDA, DHHS, or the Federal Government
The expressions SUV or Suv or Su(var) are shorter versions for “suppression of variegation”. It is discussed below under “some examples of epigenetic phenomena”.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
REFERENCES
- Aagaard-Tillery KM, Grove K, Bishop J, Ke X, Fu Q, McKnight R, Lane RH. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. J. Mol. Endocrinol. 2008;41:91–102. doi: 10.1677/JME-08-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnouti Y, Patrick JS, Klaassen CD. Tissue distribution and ontogeny of organic cation transporter in mice. Drug. Metab. Disp. 2006;34:477–482. doi: 10.1124/dmd.105.006932. [DOI] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- Brown SE, Szyf M. Dynamic epigenetic states of ribosomal RNA promoters during the cell cycle. Cell Cycle. 2008;7:382–390. doi: 10.4161/cc.7.3.5283. [DOI] [PubMed] [Google Scholar]
- Burdge GC, Slater-Jefferies J, Torrens C, Phillips ES, Hanson MA, Lillycrop KA. Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br. J. Nutr. 2007;97:435–439. doi: 10.1017/S0007114507352392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callinan PA, Feinberg AP. The emerging science of epigenomics. Hum. Mol. Genet. 2006;15:R95–R101. doi: 10.1093/hmg/ddl095. [DOI] [PubMed] [Google Scholar]
- Cheng RY, Hockman T, Crawford E, Anderson LM, Shiao YH. Epigenetic and gene expression changes related to transgenerational carcinogenesis. Mol. Carcinogen. 2004;40:1–11. doi: 10.1002/mc.20022. [DOI] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, Sugarbaker DJ, Yeh RF, Wiencke JK, Kelsey KT. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri S. Epigenetic regulation of gene and genome expression. In: Choudhuri S, Carlson DB, editors. Genomics: Fundamentals and Applications. Vol. 2009. NY: Informa Healthcare; 2009a. pp. 101–128. [Google Scholar]
- Choudhuri S. Lesser known relatives of miRNA. Biochim. Biophys. Res. Commun. 2009b;388:177–180. doi: 10.1016/j.bbrc.2009.08.039. [DOI] [PubMed] [Google Scholar]
- Choudhuri S. Small non-coding RNAs: biogenesis, function and emerging significance in toxicology. J. Biochem. Mol. Toxicol. 2010;24:195–216. doi: 10.1002/jbt.20325. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews D, Gore AC, Hsu TS, Dangleben NL, Spinetta M, Schallert T, Anway MD, Skinner MK. Transgenerational epigenetic imprints on mate preference. Proc. Natl. Acad. Sci. USA. 2007;104:5942–5946. doi: 10.1073/pnas.0610410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SH, Meehan RR, Nan X, Bird A. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat. Genet. 1997;16:256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- Cui YJ, Cheng X, Weaver YM, Klaassen CD. Tissue distribution, gender-divergent expression, ontogeny, and chemical induction of multidrug resistance transporter genes (Mdr1a, Mdr1b, Mdr2) in mice. Drug Metab. Dispos. 2009;37:203–210. doi: 10.1124/dmd.108.023721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M. Emerging roles of the SUMO pathway in mitosis. Cell Div. 2008;3:5. doi: 10.1186/1747-1028-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Bannister AJ, Göttgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bustos C, Ramos E, Young JM, Tran RK, Menzel U, Langford CF, Eichler EE, Hsu L, Henikoff S, Dumanski JP, Trask BJ. Tissue-specific variation in DNA methylation levels along human chromosome 1. Epigenetics Chromatin. 2009;2:7. doi: 10.1186/1756-8935-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding YF, Zhu C. The role of microRNAs in copper and cadmium homeostasis. Biochem. Biophys. Res. Commun. 2009;386:6–10. doi: 10.1016/j.bbrc.2009.05.137. [DOI] [PubMed] [Google Scholar]
- Douet V, Heller MB, Le Saux O. DNA methylation and Sp1 binding determine the tissue-specific transcriptional activity of the mouse Abcc6 promoter. Biochem. Biophys. Res. Commun. 2007;354:66–71. doi: 10.1016/j.bbrc.2006.12.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS. Neomorphic agouti mutations in obese yellow mice. Nat. Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- Duncan IW. Transvection effects in Drosophila. Annu. Rev. Genet. 2002;36:521–556. doi: 10.1146/annurev.genet.36.060402.100441. [DOI] [PubMed] [Google Scholar]
- Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- Edwards TM, Myers JP. Environmental exposures and gene regulation in disease etiology. Environ. Health Perspect. 2007;115:1264–1270. doi: 10.1289/ehp.9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Risques RA, Toyota M, Capella G, Moreno V, Peinado MA, Baylin SB, Herman JG. Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res. 2001;61:4689–4692. [PubMed] [Google Scholar]
- Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat. Rev. Genet. 2004;5:446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. A brief history of epigenetics. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor, NY: CSHL Press; 2007. pp. 15–22. [Google Scholar]
- Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, Corrocher R, Selhub J. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc. Natl. Acad. Sci. USA. 2002;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishiro H, Okugaki S, Yasumitsu S, Enomoto S, Himeno S. Involvement of DNA hypermethylation in down-regulation of the zinc transporter ZIP8 in cadmium-resistant metallothionein-null cells. Toxicol. Appl . Pharmacol. 2009;241:195–201. doi: 10.1016/j.taap.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Galvão TC, Thomas JO. Structure-specific binding of MeCP2 to four-way junction DNA through its methyl CpG-binding domain. Nucleic Acids Res. 2005;33:6603–6609. doi: 10.1093/nar/gki971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Dominguez M, Reyes JC. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim. Biophys. Acta. 2009;1789:451–459. doi: 10.1016/j.bbagrm.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Govindarajan B, Klafter R, Miller MS, Mansur C, Mizesko M, Bai X, LaMontagne K, Jr, Arbiser JL. Reactive oxygen-induced carcinogenesis causes hypermethylation of p16(Ink4a) and activation of MAP kinase. Mol. Med. 2002;8:1–8. [PMC free article] [PubMed] [Google Scholar]
- Hammons GJ, Yan-Sanders Y, Jin B, Blann E, Kadlubar FF, Lyn-Cook BD. Specific site methylation in the 5'-flanking region of CYP1A2 interindividual differences in human livers. Life Sci. 2001;69:839–845. doi: 10.1016/s0024-3205(01)01175-4. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Abbott C, McQueen H, Chambers D, Cross S, Bird A. Genomic structure and chromosomal mapping of the murine and human Mbd1, Mbd2, Mbd3, and Mbd4 genes. Mamm. Genome. 1999a;10:906–912. doi: 10.1007/s003359901112. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999b;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Comai L. Trans-sensing effects: the ups and downs of being together. Cell. 1998;93:329–332. doi: 10.1016/s0092-8674(00)81161-7. [DOI] [PubMed] [Google Scholar]
- Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007;22:91–103. doi: 10.1093/mutage/gel068. [DOI] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB. Incidence and functional consequences of hMLH1 promoter in colorectal carcinoma. Proc. Natl. Acad. Sci. USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudder A, Novak RF. miRNAs: effectors of environmental influences on gene expression and disease. Toxicol. Sci. 2008;103:228–240. doi: 10.1093/toxsci/kfn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP. CpG islands- ‘A rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Imai S, Kikuchi R, Kusuhara H, Yagi S, Shiota K, Sugiyama Y. Analysis of DNA methylation and histone modification profiles of liver-specific transporters. Mol. Pharmacol. 2009;75:568–576. doi: 10.1124/mol.108.052589. [DOI] [PubMed] [Google Scholar]
- Inawaka K, Kawabe M, Takahashi S, Doi Y, Tomigahara Y, Tarui H, Abe J, Kawamura S, Shirai T. Maternal exposure to anti-androgenic compounds, vinclozolin, flutamide and procymidone, has no effects on spermatogenesis and DNA methylation in male rats of subsequent generations. Toxicol. Appl. Pharmacol. 2009;237:178–187. doi: 10.1016/j.taap.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Issa JP. CpG island methylator phenotype in cancer. Nat. Rev. Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- Jin B, Park DW, Nam KW, Oh GT, Lee YS, Ryu DY. CpG methylation of the mouse CYP1A2 promoter. Toxicol. Lett. 2004;152:11–18. doi: 10.1016/j.toxlet.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Métivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- Keppler BR, Archer TK. Chromatin-modifying enzymes as therapeutic targets - Part 1. Expert Opin. Ther. Targets. 2008a;12:1301–1312. doi: 10.1517/14728222.12.10.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler BR, Archer TK. Chromatin-modifying enzymes as therapeutic targets – Part 2. Expert Opin. Ther. Targets. 2008b;12:1457–1467. doi: 10.1517/14728222.12.11.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi R, Kusuhara H, Hattori N, Shiota K, Kim I, Gonzalez FJ, Sugiyama Y. Regulation of the expression of human organic anion transporter 3 by hepatocyte nuclear factor 1alpha/beta and DNA methylation. Mol. Pharmacol. 2006;70:887–896. doi: 10.1124/mol.106.025494. [DOI] [PubMed] [Google Scholar]
- Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, Matsumoto T, Fujiyama S, Shirode Y, Yamaoka I, Kitagawa H, Takeyama K, Shibuya H, Ohtake F, Kato S. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–1012. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- Lerach S, Zhang W, Bao X, Deng H, Girton J, Johansen J, Johansen KM. Loss-of-function alleles of the JIL-1 kinase are strong suppressors of position effect variegation of the wm4 allele in Drosophila. Genetics. 2006;173:2403–2406. doi: 10.1534/genetics.106.059253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bird AP. DNA methylation in mammals. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor, NY: CSHL Press; 2007. pp. 341–356. [Google Scholar]
- Li Y, Cui Y, Hart SN, Klaassen CD, Zhong XB. Dynamic patterns of histone methylation are associated with ontogenic expression of the Cyp3a genes during mouse liver maturation. Mol. Pharmacol. 2009;75:1171–1179. doi: 10.1124/mol.108.052993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J. Nutr. 2005;135:1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, Huang TC, Unterberger A, Suderman M, Ernst C, Meaney MJ, Turecki G, Szyf M. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS One. 2008;3:e2085. doi: 10.1371/journal.pone.0002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memili E, Hong YK, Kim DH, Ontiveros SD, Strauss WM. Murine Xist RNA isoforms are different at their 3' ends: a role for differential polyadenylation. Gene. 2001;266:131–137. doi: 10.1016/s0378-1119(01)00353-5. [DOI] [PubMed] [Google Scholar]
- Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Morris JR, Chen J-L, Geyer PK. Two modes of transvection: enhancer action in trans and bypass of a chromatin insulator in cis. Proc. Natl. Acad. Sci. USA. 1998;95:10740–10745. doi: 10.1073/pnas.95.18.10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Nikitina T, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Grigoryev SA, Woodcock CL. MeCP2-chromatin interactions include the formation of chromatosome-like structures and are altered in mutations causing Rett syndrome. J. Biol. Chem. 2007;282:28237–28245. doi: 10.1074/jbc.M704304200. [DOI] [PubMed] [Google Scholar]
- Nishigaki M, Aoyagi K, Danjoh I, Fukaya M, Yanagihara K, Sakamoto H, Yoshida T, Sasaki H. Discovery of aberrant expression of R-RAS by cancer-linked DNA hypomethylation in gastric cancer using microarrays. Cancer Res. 2005;65:2115–2124. doi: 10.1158/0008-5472.CAN-04-3340. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 1998;26:2536–2540. doi: 10.1093/nar/26.11.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okino ST, Pookot D, Li LC, Zhao H, Urakami S, Shiina H, Igawa M, Dahiya R. Epigenetic inactivation of the dioxin-responsive cytochrome P4501A1 gene in human prostate cancer. Cancer Res. 2006;66:7420–7428. doi: 10.1158/0008-5472.CAN-06-0504. [DOI] [PubMed] [Google Scholar]
- Pan YZ, Gao W, Yu AM. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab . Dispos. 2009;37:2112–2117. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauler FM, Barlow DP. Imprinting mechanisms– it only takes two. Genes Dev. 2006;20:1203–1206. doi: 10.1101/gad.1437306. [DOI] [PubMed] [Google Scholar]
- Pirotta V. Transvection and chromosomal trans-interaction effects. Biochim. Biophys Acta. 1999;1424:M1–M8. doi: 10.1016/s0304-419x(99)00019-0. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reamon-Buettner SM, Mutschler V, Borlak J. The next innovation cycle in toxicogenomics: environmental epigenetics. Mutat. Res. 2008;659:158–165. doi: 10.1016/j.mrrev.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Reinberg D. Epigenetics: From phenomenon to field. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Vol. 2007. Cold Spring Harbor, NY: CSHL Press; 2007. pp. 1–13. [Google Scholar]
- Riggs AD, Martienssen RA, Russo VEA. Introduction. In: Russo VEA, et al., editors. Epigenetic Mechanisms of Gene Regulation. Vol. 1996. Cold Spring Harbor, NY: CSHL Press; 1996. pp. 1–4. [Google Scholar]
- Rodriguez-Antona C, Gomez A, Karlgren M, Sim SC, Ingelman-Sundberg M. Molecular genetics and epigenetics of the cytochrome P450 gene family and its relevance for cancer risk and treatment. Hum. Genet. 2010;127:1–17. doi: 10.1007/s00439-009-0748-0. [DOI] [PubMed] [Google Scholar]
- Sawada N, Sakaki T, Kitanaka S, Kato S, Inouye K. Structure-function analysis of CYP27B1 and CYP27A1. Studies on mutants from patients with vitamin D-dependent rickets type I (VDDR-I) and cerebrotendinous xanthomatosis (CTX) Eur. J. Biochem. 2001;268:6607–6615. doi: 10.1046/j.0014-2956.2001.02615.x. [DOI] [PubMed] [Google Scholar]
- Schneider S, Kaufmann W, Buesen R, van Ravenzwaay B. Vinclozolin-the lack of a transgenerational effect after oral maternal exposure during organogenesis. Reprod. Toxicol. 2008;25:352–360. doi: 10.1016/j.reprotox.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Schnekenburger M, Peng L, Puga A. HDAC1 bound to the Cyp1a1 promoter blocks histone acetylation associated with Ah receptor-mediated trans-activation. Biochim. Biophys. Acta. 2007;1769:569–578. doi: 10.1016/j.bbaexp.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr CJ, Levorse JM, Tilghman SM. CTCF maintains differential methylation at the Igf 2/H19 locus. Nat. Genet. 2003;33:66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- Sengupta SR, Das NK, Datta PK. Pathogenesis, clinical features and pathology of chronic arsenicosis. Indian J. Dermatol. Venereol . Leprol. 2008;74:559–570. [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah YM, Morimura K, Yang Q, Tanabe T, Takagi M, Gonzalez FJ. Peroxisome proliferator-activated receptor alpha regulates a microRNA-mediated signaling cascade responsible for hepatocellular proliferation. Mol. Cell Biol. 2007;27:4238–4247. doi: 10.1128/MCB.00317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kelly TK, Jones PA. Epigenetics in Cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Ahuja N, Shen Y, Habib NA, Toyota M, Rashid A, Issa JP. DNA methylation and environmental exposures in human hepatocellular carcinoma. J. Natl. Cancer Inst. 2002;94:755–761. doi: 10.1093/jnci/94.10.755. [DOI] [PubMed] [Google Scholar]
- Skinner MK, Anway MD, Savenkova MI, Gore AC, Crews D. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS One. 2008:e3745. doi: 10.1371/journal.pone.0003745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol. Metab. 2010;21:214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Smith IM, Glazer CA, Mithani SK, Ochs MF, Sun W, Bhan S, Vostrov A, Abdullaev Z, Lobanenkov V, Gray A, Liu C, Chang SS, Ostrow KL, Westra WH, Begum S, Dhara M, Califano J. Coordinated activation of candidate proto-oncogenes and cancer testes antigens via promoter demethylation in head and neck cancer and lung cancer. PLoS One. 2009;4:e4961. doi: 10.1371/journal.pone.0004961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Straussman R, Nejman D, Roberts D, Steinfeld I, Blum B, Benvenisty N, Simon I, Yakhini Z, Cedar H. Developmental programming of CpG island methylation profiles in the human genome. Nat. Struct. Mol. Biol. 2009;16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- Toyota M, Issa JP. CpG island methylator phenotypes in aging and cancer. Semin. Cancer Biol. 1999;9:349–357. doi: 10.1006/scbi.1999.0135. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66:9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell. Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- Wade PA. Methyl CpG-binding proteins and transcriptional repression. Bioessays. 2001;23:1131–1137. doi: 10.1002/bies.10008. [DOI] [PubMed] [Google Scholar]
- Wakefield RI, Smith BO, Nan X, Free A, Soteriou A, Uhrin D, Bird AP, Barlow PN. The solution structure of the domain from MeCP2 that binds to methylated DNA. J. Mol. Biol. 1999;291:1055–1065. doi: 10.1006/jmbi.1999.3023. [DOI] [PubMed] [Google Scholar]
- Watt PM, Kumar R, Kees UR. Promoter demethylation accompanies reactivation of the HOX11 proto-oncogene in leukemia. Genes Chromosomes Cancer. 2000;29:371–377. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1050>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Wise TL, Pravtcheva DD. The undermethylated state of a CpG island region in igf2 transgenes is dependent on the H19 enhancers. Genomics. 1999;60:258–271. doi: 10.1006/geno.1999.5921. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Smith LE, Feng Z, Tang M, Lee CS, Pfeifer GP. Methylated CpG dinucleotides are the preferential targets for G-to-T transversion mutations induced by benzo[a]pyrene diol epoxide in mammalian cells: similarities with the p53 mutation spectrum in smokingassociated lung cancers. Cancer Res. 2001;61:7110–7117. [PubMed] [Google Scholar]
- Zhang B, Pan X. RDX induces aberrant expression of microRNAs in mouse brain and liver. Environ. Health Perspect. 2009;117:231–240. doi: 10.1289/ehp.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Chen WD, Qin X, Lee K, Liu L, Markowitz SD, Gerson SL. MMTV promoter hypomethylation is linked to spontaneous and MNU assciated c-neu expression and mammary carcinogenesis in MMTV c-neu transgenic mice. Oncogene. 2001;20:6009–6017. doi: 10.1038/sj.onc.1204830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Absence of H3K27me3 and DNAme around the Ugt2 and Ugt3 gene loci during liver development. (A) H3K27me3 around the Ugt2 gene locus from day -2 to day 45 of age. (B) H3K27me3 around the Ugt3 gene locus from day -2 to day 45 of age. (C) DNAme around the Ugt2 gene locus from day -2 to day 45 of age. (D) DNAme around the Ugt3 gene locus from day -2 to day 45 of age. ChIP-on-chip data were visualized by the Affymetrix Integrated Genome Browser (IGB). Solid lines through the signal enrichment peaks indicate the threshold value (4.0-fold compared to input background) for enriched intervals. Asterisks (*) indicate the peak center.