Abstract
Motivation: Recently developed profile–profile methods rival structural comparisons in their ability to detect homology between distantly related proteins. Despite this tremendous progress, many genuine relationships between protein families cannot be recognized as comparisons of their profiles result in scores that are statistically insignificant.
Results: Using known evolutionary relationships among protein superfamilies in SCOP database, support vector machines were trained on four sets of discriminatory features derived from the output of HHsearch. Upon validation, it was shown that the automatic classification of all profile–profile matches was superior to fixed threshold-based annotation in terms of sensitivity and specificity. The effectiveness of this approach was demonstrated by annotating several domains of unknown function from the Pfam database.
Availability: Programs and scripts implementing the methods described in this manuscript are freely available from http://hhsvm.dlakiclab.org/.
Contact: mdlakic@montana.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Predicting protein functions by homology is still the only practical way of annotation that can keep up with the ever-increasing number of known protein sequences. Pairwise sequence-sequence similarity search tools such as FASTA (Pearson and Lipman, 1988) and BLAST (Altschul et al., 1990) are still the workhorses of genome annotations, in particular for sequences that share more than 30% identity. However, the performance of these methods is substandard for homologous proteins sharing <30% identity (Brenner et al., 1998), prompting the development of profile-based tools such as PSI-BLAST (Altschul et al., 1997), HMMer (Eddy, 1998) and SAM (Karplus et al., 1998). These profile-sequence methods detect many additional remote homologues missed by pairwise methods (Park et al., 1998) as they take advantage of the position-specific evolutionary information derived from the alignment of family members. Finally, profile–profile methods (Ginalski et al., 2004; Sadreyev and Grishin, 2003; Söding, 2005; Yona and Levitt, 2002) further improved our ability to detect remote yet biologically important relationships.
High-scoring and statistically significant hits from profile–profile comparisons are almost always indicative of true functional and structural relationships (Söding, 2005). Nevertheless, many biologically meaningful similarities between distantly related protein families are either completely missed, or have statistically insignificant scores that require further evidence before the homology can be inferred with certainty. Skilled practitioners of profile methods can recognize many of these additional homologues from alignments and similarities in secondary structures. This approach, however, is time-consuming and usually requires specific knowledge of proteins families in question. The whole process was recently automated by training support vector machines (SVMs) to recognize both true and false homologues from the PSI-BLAST output, with results often outperforming profile–profile methods (Shah et al., 2008).
It is intuitively clear that machine learning approaches could be trained to analyze the output of profile–profile comparisons and recognize the matches with high statistical significance, as low E-values alone are usually very discriminative without any additional features. However, in classifying true versus false homologues from the list of statistically insignificant hits, human experts use additional information available in the output such as conservation of functionally important residues and similar domain organizations. Since this decision-making process is difficult to formalize in terms of defined rules, we were interested to find out whether the human expertise in the ‘twilight zone’ of protein similarities could be replaced by discriminative machine learning approaches trained on defined sets of true and false homologues. To test this, we decided to use SVMs trained on the output of HHsearch (Söding, 2005), one of the recently developed state-of-the-art methods for profile–profile comparisons.
We started by creating profiles of protein domains classified in SCOP (Andreeva et al., 2008), and then used HHsearch to compare each of these profiles with the entire database. Known SCOP relationships between these proteins were used to create the sets of true and false homologues, and four SVMs trained on different groups of discriminatory features were tested for their ability to correctly classify the hits from the HHsearch output. We show that the best SVM classifier significantly outperforms automatic classifications based on fixed significance thresholds, both in terms of sensitivity and specificity. More importantly, the speed of classification and the availability of probability estimates for each prediction make this approach suitable for automatic parsing of the large number of profile outputs. This is illustrated by analyzing the outputs of all profiles from the Pfam database (Finn et al., 2008). We highlight several persuasive new relationships between protein families of unknown functions and the clan of PD-(D/E)xK nucleases (Knizewski et al., 2007).
2 METHODS
2.1 Training and testing datasets
SCOP classification at the superfamily level was used as a gold standard, because this is where the structural similarity between distantly related proteins can still be attributed to common ancestry. It is well known, however, that profile-based methods in some cases can identify relatedness between structurally divergent proteins classified into different SCOP superfamilies (Cheng et al., 2008; Madera and Gough, 2002; Reid et al., 2007; Söding, 2005). These exceptions were compiled by Dr Julian Gough as a set of rules which is meant to augment the SCOP classification at the superfamily level (http://www.supfam.org/SUPERFAMILY/ruleset_1.69.html). Based on these rules, all protein pairs in SCOP can be scored as related, ambiguous, or unrelated, and we eliminated all ambiguous pairs from the analysis to avoid complications in training and data interpretation.
SCOP database (v1.69) was clustered at 95% identity and the resulting 11 944 sequences were used as queries for the modified Target2K procedure (Karplus et al., 2003). Briefly, this procedure uses PSI-BLAST instead of WU-BLAST to search the protein database and automatically builds an alignment of identified homologues. Predicted secondary structure by PSI-PRED (Jones, 1999) was subsequently added to the alignments to generate profile hidden Markov models (HMMs) (Söding, 2005). This database of profile HMMs is likely to be more accurate in terms of starting alignments than the similar database distributed on the HHpred server (Söding et al., 2005). In both cases PSI-BLAST is used to collect remote homologues, but Target2K procedure realigns all identified sequences using SAM (Karplus et al., 2003). In order to maximize the number of detected homologues in the ‘twilight zone’ of similarities (Rost, 1999), HHsearch was run with E-value threshold of 100, which corresponds approximately to the P-value of 1×E−02). The probability of correct hits (switch −p) was set at 0.001 since HHsearch default threshold of 20 takes precedence over E-value and truncates the output at E-values smaller than 100.
Consensus sequences extracted from HMMs in Pfam 23.0 (Finn et al., 2008) were used as inputs for the modified Target2K procedure as described above. This group of profiles was compared in all-vs-all fashion using HHsearch, and the outputs were processed using SVMs. We were looking in particular for results that would help make sense of Pfam families presently annotated as domains of unknown function.
2.2 Training and testing SVMs
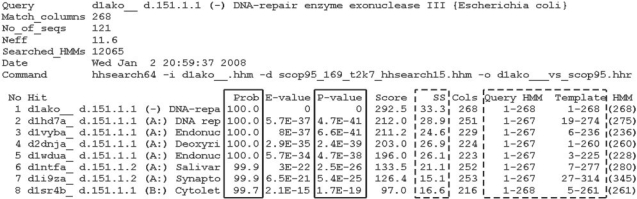
The output of HHsearch contains many useful discriminatory features, yet most users will rely on E-values (or P-values) and probabilities which indicate the likelihood of the correct match (full-line boxes in Fig. 1). These numbers provide statistical measures that are easy to understand even without looking at the underlying alignments generated during the scoring procedure. We tested several different SVMs in order to determine what features provide best discrimination between positive and negative samples. First SVM, termed HHsvm1, was trained using only P-values and probabilities of the true match as estimated by HHsearch (Söding, 2005). The reason we did not use E-values is because they change with the size of the database used for searching, while P-values are always consistent as they depend only on the internal calibration of HMMs. The second SVM, named HHsvm2, was trained by amplifying HHsvm1 with the secondary structure scores and coverage fractions of the query and its matches (columns boxed by dashed lines in Fig. 1). These features are available by directly parsing the output of HHsearch. Finally, we created HHsvm3 from HHsvm2 by adding average information contents of the complete query and its match, as well as average information contents of the query and hits over their aligned regions. Positional information content was calculated separately from profile HMMs and HHsearch alignments were used to determine the profile positions for averaging.
Fig. 1.
Opening lines of a typical HHsearch output file. Column Prob contains probabilities for the correct match as estimated by HHsearch (Söding, 2005). Values extracted from columns boxed by full lines were used for HHsvm1 training after calculating—log of the P-value (P-value of 0 was assumed to be 1×E−50). Values in columns boxed by dashed lines were used for HHsvm2 training. Query HMM and Template HMM values were converted into fractions of the full profile length.
All extracted numbers were scaled uniformly to fit the range from −1 to 1. A wide range of C and γ parameters for the radial basis function (RBF) kernel was probed first on a small number of training examples using the grid search and 5-fold validation (Chang and Lin, 2001). After finding the narrow range of parameters that resulted in best training, the grid search was repeated in finer steps using larger number of training examples. Finally, the whole dataset containing ∼1.7 million data points was divided in two halves by stratified sampling, which prevents rare target values from being excluded and ensures that proportions of classes in training and testing data are close to overall class fractions (Chang and Lin, 2001). One half was trained with C and γ parameters from the grid search that gave the best 5-fold validation accuracy, and the remaining half was used for testing.
After experimenting extensively with the RBF kernel, we tried the SVM formulation of an infinite ensemble framework. Infinite ensemble SVMs typically perform similarly to other popular SVM kernels yet train faster as only parameter C has to be optimized (Lin and Li, 2008). Somewhat surprisingly, the perceptron kernel within the infinite ensemble SVM framework consistently outperformed the RBF kernel in all quantitative aspects of our tests. This kernel was therefore used in our work and results obtained on the testing set are reported in Table 1. For the sake of comparison, we provide the summary of RBF kernel classifications in Supplementary Table 1.
Table 1.
A comparison of homology assignments using fixed P-value thresholds and HHsvm classifiers trained using the perceptron kernel
| Method | Accuracy | Sensitivity 1 | Specificity 1 | Sensitivity 0 | Specificity 0 |
|---|---|---|---|---|---|
| Fixed E−04 | 95.06 | 93.52 | 97.65 | 97.08 | 92.01 |
| HHsvm1 | 95.31 | 93.59 | 98.02 | 97.54 | 92.11 |
| HHsvm2 | 96.30 | 95.27 | 98.14 | 97.65 | 94.06 |
| HHsvm3 | 98.28 | 97.98 | 98.97 | 98.67 | 97.40 |
| HHsvm4 | 98.68 | 98.43 | 99.22 | 98.99 | 97.98 |
| HHsvm4-0.95 | 99.65 | 99.49 | 99.91 | 99.87 | 99.30 |
Row ‘Fixed E−04’ has the parameters for fixed P-value threshold of 1×E−04. HHsvm1 to HHsvm4 are for SVM classifiers as described in the text. Row HHsvm4-0.95 contains the parameters for HHsvm4 predictions made with probabilities ≥0.95.
The overall accuracy of classification was defined as (TP + TN)/(P + N), where TP and TN are numbers of entries that were correctly classified as positive and negative, respectively, while P and N are numbers of known positive and negative entries, respectively. The overall accuracy may be misleading for imbalanced datasets if the more abundant entries are classified much better. Since our training dataset had more homologous than non-homologous pairs, in addition to overall accuracy we show classification measures for individual categories. Sensitivity was defined as TP/P for homologous proteins (sensitivity 1) and TN/N for unrelated proteins (sensitivity 0). Specificity reflects the correctness of predictions and was calculated as TP/(TP + FP) for related proteins (specificity 1) and TN/(TN + FN) for unrelated proteins (specificity 0); FP stands for entries that were falsely classified as homologous and FN for those that were incorrectly classified as non-homologous.
3 RESULTS
3.1 Threshold-based assignment of HHsearch hits
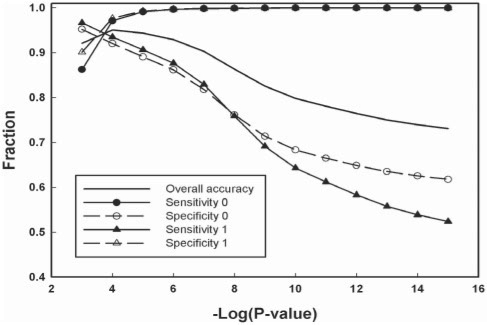
As a baseline for comparison with our method, we used the classification from fixed significance thresholds. We chose P-value thresholds instead of E-values because for HHsearch the former do not depend on the size of profile database and can be compared between different databases. Figure 2 shows overall accuracy, sensitivity and specificity as functions of the −log(P-value). As expected, the sensitivity 1 (true homologues) and sensitivity 0 (unrelated proteins) show opposite trends by decreasing and increasing, respectively, with lower P-value thresholds [higher −log(P-value)]. For both homologous and non-homologous protein pairs, the improvement in sensitivity is coupled with deterioration in specificity, and vice versa. It is therefore important to identify the threshold that offers good balance of high sensitivity and specificity, which for the SCOP dataset evaluated here coincides with highest overall accuracy at P-value of 1×E−04 (Fig. 2 and Table 1). When the threshold is identified that ensures high specificity, it is usually up to human experts to improve the sensitivity by inspecting the hits that fall beyond the threshold. As discussed previously, this approach can and does yield important findings, yet it is time-consuming and requires specific knowledge.
Fig. 2.
Classification parameters for the entire SCOP 1.69 database based on fixed P-value thresholds (10−03 to 10−15) from HHsearch. Values are plotted as a fraction of 1 (perfect classification). Accuracy, sensitivity and specificity are defined in ‘Methods’ section.
3.2 Using SVMs for classification of HHsearch hits without fixed significance thresholds
SVMs were chosen because of their ability to generalize the rules learned from a set of known examples so as to be accurate even on data not seen during the training (Vapnik, 1995). Our initial testing runs suggested that several SVM implementations achieve similar classifications, yet we used the LIBSVM library (Chang and Lin, 2001) because its classifications include probability estimates (Platt, 2000). While experimenting with various SVM kernels, we found that the perceptron kernel SVMs based on an infinite ensemble framework (Lin and Li, 2008) outperformed the more popular RBF kernel for all tested datasets. Given that SVMs with this kernel also train faster, the perceptron kernel became a clear choice for the work described here.
3.2.1 HHsvm1
First SVM, termed HHsvm1, was trained using P-values and probabilities for each match to be true positive (Söding, 2005) (relevant columns from the HHsearch output are boxed by full lines in Fig. 1). This SVM is meant to mimic a casual user who relies only on statistical evidence to decide whether a match reported by HHsearch is a true positive or not. As shown in Table 1, using these two features in the decision-making process results in slightly better classification compared to fixed P-value threshold of 1×E−04. It is important to note that HHsvm1 is already an excellent classifier based on its ROC curve (Supplementary Fig. 1).
3.2.2 HHsvm2
Our results with HHsvm1 indicated that SVMs can be very effective in classifying the output of HHsearch, and further improvements in their accuracy could be expected after including more informative features in the training process. In continuing to emulate the classification by human experts, we next considered the quality of the match between predicted secondary structures of query and each of identified hits. To a trained eye, the visual inspection of predicted secondary structures is usually one of strongest indicators of remote homology, and HHsearch even uses its secondary structure score to derive the probability of the correct match (Söding, 2005). Similarly, it is more likely that the match represent a true homologue if it can be aligned with the larger part of the query, so we also included the fraction of query aligned with the hit and vice versa. These three features (boxed by dashed lines in Fig. 1) were combined with HHsvm1 to produce the second SVM named HHsvm2. In terms of predictive ability, HHsvm2 was better than HHsvm1 (Supplementary Fig. 1). More importantly, the improvement was seen for true homologues and true negatives in terms of better sensitivity and specificity (Table 1).
3.2.3 HHsvm3
Yet another way of recognizing remote homology is by observing similarities in functionally important parts of proteins such as catalytic sites or interacting surfaces. In many cases the similarity only persists in these regions and may not give rise to statistically significant alignments, and visual inspection is sometimes the only way to tease out these alignments from the group of statistically insignificant alignments between unrelated proteins. This process is difficult to generalize for the purposes of machine learning because functionally important residues cannot always be reliably predicted despite recent advances in the field (Fischer et al., 2008). Our rationale was that the positional conservation of amino acids is usually more similar between evolutionarily conserved parts of proteins, even if the remaining parts of proteins have diverged. This conservation can be expressed as the average information content (Schneider and Stephens, 1990), either within the part of the profile that was aligned or over the whole profile length. The information content per residue is shown only qualitatively in the HHsearch output by lower- or upper-case letters in consensus sequence, but it can be easily calculated directly from profile HMMs used for comparison.
Positional information content for all residues in the profile was summed and divided by profile length to get a single number representing average information content. The procedure was repeated for the aligned part of each profile to derive average information content per alignment. These two numbers were calculated for the query and each hit, and were added to the features used for HHsvm2 training. This SVM was named HHsvm3 and it improved upon HHsvm2 in all quantitative measures of classification (Table 1 and Supplementary Fig. 1). It was particularly satisfying that the 2% improvement in overall accuracy was coupled with similar improvements in the specificity of predictions. The robust performance of HHsvm3 clearly indicated that our approach based on average information content is appropriate for delineating relevant from irrelevant alignments even when both are statistically insignificant, and even without the benefit of knowing the functionally important residues. It is possible that other approaches exploiting the information in HHsearch output and profile HMMs can further improve the SVM classification.
3.2.4 HHsvm4
After establishing HHsvm3 as the most accurate classifier, we decided to try recursive feature elimination in order to find out whether a better classification can be achieved. In all but one case, eliminating any single feature or any two features together resulted in deteriorated performance compared to HHsvm3. However, when the probability of the correct match was removed from training, the resulting classifier, HHsvm4, was better (Table 1; also compare ROC curves in Supplementary Fig. 1). In retrospect, we found that removing the same feature also improves the HHsvm2 classifier, but not HHsvm1 classifier. We conclude that, starting with HHsvm2, the inclusion of secondary structure scores and aligned fractions of the query and hit already provided more useful information than the probability of the correct match, making this feature detrimental to the overall performance. Interestingly, removing P-value from HHsvm3 instead of true match probability resulted in only slightly decreased performance (data not shown). This was somewhat surprising given that most practitioners of remote homology detection would probably rate P-value as the most useful attribute, yet this finding underscores the value of information that can be extracted from a carefully chosen set of features by machine learning.
3.3 Automatic classification using HHsvm predictions with high probabilities
The speed (>10 000 decisions/min) and accuracy of HHsvm4 classifications already bode well for its use in automated pipelines. SVM probabilities attached to each decision provide an additional safeguard against incorrect classification. When the premium is placed on quality annotations with little or no human intervention, the increased stringency can be achieved by using only high-probability classifications. As shown in Table 1, considering only classifications with probability ≥0.95 increases the overall accuracy by 1% and brings it well above 99% (row HHsvm4-0.95). The increased stringency would ultimately be detrimental if the fraction of high-probability predictions was low, yet in our test set 94.5% of predictions had the probability ≥0.95. Finally, the specificity of classifications for true positives reached 99.91%, ensuring that even fewer false positives would be assigned to this category.
3.4 HHsvm classifications in the ‘twilight zone’ of sequence identity
Sequence and profile alignments with relatively high P-values usually indicate the lack of homology, yet in some cases they occur when true homologues share low sequence identity. Because of this inherent difficulty in determining homology from alignments alone, low-identity matches (25–30%) are considered to be in the ‘twilight zone’ (Rost, 1999) and usually require evaluation by human experts and subsequent experimental validation. Given that threshold-based classification at P-value ≤1×E−04 is already very solid (Table 1), the most significant improvements in HHsvm classifications were expected for low-identity matches with P-values >1×E−04.
To evaluate the ability of HHsvm4 to correctly classify low-identity alignments, we created test datasets based on identity thresholds of 10, 20, 30, 40, 50, 70 and 90% (SCOP10 to SCOP90 in Table 2). For example, alignments in SCOP10 dataset come only from SCOP sequences that have ≤10% identity. This dataset contains the smallest number of alignments and its true positives are most difficult to classify using only P-values. Conversely, SCOP90 contains the largest number of alignments, most of which are relatively easy to classify from P-values (Table 2). HHsvm4 classification outperforms threshold-based assignments both in terms of sensitivity and specificity and across the whole range of sequence identities. Strikingly, the biggest improvement is seen in the most difficult group below 10% identity, where HHsvm4 comes ahead by 10% in its ability to correctly identify true evolutionary relationships. Just as importantly, HHsvm4 classifications steadily improve in all categories with increasing identity thresholds, whereas threshold-based assignments show declining specificity for true negatives when sequences with 50% identity and above are added to the dataset (Table 2).
Table 2.
Homology assignments at different levels of sequence identity using fixed P-value thresholds (1×E−04) versus HHsvm4
| Dataset | Accuracy | Sensitivity 1 | Specificity 1 | Sensitivity 0 | Specificity 0 |
|---|---|---|---|---|---|
| SCOP10 | 93.21/96.36 | 80.07/90.09 | 90.51/94.47 | 97.35/98.34 | 93.93/96.92 |
| SCOP20 | 93.42/96.58 | 82.15/91.14 | 90.86/95.10 | 97.21/98.42 | 94.17/97.05 |
| SCOP30 | 93.74/97.14 | 85.48/93.55 | 92.09/96.34 | 97.06/98.57 | 94.34/97.44 |
| SCOP40 | 94.02/97.53 | 86.88/94.85 | 93.23/97.05 | 97.20/98.72 | 94.34/97.74 |
| SCOP50 | 93.80/97.66 | 86.70/95.27 | 93.54/97.40 | 97.16/98.80 | 93.91/97.78 |
| SCOP70 | 93.81/97.96 | 87.80/96.30 | 94.84/98.07 | 97.26/98.91 | 93.28/97.90 |
| SCOP90 | 94.80/98.51 | 92.32/98.04 | 97.39/99.03 | 97.40/98.99 | 92.36/97.97 |
SCOP datasets clustered between 10% (SCOP10) and 90% (SCOP90) sequence identity were evaluated. In each column values calculated for fixed P-value threshold of 1×E−04 and HHsvm4 classifications are shown on left and right side, respectively.
3.5 Using HHsvm to annotate protein domains of unknown function as PD-(D/E)xK nucleases
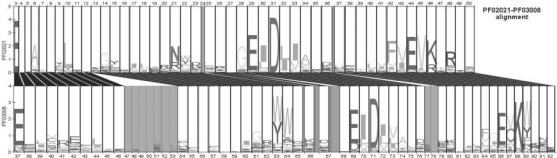
In recent years, related protein families in Pfam database have been grouped into clans (Finn et al., 2008). We decided to test the utility of HHsvm4 by trying to expand the membership of a well-characterized PD-(D/E)xK clan (Pfam designation CL0236), which contains a diverse group of nucleases (Aravind et al., 2000; Knizewski et al., 2007). PD-(D/E)xK clan has 27 annotated protein families and HHsearch was used to compare each of their profiles with all families in Pfam 23.0. The resulting HHsearch outputs were then processed by HHsvm4, and all hits with SVM probabilities >0.5 were set aside for further analysis. We constructed pairwise HMM logos (Schuster-Bockler and Bateman, 2005) for all matches outside of the PD-(D/E)xK clan and searched for conserved residues, particularly the metal-chelating acidic residues and the catalytic lysine (Aravind et al., 2000).Figure 3 illustrates one example of this conservation of critical residues between a known (PF02021) and predicted (PF03008) member of the PD-(D/E)xK clan. Four additional pairwise HMM logos between known and predicted nucleases are shown in Supplementary Figure 2.
Fig. 3.
Pairwise HMM logo (Schuster-Bockler and Bateman, 2005) of protein families PF02021 and PF03008 from Pfam 23.0 (Finn et al., 2008). PF02021 is annotated as a member of the PD-(D/E)xK clan (CL0236), while PF03008 (also known as DUF234) has no informative annotation. Despite low sequence identity, the alignment of conserved acidic residues and the lysine which are required for catalysis (Aravind et al., 2000; Knizewski et al., 2007) supports their relationship; it is also strongly supported by HHsvm4 (Supplementary Table 2).
We identified many convincing relationships between at least one of the 27 families from CL0236 and 15 other families in Pfam 23.0. A complete list of all additional families predicted to be PD-(D/E)xK nucleases is provided as Supplementary Table 2, along with P-values and SVM probabilities with the known PD-(D/E)xK match that provided the most persuasive evidence.
4 DISCUSSION AND CONCLUSIONS
At the onset of this work, we were reasonably confident that SVMs can be trained to correctly classify statistically significant hits from the HHsearch output. Indeed, an SVM trained on just two features already classifies in a manner that is quantitatively similar to assignments based on fixed thresholds of P-values. However, the real power of SVMs is realized after they are trained on larger sets of features, as this elevates their classification ability into something resembling the expertise of human annotators (Table 1). Overall, all SVMs achieve excellent classification both in terms of sensitivity and specificity, which is reflected in their ROC curves (Supplementary Fig. 1). HHsvm4 improvements are particularly impressive when classifying true homologues with ≤10% sequence identity, as these matches are challenging even for human experts. In addition to accuracy, HHsvm4 delivers more than ten thousand classifications per minute, which is orders of magnitude faster than comparable annotation by human experts. This feature makes HHsvm4 very suitable for large-scale analyses.
We used HHsvm4 to explore relationships between known members of the PD-(D/E)xK clan (Finn et al., 2008; Knizewski et al., 2007) and all other Pfam families presently lacking clan annotation. Following high-probability classifications with manual inspection of pairwise HMM logos (Schuster-Bockler and Bateman, 2005), we assigned 15 new Pfam families to the PD-(D/E)xK clan. Four of these assignments (PF04556, PF09019, PF09254 and PF09563) have already been made in earlier publication (Orlowski and Bujnicki, 2008), and one (PF04257) is a multi-domain family that may belong to several clans. Some of these protein families are already annotated as nucleases or recombinases (Supplementary Table 2), and their association with the PD-(D/E)xK clan further clarifies their function. Finally, at least five putative PD-(D/E)xK clan members lack meaningful functional annotations, and these predictions will hopefully help their future experimental characterizations.
Like any machine-based approach, HHsvm4 works best when the new data to be classified is treated the same way as the training sample. This means that profiles should be created using modified Target2K procedure tuned to identify and align distantly related homologues at the superfamily level. Target2K is part of the SAM package (http://compbio.soe.ucsc.edu/sam.html) and our modifications are easy to implement. HHsvm4 could be used to classify matches from HMM profiles distributed on the HHpred server (http://toolkit.tuebingen.mpg.de/hhpred), but the overall accuracy of classification was lower (data not shown). These profiles were created from less stringent multiple alignments derived directly from pairwise PSI-BLAST alignments between the query and its matches, and we suspect that this caused the drop in performance.
It is important to point out that 98.68% accuracy by HHsvm4 (Table 1) does not correspond to all true homologues in this SCOP dataset. Instead, it describes the performance of HHsvm4 for the true hits identified in the HHsearch output at E-value ≤100 (P ≤0.01), which included only 88.2% of all homologous pairs. Increasing the sensitivity of HHsearch, at least to the point where more distantly related proteins can be assigned P-values lower than 0.01, may further improve the overall performance of HHsvm4.
In conclusion, we show that SVMs trained on a well-defined dataset can be used with confidence in assessing even distant relationships between proteins from the HHsearch output. Combining the outputs of different profile methods and the addition of other informative features will be new avenues for potential improvements of this approach.
Supplementary Material
ACKNOWLEDGEMENTS
Most computations were done using the computer cluster at Montana State University INBRE Bioinformatics Core Facility, which is supported by grants P20-RR16455 (NIH) and IGERT DGE-0654336 (NSF). The author is grateful to Johannes Söding for providing early versions of the HHpred suite of programs and for explaining the HHM file format, and to Gary Orser for the computer maintenance at the INBRE Bioinformatics Core Facility.
Conflict of interest: none declared.
REFERENCES
- Altschul SF, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, et al. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, et al. Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner SE, et al. Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc. Natl Acad. Sci. USA. 1998;95:6073–6078. doi: 10.1073/pnas.95.11.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Lin C. LIBSVM: a library for support vector machines. 2001 http://www.csie.ntu.edu.tw/∼cjlin/libsvm. [Google Scholar]
- Cheng H, et al. Discrimination between distant homologs and structural analogs: lessons from manually constructed, reliable data sets. J. Mol. Biol. 2008;377:1265–1278. doi: 10.1016/j.jmb.2007.12.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JD, et al. Prediction of protein functional residues from sequence by probability density estimation. Bioinformatics. 2008;24:613–620. doi: 10.1093/bioinformatics/btm626. [DOI] [PubMed] [Google Scholar]
- Ginalski K, et al. Detecting distant homology with Meta-BASIC. Nucleic Acids Res. 2004;32:W576–W581. doi: 10.1093/nar/gkh370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Karplus K, et al. Hidden Markov models for detecting remote protein homologies. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- Karplus K, et al. Combining local-structure, fold-recognition, and new fold methods for protein structure prediction. Proteins. 2003;53(Suppl. 6):491–496. doi: 10.1002/prot.10540. [DOI] [PubMed] [Google Scholar]
- Knizewski L, et al. Realm of PD-(D/E)XK nuclease superfamily revisited: detection of novel families with modified transitive meta profile searches. BMC Struct. Biol. 2007;7:40. doi: 10.1186/1472-6807-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H-T, Li L. Support vector machinery for infinite ensemble learning. J. Mach. Learn. Res. 2008;9:285–312. [Google Scholar]
- Madera M, Gough J. A comparison of profile hidden Markov model procedures for remote homology detection. Nucleic Acids Res. 2002;30:4321–4328. doi: 10.1093/nar/gkf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski J, Bujnicki JM. Structural and evolutionary classification of Type II restriction enzymes based on theoretical and experimental analyses. Nucleic Acids Res. 2008;36:3552–3569. doi: 10.1093/nar/gkn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, et al. Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J. Mol. Biol. 1998;284:1201–1210. doi: 10.1006/jmbi.1998.2221. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt J. Probabilities for SV machines. In: Bartlett PS, et al., editors. Advances in Large Margin Classifiers. Cambridge, MA: MIT Press; 2000. pp. 61–74. [Google Scholar]
- Reid AJ, et al. Methods of remote homology detection can be combined to increase coverage by 10% in the midnight zone. Bioinformatics. 2007;23:2353–2360. doi: 10.1093/bioinformatics/btm355. [DOI] [PubMed] [Google Scholar]
- Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- Sadreyev R, Grishin N. COMPASS: a tool for comparison of multiple protein alignments with assessment of statistical significance. J. Mol. Biol. 2003;326:317–336. doi: 10.1016/s0022-2836(02)01371-2. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Bockler B, Bateman A. Visualizing profile-profile alignment: pairwise HMM logos. Bioinformatics. 2005;21:2912–2913. doi: 10.1093/bioinformatics/bti434. [DOI] [PubMed] [Google Scholar]
- Shah AR, et al. SVM-HUSTLE–an iterative semi-supervised machine learning approach for pairwise protein remote homology detection. Bioinformatics. 2008;24:783–790. doi: 10.1093/bioinformatics/btn028. [DOI] [PubMed] [Google Scholar]
- Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Söding J, et al. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik VN. The Nature of Statistical Learning Theory. New York: Springer; 1995. [Google Scholar]
- Yona G, Levitt M. Within the twilight zone: a sensitive profile-profile comparison tool based on information theory. J. Mol. Biol. 2002;315:1257–1275. doi: 10.1006/jmbi.2001.5293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.