Abstract
Oxalate is widely distributed in the plant kingdom. While excess oxalate in food crops is detrimental to animal and human health, it may play various functional roles in plants, particularly for coping with environmental stresses. Understanding its biosynthetic mechanism in plants, therefore, becomes increasingly important both theoretically and practically. However, it is still a matter of debate as to what precursor and pathway are ultimately used for oxalate biosynthesis in plants. In this study, both physiological and molecular approaches were applied to address these questions. First, it was observed that when glycolate or glyoxylate was fed into detached leaves, both organic acids were equally effective in stimulating oxalate accumulation. In addition, the stimulation could be completely inhibited by cysteine, a glyoxylate scavenger that forms cysteine–glyoxylate adducts. To verify the role of glyoxylate further, various transgenic plants were generated, in which several genes involved in glyoxylate metabolism [i.e. SGAT (serine-glyoxylate aminotransferase), GGAT (glutamate-glyoxylate aminotransferase), HPR (hydroxypyruvate reductase), ICL (isocitrate lyase)], were transcriptionally regulated through RNAi or over-expression. Analyses on these transgenic plants consistently revealed that glyoxylate acted as an efficient precursor for oxalate biosynthesis in rice. Unexpectedly, it was found that oxalate accumulation was not correlated with photorespiration, even though this pathway is known to be a major source of glyoxylate. Further, when GLDH (L-galactono-1,4-lactone dehydrogenase), a key enzyme gene for ascorbate biosynthesis, was down-regulated, the oxalate abundance remained constant, despite ascorbate having been largely reduced as expected in these transgenic plants. Taken together, our results strongly suggest that glyoxylate rather than ascorbate is an efficient precursor for oxalate biosynthesis, and that oxalate accumulation and regulation do not necessarily depend on photorespiration, possibly due to the occurrence of the anaplerotic reaction that may compensate for glyoxylate formation in rice.
Keywords: Ascorbate, glycolate, glyoxylate, oxalate, rice
Introduction
Oxalate, the simplest dicarboxylic acid, can be found in a wide variety of plants and may constitute as much as 3–10% of plant dry mass (Nakata, 2003; Franceschi and Nakata, 2005). This value in rice is generally between 3–6%, depending on growth stage and culture conditions (Libert and Franceschi, 1987; Ji and Peng, 2005; Xu et al., 2006). While oxalate used to be considered as an inert metabolic product, a growing body of evidence has shown that it may play various functional roles in plants. Certain plants, such as buckwheat, taro, and rice, may exude and/or accumulate internal oxalate to cope with aluminium or lead toxicity (Ma et al., 1997; Ma and Miyasaka, 1998; Yang et al., 2000; Morita et al., 2008). Reportedly, oxalate was also able to detoxify other hazardous metals in plants, such as strontium (Franceschi and Schueren, 1986), cadmium (Choi et al., 2001), and copper (Mazen and EI Maghraby, 1997; Mijovilovich et al., 2009). In addition, oxalate quenches the oxidative burst during pathogen attack in plants (Cessna et al., 2000), scavenges excreted phytotoxins from weeds (Weir et al., 2006), and is involved in the plant programmed cell death (Kim et al., 2008; Errakhi et al., 2008). Oxalate may regulate stomatal aperture by binding calcium ions in the vicinity of the guard cells (Ruiz and Mansfield, 1994). Korth et al. (2006) found that calcium oxalate crystals acted as an effective defence against chewing insects. Understanding how plants biosynthesize and regulate oxalate is, therefore, of both theoretical and practical significance.
Three pathways for oxalate biosynthesis have been proposed in plants, although none of them have been proven conclusively. Oxidation of glycolate/glyoxylate during photorespiration has long been considered as a biosynthetic pathway for oxalate in plants (Libert and Franceschi, 1987; Fujii et al., 1993; Nakata, 2003; Franceschi and Nakata, 2005). This hypothesis is mainly based on the evidence that isotope-labelled glycolate/glyoxylate was incorporated into oxalate and that glycolate oxidase (GLO) was able to catalyse the oxidation of glyoxylate to oxalate in vitro (Richardson and Tolbert, 1961; Xu et al., 2006). However, this pathway has been continuously challenged, particularly in recent years. For instance, Raven et al. (1982) showed that 18O2 incorporation into glycolate did not extend to oxalate. Oxalate accumulates even in the dark or in callus, where presumably no GLO or photorespiration occurs (Franceschi, 1987). More recently, a number of isotope-labelling studies have demonstrated that the 1 and 2 carbons of the 14C-labelled L-ascorbate gave rise to oxalate in Lemna minor, Yucca torreyii, and Pistia stratiotes (Franceschi, 1987; Horner et al., 2000; Keates et al., 2000; Kostman et al., 2001; Franceschi and Nakata, 2005). The 5-carbon ascorbate analogue, erythorbic acid, and precursors of ascorbic acid labelled at the 1 carbon also gave rise to oxalate (Keates et al., 2000; Kostman et al., 2001; Franceschi and Nakata, 2005). Green and Fry (2005) showed that the ascorbate catabolic pathway operates extracellularly to produce oxalate in plant cells. Oxaloacetate breakdown was reported to be the third source for oxalate in plants, which was presumably catalysed by an oxaloacetase. While this enzyme was once reported in beetroot and spinach crude preparations (Chang and Beevers, 1968), no relevant follow-up studies have confirmed their results. We have tried for years to identify this enzyme from various plants and have not been successful (X Peng, unpublished data). Thus, the mechanisms underlying oxalate biosynthesis and regulation are still undetermined in plants.
In this study, in addition to the physiological dissection, various transgenic plants were generated, which had up-regulated SGAT (serine-glyoxylate aminotransferase), GGAT (glutamate-glyoxylate aminotransferase), and ICL (isocitrate lyase), and down-regulated HPR (hydroxypyruvate reductase) and GLDH (L-galactono-1,4-lactone dehydrogenase). Analyses on these transgenic plants revealed that glycolate/glyoxylate rather than ascorbate is the efficient precursor for oxalate biosynthesis, but oxalate accumulation and regulation do not necessarily depend on photorespiration, possibly due to the existence of an anaplerotic reaction which may compensate for the glyoxylate formation in rice. This finding laid a foundation for being able to bypass more photorespiratory glyoxylate into oxalate through molecular regulation. Such a metabolic engineering would hopefully achieve a win–win scenario, i.e. to reduce the carbon loss due to photorespiration and, meanwhile, to improve plant stress resistance by increasing the oxalate accumulation.
Materials and methods
Plant materials
Two rice cultivars (Oryza sativa L.) were used: Xiangzhongxian 2 (XX2) for the physiological studies and Zhonghua 11 (ZH 11) for generating the transgenic lines.
Growth conditions and treatments
Germinated seeds of XX2 and the transgenic plants were pre-grown with complete Kimura B nutrient solution (Yoshida et al., 1976) in a greenhouse under natural conditions. After reaching the four-leaf stage, the seedlings were transferred to a growth chamber for the different treatments [chamber conditions; 30/25 °C temperature (day/night), ∼60% relative humidity, 100–800 μmol m−2 s−1 light intensity as specified in the figure legends, and 14/10 h photoperiod (day/night)]. For the different light treatments, four-leaf age seedlings grown with complete Kimura B were first grown in sole ammonium-N (3 mM) Kimura B solution for an additional 3 d, then transferred to either sole nitrate or sole ammonium and grown under different light intensities (100–800 μmol m−2 s−1) for various times (0–72 h). For air or high CO2 treatment, the seedlings were grown at 400 μmol m−2 s−1 light either in normal air or 0.5% CO2. The high CO2 treatment was realized by introducing pure CO2 gas into the growth chamber, controlled by a rotameter and monitored by an infrared gas analyser. For the exogenous feeding experiment, the second leaf from the top was cut at 13 cm length in water. The detached leaf was then dipped into the treatment solutions contained in 5 ml centrifuge tubes. The treatment solution was made by adding the specified substances into sole ammonium-N Kimura B solution. The tubes were placed in a growth chamber under 30/25 °C temperature (day/night), ∼60% relative humidity, 50 μmol m−2 s−1 light, and a 12/12 h photoperiod (day/night). After 48 h treatment, the leaf was subjected to analysis. Some experiments with different treatments are specified in the figure legends.
Generation of transgenic plants
Construction of the vectors:
The overexpression vectors for SGAT, GGAT, and ICL: the complete cDNA sequences for SGAT, GGAT, and ICL (Accession nos AK064774, AK067732, and AK063353, respectively), were cloned by RT-PCR with upstream primers 5′-AGCTGAGGATCCGAGATGGCGGACTACGTGTAT-3′ (SGAT), 5′-ATTTGGATCCTGCGGCAAGATGTT-CGG-3′ (GGAT), and 5′-GGCCGAAGCTTTCTTGGTTATCATGTCGT-3′ (ICL), and with downstream primers 5′-AAACTGACGCGTGAACCATCGCTGCTGAGAAT-3′ (SGAT), 5′-CTTGTTACGCGTTCACATCCTGGAGTAGCCAT-3′ (GGAT), and 5′-TATATACGCGTGCTCCTTGGCTGAAGTCC-3′ (ICL). Then the sequences were inserted into the vector between BamHI and MluI restriction sites (the vector named as pYLox.5 was kindly provided by Dr Yao-Guang Liu, College of Life Sciences, South China Agricultural University). First, PCR with specific primers and cutting with restriction enzymes showed that the target fragment had been correctly ligated.. DNA sequencing finally confirmed the correct orientation and 100% cDNA identity to that reported in the GeneBank.
The RNAi vectors for HPR1 and GLDH: the cDNA fragments (1238 bp for HPR1 and 554 bp for GLDH) were cloned by RT-PCR with upstream primers 5′-GCAAAAGCTTCGAAAGCAAAACAAGAAATGGC-3′ (HPR1) and 5′-ATTTGAGCTCTTCGTCCCTGTTCTGCCG-3′ (GLDH); and with downstream primers 5′-GAGAGAACGCGTGAGTATGATCTTGAACCGACA-3′ (HPR1) and 5′-TAATAAGCTTGCGTATCCCGGCCTGCAC-3′ (GLDH). The sequences were then inserted into the vector pYLRNAi.5 (kindly provided by Dr Yao-Guang Liu, College of Life Sciences, South China Agricultural University) between BamHI and SacI restriction sites. Both PCR with the specific primers and restriction enzyme cutting verified that the fragment had been correctly inserted into the vector. This first-round- ligated vector was then used as the template to amplify a second sequence with two unique restriction sites in both ends (RNAi-MluI: 5′-CACCCTGACGCGTGGTGTTACTTCTGAAGAGG-3′; RNAi-PstI: 5′-ACTAGAACTGCAGCCTCAGATCTACCATGGTCG-3′). The second sequence was subsequently cloned at MCS2 between PstI and MluI, resulting in an opposite orientation in contrast to the sequence in MCS1. Restriction enzyme cutting showed that the second target fragment had been correctly inserted into the vector. Finally, the DNA sequencing further confirmed the correct orientation and 100% cDNA identity identical to that reported in the GenBank
Transformation of the genes into rice:
The various vectors constructed as described above were transformed into rice callus by Agobacterium-mediated infection (strain EHA105). The seeds harvested from the positive independent T0 lines were germinated and grown in complete Kimura B nutrient solution, then transferred to normal soil conditions to grow until the seeds were harvested. T2 or T3 seeds were used for the functional analysis in this study.
Semi-quantitative PCR analysis of gene expressions:
The optimal number of PCR cycles was first tested gene by gene during semi-quantitative PCR analysis. The PCR was performed with PTC-200 (Bio-Rad, Hercules CA, USA), and the PCR products were separated on 1% (w/v) agarose gels and visualized by Goldview staining. Sequences of the primers for the semi-quantitative RT-PCR were listed in Supplementary Table S1 available at JXB online.
Assay of enzyme activities
SGAT and GGAT
50 mg of leaves were homogenized in 1 ml 50 mM K-phosphate (pH 7.4) at 4 °C, and the homogenate was then centrifuged at 12 000 rpm and 4 °C for 30 min. The supernatant was used as an enzyme extract. The reaction mixture (500 μl) contained 20 mM L-serine for SGAT or L-glutamate GGAT, 5 mM glyoxylate, 10 μM pyridoxal-5-phosphate (PAL), and appropriate enzyme extract. The reaction was started by the addition of glyoxylate and conducted at 30 °C for 20 min. The reaction was terminated by adding 100 μl of 20% TCA. After a centrifugation, the supernatant was derived with dinitroflurobenzene at 60 °C for 1 h. The amino acid derivatives were then separated on a C-18 column equipped with an HPLC system (Waters-2695, USA) and the amount of glycine produced was detected to measure the SGAT and GGAT activity.
HPR:
100 mg of leaves was homogenized in 1 ml extraction buffer (10 mM TRIS-HCl, 1 mM EDTA, 2 mM MgCl2, and 1 mM β-mercaptoethanol, pH 7.5) at 4 °C, then the homogenate was centrifuged at 12 000 rpm and 4 °C for 20 min. The supernatant was used for HPR activity assay. 1 ml of reaction mixture contained the extraction buffer as described above, 0.2 mM NADH or NADPH, 0.5 mM hydroxypyruvate, and the appropriate enzyme extract. The reaction was started by the addition of hydroxpyruvate, and the oxidation of NADH or NADPH was spectrophotometrically detected at 340 nm.
ICL:
100 mg of leaves was homogenized in 0.8 ml 50m M TRIS-HCl buffer (pH 7.8) containing 10 mM β-mecaptoethanol. The homogenate was then centrifuged at 12 000 rpm and 4 °C for 15 min. The supernatant was used for ICL activity assay. The activity was determined according to the method of Ranaldi et al. (2000) with some modifications. The reaction mixture contained 25 mM TRIS-HCl buffer (pH 7.8), 0.75 mM MgCl2,, 6 mM L-Cys, 0.033% phenylhydrazine-HCl (w/v), 2.4 mM DL-isocitrate, and appropriate enzyme extract. The enzyme extract was finally added to start the reaction. The reaction was conducted at 30 °C for 15 min, then terminated by adding 0.1 ml of 2 M HCl. The mixture was placed on ice for 5 min, then 0.5 ml of concentrated HCl was added to the mixture. The mixture was kept on ice for another 5 min. Finally, 0.1 ml of 1.65% K3Fe(CN)6 (w/v) was added to develop the red colour (20 min in the dark) and spectrophotometrically detected at 550 nm..
Quantification of organic acids
Oxalate and glyoxylate were determined after Xu et al. (2006). For the determination of ascorbic acid, 0.1 g of fresh leaves was homogenized in 1 ml 6% TCA solution and the homogenate was centrifuged at 12 000 rpm and 4 °C for 10 min. The supernatant was used for ascorbate analysis according to Kampfenkel et al. (1995).
Measurement of photorespiratory rates
The photorespiratory rate (Rp) was estimated by measuring the differences in net photosynthetic rates (Pn) between under 2% O2 air system (2% O2, air CO2, balance with N2) and under normal air (Sharkey, 1988).
Protein determination
Protein content was determined according to Bradford (1976).
Results
Relationship between oxalate accumulation and photorespiration
While it has been long noticed that oxalate content is higher in nitrate-grown plants than ammonium-grown ones, its mechanism is still a matter of debate (Tian et al., 2008). To understand the link between oxalate accumulation and photorespiration further, the response of oxalate accumulation to different light intensities was determined first under nitrate-N conditions, based on the fact that photorespiration is light-dependent and usually increases as light intensity increases (Brown and Morgan, 1980; Lin et al., 2000; Zhang et al., 2009). In one set of experiments, different light intensities ranging from 100–800 μmol m−2 s−1 were used for plant growth. No significant effects were observed on oxalate accumulation under nitrate-N; however, low light (i.e. 100 μmol m−2 s−1) did slow down the oxalate decrease under ammonium-N (Fig. 1A). It is well known that high CO2 can effectively suppress photorespiration (Somerville, 2001). When plants were grown in 0.5% CO2 for various times, the oxalate accumulation under nitrate-N was unaffected as compared with that under normal air (0.04% CO2) (Fig. 1B). The photorespiratory rate (Rp) was even slightly lower for nitrate-fed leaves than that for ammonium-fed ones (Fig. 1C), in contrast to the oxalate content that was much higher in nitrate-fed leaves than that in ammonium-fed ones (Fig. 1A). The results consistently suggest that oxalate accumulation is not necessarily dependent upon photorespiration in rice.
Fig. 1.
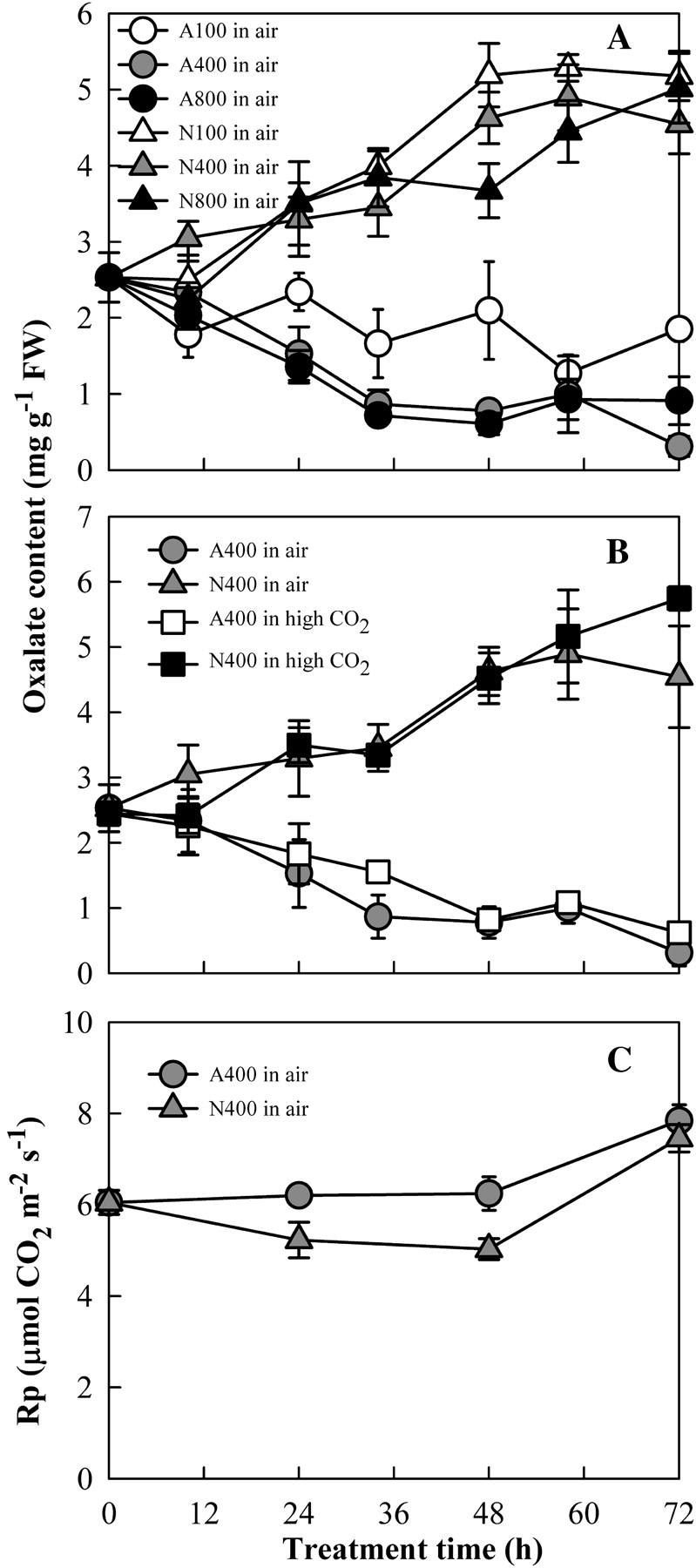
Relationship between oxalate accumulation and photorespiration. Four-leaf age seedlings grown in complete Kimura B nutrient solution were treated for 72 h under different conditions. (A) Oxalate accumulation for nitrate- and ammonium-fed leaves in air under different light intensities; (B) oxalate accumulation for nitrate- and ammonium-fed leaves at 400 μmol m−2 s−1 light intensity in air and high CO2 (0.5%); and (C) photorespiartory rate (Rp) for nitrate- and ammonium-fed leaves in air at 400 μmol m−2 s−1. N and A indicate that plants were grown in the sole nitrate and ammonium nutrient solution, respectively. The numbers 100, 400, and 800 following both N and A indicate that the plants were grown at 100, 400, and 800 μmol m−2 s−1 light intensity, respectively.
Effect of glycolate/glyoxylate on oxalate accumulation
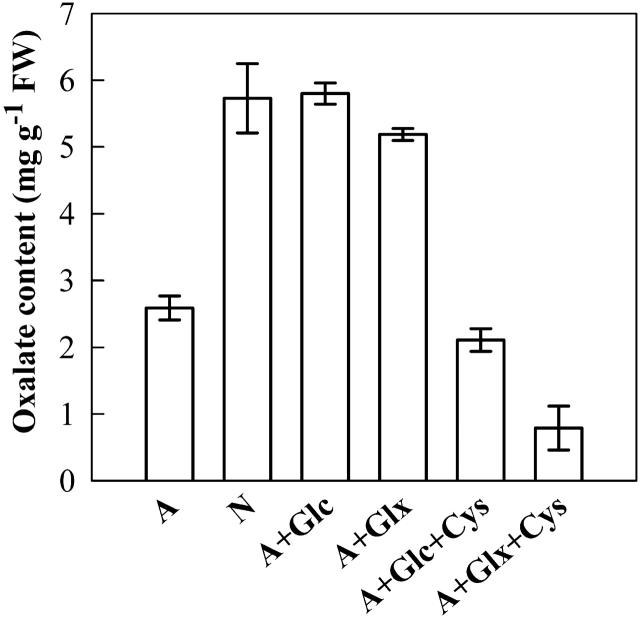
It was noticed previously that glycolate was able effectively to restore the ammonium-decreased oxalate accumulation and that glyoxylate was less effective in this aspect (Xu et al., 2006). In this study, the effects of glycolate and glyoxylate were tested further using a different approach. When glycolate or glyoxylate was fed into detached leaves by dipping the cut end into the same solution as formerly used for the intact plants (Xu et al., 2006), the two substances were equally effective in promoting oxalate accumulation (Fig. 2). Interestingly, the promoting effect was totally inhibited when cysteine was fed together with glycolate or glyoxylate (Fig. 2).
Fig. 2.
Effects of glycolate, glyoxylate, and cysteine on oxalate accumulation. Detached leaves were treated for 48 h as follows: sole ammonium-N Kimura B solution (designated as A), sole nitrate-N Kimura B solution (designated as N), 5 mM glycolate in A (A+Glc), 5 mM glyoxylate in A (A+Glx), 5 mM glycolate and 5 mM cysteine in A (A+Glc+Cys), 5 mM glyoxylate and 5 mM cysteine in A (A+Glx+Cys).
Modulation of the genes involved in glyoxylate metabolism and the effects on oxalate accumulation
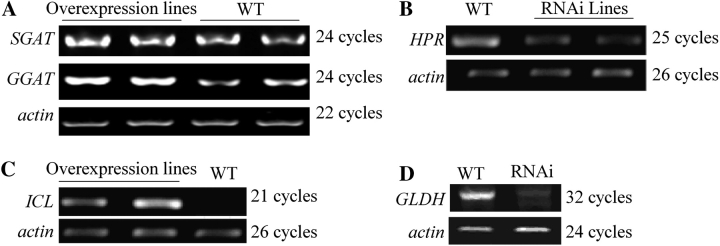
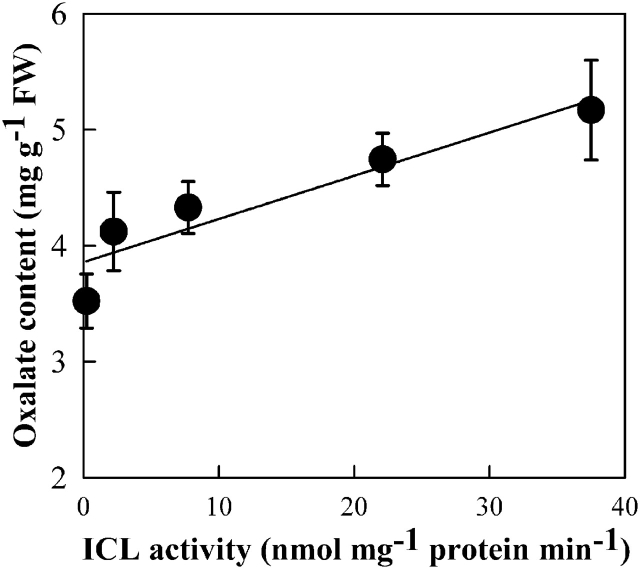
In order to confirm whether glyoxylate is involved in oxalate biosynthesis, various transgenic lines were generated in which several glyoxylate metabolism related genes were either over-expressed or down-regulated. SGAT and GGAT encode Ser:glyoxylate aminotransferease (SGAT) and Glu:glyoxylate aminotransferease (GGAT), respectively. These two enzymes catalyse the conversion of glyoxylate into glycine (Liepman and Olsen, 2001; Igarashi et al., 2006). ICL codes for isocitrate lyase (ICL), which catalyses the production of glyoxylate by splitting isocitrate. Hydroxypyruvate reductase (HPR) is reportedly able to reduce either hydroxypyruvate or glyoxylate (Givan and Kleczkowski, 1992). SGAT and GGAT were transcriptionally up-regulated (Fig. 3A) and the enzyme activities were correspondingly increased in the over-expression lines (Fig. 4A, B). Analyses of gloxylate and oxalate contents in these plants showed that oxalate accumulation was significantly positively correlated with glyoxylate content (P <0.01) (Fig. 4C) but significantly negatively correlated with the enzyme activities (P <0.01) (Fig. 4A, B). When HPR-1 expression was suppressed (Fig. 3B), NADH-HPR activity was almost completely lost and NADPH-HPR was partially reduced (Table 1). These HPR-1-interfered plants can grow normally in air but both glyoxylate and oxalate levels were significantly increased in these plants (Table 1). This result is well in agreement with that of Kleczkowski et al. (1990). ICL gene expression and activity were very low in WT plants (Figs 3C, 5) and dramatically up-regulated in the over-expression lines (Figs 3C, 5). Oxalate also significantly increased as ICL activity was up-regulated (Fig. 5).
Fig. 3.
RT-PCR analysis of transcriptional expression of genes from the different transgenic plants. (A) SGAT and GGAT over-expression lines; (B) HPR-1 RNAi lines; (C) ICL over-expression lines; (D) GLDH RNAi lines.
Fig. 4.
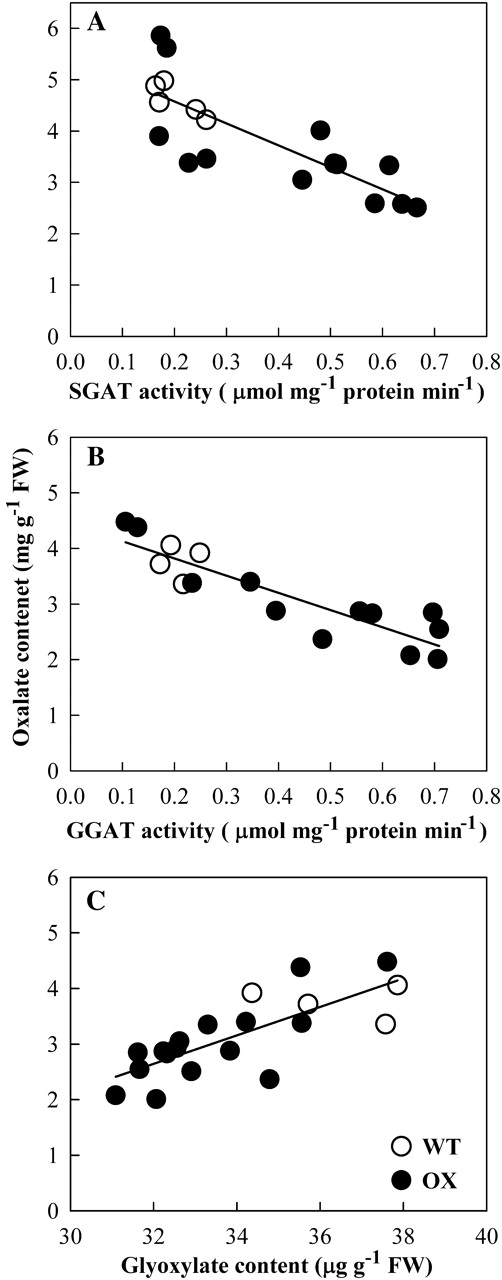
Oxalate accumulation in SGAT- and GGAT-over-expressed plants and its correlation with glyoxylate content. Four-leaf age seedlings grown in complete Kimura B nutrient solution were transferred to the sole nitrate-N Kimura B solution for 3 d, then the leaves were sampled for the analysis. Two independent RNAi lines were used for the analysis. Each data point represents the enzyme activity and the corresponding oxalate content in various individual plants. (A) Changes in oxalate accumulation as SGAT activity was up-regulated; (B) changes in oxalate accumulation as GGAT activity was up-regulated; and (C) the correlation between oxalate accumulation and glyoxylate content in the transgenic plants (y=0.2557x–5.5378, R2=0.5826, n=20, P=0.00009031).
Table 1.
Effect of HPR-1 down-regulation on glyoxylate and oxalate accumulation
| Activity | Content | |||
| (μmol mg−1 protein min−1) |
(μg g−1 FW) |
(mg g−1 FW) |
||
| NADH-HPR | NADPH-HPR | Glyoxylate | Oxalate | |
| WT | 1.016±0.215 | 0.017±0.003 | 20.296±3.860 | 3.278±0.220 |
| HPR-1 | 0.048±0.005 | 0.0108±0.001 | 40.170±2.879 | 4.830±0.746 |
| Relative (%) | –95 | –37 | +98 | +47 |
The four-leaf age seedlings grown in the Kimura B complete nutrient solution were transferred to the sole nitrate-N Kimura B solution for 3 d, then the leaves were sampled for the analyses.
Fig. 5.
Changes in oxalate accumulation as ICL activity was up-regulated in the ICL over-expressed plants. Four-leaf age seedlings grown in the complete Kimura B nutrient solution were transferred to the sole nitrate-N Kimura B solution for 3 d, then the leaves were sampled for the analysis. Four independent RNAi lines were used for the analysis. The ICL activity was first determined for each individual plant, then those individuals with similar ICL activities were pooled and sampled for measuring the corresponding oxalate content. Each data point represents means ±SE of three replicates.
Modulation of GLDH and its effect on ascorbate and oxalate accumulation
While conversion of ascorbate to oxalate was well documented, its biochemical basis has so far not been established (Franceshi and Nakata, 2005). It was noticed previously that when ascorbate was fed into rice in the same manner as glycolate, only slight increases occurred in oxalate levels, in contrast to the effect of glycolate (Xu et al., 2006). In order to define the correlation between ascorbate and oxalate in rice further, transgenic lines were constructed that had down-regulated GLDH. GLDH encodes L-galactono-1,4-lactone dehydrogenase (GLDH), a key enzyme for ascorbate biosynthesis (Wheeler et al., 1998). The GLDH transcripts were much lower in the interfered plants than in WT (Fig. 3D). Reduced ascorbate, dehydroascorbate, and the total content were all markedly altered as expected in the transgenic plants. For instance, the total ascorbate was decreased by up to 85% in the interfered plants. However, oxalate levels were not significantly changed in these transgenic plants (Fig. 6). These data strongly suggest that oxalate accumulation and regulation are independent of ascorbic acid levels in rice plants.
Fig. 6.
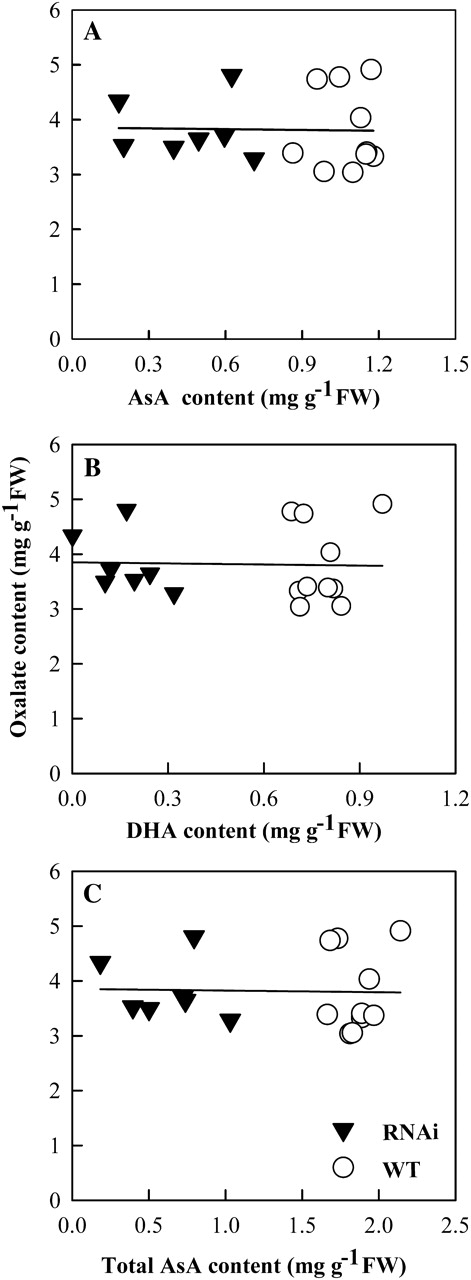
The correlation between oxalate accumulation and ascorbate content in the GLDH down-regulated plants. Four-leaf age seedlings grown in the complete Kimura B nutrient solution were transferred to the sole nitrate-N Kimura B solution for 3 d, then the leaves were sampled for the analysis. One independent RNAi line was used for the analysis. Each data point represents ascorbate and the corresponding oxalate content in various individual plants. (A) Changes in oxalate accumulation as the internal reduced ascorbate (AsA) content was regulated; (B) changes in oxalate accumulation as the internal dehydroascorbate (DHA) content was regulated; and (C) changes in oxalate accumulation as the total ascorbate content was regulated.
Discussion
Glyoxylate rather than ascorbate is an efficient precursor for oxalate biosynthesis
It had previously been observed that oxalate content increased when plants were fed with glycolate or glyoxylate through roots, with the former being more efficient than the latter (Xu et al., 2006). It was hypothesized that a glycolate dehydrogenase exists in plants, similar to the one identified from human liver that catalyses a direct oxidation of glycolate into oxalate without forming glyoxylate as an intermediate (Fry and Richardson, 1979). If this is true in rice, it is reasonable that glycolate could be more effective than glyoxylate in stimulating oxalate accumulation. However, our long-term effort to identify this enzyme has not been successful (Ji, 2004; X Peng, unpublished data). In addition, it was noted that oxalate abundance was not increased when glycolate was highly accumulated in the GLO-suppressed plants (Xu et al., 2006, 2009), indirectly negating the existence in rice of a human-like glycolate dehydrogenase. In this study, it is shown further that if glycolate or glyoxylate was fed into detached leaves, both substances were equally effective in stimulating oxalate accumulation (Fig. 2). This implies that the lowered effectiveness for glyoxylate to stimulate oxalate accumulation in intact plants may be caused by its less efficient acquisition and transportation, since, in detached leaves, the resistance for acquisition and transportation has been largely reduced. In addition, it has been reported that glycolate could be more effectively taken up into the human HepG2 cells than glyoxylate (Baker et al., 2004). Moreover, glyoxylate is known to be susceptible to non-enzymatic attack by certain amino acids such as lysine, arginine or cysteine (Dutta et al., 2007), causing the cysteine inhibition on the urinary oxalate excretion under hyperoxaluric conditions (Bais et al., 1991; Baker et al., 1998). Similarly, cysteine abolished both glycolate- and glyoxylate-promoted oxalate accumulations in detached leaves (Fig. 2). These data indicate that glyoxylate is involved in oxalate biosynthesis and may mediate the effect of glycolate in rice.
To define further whether glyoxylate is involved in oxalate accumulation, various transgenic plants were generated, in which several genes involved in glyoxylate metabolism were molecularly regulated. SGAT and GGAT catalyse the conversion of glyoxylate into glycine in plants (Liepman and Olsen, 2001; Igarashi et al., 2006). When these two genes were over-expressed and the enzyme activities increased as expected (Figs 3A, 4), both oxalate and glyoxylate contents were decreased with a significant and positive correlation (Fig. 4). Similar results have been reported in human and animals showing that oxalate accumulation was stimulated due to the deficiency in peroxisomal Ala:glyoxylate aminotransferease (AGT) (Baker et al., 2004; Mdluli et al., 2005; Salido et al., 2006). HPR is known to have activity to reduce both hydroxypyruvate and glyoxylate (Givan and Kleczkowski, 1992). Both oxalate and glyoxylate contents were increased when HPR was suppressed (Fig. 3B; Table 1). ICL codes for isocitrate lyase, which catalyses the production of glyoxylate. Oxalate increased as the ICL activity was up-regulated in the over-expression lines (Figs 3C, 5). All of these results further confirm that glyoxylate is an efficient precursor for oxalate biosynthesis in rice.
The conversion of ascorbate to oxalate has been noticed for a long time (Yang and Loewus, 1975), and stronger evidence has emerged more recently (Saito, 1996; Keates et al., 2000; Kostman et al., 2001; Green and Fry, 2005). However, all of the data were obtained by using isotope-labelling techniques. In this study, transgenic plants were constructed that had down-regulated GLDH (Fig. 3D), a key enzyme gene for ascorbate biosynthesis (Wheeler et al., 1998).The ascorbate contents were markedly altered as expected in these transgenic plants, but the oxalate levels were unchanged (Fig. 6). Since the RNAi construct was inserted into the genome in a three-copy fashion (data not shown), the generations were segregated, leading to a wide spread of ascorbate concentrations. It was earlier observed that no correlation exists between oxalate and ascorbate contents, either in different growth stages of tobacco or various plant species (Li and Peng, 2006), and that feeding rice with ascorbate did not significantly promote oxalate accumulation, in contrast to glycolate (Xu et al., 2006). Similar results were also recently presented by Proietti et al. (2009). Therefore, it is considered that oxalate accumulation and regulation are independent of ascorbic acid levels in rice plants.
The next question is, which enzyme(s) catalyses the oxidation of glyoxylate into oxalate. It was recently proved that GLO is not involved in oxalate accumulation and regulation in rice (Xu et al., 2006), even though this enzyme has long shown to be able to catalyse the oxidation of glyoxylate into oxalate in vitro, in addition to its major catalytic activity on the conversion of glycolate to glyoxylate (Richardson and Tolbert, 1961; Xu et al., 2006). It was therefore speculated that an as yet unidentified enzyme exists which is responsible for catalysing the conversion of glyoxylate into oxalate. This novel enzyme is probably a dehydrogenase that is metabolically linked with or regulated by nitrate reduction (Tian et al., 2008). Lactate dehydrogenase had a strong possibility to be the candidate since this enzyme purified from animals and microbes exhibited catalytic activity on the oxidation of glyoxylate into oxalate (Baker et al., 2004; Mdluli et al., 2005). However, the lactate dehydrogenase purified from various plants showed no activity on glyoxylate (Sugiyama and Taniguchi, 1997; data not shown). A glyoxylate dehydrogenase has been reported in microbes and animals (Quayle and Taylor, 1961; Balmforth and Thomson, 1984; Tokimatsu et al., 1998), and whether such an enzyme exists in plants remains to be identified.
The glyoxylate-dependent oxalate accumulation does not necessarily depend on photorespiration
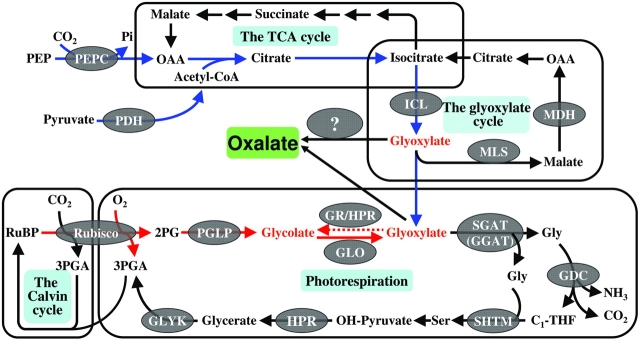
The photorespiratory pathway is known to be a major source of glyoxylate in plants. Now that glyoxylate is considered to be an efficient precursor for oxalate biosynthesis, there should be a correlation between oxalate accumulation and photorespiration. However, lines of evidence to indicate that oxalate accumulation is not necessarily dependent on photorespiration are shown here. First, oxalate accumulation was not dependent on light intensity (Fig. 1A) which is known to be a key factor regulating photorespiration. Secondly, high CO2 failed to reduce the oxalate accumulation (Fig. 1B). Third, there was no correlation between photorespiratory rates (Rp) and oxalate accumulation (Fig. 1C). In support, oxalate can be accumulated in callus, where there is supposedly no photorespiration (Franceschi, 1987). It was also noted recently that suppression of GLO, a key enzyme in photorespiration, had no impact on oxalate accumulation in rice (Xu et al., 2006). It is then curious as to how oxalate accumulation can not be correlated with photorespiration or glycolate oxidase since its glyoxylate-dependence has been considered? Several possibilities are proposed here. First, the photorespiratory glyoxylate could not be efficiently utilized for oxalate biosynthesis due to cellular compartmentalization. If oxalate is not biosynthesized in peroxisomes where photorespiratory glyoxylate production occurs, the channelling of glyoxylate out of peroxisomes, rather than its formation, becomes the limiting factor in regulating oxalate accumulation. The second possibility is that there may exist an anaplerotic reaction to supplement the glyoxylate formation once the photorespiratory source is disrupted. This possibility has been supported by our recent data. Xu et al. (2009) found that although GLO activity was dramatically reduced there were minimal changes for the product glyoxylate, and for some other downstream metabolites or genes. The Rp measurement using the low O2 (1%) method showed that Rp was not significantly reduced even if GLO was dramatically suppressed (data not shown). Further analyses revealed that ICL and MLS (malate synthase), two key enzyme genes for the glyoxylate cycle, were highly up-regulated, indicating that the glyoxylate cycle may serve as anaplerotic reactions to compensate for the glyoxylate formation, particularly when the glycolate oxidation pathway is disrupted (Xu et al., 2009). Actually, similar results were earlier reported by Zelitch (1973, 1988). Moreover, the increased oxalate in the ICL over-expression lines (Fig. 5) further support that the glyoxylate from the glyoxylate cycle can be effectively utilized for oxalate biosynthesis in rice. Based on these results, a hypothetical pathway is proposed, illustrating the possible metabolic interactions for regulating glycolate/glyoxylate in relation to oxalate biosynthesis in rice (Fig. 7).
Fig. 7.
A hypothetical ananplerotic pathway for glycolate/glyoxylate in relation to oxalate formation. Glycolate/glyoxylate is produced during photorespiration (red arrows) or can be compensated via ananplerotic reactions related to the glyoxylate cycle (blue arrows). PEPC, PEP carboxylase; PDH, pyruvate dehydrogenase; MDH, malate dehydrogenase; GR, glyoxylate reductase; GGAT, Glu:glyoxylate aminotransferase; SHMT, Ser hydroxymethyltransferase; OAA, oxaloacetate; RuBP, ribulose-1,5-bisphosphate; 2PG, 2-phosphoglycolate; C1-THF, C1-tetrahydrofolate; OH-pyruvate, hydroxypyruvate; 3PGA, 3-phosphoglycerate.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. The primer sequences used for quantifying the gene expression.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (30971710, 30870184) and the National Basic Research Program of China (973 Program) (2009CB118504). This work was also partially supported by the National Institutes of Health (SCORE award no. S06 GM52588) to ZHH. We thank Dr Colin Leasure (Department of Biology, San Francisco State University) for reading this manuscript.
References
- Bais R, Rofe AM, Conyers RA. The inhibition of metabolic oxalate production by sulfhydryl compounds. Journal of Urology. 1991;145:1302–1305. doi: 10.1016/s0022-5347(17)38619-6. [DOI] [PubMed] [Google Scholar]
- Baker PRS, Cramer SD, Kennedy M, Assimos DG, Holmes RP. Glycolate and glyoxylate metabolism in HepG2 cells. American Journal of Physiology- Cell Physiology. 2004;287:C1359–C1365. doi: 10.1152/ajpcell.00238.2004. [DOI] [PubMed] [Google Scholar]
- Baker PW, Rofe AM, Bais R. The effect of (l)-cysteine and (l)-2-oxothiazolidine-4-carboxylic acid (OTZ) on urinary oxalate excretion: studies using a hyperoxaluric rat model. Journal of Urology. 1998;159:2177–2181. doi: 10.1016/S0022-5347(01)63301-9. [DOI] [PubMed] [Google Scholar]
- Balmforth AJ, Thomson A. Isolation and characterization of glyoxylate dehydrogenase from the fungus Sclerotium rolfsii. Biochemical Journal. 1984;218:113–118. doi: 10.1042/bj2180113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brown RH, Morgan JA. Photosynthesis of grass species differing in carbon dioxide fixation pathways. vi. Differential effects of temperature and light intensity on photorespiration in C3, C4, and intermediate species. Plant Physiology. 1980;66:541–544. doi: 10.1104/pp.66.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cessna SG, Sears VE, Dickman MB, Low PS. Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. The Plant Cell. 2000;12:2191–2199. doi: 10.1105/tpc.12.11.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Beevers H. Biogenesis of oxalate in plant tissues. Plant Physiology. 1968;43:1821–1828. doi: 10.1104/pp.43.11.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YE, Harada E, Wada M, Tsuboi H, Morita Y, Kusano T, Sano H. Detoxification of cadmium in tobacco plants: formation and active excretion of crystal containing cadmium and calcium through trichomes. Planta. 2001;213:45–50. doi: 10.1007/s004250000487. [DOI] [PubMed] [Google Scholar]
- Dutta U, Cohenford MA, Guha M, Dain JA. Non-enzymatic interactions of glyoxylate with lysine, arginine, and glucosamine: a study of advanced non-enzymatic glycation like compounds. Bioorganic Chemistry. 2007;35:11–24. doi: 10.1016/j.bioorg.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Errakhi R, Meimoun P, Lehner A, Vidal G, Briand J, Corbineau F, Rona JP, Bouteau F. Anion channel activity is necessary to induce ethylene synthesis and programmed cell death in response to oxalic acid. Journal of Experimental Botany. 2008;59:3121–3129. doi: 10.1093/jxb/ern166. [DOI] [PubMed] [Google Scholar]
- Franceschi VR. Oxalic acid metabolism and calcium oxalate formation in Lemna minor L. Plant, Cell and Environment. 1987;10:397–406. [Google Scholar]
- Franceschi VR, Nakata PA. Calcium oxalate in plants: formation and function. Annual Review of Plant Biology. 2005;56:41–71. doi: 10.1146/annurev.arplant.56.032604.144106. [DOI] [PubMed] [Google Scholar]
- Franceschi VR, Schueren AM. Incorporation of strontium into plant calcium oxalate crystals. Protoplasma. 1986;130:199–205. [Google Scholar]
- Fry DW, Richardson KE. Isolation and characterization of glycolic acid dehydrogenase from human liver. Biochimica et Biophysica Acta. 1979;567:482–491. doi: 10.1016/0005-2744(79)90134-7. [DOI] [PubMed] [Google Scholar]
- Fujii N, Watanabe M, Watanabe Y, Shimada N. Rate of oxalate biosynthesis from glycolate and ascorbic acid in spinach leaves. Journal of Plant Nutrition and Soil Science. 1993;39:627–634. [Google Scholar]
- Givan CV, Kleczkowski LA. The enzymic reduction of glyoxylate and hydroxypyruvate in leaves of higher plants. Plant Physiology. 1992;100:552–556. doi: 10.1104/pp.100.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MA, Fry SC. Vitamin C degradation in plant cells via enzymatic hydrolysis of 4- O-oxalyl-l-threonate. Nature. 2005;433:83–87. doi: 10.1038/nature03172. [DOI] [PubMed] [Google Scholar]
- Horner HT, Kausch AP, Wagner BL. Ascorbic acid: a precursor of oxalate in crystal idioblasts of Yucca torreyi in liguid root culture. International Journal of Plant Science. 2000;161:861–868. [Google Scholar]
- Igarashi D, Tsuchida H, Miyao M, Ohsumi C. Glutamate: glyoxylate aminotransferase modulates amino acid content during photorespiration. Plant Physiology. 2006;142:901–910. doi: 10.1104/pp.106.085514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Peng X. Oxalate accumulation as regulated by nitrogen forms and its relation to photosynthesis in rice. Journal of Integrative Biology. 2005;47:831–838. [Google Scholar]
- Kampfenkel K, Motagu MV, Inzé D. Extraction and determination of ascorbate and dehydroascorbate from plant tissue. Analytical Biochemistry. 1995;225:165–167. doi: 10.1006/abio.1995.1127. [DOI] [PubMed] [Google Scholar]
- Keates SE, Tarlyn NM, Loewus FA, Franceschi VR. L-Ascorbic acid and l-galactose are sources for oxalic acid and calcium oxalate in Pistia stratiotes. Phytochemistry. 2000;53:433–440. doi: 10.1016/s0031-9422(99)00448-3. [DOI] [PubMed] [Google Scholar]
- Kim KS, Min JY, Dickman MB. Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Molecular Plant–Microbe Interactions. 2008;21:605–612. doi: 10.1094/MPMI-21-5-0605. [DOI] [PubMed] [Google Scholar]
- Kleczkowski LA, Edwards GE, Blackwell RD, Lea PJ, Givan CV. Enzymology of the reduction of hydroxypyruvate and glyoxylate in a mutant of barley lacking peroxisomal hydroxypyruvate reductase. Plant Physiology. 1990;94:819–825. doi: 10.1104/pp.94.2.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth KL, Doege SJ, Park SH, Goggin FL, Wang Q, Gomez SK, Liu G, Jia L, Nakata PA. Medicago truncatula mutants demonstrate the role of plant calcium oxalate crystals as an effective defense against chewing insects. Plant Physiology. 2006;141:188–195. doi: 10.1104/pp.106.076737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostman TA, Tarlyn NM, Loewus FA, Franceschi VR. Biosynthesis of l-ascorbic acid and conversion of carbons 1 and 2 of l-ascorbic acid to oxalic acid occurs within individual calcium oxalate crystal idioblasts. Plant Physiology. 2001;125:634–640. doi: 10.1104/pp.125.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BS, Peng XX. Relationship between oxalate accumulation and ascorbate content in plant leaves. Plant Physiology Communications. 2006;42:31–33. [Google Scholar]
- Libert B, Franceschi VR. Oxalate in crop plants. Journal of Agricultural and Food Chemistry. 1987;35:926–936. [Google Scholar]
- Liepman AH, Olsen LJ. Peroxisomal alanine: glyoxylate aminotransferase (AGT1) is a photorespiratory enzyme with multiple substrates in Arabidopsis thaliana. The Plant Journal. 2001;25:487–498. doi: 10.1046/j.1365-313x.2001.00961.x. [DOI] [PubMed] [Google Scholar]
- Lin Z, Peng C, Sun Z, Lin G. Effect of light intensity on partitioning of photosynthetic electron transport to photorespiration in four subtropical forest plants. Science in China Series C: Life Sciences. 2000;43:347–354. doi: 10.1007/BF02879298. [DOI] [PubMed] [Google Scholar]
- Ma JF, Zheng SJ, Hiradate S, Matsumoto H. Detoxifying aluminum with buckwheat. Nature. 1997;390:569–570. [Google Scholar]
- Ma Z, Miyasaka SC. Oxalate exudation by taro in response to Al. Plant Physiology. 1998;118:861–865. doi: 10.1104/pp.118.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazen AMA, EI Maghraby OMO. Accumulation of cadmium, lead and strontium, and a role of calcium oxalate in water hyacinth tolerance. Biologia Plantarum. 1997;40:411–417. [Google Scholar]
- Mdluli K, Booth MP, Brady RL, Rumsby G. A preliminary account of the properties of recombinant human glyoxylate reductase (GRHPR), LDHA, and LDHB with glyoxylate, and their potential roles in its metabolism. Biochimica et Biophysica Acta. 2005;1753:209–216. doi: 10.1016/j.bbapap.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Mijovilovich A, Leitenmaier B, Meyer-Klaucke W, Kroneck PM, Gotz B, Kupper H. Complexation and toxicity of copper in higher plants (II): different mechanisms for Cu versus Cd detoxification in the Cu-sensitive Cd/Zn hyperaccumulator Thlaspi caerulescens (Ganges ecotype) Plant Physiology. 2009;151:715–731. doi: 10.1104/pp.109.144675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita A, Yanagisawa O, Takatsu S, Maeda S, Hiradate S. Mechanism for the detoxification of aluminum in roots of tea plant (Camellia sinensis (L.) Kuntze) Phytochemistry. 2008;69:147–153. doi: 10.1016/j.phytochem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Nakata PA. Advances in our understanding of calcium oxalate crystal formation and function in plants. Plant Science. 2003;164:901–909. [Google Scholar]
- Proietti S, Moscatello S, Famiani F, Battistelli A. Increase of ascorbic acid content and nutritional quality in spinach leaves during physiological acclimation to low temperature. Plant Physiology and Biochemistry. 2009;47:717–723. doi: 10.1016/j.plaphy.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Quayle JR, Taylor GA. Carbon assimilation by Pseudomonas oxalaticus (OX1). 5. Purification and properties of glyoxylate dehydrogenase. Biochemical Journal. 1961;78:611–615. doi: 10.1042/bj0780611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranaldi F, Vanni P, Giachetti E. Multisite inhibition of Pinus pinea isocitrate lyase by phosphate. Plant Physiology. 2000;124:1131–1138. doi: 10.1104/pp.124.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA, Griffiths H, Glidewell SM, Preston T. The mechanism of oxalate biosynthesis in higher plants: investigations with the stable isotopes 18O and 13C. Proceedings of the Royal Society of London, Series B-Biological Sciences. 1982;216:87–101. [Google Scholar]
- Richardson KE, Tolbert NE. Oxidation of glyoxylic acid to oxalic acid by glycolate oxidase. Journal of Biological Chemistry. 1961;236:1280–1284. [PubMed] [Google Scholar]
- Ruiz lP, Mansfield TA. A postulated role for calcium oxalate in the regulation of calcium ions in the vicinity of stomatal guard cells. New Phytologist. 1994;127:473–481. [Google Scholar]
- Saito K. Formation of l-ascorbic acid and oxalate acid from d-glucosone in Lemna minor. Phytochemistry. 1996;41:145–149. [Google Scholar]
- Salido EC, Li XM, Lu Y, Wang X, Santana A, Roy-Chowdhury N, Torres A, Shapiro LJ, Roy-Chowdhury J. Alanine-glyoxylate aminotransferase-deficient mice, a model for primary hyperoxaluria that responds to adenoviral gene transfer. Proceedings of the National Academy of Sciences, USA. 2006;103:18249–18254. doi: 10.1073/pnas.0607218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey TD. Estimating the rate of photorespiration in leaves. Physiologia Plantarum. 1988;73:147–152. [Google Scholar]
- Somerville CR. An early Arabidopsis demonstration. Resolving a few issues concerning photorespiration. Plant Physiology. 2001;125:20–24. doi: 10.1104/pp.125.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N, Taniguchi N. Evaluation of the role of lactate dehydrogenase in oxalate synthesis. Phytochemistry. 1997;44:571–574. doi: 10.1016/s0031-9422(96)00629-2. [DOI] [PubMed] [Google Scholar]
- Tian H, Jiang L, Liu E, Zhang J, Liu F, Peng X. Dependence of nitrate-induced oxalate accumulation on nitrate reduction in rice leaves. Physiologia Plantarum. 2008;133:180–189. doi: 10.1111/j.1399-3054.2008.01052.x. [DOI] [PubMed] [Google Scholar]
- Tokimatsu T, Nagai Y, Hattori T, Shimada M. Purification and characteristics of a novel cytochrome c dependent glyoxylate dehydrogenase from a wood-destroying fungus Tyromyces palustris. FEBS Letters. 1998;437:117–121. doi: 10.1016/s0014-5793(98)01104-1. [DOI] [PubMed] [Google Scholar]
- Weir TL, Bais HP, Stull VJ, Callaway RM, Thelen GC, Ridenour WM, Bhamidi S, Stermitz FR, Vivanco JM. Oxalate contributes to the resistance of Gaillardia grandiflora and Lupinus sericeus to a phytotoxin produced by Centaurea maculosa. Planta. 2006;223:785–795. doi: 10.1007/s00425-005-0192-x. [DOI] [PubMed] [Google Scholar]
- Wheeler GL, Jones MA, Smirnoff N. The biosynthetic pathway of vitamin C in higher plants. Nature. 1998;393:365–369. doi: 10.1038/30728. [DOI] [PubMed] [Google Scholar]
- Xu H, Zhang J, Zeng J, Jiang L, Liu E, Peng C, He Z, Peng X. Inducible antisense suppression of glycolate oxidase reveals its strong regulation over photosynthesis in rice. Journal of Experimental Botany. 2009;60:1799–1809. doi: 10.1093/jxb/erp056. [DOI] [PubMed] [Google Scholar]
- Xu HW, Ji XM, He ZH, Shi WP, Zhu GH, Niu JK, Li BS, Peng XX. Oxalate accumulation and regulation is independent of glycolate oxidase in rice leaves. Journal of Experimental Botany. 2006;57:1899–1908. doi: 10.1093/jxb/erj131. [DOI] [PubMed] [Google Scholar]
- Yang JC, Loewus FA. Metabolic conversion of l-ascorbic acid to oxalic acid in oxalate-accumulating plants. Plant Physiology. 1975;56:283–285. doi: 10.1104/pp.56.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YY, Jung JY, Song WY, Suh HS, Lee Y. Identification of rice varieties with high tolerance or sensitivity to lead and characterization of the mechanism of tolerance. Plant Physiology. 2000;124:1019–1026. doi: 10.1104/pp.124.3.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA. Laboratory manual for physiological studies of rice. Manila. Philippines: International Rice Research Institute; 1976. [Google Scholar]
- Zelitch I. Alternate pathways of glycolate synthesis in tobacco and maize leaves in relation to rates of photorespiration. Plant Physiology. 1973;51:299–305. doi: 10.1104/pp.51.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelitch I. Synthesis of glycolate from pyruvate via isocitrate lyase by tobacco leaves in light. Plant Physiology. 1988;86:463–468. doi: 10.1104/pp.86.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JL, Meng LZ, Cao KF. Sustained diurnal photosynthetic depression in uppermost-canopy leaves of four dipterocarp species in the rainy and dry seasons: does photorespiration play a role in photoprotection? Tree Physiology. 2009;29:217–228. doi: 10.1093/treephys/tpn018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.