Abstract
MicroRNAs (miRNAs) act in post-transcriptional gene silencing and are proposed to function in a wide spectrum of pathologies, including cancers and viral diseases. Currently, to our knowledge, no detailed mechanistic characterization of small molecules that interrupt miRNA pathways have been reported. In screening a small chemical library, we identified compounds that suppress RNA interference activity in cultured cells. Two compounds were characterized; one impaired Dicer activity while the other blocked small RNA-loading into an Argonaute 2 (AGO2) complex. We developed a cell-based model of miRNA-dependent tumorigenesis, and using this model, we observed that treatment of cells with either of the two compounds effectively neutralized tumor growth. These findings indicate that miRNA pathway-suppressing small molecules could potentially reverse tumorigenesis.
Keywords: Cancer, Diseases/Cancer/Carcinogenesis, RNA/Interference/RNAi, RNA/MicroRNA, Carcinogenesis, Adult T-cell Leukemia, HTLV
Introduction
MicroRNAs (miRNAs)3 are small non-coding RNAs that regulate the expression of many genes and are involved in a variety of cellular processes, including development, cell proliferation, apoptosis, immunity, hematopoiesis, tumorigenesis, and viral infection (1–4). Primary miRNAs are transcribed and subsequently processed to precursor miRNAs (pre-miRNAs), which are exported into the cytoplasm and cleaved by Dicer into ∼23 nucleotide (nt) RNA duplexes (5–7). These small RNAs are loaded onto an effector complex, termed the RNA-induced silencing complex (RISC), which is constituted by the argonaute (AGO) protein as a central core component (7–11) together with other regulatory proteins such as TRBP, PACT, MOV10, Gemin3/4, GW182, TNRC6B, and RNA helicase A (RHA) (12–19). Once RISC is loaded with a guide miRNA or siRNA, the mi-RISC or si-RISC can recognize and anneal to target mRNAs to down-regulate their expression (20, 21).
Dysregulated miRNAs contribute to disease pathogenesis (22, 23). For example, an miRNA cluster miR-17–92 is often up-regulated in human B-cell lymphomas, leading to accelerated disease progression (24). miRNA-155, overexpressed in several types of B-cell lymphomas and solid tumors (25, 26), can also contribute to B-cell malignancy (27). miR-21 is augmented in various tumors and exhibits pro-tumorigenic activity (26, 28, 29). Other examples include the oncological roles played by miR-93 and miR-130b in human T-cell leukemia virus type-I (HTLV-I)-infected adult T-cell leukemia (30), the role of let-7, miR-15, and miR-16 as tumor suppressors (31, 32), and the contributions of RISC components such as AGO proteins (33–35), Dicer (36, 37), and TRBP (38) to cellular proliferation. The variegated actions of miRNAs in cell growth are likely cell-type- and context-specific. Until now, there is no well characterized demonstration that chemical manipulation to activate or inactivate miRNA pathways can influence miRNA-related tumorigenesis.
Here, we screened a chemical compound library for moieties that modulate miRNA-mediated gene silencing in cells. Two compounds with relatively non-cytotoxic profiles that suppress miRNA-RISC activity, polylysine (PLL) and trypaflavine (TPF), were identified. In a miRNA-dependent cellular transformation model, treatment of cells with PLL and TPF effectively attenuated the cells' tumorigenic properties.
EXPERIMENTAL PROCEDURES
Reagents and Compound Screening
530 compounds were screened (supplemental Table S1). The compounds included some whose bioactivities and pharmacological properties have been well characterized. For most of the compounds, we examined each at six different concentrations (0.041, 0.12, 0.37, 1.1, 3.3, and 10 μm). An initial screening was conducted by transfection with pRS-shLuc, pRL-TK, and pGL3 promoter-plasmids, which express sh-Fluc (shRNA targeting firefly luciferase mRNA) or Rluc (Renilla luciferase expression vector), or SV40 promoter-driven Fluc (firefly luciferase), respectively, into 293T cells (supplemental Figs. S1 and S2). This screening identified 19 compounds (3.6% of the total number of compounds tested) that augmented the ratio of Fluc/Rluc more than 2-fold, and 3 compounds (0.57% of the total compounds) that decreased Fluc/Rluc by 2-fold or more. Next, two follow-up screenings to cull false positives from the initial screening were performed (supplemental Fig. S3). The first follow-up employed pCMV-luc, encoding a CMV promoter-driven Fluc, instead of the SV40 promoter-driven Fluc used in the initial screening. In the second follow-up, a pRS vector expressing sh-GFP in place of pRS-shLuc was used. Compounds that showed positive results in the initial and second screenings, but negative or unchanged results in the third screening were sought. Based on these criteria, two candidates, PLL and TPF, from our pool of 530 compounds were identified (supplemental Fig. S4). The PLL (poly-l-lysine hydrobromide, molecular weight 4,000–15,000 (Sigma-Aldrich, P6516)) and TPF (3,6-diamino-10-methylacridinium chloride) used in this study were purchased from Sigma-Aldrich.
Plasmid Construction
The expression vectors for Fluc and Rluc were the pGL3-plasmid (Promega, Madison, WI) and the pRL-TK plasmid (Promega), respectively. Those for sh-Fluc, sh-GFP, and their corresponding empty vector were pRS-shLuc, pRS-shGFP, and pRS control plasmid (OriGene), respectively. pIRESneo-FLAG/HA-AGO2 was provided from Drs. Thomas Tuschl and Gunter Meister at the Rockefeller University through Addgene Inc. (7). pCA-FLAG·AGO2 and pCA-HA-AGO2 were prepared by inserting a PCR product using 5′-GTTGAATTCATGTACTCGGGAG-3′ and 5′-GTTGCGGCCGCTCAAGCAAAGTAC-3′ as primers and pIRESneo-FLAG/HA-AGO2 as a template. pFLAG-TRBP was generated based on the TRBP expression plasmid as described previously (39). pcDNA-FLAG-RHA was provided from Dr. Chee-Gun Lee at the University of Medicine and Dentistry of New Jersey. The expression plasmids for miR-93 and miR-130b were described previously (30, 40).
Cell Culture and Transfection
293T and HeLa cells were cultured with DMEM (Invitrogen) supplemented with 10% fetal bovine serum and l-glutamine (Invitrogen, growth medium). 293T(FLAG·AGO2) cells were established by transfection with pCA-FLAG·AGO2 and pRS vector into 293T cells followed by selection with 1 μg/ml puromycin for 3 weeks. These cells were maintained with the above growth medium supplemented with 1 μg/ml puromycin. 3T3-miR-93, 3T3-miR-130b, 3T3-Ras, and 3T3-control cells were established by transduction of miR-93-, miR-130b-, Ras-, or empty vector-packaged feline immunodeficiency virus into NIH3T3 cells followed by selection with G418. HTLV-I-infected cells and MT1, MT4, and ED cells were cultured in RPMI 1640 (Invitrogen) supplemented with 10% fetal bovine serum and l-glutamine. PBMCs were isolated from healthy donors and cultured with the above medium supplemented with 2 μg/ml phytohemagglutinin and 20 units/ml interleukin-2.
Plasmid transfection into cells was performed using TurboFectin 8.0 (OriGene), Lipofectamine 2000 (Invitrogen), and FuGENE 6 transfection reagent (Roche Applied Science) according to the manufacturer's protocol. Small RNAs were transfected with Lipofectamine 2000 and Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's protocol.
Antibodies
Antibodies used in this study were anti-Fluc (PM016, MBL), anti-Rluc (MAB4410, Millipore), anti-actin (Sigma), anti-FLAG (M2, Sigma), anti-HA (HA-7, Sigma), and anti-RHA (anti-DHX9 A300–855A, Bethyl Laboratories) antibodies.
Luciferase Assay
Luciferase activity was quantified using the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer's protocol.
Cell Proliferation Assay
Cell proliferation was examined by MTT assay using the Cell Proliferation Kit II (XTT, Roche Applied Science) according to the manufacturer's protocol.
Immunoblot Analysis
Immunoblots were performed as described previously (41).
Recovery and Quantification of Small RNA
Small RNA fraction was recovered with an mirVana miRNA isolation kit (Ambion) or TRIzol (Invitrogen). siRNA and miRNA were quantified using a QuantiMir RT kit (System Biosciences) and a TaqMan MicroRNA Assay kit (Applied Biosystems) by following the manufacturers' protocol. Real-time PCR analysis was done as described previously (30). The antisense primers used for the real-time PCR followed the procedure with QuantiMir RT kit and are 5′-GCTGGAGTACAACTACAACAGCC-3′, 5′-TAGCAGCACGTAAATATTGGCG-3′, 5′-CAAAGTGCTTACAGTGCAGGTAG-3′, 5′-TGAGGTAGTAGGTTGTATAGTT-3′, 5′-TAGCTTATCAGACTGATGTTGA-3′, 5′-TGTAAACATCCTCGACTGGAAG-3′, and 5′-TAGGTAGTTTCATGTTGTTGGG-3′ for detection of si-GFP, miR-16, miR-17, let-7a, miR-21, miR-30a, and miR-196a, respectively.
In Vitro Dicer Assay
The enzymatic activity of Dicer was studied in vitro with Turbo Dicer siRNA generation kit (Genlantis). For preparing a substrate RNA, the control GFP plasmid was amplified by PCR reaction. The PCR product was transcribed using TurboScript T7 transcription kit (Genlantis) in the presence of [α-32P]UTP. The resulting substrate RNA was reacted with a recombinant Dicer by following the manufacturer's protocol. After electrophoresis in a denaturing gel, labeled RNAs were visualized with an imaging analyzer FLA-7000 (Fujifilm).
Immunoprecipitation Analysis
Immunoprecipitation (IP) for recovering AGO2-IP fraction was performed using the IP buffer comprising 50 mm HEPES, 150 mm NaCl, 0.5% Nonidet P-40, 10% glycerol, 1.5 mm MgCl2, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, protease inhibitor (Complete, Roche), and 100 units/ml SUPERase-In RNase inhibitor (Ambion). After lysing cells, 2 μg of anti-FLAG antibody was added to isolate the AGO2 complex. In co-immunoprecipitation analysis for protein binding, cells were lysed with the IP buffer without SUPERase-In. 2 μg of anti-FLAG, anti-HA, and anti-RHA antibodies was used for the immunoprecipitation.
RNA Precipitation Analysis
Unless otherwise indicated, the small RNAs used for RNA precipitation assay were biotinylated (bio-) with si-GFP; the RNAs were prepared by annealing the two strands 5′-CUUGAAGAAGUCGUGCUGCdTdT-bio-3′ and 5′-GCAGCACGACUUCUUCAAGdTdT-3′ (Dharmacon).
293T(FLAG·AGO2) (42) cells that were treated with TPF or PLL, or not treated with TPF or PLL, were lysed and incubated with bio-si-GFP. The RNA·protein complex was recovered by precipitation with streptavidin-Sepharose (GE healthcare) (streptavidin precipitation fraction) or without precipitation to detect the total FLAG·AGO2 complex in the lysate. Where indicated, 293T(FLAG·AGO2) cells were transfected with bio-si-GFP and treated with TPF or were untreated. These cells were then lysed and recovered with streptavidin-Sepharose (streptavidin precipitation, upper panel) or directly recovered without beads precipitation. Recovered AGO2 complex was detected with anti-FLAG antibody.
Colony Formation in Soft Agar
The base and the top agar were prepared with 0.5% agar (bacto-agar, DIFCO)/DMEM/F-12 (Invitrogen) and 0.35% agar (agar noble, DIFCO)/DMEM/F-12, respectively. After laying a base agar, 7 × 102 cells were mixed with a top agar solution to spread over the base agar in a 6-well plate. Cells were observed for 1 week for colony formation. Cells were pretreated with PLL or TPF or were untreated for 3 days before being subjected to the soft agar assay. The agar used in this assay was supplemented with PLL or TPF or was unsupplemented.
In Vivo Implantation of Cells into Nude Mice
2 × 106 cells were suspended in 100 μl of DMEM/2% fetal bovine serum/25% Matrigel (BD Biosciences). The cell suspension was injected into 5- to 8-week-old female nude mice (BALBNU-M-F (C.Cg/AnNTac-FoxnlnuNE9), Taconic) subcutaneously in the neck region according to the protocol approved by the NIAID, National Institutes of Health animal care and use committee. Three mice per group were used for the experiment. At 2 weeks after implantation of cells, the mice were observed for tumor formation. Tumor weight was measured after dissection.
RESULTS
Identification of Compounds That Suppress RNAi
miRNA and shRNA share some common steps in biogenesis and activity. To measure the gene-silencing activities of miRNAs and shRNAs in a cell-based assay, we employed a firefly luciferase (Fluc) mRNA-targeting shRNA (sh-Fluc), which is processed inside cells to a Fluc-specific siRNA (supplemental Fig. S1). Cells transfected with sh-Fluc silenced a Fluc reporter, but not a control Renilla luciferase (Rluc) reporter (supplemental Fig. S2A). This sh-Fluc-induced gene silencing was less effective if cell endogenous AGO2 was first knocked down (supplemental Fig. S2B), verifying that the observed effect was indeed due to RNAi. Using this assay, we screened a library of 530 compounds containing many previously characterized drugs (supplemental Table S1).
Our initial screening netted 19 compounds that increased, and 3 compounds that decreased, the Fluc/Rluc ratio in cells transfected with Fluc, Rluc, and sh-Fluc. This first screening used a SV40 promoter-driven Fluc plasmid. In a second screening, we employed a Fluc expression plasmid driven by a CMV promoter (supplemental Fig. S3); finally, as a negative control, we performed a third screening in which Fluc and Rluc expression plasmids were targeted with an irrelevant shRNA (an shRNA that targets GFP (sh-GFP)) in place of sh-Fluc (supplemental Fig. S3). Collectively, the goal of these assays was to identify compounds that modulate specific sh-Fluc targeting of Fluc (as measured by the Fluc/Rluc ratio) normalized against any nonspecific sh-GFP effect on the Fluc/Rluc ratio (supplemental Fig. S3, right column).
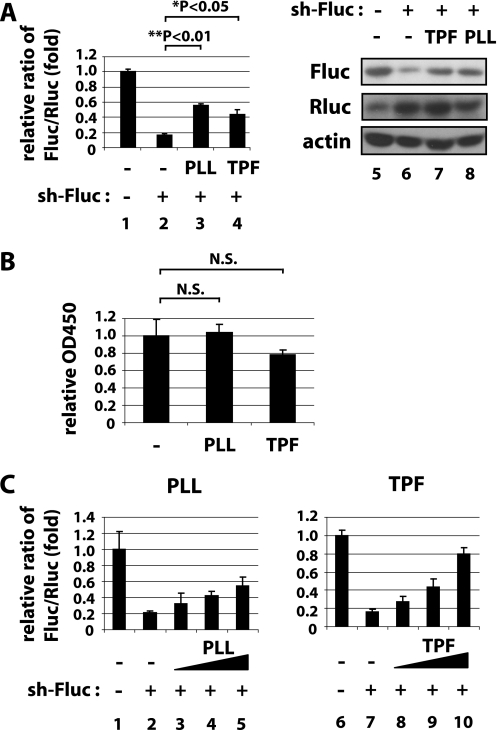
Among the 530 samples after the three screenings, two compounds, PLL and TPF (supplemental Fig. S4), were found to suppress significantly the specific Fluc/Rluc ratio while minimally perturbing the assay that used a negative control sh-GFP (supplemental Fig. S3). We next verified that treatment of cells with PLL or TPF did reduce specific sh-Fluc targeting of Fluc (Fig. 1A, left, graph of luciferase assay; right, Western blotting), but did not elicit significant cytotoxicity (Fig. 1B). In immunoblotting for luciferase protein in sh-Fluc-transfected cells treated without (Fig. 1A, lane 6) or with TPF (Fig. 1A, lane 7) or PLL (Fig. 1A, lane 8), we observed increased Fluc protein in the PLL- and TPF-treated cells (Fig. 1A, compare lane 6 to lanes 7 and 8 of the upper panel), and we found that both PLL (Fig. 1C, lanes 1–5) and TPF (Fig. 1C, lanes 6–10) exhibited dose-dependent reversal of sh-Fluc-mediated silencing of Fluc (Fig. 1C).
FIGURE 1.
PLL and TPF suppress shRNA-mediated gene silencing. A, 293T cells were transfected with expression plasmids for firefly (Fluc) and Renilla luciferase (Rluc) together with a shRNA that targets Fluc (sh-Fluc, lanes 2–4) or a control irrelevant shRNA (sh-control, lane 1). Sh-Fluc is designed to silence Fluc, but not Rluc. In parallel, cells were treated with 3.3 μm PLL (lane 3), 3.3 μm TPF (lane 4), or were untreated (lanes 1 and 2). In the left graph, luciferase activities were quantified at 24 h post treatment, and Fluc/Rluc ratios are graphed based on the averages from three independent experiments. In the right panels, Fluc (top), Rluc (middle), and actin (bottom) as an internal control were immunoblotted. Effective silencing of Fluc by sh-Fluc was seen in lane 2 while treatment with PLL (lane 3) or TPF (lane 4) reduced the effectiveness of silencing. B, MTT assays for cell viability of control-treated or PLL- or TPF-treated cells. C, dose-dependent suppression of shRNA silencing by PLL (left) and TPF (right). 293T cells were transfected with plasmids as in A and then treated with escalating amounts of PLL (0, 0, 0.31, 1.3, and 5.0 μm in lanes 1–5) or TPF (0, 0, 1.5, 3.3, and 7.5 μm in lanes 6–10). Luciferase activities were quantified as in A.
PLL and TPF Affect Different Mechanistic Steps
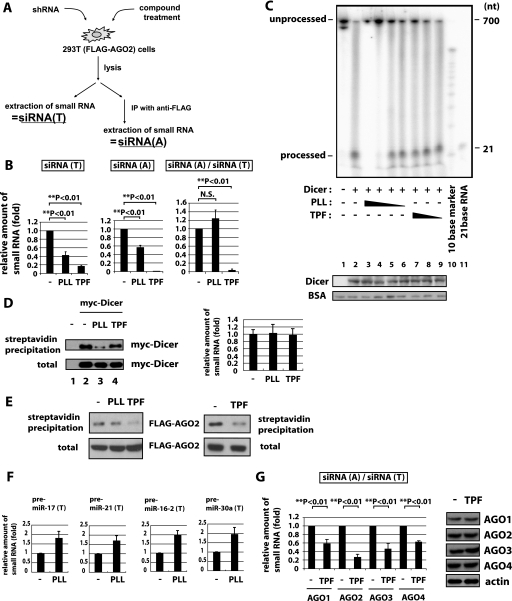
How do PLL and TPF affect shRNA-mediated silencing? Suppression of silencing can occur at several steps. Among others, these steps include Dicer processing of shRNA/pre-miRNA to siRNA/miRNA, the incorporation of guide siRNA/miRNA into RISC, the recognition of target RNA by guide RNA, and the suppression of protein expression by mi- or si-RISC. To investigate these mechanisms, we used a 293T cell line with a stably integrated FLAG·AGO2 expression vector (293T(FLAG·AGO2), Fig. 2A) (42). This 293T(FLAG·AGO2) cell permits the recovery of AGO2 complexes using anti-FLAG monoclonal antibody. An shRNA when transfected into 293T(FLAG·AGO2) cells is expected to be processed by Dicer to siRNA; this siRNA can enter either the total small RNA (siRNA(T)) pool and/or the AGO2-associated small RNA (siRNA(A)) pool (Fig. 2A). One could then query if PLL or TPF treatment changes the relative amounts of shRNA-produced small RNA in the siRNA(T) and/or siRNA(A) fractions. Mechanistically, if PLL or TPF affects Dicer function, then the amount of siRNA in siRNA(T) might be expected to be reduced. On the other hand, the amount of siRNA in the siRNA(T) fraction could also be decreased if Dicer function is not affected, but some other process perturbs siRNA stability. Interestingly, sh-GFP-transfected cells treated with either PLL or TPF showed clearly decreased amounts of GFP siRNA in the siRNA(T) fraction (Fig. 2B, left graph).
FIGURE 2.
PLL and TPF inhibit small RNA biogenesis/silencing in different ways. A, schematic representation of the experimental procedure for recovering shRNA produced small RNAs in AGO2-IP and total cell fractions. 293T(FLAG·AGO2) cells after shRNA transfection and treatment with compounds were lysed and equally divided and then immediately extracted for total small siRNAs (siRNA(T); left). The lysate was also subjected to immunoprecipitation with anti-FLAG followed by the extraction of small AGO2-associated siRNAs (siRNA(A); right). B, siRNA(T) and siRNA(A) were extracted as described in A from 293T(FLAG·AGO2) cells treated with PLL, or TPF, or untreated cells (−). The amount of processed sh-GFP (si-GFP) was measured by real-time RT-PCR. The relative ratio of si-GFP in sRNA(T) (si-GFP(T), left) and that in sRNA(A) (si-GFP(A), center) from control-, PLL-, or TPF-treated cells was calculated using cells without treatment (−) set as 1. To quantify AGO2 association of si-GFP, the si-GFP(A) value was normalized by dividing with the si-GFP(T) value and graphed (right). The association of si-GFP with AGO2 (siRNA(A)/siRNA(T)) in cells treated with TPF was reduced compared with control (−) or PLL treated cells. C, in vitro Dicer assay. Various amounts of PLL (8, 4, 2, and 1 μm in lanes 3–6) or TPF (100, 10, and 1 μm in lanes 7–9) were added to the reactions. The substrate (unprocessed) and processed 32P-labeled RNAs were visualized. The presence of a “processed” band measures the Dicer enzymatic activity. The lower panel shows the amount of immunoblotted Dicer protein in each lane. Bovine serum albumin carrier protein was also blotted in each sample as a loading control. The addition of PLL, but not TPF, decreased Dicer-mediated processing of the ∼700 nt to the ∼21 nt band. D, in vitro RNA binding assay. 293T cells overexpressing myc-Dicer were treated with PLL (lane 3), TPF (lane 4), or untreated (lane 2). 293T cells, which do not express myc-Dicer, were used as a negative control (lane 1). Lysates from these cells were incubated with biotinylated pre-let-7a RNA and then precipitated with streptavidin-Sepharose. In the left panel, myc-Dicer was detected in the precipitates by immunoblotting (top). Total myc-Dicer in each sample was measured by immunoblotting at the bottom (total). In the right panel, total pre-let7a RNA in the samples used for lanes 2–4 of the left panel was quantified by real-time RT-PCR in three replicates. E, siRNA association with AGO2 after PLL or TPF treatment. 293T(FLAG·AGO2) cells treated with PLL or TPF or untreated (−) were lysed and mixed with biotinylated si-GFP RNA (left panels). si-GFP-containing complexes were captured with streptavidin beads (top) or left unprecipitated (bottom) and detected for FLAG·AGO2 by immunoblotting. siRNA association with AGO2 was reduced in lysates from cells treated with TPF but not PLL. Separately, an RNA precipitation assay with transfected RNA was performed (right panels). 293T(FLAG·AGO2) cells were transfected with biotinylated si-GFP, and cells were treated with TPF or were untreated. The cells were lysed and captured with streptavidin beads (top) or left unprecipitated (bottom) for detection of FLAG·AGO2. F, pre-miR-17, -21, -16–2, and -30a in total RNA fraction were quantified by real-time RT-PCR in 293T cells treated with PLL or untreated (−). G, 293T cells overexpressing FLAG·AGO1, -2, -3, or -4 were transfected with sh-GFP and treated with TPF or untreated (−). Left, the ratios of siRNA(A)/siRNA(T) were quantified as in Fig. 2B. Right, AGO1, -2, -3, or -4 expression was immunoblotted using anti-FLAG antibody. Anti-actin was used as a loading control.
Dicer activity has been reported to be reduced directly by highly basic polypeptides such as the human immunodeficiency virus-1 Tat protein (43), which suppresses RNA silencing in mammalian cells (44). Because PLL is also a highly basic polypeptide, we reasoned that its effect in Fig. 2B could result from a direct repression of Dicer activity. To address this possibility, we performed an in vitro Dicer assay. Purified recombinant Dicer was added to a 32P-labeled double-stranded GFP RNA of 700 nucleotides, and this incubation led to the expected cleavage of GFP-double strand RNA to 21 nucleotide products. We then asked how PLL would affect Dicer activity. Indeed, when PLL was added to the reaction, it reduced Dicer-mediated RNA-processing (compare the 21-nt band in lanes 2–6), without affecting the stability of the Dicer protein (Fig. 2C, bottom Western blot Dicer, see lanes 2–9) or the stability of the added carrier bovine serum albumin protein (Fig. 2C, bottom bovine serum albumin, see lanes 1–9). By contrast, the addition of TPF to the same assays did not change Dicer-mediated processing of RNA (Fig. 2C, compare the 21-nt band in lane 2 to the band in lanes 7–9). A similar decrease in Dicer's in vitro processing of RNA by PLL, but not TPF, was seen using pre-let-7a instead of double strand GFP RNA as a Dicer substrate (supplemental Fig. S5A).
The reduction of Dicer-mediated RNA processing by PLL (Fig. 2C and supplemental Fig. S5A) could result from two possible steps: a decreased association of Dicer with substrate RNA or a direct impairment of the enzymatic activity of Dicer. To gain insights into these mechanisms, we examined Dicer-RNA association using an in vitro RNA precipitation assay. We incubated biotinylated pre-let-7a RNA as a probe with lysate from Dicer-overexpressing 293T cells that were treated with PLL or TPF or untreated. Streptavidin-captured RNA co-purified Dicer protein (Fig. 2D, left), and the streptavidin-beads captured equal amounts of input biotinylated pre-let-7a RNA in each of the control reactions incubated without lysates (Fig. 2D, right). The amount of RNA-associated Dicer was decreased by treatment of the cells with PLL (Fig. 2D, upper left, compare lane 3 to 2), but not by TPF (Fig. 2D, upper left, compare lane 4 to 2). Neither PLL nor TPF treatment affected the Dicer level in the total cell lysates (Fig. 2D, lower left panel, total). These findings are consistent with the interpretation that PLL treatment reduces the formation of Dicer·RNA complexes. In separate in vitro RNA precipitation assays, we observed that treatment with PLL, but not TPF, also decreased RNA association with Drosha (supplemental Fig. S5B). This latter PLL activity could also influence the amount of processed miRNA that enters the miRNA(T) pool.
The above results suggest that PLL, but not TPF, perturbs the association of Dicer with RNA. If TPF did not influence Dicer's processing of pre-miRNA to miRNA (supplemental Fig. S5A), or its processing of shRNA to siRNA (Fig. 2C), how would it modulate shRNA-mediated silencing (Fig. 1)? Upon closer inspection, we noted a dramatically larger reduction in processed sh-GFP (i.e. GFP-siRNA) in the AGO2-associated siRNA(A) fraction (Fig. 2B, middle graph) in TPF-treated versus PLL-treated cells. By calculating the ratio (i.e. siRNA(A)/siRNA(T)) of GFP siRNA in the AGO2-associated siRNA(A) fraction versus that in the total siRNA(T) fraction as a normalized measure of the amount of GFP-siRNA loaded into the AGO2 complex, we found a statistically significant decrease in this ratio when comparing mock treated cells to TPF-treated cells, but no statistically significant decrease when comparing mock treated cells to PLL-treated cells (Fig. 2B, right graph).
There can be several ways to interpret a change in the siRNA(A)/siRNA(T) ratio. One interpretation, which does not exclude others, is that TPF primarily affects the association or loading of processed siRNAs into an AGO2 complex, whereas PLL affects the association of substrate RNAs with Dicer and/or Drosha. Accordingly, the latter (PLL), but not the former (TPF), could directly affect the processing of pre-miRNA/shRNA to miRNA/siRNA. Thus, PLL treatment affects the abundance of miRNAs/siRNAs that enter the miRNA(T)/siRNA(T) fraction without affecting how the RNAs in the miRNA(T)/siRNA(T) fraction are loaded onto AGO2 (i.e. the miRNA(A)/siRNA(A) fraction). By contrast, TPF treatment would not affect directly the biogenesis of miRNA/siRNA that enters the miRNA(T)/siRNA(T) fraction, but rather influences the efficiency that miRNA/siRNA associates with an AGO2 complex (i.e. the miRNA(A)/siRNA(A) fraction). One should note that preventing si-/miRNA loading to AGO2 could destabilize the RNA. In this respect, an indirect consequence of TPF treatment could be a reduction in the measured miRNA(T) or siRNA(T) values (see “Discussion”).
To investigate how TPF might influence the miRNA(A)/siRNA(A) value, we performed a precipitation assay using streptavidin beads and biotinylated si-GFP RNA to investigate RNA·AGO2 complexes after PLL or TPF treatment (Fig. 2E). The results showed that cells treated with TPF exhibited reduced ability to form siRNA·AGO2 complexes (Fig. 2E, TPF), whereas cells treated with PLL were minimally affected in these complexes (Fig. 2E, left, PLL). Collectively, the results in Fig. 2 (D and E) suggest that PLL treatment reduces pre-miRNA association with Dicer while TPF treatment reduces siRNA/miRNA association with AGO2.
Two additional experiments were performed to clarify the effects of PLL and TPF treatment. To assess the activity of PLL on pre-miRNA-Dicer interaction, we checked the abundance of four cell endogenous pre-miRNAs before and after PLL treatment. Indeed, the amount of pre-miR-17, -21, -16–2, and -30a quantified in the pre-miRNA(T) fraction (total RNA isolated by TRIzol, and pre-miRNA measured by quantitative RT-PCR) was higher in PLL- treated versus untreated cells (Fig. 2F). We also checked how TPF might affect siRNA association with the four known types of mammalian AGO proteins. We transfected 293T cells with epitope-tagged AGO1, -2, -3, and -4 expression plasmids with sh-GFP and measured AGO-associated GFP-siRNA in cells with or without TPF treatment. When normalized to the total small RNA fraction siRNA(T), TPF treatment decreased AGO1, -2, -3, and -4 associated siRNAs as measured by the siRNA(A)/siRNA(T) ratios (Fig. 2G, left). The TPF treatment did not destabilize the AGO proteins as evidenced by Western blotting (Fig. 2G, right). Additional studies are needed to clarify the implications of the TPF effect on the functions of AGO1, -3, and -4.
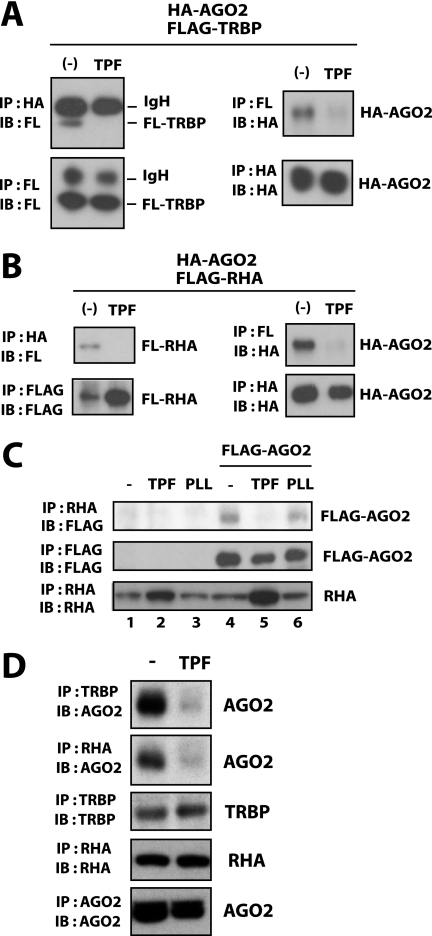
Elsewhere, TRBP and RHA have been reported as cofactors that regulate RNA association with AGO2 (12, 19). We extended our exploration of the TPF mechanism by asking if this treatment could affect TRBP and/or RHA activity. TPF treatment of 293T cells reduced the association of transfected FLAG-TRBP or FLAG-RHA with HA-AGO2 (Fig. 3, A and B). We also found that TPF, but not PLL treatment disrupted FLAG·AGO2 association with cell endogenous RHA (Fig. 3C, lane 5). Interestingly, whereas the interaction between cell endogenous AGO2 and TRBP or RHA was reduced following TPF treatment (Fig. 3D), the interaction between Dicer and AGO2 was unaffected by TPF treatment (supplemental Fig. S5C). Taken together, the collective data suggest that TPF acts differently from PLL, and that one of its effects is through influencing the association of TRBP and AGO2 and/or RHA and AGO2. By disturbing these interactions, TPF potentially affects the loading of miRNAs/siRNAs into AGO2-RISC.
FIGURE 3.
TPF disrupts the association of TRBP and RHA with AGO2. A and B, 293T cells were transfected with HA-tagged AGO2 (HA-AGO2) and FLAG-tagged TRBP (FLAG-TRBP in A) or RHA (FLAG-RHA in B). At 24 h post transfection, cells were treated without or with TPF. After a further 24 h, cells were lysed and immunoprecipitated with anti-FLAG or anti-HA. The immunoprecipitates were blotted with anti-FLAG or anti-HA as indicated. In the left panels of A, the recovered bands for the IgG heavy chain (IgH) as well as for FLAG-TRBP are indicated. Recovery of HA-AGO2 is shown in the right panels. In B, recoveries of FLAG-RHA and HA-AGO2 are shown. C, 293T cells transfected with FLAG·AGO2 (lanes 4–6) or the corresponding empty vector (lanes 1–3) were treated with TPF (lanes 2 and 5), PLL (lanes 3 and 6), or were untreated (lanes 1 and 4). Cells were lysed and immunoprecipitated with anti-RHA (top and bottom) or anti-FLAG (middle). The immunoprecipitates were blotted with anti-FLAG (top and middle) or anti-RHA (bottom). D, after 24-h treatment with TPF or untreated (−), 293T cells were lysed and immunoprecipitated with anti-TRBP, anti-RHA, or anti-AGO2 as indicated. The immunoprecipitates were blotted with anti-AGO2, anti-TRBP, or anti-RHA. Endogenous AGO2 co-precipitated with TRBP or RHA was decreased in cells treated with TPF.
Next, we wished to exclude that the observed PLL and TPF effects might be due to nonspecific influences on the general stability of small RNAs. We thus checked how PLL or TPF might affect the ambient levels of two small nuclear RNAs. Neither PLL nor TPF significantly changed the intracellular levels of ACA17 and ACA19 small nuclear RNAs (supplemental Fig. S5D). These results indicate that the effects of PLL and TPF on siRNA/miRNA/Dicer/AGO2 interactions are unlikely from generalized nonspecific activities on small RNAs. Because TPF affected miRNA association with AGO2, we also checked to see if this compound influenced the stability of cell endogenous AGO2. No effect of TPF on AGO2 stability was observed (supplemental Fig. S5E).
Effects of PLL and TPF on Cell Endogenous miRNA
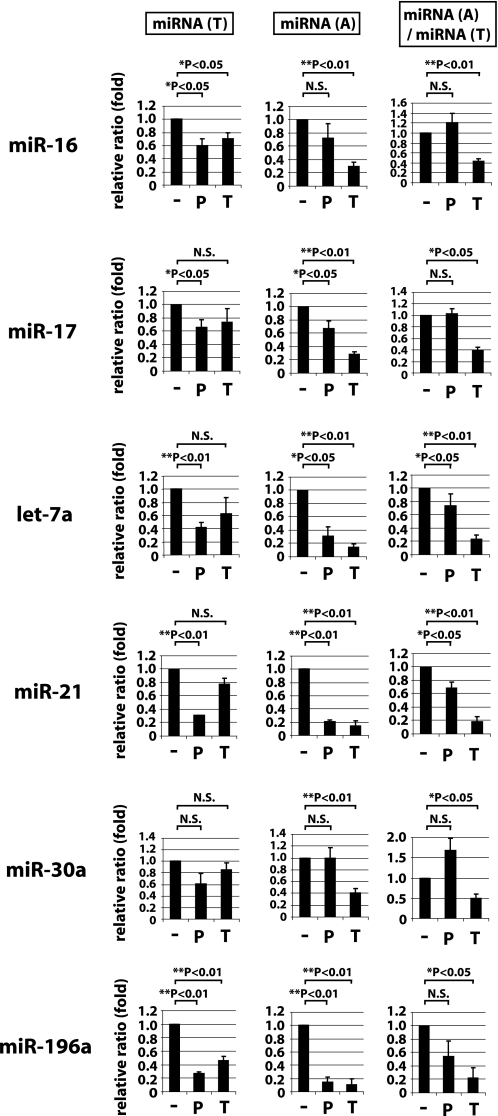
Many of the above experiments examined PLL and TPF effects on overexpressed transfected shRNAs. We also wished to understand their effects on cell endogenous miRNAs. We selected six cellular miRNAs, miR-16, -17, -21, -30a, -196a, and let-7a, and asked how PLL or TPF treatment would affect their expression (Fig. 4). Treated and untreated cells were fractionated and measured for total small RNAs (miRNA(T), Fig. 4, left column) and AGO2-associated small RNAs (miRNA(A), Fig. 4, middle column) after 2 days of treatment with compound, and we calculated the miRNA(A)/miRNA(T) ratios (Fig. 4, right column). Overall, PLL had a greater effect than TPF on reducing the miRNA(T) (Fig. 4, left column), whereas TPF had a larger effect than PLL on reducing miRNA(A) (Fig. 4, middle column). When these two values were integrated, we observed that TPF, but not PLL, compared with mock treated cells consistently decreased the miRNA(A)/miRNA(T) ratio in a statistically significant manner (Fig. 4, right column). These findings from cell endogenous miRNAs agree in part with the results from transfected shRNAs (Fig. 2B) that TPF predominantly affects miRNA(A), whereas PLL primarily decreases the miRNA(T) (Fig. 4, left column). The miRNA results are consistent with an interpretation that PLL affects Dicer-mediated processing of pre-miRNA to miRNA while TPF affects the association of processed miRNA with an AGO2 complex.
FIGURE 4.
Effect of PLL or TPF treatments on the abundance of cell endogenous miRNAs. 293T(FLAG·AGO2) cells treated with 5 μm PLL (P), 10 μm TPF (T), or untreated (−) for 2 days were recovered for small RNA as described in Fig. 2A. Total miRNA or AGO2-associated miRNA fractions are termed miRNA(T) and miRNA(A), respectively. The amounts of miR-16, -17, let-7a, miR-21, -30a, and -196a were quantified using real-time RT-PCR.
We next examined if PLL and TPF treatment could functionally inhibit the silencing activity of miRNAs. To test this point, we employed a construct for luciferase fused with three repeats of let-7a target sequences positioned in the downstream untranslated region. We transfected this plasmid together with a let-7a expression plasmid into HeLa cells. The luciferase activity was reduced by the overexpression of let-7a in a dose-dependent manner (supplemental Fig. S6, lanes 1–4), reflecting the silencing activity of let-7a. Treatment with PLL or TPF attenuated this silencing activity (supplemental Fig. S6, compare the difference between lanes 1 and 4 with those between lanes 5 and 6 (for PLL), or with those between lanes 7 and 8 (for TPF)). These results support that PLL or TPF treatment inhibits the silencing function of miRNAs.
PLL and TPF Treatment Reversed miRNA-dependent Tumorigenesis
AGO2 and Dicer overexpression have been correlated with the development of several types of cancer (33, 37, 45). In settings where cell growth and cellular transformation may be miRNA-dependent, we reasoned that treatment of cells with PLL or TPF could change the proliferative phenotype. Previously, we had found that the overexpression of miR-93 and miR-130b impacts leukemogenesis (30). Extending those findings, we established NIH3T3 cells that stably overexpress miR-93 (3T3-miR-93) or miR-130b (3T3-miR-130b) or Ras (3T3-Ras) and found that these cells, different from parental NIH3T3 cells, formed significant anchorage-independent colonies in soft agar (supplemental Fig. S7A). Moreover, the implantation of 3T3-miR-93, 3T3-miR-130b, or 3T3-Ras cells, but not parental NIH3T3 cells, into nude mice produced gross in vivo tumors (supplemental Fig. S7B) providing a model of miRNA-dependant cell growth and tumorigenesis.
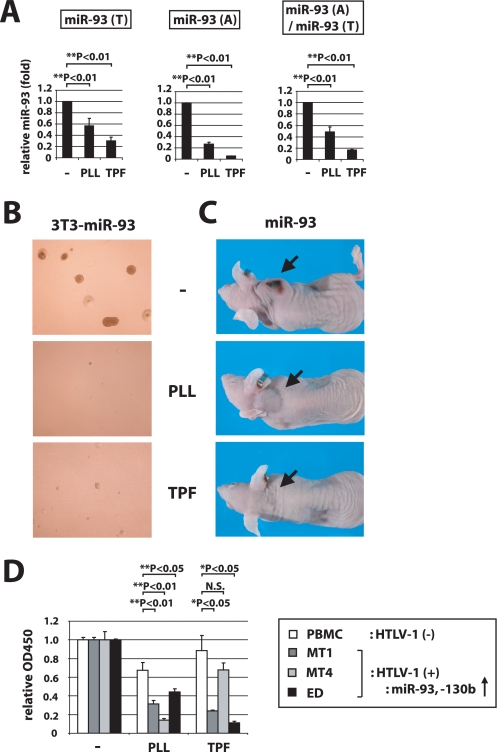
We investigated the consequence of PLL or TPF treatment of 3T3-miR-93 and 3T3-miR-130b cells. In assays for miR-93 and miR-130b, TPF treatment most significantly decreased the miRNA(A)/miRNA(T) ratio while PLL treatment decreased significantly the level of assayed small RNA in the miRNA(T) pool (Fig. 5A and supplemental Fig. S8B). Treatment with PLL or TPF also significantly abrogated the ability of 3T3-miR-93 and 3T3-miR-130b cells to form colonies in soft agar (Fig. 5B and supplemental Fig. S8C), but did not affect the baseline colony numbers seen with 3T3 control cells (supplemental Fig. S8D). As a control experiment, the colony forming ability of 3T3-miR-93 cells was decreased by transfection with antagonizing oligonucleotide for miR-93 (supplemental Fig. S8E). These PLL and TPF effects were not due to general cytotoxicity, because PLL or TPF treatment of cells in tissue culture for 3, 5, and 7 days using the same conditions elicited no perturbation of cell growth (supplemental Fig. S8F). TPF- and PLL-treated 3T3-miR-93 cells also showed significant reduction in their ability to form tumors when implanted into nude mice (Fig. 5C and supplemental Fig. S8G).
FIGURE 5.
PLL and TPF treatments suppress the tumorigenic activity of miR-93 overexpressing NIH3T3 cells. A, 3T3-miR-93 cells transfected with FLAG·AGO2 were treated with PLL or TPF, or were untreated (−) for 3 days and then recovered for small RNA as described in Fig. 4. miR-93 in miRNA(T) and miRNA(A) was quantified, and the ratios of miRNA(A)/miRNA(T) are also shown. B, 3T3-miR-93 cells treated with 2 μm PLL, 1 μm TPF, or untreated were observed in a soft agar growth assay. Representative images are shown. C, 3T3-miR-93 cells were pretreated without or with PLL or TPF for 3 days and subsequently implanted into nude mice subcutaneously for observation of tumor formation. Treated cells did not form tumors in mice. Representative pictures of mice are shown. D, cell growth of HTLV-I-infected leukemic cell lines and normal PBMCs upon PLL and TPF treatment. Cell proliferation was examined by MTT assay on HTLV-1-cells, MT1, MT4, and ED, or on normal PBMC treated with 3 μm PLL, 3 μm TPF, or untreated (−) for 4 days. MT1, MT4, and ED cells all overexpress miR-93 and miR-130b. The A450 value for the untreated sample was set as 1 for each cell type. Treatment with PLL and TPF reduced the growth of HTLV-1-cells compared with normal PBMCs.
We next examined the compounds' effect by testing a physiological tumor cell model. Previously, we have reported that HTLV-1-infected cells overexpress miR-93 and -130b at amounts 20 to 4000 times higher than those in normal peripheral blood mononuclear cells (PBMC) and that these miRNAs play pivotal roles in virus-mediated leukemogenesis (30, 46). To ask how miRNA inhibition might affect HTLV-1 tumor maintenance, we tested PLL and TPF treatment of adult T-leukemic cell lines (MT-1, MT-4, and ED) and normal PBMCs. The effect of treatment on cell growth was checked by MTT assay. As shown in Fig. 5D, PBMC viability was minimally affected by PLL and TPF, whereas adult T-cell leukemia cells were significant reduced in cell viability. These results support a role played by miRNA-dysregulation in certain leukemogenic events and illustrate that chemical inhibition of miRNA pathways can in selected settings reverse oncogenic miRNA (oncomiR)-mediated cellular proliferation.
DISCUSSION
Here, we have identified PLL and TPF as inhibitors of miRNA-mediated silencing function. To date, there has been a single report of the screening of compounds that suppressed miR-21 activity; however, in that study, the mechanistic action and cellular target(s) of the compounds were not clarified (47). In our work, we have investigated in detail two chemical compounds that suppress small RNA-mediated gene silencing. We note with interest that others with complementary goals have screened for compounds that activate, rather than suppress, RNA-mediated silencing (48).
In our analyses, PLL and TPF act differently. PLL inhibited Dicer-mediated processing (Fig. 2C), of pre-miRNA/shRNA to miRNA/siRNA. PLL treatment of cells also reduced mature miRNA(T) levels (Fig. 4) while increasing the amount of unprocessed pre-miRNAs (Fig. 2F). The PLL mechanism appears to be an inhibition of the association of pre-miRNA with Dicer (Fig. 2D). It is currently unclear how PLL reduces the interaction of RNA substrates with Dicer (Fig. 2D) or Drosha (supplemental Fig. S5B). Further efforts to clarify the structural bases for these effects are needed, but are outside of the scope of the current study.
By contrast, TPF appears to act by reducing the association of siRNA/miRNA with AGO2 (Fig. 2E). This inhibitory effect of TPF may be due to its ability to disrupt the protein-protein association between TRBP and AGO2 (Fig. 3A) and between RHA and AGO2 (Fig. 3B). Previous reports in the literature have suggested that a failure of siRNA/miRNA association with AGO2 leads to the destabilization of these small RNAs, decreasing their intracellular amounts (49, 50). Thus, although TPF primarily affects the loading of si-/miRNAs into the AGO2 complex as reflected by its reduction of siRNA(A) (Fig. 2B) and miRNA(A) (Fig. 4) values, this reduced loading of si-/miRNA to AGO2 can in some settings destabilize the not loaded small RNAs, leading to an indirect reduction in miRNA(T) or siRNA(T) values (Fig. 2B). We should add that these mechanistic interpretations represent our operational observations of the effects of PLL and TPF on transiently transfected shRNA (Fig. 2) and on cell endogenous miRNAs (Fig. 4) in 293T cells. TPF and PLL effects on oncomiRs in cells that are selected for transformation (Fig. 5) may be more complex due to additional secondary changes that could occur during transformation.
It remains unknown how TPF disrupts the association of AGO2 with TRBP or RHA. In our experiments, TPF did not affect the subcellular localizations of AGO2, TRBP, or RHA (data not shown). We also did not observe a TPF effect on the half-life of AGO2 (supplemental Fig. S5E). At this juncture, we do not have the capability to fully resolve the definitive TPF mechanism. Elsewhere, it has been reported that TRBP stabilizes the Dicer·AGO2 complex and enhances the efficiency of RNA transfer from Dicer to AGO2 (51). According to this scenario, TPF may decrease the efficiency of RNA transfer by reducing TRBP-AGO2 association (Fig. 3A). Alternatively, a second possibility is that TPF directly interrupts the binding of AGO2 with RNA, which could then affect the formation of an AGO2·TRBP/RHA complex (Fig. 3B).
Our two compounds can reverse the tumorigenicity of miR-93- and miR-130b- overexpressing cells (Fig. 5 and supplemental Fig. S8). We believe that treatment with these two compounds affects general miRNA activities, rather than only the specific activities of miR-93 or miR-130b. However, in settings where tumorigenesis is dependent on the overexpression of oncomiRs, reducing the activities of these oncomiRs, albeit accompanied by commensurate reductions in other cellular miRNAs, may be therapeutic (Fig. 5 and supplemental Fig. S8). Of interest, the tumorigenicity of Ras-overexpressing NIH3T3 cells was also decreased by compound treatment to ∼50–60% of untreated cells, although this value appears not to be statistically significant (supplemental Fig. S8D). The recent report that Ras-induced cellular transformation also includes an miRNA-dependent mechanism (52) would seem to agree with our observed decrease in tumorigenicity.
As a final thought, oncomiRs such as the miR-17–92 cluster, miR-155, and miR-21 play important roles in several cancers (22, 23, 53). It has not been examined whether PLL and TPF reverse the tumorigenic influence of these miRNAs. Additional investigation in this direction could be of future utility to further establish the relevance of chemical inhibition of miRNA pathways in tumor reversion. Going forward, further screenings for small molecules that more specifically inhibit the biological effects of oncomiRs may be scientifically informative.
Supplementary Material
Acknowledgments
We are grateful to Alicia Buckler-White and Ronald Plishka for sequencing and analyses. We also thank Andrew Dayton and K.T.J.'s laboratory members for critical readings of the manuscript. pIRESneo-FLAG/HA-AGO2 (corrected) and pcDNA-FLAG-RHA were provided by Drs. Thomas Tuschl and Gunter Meister at The Rockefeller University and Dr. Chee-Gun Lee at the University of Medicine and Dentistry of New Jersey, respectively.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01 GM087738-01A1 (to R. S. H.) and by intramural funds from NIAID, NIH and the Intramural AIDS Targeted Antiviral Program from the office of the Director, NIH (to K.-T. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8 and Table S1.
- miRNA
- microRNA
- RNAi
- RNA interference
- pre-miRNA
- precursor miRNA
- nt
- nucleotide(s)
- RISC
- RNA-induced silencing complex
- AGO
- argonaute
- RHA
- RNA helicase A
- HTLV-I
- human T-cell leukemia virus, type I
- PLL
- polylysine
- TPF
- trypaflavine
- DMEM
- Dulbecco's modified Eagle's medium
- shRNA
- short hairpin RNA
- Fluc
- firefly luciferase
- Rluc
- Renilla luciferase
- sh-Fluc
- shRNA targeting firefly luciferase mRNA
- oncomiR
- oncogenic mRNAs
- PBMC
- peripheral blood mononuclear cell
- CMV
- cytomegalovirus
- GFP
- green fluorescent protein
- sh-GFP
- shRNA that targets GFP
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- IP
- immunoprecipitation
- RT
- reverse transcription
- si-GFP
- siRNA targeting GFP.
REFERENCES
- 1.Baltimore D., Boldin M. P., O'Connell R. M., Rao D. S., Taganov K. D. (2008) Nat. Immunol. 9, 839–845 [DOI] [PubMed] [Google Scholar]
- 2.Garofalo M., Condorelli G., Croce C. M. (2008) Curr. Opin. Pharmacol. 8, 661–667 [DOI] [PubMed] [Google Scholar]
- 3.O'Connell R. M., Rao D. S., Chaudhuri A. A., Boldin M. P., Taganov K. D., Nicoll J., Paquette R. L., Baltimore D. (2008) J. Exp. Med. 205, 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soifer H. S., Rossi J. J., Saetrom P. (2007) Mol. Ther. 15, 2070–2079 [DOI] [PubMed] [Google Scholar]
- 5.Grishok A., Pasquinelli A. E., Conte D., Li N., Parrish S., Ha I., Baillie D. L., Fire A., Ruvkun G., Mello C. C. (2001) Cell 106, 23–34 [DOI] [PubMed] [Google Scholar]
- 6.Hutvágner G., McLachlan J., Pasquinelli A. E., Balint E., Tuschl T., Zamore P. D. (2001) Science 293, 834–838 [DOI] [PubMed] [Google Scholar]
- 7.Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. (2004) Mol. Cell 15, 185–197 [DOI] [PubMed] [Google Scholar]
- 8.He L., Hannon G. J. (2004) Nat. Rev. Genet 5, 522–531 [DOI] [PubMed] [Google Scholar]
- 9.Hutvágner G., Zamore P. D. (2002) Science 297, 2056–2060 [DOI] [PubMed] [Google Scholar]
- 10.Liu J., Carmell M. A., Rivas F. V., Marsden C. G., Thomson J. M., Song J. J., Hammond S. M., Joshua-Tor L., Hannon G. J. (2004) Science 305, 1437–1441 [DOI] [PubMed] [Google Scholar]
- 11.Pillai R. S., Artus C. G., Filipowicz W. (2004) RNA 10, 1518–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chendrimada T. P., Gregory R. I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. (2005) Nature 436, 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haase A. D., Jaskiewicz L., Zhang H., Lainé S., Sack R., Gatignol A., Filipowicz W. (2005) EMBO Rep. 6, 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J. C., Fritzler M. J., Chan E. K. (2005) Nat. Cell Biol. 7, 1267–1274 [DOI] [PubMed] [Google Scholar]
- 15.Lee Y., Hur I., Park S. Y., Kim Y. K., Suh M. R., Kim V. N. (2006) EMBO J. 25, 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J., Rivas F. V., Wohlschlegel J., Yates J. R., 3rd, Parker R., Hannon G. J. (2005) Nat. Cell Biol. 7, 1261–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meister G., Landthaler M., Peters L., Chen P. Y., Urlaub H., Lührmann R., Tuschl T. (2005) Curr. Biol. 15, 2149–2155 [DOI] [PubMed] [Google Scholar]
- 18.Mourelatos Z., Dostie J., Paushkin S., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G. (2002) Genes Dev. 16, 720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robb G. B., Rana T. M. (2007) Mol. Cell 26, 523–537 [DOI] [PubMed] [Google Scholar]
- 20.Bartel D. P. (2009) Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leung A. K., Sharp P. A. (2006) Cold Spring Harb. Symp. Quant. Biol. 71, 29–38 [DOI] [PubMed] [Google Scholar]
- 22.Chang T. C., Mendell J. T. (2007) Annu. Rev. Genomics Hum. Genet 8, 215–239 [DOI] [PubMed] [Google Scholar]
- 23.Deng S., Calin G. A., Croce C. M., Coukos G., Zhang L. (2008) Cell Cycle 7, 2643–2646 [DOI] [PubMed] [Google Scholar]
- 24.He L., Thomson J. M., Hemann M. T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S. W., Hannon G. J., Hammond S. M. (2005) Nature 435, 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eis P. S., Tam W., Sun L., Chadburn A., Li Z., Gomez M. F., Lund E., Dahlberg J. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Costinean S., Zanesi N., Pekarsky Y., Tili E., Volinia S., Heerema N., Croce C. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7024–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan J. A., Krichevsky A. M., Kosik K. S. (2005) Cancer Res. 65, 6029–6033 [DOI] [PubMed] [Google Scholar]
- 29.Iorio M. V., Ferracin M., Liu C. G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M., Ménard S., Palazzo J. P., Rosenberg A., Musiani P., Volinia S., Nenci I., Calin G. A., Querzoli P., Negrini M., Croce C. M. (2005) Cancer Res. 65, 7065–7070 [DOI] [PubMed] [Google Scholar]
- 30.Yeung M. L., Yasunaga J., Bennasser Y., Dusetti N., Harris D., Ahmad N., Matsuoka M., Jeang K. T. (2008) Cancer Res. 68, 8976–8985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cimmino A., Calin G. A., Fabbri M., Iorio M. V., Ferracin M., Shimizu M., Wojcik S. E., Aqeilan R. I., Zupo S., Dono M., Rassenti L., Alder H., Volinia S., Liu C. G., Kipps T. J., Negrini M., Croce C. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson S. M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K. L., Brown D., Slack F. J. (2005) Cell 120, 635–647 [DOI] [PubMed] [Google Scholar]
- 33.Adams B. D., Claffey K. P., White B. A. (2009) Endocrinology 150, 14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koesters R., Adams V., Betts D., Moos R., Schmid M., Siermann A., Hassam S., Weitz S., Lichter P., Heitz P. U., von Knebel Doeberitz M., Briner J. (1999) Genomics 61, 210–218 [DOI] [PubMed] [Google Scholar]
- 35.Qiao D., Zeeman A. M., Deng W., Looijenga L. H., Lin H. (2002) Oncogene 21, 3988–3999 [DOI] [PubMed] [Google Scholar]
- 36.Kumar M. S., Lu J., Mercer K. L., Golub T. R., Jacks T. (2007) Nat. Genet. 39, 673–677 [DOI] [PubMed] [Google Scholar]
- 37.Mudhasani R., Zhu Z., Hutvagner G., Eischen C. M., Lyle S., Hall L. L., Lawrence J. B., Imbalzano A. N., Jones S. N. (2008) J. Cell Biol. 181, 1055–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benkirane M., Neuveut C., Chun R. F., Smith S. M., Samuel C. E., Gatignol A., Jeang K. T. (1997) EMBO J. 16, 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gatignol A., Buckler C., Jeang K. T. (1993) Mol. Cell. Biol. 13, 2193–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gou D., Zhang H., Baviskar P. S., Liu L. (2007) Physiol. Genomics 31, 554–562 [DOI] [PubMed] [Google Scholar]
- 41.Watashi K., Khan M., Yedavalli V. R., Yeung M. L., Strebel K., Jeang K. T. (2008) J. Virol. 82, 9928–9936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeung M. L., Houzet L., Yedavalli V. S., Jeang K. T. (2009) J. Biol. Chem. 284, 19463–19473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennasser Y., Le S. Y., Benkirane M., Jeang K. T. (2005) Immunity 22, 607–619 [DOI] [PubMed] [Google Scholar]
- 44.Qian S., Zhong X., Yu L., Ding B., de Haan P., Boris-Lawrie K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiosea S., Jelezcova E., Chandran U., Acquafondata M., McHale T., Sobol R. W., Dhir R. (2006) Am J. Pathol. 169, 1812–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouzar A. B., Willems L. (2008) Retrovirology 5, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gumireddy K., Young D. D., Xiong X., Hogenesch J. B., Huang Q., Deiters A. (2008) Angew Chem. Int. Ed. Engl. 47, 7482–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shan G., Li Y., Zhang J., Li W., Szulwach K. E., Duan R., Faghihi M. A., Khalil A. M., Lu L., Paroo Z., Chan A. W., Shi Z., Liu Q., Wahlestedt C., He C., Jin P. (2008) Nat. Biotechnol. 26, 933–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diederichs S., Haber D. A. (2007) Cell 131, 1097–1108 [DOI] [PubMed] [Google Scholar]
- 50.O'Carroll D., Mecklenbrauker I., Das P. P., Santana A., Koenig U., Enright A. J., Miska E. A., Tarakhovsky A. (2007) Genes Dev. 21, 1999–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H. W., Noland C., Siridechadilok B., Taylor D. W., Ma E., Felderer K., Doudna J. A., Nogales E. (2009) Nat. Struct. Mol. Biol. 16, 1148–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Talotta F., Cimmino A., Matarazzo M. R., Casalino L., De Vita G., D'Esposito M., Di Lauro R., Verde P. (2009) Oncogene 28, 73–84 [DOI] [PubMed] [Google Scholar]
- 53.Medina P. P., Slack F. J. (2008) Cell Cycle 7, 2485–2492 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.