Abstract
In order to study the intragenic profiles of active transcription, we determined the relative levels of active RNA polymerase II present at the 3′- and 5′-ends of 261 yeast genes by run-on. The results obtained indicate that the 3′/5′ run-on ratio varies among the genes studied by over 12 log2 units. This ratio seems to be an intrinsic characteristic of each transcriptional unit and does not significantly correlate with gene length, G + C content or level of expression. The correlation between the 3′/5′ RNA polymerase II ratios measured by run-on and those obtained by chromatin immunoprecipitation is poor, although the genes encoding ribosomal proteins present exceptionally low ratios in both cases. We detected a subset of elongation-related factors that are important for maintaining the wild-type profiles of active transcription, including DSIF, Mediator, factors related to the methylation of histone H3-lysine 4, the Bur CDK and the RNA polymerase II subunit Rpb9. We conducted a more detailed investigation of the alterations caused by rpb9Δ to find that Rpb9 contributes to the intragenic profiles of active transcription by influencing the probability of arrest of RNA polymerase II.
INTRODUCTION
In the last decade, the importance of transcription elongation regulation has been brought into focus. Many factors have been associated with this key step of gene expression, and it has been proved that several biological processes are connected to this transcription phase, including response to stress, development and viral infections (1,2).
Chromatin immunoprecipitation (ChIP) (3,4) using antibodies against different phosphorylated forms of RNA polymerase II (RNA pol II) (5) enables the measurement of elongation rates and processivity (6). Besides, the combination of RNA pol II ChIP with DNA arrays and massive sequencing has provided pictures of the distribution of RNA pol II in several genomes (7). Studying transcription elongation in vivo has also involved the use of other techniques, including the depletion of the intracellular pools of ribonucleotide triphosphates by drugs like 6-azauracile (8) and mycophenolic acid (9), or the comparison of reporter genes of different lengths (10).
One of the drawbacks of the ChIP of RNA pol II is its lack of specificity against the active, elongation-competent form of the polymerase. In vitro studies have shown that RNA pol II often becomes arrested during elongation in the chromatin context (11), while molecular modeling has suggested that backtracking during elongation is indeed a frequent phenomenon in vivo (12).
The run-on technique has proved highly appropriate to deal with these issues. It enables the measurement of the density of actively transcribing RNA polymerases by labelling nascent mRNA in the presence of high salt and sarkosyl, which inhibits a new round of transcription initiation without affecting the elongation reaction (13). Global transcription analyses have been carried out by combining run-on with either DNA arrays hybridization (14) or massive sequencing (15). Using this genomic run-on (GRO) approach, we have recently shown that some functional gene categories are controlled at the elongation step by modulating the fraction of RNA polymerases that become inactive during transcription (16).
In the present work, we have used the run-on technique and a new type of custom-developed DNA arrays to quantitatively analyse the intragenic distribution of active RNA pol II. By probing run-on preparations with the DNA sequences of the two ends of a broad set of genes, we found that the 3′/5′ ratio of actively transcribing polymerases is gene-specific. Among the tested genes, those encoding structural ribosomal proteins (RP) showed the lowest 3′/5′ run-on ratios. We also measured these ratios under several conditions and in mutant backgrounds, and we detected a clear influence of some elements of the transcriptional machinery on the intragenic distribution of active RNA pol II.
MATERIALS AND METHODS
Strains and media
All the strains, except W303 (MAT a ade2-1 can1-100 ura3-1 leu2-3,112 his3-11,15 trp1-1) (17) and its derivatives SET1-myc and set1AA-myc (18), were purchased from EUROSCARF and were isogenic to BY4741 (MAT a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) with the corresponding gene substituted by a KANMX4 cassette.
Rich (YPD) and minimal (SC) media were used as previously described (19). Osmotic shock was produced by adding NaCl at a final concentration of 0.5 M.
Macroarray fabrication
DNA macroarrays were produced in the Sección de Chips de DNA-S.C.S.I.E of the University of Valencia following the procedure described in (20). The double strand DNA 300-bp-length probes obtained by PCR were printed onto positively charged nylon membranes using a BioGridTM robot (BioRobotics). Later, 20–30 ng of DNA were spotted on each position.
Some of the probes were amplified using genomic DNA as a template, while some others were amplified using plasmids containing the individual ORFs (20). The primers used (Supplementary Table SI) are available upon request.
Slot-blotted membranes
A Hybond N+ membrane (Amersham) was placed into a PR600 Slot Blot device (Hoefer Scientific Instruments). Each well of the device was loaded with 500 ng of dsDNA probes diluted in 0.5 ml 0.4 M NaOH 200 mM EDTA, previously boiled to denaturalize the DNA. The solution was forced through the membrane by applying a vacuum pump to the slot blot device. The membrane was then washed by rinsing it in 0.6 M NaCl 60 mM Na citrate (SSC 2×). Probes were obtained by PCR using the primers listed in Supplementary Table SII.
Run-on
Run-on assays were performed as described in (21) with minor modifications. Shortly, 50 ml of yeast culture were collected at OD600 0.5 by centrifugation at 4°C. Cells were washed in 5 ml cold TMN solution (10 mM Tris–HCl pH 7.4, 5 mM MgCl2, 10 mM NaCl), centrifuged again and resuspended in 950 µl cold H2O. Next, 50 µl of 10% sarkosyl were added at a final concentration of 0.5%. Cells were incubated in this solution for 20 min at 4°C for permeabilization purposes. Afterward, cells were centrifuged and the supernatant was completely removed. The transcription reaction was performed in 150 µl of transcription buffer (50 mM Tris–HCl pH 7.9, 100 mM KCl, 5 mM MgCl2, 1 mM MnCl2, 2 mM dithiothreitol, ATP, GTP and CTP 0.5 mM each and 100 µCi alpha-33P] UTP (3000 Ci/mmol)). The mix was incubated for 3 min at 30°C. The reaction was stopped by adding 1 ml cold TMN solution. RNA was immediately extracted following the acid-phenol protocol (22).
ChIP
ChIP experiments were performed as previously described (23) with minor modifications. Shortly, 50 ml of yeast culture were collected at OD600 0.5. Crosslinking was performed by adding formaldehyde to 1% to the culture and by incubating at room temperature for 15 min. Then 2.5 ml 2.5 M glycine were added and the culture was incubated for 5 min. Cells were then harvested and washed four times with 25 ml Tris–saline buffer (150 mM NaCl 20 mM Tris–HCl pH 7.5) at 4°C. Cell breakage was performed in 300 µl of lysis buffer (see the above reference) with glass beads, and cell extracts were sonicated in a Bioruptor sonicator (Diagenode) for 30 min in 30 s on/30 s off cycles (chromatin is sheared into an average size of 300 bp). Immunoprecipitation was performed with magnetic beads coated with pan anti-IgG antibodies (Dynal), which were incubated with the 8WG16 monoclonal antibody beforehand.
Finally, 25 µl real-time PCR were performed to quantify immunoprecipitation using a dilution of 1:1500 for the input samples and another of 1:10 for the immunoprecipitated samples. Immunoprecipitation was defined as the ratio of each specific probe product in relation to that of a non transcribed region (chromosome V, coordinates 9716 to 9863). The primers used are listed in Supplementary Table SIII.
For ChIP on chip purposes, the immunoprecipitated DNA was amplified and labelled following the procedure described in (24).
Genomic DNA labeling
First, 50–20 ng of yeast genomic DNA was diluted in 35 µl H2O and then boiled. Afterward, 5 µl of the 10× random hexanucleotides mix (ROCHE), 5 µl of a solution containing dATP, dGTP, dTTP, 0.5 mM each, 20 µCi [α-32P]dCTP (3000 Ci/mmol) and 2 U of Klenow polymerase were added to a final volume of 50 µl. The reaction mix was incubated for 1 h at 37°C. Non incorporated nucleotides were eliminated by purifying the sample through a Sephadex G-50 column.
Hybridization
Radio-labelled RNA from the run-on was fragmented and denatured prior to hybridization by adding NaOH to the sample at a final concentration of 50 mM and by incubating for 5 min on ice. Fragmentation was stopped by adding HCl to a concentration of 50 mM. Genomic DNA was denatured before hybridization by boiling it for 5 min. Hybridization with labelled RNA (run-on) or DNA (ChIP or genomic DNA) samples was performed as described in (16). Membranes were exposed in Fuji BAS screens for 5–7 days and were developed with a FUJIX FLA3000 device. Signals were quantified using the Array Vision software, version 8.0 (Imaging Research Inc.).
Normalizations, quality filters and statistical analysis
Only those spots with a signal 1.3 times over the background were considered. After this filtering and background subtraction, the average signal of the two replicas of each probe was only obtained if both values were over the threshold value. For RNA hybridizations, this value was corrected by the number of uracils present in the coding strand of the probe. Then the 3′/5′ ratio was obtained. Similarly, genomic DNA signals were corrected for the numbers of cytidines present in the probes. The run-on ratio was divided by the genomic DNA ratio to correct probe to probe differences in DNA amount and hybridization efficiency. Finally, the log2 of this ratio was obtained. At least three replicas of each experiment were performed.
The comparisons made of the results obtained for each strain or condition were analysed with a Student’s two-tailed paired t-test (25).
RESULTS
The 3′/5′ run-on ratio as a parameter reflecting the intragenic distribution of active RNA pol II
In order to determine the intragenic distribution of transcriptionally active RNA pol II in a large number of genes, we designed a DNA macroarray containing probes of about 300 bp corresponding to the 5′ and 3′ ends of 377 S. cerevisiae ORFs (Supplementary Table SI). We also designed a second array containing similar probes for a subset of 76 highly expressed genes (26) (Supplementary Table SI). We used these membranes for the hybridization done with the labelled RNA via the run-on technique so that the signal obtained in each probe was proportional to the density of the active RNA pol II present in this particular piece of the genome. Then we divided the signal obtained in the 3′ probe by the signal obtained in the 5′ probe of each transcriptional unit. This ratio was used as a parameter to reflect the intragenic distribution of the transcriptionally active, i.e. transcriptionally competent, RNA pol II in the genes represented in the membrane (see ‘Materials and Methods’ section for the normalization and quality control procedures). Log2 scale is used; therefore, positive values represent a higher density of active RNA polymerase present at the 3′-end than at the 5′-end, while negative values mean that more active RNA polymerases are present at the 5′-end than at the 3′-end.
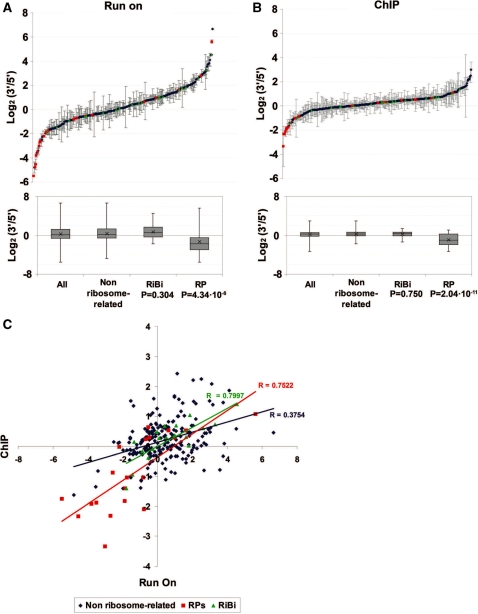
First, we tested the intragenic distribution of transcriptionally active RNA pol II in a BY4741 wild-type strain. The results of 261 transcriptional units were obtained after the quality controls and normalizations (see ‘Materials and Methods’ section). We did not obtain consistent signals (repetitively over the background threshold) for at least one of the two probes of the other 116 genes represented in the array. The average results are depicted in Figure 1A as a dot plot in a log2 scale with the standard deviations shown as error bars. The 3′/5′ ratio range covered more than 12 log2 units (from –5.49 in RPL25 to 6.66 in YRF1-2; Supplementary Table SIV) and adapted to a normal distribution (Figure 1A, Supplementary Figure S2A and B). Conventional run-on assays of five highly expressed genes confirmed that the intragenic distribution of active RNA pol II was not uniform (Supplementary Figure S2C). Additional evidence indicating that the dispersion of the 3′/5′ results did not happen by chance was obtained by randomly pairing the 3′ probes signal with the 5′ probes signal of a particular experiment. The results of five random distributions were much more dispersed than the results of the real experiment (Supplementary Figure S3). Therefore, in spite of the background noise that the use of single probes might have introduced in these results, we conclude that the distribution of active transcription is gene-specific.
Figure 1.
(A) Average 3′/5′ ratios of the run-on signals obtained from a wild-type BY4741 strain for the transcriptional units present in the 377 genes array. RP genes are represented in red and RiBi genes in green. Error bars represent standard deviation. The box-and-whiskers diagrams represent the minimum, the maximum, the average (cross), the median and the 25th and the 75th percentiles of the average 3′/5′ run-on ratios for the indicated gene categories. The P-values of the Student’s two-tailed t-test for those samples with unequal variances are shown. (B) The same as in A but for the ChIP signals obtained with the 8WG16 antibody. (C) Scatter plot of the average 3′/5′ run-on ratios in A (X-axis) versus the average 3′/5′ ChIP ratios in B (Y-axis). Linear regressions and their corresponding Pearson’s correlation coefficients (R) are shown.
In order to understand the cause of these differences in the 3′/5′ run-on ratios, we studied whether there was a correlation between the 3′/5′ ratios and several gene properties. We found no correlation with either the G + C content (Supplementary Figure S4A) or the distance to the nearest telomere (Supplementary Figure S4B), although the subtelomeric YRF1 genes exhibited the highest 3′/5′ ratios (Supplementary Table SIV). Since the 5′ probes were situated inside the ORFs, we looked for a possible correlation between the 3′/5′ run-on ratios and the length of the 5′ UTRs. By using the data of the transcription start sites published by Zhang and Dietrich (27), we found no correlation between these two parameters (Supplementary Figure S4C). The data also dismissed a significant influence of overlapping cryptic unstable transcripts (CUTs) or stable unannotated transcripts (SUTs) (28) on the 3′/5′ run-on ratios (Supplementary Figure S4D and E). We observed a slightly positive correlation of the 3′/5′ run-on ratios with the length of the transcription unit (Supplementary Figure S4F) and a weak negative correlation with the expression level according to Holstege et al. (26) (Supplementary Figure S4G). We also found that the average 3′/5′ ratios of intron-containing genes were slightly lower than those of intron-less genes (Supplementary Figure S4H).
These results suggest that short, highly expressed, intron-containing genes should have the lowest average 3′/5′ run-on ratios. The functional group of genes that best fitted these three features is that of the RP genes. The positive correlation between gene length and the 3′/5′ run-on ratio, and the negative correlation between this and the expression level, diminished when the RP genes were excluded (Supplementary Figure S4F and G), while the average 3′/5′ run-on ratio of the intron-containing genes, once non-RP genes were excluded, was even more significantly different to intron-less genes (Supplementary Figure S4H). Accordingly, we found an overrepresentation of the RP genes in the minimal values of the 3′/5 ratios distribution (Figure 1A; Supplementary Figure S2A), where the average 3′/5′ ratio (−1.351 log2 units) was significantly lower than that of those genes without a functional link to the ribosomes (0.375 log2 units; P = 4.34 × 10–6; Figure 1A). The low 3′/5′ run-on ratio is a very specific feature of those genes encoding structural RPs since other ribosome-related genes belonging to the RiBi regulon (29) displayed a scattered distribution of 3′/5′ ratios, while their average ratio (0.74 log2 units) did not show a statistically significant difference to the non ribosome-related genes (P = 3.04 × 10–1; Figures 1A and S2A). Gene ontology analysis of the data, using the Fatigo application from Babelomics (30), revealed that RP-related categories were the only ones exhibiting a significantly biased distribution of active transcription. ‘Structural constituent of ribosomes’ was the item showing the highest adjusted P-value (2.68 × 10–6).
Run-on assays measure the density of the actively elongating RNA polymerases present in a given sequence, but it does not detect enzymes that are not competent for elongation, because either they have not yet initiated, have already terminated, or have been arrested and backtracked during elongation (13). In order to test whether the different run-on profiles of the genes related to the different proportions of active RNA pol II (active versus total), we performed ChIP on chip experiments using the 8WG16 antibody which recognizes the carboxy-terminal domain of the biggest RNA pol II subunit (5), and the same 5–3′ macroarrays we used in the run-on experiments; a protocol that we called RPCC (RNA Pol ChIP on Chip) in a previous paper (16). The 3′/5′ ChIP ratios extended between –3.35 (RPL24B) and 2.99 (GAL7), a considerably shorter range than the one we found for the run-on ratios (Figure 1B). Of the analysed genes, 80% showed ChIP ratios close to 1 (between 0.5 and 2; Supplementary Figure S2B), whereas only 53% exhibited run-on ratios within this range (Supplementary Figure S2A). The biological meaning of this difference is uncertain and it might be related to the shorter dynamic range of RPCC vs GRO (16).
RP genes were also overrepresented in the lowest part of the 3′/5′ ChIP ratios distribution and the average ChIP ratio for RP genes (−0.91 log2 units) even differed more significantly from the non ribosomal genes (0.28 log2 units; P = 2.04 × 10–11) than the average run-on ratio (Figure 1B). However, the overall correlation between run-on and ChIP ratios was poor (Figure 1C) and exhibited a Pearson’s coefficient of only 0.375. These results suggest that the 3′/5′ run-on ratio is not just a direct consequence of the distribution of total RNA Pol II within the genes, but reflects a combination of this parameter with others that contribute to set the proportion of active enzyme along the gene. We also measured the distribution of RNA pol II by ChIP and quantitative PCR in the individual genes tested before. The difference between the run-on and ChIP profiles was clear in most cases (Supplementary Figure 2C). We conclude that RNA pol II has the tendency to become transcriptionally inactive by becoming arrested during elongation in certain gene contexts.
The 3′/5′ run-on ratios are not globally affected by nutritional conditions or stress, but depend on the genetic background
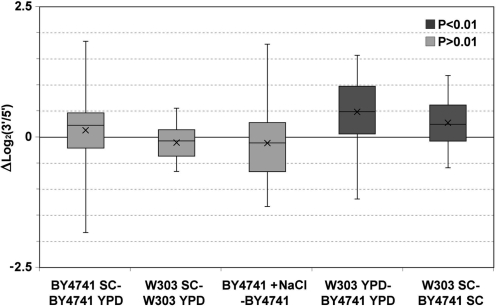
In spite of the low overall correlation between the ChIP and run-on ratios, some functional categories, like RP and RiBi, presented more significant correlations (Figure 1D). This suggests that the distribution of active RNA pol II within a particular gene could be affected by its regulatory mechanisms. To test this hypothesis, we investigated whether the run-on ratios of a subset of 76 highly expressed genes globally respond to physiological changes. First, we compared the distribution of the RNA pol II in those cells grown in a rich medium (YPD) and in a minimal medium (SC) for two wild-type strains: BY4741 and W303 (Supplementary Figure S5A and B). In all the comparisons made in this work, we did not find a uniform homogenous behaviour of all the genes represented in the array as there was always a fraction of genes that increased the ratios while another fraction decreased them. Therefore, the same absolute value of average increment can reflect a modest, but very general effect on the gene population, or a stronger effect in a subpopulation of genes. To display this information, we presented the data using box-and-whisker diagrams in which the minimum, the maximum, the average, the median and the 25th and 75th percentiles are all represented. To test whether the two data distributions significantly differed, we performed Student’s two-tailed t-tests for paired samples. In BY4741, there was an apparently slight displacement of active polymerases toward the 3′ end of the transcription units in the minimal medium (Figure 2). The average log2 (3′/5′ ratio) of the minimal medium was −0.29 and −0.42 for the rich medium (Table 1). A small shift toward the 5′ end was also observed in W303 (with averages of 0.09 and −0.01 for the rich and the minimal mediums, respectively). However, the level of confidence that we set for this work (P <0.01) indicates that these changes are not statistically significant (Figure 2, Table 1).
Figure 2.
Box-and-whiskers diagrams representing the increments in the 3′/5′ run-on ratios for the transcription units present in the array of highly expressed genes in the indicated strains and conditions. The diagrams summarize the data represented as scatter plots in the Supplementary Figure S5. Dark gray indicates a significant P-value.
Table 1.
Average change of 3′/5′ ratios in all mutants and conditions tested
| Query condition/mutant | Control | Query log2 (3′/5′ ratio) average | Control log2 (3′/5′ ratio) average | Difference (mutant-control) | Student’s t-test P-value | Number of genes analysed |
|---|---|---|---|---|---|---|
| BY4741-SC | BY4741-YPD | −0.29 | −0.42 | 0.13 | 1.56 × 10−1 | 50 |
| W303-SC | W303-YPD | −0.01 | 0.09 | −0.11 | 2.13 × 10−2 | 43 |
| BY4741-YPD + NaCl 0.5M | BY4741-YPD | −0.41 | −0.29 | −0.12 | 2.03 × 10−1 | 54 |
| W303-YPD | BY4741-YPD | 0.15 | −0.33 | 0.48 | 2.39 × 10−6 | 46 |
| W303-SC | BY4741-SC | −0.06 | −0.34 | 0.28 | 5.25 × 10−5 | 47 |
| bre1Δ | BY4741 | −0.55 | −0.23 | −0.32 | 7.93 × 10−3 | 55 |
| bur2Δ | BY4741 | −0.01 | −0.34 | 0.33 | 3.00 × 10−3 | 49 |
| ccr4Δ | BY4741 | 0.22 | −0.43 | 0.65 | 3.70 × 10−4 | 36 |
| cdc73Δ | BY4741 | −0.39 | −0.23 | −0.16 | 1.50 × 10−1 | 52 |
| dst1Δ | BY4741 | −0.52 | −0.31 | −0.21 | 8.61 × 10−3 | 45 |
| fes1Δ | BY4741 | −0.84 | −0.47 | −0.37 | 4.17 × 10−3 | 38 |
| fun12Δ | BY4741 | −0.07 | −0.40 | 0.32 | 5.02 × 10−3 | 50 |
| med2Δ | BY4741 | −0.85 | −0.38 | −0.47 | 3.17 × 10−4 | 38 |
| not5Δ | BY4741 | −0.29 | −0.43 | 0.14 | 3.97 × 10−1 | 25 |
| rpb4Δ | BY4741 | −0.62 | −0.35 | −0.27 | 1.53 × 10−2 | 54 |
| rpb9Δ | BY4741 | 0.76 | −0.27 | 1.03 | 1.68 × 10−12 | 47 |
| rtf1Δ | BY4741 | −0.59 | −0.34 | −0.25 | 6.47 × 10−2 | 27 |
| set1Δ | SET1-myc (W303) | 0.14 | 0.19 | −0.05 | 6.80 × 10−1 | 48 |
| set1-AA-myc | SET1-myc (W303) | −0.19 | 0.21 | −0.40 | 1.15 × 10−4 | 42 |
| spt4Δ | BY4741 | −0.59 | −0.28 | −0.31 | 8.38 × 10−4 | 50 |
| spt7Δ | BY4741 | −0.33 | −0.48 | 0.15 | 2.97 × 10−1 | 37 |
| spt20Δ | BY4741 | −0.05 | −0.21 | 0.16 | 2.72 × 10−1 | 32 |
| ssd1Δ(BY4741) | BY4741 | 0.11 | −0.28 | 0.39 | 6.30 × 10−3 | 57 |
| ssd1Δ(BY4741) | ssd1-d (W303) | 0.09 | 0.15 | −0.06 | 6.87 × 10−1 | 47 |
| 377 array: | ||||||
| rpb9Δ (377 array) | BY4741 (377 array) | 0.91 | 0.25 | 0.66 | 5.14 × 10−31 | 261 |
| rpb9ΔChIP | BY4741 ChIP | 0.08 | 0.19 | −0.11 | 4.92 × 10−5 | 324 |
| BY4741 run on | BY4741 ChIP | 0.29 | 0.12 | 0.17 | 5.46 × 10−2 | 250 |
| rpb9Δrun on | rpb9ΔChIP | 0.93 | 0.03 | −0.90 | 2.93 × 10−19 | 250 |
We then investigated the effect of osmotic stress on the intragenic distribution of active RNA pol II. We performed run-on assays on a BY4741 strain with or without adding NaCl at a final concentration of 0.5 M and after a 20-min incubation. No general change was observed in the 3′/5′ ratios (Supplementary Figure S5C). The difference between the averages of both conditions was only 0.12 log2 units (−0.29 and −0.41 for the non-treated and the NaCl-treated cultures, respectively), with no statistical significance (Figure 2, Table 1). We conclude that the physiological stimuli tested do not have a general effect on the intragenic distribution of active RNA pol II.
We then compared the 3′/5′ run-on ratios of the two genetic backgrounds studied. The general distribution in both strains was conserved. Those genes with a high 3′/5′ run-on ratio in the BY4741 background also showed a high ratio in the W303 background and the transcription units with a low 3′/5′ run-on ratio in BY4741 also presented a low one in W303 (Supplementary Figure S5D). The distribution of the cells grown in the YPD medium showed a high Pearson’s correlation coefficient (r = 0.806). However, the average distribution of active polymerases shifted to the 3′-end in W303 in relation to BY4741. The average 3′/5′ ratios were −0.33 log2 units for BY4741 and 0.15 for W303, with a very significant statistical difference (P = 2.39 × 10–6; Figure 2, Table 1). Very similar results were obtained in the minimal medium (Figure 2, Supplementary Figure S5E). In this case, the correlation between the two series of data was even higher (r = 0.902) and the difference between the averages of both strains was 0.28 logarithmic units, while the P-value in the Student’s t-test was 5.25 × 10–5 (Table 1). We conclude that the genetic background has a general influence on the intragenic distribution of active RNA pol II.
Genetic analysis of the intragenic distribution of active transcription
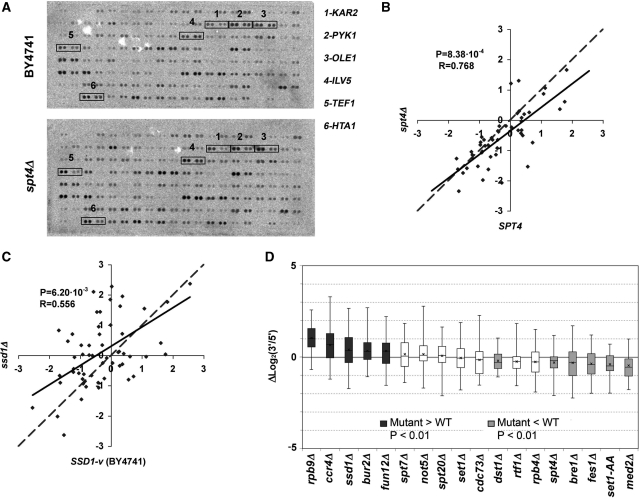
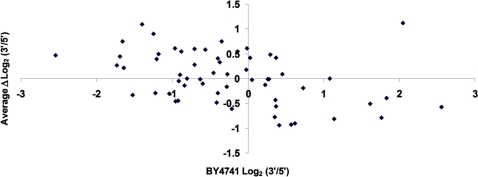
In order to test whether a single-gene mutation can alter the intragenic distribution of active pol II, we performed a series of run-on assays with an spt4Δ mutant, which lacks one of the two subunits of the well-known transcription elongation factor DSIF (31–35). Figure 3A shows a typical run-on hybridization experiment to compare a wild-type BY4741 with an isogenic spt4Δ mutant. A visual inspection reveals that the 3′/5′ ratio in several genes is lower in the mutant than in the wild type. The quantification of the ratio in both strains confirmed this observation (Figure 3B). The statistical analysis of the data showed the two distributions to be different (P = 8.38 × 10–4) with a log2 average difference of −0.31 units (Table 1).
Figure 3.
(A) Image of a typical hybridization of the array of highly expressed genes with a run-on preparation from the BY4741 wild-type strain (top) and an isogenic spt4Δ mutant (bottom). See Supplementary Figure S1 and Supplementary Table SI for probe keys. The signals of six transcriptional units, in which changes in the 3′/5′ ratio between the wild type and the mutant can be easily seen, are squared. (B) Scatter plot of the run-on 3′/5′ ratios obtained for a wild-type BY4741 strain (X-axis) and an spt4Δ mutant (Y-axis). The dashed line represents a perfect match for the X and Y values. The continuous line represents linear regression. The distance between the two lines reflects the average effect of the mutant. P-value for the Student’s two-tailed t-test for paired samples (P) and Pearson’s coefficient (R) are shown. The lower the Pearson’s coefficient of a mutant, the more gene-specific its effect on the 3′/5′ ratios. (C) The same as in (B) but for the ssd1Δ mutant. (D) Box-and-whiskers diagrams representing the increments in the 3′/5′ run-on ratios for the transcription units present in the array of highly expressed genes, in the indicated mutant, related to the corresponding isogenic wild types. The boxes representing the mutants with a significantly higher P-value are shown in dark gray, those with a significantly lower value are represented in light gray, and those with no statistically significant difference are represented in white.
Given this result, we wondered whether the difference observed between BY4741 and W303 might be due to a single mutation. One of the genetic specificities of the W303 background is the SSD1 gene, which is involved in several aspects of transcription and mRNA biogenesis (36–41).The BY4741 strain bears the dominant SSD1-v allele, whereas the W303 background possesses the ssd1-d allele that encodes a truncated form of the Ssd1 protein (42). We analysed the 3′/5′ run-on ratios in an ssd1Δ strain (BY4741 background). The results obtained showed a clear shift of the run-on signal toward the 3′-end in ssd1Δ (a difference of 0.39 log2 units and a P-value of 6.20 × 10–3) (Figure 3C and D). A comparison between the ssd1Δ mutant in the BY4741 background and the wild-type W303 strain revealed no significant difference, with average ratios of 0.09 and 0.15 log2 units, respectively, and a P-value of 6.87 × 10–1 (Supplementary Figure S6A). We conclude that the ssd1 mutation in W303 is the most likely reason for its different RNA pol II intragenic distribution, as compared with BY4741.
These results might suggest that any alteration of the RNA pol II machinery has a significant unspecific impact on the intragenic profiles of active transcription. Alternatively, Spt4 and Ssd1 might belong to a more select group of elongation factors that are involved in shaping these profiles. In order to test these hypotheses, we decided to carry out a more systematic genetic analysis of the intragenic distribution of active RNA pol II. We analysed the 3′/5′ run-on ratios in a collection of mutants defective for the elements involved in RNA pol II transcription elongation. First, we studied mutants lacking one of the two non essential subunits of the RNA pol II, namely Rpb4 and Rpb9 (43,44). On the one hand, the deletion of RPB4 resulted in a slight shift of the active RNA pol II distribution toward the 5′-end of −0.27 log2 units (Figures 3D and Supplementary S6B); however, this shift was not statistically significant (P = 1.53 × 10–2; Table 1). On the other hand, the deletion of RPB9 led to a very marked change in the distribution of active RNA pol II in the opposite direction (Figures 3D and Supplementary S6C). The average 3′/5′ ratio in the rpb9Δ mutant was 0.76 log2 units, while it was − 0.27 in the wild type, yielding an extremely significant P-value of 1.68 × 10–12 (Table 1).
The next mutant we studied was bur2Δ. It lacks a component of the BUR cyclin-kinase complex, which phosphorylates not only the carboxy-terminal domain of the DSIF component Spt5 (45,46), but possibly the CTD of Rpb1 (47–49). The deletion of BUR2 led to a general shift of active RNA pol II toward the 3′-end of the transcription units (Figure 3D and Supplementary S6D), presenting a statistically significant average difference of 0.33 log2 units (P = 3.00 × 10–3; Table 1).
The lack of TFIIS/Dst1, a cleavage factor which rescues RNA pol II from arrested situations (50–52), led to an accumulation of the run-on signal toward the 5′-end of the transcribed regions (Figures 3D and Supplementary S6E). This alteration is statistically significant according to the Student’s t-test (P = 8.61 × 10–3; Table 1) despite being slight (an average log2 difference of −0.21 units).
We have recently published the findings that those mutations lacking subunits of the Mediator (53) and the CCR4-Not complex (54) produce transcription elongation defects both in vitro and in vivo (55). The med2Δ mutant, lacking a subunit of the Mediator tail domain, produced a clearly significant (P = 3.17 × 10–4) displacement of the run-on signal toward the 5′-end with an average difference of −0.47 log2 units (Figures 3D and Supplementary S6F, Table 1). With regard to the CCR4–Not complex, we studied ccr4Δ and not5Δ. Although the not5Δ mutant presented no significant differences with the wild-type strain (Figures 3D and S6G, Table 1), the ccr4Δ mutant was clearly affected (Figures 3D and Supplementary S6H) and showed a general accumulation of the run-on signal at the 3′-ends of the transcription units (increment of 0.65 log2 units, P = 3.70×10–4; Table 1).
Next, we decided to analyse some of the mutations affecting the chromatin-related factors involved in the covalent modifications of histones. We chose spt7Δ and spt20Δ which lack subunits of SAGA (56,57); cdc73Δ and rtf1Δ which lack subunits of the PAF complex (58,59); set1Δ which lacks the catalytic subunit of the COMPASS H3-K4 methylase complex (60,61); and bre1Δ which lacks the E3 ubiquitin ligase involved in H2B ubiquitination (62). We found no significant difference in the two SAGA mutants when compared with the wild type (Figures 3D and Supplementary S6I-J, Table 1). The same happened with cdc73Δ, rtf1Δ and set1Δ (Figures 3D and Supplementary S6K-M, Table 1). In contrast, bre1Δ presented a significant decrease in the average log2 (3′/5′ ratio) when compared with the wild type (−0.55 versus −0.23 with a P-value of 7.93 × 10–3) (Figures 3D and Supplementary S6N and Table 1).
H2B ubiquitination and H3-K4 methylation are closely connected as the former is a covalent modification required for H3-K4 di- and tri-methylation (18,63). Since set1Δ abolishes any kind of H3-K4 methylation and bre1Δ only eliminates H3-K4 di- and tri-methylation, we tested set1AA, a point mutation in an RRM domain of Set1 that almost abolishes H3-K4 tri-methylation, but accumulates significant levels of dimethylation at the 5′-end of the transcribed region (64). The analysis of the set1AA mutant shows a clear shift of the run-on signal toward the 5′-end (increment of −0.40 log2 units, P = 1.15 × 10–4; Figures 3D and Supplementary S6O, Table 1), which suggests that the imbalance between tri- and mono-/di-methylation in the transcribed regions brings about an aberrant intragenic distribution of active RNA pol II.
Finally, we investigated the consequences of two mutations related to translation, fun12Δ and fes1Δ, which somehow cause transcription elongation-related phenotypes (55). FUN12 encodes a translation initiation factor (65). Its deletion brought about an important change in the 3′/5′ run-on, and an average 3′/5′ratio of −0.07 log2 units was noted as opposed to the −0.40 log2 units of the wild type (P = 5.02 × 10–3; Figures 3D and Supplementary S6P, Table 1). The last factor studied, Fes1, is the nucleotide exchange factor of the Ssa1 and Ssb1 chaperones (66,67), both of which are associated with elongating ribosomes. The effect of this mutation on the active RNA pol II distribution had the opposite effect (Figures 3D and Supplementary S6Q, Table 1) since it produced a lower average 3′/5′ ratio than the wild type (−0.84 versus −0.47 log2 units, P = 3.17 × 10–4). These results reinforce the connection of these two translation-related proteins to transcription elongation.
The alterations of the run-on profiles caused by rpb9Δ do not involve major changes in the distribution of total RNA pol II
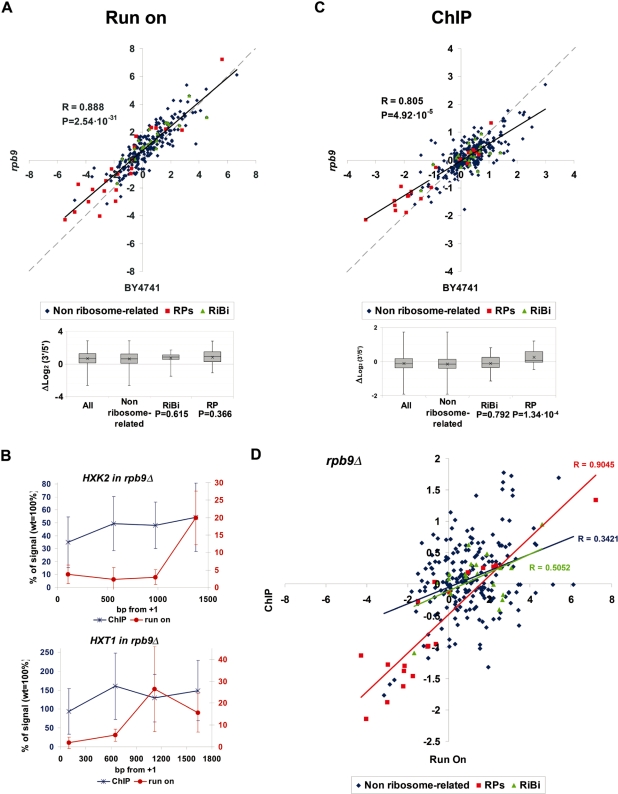
The genetic analysis carried out with the array of 76 highly expressed genes indicates that the intragenic profiles of active transcription do not respond to all the alterations of the transcriptional machinery, but to a subset of elongation-related factors. In order to do an in-depth investigation into rpb9Δ, which presented the most extreme alteration in the 3′/5′ run-on ratio, we repeated the analysis using the extended array that we had previously used for characterizing the 3′/5′ run-on ratios in the wild type. The results confirmed that the shift in the run-on ratios produced by rpb9Δ was highly significant (0.66 log2 units in average, P = 2.54 × 10–31) (Figure 4A). The effect is general (r = 0.888), including both the RP (0.80 log2 units in average) and the RiBi genes (0.73 log2 units; Figure 4A). We also analysed some individual genes in detail. The results confirmed a stronger decrease of active RNA pol II density at the 5′-end of HXK2 and HXT1 in rpb9Δ in relation to the wild type than at the 3′ end (Figure 4B). In contrast, we detected neither comparable changes in the distribution of total RNA pol II by ChIP along these two genes (Figure 4B) nor general changes in the 3′/5′ ratios of the total RNA pol II for the genes present in the array. As shown in Figure 4C, the variation in the ChIP ratios produced by rpb9Δ was in the opposite direction to the change in run-on ratios and much less significant than this. The RP genes were the exception, since their average 3′/5′ ratio slightly increased (0.26 log2 units), unlike the slight decrease noted (−0.14 log2 units) of non ribosome-related genes (P = 1.34 × 10–4; Table 1). This coordinated shift of run-on and ChIP ratios for the RP genes originated a higher correlation between run-on and ChIP ratios for this regulon, whereas the correlation for the rest of the genes did not increase (non ribosome-related), or even decreased (RiBi) (compare Figures 1C and 4D). We conclude that rpb9Δ globally alters the intragenic distribution of active RNA pol II by changing the proportion of enzymes that become transiently arrested at the 5′-end of the transcribed region.
Figure 4.
(A) Scatter plot of the run-on 3′/5′ ratios obtained for a wild-type BY4741 strain (X axis) and an rpb9Δ mutant (Y axis) using the array of 377 genes. The box-and-whiskers diagrams summarize the rpb9Δ/wild-type differences for the genes belonging to the indicated gene categories. The effect of rpb9Δ on RP and RiBi genes, relative to non ribosome-related genes, was challenged with the Student’s two-tails t-test for samples with unequal variances; P-values are shown underneath. The other details are as in Figure 3B. (B) Comparison of the run-on profile (red) and the RNA pol II ChIP profile (blue) of the rpb9Δ mutant (relative to the wild-type profile) in HXK2 and HXT1. Error bars represent standard deviation. (C) The same as in (A) but for the RNA pol II 3′/5′ ratios. (D) Scatter plot of the average run-on and ChIP 3′/5′ ratios of rpb9Δ corresponding to the genes represented in the 377 genes array. Linear regressions and their corresponding Pearson’s coefficients are shown.
DISCUSSION
The intragenic distribution of active RNA pol II is gene-specific
The use of the run-on technique in combination with DNA arrays has allowed us to study the distribution of transcriptionally competent polymerases in a broad set of yeast genes. The results obtained reveal that the intragenic distribution of active transcription varies vastly from gene to gene. CUTs or SUTs overlapping the probes do not explain this variation, since the number of genes affected in the array was very low (32 out of 377). Even after excluding 40 additional genes located in the vicinity of CUTs and SUTs (<200 bp away from any of the probes present in the array) the run-on ratios were clearly gene-specific (Supplementary Figure S4E). The effect of the tested mutants on the run-on ratios was also unaffected by the exclusion of CUTs and SUTs overlapping genes (Supplementary Figure S6R). The intragenic distribution of active transcription does not correlate extensively with any of the other parameters that we analysed (length, G + C content, expression level, presence of introns). It seems to be an intrinsic feature of each transcription unit as it is roughly conserved in all the mutants and under all study conditions (Supplementary Figure S7A).
When we compare the distributions of RNA pol II determined by run-on and ChIP, we observe a much more uniform profile by ChIP, with the majority of genes showing 3′/5′ ChIP ratios close to 1. The detailed analysis of individual genes shows an almost constant distribution of total RNA pol II along their bodies, whereas their run-on profiles for each gene are markedly different and show considerable fluctuations along the transcription units. The more homogenous patterns produced by ChIP may not be just a consequence of the lower resolution of this technique since the RP genes (generally short and, therefore, more sensitive to cross-contamination between the 3′ and 5′ ChIP signals) presented the most biased ChIP ratios. We rather think that the simplest explanation for this phenomenon is the existence of arrested, transcriptionally inactive polymerases along the transcribed regions, which do not produce a run-on signal despite being detected by ChIP.
Although pausing and arrest seem to be common phenomena in each transcription cycle in vivo (68), we show that the tendency of RNA pol II to become inactively arrested is variable among genes and likely influenced by the genomic environment. It is possible that nucleosome positioning, a genomic feature that is considerably gene-specific (69), may increase the probability of RNA pol II becoming arrested. This hypothesis is in good agreement with the conclusions drawn from in vitro experiments that present a high tendency of RNA pol II to backtrack in front of a nucleosome (11).
By considering the whole gene as the unit of analysis, we have recently published that a significant proportion of the total RNA pol II that is engaged in transcription under standard growth conditions is not detectable in a GRO assay (16). We showed that this accumulation of inactive RNA pol II is especially relevant in the RP genes (16), which, according to the present work, exhibit the most 5′-biased distribution of total RNA pol II. It is appealing in this respect that the transcribed regions of the RP genes show some specific chromatin features, including a significantly lower 5′ nucleosome spacing than the rest of the genome (70). Transcription through these highly packaged genes would involve a higher probability of pausing and arrest and a subsequent 5′ bias in the distribution of total RNA pol II. However, Weiner et al. (70) have provided evidence of transcriptional activity modulating chromatin packaging, and not vice versa. Accordingly, we failed to detect a significant quantitative correlation between the 3′/5′ run-on ratios and the corresponding ratios of sensitivity to micrococcal nuclease when we used the data kindly provided by O. Rando (results not shown). Therefore, if nucleosome positioning plays a role in restricting RNA pol II activity, additional chromatin elements should be involved. The genetic analysis carried out in the second part of this work sheds some light in this respect (see subsequently).
Gene specificity is not contradictory to coordinated regulation. The RP genes exhibit an exceptional positive correlation between ChIP and the run-on ratios in both the wild type and rpb9Δ. This fact suggests that, despite their differences, the intragenic distributions of total and active RNA pol II can be somehow interdependent and that there should be a mechanism connecting them. This mechanism might be characteristic of each regulon. In this sense, and unlike the RP regulon, the RiBi one shows a higher ChIP-run-on correlation in the wild type than in rpb9Δ, which indicates that Rpb9 plays a more prominent role in linking the intragenic distributions of total and active RNA pol II in the RiBi genes. We have previously shown that the proportion of active versus total RNA pol II during elongation is differentially controlled in response to the carbon source in three regulons (RP, RiBi and mitochondrial), which represent >10% of yeast genes (16). Accordingly, the gene-specific profiles of active and total RNA pol II might be the consequence of the local effects caused by the regulatory mechanisms controlling transcription at the elongation level.
Transcription elongation factors control the intragenic distribution of active RNA pol II
We have hereby shown that some single mutations can have general effects on the intragenic profiles of active transcription, whereas other mutations, including those affecting important general transcription factors like SAGA or the Paf complex, do not significantly affect the 3′/5′ run-on ratios. All the tested mutations were viable and, therefore, their average effects on the 3′/5′ run-on ratios were modest (1 log2 units at the most; Table 1). These moderate alterations of the active RNA pol II profiles are fully compatible with normal cell growth under laboratory conditions. W303, for instance, which has been considered the reference strain in many gene-expression studies, displays significantly higher 3′/5′ run-on ratios than BY4741. We have shown that this difference is likely due to the presence in W303 of a recessive allele (ssd1-d) that encodes a truncated form of Ssd1 lacking an RNAse II-like domain (42). Ssd1 participates in mRNA processing (39,40), binds RNA (38) and physically interacts with the hyperphosphorylated form of the RNA pol II CTD (71). Our results indicate an important role of Ssd1 during transcription elongation and help explain why some gene expression-related phenotypes are specific of the W303 background (37,41).
The results of the genetic analysis that we have carried out confirm the involvement of some protein factors in transcription elongation (Ccr4, Fun12 and Fes1), whose molecular connection with the transcriptional machinery is uncertain (55). In other cases, previous knowledge on the factors function allows approaching the molecular basis of this phenomenon. This is the case of bre1Δ and set1AA, producing a shift in the distribution of active pol II toward the 5′-end of the genes. The deletion of BRE1 abolishes H2B ubiquitynation and, subsequently, H3-K4 di- and trimethylation (18)(63), whereas set1AA causes a defect in H3-K4 trimethylation without eliminating dimethylation (64). In contrast, the deletion of SET1, which abolishes all forms of H3-K4 methylation, does not produce any significant shift in the intragenic localization of active RNA pol II. We propose that the balance between the different forms of H3-K4 methylation (mono-, di- and tri-methylation) regulates RNA pol II activity during transcription elongation either directly or by affecting additional chromatin modifications (72).
The other mutants that produce general shifts in the 3′/5′ run-on ratios encode general transcription factors that interact directly with RNA pol II. Of these, the deletion of the polymerase subunit Rpb9 is particularly striking. The comparison of the run-on results with those produced by ChIP indicates the presence of transcriptionally incompetent polymerases in the 5′ half of the genes studied, suggesting a higher tendency of Rpb9-defective polymerases to become arrested in that particular region. Such arrested polymerases would need TFIIS to resume transcription, which is consistent with the synthetic lethality exhibited by rpb9Δ and dst1Δ (73). However, dst1Δ does not produce a 3′-biased profile of active transcription, like rpb9Δ, but a weaker bias toward the 5′-end. This result indicates that wild-type RNA pol II does not tend to become preferentially inactive at the 5′-end, as it does in the absence of Rpb9, but inactivates more modestly at the 3′-end, where it may be efficiently compensated by the action of Dst1.
The deletion of Bur2 cyclin-kinase also displaces the distribution of active RNA pol II toward the 3′-end. The main function of Bur2 is to phosphorylate the Spt5 subunit of yDSIF (45,46). This phosphorylation is essential for overcoming the early elongation pause. So it is conceivable that a lack of phosphorylation in Spt5 produces a higher frequency of RNA pol II arrest at the 5′-end, hence lowering the run-on signal at this region. This is in agreement with the results obtained by Chu et al. (74) in PMA1, where bur2Δ has been seen to bring about a decrease in histone H4-K36 trimethylation, a chromatin mark of transcription elongation, at the 5′ of this gene without affecting the total amount of RNA pol II present in this region. If this hypothesis is true, and given that spt4Δ produces a bias of active transcription in the opposite orientation, the two subunits of yDSIF would play distinct complementary roles during elongation. Although we favour this interpretation, we cannot rule out that the deletion of Bur2 produces a perturbation in the intragenic distribution of active RNA pol II as it affects the phosphorylation status of the Rpb1 CTD (49).
The last intriguing finding of our genetic analysis of active RNA polymerase distribution is the marked 5′-bias produced by med2Δ. The Med2 protein belongs to the Mediator complex, which was originally described for its role in the regulation of the pre-initiation-complex assembly. Following a completely different experimental approach, we have already linked the Mediator complex with the elongation phase (55). Human Mediator has also been connected to post-initiation transcription since it was found to be involved in the DSIF-dependent activation of transcription (75). This fact enables a mechanism by which Mediator might indirectly modulate the intragenic distribution of active RNA pol II.
As part of the general tendency of each gene to maintain a characteristic 3′/5′ run-on ratio in all backgrounds and under all conditions, there is a certain degree of overlapping among the genes when comparing all the average ratios obtained for the 18 mutants tested (Supplementary Figure S7A). As expected, the clustering analysis of this data recapitulates the functional relationship of some mutants (set1AA and set1Δ) or their average effect on the run-on ratios (rpb9Δ, ccr4Δ and ssd1Δ) (Supplementary Figure S9C). With some minor exceptions, all the genes present in the array were influenced in their 3′/5′ run-on ratios by the mutants tested, exhibiting a variation of at least 1 log2 units in one of the mutant strains (Supplementary Figure S7B). Some genes tended to shift their run-on ratio toward the 3′ end in the mutants analysed, whereas others did so toward the 5′-end (Supplementary Figure S7B). When we plot the average mutant-wild-type increment in the 3′/5′ run-on ratio for each gene versus its 3′/5′ run-on ratio in the wild type, we see that most genes exhibiting significantly biased distributions of active RNA pol II in the wild type tend to equilibrate them in the mutant backgrounds (Figure 5). We can conclude that the default situation is the unbiased distribution of active transcription and that transcription elongation factors are collectively involved in shaping the profile of active RNA pol II along the transcribed region. Accordingly, gene-specific profiles would be the result of combining local chromatin with the action of the transcription elongation machinery in response to gene-specific regulatory signals. The recent development of more sensitive high-resolution methods of genomic run-on (15) should facilitate the test of this model.
Figure 5.
Scatter plot representing the average increment of the 3′/5′ run-on ratio produced on each gene by the 18 mutation tested (Y-axis) against the run-on 3′/5′ ratio exhibited by that gene in the wild-type BY4741 strain (X-axis). Spearmann’s rank correlation ρ = –0,428; P = 8.09 × 10–4.
FUNDING
Spanish Ministry of Education and Science (BFU2007-67575-CO3-01/BMC to J.E.P-O. and BFU2007-67575-CO3-02/BMC to S.C.); Regional Valencian Government (ACOMP2009/368 to J.E.P.-O.); Regional Andalusian Government (P07-CVI02623 to S.C.). A.R-G. was covered by a fellowship from the University of Seville. Funding for open access charge: Andalusian Government.
Conflict of interest statement. None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank O. Rando for sharing data, F. Posas, E. de Nadal and the members of our labs for comments and advice, and Helen Warburton for English corrections.
REFERENCES
- 1.Price DH. Poised polymerases: on your mark…get set…go! Mol. Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Zhou Q, Yik JH. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hecht A, Grunstein M. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 1999;304:399–414. doi: 10.1016/s0076-6879(99)04024-0. [DOI] [PubMed] [Google Scholar]
- 4.Hecht A, Strahl-Bolsinger S, Grunstein M. Mapping DNA interaction sites of chromosomal proteins. Crosslinking studies in yeast. Methods Mol. Biol. 1999;119:469–479. doi: 10.1385/1-59259-681-9:469. [DOI] [PubMed] [Google Scholar]
- 5.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 7.Gilchrist DA, Fargo DC, Adelman K. Using ChIP-chip and ChIP-seq to study the regulation of gene expression: Genome-wide localization studies reveal widespread regulation of transcription elongation. Methods. 2009;48:398–408. doi: 10.1016/j.ymeth.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Exinger F, Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 9.Glesne DA, Collart FR, Huberman E. Regulation of IMP dehydrogenase gene expression by its end products, guanine nucleotides. Mol. Cell Biol. 1991;11:5417–5425. doi: 10.1128/mcb.11.11.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morillo-Huesca M, Vanti M, Chavez S. A simple in vivo assay for measuring the efficiency of gene length-dependent processes in yeast mRNA biogenesis. FEBS J. 2006;273:756–769. doi: 10.1111/j.1742-4658.2005.05108.x. [DOI] [PubMed] [Google Scholar]
- 11.Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Mol. Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 12.Voliotis M, Cohen N, Molina-Paris C, Liverpool TB. Fluctuations, pauses, and backtracking in DNA transcription. Biophys. J. 2008;94:334–348. doi: 10.1529/biophysj.107.105767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirayoshi K, Lis JT. Nuclear run-on assays: assessing transcription by measuring density of engaged RNA polymerases. Methods Enzymol. 1999;304:351–362. doi: 10.1016/s0076-6879(99)04021-5. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Martinez J, Aranda A, Perez-Ortin JE. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell. 2004;15:303–313. doi: 10.1016/j.molcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pelechano V, Jimeno-Gonzalez S, Rodriguez-Gil A, Garcia-Martinez J, Perez-Ortin JE, Chavez S. Regulon-specific control of transcription elongation across the yeast genome. PLoS Genet. 2009;5:e1000614. doi: 10.1371/journal.pgen.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 18.Dehe PM, Pamblanco M, Luciano P, Lebrun R, Moinier D, Sendra R, Verreault A, Tordera V, Geli V. Histone H3 lysine 4 mono-methylation does not require ubiquitination of histone H2B. J. Mol. Biol. 2005;353:477–484. doi: 10.1016/j.jmb.2005.08.059. [DOI] [PubMed] [Google Scholar]
- 19.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Sprong Harbor, NY: Cold Sprong Harbor Laboratory Press; 1990. [Google Scholar]
- 20.Alberola TM, Garcia-Martinez J, Antunez O, Viladevall L, Barcelo A, Arino J, Perez-Ortin JE. A new set of DNA macrochips for the yeast Saccharomyces cerevisiae: features and uses. Int. Microbiol. 2004;7:199–206. [PubMed] [Google Scholar]
- 21.Akhtar A, Faye G, Bentley DL. Distinct activated and non-activated RNA polymerase II complexes in yeast. EMBO J. 1996;15:4654–4664. [PMC free article] [PubMed] [Google Scholar]
- 22.Kohrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- 23.Jimeno-Gonzalez S, Gomez-Herreros F, Alepuz PM, Chavez S. A gene-specific requirement for FACT during transcription is related to the chromatin organization of the transcribed region. Mol. Cell Biol. 2006;26:8710–8721. doi: 10.1128/MCB.01129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosaleny LE, Ruiz-Garcia AB, Garcia-Martinez J, Perez-Ortin JE, Tordera V. The Sas3p and Gcn5p histone acetyltransferases are recruited to similar genes. Genome Biol. 2007;8:R119. doi: 10.1186/gb-2007-8-6-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berry G, Armitage P. Statistical Methods in Medical Research. 3rd ed. Oxford: Blackwell Science; 1994. [Google Scholar]
- 26.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, Dietrich FS. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res. 2005;33:2838–2851. doi: 10.1093/nar/gki583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Shahrour F, Minguez P, Tarraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 2006;34:W472–W476. doi: 10.1093/nar/gkl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindstrom DL, Hartzog GA. Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics. 2001;159:487–497. doi: 10.1093/genetics/159.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rondon AG, Garcia-Rubio M, Gonzalez-Barrera S, Aguilera A. Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J. 2003;22:612–620. doi: 10.1093/emboj/cdg047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kebaara B, Nazarenus T, Taylor R, Atkin AL. Genetic background affects relative nonsense mRNA accumulation in wild-type and upf mutant yeast strains. CURR. Genet. 2003;43:171–177. doi: 10.1007/s00294-003-0386-3. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, McIntosh KB, Rudra D, Schawalder S, Shore D, Warner JR. Fine-structure analysis of ribosomal protein gene transcription. Mol. Cell Biol. 2006;26:4853–4862. doi: 10.1128/MCB.02367-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uesono Y, Toh-e A, Kikuchi Y. Ssd1p of Saccharomyces cerevisiae associates with RNA. J. Biol. Chem. 1997;272:16103–16109. doi: 10.1074/jbc.272.26.16103. [DOI] [PubMed] [Google Scholar]
- 39.Fortes P, Kufel J, Fornerod M, Polycarpou-Schwarz M, Lafontaine D, Tollervey D, Mattaj IW. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol. Cell Biol. 1999;19:6543–6553. doi: 10.1128/mcb.19.10.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luukkonen BG, Seraphin B. A conditional U5 snRNA mutation affecting pre-mRNA splicing and nuclear pre-mRNA retention identifies SSD1/SRK1 as a general splicing mutant suppressor. Nucleic Acids Res. 1999;27:3455–3465. doi: 10.1093/nar/27.17.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stettler S, Chiannilkulchai N, Hermann-Le Denmat S, Lalo D, Lacroute F, Sentenac A, Thuriaux P. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol. Gen. Genet. 1993;239:169–176. doi: 10.1007/BF00281615. [DOI] [PubMed] [Google Scholar]
- 42.Jorgensen P, Nelson B, Robinson MD, Chen Y, Andrews B, Tyers M, Boone C. High-resolution genetic mapping with ordered arrays of Saccharomyces cerevisiae deletion mutants. Genetics. 2002;162:1091–1099. doi: 10.1093/genetics/162.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woychik NA, Lane WS, Young RA. Yeast RNA polymerase II subunit RPB9 is essential for growth at temperature extremes. J. Biol. Chem. 1991;266:19053–19055. [PubMed] [Google Scholar]
- 44.Woychik NA, Young RA. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol. Cell Biol. 1989;9:2854–2859. doi: 10.1128/mcb.9.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wood A, Shilatifard A. Bur1/Bur2 and the Ctk complex in yeast: the split personality of mammalian P-TEFb. Cell Cycle. 2006;5:1066–1068. doi: 10.4161/cc.5.10.2769. [DOI] [PubMed] [Google Scholar]
- 46.Zhou K, Kuo WH, Fillingham J, Greenblatt JF. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl Acad. Sci. USA. 2009;106:6956–6961. doi: 10.1073/pnas.0806302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao S, Neiman A, Prelich G. BUR1 and BUR2 encode a divergent cyclin-dependent kinase-cyclin complex important for transcription in vivo. Mol. Cell Biol. 2000;20:7080–7087. doi: 10.1128/mcb.20.19.7080-7087.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murray S, Udupa R, Yao S, Hartzog G, Prelich G. Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol. Cell Biol. 2001;21:4089–4096. doi: 10.1128/MCB.21.13.4089-4096.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu H, Hu C, Hinnebusch AG. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell. 2009;33:752–762. doi: 10.1016/j.molcel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kipling D, Kearsey SE. Function of the S. cerevisiae DST1/PPR2 gene in transcription elongation. Cell. 1993;72:12. doi: 10.1016/0092-8674(93)90044-q. [DOI] [PubMed] [Google Scholar]
- 51.Awrey DE, Weilbaecher RG, Hemming SA, Orlicky SM, Kane CM, Edwards AM. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J. Biol. Chem. 1997;272:14747–14754. doi: 10.1074/jbc.272.23.14747. [DOI] [PubMed] [Google Scholar]
- 52.Fish RN, Kane CM. Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 53.Bjorklund S, Gustafsson CM. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 2005;30:240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 54.Collart MA. Global control of gene expression in yeast by the Ccr4-Not complex. Gene. 2003;313:1–16. doi: 10.1016/s0378-1119(03)00672-3. [DOI] [PubMed] [Google Scholar]
- 55.Gaillard H, Tous C, Botet J, Gonzalez-Aguilera C, Quintero MJ, Viladevall L, Garcia-Rubio ML, Rodriguez-Gil A, Marin A, Arino J, et al. Genome-wide analysis of factors affecting transcription elongation and DNA repair: a new role for PAF and Ccr4-not in transcription-coupled repair. PLoS Genet. 2009;5:e1000364. doi: 10.1371/journal.pgen.1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daniel JA, Grant PA. Multi-tasking on chromatin with the SAGA coactivator complexes. Mutat Res. 2007;618:135–148. doi: 10.1016/j.mrfmmm.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Timmers HT, Tora L. SAGA unveiled. Trends Biochem Sci. 2005;30:7–10. doi: 10.1016/j.tibs.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 58.Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi X, Chang M, Wolf AJ, Chang CH, Frazer-Abel AA, Wade PA, Burton ZF, Jaehning JA. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell Biol. 1997;17:1160–1169. doi: 10.1128/mcb.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dehe PM, Geli V. The multiple faces of Set1. Biochem. Cell Biol. 2006;84:536–548. doi: 10.1139/o06-081. [DOI] [PubMed] [Google Scholar]
- 61.Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 62.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 63.Shahbazian MD, Zhang K, Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell. 2005;19:271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 64.Tresaugues L, Dehe PM, Guerois R, Rodriguez-Gil A, Varlet I, Salah P, Pamblanco M, Luciano P, Quevillon-Cheruel S, Sollier J, et al. Structural characterization of Set1 RNA recognition motifs and their role in histone H3 lysine 4 methylation. J. Mol. Biol. 2006;359:1170–1181. doi: 10.1016/j.jmb.2006.04.050. [DOI] [PubMed] [Google Scholar]
- 65.Choi SK, Lee JH, Zoll WL, Merrick WC, Dever TE. Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science. 1998;280:1757–1760. doi: 10.1126/science.280.5370.1757. [DOI] [PubMed] [Google Scholar]
- 66.Dragovic Z, Shomura Y, Tzvetkov N, Hartl FU, Bracher A. Fes1p acts as a nucleotide exchange factor for the ribosome-associated molecular chaperone Ssb1p. Biol. Chem. 2006;387:1593–1600. doi: 10.1515/BC.2006.198. [DOI] [PubMed] [Google Scholar]
- 67.Kabani M, Beckerich JM, Brodsky JL. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell Biol. 2002;22:4677–4689. doi: 10.1128/MCB.22.13.4677-4689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu. Rev. Biochem. 2009;78:245–271. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Phatnani HP, Jones JC, Greenleaf AL. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry. 2004;43:15702–15719. doi: 10.1021/bi048364h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hemming SA, Jansma DB, Macgregor PF, Goryachev A, Friesen JD, Edwards AM. RNA polymerase II subunit Rpb9 regulates transcription elongation in vivo. J. Biol. Chem. 2000;275:35506–35511. doi: 10.1074/jbc.M004721200. [DOI] [PubMed] [Google Scholar]
- 74.Chu Y, Simic R, Warner MH, Arndt KM, Prelich G. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 2007;26:4646–4656. doi: 10.1038/sj.emboj.7601887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malik S, Barrero MJ, Jones T. Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. Proc. Natl Acad. Sci. USA. 2007;104:6182–6187. doi: 10.1073/pnas.0608717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.