Abstract
Circadian oscillators provide rhythmic temporal cues for a range of biological processes in plants and animals, enabling anticipation of the day/night cycle and enhancing fitness-associated traits. We have used engineering models to understand the control principles of a plant’s response to seasonal variation. We show that the seasonal changes in the timing of circadian outputs require light regulation via feed-forward loops, combining rapid light-signaling pathways with entrained circadian oscillators. Linear time-invariant models of circadian rhythms were computed for 3,503 circadian-regulated genes and for the concentration of cytosolic-free calcium to quantify the magnitude and timing of regulation by circadian oscillators and light-signaling pathways. Bioinformatic and experimental analysis show that rapid light-induced regulation of circadian outputs is associated with seasonal rephasing of the output rhythm. We identify that external coincidence is required for rephasing of multiple output rhythms, and is therefore important in general phase control in addition to specific photoperiod-dependent processes such as flowering and hypocotyl elongation. Our findings uncover a fundamental design principle of circadian regulation, and identify the importance of rapid light-signaling pathways in temporal control.
Keywords: circadian rhythms, photoperiod, systems identification, linear time-invariant systems, external coincidence
In plants and animals, the timing of gene expression during the day is maintained by circadian oscillators that comprise networks of interlocking transcriptional negative feedback loops (1–4). In Arabidopsis thaliana, a negative feedback loop between CIRCADIAN CLOCK ASSOCIATED 1 (CCA1)/LATE ELONGATED HYPOCOTYL (LHY), and TIMING OF CAB2 EXPRESSION 1 (TOC1) forms part of the circadian clock (3). CCA1/LHY dimers bind directly to a conserved evening element (EE) promoter motif found in a large subset of circadian-regulated transcripts, including TOC1, conferring rhythmic expression (5). The CCA1/LHY-TOC1 loop is one of at least three interlocking negative-feedback loops that form the circadian oscillator (6). It is proposed that this complexity provides robustness and multiple entry points for light input, allowing entrainment and adaptation to changing photoperiod (7, 8). Light input is mediated by the red-light receptors PHYTOCHROME A/B/D/E, the blue-light receptors CRYPTOCHROME 1/2, and the light oxygen voltage (LOV) domain proteins ZEITLUPE (ZTL) and FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1) (4). Photoperception pathways may also directly regulate circadian outputs; for example, expression of CHLOROPHYLL A/B BINDING PROTEIN2 (CAB2) is controlled by both the circadian oscillator and phytochrome signaling pathways (9), and light-induced transcription of CAB2 is gated by the circadian oscillator (10, 11), indicating interactions between the two regulatory modules.
Many circadian outputs adapt their phase to seasonal changes in photoperiod (12). A change in the timing of peak activity (or phase shift) is required to synchronize time-of-day-specific processes (e.g., those with peak activity in the middle of the photoperiod) with the changing day-length of the external environment. Two models describing mechanisms by which the circadian clock contributes to photoperiod-dependent phasing of an output rhythm have been proposed. An “external coincidence” model proposes that the phase of an activity is the result of coincidence between the phase of the oscillator and that of the external light and dark cycle (13). Alternatively, an “internal coincidence” model proposes that the role of light is to differentially entrain at least two rhythms (e.g., the morning and evening loops of the central oscillator) (6) and that the phase of the output is dependent on the phase relationship between the two oscillators (8, 14). Examples of both internal and external coincidence mechanisms are found in the photoperiodic control of flowering time in Arabidopsis. Expression of the floral regulator CONSTANS (CO) requires formation of a complex between GIGANTEA (GI) and FKF1. The formation of this complex is an example of both external and internal coincidence. The expression of GI and FKF1 are both circadian regulated, and the expression peaks have the same phase in long days but are phased differently in short days (15) and can therefore only form a complex and promote CO transcription under long photoperiods. The next stage of the pathway requires external coincidence, because FKF1 can act as a blue light receptor, light favors the FKF-GI complex formation, and coincidence between CO transcription and light-dependent stabilization of CO protein in long days (16) results in transcription of the key floral regulator FLOWERING LOCUS T (FT).
We found that oscillations in the concentration of free Ca2+ ions in the cytosol ([Ca2+]cyt) are regulated both by the circadian oscillator and light signaling (17), and that the phase of circadian [Ca2+]cyt oscillations changes in response to photoperiod (18). We were therefore interested to test whether this dual regulation by light and the clock might be related to phase control. Using a combination of reverse engineering and experimentation, we tested the hypothesis that appropriate phasing of circadian outputs, such as [Ca2+]cyt oscillations, is a result of external coincidence.
We used linear time-invariant (LTI) models to describe the regulation of circadian outputs by light and the circadian oscillator to overcome the algorithmic and combinatorial challenges of fitting detailed nonlinear systems to limited data. Systems identification techniques enable efficient estimation of parameters in LTI models from input-output data (19), which, in principle, contain any number of hypothetical components or hidden variables. Approximation to a LTI system allowed access to a wealth of analytical tools not applicable to nonlinear models, permitting dissection of the control principles of the circadian network. We show that LTI models can be used to approximate nonlinear biological phenomena by testing hypotheses about the network controlling [Ca2+]cyt and gene expression. We also demonstrate that Bode analysis (20), in which the response of an LTI system to sinusoidal inputs of various frequencies is measured, can be used to estimate the relative contribution of individual regulatory pathways to overall control of a biological process regulated by multiple inputs. Our data suggest that rapid light input is required for photoperiod-dependent changes in the phase of the daily rhythms in Arabidopsis, providing evidence of external coincidence as a mechanism of global phase control.
Results
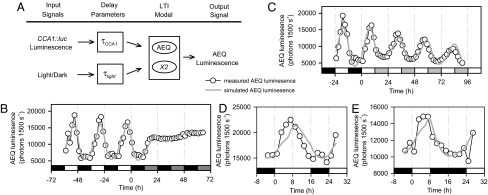
We derived mathematical models using LTI systems identification (19, 21, 22) on input-output data sets (Fig. 1A), with [Ca2+]cyt (output) measured as AEQUORIN (AEQ) bioluminescence (Fig. 1B and SI Appendix, Fig. S1B). CCA1 is required for circadian oscillations of [Ca2+]cyt (17). Therefore, CCA1 promoter activity measured as CCA1::luciferase (CCA1::luc) (23) luminescence represented input from the circadian clock (SI Appendix, Fig. S1A). Light input was incorporated on two levels, either via the transcriptional activation of CCA1 and hence implicitly encapsulated by CCA1::luc activity (24, 25), or through an explicit independent pathway. The timescales of regulation by CCA1 and light were considered by introducing delay parameters (τCCA1 and τlight). Model parameters were estimated using a single experiment of input-output measurements in 12-h light/12-h dark cycles (12L/12D), followed by transfer to constant dark (DD; Fig. 1B), then cross-validated by comparing model-predicted outputs with input-output data (that had not been used for model identification) in constant light and cycles of long and short photoperiods (LL, 16L/8D, and 8L/16D, respectively; Fig. 1 C–E and SI Appendix, Figs. S1–S3). An optimal model predicted key features of the experimental data (Fig. 1 C–E) that were not overt in the training data (Fig. 1B), such as rephasing between 16L/8D (measured peaks 7.0 ± 0.5 h after dawn, n = 24; simulated peaks 8.95 h after dawn) and 8L/16D (peaks 4.9 ± 0.4 h after dawn, n = 22; simulated peaks 7.57 h after dawn; Fig. 1). The CCA1-independent light-input pathway might involve alterations to cellular physiology, e.g., ion channel activation, because the optimal model indicated that light rapidly affects [Ca2+]cyt (τlight = 0 h; SI Appendix, Fig. S4A), whereas a sequence of molecular events probably link CCA1 activity to [Ca2+]cyt (τCCA1 = 5.2 h; SI Appendix, Fig. S2B). When selecting the optimal model, a range of complexity was considered by providing hidden variables, which enable increasingly rich dynamics with the cost of increased uncertainty (SI Appendix). Biologically, hidden variables represent cellular components that interact with modeled output process and are required for the behaviors observed, but are not included explicitly in the model definition. The optimal model for [Ca2+]cyt incorporated a single hidden variable (termed X2; Fig. 1).
Fig. 1.
Architecture and validation of a dual-input linear systems model of the circadian [Ca2+]cyt oscillation. (A) Schematic for a mathematical model of circadian and light/dark regulation of [Ca2+]cyt. A linear time-invariant (LTI) model incorporates inputs from the circadian clock (CCA1::luc) and an independent light/dark pathway, which regulate a hidden variable X2 and [Ca2+]cyt (AEQ luminescence). Light input to the model was assumed to be binary (light = 1; dark = 0). (B) Estimation of the model with wild-type (Ws-0) AEQ luminescence data obtained under LD-DD cycles. (C–E) Cross-validation of the model with wild-type (Ws-0) AEQ luminescence data obtained under (C) LD-LL, (D) 16L/8D, and (E) 8L/16D cycles. Open circles indicate experimental data measuring AEQ luminescence, and gray lines simulated model outputs. Simulations were scaled to overlap the data points using linear regression. Open bars indicate light, closed bars dark, and gray bars subjective light or dark, as appropriate. [B and C, data re-plotted from ref. 17 (Copyright American Society of Plant Biologists).]
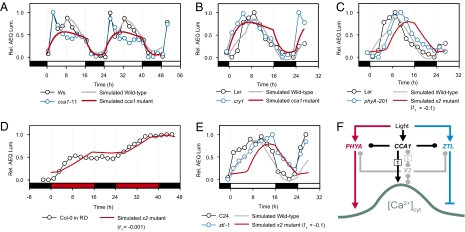
To identify pathway(s) that contribute to the hidden variable X2, and to interpret the model in a cellular context, we simulated the effects of CCA1 and X2 mutations (see SI Appendix for simulation methods) and compared the results with experimental data (Fig. 2 and SI Appendix, Figs. S4–S7). The simulated [Ca2+]cyt oscillation of a cca1 null model (CCA1 input = 0) in 16L/8D had dynamics consistent with experimental data from cca1-11 (Fig. 2A and SI Appendix, Fig. S4 A and B), in that the main peak of [Ca2+]cyt in the middle of the day was absent in the cca1 null. The model also predicted a large reduction in amplitude of the oscillation in the cca1 null (SI Appendix, Fig. S4 C and E), but this was not tested because of the difficulty in calibrating AEQ luminescence to [Ca2+]cyt in long-term experiments (17). The early [Ca2+]cyt peak in this accession of Wassilewskija (Ws) was not encapsulated in the wild-type simulation because the model was derived using data from a Ws accession that lacked this peak; analysis of a number of Arabidopsis accessions revealed diversity in early morning [Ca2+]cyt dynamics (SI Appendix, Fig. S8) (17).
Fig. 2.
Dual light and circadian control is required for appropriate shape and phasing of the daily [Ca2+]cyt oscillation. (A and B) Simulated cca1 null mutant compared with measurements of AEQ luminescence (A) cca1-11 and (B) cry1 in 16L/8D cycles. (C) Simulated x2 mutant (t1 = −0.1) compared with measurements of AEQ luminescence in phyA-201. (D) Simulated x2 mutant (t1 = −0.001) compared with AEQ luminescence in Col-0 grown under red/dark (RD) cycles. (E) Simulated x2 mutant (t1 = −0.1) compared with measurements of AEQ luminescence in ztl-1. (A–E) AEQ measurements (open circles) were normalized for clarity, indicating wild-type (black) and mutant (blue) Arabidopsis. Furthermore, to determine whether a simulated mutation had dynamics that were consistent with specific genetic lesions measured with the scale-free reporter AEQ, the simulations were rescaled to overlap the measured data. More specifically, simulations (solid lines) were scaled to overlap the data points using linear regression, comparing simulated wild type (gray) and mutants (red). In abscissa, white bars indicate R+B light, red bars for red light and black bars for dark. (F) The shape, phase, and amplitude of the [Ca2+]cyt oscillation is regulated by the circadian oscillator (via CCA1; black arrows) through a pathway of duration τ h, and through the photoreceptors PHYA (red arrows) and ZTL (blue arrows). The hypothetical component X2 (gray symbols and arrows) might represent PHYA and ZTL in the temporal separation of red and blue light input to the oscillation. Arrow endings indicate known activation (→) or inactivation (|), or an unknown causal relationship (●).
Functionally distinct roles of the photoreceptors CRY1 and PHYA were identified by comparing the simulated effects of removing either CCA1 or the hidden variable X2 with experimental data from photoreceptor null mutations (Fig. 2 B and C and SI Appendix, Figs. S5 and S6). The [Ca2+]cyt oscillation in the cry1 mutant (26) had an early peak, similar to the simulated cca1 mutant (Fig. 2B and SI Appendix, Fig. S6), indicating that CRY1 might be responsible for light input to the circadian oscillator. Because X2 is a hidden variable, it was not possible to simulate its absence in a unique way. A family of simulations is parameterized by a variable t1 (see SI Appendix for further details), each of which was compared with measured data. In 16L/8D, simulated x2 mutations caused a lag of ≈4 h between dawn and the [Ca2+]cyt increase (t1 < −0.001; SI Appendix, Fig. S6C), and most simulations predicted a phase delay in 16L/8D (t1 > −1; Fig. 2C and SI Appendix, Fig. S1C). Only the oscillation in phyA-201 (27) reproduced these behaviors, having a significant delay to the increase in [Ca2+]cyt after dawn, and peaking 2 h later than wild type (Fig. 2C and SI Appendix, Figs. S5A and 6C). Therefore, PHYA can be considered a candidate for part of X2.
X2 also describes blue light (BL) input at the end of the photoperiod. In red-light/dark cycles, [Ca2+]cyt progressively increased during each circadian period, similar to a simulated x2 mutant in 16L/8D cycles (t1 = −0.001; Fig. 2D and SI Appendix, Fig. S6C). This indicates BL is required for the decrease in [Ca2+]cyt during the afternoon, and dark-mediated repression of [Ca2+]cyt requires prior BL signaling. Simulated x2 mutants failed to decrease [Ca2+]cyt in anticipation of dusk in 16L/8D (t1 ≥ −0.01; SI Appendix, Fig. S6C), a pattern also observed in the clock-associated BL receptor mutant ztl-1 (Fig. 2E and SI Appendix, Fig. S7D). X2 therefore describes the temporal separation of wavelength-dependent light signaling (Fig. 2F).
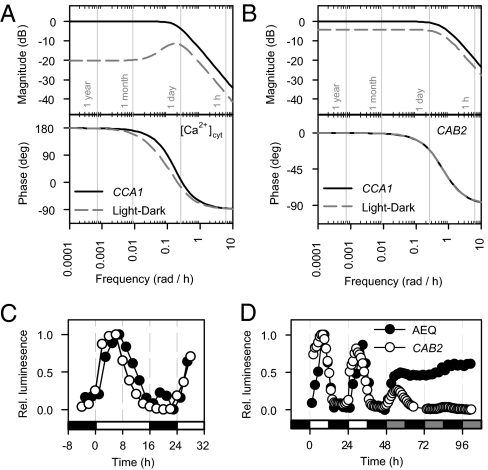
The model demonstrated that oscillations of [Ca2+]cyt are dual controlled by rapid light signals and circadian inputs, and consequently that the network connecting light to [Ca2+]cyt is an incoherent feed-forward loop (Fig. 2F) (7, 28). To determine the relative importance and temporal characteristics of each input pathway, we used Bode analysis to investigate the frequency response of [Ca2+]cyt oscillations (Fig. 3A; see SI Appendix for details) (20, 21). Bode analysis describes the response of a LTI system to purely sinusoidal inputs over a range of frequencies, in terms of the relative amplitude (magnitude) and phase. The degree of amplification of the input signal is measured by its effect on the amplitude of the output in decibel (dB) units. Biologically, this can be considered as estimating how much effect a periodic signal has on a biological system, and allows multiple inputs to be considered. Using sinusoids to analyze rhythmic processes is common in engineering applications, and also forms the basis of algorithms such as BRASS (29), which is used to estimate circadian periods. The CCA1 (circadian) input pathway dominated the magnitude response at all frequencies, and was ≈7 dB greater than that from the light input pathway at the frequency corresponding to a 24-h period. However, at lower frequencies (long period oscillations, e.g., annual cycles), the light input pathway contributed much less than the CCA1 pathway (20 dB difference; Fig. 3A), indicating that light input only modulates [Ca2+]cyt over faster variations in light availability.
Fig. 3.
The frequency responses of [Ca2+]cyt and CAB2 to the central oscillator and light-dark cycles are similar. (A and B) Bode analysis of (A) [Ca2+]cyt and (B) CAB2 LTI models. Bode magnitude and phase angle frequency responses plotted for a range of driving sinusoidal signals with various frequencies for the CCA1 input (solid lines) and the independent light-dark pathway (broken lines). A driving frequency of π/12 rad h−1 (0.261 rad h−1) corresponds to a 24-h period. Magnitude is obtained on a log scale in decibels (dB) and represents the input-output gain (i.e., amplitude ratio) at each frequency. Phase is obtained in degrees and represents the input-output phase difference at each frequency. 180 degrees corresponds to a 12-h phase change (positive = advance, negative = delay). (C and D) Comparison of [Ca2+]cyt (AEQ luminescence; ●) and CAB2 (CAB2::luc luminescence; ○) oscillations in (C) 16L/8D and (D) 12L/12D followed by DD. In abscissa, open bars indicate light, closed bars dark, and shaded bars subjective light.
We wished to assess whether our finding that regulation of [Ca2+]cyt requires both a rhythm-generating oscillator and a rapid feed-forward light signaling pathway is broadly applicable to other circadian outputs. We therefore constructed a similar model and Bode analysis of CAB2 promoter activity (Fig. 3B and SI Appendix, Fig. S9) (30) because CAB2 is also regulated by light and the circadian oscillator (SI Appendix, Table S1) (10), and has similar dynamics to the [Ca2+]cyt oscillation in 12L/12D (Fig. 3 C and D). The input delay of the circadian clock-dependent pathway regulating the CAB2 promoter was shorter than in the model for [Ca2+]cyt ([Ca2+]cyt: 5.2 h, CAB2: 0.6 h), consistent with the direct regulation of the CAB2 promoter by clock components (31). Similar to the [Ca2+]cyt oscillation, a CCA1-independent light input pathway was required to describe the regulation of the CAB2 promoter, operating through a rapid pathway (no input delay) at 4 dB less than the regulation by the circadian clock (Fig. 3B).
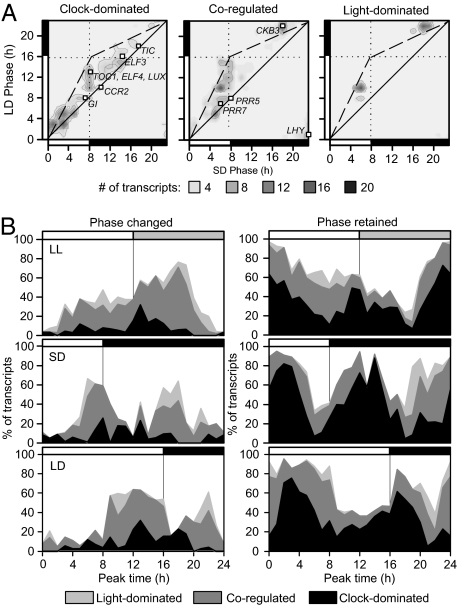
We hypothesized that the incorporation of rapid light input pathways is associated with correct circadian timing, because removal of ZTL or PHYA function altered circadian phase of [Ca2+]cyt, similar to simulated removal of light input (Fig. 2). We also hypothesized that regulation by rapid light signaling is not associated with specific biological function because both [Ca2+]cyt and CAB2 are dual controlled but involved in different biological processes. To test our hypotheses, we constructed similar models and Bode analyses for 3,503 rhythmic transcripts (32) from published microarray data (5, 33) using either CCA1 or TOC1 transcript abundance data as a proxy for the circadian oscillator. The light input was assumed to be rapid (no input delay) in this analysis to specifically investigate whether a general role exists for rapid light-induced events at the circadian subgenome level. A minimal performance criterion was used to determine suitability of the derived models (rw > 0.75; see equation 7 in SI Appendix), which favored CCA1-driven models (CCA1: 1,083, TOC1 – 460 of 3,503 models; SI Appendix, Fig. S10A). Of the 460 transcripts with high-performing TOC1-driven models, the majority (379 transcripts; 82.3%) also had high-performing CCA1-driven models. This indicated that performance was not a consequence of our choice of proxy for the circadian oscillator, but instead a property of the dynamics we intended to describe. Higher suitability of CCA1-driven models might be ascribable to CCA1 having a larger output connectivity than TOC1 within the central oscillator network (6) and/or direct binding of CCA1 to a large number of circadian-regulated gene promoters (3, 5). CCA1-driven models were then analyzed for the relative magnitudes of each input pathway (SI Appendix, Fig. S10). Transcripts were considered to be coregulated if the difference in the Bode magnitude of the two input pathways was less than 7 dB, and clock or light-dominated otherwise (Dataset S1). The 7-dB threshold was chosen because this matched the magnitude difference in the [Ca2+]cyt model, which was found to be coregulated experimentally (Fig. 2). The regulation of TOC1, GI, LUX ARRHYTHMO (LUX), TIME FOR COFFEE (TIC), and EARLY FLOWERING 3 and 4 (ELF3/4) were predicted to be clock dominated, and LHY, CASEIN KINASE II BETA CHAIN 3 (CKB3), and PSEUDO RESPONSE REGULATORs 5 and 7 (PRR5/7) were coregulated (Fig. 4A). This largely agrees with published experimental evidence, which suggests LHY, PRR5/7, and GI are transcriptionally regulated by light, whereas the other genes are thought not to be light regulated at the transcript level (4).
Fig. 4.
Transcripts undergoing photoperiod-dependent phase changes are associated with light regulation. Experimentally determined oscillation phase analyzed for 1,083 rhythmic transcripts with high-performing models (rw > 0.75), when categorized by dominant input pathway(s). (A) Heat map of the relationship between phases in 8L/16D and 16L/8D, indicating phase invariance (along solid line) or photoperiod-dependent phase changes (along broken line). Specific circadian-associated genes indicated by open squares. (B) Histographs represent the model classifications of rhythmically expressed transcripts, which are dominated by light-input pathways (light gray regions), by clock-associated pathways (black regions) or coregulated (dark gray regions), binned according to their measured time of maximal expression in LL (Top), 8L/16D (Middle), or 16L/8D (Bottom). The reported value is defined as 100 × (number of category-specific transcripts which peak at the specified time)/(total number of transcripts which peak at the specified time). Transcripts are classified by whether they change phase by at least 4 h (Left) or retain phase between 8L/16D and 16L/8D (Right).
Coregulated, light-dominated, and clock-dominated model classes were not associated with particular biological functions (SI Appendix, Fig. S11). In contrast, there was a strong association between model class and the phase of peak accumulation of transcript reported in the DIURNAL database (http://diurnal.cgrb.oregonstate.edu/) (34). Clock-dominated transcripts generally peaked in the early morning in both 8L/16D and 16L/8D, whereas coregulated or light-dominated transcripts peaked in the second half of the photoperiod or in the middle of the night (Fig. 4A). There was a significant relationship between the dominant regulatory pathway and rephasing of a transcript between 8L/16D and 16L/8D (>4 h); only 17.6% of clock-dominated transcripts changed phase between 8L/16D and 16L/8D, compared with 47.9% of coregulated and 54.6% of light-dominated transcripts (χ2 statistic: 6.2 × 10−6). Clock-dominated transcripts were therefore associated with morning expression independent of the external photoperiod, whereas coregulated and light-dominated transcripts peaked later in the day/night and had altered peak times between 8L/16D and 16L/8D (Fig. 4B).
Discussion
We used reverse engineering with simple linear time-invariant models to form a global view of the responses of rhythmic processes to changing photoperiod. Our approach was based on dynamical analyses of behavior rather than detailed biochemical investigation. It is computationally light and therefore suited to the reconstruction of networks in systems where information is limited and to genome-scale analyses. Our approach differs from the formulation of the majority of mathematical models, which describe circadian oscillators using ordinary differential equations based on mass-action kinetics with multiple nonlinear terms and kinetic rate parameters (6, 35, 36). Nonlinear models have proven powerful in the modeling of the Arabidopsis oscillator, providing descriptions of processes such as transcription, translation, nuclear-cytoplasmic transport, and degradation, and helped in identifying new oscillator components (6, 36). However, obtaining estimates for the kinetic parameter rates in nonlinear systems is not trivial, and prior knowledge of the underlying components is often required to describe the processes mathematically. The use of linear models to approximate nonlinear and stochastic dynamics provided us with many advantages, including automatic parameter estimation, automatic provision of latent/hidden variables, and Bode analysis. Most strikingly, we were able to derive LTI models for the circadian regulation of 3,503 genes, a process which would currently be impractical using nonlinear approaches. Systems identification techniques provided 1,083 high-performing linear models on which we based our analysis of photoperiodic phase adjustment. It is likely that the remaining transcripts require either higher complexity linear or nonlinear systems to accurately capture their dynamical behavior.
We have identified that correct daily regulation of multiple biological rhythms, independent of function, requires both a rhythm-generating oscillator and rapid light-signaling pathways. Our analyses show that rapid, light-activated feed-forward loops modulate both oscillator components and circadian outputs. The identified incoherent feed-forward loop structure is often associated with noisy step-like information processing (28), such as occurs in light signaling. The rapid timescale over which light-signaling feed-forward loops operate precludes a cascade of transcriptional activation/inactivation, such as found within the transcriptional feedback loops of the circadian oscillator. Rapid light signaling might include changes in second messengers or posttranslational modifications, which could target components that are or are not part of the oscillator. We found that there is, at least for the regulation of [Ca2+]cyt, modulation of the rapid light-signaling pathways by the circadian oscillator, with red light increasing [Ca2+]cyt in the morning, through PHYA, and blue light, acting through ZTL being required for a decrease in [Ca2+]cyt later in the cycle.
Photoperiodic responses such as floral initiation and hypocotyl elongation are produced through external coincidence, with the combination of a specifically phased rhythm and light availability determining whether a downstream component is activated (37, 38). Our data suggest an additional example of external coincidence, with rapid light-signaling pathways and the rhythm-generating oscillator acting over different timescales to confer photoperiod-dependent rephasing of circadian outputs. Rhythmic outputs are the targets of multiple light-signaling pathways, allowing photoperiod adjustment at the level of the individual output rather than solely through the circadian oscillator. Our data suggest that the correct phasing of rhythmic behavior requires rapid light-signaling pathways that act in concert with the circadian oscillator, and that this is a widely used mechanism of phase control. Therefore, previously identified internal coincidence mechanisms confer only one part of the response of the Arabidopsis circadian system to changing photoperiod (8); additional rapid light-signaling pathways acting through external coincidence mechanisms are also required for accurate timing of circadian outputs.
Materials and Methods
Luminescence Imaging.
For luminescence imaging, plants were grown on 1/2 MS nutrient media (without sucrose) under 100 μmol m−2·s−1 mixed light in the appropriate photoperiodic regime for ≈10 days before imaging (18). Seedlings were then imaged using a Photek ICCD 225 photon-counting camera (18) for imaging under R+B, or similarly imaged using a Photek high-resolution photon-counting system for analysis under single wavelengths of light (red, 660 nm, 70–90 μmol m−2·s−1; blue, 470 nm, 70–90 μmol m−2·s−1).The cca1-11 mutant and the specific Ws background accession for this mutant were a gift from Seth Davis (Max-Planck Institute for Plant breeding Research, Cologne, Germany) (39), a Col-0 accession was a gift from Takeshi Mizuno, Nagoya, Japan (40), and a C24 accession was a gift from Laszlo Kozma-Bognar, Szeged, Hungary (41). These lines were transformed with pGIFAEQ (42). Other lines and transformation protocols used were described previously (17).
Bioinformatic Analysis.
Data describing the rhythmic regulation of abundance in 8L/16D and 16L/8D for the 3,503 transcripts used for the generation of models was obtained from the DIURNAL database (http://diurnal.cgrb.oregonstate.edu/) (34). Of the total 3,503 transcripts, an estimate of circadian phase in both photoperiods was available for 1,845 transcripts (Dataset S1). Of the 1,083 transcripts passing a modeling performance threshold (SI Appendix), an estimate of circadian phase in both photoperiods was available for 925. Transcripts were defined as changing phase between 8L/16D and 16L/8D if a phase difference of 4 h or more existed between the two day lengths. For the generation of the phase profiles represented in Fig. 4, the 925 transcripts exceeding the modeling performance threshold with 8L/16D and 16L/8D data available were binned according to their times of maximal abundance in 8L/16D, 16L/8D, and LL (using published phase data in ref. 32), respectively, and then further categorized by the input regulatory structure identified in the modeling and also according to whether the transcript changed phase between 8L/16D and 16L/8D.
Supplementary Material
Acknowledgments
Funding was provided by the Biotechnology and Biological Sciences Research Council (BBSRC) UK (A.A.R.W., J.M.G., N.D., F.C.R., and H.M.B.), Engineering and Physical Sciences Research Council UK (A.A.R.W., J.M.G., and G.-B.S.), and the Isaac Newton Trust (A.A.R.W. and J.M.G.). K.E.H. was funded by a BBSRC Doctoral Training Grant award, and C.T.H. was a CAPES Brazil Scholar.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001429107/-/DCSupplemental.
References
- 1.Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 2.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alabadí D, et al. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 4.Gardner MJ, Hubbard KE, Hotta CT, Dodd AN, Webb AAR. How plants tell the time. Biochem J. 2006;397:15–24. doi: 10.1042/BJ20060484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harmer SL, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 6.Locke JCW, et al. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol. 2006;2:59. doi: 10.1038/msb4100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Troein C, Locke JCW, Turner MS, Millar AJ. Weather and seasons together demand complex biological clocks. Curr Biol. 2009;19:1961–1964. doi: 10.1016/j.cub.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 8.Salazar JD, et al. Prediction of photoperiodic regulators from quantitative gene circuit models. Cell. 2009;139:1170–1179. doi: 10.1016/j.cell.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 9.Anderson SL, Kay SA. Functional dissection of circadian clock-regulated and phytochrome-regulated transcription of the Arabidopsis CAB2 gene. Proc Natl Acad Sci USA. 1995;92:1500–1504. doi: 10.1073/pnas.92.5.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Millar AJ, Kay SA. Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:15491–15496. doi: 10.1073/pnas.93.26.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McWatters HG, Bastow RM, Hall A, Millar AJ. The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature. 2000;408:716–720. doi: 10.1038/35047079. [DOI] [PubMed] [Google Scholar]
- 12.Michael TP, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008;4:e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bünning E. Die endogene tagesrhythmik als grundlage der photoperiodischen reaktion. Ber Deut Bot Ges. 1936;54:590–607. [Google Scholar]
- 14.Pittendrigh CS, Minis DH. The entrainment of circadian oscillations by light and their role as photoperiodic clocks. Am Nat. 1964;98:261–294. [Google Scholar]
- 15.Sawa M, Kay SA, Imaizumi T. Photoperiodic flowering occurs under internal and external coincidence. Plant Signal Behav. 2008;3:269–271. doi: 10.4161/psb.3.4.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanovsky MJ, Kay SA. Living by the calendar: How plants know when to flower. Nat Rev Mol Cell Biol. 2003;4:265–275. doi: 10.1038/nrm1077. [DOI] [PubMed] [Google Scholar]
- 17.Xu X, et al. Distinct light and clock modulation of cytosolic free Ca2+ oscillations and rhythmic CHLOROPHYLL A/B BINDING PROTEIN2 promoter activity in Arabidopsis. Plant Cell. 2007;19:3474–3490. doi: 10.1105/tpc.106.046011. Available at www.plantcell.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love J, Dodd AN, Webb AAR. Circadian and diurnal calcium oscillations encode photoperiodic information in Arabidopsis. Plant Cell. 2004;16:956–966. doi: 10.1105/tpc.020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ljung L. Systems Identification: Theory for the User. Englewood Cliffs, NJ: Prentice Hall; 1999. [Google Scholar]
- 20.Dorf RC, Bishop RH. Modern Control Systems. Englewood Cliffs, NJ: Prentice Hall; 2007. [Google Scholar]
- 21.Mettetal JT, Muzzey D, Gómez-Uribe C, van Oudenaarden A. The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science. 2008;319:482–484. doi: 10.1126/science.1151582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett MR, et al. Metabolic gene regulation in a dynamically changing environment. Nature. 2008;454:1119–1122. doi: 10.1038/nature07211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salomé PA, McClung CR. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell. 2005;17:791–803. doi: 10.1105/tpc.104.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devlin PF, Kay SA. Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell. 2000;12:2499–2510. doi: 10.1105/tpc.12.12.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 26.Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 27.Nagatani A, Reed JW, Chory J. Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 1993;102:269–277. doi: 10.1104/pp.102.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milo R, et al. Network motifs: Simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 29.Plautz JD, et al. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- 30.Millar AJ, Carré IA, Strayer CA, Chua NH, Kay SA. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science. 1995;267:1161–1163. doi: 10.1126/science.7855595. [DOI] [PubMed] [Google Scholar]
- 31.Carré IA, Kay SA. Multiple DNA-protein complexes at a circadian-regulated promoter element. Plant Cell. 1995;7:2039–2051. doi: 10.1105/tpc.7.12.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwards KD, et al. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell. 2006;18:639–650. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanovsky MJ, Kay SA. Molecular basis of seasonal time measurement in Arabidopsis. Nature. 2002;419:308–312. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]
- 34.Mockler TC, et al. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 35.Leloup J-C, Goldbeter A. Toward a detailed computational model for the mammalian circadian clock. Proc Natl Acad Sci USA. 2003;100:7051–7056. doi: 10.1073/pnas.1132112100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Locke JCW, et al. Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol. 2005;1:2005.0013. doi: 10.1038/msb4100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imaizumi T, Kay SA. Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci. 2006;11:550–558. doi: 10.1016/j.tplants.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 39.Ding Z, Millar AJ, Davis AM, Davis SJ. TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell. 2007;19:1522–1536. doi: 10.1105/tpc.106.047241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamichi N, et al. The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol. 2005;46:609–619. doi: 10.1093/pcp/pci061. [DOI] [PubMed] [Google Scholar]
- 41.Kevei E, et al. Arabidopsis thaliana circadian clock is regulated by the small GTPase LIP1. Curr Biol. 2007;17:1456–1464. doi: 10.1016/j.cub.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 42.Kaplan B, et al. Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell. 2006;18:2733–2748. doi: 10.1105/tpc.106.042713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.