Abstract
The variation of expression pattern exhibited by a transgene as a result of random integration, known as position effect, is, among other mechanisms, a particular challenge to reverse genetics. We present a strategy to counteract position effect in Arabidopsis thaliana by flanking the transgenes with the gypsy insulator from Drosophila melanogaster. In addition, Suppressor of Hairy-wing [Su(Hw)], the binding protein of the gypsy insulator, was coexpressed. Results indicated that the gypsy insulators could efficiently improve the expression levels of reporter genes driven by various kinds of promoters by 8- to 13-fold. Coexpression of the Su(Hw) protein led to a more uniform expression level of transgenes, as the coefficient of variation of expression levels was reduced further. The gypsy-Su(Hw) system enhanced expression levels, but did not alter the specificity of promoter activities, as experimentally evidenced by the promoters of the PIN and the AFB gene families. Interestingly, the gypsy insulator was also able to improve the expression of a selectable marker gene outside the insulated region, which facilitated the screen of transformants. Our system will likely decrease the number of lines that experimenters need to create and examine for a given transgene by contributing to relatively high and precise expression of transgenes in plants. Certain features of the gypsy insulator in Arabidopsis also provide new perspectives on the insulator field.
GENETIC engineering has become a routine technique for studying gene function and regulation of complex physiological networks. In plant transgenesis, the transgene is usually integrated randomly into the host genome. The expression level of the transgene may vary among independent lines due to a number of factors including the transgene copy number, RNA silencing, and transgene insertion site (Butaye et al. 2004; De Bolle et al. 2007; De Paepe et al. 2009). The variation of transgene expression has complicated phenotype characterization and necessitates the analysis of multiple transgenic lines.
Recent studies have identified sequence-specific mRNA decay by post-transcriptional gene silencing (PTGS) as the major cause in the variation of the expression level of transgenes driven by strong promoters in plants. RNA silencing was triggered if the transcript level of a transgene surpassed a gene-specific threshold (Schubert et al. 2004). Improperly terminated, unpolyadenylated mRNA from transgene transcriptions generated by 3′ readthrough was subjected to RNA-DEPENDENT RNA POLYMERASE6 (RDR6)-mediated RNA silencing (Luo and Chen 2007).
Variation of transgene expression induced by the copy number and the homology-based PTGS could be minimized by several approaches (Butaye et al. 2005). Screening for single-copy T-DNA transformants greatly enriches for stable and high transgene expression because PTGS is thought to be particularly triggered by multiple, complexly arranged copies of transgenes (De Buck et al. 2004; Nagaya et al. 2005). In addition, the use of PTGS mutant backgrounds as the target of transformation has been proven to be of high value for a stable and high expression level of transgenes (Butaye et al. 2004; De Bolle et al. 2007). Recently, it has been shown that the CRE/loxP recombination system is capable of resolving complex T-DNA loci into single T-DNA inserts and producing a high frequency of single-copy T-DNA transformants (De Buck et al. 2007; De Paepe et al. 2009).
The variation of transgene expression induced by the insertion site itself is known as position effect. The surrounding host chromatin structure as well as the negative and positive regulatory elements in adjacent loci are implicated in mediating position effects (Kellum and Schedl 1991; Peach and Velten 1991; Gelvin and Kim 2007). Although a great deal is known about position effects in Drosophila, there is conflicting evidence as to whether or not they exist in plants (Matzke and Matzke 1998; De Buck et al. 2004; Nagaya et al. 2005). It has been suggested that pronounced position effects should never be detected in stable transformants that have undergone antibiotic selection, as negative position effects should also suppress the expression of the resistance marker (Kim et al. 2007). However, several lines of evidence have suggested that position effects should exist in plants. As in Drosophila, large-scale enhancer trapping using transgenes equipped with minimal promoters has provided a powerful approach for identifying tissue- and stage-specific gene expression in Arabidopsis (Campisi et al. 1999; Laplaze et al. 2005; Liu et al. 2005). Many of the proteins required for the machinery of the position effects are conserved from Drosophila to Arabidopsis (Henderson and Jacobsen 2007). Genome-wide transposon tagging revealed location-dependent effects on transcription and chromatin organization in Arabidopsis (Rosin et al. 2008). An example of a position effect that is associated with the reversal of epigenetic silencing has recently been documented in maize (Singh et al. 2008).
Genes subjected to reverse genetic studies may have very subtle phenotypes that are masked by “noise” from the position effect and fall beneath the threshold of detection. In promoter analysis, position effects can be substantially deleterious if the expression of the reporter gene is influenced by particular spatial and temporal factors.
Various techniques have been developed to overcome the confounding potential of position effects. The ability of matrix attachment regions (MARs) to anchor DNA to the nuclear matrix has made MARs interesting candidates as transgene boundary elements in animals (Stief et al. 1989; Namciu et al. 1998; Sgourou et al. 2009) and plants (Spiker and Thompson 1996; Holmes-Davis and Comai 1998; Halweg et al. 2005). Several models have been proposed to explain the function of MARs. The loop-domain model suggested that MARs could induce transgenes to form independent loops that create protective environments for them (Spiker and Thompson 1996). Another model suggested that MARs could inhibit transcriptional read-out and thus prevent RNA silencing of transgenes (Mlynarova et al. 2003).
MARs have not been widely applied in animal and plant transgenesis. They often consist of long DNA fragments that are not amenable to processing via the traditional recombinant DNA systems. The limited knowledge about protein factor(s) that interact with MARs precludes their wide application in transgenesis (De Bolle et al. 2007). Furthermore, while MARs have been used in conjunction with the 35S CaMV promoter in plants to achieve high-level expression of transgenes, it is unclear whether MARs could guarantee the precise expression of transgenes driven by other promoters. Two recent reports revealed negative results of MARs in plant transgenesis (De Bolle et al. 2007; Li et al. 2008).
Insulators are another type of DNA boundary elements that have been attractive candidates for protection against positional effects (Bell et al. 2001; Gerasimova and Corces 2001). One type of insulator element is thought to establish independent domains that block enhancer–promoter communication whereas a second type is postulated to create a barrier against the spread of heterochromatin. Some insulators are composite elements with separable activities, while others employ a single mechanism to confer both properties (Kuhn and Geyer 2003; Gaszner and Felsenfeld 2006). Insulators that have been well studied in transgenic animals include the gypsy and scs/scs′ insulators from Drosophila (Barolo et al. 2000; Sarkar et al. 2006; Markstein et al. 2008) and the β-globin HS4 insulator from chicken (Wang et al. 2009).
The gypsy insulator, originally identified from the gypsy retrotransposon, is one of the best-characterized insulators (Gdula et al. 1996; Bell et al. 2001; Kuhn and Geyer 2003). Studies have revealed that it contains a cluster of binding sites for the Suppressor of Hairy-wing [Su(Hw)] protein, a zinc-finger DNA-binding protein (Harrison et al. 1993; Gaszner and Felsenfeld 2006). The gypsy insulator is believed to regulate gene expression by establishing higher-order domains of chromatin structure and blocking the interference of nearby enhancers or repressors (Gaszner and Felsenfeld 2006; Bushey et al. 2008).
In a pioneering study, the insulator element ArsI (arylsulfatase) from sea urchin (Hemicentrotus pulcherrimus) was shown to suppress variations of transgene expression in cultured tobacco cells (Nagaya et al. 2001). However, reports concerning the exploitation of insulators in plant transgenesis are limited. Here, we have introduced the gypsy insulator and its binding protein Su(Hw) into Arabidopsis. The transgenic vectors were constructed using GATEWAY technology based on a site-specific recombination cloning system. We found that the gypsy insulators improved the expression level of transgenes driven by various promoters. The gypsy-Su(Hw) system reduced position effects and mediated the specificity of promoter activities. Our strategy can be used to guarantee high and precise expression of transgenes in plants. Certain features of the gypsy insulator in Arabidopsis also provide new perspectives on the insulator field.
MATERIALS AND METHODS
Materials:
All transgenic experiments used accession Columbia-0 of Arabidopsis thaliana provided by the Arabidopsis Biological Resource Center (ABRC).
Chemicals and original vectors:
Clones that contained sequences of Su(Hw) and the gypsy insulator of Drosophila were purchased from the Drosophila Genomics Resource Center. GATEWAY-compatible destination vectors (Karimi et al. 2002) for promoter analysis (pHGWFS7) and overexpression (pH7FWG2.0, pK7WG2D.1) were ordered from the Department of Plant Systems Biology, VIB-Ghent University (Ghent, Belgium). The vector pAVA319 contained a translation leader (TL) and was obtained from ABRC. The TOPO cloning and LR reaction kits were purchased from Invitrogen (http://www.invitrogen.com).
Construction of entry vectors:
Entry vectors for promoter analysis of Silent Information Regulator 2 (SIRT2), the Pinform (PIN) gene family, and the Auxin Receptor F-box (AFB) gene family and for overexpression of Su(Hw) were constructed using the pENTR/D-TOPO kits. The PCR primers for construction of entry vectors are listed in the supporting information, Table S1. Each entry clone was confirmed by DNA sequencing.
Construction of destination vectors containing insulators:
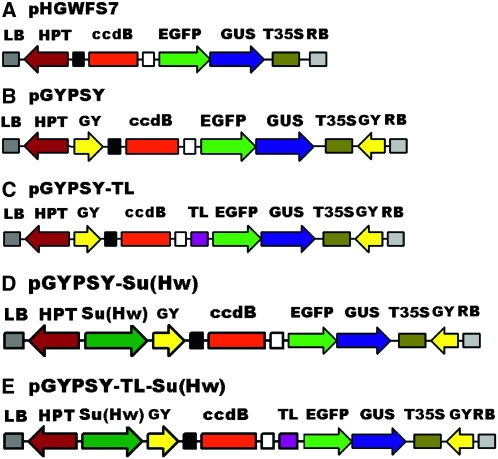
The gypsy insulator (395 bp) was amplified from the pH-Stinger vector (Barolo et al. 2000), with primers indicated in Table S1. First, one copy of the gypsy insulator, named I2, was inserted at the SacI/SpeI sites of pHGWFS7 to obtain pHGWFS7-I2. Then, pHGWFS7-I2 was divided into a long and a short fragment by AatII digestion. Another gypsy insulator with BstXI sites introduced at both ends was inserted in the AatII site of the long fragment. This transition vector was ligated to the short fragment of pHGWFS7-I2 to produce the pGYPSY vector with gypsy insulators flanking both sides of the EGFP-GUS reporter gene (Figure 1B). The TL from the vector pAVA319 was cloned into pGYPSY at the SacII site to produce the pGYPSY-TL vector (Figure 1C). Finally, we inserted the Su(Hw) overexpression cassette, driven by the 35S CaMV promoter, into the SacI/XhoI site of pGYPSY and pGYPSY-TL to obtain pGYPSY-Su(Hw) and pGYPSY-TL-Su(Hw) (Figure 1, D and E). The PCR primers used for the construction of destination vectors are listed in Table S2.
Figure 1.—
Schematic overview of the promoter analysis vectors used in this study. The positions and relative orientations of the selectable marker gene hygromycin phosphotransferase (HPT) and the reporter gene EGFP-GUS are shown with respect to the right border (RB) and the left border (LB) of the T-DNA regions. The ccdB region was expected to be replaced by a promoter sequence by a LR reaction of the GATEWAY system. (A) The pHGWFS7 vector (Karimi et al. 2002). (B) pGYPSY harbors gypsy insulators (GY) at both ends of the EGFP-GUS expression cassette based on the pHGWFS7. (C) pGYPSY-TL with a translation leader (TL) inserted into pGYPSY. (D) pGYPSY-Su(Hw) with an overexpression cassette of Su(Hw), the binding protein of the gypsy insulator, inserted into pGYPSY. (E) pGYPSY-TL-Su(Hw) with a Su(Hw) overexpression cassette inserted into pGYPSY-TL.
Construction of expression vectors:
The LR reaction was conducted to generate different expression vectors. The system contains entry vectors of interest, their corresponding destination vectors, and the LR recombinant enzyme.
Transformation and selection of transformed plants:
The selected expression vectors were transformed into Agrobacterium tumefaciens (GV3101) via electroporation. Arabidopsis plants were transformed using the vacuum infiltration method of Bechtold et al. (1993). Seeds collected from infiltrated plants were cultured on B5 medium containing hygromycin (12 mg/L) or kanamycin (30 mg/L). To estimate the number of transgene integration loci in the T1 generation, about 100–150 seeds were plated on a hygromycin medium to determine the segregation of the selectable marker gene HPT (hygromycin phosphotransferase). Transgenic lines whose progeny showed Mendelian characteristics (3:1 segregation, χ2 test) were termed “single-locus lines” (Forsbach et al. 2003). The single-locus lines were then subjected to Southern blot analysis to reveal the single-copy lines. Genomic DNA was digested by SacI and hybridized with the HPT probe. The DNA probe was labeled and detected using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics). During the development of the gypsy-Su(Hw) system in Arabidopsis, single-copy lines were used for the quantitative analysis of the promoter activities of SIRT2, 35S, PIN1, and PIN2. During the extension of the gypsy-Su(Hw) system in Arabidopsis, single-locus lines were used for the qualitative analysis.
Seed germination and plant growth measurement:
Arabidopsis seeds were surface-sterilized and cultured aseptically on 9-cm petri dishes containing Gamborg's B5 medium with 1% (w/v) sucrose and 1% (w/v) agar. The plates were maintained at 4° for 2 days and then transferred to a culture room (23°, 100 μm m−2·s−1 irradiance with a 16-hr photoperiod, 30–40% relative humidity). After 6 days, the plates were digitally photographed. Root and hypocotyl length was measured using magnified images. Statistical analyses of the data were performed using Microsoft Excel and Student's t-test.
Histochemical GUS staining and quantitative analysis:
Six-day-old seedlings containing the reporter gene GUS (β-glucuronidase) were collected and stained at specific times at 37° in X-glucuronide (5-bromo-4-chloro-3-indolyl-β-d-glucuronic) dissolved in 10 mm EDTA, 1 mm potassium hexacyanoferrate, 0.1% Triton X-100, and 100 mm phosphate buffer, pH 7.0. The GUS images were examined under a Nikon Eclipse 80i microscope with DXM1200 CCD camera and EclipseNet software (http://www.laboratory-imaging.com). Quantitative GUS activity was measured through the detection of the cleavage of 4-methylumbelliferyl-β-d-glucuronide (MUG) into 4-methylumbelliferon by fluorometric assay and calculated as nanomoles of 4-methylumbelliferon per minute and per milligram of total soluble proteins (Jefferson et al. 1987). The protein extraction solution contained 100 mm PBS, pH 7.0, 10 mm EDTA, 10 mm β-mercaptoethanol, 0.1% Triton X-100, and 140 μm PMSF. Protein concentrations were determined by methods described by Bradford (1976). The GUS reaction solution contained 1 mm MUG in the protein extraction solution. The reaction was carried out in 37° for 20 min. Fluorescence was measured using a spectrofluorophotometer (RF-5301PC, Shimadzu) at 460 nm with excitation at 355 nm.
Enhanced green fluorescent protein visualization:
Enhanced green fluorescent protein (EGFP) fluorescence and differential-interference-contrast images were visualized using a fluorescent microscope (Nikon Eclipse 80i). Band-pass filter sets for EGFP (excitation at 460–500 nm, emission at 510–550 nm) were used. Seedlings were mounted in water.
RESULTS
Construction of T-DNA vectors consisted of gypsy insulators:
To counteract position effects in the transgenesis of Arabidopsis, T-DNA destination vectors containing gypsy insulators of Drosophila were constructed. The procedure has been detailed in materials and methods, and the schematic overview of these vectors is illustrated in Figure 1. The original vector for promoter analysis, pHGWFS7, employed a fused EGFP and GUS as its reporter gene and the HPT driven by the nopaline synthase promoter (NOS) as its selectable marker (Figure 1A; Karimi et al. 2002). In the pGYPSY vector, the expression cassette of the GUS-EGFP gene was located between the two gypsy insulators, whereas the expression cassette of the hygromycin marker gene was outside the insulated region (Figure 1B), with the intention of reducing the effects of the enhancers of the selectable marker gene on the promoters under analysis. On the basis of the pGYPSY vector, the pGYPSY-TL vector was constructed to include a translation leader to the reporter gene in anticipation of the benefit to the expression of the fusion protein (Figure 1C). Su(Hw) is the binding protein of the gypsy insulator, and it is critical to efficient activity in Drosophila (Kuhn and Geyer 2003). To explore the function of Su(Hw) in Arabidopsis, a Su(Hw) overexpression cassette driven by the 35S CaMV promoter was introduced into pGYPSY and pGYPSY-TL vectors, resulting in two vectors pGYPSY-Su(Hw) and pGYPSY-TL-Su(Hw) (Figure 1, D and E).
The gypsy insulator boosted the expression of reporter genes with different promoters in Arabidopsis:
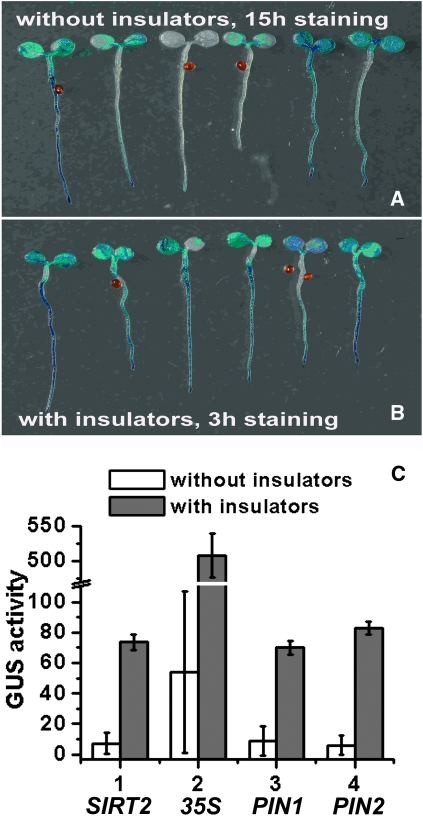
The SIRT gene family encodes nicotinamide adenine dinucleotide (NAD+)-dependent protein deacetylases evolutionarily conserved from Archaebacteria to humans (Hunt et al. 2004). Arabidopsis has two SIRT homologs: AtSIRT1 (AT5G55760) and AtSIRT2 (AT5G09230). We aimed to study the expression pattern of AtSIRT2 using promoter reporter vectors pHGWFS7-SIRT2 (the control) and pGYPSY-SIRT2 (including the gypsy insulators). Single-copy and homozygous transgenic lines were studied using histochemical staining and quantitative analysis. In the GUS staining assay, results demonstrated that the expression level of β-glucuronidase in transgenic lines without the gypsy insulators varied among the lines tested even after staining for 15 hr, which presented a substantial technical challenge to interpreting results and limited its efficacy as a reporter system (Figure 2A). In contrast, when the gypsy insulator was introduced, the staining pattern was more reproducible among the lines, and the threshold of detection was reached in only 3 hr (Figure 2B; Figure S1). A quantitative fluorometric assay revealed that GUS activities in transgenic lines with insulators were 10.20 ± 0.71-fold higher than in the uninsulated ones (Figure 2C, n = 15).
Figure 2.—
The gypsy insulator improved the expression of reporter genes with different promoters. (A) Histochemical GUS staining indicated the promoter activity of Silent Information Regulator 2 (AtSIRT2) in transgenic lines lacking the gypsy insulators (pHGWFS7-SIRT2). The expression level varied among the lines tested, even after staining for 15 hr. Six lines are shown. (B) Histochemical GUS staining indicated the promoter activity of AtSIRT2 in transgenic lines including the gypsy insulators (pGYPSY-SIRT2). The staining pattern was less variable (staining for 3 hr). Six lines are shown. (C) Quantitative GUS assay of promoter activities of AtSIRT2, 35S cauliflower mosaic virus (CaMV), Pinform1 (PIN1; auxin efflux carrier), and Pinform2 (PIN2) in transgenic lines with (using vector pGYPSY) or without (using vector pHGWFS7) the gypsy insulators. The GUS activity was calculated as nanomoles of 4-methylumbelliferone per minute per milligram of protein. Each value was the mean of 15 single-copy transgenic lines. Error bars represent standard deviation.
To examine whether the effectiveness of the gypsy insulator was dependent on the type of promoter, various well-characterized promoters were tested. The 35S CaMV promoter remains the most frequently used promoter in plant transformation. The PIN1 promoter drives an auxin efflux carrier gene important for proper embryogenesis, vascular bundle formation, and many other developmental processes, while the PIN2 promoter drives another auxin efflux carrier gene that affects gravitropism of Arabidopsis roots (Paponov et al. 2005). The 35S CaMV promoter is constitutive, the PIN1 promoter is tissue-specific, and the PIN2 promoter is tissue-specific and inducible. Quantitative analysis of the 35S, PIN1, and PIN2 promoter activities alone or in the presence of insulators revealed that the magnitude of improvement by the gypsy insulators was 9.41 ± 0.58, 8.13 ± 0.56, and 13.7 ± 0.69, respectively (Figure 2C). These data suggested that the gypsy insulator might improve the expression of reporter genes driven by different types of promoters.
The variability of transgene expression was further reduced by coexpression of Su(Hw):
The gypsy insulator contains 12 binding sites for Su(Hw), a protein factor proven to be pivotal for proper insulation function in Drosophila (Gdula et al. 1996; Bell et al. 2001; Kuhn and Geyer 2003). However, there is no homolog of Su(Hw) in the Arabidopsis genome. Hence, we investigated the effects of Su(Hw) coexpression upon the efficacy of the gypsy insulator in the transgenesis of Arabidopsis.
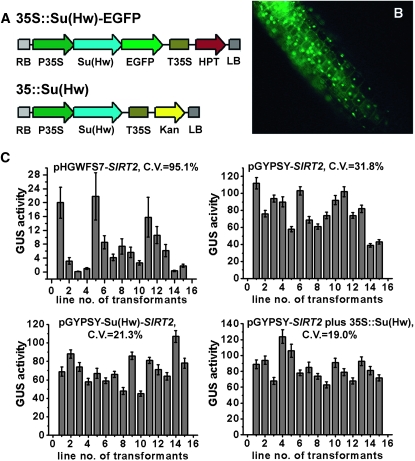
Initially, we overexpressed the Su(Hw)–EGFP fusion construct in conjunction with the 35S promoter (Figure 3A) to verify the subcellular localization of the Su(Hw) protein in Arabidopsis. As shown in Figure 3B, EGFP fluorescence was detected in the nucleus, indicating that the localization pattern of Su(Hw) protein in plant was identical to that observed in Drosophila (Harrison et al. 1993).
Figure 3.—
Variability of reporter gene expression driven by AtSIRT2 (Silent Information Regulator 2) promoter was reduced when Su(Hw) was expressed in a cis or a trans manner. (A) Schematic for the vectors that overexpress the Su(Hw)∷GFP fusion and the Su(Hw) proteins. (B) Nuclear localization of the Su(Hw)∷GFP fusion in the hypocotyls of 6-day-old transgenic seedlings. (C) Comparison of GUS activities in four classes of transgenic lines: pHGWFS7-SIRT2, the control; pGYPSY-SIRT2, with the gypsy insulators; pGYPSY-Su(Hw)-SIRT2, the Su(Hw) protein coexpressed in the cis mode; and pGYPSY-SIRT2 plus 35S∷Su(Hw), the Su(Hw) protein coexpressed in the trans mode. The coefficient of variation of each treatment is indicated. The GUS activity was calculated as nanomoles of 4-methylumbelliferone per minute per milligram of protein. Each value was the mean of 15 single-copy transgenic lines. Error bars represent standard deviation.
We then investigated the role played by Su(Hw) in two different modes of expression. In the cis expression mode, the T-DNA contained a promoter analysis region and a Su(Hw) overexpression cassette [pGYPSY-Su(Hw), Figure 1D]. In the trans expression mode, one T-DNA contained a promoter analysis region with hygromycin resistance as the selection marker (pGYPSY, Figure 1B), while the other T-DNA contained a Su(Hw) overexpression cassette with kanamycin resistance as the selection marker (Figure 3A). In the latter case, homozygous Arabidopsis plants overexpressing Su(Hw) were generated first and provided the transgenic backgrounds for the second transformation.
Four classes of transgenic lines (n = 15 each) were created for promoter analysis of AtSIRT2. As shown in Figure 3C, the GUS activities for uninsulated lines (pHGWFS7-SIRT2) were lower and more variable than that observed for the insulated lines (pGYPSY-SIRT2). The variation of GUS activities was reduced when the Su(Hw) protein was overexpressed in the cis [pGYPSY-Su(Hw)-SIRT2] or the trans modes [pGYPSY-SIRT2 plus 35S∷Su(Hw)] (Figure 3C). The coefficient of variation (standard deviation/mean × 100%) for GUS activities of the four classes of transgenic lines was 95.1%, 31.8%, 21.3%, and 19.0%, respectively, indicating that the gypsy insulator acting with the Su(Hw) was able to reduce the coefficient of variation by approximately fivefold.
The gypsy-Su(Hw) system does not alter the specificity of promoter activities of the PIN and AFB gene families:
The above results demonstrated that the gypsy insulator and its binding protein Su(Hw) were able to boost the expression of transgenes and reduce the variability of the expression of transgenes in Arabidopsis. We next sought to determine whether this system altered the specificity of plant promoter activities.
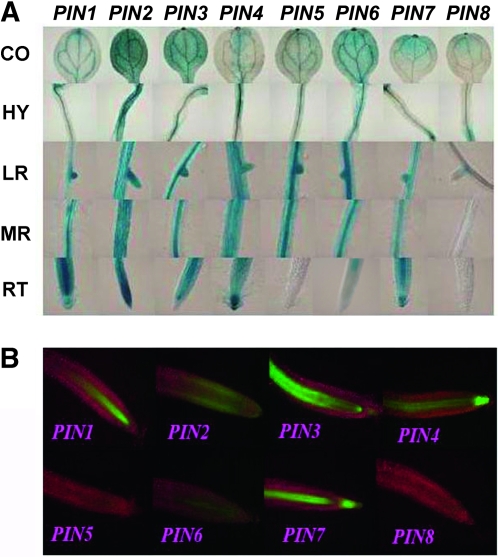
There are eight members of the PIN family encoding auxin efflux carriers that are crucial to polar auxin transport (Blilou et al. 2005). We constructed a set of promoter analysis vectors for this gene family, from PIN1 to PIN8, using the pGYPSY-TL-Su(Hw) construct, where the gypsy insulators flanked the transgenes and the Su(Hw) protein was expressed in the cis manner. In 6-day-old seedlings, GUS activity in cotyledon, hypocotyl, lateral root, and primary root and EGFP signal in root tips were examined (Figure 4A). As expected, transgene expression variability was minimal among different transgenic lines for all eight promoters, which facilitated the comparison of expression patterns of the different promoters. In root tips, PIN1 was strongly expressed in vascular tissue, whereas PIN2 was expressed in the outer cortical cells and epidermis cells. PIN3 was transcribed in the vascular cells and notably at the basal end of the provascular cells. PIN4 was restricted to the vascular cells and principally to the quiescent center and auxin peak region. PIN7 was present in the vascular cells and was strongly expressed in the columella cells. PIN6 was weakly expressed in provascular cells, whereas PIN5 and PIN8 were undetectable in root tips (Figure 4B).
Figure 4.—
The gypsy-Su(Hw) system did not alter the specificity of promoter activities of the Pinform (PIN; auxin efflux carrier) gene family. Promoter analysis lines for this gene family were generated using the pGYPSY-TL-Su(Hw) construct where the gypsy insulators flanked the transgenes and the Su(Hw) protein was expressed in a cis manner. (A) The GUS staining pattern in cotyledon (CO), hypocotyl (HY), lateral root (LR), mature root (MR), and root tip (RT) of 6-day-old transgenic seedlings. (B) The EGFP fluorescence images for the expression patterns of the PIN family promoters in root tips of 6-day-old Arabidopsis seedlings.
The TIR1/AFB gene family encodes proteins related to auxin responses (Dharmasiri et al. 2005a). As a subunit of the E3 ubiquitin ligase SCFTIR1, TIR1 functions as a receptor for plant hormone auxin. After binding of auxin to TIR1, ubiquitination and degradation of AUX/IAA proteins, a class of transcriptional repressors in auxin signaling, is triggered (Dharmasiri et al. 2005b; Kepinski and Leyser 2005). Using the gypsy-Su(Hw) system, we generated a set of promoter analysis lines for the TIR1/AFB gene family.
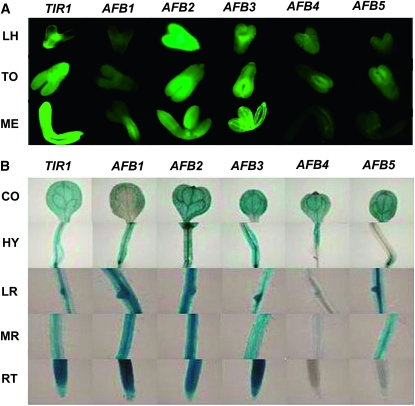
The expression patterns of the TIR1/AFB gene family members during certain stages of embryogenesis were examined. As shown in Figure 5A, at the late heart stage, the TIR1 promoter exhibited a stronger expression pattern in shoot primordium than in root primordium. In contrast, at the torpedo stage, it exhibited a stronger expression in root primordium than that in shoot primordium. At the mature embryo stage, TIR1 was strongly expressed throughout the whole embryo. AFB1 was expressed in the provascular bundle at the mature embryo stage, and its expression at the heart and torpedo stages was low. In contrast, AFB2 was highly expressed in precotyledons from the heart stage to the mature embryo stage. At the late heart and torpedo stages, AFB3 promoter produced striking EGFP signal in shoot primordium and provascular bundles, while at the mature embryo stage, its expression was more widespread, similar to that observed for the TIR1 promoter. For both AFB4 and AFB5 promoters, weak signals on the paraxial side of two precotyledons at the torpedo stage were detected (Figure 5A).
Figure 5.—
The gypsy-Su(Hw) system facilitated the examination of the promoter activities of the Transport Inhibitor Response 1/Auxin Receptor F box (TIR1/AFB) gene family. Promoter analysis lines for this gene family were generated using the pGYPSY-TL-Su(Hw) construct in which the gypsy insulators flanked the transgenes, and the Su(Hw) protein was expressed in a cis manner. (A) The EGFP images of the expression pattern of the TIR1/AFB gene family during embryogenesis at the late heart (LH), torpedo (TO), and mature embryo (ME) stages. (B) The GUS images of the expression pattern of the TIR1/AFB gene family in cotyledon (CO), hypocotyl (HY), lateral root (LR), mature root (MR), and root tip (RT) of 6-day-old seedlings.
We then examined the expression patterns of these TIR1/AFB genes in 6-day-old seedlings using GUS staining (Figure 5B). For all six promoters, variation of expression pattern among different transgenic lines was less detectable, which indicated that position effects were largely reduced. Taking advantage of higher intensity signals and lower variation, the more subtle differences in expression features of TIR1/AFB genes in the cotyledon, hypocotyl, lateral root, mature root region, and root tip were readily discernible (Figure 5B).
The gypsy insulator improved the expression of a selectable marker gene outside the insulated region:
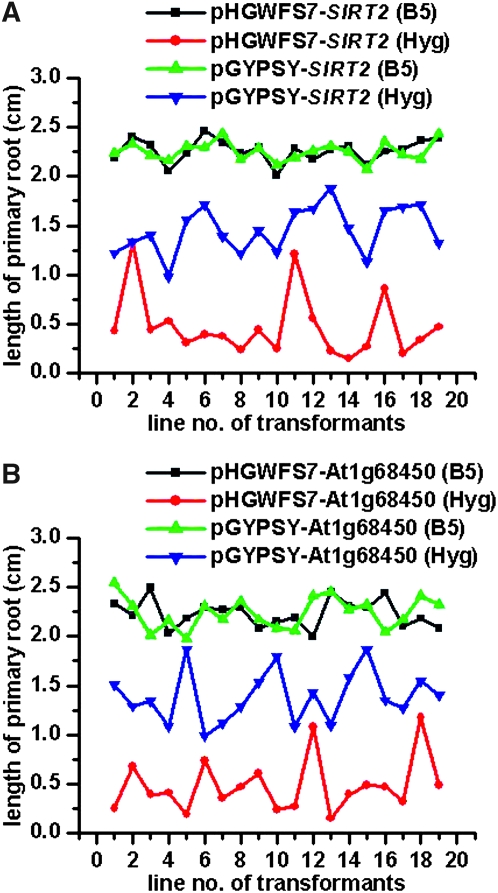
The expression cassette of the hygromycin marker was outside the insulated region, although its NOS promoter was adjacent to one of the insulators (Figure 1B). We observed an improved hygromycin resistance in transgenic plants with the gypsy insulators. To characterize the hygromycin resistance capacity, we measured the primary root length of 6-day-old seedlings. Data indicated that the mean values of root length for transgenic lines with or without insulators were similar when germinated on medium without antibiotics (with insulators: 2.25 ± 0.09 cm, n = 19; without insulators: 2.26 ± 0.11 cm, n = 19; Figure 6A). However, as seedlings grown in antibiotic-containing medium, the root length for the insulated lines (1.45 ± 0.23 cm, n = 19) was on average threefold greater than that measured for the uninsulated lines (0.47 ± 0.32 cm, n = 19, Figure 6A). Root length of transgenic lines without insulators fluctuated more greatly than their counterparts in response to antibiotic exposure: the coefficient of variation for uninsulated and insulated lines was 68.1% and 15.8%, respectively.
Figure 6.—
Transgenic plants with gypsy insulators exhibited improved hygromycin resistance. The promoter analysis vectors pHGWFS7 (control) and pGYPSY (with gypsy insulators) included a hygromycin resistance marker. Single-locus transgenic lines were cultured on hygromycin (Hyg) or control (B5) medium. (A) Comparison of pGYPSY-SIRT2 with pHGWFS7-SIRT2. Silent Information Regulator 2 (SIRT2) is a promoter sequence of AT5G09230. (B) Comparison of pGYPSY-At1g68450 with pHGWFS7-At1g68450. At1g68450 is a protein-coding sequence of the corresponding gene. The root lengths of 6-day-old seedlings were measured. Each treatment was carried out on 19 transgenic lines. Error bars represent standard deviation.
To rule out the possibility that increased hygromycin resistance in insulated transgenic lines was due to activities of the AtSIRT2 promoter, we introduced a protein coding sequence of At1g68450 (a VQ-domain-containing protein with unknown function). Results for pHGWFS7-At1g68450 and pGYPSY-At1g68450 constructs are shown in Figure 6B. Again, when seedlings were grown on antibiotic-free medium, the root length of transgenic lines with insulators was similar to that of uninsulated ones (with insulators: 2.24 ± 0.16 cm, n = 19; without insulators: 2.23 ± 0.14 cm, n = 19), but exhibited an approximately threefold difference on antibiotic-infused medium (with insulators: 1.39 ± 0.26 cm, n = 19; without insulators: 0.48 ± 0.28 cm, n = 19). All data considered, the gypsy insulator improved the expression of a selectable marker outside the insulated region.
DISCUSSION
To overcome position effects in the transgenesis of Arabidopsis, we adopted the gypsy insulator of Drosophila together with its binding protein Su(Hw) in Arabidopsis. The transgenic vector system described here has several features. First, previous approaches using animal MARs or insulators to create transgenic plants have focused on cis-acting elements only. Our transgenic vectors have been combined with relevant cis-acting elements with their cognate trans-acting factors. Second, previous studies dealt with strong promoters and were concerned with the expression level only. Our goal was to maximize the expression level and also the specificity of promoter activities. Finally, the GATEWAY high-throughput system was employed, allowing for the construction of T-DNA vectors with high speed and reliability.
We found that, in Arabidopsis, the gypsy insulators were able to boost the expression of reporter genes driven by various kinds of promoters, such as AtSIRT2, 35S, PIN1, and PIN2, by ∼8- to 13-fold (Figure 2). To ensure proper function of the gypsy insulator in Arabidopsis, the Su(Hw) protein was overexpressed in a cis or trans manner (Figure 3A). Interestingly, the Su(Hw) co-expression led to a more uniform expression level of a transgene, as the coefficient of variation of observed expression levels was reduced further (Figure 3C), but not to a general increase in the expression level.
We also found that the gypsy-Su(Hw) system reduced position effect but did not alter the specificity of promoter activities of the PIN and the TIR1/AFB gene family (Figures 4 and 5). Promoter reporter lines and immunocytochemical analysis data for PIN1, PIN2, PIN3, PIN4, and PIN7 have been previously documented (Blilou et al. 2005), as has TIR1, AFB1, AFB2, and AFB3 (Dharmasiri et al. 2005a). During the writing of this article, the expression pattern of PIN5 was reported (Mravec et al. 2009). Our results for the promoter activities of the PIN and the AFB gene families were in agreement with the published data. This suggested that the gypsy insulators did not act as tissue-specific or time-specific enhancers directly and that they were applicable to precise gene expression in Arabidopsis. The enhanced gene expression properties and the specificity of promoter activities make our transgenic approach a promising candidate technique for effective plant reverse genetics.
The mechanism mediating the function of the gypsy insulator has not yet been completely elucidated, but several models have been proposed in Drosophila. The transcriptional model and the structural model are two of the most discussed models (Kuhn and Geyer 2003; Gaszner and Felsenfeld 2006; Bushey et al. 2008). The former emphasized the competition between insulators and enhancers/repressors, and the latter stressed the formation of a chromatin loop with the help of insulators. In addition, the gypsy insulator is believed to facilitate the assembly of the transcriptional complex to improve the expression of transgenes in Drosophila (Golovnin et al. 2005). As insulators are known to block both the repressive and the activating influences of the surrounding environment, the pervasive boosting effects of gypsy insulators observed in Drosophila transgenesis suggest that, on balance, transgenes in Drosophila are under repressive influences (Markstein et al. 2008).
In Drosophila, the Su(Hw) protein is necessary for chromatin loop formation and enhancer/repressor blockage (Gerasimova and Corces 2001; Kuhn and Geyer 2003). In Arabidopsis, the gypsy insulators themselves can reduce transgene variations efficiently by minimizing those transgenic lines with low-level expression (Figure 2). These findings implied that an Arabidopsis protein was binding to the gypsy insulator and was providing insulator activity. However, expression of the Su(Hw) protein had some effects. The variability in transgene expression among individual lines was reduced further (Figure 3C). Perhaps the Su(Hw) protein of Drosophila acted as a better insulator protein than the Arabidopsis counterpart. Since there is no homolog of Su(Hw) in the Arabidopsis genome, further investigation using the yeast one-hybrid technique to identify the putative protein factor(s) recruited by the gypsy insulator promises to clarify the mechanism. A better understanding of these processes would provide a new perspective on the function of the gypsy insulator and enhance the insulator field in general.
The chromatin loop model cannot explain all of the findings from our studies. The insulator improved the expression of a selectable marker gene, although its expression cassette was not located between the two insulators (Figure 6). It would be helpful to know if protection by the insulators represents a block of a spread of silencing or some other effect of the gypsy insulators as discussed below.
Position-effect variegation is a well-documented concept in Drosophila, but very little is known about it in Arabidopsis, where the 35S promoter of virus origin has been extensively employed in transgenesis. The variation of the expression level of a transgene driven by this strong and constitutive promoter has been attributed largely to a gene-specific RNA-silencing mechanism (Schubert et al. 2004; Luo and Chen 2007). The impact of position effects on the variation of the expression pattern of a transgene driven by a tissue- or stage-specific plant promoter was less documented. Here, we show that, similar to that in Drosophila, the gypsy insulator, together with its binding protein Su(Hw), facilitate high and precise expression of transgenes in Arabidopsis. Since we had not directly detected the epigenetic status of the transgenes with and without the gypsy insulators, interpretations other than the shielding of position effects could not be excluded. The gypsy insulator upstream from the promoter region of transgenes might facilitate the assembly of the transcriptional complex, as suggested in Drosophila by Golovnin et al. (2005). Increasing evidence suggests that insulators have evolved from promoters (Raab and Kamakaka 2010). The gypsy insulator downstream from the terminator region of transgenes might ensure correct termination of reporter transcripts, because it was documented that RNA silencing triggered by mRNA 3′ readthrough was a major cause of reduced expression of transgenes in plants (Luo and Chen 2007). However, terminator activity for the gypsy insulator was recently tested in Drosophila, where it was found not to function (Silicheva et al. 2010). It is also possible that transgenes with gypsy insulators integrated in distinct genomic regions that might be more permissive to gene expression. Further work is required to address these questions.
Acknowledgments
We thank the Flanders Interuniversity Institute for Biotechnology-Ghent University for the original vectors. We are also grateful to the Arabidopsis Biological Resource Center (ABRC) and Drosophila Genomics Resource Center, respectively, for the distribution of Arabidopsis and Drosophila materials. This work was supported by funding from the National Natural Science Foundation of China (grant nos. 30470923, 30400036, and 60533050) and the Natural Science Foundation of Zhejiang Province (grant no. R304098). Transgenic lines generated in the present article have been donated to ABRC.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.117960/DC1.
References
- Barolo, S., L. A. Carver and J. W. Posakony, 2000. GFP and β-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. BioTechniques 29 726–732. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., J. Ellis and G. Pelletier, 1993. In planta Agrobacterium-mediated gene-transfer by infiltration of adult Arabidopsis thaliana plants. Compt Rend Acad Sci III 316 1194–1199. [Google Scholar]
- Bell, A. C., A. G. West and G. Felsenfeld, 2001. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science 291 447–450. [DOI] [PubMed] [Google Scholar]
- Blilou, I., J. Xu, M. Wildwater, V. Willemsen, I. Paponov et al., 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433 39–44. [DOI] [PubMed] [Google Scholar]
- Bradford, M. M., 1976. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Bushey, A. M., E. R. Dorman and V. G. Corces, 2008. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol. Cell 32 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butaye, K. M., I. J. Goderis, P. F. Wouters, J. M. Pues, S. L. Delauré et al., 2004. Stable high-level transgene expression in Arabidopsis thaliana using gene silencing mutants and matrix attachment regions. Plant J. 39 440–449. [DOI] [PubMed] [Google Scholar]
- Butaye, K. M. J., B. P. A. Cammue, S. L. Delauré and M. F. C. De Bolle, 2005. Approaches to minimize variation of transgene expression in plants. Mol. Breed. 16 79–91. [Google Scholar]
- Campisi, L., Y. Yang, Y. Yi, E. Heilig, B. Herman et al., 1999. Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 17 699–707. [DOI] [PubMed] [Google Scholar]
- De Bolle, M. F., K. M. Butaye, I. J. Goderis, P. F. Wouters, A. Jacobs et al., 2007. The influence of matrix attachment regions on transgene expression in Arabidopsis thaliana wild type and gene silencing mutants. Plant Mol. Biol. 63 533–543. [DOI] [PubMed] [Google Scholar]
- De Buck, S., P. Windels, M. D. Loose and A. Depicker, 2004. Single-copy T-DNAs integrated at different positions in the Arabidopsis genome display uniform and comparable beta-glucuronidase accumulation levels. Cell. Mol. Life Sci. 61 2632–2645. [DOI] [PubMed] [Google Scholar]
- De Buck, S., I. Peck, C. D. Wilde, G. Marjanac and J. Nolf, 2007. Generation of single-copy T-DNA transformants in Arabidopsis by the CRE/loxP recombination-mediated resolution system. Plant Physiol. 145 1171–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe, A., S. De Buck, K. Hoorelbeke, J. Nolf, I. Peck et al., 2009. High frequency of single-copy T-DNA transformants produced by floral dip in CRE-expressing Arabidopsis plants. Plant J. 59 517–527. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., S. Dharmasiri and M. Estelle, 2005. a The F-box protein TIR1 is an auxin receptor. Nature 435 441–445. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., S. Dharmasiri, D. Weijers, E. Lechner, M. Yamada et al., 2005. b Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9 109–119. [DOI] [PubMed] [Google Scholar]
- Forsbach, A., D. Schubert, B. Lechtenberg, M. Gils and R. Schmidt, 2003. A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant Mol. Biol. 52 161–176. [DOI] [PubMed] [Google Scholar]
- Gaszner, M., and G. Felsenfeld, 2006. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7 703–713. [DOI] [PubMed] [Google Scholar]
- Gdula, D. A., T. I. Gerasimova and V. G. Corces, 1996. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc. Natl. Acad. Sci. USA 93 9378–9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin, S. B., and S. I. Kim, 2007. Effect of chromatin upon Agrobacterium T-DNA integration and transgene expression. Biochim. Biophys. Acta 1769 410–421. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T. I., and V. G. Corces, 2001. Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu. Rev. Genet. 35 193–208. [DOI] [PubMed] [Google Scholar]
- Golovnin, A., E. Melnick, A. Mazur and P. Georgiew, 2005. Drosophila Su(Hw) insulator can stimulate transcription of a weakened yellow promoter over a distance. Genetics 170 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halweg, C., W. F. Thompson and S. Spiker, 2005. The Rb7 matrix attachment region increases the likelihood and magnitude of transgene expression in tobacco cells: a flow cytometric study. Plant Cell 17 418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, D. A., D. A. Gdula, R. S. Coyne and V. G. Corces, 1993. A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Genes Dev. 7 1966–1978. [DOI] [PubMed] [Google Scholar]
- Henderson, I. R., and S. E. Jacobsen, 2007. Epigenetic inheritance in plants. Nature 447 418–424. [DOI] [PubMed] [Google Scholar]
- Holmes-Davis, R., and L. Comai, 1998. Nuclear matrix attachment regions and plant gene expression. Trends Plant Sci. 3 91–97. [Google Scholar]
- Hunt, L., F. Lerner and M. Ziegler, 2004. NAD-new roles in signaling and gene regulation in plants. New Phytol. 163 31–44. [DOI] [PubMed] [Google Scholar]
- Jefferson, R. A., T. A. Kavanagh and M. W. Bevan, 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M., D. Inze and A. Depicker, 2002. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kellum, R., and P. Schedl, 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64 941–950. [DOI] [PubMed] [Google Scholar]
- Kepinski, S., and O. Leyser, 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451. [DOI] [PubMed] [Google Scholar]
- Kim, S. I., Veena, S. B. Gelvin, 2007. Genome-wide analysis of Agrobacterium T-DNA integration site in Arabidopsis genome generated under non-selective conditions. Plant J. 51 779–791. [DOI] [PubMed] [Google Scholar]
- Kuhn, E. J., and P. K. Geyer, 2003. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 15 259–265. [DOI] [PubMed] [Google Scholar]
- Laplaze, L., B. Parizot, A. Baker, L. Ricaud, A. Martinière et al., 2005. GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J. Exp. Bot. 56 2433–2442. [DOI] [PubMed] [Google Scholar]
- Li, J., A. M. Brunner, R. Meilan and S. H. Strauss, 2008. Matrix attachment region elements have small and variable effects on transgene expression and stability in field-grown Populus. Plant Biotechnol. J. 6 887–896. [DOI] [PubMed] [Google Scholar]
- Liu, P., N. Koizuka, T. M. Homrichhausen, J. R. Hewitt, R. C. Martin et al., 2005. Large-scale screening of Arabidopsis enhancer-trap lines for seed germination-associated genes. Plant J. 41 936–944. [DOI] [PubMed] [Google Scholar]
- Luo, Z., and Z. Chen, 2007. Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell 19 943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein, M., C. Pitsouli, C. Villalta, S. E. Celniker and N. Perrimon, 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke, A. J. M., and M. A. Matzke, 1998. Position effects and epigenetic silencing of plant transgenes. Curr. Opin. Plant Biol. 1 142–148. [DOI] [PubMed] [Google Scholar]
- Mlynarova, L., A. Hricova, A. Loonen and J. P. Nap, 2003. The presence of a chromatin boundary appears to shield a transgene in tobacco from RNA silencing. Plant Cell 15 2203–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec, J., P. Skupa, A. Bailly, K. Hoyerova, P. Krecek et al., 2009. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459 1136–1140. [DOI] [PubMed] [Google Scholar]
- Nagaya, S., K. Yoshida, K. Kato and A.A. Shinmyo, 2001. An insulator element from the sea urchin Hemicentrotus pulcherrimus suppresses variation in transgene expression in cultured tobacco cells. Mol. Genet. Genomic 265 405–413. [DOI] [PubMed] [Google Scholar]
- Nagaya, S., K. Kato, Y. Ninomiya, R. Horie, M. Sekine et al., 2005. Expression of randomly integrated single complete copy transgenes does not vary in Arabidopsis thaliana. Plant Cell Physiol. 46 438–444. [DOI] [PubMed] [Google Scholar]
- Namciu, S. J., K. B. Blochlinger and R. E. K. Fournier, 1998. Human matrix attachment regions insulate transgene expression from chromosomal position effects in Drosophila melanogaster. Mol. Cell. Biol. 18 2382–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov, I. A., W. D. Teale, M. Trebar, I. Blilou and K. Palme, 2005. The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci. 10 170–177. [DOI] [PubMed] [Google Scholar]
- Peach, C., and J. Velten, 1991. Transgene expression variability (position effect) of CAT and GUS reporter genes driven by linked divergent T-DNA promoters. Plant Mol. Biol. 17 49–60. [DOI] [PubMed] [Google Scholar]
- Raab, J. R., and R. T. Kamakaka, 2010. Insulators and promoters: closer than we think. Nat. Rev. Genet. 11 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin, F. M., N. Watanabe, J. L. Cacas, N. Kato, J. M. Arroyo et al., 2008. Genome-wide transposon tagging reveals location-dependent effects on transcription and chromatin organization in Arabidopsis. Plant J. 55 514–525. [DOI] [PubMed] [Google Scholar]
- Sarkar, A., A. Atapattu, E. J. Belikoff, J. C. Heinrich, X. Li et al., 2006. Insulated piggybac vectors for insect transgenesis. BMC Biotechnol. 6: 27. [DOI] [PMC free article] [PubMed]
- Schubert, D., B. Lechtenberg, A. Forsbach, M. Gils, S. Bahadur et al., 2004. Silencing in Arabidopsis T-DNA transformants: the predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell 16 2561–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgourou, A., S. Routledge, D. Spathas, A. Athanassiadou and M. N. Antoniou, 2009. Physiological levels of HBB transgene expression from S/MAR element-based replicating episomal vectors. J. Biotechnol. 143 85–94. [DOI] [PubMed] [Google Scholar]
- Silicheva, M., A. Golovnin, E. Pomerantseva, A. Parshikov, P. Georgiev et al., 2010. Drosophila mini-white model system: new insights into positive position effects and the role of transcriptional terminators and gypsy insulator in transgene shielding. Nucleic Acids Res. 38 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, J., M. Freeling and D. Lisch, 2008. A position effect on the heritability of epigenetic silencing. PloS Genet. 4 e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiker, S., and W. F. Thompson, 1996. Nuclear matrix attachment regions and transgene expression in plants. Plant Physiol. 110 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stief, A., D. M. Winter, W. H. Stratiing and A. E. Sippel, 1989. A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature 341 343–345. [DOI] [PubMed] [Google Scholar]
- Wang, X., L. Li, S. Ding, X. Huang, J. Zhang et al., 2009. Chicken HS4 insulator significantly improves baculovirus-mediated foreign gene expression in insect cells by modifying the structure of neighbouring chromatin in virus minichromosome. J. Biotechnol. 142 193–199. [DOI] [PubMed] [Google Scholar]