Abstract
LINE-1 (L1) retrotransposons represent the most abundant family of non-LTR retrotransposons in virtually all mammals. The only currently known exception is Platypus, where it is found only in low copy numbers. Autonomous L1s encode two proteins, ORF1p and ORF2p, both of which are required for the transposition of L1s. L1 replicative machinery is also involved in the trans-mobilization of non-autonomous retrotransposons, such as diverse short interspersed repetitive elements (SINEs) and processed pseudogenes. Here, we focus on a unique category of “half -L1” elements (HAL1s), which encode ORF1p but not ORF2p. HAL1s are present both in placental mammals and marsupials. We demonstrate that HAL1s originated independently several times during the evolution of mammals. The youngest mammalian HAL1 elements analyzed in this paper were identified in the guinea pig genome. Our analysis strongly suggests that HAL1-encoded ORF1p is essential for the transposition of HAL1s and indicates that the evolution of ORF1p in HAL1s is faster than in L1s. The implications of HAL1 for the evolution of L1 elements and the host genomes are discussed.
Keywords: LINE-1, HAL1, non-LTR retrotransposon, evolution
1. Introduction
Long interspersed element 1 (LINE-1 or L1) is a dominant clade of non-LTR retrotransposons in mammalian genomes (Lander et al., 2001; Waterston et al., 2002; Kapitonov et al., 2009). The vast majority of mammalian L1 elements are 5’-truncated or internally rearranged, and unable to retrotranspose. Complete L1 elements contain two open reading frames (ORFs). ORF1 encodes a ~40-KD protein (ORF1p or p40) with RNA binding (Hohjoh and Singer, 1996) and nucleic acid chaperone activity (Martin and Bushman, 2001). Vertebrate ORF1p consist of three domains: N-terminal coiled-coil (CC) domain, the central noncanonical RNA-recognition-motifs (RRMs) and the C-terminal domain (CTD). The second and third domains are critical for its RNA binding function (Januszyk et al., 2007; Khazina and Weichenrieder, 2009). The ORF2-encoded protein (ORF2p) contains endonuclease (EN) and reverse transcriptase (RT) domains (Mathias et al., 1991; Feng et al., 1996). Both ORF1p and ORF2p proteins are required for the retrotransposition of L1 elements (Moran et al., 1996), in a process termed target site-primed reverse transcription (TPRT) (Luan et al., 1993). The two proteins interact cis-preferentially with the L1 RNA transcripts (Wei et al., 2001; Kulpa and Moran, 2006), and form cytoplasmic ribonucleoprotein particles (RNPs) as the intermediate in retrotransposition (Kulpa and Moran, 2005; Basame et al., 2006).
L1-encoded enzymes are also responsible for the trans-mobilization of a variety of other non-autonomous retroelements, such as Alu and other SINE elements or processed pseudogenes (Jurka, 1997; Esnault et al., 2000; Wei et al., 2001; Dewannieux et al., 2003). The ORF1p protein is not required for the mobilization of non-autonomous Alu elements, but it can enhance the efficiency of their transposition (Hulme et al., 2007; Wallace et al., 2008). In addition, by means of template switching L1 can mobilize other types of cellular RNAs, such as U6 snRNA (Buzdin et al., 2002; Buzdin et al., 2003; Gilbert et al., 2005). In contrast to these non-autonomous retrotransposons, “half-L1” elements (HAL1s) encode a single protein homologous to L1-encoded ORF1p (Smit, 1999). HAL1 elements were first reported in the human genome over a decade ago (Smit, 1999). However, these ancient HAL1s are represented by highly mutated sequence remnants and their biological properties were not studied. Since then, additional HAL1 sequences were deposited in Repbase (Jurka et al., 2005), but details of their origin and evolution were not studied, although artificial constructs similar to HAL1 have been used in several experimental studies (Wei et al., 2001; Garcia-Perez et al., 2007). The HAL1 elements deposited in Repbase (L1-2_Cho, L1-3_ME, L1-3A_ME, L1-2a_MD, L1N1_MD, L1-1N_Tbel) were usually annotated as L1-like elements. This is partly due to the fact that these HAL1 elements are relatively old and difficult to analyze in terms of their biological properties, and no separate annotation procedure was established for HAL1 elements. Our studies combined with the previously reported data show that HAL1-like elements are widespread in mammalian genomes. In this paper we focus on the guinea pig genome, in which both old and young (active) HAL1 elements are present. The ORF1s of the youngest guinea pig HAL1 elements appear to be most intact, implying that they are essential for HAL1 retrotransposition. Our analysis indicates that the HAL1-encoded ORF1p proteins evolve faster than L1-encoded ORF1p. The implications of the HAL1 studies for a broader understanding of other non-LTR retrotransposons and the host genomes are discussed.
2. Materials and methods
HAL1 and L1 repeats were identified by a systematic screening of available mammalian genomes (http://en.wikipedia.org/wiki/List_of_sequenced_eukaryotic_genomes), as a part of the development of Repbase (Jurka et al., 2005), using approaches similar to those described previously (Bao and Eddy, 2002). The full-length or partial consensus sequences for each L1 or HAL1 subfamily were constructed from ~10–30 elements. The copy numbers of each L1 or HAL1 subfamily in each particular genome were determined using BLASTN results with the consensus sequences as query. To avoid overestimating, only the hits corresponding to the 3’-end of the 3’-UTR (> 150-bp length, > 68% identity) were counted. The possibility of positive selection for ORF1p, measured by the ratio of non-synonymous to synonymous substitution rates (Ka/Ks), was estimated for each pair of most-closely related subfamilies. Ka/Ks values were calculated using the KaKs_Calculator program (Zhang et al., 2006). Phylogenetic trees based on the ORF1 or RT region were built with the Mega 4.0 program package (Tamura et al., 2007) using neighbor joining (NJ) method, Kimura 2-parameter distance, and 1,000 bootstrap replicates. The sequence alignments of ORF1 and the RT region are shown in Supplementary Files S2 and S3, respectively.
The age of any particular L1 or HAL1 subfamily was estimated by calculating the average pairwise divergence between each element and all other elements in the subfamily (Khan et al., 2006). Specifically, for each subfamily, ~100–200 elements were randomly collected and were aligned by CLUSTALW (Larkin et al., 2007). The average pair-wise divergence (and standard error) of each subfamily was calculated for the 3’-UTR ~500-bp portion ~50-bp downstream from the ORF1 (for HAL1 elements) or ORF2 (for L1 elements); CpG dinucleotides and the highly mutable poly-purine tracts were not included in the calculation. Divergence was calculated using the Kimura 2-parameter correction installed in Mega program package (Tamura et al., 2007). Because no estimate of the DNA substitution rate in guinea pig was available, we used the mouse DNA substitution rate of 0.34%/Myr (Waterston et al., 2002) as a close approximation when calculating the age of guinea pig L1 or HAL1 sequences. All HAL1 and L1 families described here (Supplementary data) were deposited in Repbase (Jurka et al., 2005).
3. Results and Discussion
3.1. HAL1 elements from Guinea pig
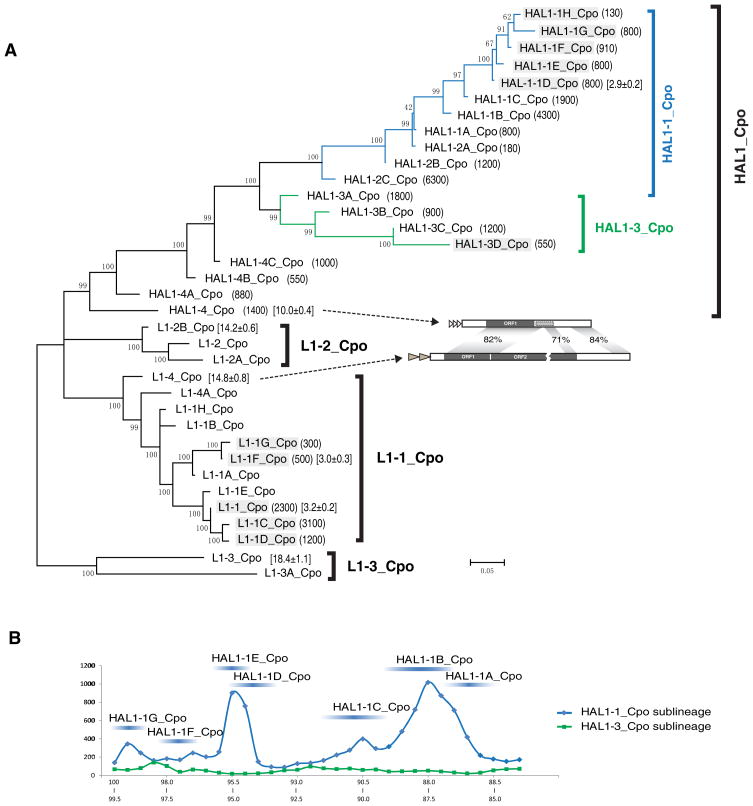
In addition to the complete ~6–7 kb long L1 retrotrotransposons, a large number of ~3–4 kb non-LTR retrotransposons were identified in the guinea pig (Cavia porcellus) genome. Like L1 retrotransposons, these elements typically end with 3’-poly(A) tails and are flanked by 8–16 bp target site duplications (TSDs) (data not shown). Based on the overall sequence similarity or different 5’- or 3’-terminal sequences, or both, these elements were grouped into 19 subfamilies (Fig. 1, 2, Supplementary File S1), representing an estimated 26,000 or more copies (Fig. 1). Analogously, the L1 elements were grouped into 16 subfamilies (Fig. 1, 2). Like the previously identified HAL1 element (HAL1) (Smit, 1999), each of the 19 subfamilies contains a single open reading frame (ORF) homologous to the ORF1 of L1 elements. Therefore, these 19 subfamilies hereafter are referred to as HAL1_Cpo elements, and the ORF is also called ORF1. For example, the ORF1 of HAL1-4_Cpo subfamily shares 82% sequence identity with the ORF1 of L1-4_Cpo subfamily (Fig. 1A). Remarkably, in some HAL1_Cpo subfamilies, such as HAL1-1H_Cpo and HAL1-1G_Cpo, most elements are less than 1% divergent from the consensus, and some elements are even identical to each other, indicating their recent transpositional activity.
Fig. 1. Phylogeny and amplification dynamics of the HAL1 elements from guinea pig.
(A) Phylogenetic tree of the HAL1 and L1 subfamilies. The tree is based on the alignment of the nucleotide sequences of the ORF1 (Supplementary File S2). The three L1 lineages and one HAL1 Lineage are indicated on the right. The two HAL1 sub-lineages are marked in blue and green. The copy numbers of the subfamilies are included in parentheses. Average divergences from the consensus sequences (and standard errors) are shown in brackets (see Methods). The subfamilies expanding during the last ~9 Myr are shaded by gray boxes. The structure comparison between L1-4_Cpo and HAL1-4_Cpo is shown. (B) The HAL1 abundance profile of the HAL1-1_Cpo (blue) and HAL1-3_Cpo (green) sub-lineages in the last ~12 Myrs. The X-axis indicates percentage of identity between ORF1 sequences from individual HAL1 elements and the corresponding consensus sequences of the HAL1-1G_Cpo (blue) and HAL1-3D_Cpo (green) subfamilies. The Y-axis indicates cumulative numbers of elements for each increment of 0.5% sequence identity. The corresponding ranges for the subfamilies of the HAL1-1_Cpo sub-lineage are indicated.
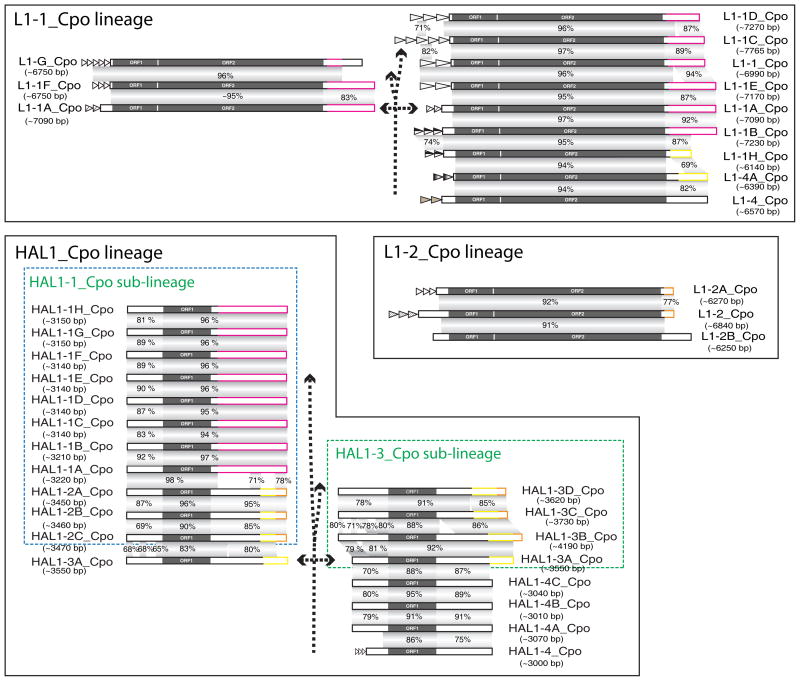
Fig. 2. The structure of L1 and HAL1 elements in the guinea pig genome.
Structure and evolutionary variations in: L1-1_Cpo lineage, L1-2_Cpo lineage and HAL1_Cpo lineage. Elements are arranged according to their evolutionary relationship shown in Figure 1A (dashed branching arrows): from the oldest (bottom) to the youngest subfamily (top). For the purpose of illustration, certain elements near to the branch point are depicted twice (double headed arrows). Tandem repeats in the promoter region are indicated by triangles. The inferred swapped regions are highlighted by different colors and sequence similarities between the regions are shown in Supplementary Fig. S3.
Phylogenetic analysis revealed that the HAL1_Cpo elements belong to one lineage (HAL1_Cpo lineage), including two sub-lineages: HAL1-1_Cpo sub-lineage and HAL1-3_Cpo sub-lineage, whereas L1 elements belong to three L1 lineages: L1-1_Cpo lineage, L1-2_Cpo lineage and L1-3_Cpo lineage (Fig. 1A). L1-3_Cpo is the oldest of the three L1 lineages, as confirmed by the phylogeny of the reverse transcriptase (RT) (Supplementary Figure S1). Based on the sequence divergence, L1-1_Cpo is likely to be the only L1 lineage that contains active subfamilies, including L1-1C_Cpo and L1-1D_Cpo subfamilies (divergence < 1%). While the HAL1-Cpo lineage possibly originated from the common ancestor of the L1-1_Cpo and L1-2_Cpo lineages, the structure and sequence analyses reveal that the HAL1_Cpo lineage is more closely related to the L1-1_Cpo lineage than to the L1-2_Cpo lineage (Fig. 1A). The ORF1 sequence of HAL1-4_Cpo (the oldest HAL1 element) shares a greater identity with L1-4_Cpo than with the L1-2B_Cpo subfamily consensus (82% vs 79%). Furthermore, there is ~84% sequence identity between the 478-bp 3’-end of L1-4_Cpo and the corresponding region of the HAL1-4_Cpo subfamily (Fig. 1A), which is not shared between HAL1-4_Cpo and L1-2B_Cpo. Notably, while the ORF2p coding region is absent in all HAL1_Cpo elements, small partial ORF2 sequences are present in some old subfamilies. For example, the ~200-bp 5’ and ~150-bp 3’ terminal sequences of the L1-4_Cpo ORF2 region are present in HAL1-4_Cpo sequences (Fig. 1A). Taken together, this suggests that HAL1-4_Cpo originated from an ancient L1-4_Cpo-like element by deleting a part of the ORF2 region.
To determine when the first HAL1-Cpo element appeared in guinea pig, we estimated the ages of the oldest subfamilies in the L1 and HAL1 lineages. Age estimates were based on average pairwise divergence and were calculated using the average DNA substitution rate of 0.34%/Myr for mouse (Waterston et al., 2002), although there may be differences between the two rodent species (Fieldhouse et al., 1997). By this estimate the L1-3_Cpo subfamily is ~54 Myr old, the L1-4_Cpo subfamily is ~44 Myr old, the L1-2B_Cpo subfamily is ~42 Myr old, and the HAL1-4_Cpo is ~29 Myr old (Fig. 1A). Given the possible recent expansion of the HAL1-4_Cpo subfamily and the different evolutionary rate between HAL1 and L1 elements (discussed below), 29 Myr is likely to be an underestimate for the HAL1_Cpo lineage. Therefore, taking into account the close relationship between HAL1-4_Cpo and L1-4_Cpo (Fig. 1A), we assume that the HAL1_Cpo lineage originated in the guinea pig genome some ~ 29–44 Myr ago. Again, this rough estimate is likely to be adjusted if more accurate substitute rates in guinea pig become available.
3.2. Strong conservation of intact ORF1 in HAL1s
To evaluate the biological significance of the ORF1 in HAL1 elements, we analyzed the preservation of an intact ORF1 in each subfamily and determined that younger subfamilies show a higher percentage of elements retaining intact ORF1 than the older ones. For example, in the relatively young subfamilies HAL1-1H_Cpo, HAL1-1G_Cpo and HAL1-3D_Cpo, the percentages of intact ORF1s are 89%, 65% and 60%, respectively. In older subfamilies such as HAL1-1C_Cpo, HAL1-1B_Cpo and HAL1-3C_Cpo the respective percentages are 13%, 5% and 10%. Analogously, the percentages of intact ORF1s in active L1-1C_Cpo and L1-1D_Cpo subfamilies are 65% and 61%, respectively, and only 18% in the older L1-1_Cpo subfamily. Thus, given the relatively long evolutionary history of HAL1_Cpo elements, the apparent preservation of intact ORF1s in younger (active) elements strongly indicates HAL1-encoded ORF1p is needed in cis for the replication of HAL1_Cpo elements. This conclusion based on evolutionary analysis is consistent with the available experimental data (Wei et al., 2001; Garcia-Perez et al., 2007). Subsequently, we aligned the HAL1 ORF1p sequences with L1s ORF1p sequences in guinea pig and other mammalian species. As in the case of L1 ORF1p (Januszyk et al., 2007; Khazina and Weichenrieder, 2009), the HAL1-encoded ORF1p also shows the most conservative parts in the RRM and the CTD domains. Importantly, L1s and HAL1 ORF1p have the same conserved amino acids (Supplementary Fig. S2).
3.3. The dynamics of HAL1s in guinea pig
The 5’ untranslated regions (UTRs) of L1 elements are evolutionarily variable, as previously reported (Adey et al., 1994; Naas et al., 1998; Goodier et al., 2001; Khan et al., 2006). Also, recombination between different L1s can produce novel L1 subfamilies (Hayward et al., 1997). In guinea pig, similar dynamics were observed in both the 5’- and 3’-UTRs of L1 and HAL1 elements. The 5’- or 3’-terminal sequences of almost all HAL1_Cpo subfamilies are either inherited from an ancestral subfamily or “borrowed” from a non-ancestral L1 subfamily (Fig. 2, Supplementary Fig. S3). The most striking example is the HAL1-1A_Cpo subfamily, which acquired its 1301-bp long 3’-end sequence from the 3’-UTR of an element belonging to the L1-1B_Cpo subfamily (Fig. 2, Supplementary Fig. S3B). Other examples include HAL1-4A_Cpo subfamily (“borrowing” its 518-bp 5’-UTR from L1-3_Cpo subfamily) and HAL1-3A_Cpo subfamily (acquiring its 3’-end 637-bp sequence from L1-4A_Cpo subfamily) (Fig. 2, Supplementary Fig. S3). In the case of HAL1-2C_Cpo and HAL1-3B_Cpo subfamilies, their ~250-bp long 3’-ends are ~89% identical to the 3’-ends of L1-2_Cpo elements (Supplementary Fig. S3D). However, the 258-bp 3’-end of L1-2_Cpo itself is likely acquired from other elements (Fig. 2, Supplementary Fig. S3E). Interestingly, in the L1-2_Cpo and HAL1-2B_Cpo subfamilies the sequence junctions between the newly acquired sequence and the original sequence are located at similar positions (Supplementary Fig. S3E), suggesting a common mechanism for the sequence exchange.
In addition to the HAL1-1_Cpo sub-lineage, HAL1-3_Cpo sub-lineage also retains the capacity for transposition (members of the HAL1-3D_Cpo subfamily are >99% identical to their consensus sequence and ~60% elements contain intact ORF1s). As shown in figure 1B, however, the two sub-lineages differ significantly in their amplification profiles. Based on the fact that a single amino acid substitution, in some critical positions, can dramatically affect the efficiency of L1 transposition (Kulpa and Moran, 2005; Garcia-Perez et al., 2007; Martin et al., 2008), we assumed that the biochemical properties of ORF1p could largely account for this long-term difference. Indeed, the alignment of ORF1p suggests that some candidate substitutions may be linked to the low profile of HAL1-3_Cpo sub-lineage (Supplementary Fig. S2). For example, K284 is limited to HAL1-3_Cpo sub-lineage, while R284 is conserved in all other L1s and HAL1s.
During the last ~12 Myrs’ evolution, several master elements in the HAL1-1_Cpo sub-lineage showed elevated transpositional activity and produced several peaks (subfamilies) in the abundance profile (Fig. 1B). Of the various subfamilies generated from the master HAL1 elements, the HAL1-1B_Cpo subfamilies are of particular interest from an evolutionary point of view. During the transition from HAL1-1A_Cpo to HAL1-1B_Cpo the estimated ratios of non-synonymous to synonymous substitution rates (Ka/Ks) in the N-terminal half of their ORF1p are all greater than 1, indicating positive selection (Supplementary Fig. S2; positions 1–167). Depending on the models chosen, the Ka/Ks values vary from 1.45 to 3.46 (Supplementary Table S1). Given that the number of HAL1-1B_Cpo elements is ~4 times greater than that of the HAL1-1A_Cpo subfamily, it is tempting to speculate that the sharp increase in retrotransposition of HAL1-1B_Cpo was associated with the positively-selected amino acid substitutions. However it is also possible that other stochastic factors, such as the local chromatin context of the HAL1-1B_Cpo master gene, could account for the difference. In addition, it is worth noting that this positive selection occurred shortly, if not immediately, after the 3’-UTR of HAL1-1A_Cpo was acquired from a L1-1B_Cpo element (Fig. 2). In comparison to other sequence changes in successive subfamilies, this 3’-UTR switching is substantial and it produced two separated regions in the 3’-UTR with low sequence identity (~70% identity over ~260-bp) to its predecessor (Fig. 2), which suggests that positive selection of the HAL1-1A_Cpo ORF1p might have been triggered by large-scale sequence swaps. Positive selection has also been reported in the N-terminal half of ORF1p in some human L1 families (Boissinot and Furano, 2001; Khan et al., 2006), but no large-scale sequence changes were reported.
Analogously to SINE elements, HAL1s must compete for reverse transcriptases (RTs) encoded by other autonomous L1s, which are normally cis-preferential for their own RNA transcript. Notably, HAL1 elements were amplified with considerable success in guinea pig during the last 9 Myr or so. This period roughly overlaps with the amplification of 11 HAL1 and L1 subfamilies that are outlined in grey in Figure 1A. Overall, there are ~7,400 copies of these L1 elements, while the co-existing HAL1 elements add up to ~3,950 copies, which amount to 53% (3,950/7,400) of the retrotransposition frequency of the autonomous L1 elements. This level of mobilization efficiency in trans is considerably higher compared to available experimental data demonstrating that only 0.4–0.7 % of the HAL1-like transcripts (referred to as ORF1mneoI) are trans-mobilized by autonomous L1s in HeLa cells (Table 1 and 2 of Wei et al. 2001). The observed differences may be due to different 3’-UTR sequences used in the experiments as artificial indicator cassettes. In a recent paper it has been demonstrated that HAL1-like transcripts (ORF1mneoI) can be trans-mobilized with 50–100 times higher efficiency than ORF1-mutated HAL1-like transcripts (Figure 2A in Garcia-Perez et al. 2007). Nevertheless, it remains unclear how efficient the trans mobilization can be in vivo. In any case, the data from the guinea pig represent an example of very successful “parasite” HAL1 elements.
Table 1.
Mammalian HAL1 elements
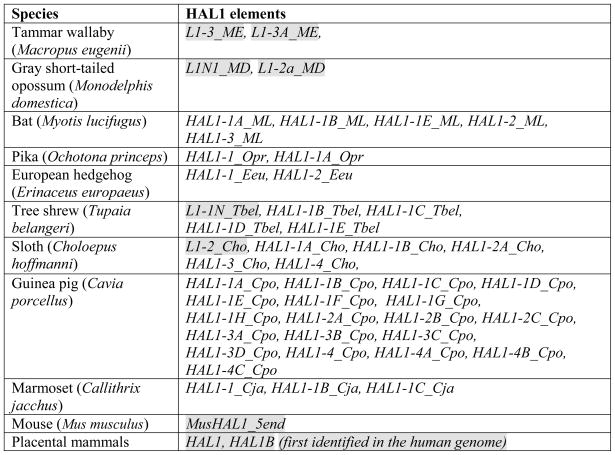 |
HAL1s which were previously deposited in Repbase are shaded (some of them were previously annotated as L1 elements). HAL1s first described in this paper are not shaded.
3.4. The accelerated evolution of ORF1p in HAL1s
Compared to autonomous L1 lineages, the HAL1 lineage in guinea pig is represented by a remarkably long branch (Fig. 1A), reflecting frequent outbursts of new subfamilies. This pattern suggests that ORF1s may evolve faster in the HAL1 lineage than in autonomous L1 elements. However, of all HAL1 subfamilies studied here, only the ORF1p of HAL1-1A_Cpo and HAL1-1B_Cpo show substitution patterns consistent with positive selection (Ka/Ks > 1), suggesting that positive selection cannot fully explain the observed accelerated evolution in the HAL1 lineage. More likely, the observed differences may be caused by different selective constraints affecting ORF1 mutations in HAL1s and in L1 elements. The ORF2 sequence is typically more conserved than the ORF1 sequence, possibly due to a stronger negative selection. Assuming that the number of random mutations in a given protein is proportional to the length of its open reading frame, a mutation in the ORF1 of a given L1 element is likely to coincide with mutations in the ORF2, which is much longer. Due to the cis-preference of ORF1p and ORF2p, a large fraction of ORF1 mutations are likely to be eliminated due to mutations that impair the function of ORF2p. Therefore, the lack of ORF2 could explain the faster evolution of ORF1 in HAL1 than in L1 elements.
3.5. Diverse HAL1 non-autonomous elements in other mammalian genomes
Several other mammalian species also carry HAL1-type families in their genomic DNA (see Table 1 for Repbase loci names). The originally reported ancient eutherian HAL1 family (Smit, 1999) is represented by ~2,100 copies in the human genome and ~2,300 copies in the horse genome, with individual HAL1 elements only ~70% identical to the consensus. In two marsupials, tammar wallaby (Macropus eugenii) and gray short-tailed opossum (Monodelphis domestica), large numbers of HAL1 elements were found (~24,000 and ~28,000, respectively), but the youngest HAL1 elements found in the M. eugenii genome are 5–6% divergent from their consensus, and in the M. domestica genome the youngest elements are ~11% divergent from the consensus sequence; for this reason they were not chosen for detailed analysis here. In addition, HAL1-like elements were identified in sloth (Choloepus hoffmanni) (~5,000 copies), marmoset (Callithrix jacchus) (~4,700 copies), bat (Myotis lucifugus) (~5,700 copies), tree shrew (Tupaia belangeri) (~6,600 copies), pika (Ochotona princeps) (~3,500 copies), mouse (Mus musculus) (~380 copies) and hedgehog (Erinaceus europaeus) (~250 copies). Notably, in the C. hoffmanni genome, three different lineages of HAL1 elements were found: HAL1-2A_Cho, HAL1-4_Cho and HAL1-1A_Cho. Most likely, these three HAL1 lineages originated independently from three different L1 retrotransposons (data not shown).
Taken together, these data suggest that different HAL1 families originated spontaneously and recurrently throughout the evolution of mammals. Once generated, HAL1s can persist in their host species for millions of years. Our data from the guinea pig genome also indicate that at certain points in their evolutionary history, HAL1s might have proliferated nearly as efficiently as L1 elements. The existence of active HAL1 elements may generate additional ORF1 proteins in the host cells, which may potentially improve the retrotransposition of other non-autonomous retrotransposons, such as Alu-like SINE elements (Wallace et al., 2008), other processed pseudogenes, or even some mutated L1s. Furthermore, given its faster evolution the HAL1-encoded ORF1p may have selective advantages over the L1-encoded ORF1p in terms of retrotransposition efficiency. Moreover, HAL1s may contribute their fast-evolving ORF1s to L1s by occasionally forming functional chimeric L1 elements, which may in turn give them selective advantages. This process may replay the initial stage in the evolutionary history of the L1 clade when an ORF1-like gene was first recruited by the non-LTR retrotransposons. In general, the L1/HAL1 system may be a relatively simple model for studies of the advantages and disadvantages of “gene consolidation” into a single replicating unit as opposed to a looser “federation” of genes working with each other. Such a model may be of interest for a broader understanding of early genome evolution (Jurka and Kapitonov, 1999).
3.6. Hypothetical mechanisms for the origin of HAL1 elements
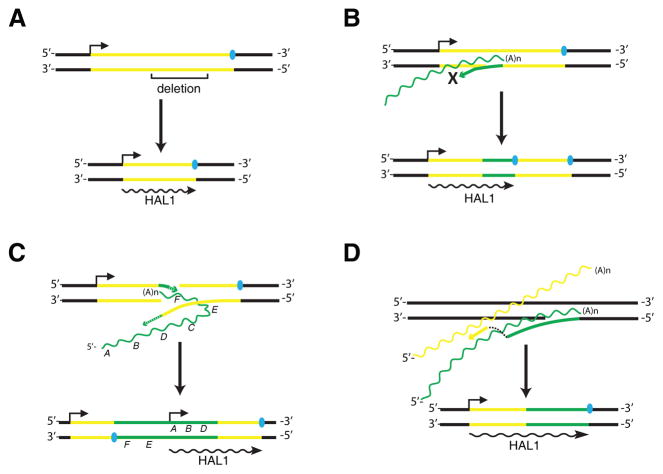
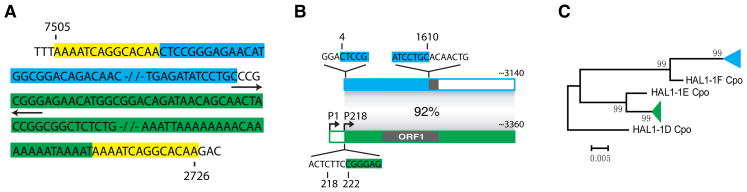
Most likely, each HAL1 element originated from a particular autonomous L1 element containing two ORFs. Theoretically, a new HAL1 element could be generated from any L1 sequence in which the ORF1p region is accidentally followed by a transcription termination signal, but the specific processes leading to this may vary. Due to the abundance of L1 retrotransposons and their retrotranspositional activities, the most accidental transcription termination signals are likely to be provided by the L1 elements. We analyzed several potential mechanisms that can explain the origins of HAL1s. The first model proposes that HAL1s originate from an L1 element affected by an internal deletion (Fig. 3A), which may occur at the DNA level or during reverse transcription of the L1 RNA (Gilbert et al., 2005). The oldest subfamily in guinea pig, HAL1-4_Cpo, was likely to be created by such process (Fig. 1A). This process is supported by experimental data (Gilbert et al., 2005). In addition, it is conceivable that the internal deletion can be introduced by splicing of the L1 transcript (Belancio et al., 2006). Another possible process may involve the insertion of the 3’-UTR of L1 elements into other L1 elements (Fig. 3B). According to our incomplete survey in guinea pig, this type of rearrangement is also relatively common (data not shown). Alternatively, it may involve insertion of a 5’-inverted L1 element into a second L1 element (Fig. 3C). The 5’-inverted L1 element could be generated by a twin-priming mechanism (Ostertag and Kazazian, 2001). Finally, template switching may also potentially explain the origin of HAL1 elements (Fig. 3D). This mechanism was invoked in the formation of U6/L1 and some other type of chimeric retrotranscripts (Buzdin et al., 2002; Buzdin et al., 2003; Garcia-Perez et al., 2007), but the case of template switching between heterologous L1 RNAs was rarely reported (Gilbert et al., 2005). In the guinea pig genome we found one HAL1/HAL1 chimeric sequence (AAKN02025597.1, position 7505-2726) flanked by 14-bp TSD, which most likely was generated by template switching (Fig. 4A). Its 1607-bp 5’- portion and 3142-bp 3’-portion originated from two different HAL1 clades (green and blue triangle, Fig. 4C), excluding the possibility of duplication during the reverse transcription. The 5’-portion covers the 5’-UTR and most of the ORF1 region, whereas the 3’-portion represents almost the entire second HAL1 element involved in the formation of the chimeric sequence (Fig. 4B). Notably, a 4-bp palindrome is present at the junction of the two HAL1 sequences, possibly generated by a self-priming process. Short palindromes or extra sequences are often observed at the 5’-ends of the newly inserted L1 elements (Gilbert et al., 2005). Apart from template switching, the origin of the chimeric element can also potentially be explained by a second model involving two independent processes: an insertion of a HAL1 element (green) at the junction between the 3'-UTR and the poly-A tract of the previously inserted HAL1 element (blue, Fig. 4), followed by an internal deletion event. As reported before, the L1/L1 chimeras are mainly generated by a process termed as non-allelic cDNA-mediated homologous recombination, triggered by single-strand annealing during TPRT (Gilbert et al., 2005). However, this process does not produce any internal deletion in the chimeric L1 element. Because template switching occurs between two RNA molecules, the newly formed HAL1 element, if any, is likely to be retrotransposition-competent like its parent L1 elements. In conclusion, even if template switching is relatively rare, it is also a possible mechanism for formation of HAL1 elements. Due to the scarcity of the sequence data and the fact that HAL1 elements are relatively old, it is difficult to determine which of the above processes contributed the most to the formation of HAL1 elements.
Fig. 3. Hypothetical schemes depicting the origins of HAL1 from L1 elements.
(A) Internal deletion. (B) Insertion of the 3’-end sequence of another L1 element. (C) Insertion of an 5’-inverted L1 element due to twin priming (Ostertag and Kazazian, 2001). (D) Template switching during reverse transcription. Bold lines indicate DNA strands. The wavy lines indicate RNA transcripts. Regions from different L1 molecules (RNA or DNA) are differentiated by yellow and blue colors. Blue solid ovals indicate the transcription stop site of the L1 or HAL1 elements. Arrows above the upper DNA strand indicate the transcription start site. The transcript of the newly formed HAL1 element is shown below (in black).
Fig. 4. Putative chimeric retrotranscript derived by template switching.
(A) The chimeric retrotranscript is from guinea pig genomic sequence contig AAKN02025597.1, positions 7505 - 2726. The 14-bp target site duplications are shaded in yellow. The 5’ and 3’ portions derived from two different HAL1 elements are shaded in blue and green. The 4-bp palindrome is indicated by the opposite arrows below the sequence. (B) Consensus sequences were constructed to represent the two parental HAL1 elements of the chimeric HAL1 shown in (A), sharing 92% sequence identity. The corresponding regions forming the chimera are shaded accordingly at position 4 to1610 of the upper element and position 222 to the 3’-end of the bottom element. The sequences around the junction are shown. The bottom (green) subfamily most likely carries two promoters: P1 and P218, starting from position 1 and 218, respectively. Either the 1607-bp 5’-end or the 3142-bp 3’- end of the chimera element were used as query sequences, and ~20 top scoring elements (by BLASTN), were used to construct their consensus sequences.(C) The 5’- and 3’-end portions of the chimeric HAL1 are clustered with other related HAL1 elements into two different clades (blue and green triangle). Each triangle contains ~20 different HAL1 sequences. HAL1-1D_Cpo, HAL1-1E_Cpo and HAL1-1F_Cpo represent consensus sequences of these subfamilies.
Supplementary Material
Acknowledgments
We thank Karolina Walichiewicz for help with editing the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Library of Medicine or the National Institutes of Health. This work was supported in part by the National Library of Medicine [P41 LM006252].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adey NB, Schichman SA, Graham DK, Peterson SN, Edgell MH, Hutchison CA., 3rd Rodent L1 evolution has been driven by a single dominant lineage that has repeatedly acquired new transcriptional regulatory sequences. Mol Biol Evol. 1994;11:778–89. doi: 10.1093/oxfordjournals.molbev.a040158. [DOI] [PubMed] [Google Scholar]
- Bao Z, Eddy SR. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12:1269–76. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basame S, Wai-lun Li P, Howard G, Branciforte D, Keller D, Martin SL. Spatial assembly and RNA binding stoichiometry of a LINE-1 protein essential for retrotransposition. J Mol Biol. 2006;357:351–7. doi: 10.1016/j.jmb.2005.12.063. [DOI] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34:1512–21. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Furano AV. Adaptive evolution in LINE-1 retrotransposons. Mol Biol Evol. 2001;18:2186–94. doi: 10.1093/oxfordjournals.molbev.a003765. [DOI] [PubMed] [Google Scholar]
- Buzdin A, et al. The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Res. 2003;31:4385–90. doi: 10.1093/nar/gkg496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzdin A, Ustyugova S, Gogvadze E, Vinogradova T, Lebedev Y, Sverdlov E. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3' terminus of l1. Genomics. 2002;80:402–6. doi: 10.1006/geno.2002.6843. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–8. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–7. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–16. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Fieldhouse D, Yazdani F, Golding GB. Substitution rate variation in closely related rodent species. Heredity. 1997;78( Pt 1):21–31. doi: 10.1038/hdy.1997.3. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez JL, Doucet AJ, Bucheton A, Moran JV, Gilbert N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 2007;17:602–11. doi: 10.1101/gr.5870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol. 2005;25:7780–95. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Du K, Kazazian HH., Jr A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11:1677–85. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward BE, Zavanelli M, Furano AV. Recombination creates novel L1 (LINE-1) elements in Rattus norvegicus. Genetics. 1997;146:641–54. doi: 10.1093/genetics/146.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. Embo J. 1996;15:630–9. [PMC free article] [PubMed] [Google Scholar]
- Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januszyk K, et al. Identification and solution structure of a highly conserved C-terminal domain within ORF1p required for retrotransposition of long interspersed nuclear element-1. J Biol Chem. 2007;282:24893–904. doi: 10.1074/jbc.M702023200. [DOI] [PubMed] [Google Scholar]
- Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci U S A. 1997;94:1872–7. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV. Sectorial mutagenesis by transposable elements. Genetica. 1999;107:239–48. [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–7. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Tempel S, Jurka J. Simple and fast classification of non-LTR retrotransposons based on phylogeny of their RT domain protein sequences. Gene. 2009 doi: 10.1016/j.gene.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazina E, Weichenrieder O. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc Natl Acad Sci U S A. 2009;106:731–6. doi: 10.1073/pnas.0809964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum Mol Genet. 2005;14:3237–48. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol. 2006;13:655–60. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Martin SL, et al. A single amino acid substitution in ORF1 dramatically decreases L1 retrotransposition and provides insight into nucleic acid chaperone activity. Nucleic Acids Res. 2008;36:5845–54. doi: 10.1093/nar/gkn554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol. 2001;21:467–75. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–10. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–27. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Naas TP, et al. An actively retrotransposing, novel subfamily of mouse L1 elements. Embo J. 1998;17:590–7. doi: 10.1093/emboj/17.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 2001;11:2059–65. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–63. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Wallace N, Wagstaff BJ, Deininger PL, Roy-Engel AM. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene. 2008;419:1–6. doi: 10.1016/j.gene.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wei W, et al. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–39. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao XQ, Wang J, Wong GK, Yu J. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 2006;4:259–63. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.