Summary
Understanding luciferase enzymology and the structure of compounds that modulate luciferase activity can be used to improve the design of luminescence-based assays. This review provides an overview of these popular reporters with an emphasis on the commonly used firefly luciferase from Photinus pyralis (FLuc). Large-scale chemical profile studies have identified a variety of scaffolds that inhibit FLuc. In some cell-based assays these inhibitors can act in a counter-intuitive way –leading to a gain in luminescent signal. Although formerly attributed to transcriptional activation, intracellular stabilization of FLuc is the primary mechanism underlying this observation. FLuc inhibition/stabilization can be complex, as illustrated by the compound PTC124, which is converted by FLuc in the presence of ATP to a high affinity multi-substrate-adduct inhibitor, PTC124-AMP. The potential influence these findings can have on drug discovery efforts is provided here.
Introduction
Technical advances in molecular biology, microtiter plate-based spectrophotometry, and automation have enabled the development of biologically-relevant assays with robust performance that can be utilized in high-throughput screening (HTS) for chemical biology and drug discovery applications. These sophisticated assays, characterized by their ability to be translated from the bench to fully-automated HTS systems while maintaining sensitivity, signal strength, and biological fidelity, often involve capturing a biological response using a single reporter that has a light-based output. While such assays greatly improve our ability to monitor intricate biological processes, incorporation of a biological reporter can influence assay results in unintended and unanticipated ways. An understanding of these reporter-related complexities is essential, and appropriate follow-up assays should be aimed at identifying reporter-related artifacts in order to validate the HTS result as well as provide an accurate interpretation of structure activity relationships (SAR) derived during subsequent probe/lead optimization efforts.
Bioluminescence is a commonly exploited detection technology used across academia and industry. An indicator of this is evidenced by the nearly 2,000 assays currently listed in the PubChem database: approximately 21% are bioluminescence and 53% are fluorescence, with the remainder using other assay formats, such as absorbance (Figure 1; see review by [Inglese et al., 2007], for an overview of different HTS assay formats). Bioluminescent assays rely on luciferase enzymes, which catalyze the oxidation of specific substrates known as luciferins to form oxyluciferin, with the concurrent emission of a photon. Although all luciferases catalyze light-emitting reactions, the luciferin substrates are structurally diverse. Luciferases from many bioluminescence-producing organisms have been cloned and isolated for the purpose of constructing bioassay systems (Fan and Wood, 2007; Hoshino, 2009), including luciferases from fireflies (Lampyridae), click beetles (Elateridae), the larvae of certain beetles known as glow worms (Phengodidae), and various marine organisms such as the sea pansy (Renillidae).
Figure 1. PubChem analysis by detection type.

Currently, of the >2,000 assays listed in PubChem, bioluminescence and fluorescence represent the two most prominent detection strategies. NCI is the NCI-60 cell viability assay panel.
This review provides a summary of commonly used luciferases and suggests recommendations for their appropriate use in common HTS assays. A particular focus is placed on the widely employed firefly luciferase, FLuc. All assay formats, including those involving FLuc, have associated artifacts which interfere with the interpretation of assay results. For this reason, we will describe our efforts to apply titration-based screening (quantitative HTS, qHTS) to identify, categorize and understand the behavior of compounds that interact with FLuc in biochemical and cell-based assays. Compounds that interfere with FLuc detection are predominantly inhibitors of the enzyme, but vary in their mode of action, from simple competitive inhibitors to unique multi-substrate adduct inhibitors, as illustrated by PTC124 (Ataluren ). Here we describe the complexities associated with reporter enzymology and how this may influence compound discovery efforts.
Overview of luciferases and uses in HTS
Luciferases employed in HTS can be divided into several major groups based on their substrate and co-factor dependence and whether the enzyme is expressed intracellularly or is secreted from cells (Table 1 and Table S1). Additionally, some luciferases are utilized for either cell- or biochemical-based HTS assays, whereas others have been developed for use in both assay formats (Table 1). One of the main reasons for the wide-spread use of luciferase in bioassays is that, unlike fluorescence, luminescence does not require excitation light energy, thus lowering the background signal to provide a sensitive assay with a high signal: background ratio (S:B). Elimination of an excitation light source also prevents interference by compound fluorescence (Simeonov et al., 2008) and fluorophore photobleaching. Therefore, luminescence assays can be very sensitive, despite a significantly weaker signal intensity compared to fluorescence. For cell based assays, the use of FLuc or Renilla luciferase (RLuc) enzymes as reporters enables the measurement of dynamic changes in reporter transcription levels as the intracellular protein half-lives of these luciferase enzymes are relatively short compared to non-enzymatic fluorescent protein reporters such as GFP (protein half-life – 26 hours [Corish and Tyler-Smith, 1999]). By comparison, the protein half-lives of Gaussia luciferase (GLuc) (Wurdinger et al., 2008) and Cypridina luciferin 2-monooxygenase (CLuc) (Nakajima et al., 2004), once secreted, are significantly longer than either FLuc or RLuc (6 days and 53 hours, respectively, in cell-culture media; Table 1), although, at this time, neither GLuc nor CLuc is commonly used in HTS.
Table 1.
Luciferases employed in chemical biology applications
| Luciferase | Species | Molecular Weight |
Peak Emission Wavelength |
ATP- dependent? |
Substrates | Secreted? | Protein half- life |
HTS Assay Type |
|---|---|---|---|---|---|---|---|---|
| Firefly (FLuc) | Photinus pyralis | 62 kDa | 550–570nm | Yes | D-luciferin/ATP | No | 3 hrs | B & C |
| Modified Firefly (Ultra-Glo) | Photuris pennsylvanica | 61 kDa | 550–570nm | Yes | D-luciferin/ATP | No | N/A | B |
| Click beetle (CBLuc) | Pyrophorus plagiophthalamus | 60 kDa | green: 537nm red:613nm | Yes | D-luciferin/ATP | No | 7 hrs | C |
| Sea pansy (RLuc) | Renilla reniformas | 36 kDa | 480nm | No | Coelenterazine | No | 4.5 hrs | B & C |
| Copepod crustacean (GLuc) | Gaussia princeps | 20 kDa | 460nm | No | Coelenterazine | Yes | 6 days (in cell media) | † |
| Ostracod crustacean (CLuc) | Cypridina noctiluca | 62 kDa | 465nm | No | Vargulin (aka Cypridina luciferin) | Yes | 53 hrs (in cell media) | † |
B = Biochemical assay format; C = Cell-based assay format
N/A = not applicable. The luciferase is not currently used in cell-based formats
= these luciferases are currently not widely used in HTS
The application of FLuc as a reporter began in the late 1980s, following its initial cloning (de Wet et al., 1985), and expression in mammalian cells (de Wet et al., 1987; de Wet et al., 1985). Found to be a sensitive reporter for biological assays (de Wet et al., 1987; Nguyen et al., 1988; Ow et al., 1986), and catalytically active as a monomer (Wood, 1995), the FLuc enzyme was optimized for expression and activity as a reporter gene in mammalian cells (Wood, 1998), though it is now widely used for biochemical applications in HTS as well. A luciferase from a different species of firefly – Photuris pennsylvanica – has also been optimized and developed for use as a reporter (though for biochemical assays only) based upon its utilization of D-luciferin (D-LH2) and ATP as substrates (e.g. Ultra-Glo ) (Fan and Wood, 2007). Typical biochemical assays that utilize either FLuc or Ultra-Glo include those that measure ATP or ADP concentrations (Singh et al., 2004; Tanega et al., 2009) or use pro-luciferin substrates to detect target protein activity (Cali et al., 2006; Fan and Wood, 2007) (Table S1).
RLuc, another commonly used luciferase reporter (Roda et al., 2009), does not share amino acid sequence similarity to FLuc (Greer and Szalay, 2002; Lorenz et al., 1991) and light production is ATP independent (Hart et al., 1979). RLuc is enzymatically active as a monomer (Matthews et al., 1977), catalyzing the oxidative decarboxylation of the luciferin coelenterazine via a dioxetane intermediate prior to conversion to coelenteramide, with the concurrent emission of blue light (Figure 2A) (Hart et al., 1978). Compared to FLuc, RLuc has a few disadvantages as an assay reporter as described in Table S1.
Figure 2. Luciferase assay configurations.
A. The dual luciferase assay protocol is illustrated. Cells are lysed with a detection reagent containing luciferase substrates (1), FLuc luminescence is measured (2), the FLuc reaction is then stopped along with the addition of the coelenterazine substrate for RLuc (3), and luminescence is again measured (4). In this case total luminescence is measured using a clear filter; the luminescence emission spectra of FLuc and RLuc are also shown (CPS, counts per second measured on a luminometer). B. CBLuc two-color dual-luciferase assay protocol is shown in which cell lysis and detection are performed with a single reagent addition (1) and green luminescence and red luminescence are then measured sequentially on a microtiter plate reader (2) using the appropriate optical filters. The luminescence emission maximums are noted and the luminescence emission spectra for both green and red CBLuc are shown below. The dotted lines on the emission spectra represent the optical filters used in (Davis et al., 2007). C. A live cell kinetic assay is depicted using a secreted luciferase (GLuc) that uses coelenterazine as the substrate. Cell culture supernatant containing the secreted GLuc is removed at different times (1) and upon mixing with the coelenterazine substrate (2) the total luminescence for each time point is measured using a clear filter (3). Also shown is the emission spectra for GLuc relative to RLuc (spectra adapted from (Tannous et al., 2005). The GLuc wavelength emission maximum is similar to RLuc, but GLuc shows brighter luminescence.
The luciferases FLuc, RLuc, and the red- or green-emitting Caribbean click beetle (CBLuc) varieties - have been configured for use in dual-luciferase assays. One such dual-luciferase assay is designed such that activity from one luciferase (typically FLuc) tracks with the target biology while the second luciferase, e.g. RLuc, is used for assay normalization (to account for cytotoxicity, differences in cell number, and transfection efficiency in the case of transiently transfected cell lines) (Stables et al., 1999). In this dual luciferase assay system, measurement of the two luciferases is sequential requiring separate substrate addition steps (Figure 2A), as the two luciferases need different substrates/buffers, and collection of the second luminescence signal requires termination of the preceding reaction (Hannah et al., 1998 ). Commercial detection reagents are available for sequential measurement of FLuc and RLuc, but non-commercial formulations have also been described (Dyer et al., 2000). While the FLuc/RLuc dual-luciferase assay is ratiometric by design, limitations to this system should be considered. For miniaturized assay volumes (< 10 μL) the two-step addition protocol is not optimal due to well volume limitations.
Additionally, as discussed in detail in the following section, either luciferase enzyme can be inhibited by small molecule compounds used in HTS. In the case of disparate enzymes such as FLuc and RLuc, which are evolutionarily unrelated and have different substrate specificities (Loening et al., 2007), it is expected that these two enzymes will have dissimilar inhibition profiles. Thus, the changes in the FLuc/RLuc luminescence ratio intended to be reflective of target modulation could actually be the result of the direct and selective inhibition of either luciferase reporter enzyme.
An alternative dual luciferase reporter-gene assay takes advantage of luciferase mutants derived from the yellow-light-emitting CBLuc. Variants of CBLuc, which share over 95% amino acid identity and catalyze the oxidation of the same substrate, D-LH2, have been engineered to emit either green or red light (Almond et al., 2003; Wood et al., 1989). The two-color dual luciferase assay requires a single substrate addition with activity from the two variant luciferases differentiated based on their shifted bioluminescent emission wavelengths (Figure 2B). Due to high amino acid sequence similarity it is expected that inhibitors would most likely affect both CBLucs similarly, that is, a chemical that inhibited the enzymatic activity of one CBLuc would inhibit the other as well, thus giving an advantage to the two color dual-luciferase assay over the FLuc/RLuc dual-luciferase assay. One disadvantage of the CBLuc assay is the, albeit partial, spectral overlap of light emitted by these luciferases. This necessitates the calculation of correction factors for the overlapping green and red emission spectra using separate cell lines that express the individual CBLuc variants separately (Davis et al., 2007).
Detection of the beetle luciferases (FLuc and CBLuc) in cell-based reporter gene assays used in HTS typically requires cell lysis, and as such, restricts these assays to a single endpoint measurement of luciferase activity. The use of luciferases that are naturally secreted, such as GLuc (Verhaegent and Christopoulos, 2002) or CLuc (Nakajima et al., 2004), appear suited for assays that could benefit from noninvasive detection of reporter response at different time points throughout the course of an experiment (Figure 2C). GLuc is monomeric, represents the smallest luciferase cloned to date (at ~20kDa), and contains a signal sequence required for efficient expression, secretion, and activity of the enzyme (Tannous et al., 2005). While GLuc produces brighter luminescence than RLuc, it decays extremely rapidly, requiring the use of injection-based readers (Tannous et al., 2005). To address this, GLuc variants have been constructed whose luminescence half-life is stable over several minutes, although the specific activity of these variants is significantly less. However, stable light emission appears achievable by adding 0.1% Triton to the supernatant obtained from GLuc-secreting cells (Maguire et al., 2009).
Structure and function of FLuc
FLuc is a member of a superfamily of adenylate-forming enzymes, which includes acyl- and aryl-CoA synthetases, and nonribosomal peptide synthetases (Gulick, 2009) and by primary sequence homology, appears to be most closely related to certain acyl-CoA ligases (~30% homology to plant 4-coumarate:CoA ligase) (Baldwin, 1996). Transfer of an adenylate group to a substrate to form an enzyme-associated intermediate is, mechanistically, a common mode of substrate activation in this enzyme family (Conti et al., 1996; McElroy et al., 1967).
FLuc is an ATP-hydrolyzing, decarboxylating 4-oxidoreductase that requires a luciferin substrate containing a hydroxy-benzothiazolyl-thiazoline-carboxylic acid structure (D-LH2), ATP, oxygen, and a metal cation (typically Mg2+) to produce light (Figure 3A i) (Marques and Esteves da Silva, 2009). Bioluminescence is produced from D-LH2 by FLuc through an SN2 nucleophilic displacement reaction in which the carboxylate on the thiazoline ring attacks the α-phosphoryl moiety of ATP (Figure 3A i). The first step in the reaction results in the release of PPi and the formation of an enzyme-bound luciferyl-adenylate, D-LH2-AMP (Fraga et al., 2004; Rhodes and McElroy, 1958). The second step of the reaction involves the oxidation of D-LH2-AMP by molecular oxygen. This leads to the production of an unstable luciferin dioxetanone which generates CO2 and an electronically excited oxyluciferin which, upon spontaneous decay to the ground state, generates photons of both green and red light (Figure 3A i) (Inouye, 2010; Marques and Esteves da Silva, 2009).
Figure 3. Bioluminescent and dark reactions catalyzed by FLuc.
A. The light reaction catalyzed by FLuc (i). The substrates D-luciferin (D-LH2) and ATP are used by FLuc to form a luciferyl-adenylate intermediate (LH2-AMP). This intermediate then undergoes nucleophilic attack by molecular oxygen, and upon subsequent displacement of AMP, an unstable dioxetanone is formed which then spontaneously breaks down to form oxylucifein, and CO2 with the emission of a photon (Marques and Esteves da Silva, 2009). (ii) Dark reactions catalyzed by FLuc are shown in the grey shaded area. One of these involves a side-reaction in which oxidation of LH2-AMP occurs to form the potent inhibitor L-AMP which can undergo pyrophosphorolysis or thiolysis to yield less potent inhibitors, L or L-CoA, respectively (Fontes et al., 1997). B) FLuc has also been reported to use certain fatty acids as substrates yielding fatty acyl-CoA metabolites. Kinetic constants for the synthesis of lineoic acid-CoA are taken from Oba et al. (2003). C. The potent inhibition observed for the novel 3,5-diaryloxadiazole, PTC124, is due to exploitation of a dark reaction catalyzed by FLuc in which PTC124 forms an adduct with AMP via its m-carboxylate. Also shown is the potential FLuc-catalyzed thiolysis of PTC124-AMP by CoASH to yield a metabolite, PTC124-CoA.
In addition to catalyzing a light-emitting reaction, several side-reactions that do not produce light, so-called “dark” reactions, are also catalyzed by FLuc. One such dark reaction involves the oxidation of D-LH2-AMP resulting in the formation of dehydro-luciferyl AMP (L-AMP) – a potent FLuc inhibitor (Figure 3A ii). L-AMP has been shown to be a tight-binding active site inhibitor (Rhodes and McElroy, 1958; Ribeiro and Esteves da Silva, 2008) (Figure 3A ii), and it has been documented that up to 20% of the D-LH2 substrate is diverted to formation of this inhibitory compound (Fraga et al., 2006). The L-AMP inhibitor is an example of a special type of enzyme inhibition in which the enzyme catalyzes the formation of its own inhibitor. This type of inhibitor is known as a multi-substrate adduct inhibitor (MAI), the affinity of which can be estimated from the product of the KDs for the two substrates (D-LH2 and ATP) and in the case of L-AMP, nanomolar inhibition is observed (Figure 3A ii). Interestingly, the flashing of fireflies as seen on a summer’s night may be attributed to the light-producing luciferase reaction interrupted intermittently by rapid and potent inhibition of the bioluminescent reaction through formation of L-AMP (Ghiradella and Schmidt, 2004). Regeneration of active luciferase within the firefly lantern is believed to occur either by reaction of L-AMP with pyrophosphate and/or CoASH, as breakdown of L-AMP has been observed when micromolar concentrations of these compounds are added to in vitro FLuc reactions (Fontes et al., 2008; Fraga et al., 2005) (Figure 3A ii). Both pyrophosphorolysis and thiolysis of L-AMP yield less potent (μM) inhibitors (L or L-CoA, respectively), thus facilitating competition by the D-LH2 substrate and regeneration of the active enzyme (Marques and Esteves da Silva, 2009). Commercial FLuc detection reagents often contain CoASH, and addition of ~100 μM CoASH to the FLuc reaction converts the bioluminescence from a flash to a glow response (Fraga et al., 2005).
FLuc catalysis utilizing CoASH as a substrate (Fraga et al., 2005) is not surprising considering that FLuc and fatty acyl-CoA synthetases belong to the same superfamily (Gulick, 2009; Oba et al., 2003). Indeed, FLuc can catalyze the formation of certain fatty acyl-CoAs (>C16; Figure 3B) from substrates such as arachidonic acid, albeit at a reduced catalytic efficiency relative to the luciferin reaction (Oba et al., 2003). Additionally, Oba and colleagues (2009) have been able to convert a fatty acyl-CoA synthetase derived from a non-luminous click beetle into a luciferase enzyme through point mutations of active site residues (Oba et al., 2009). These studies lend support to the premise that FLuc arose from an ancestral fatty acyl-CoA synthetase.
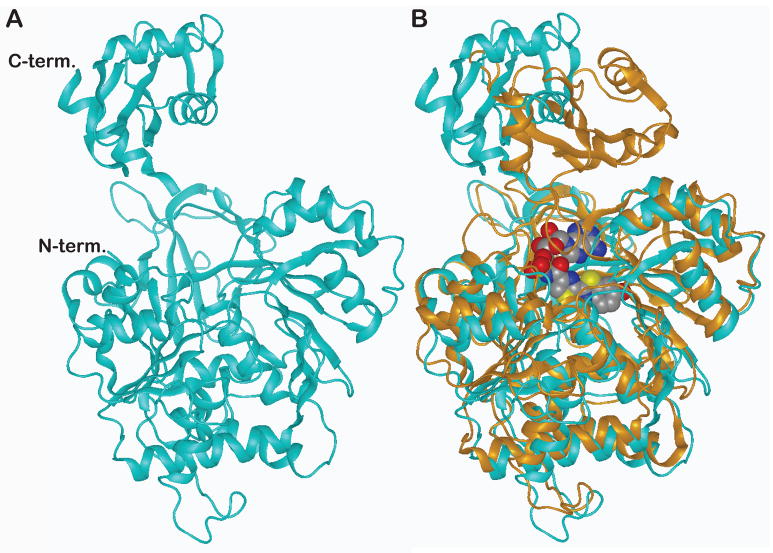
FLuc was the first enzyme in the superfamily of adenylate-forming enzymes for which the X-ray structure was determined (Conti et al., 1996). The apo FLuc structure consists of N-terminal and C-terminal domains, both containing α+β structure, that are linked by a flexible hinge region (Figure 4A) (Conti et al., 1996). The active site of the enzyme is located in the N-terminal domain near the hinge region (Conti et al., 1996). In addition, it was noted by Conti and colleagues (Conti et al., 1996) that the structure of FLuc has a novel ATP-binding motif (PDB:1LCI).
Figure 4. Structure of beetle luciferases.
A. The apo structure of FLuc as determined by Conti and colleagues (Conti et al., 1996) is shown (PDB: 1LCI) depicting the secondary α and β structural elements comprising the N- and C-terminal domains. B. Overlay of the apo FLuc structure (cyan) with the DLSA bound structure of LcrLuc (gold; PDB 2D1S). Closure of the cleft between the N- and C-terminal domains is observed in the DLSA-bound structure. The active site resides in the N-terminal domain near the cleft.
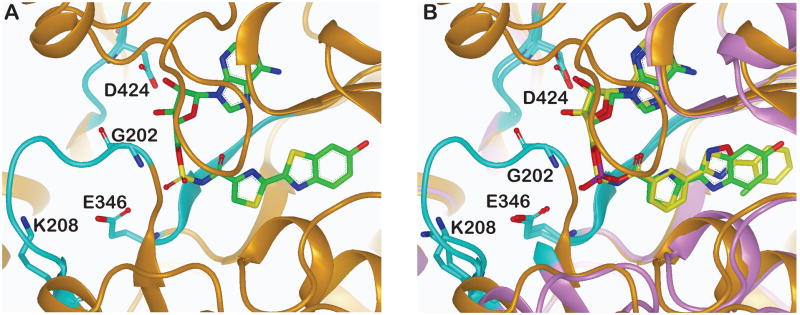
One of the most informative structures for examining the role of active site residues is the 1.3 Å resolution co-crystal structure for the firefly luciferase from Luciola cruciata (LcrLuc) bound to an analogue of the luciferyl-adenylate intermediate (5’-O-[dehydroluciferyl)-sulfamoyl]adenosine; DLSA; PDB: 2D1S) (Nakatsu et al., 2006). The structure of the DLSA-bound to beetle luciferase confirms the role of many of the invariant residues in FLuc (identified by Conti et al., 1996) that are involved with substrate binding, for example, Asp424 of LcrLuc forms H-bonds to the ribose ring of ATP (Figure 5A). Also found at the active site of LcrLuc is a nine residue peptide containing a signature sequence for this enzyme superfamily, with invariant residues Gly202 and Lys208 anchoring the ends of this peptide. A third motif is found to line the active site and contains the invariant residue Glu346 of LcrLuc. This structure confirms that the enzyme active site is lined by three regions originally identified by Conti and colleagues (Conti et al., 1996).
Figure 5. Inhibitor-bound beetle luciferase structures.
A. The LcrLuc:DLSA bound structure is shown and the three conserved motifs, originally proposed by (Conti et al., 1996) as likely candidates of the active site are shown in cyan. Several invariant residues that line the active site are highlighted (residue numbering based on LcrLuc). B. Overlay of the LcrLuc:DLSA bound structure to the FLuc:PTC124-AMP bound inhibitor complex. LcrLuc:DLSA (protein in gold; ligand in green), PTC124-AMP (protein in purple; ligand in yellow). Conserved motifs are again shown in cyan. The interactions and conformations of both ligands and invariant protein regions are highly conserved between the two structures.
Comparison of the apo FLuc (Conti et al., 1996) and LcrLuc:DLSA-bound (Nakatsu et al., 2006) structures suggest that a conformational change occurs upon substrate binding, wherein a large cleft between the N- and C-terminal domains closes (Figure 4B). Closure of this cleft is thought to increase catalytic efficiency, perhaps by water exclusion which would prevent quenching of the excited oxyluciferin product (Conti et al., 1996; Nakatsu et al., 2006). Recently, robust biosensors for HTS assays have been developed using an approach that takes advantage of this conformational change - circularly permuted reporters (Fan et al., 2008). These reporters have been used in assays to detect cAMP (and other cellular analytes), as well as intracellular protease activity (split-luciferases have also been used as intramolecular sensors; Table S1) (Binkowski et al., 2009; Fan et al., 2008).
Defining FLuc inhibitor chemotypes using quantitative HTS (qHTS)
To better understand the nature and frequency of FLuc inhibition by small molecules, we have profiled a 72K portion of the Molecular Libraries Small Molecule Repository (MLSMR; PubChem Assay Identification –AID - 411) for FLuc activity (in an assay using a commercial detection reagent formulated to measure ATP levels [Auld et al., 2008a]). Through screening at multiple compound concentrations (qHTS) (Inglese et al., 2006), potency and efficacy values were derived for every compound in this representative library. From the FLuc qHTS, we found that approximately 3% of the compounds in this screen inhibited FLuc. Potent inhibition (IC50 ≤ 10 μM), well within typical screening concentration ranges, was observed for 0.9% of the library. Major chemical series associated with FLuc inhibition included mimics of the D-LH2 substrate such as benzthiazoles, benzimidazoles, and benzoxazoles, as well as chemical series that are devoid of obvious structural similarity to either ATP or D-LH2 substrates, such as the 3,5-diaryl oxadiazoles (Figure 6A). Additional experiments confirmed that many of the benzthiazole-type compounds were competitive with D-LH2 (see Figure 6B for example).
Figure 6. Types and behavior of firefly luciferase inhibitors.
A. Potency distribution (pAC50) for four prominent chemotypes found to inhibit FLuc from profiling efforts. Shown are benzthiazoles (i), benzimidazoles (ii), benzoxazoles (iii), and 3,5-diaryl-oxadiazoles (iv). B. Comparison of FLuc and Ultra-Glo luciferase against a 2-phenylbenzothiazole luciferase inhibitor at multiple substrate concentrations. Graphs of ATP variation (main) or D-LH2 variation (inset) are shown and were varied at 0.25, 2, 25, and 250 μM, resulting in four sets of CRCs. In each case, the constant substrate was present at 250 μM. The FLuc data is shown as solid circles and the Ultra-Glo luciferase is shown as open circles. Simple benzthiazoles appear to be purely competitive with D-LH2 for either luciferase as seen from the right shift of the CRCs (decrease in potency). C. Difference in potency distribution between inhibitors identified in a commercial formulation of FLuc (PK-Light™) and a commercial formulation of Ultra-Glo (KinaseGlo™), illustrating how the source of luciferase reagent can affect in vitro assay interference. D. Comparison of FLuc and Ultra-Glo luciferase inhibition potencies for quinoline analogs assayed using KM levels of substrates. CRCs with for quinolines i (squares) and ii (circles) are shown for FLuc (solid shapes and dotted lines) and Ultra-Glo luciferase (open shapes and solid lines). Selectivity between the two luciferases is observed for such quinolines. Figures adapted from (Auld et al., 2008a; Auld et al., 2009b).
This profiling work also highlighted two key variables to consider when examining library activity against FLuc - the source of luciferase and the formulation of substrate reagents used (KM concentrations of substrates versus excess substrates typical of detection reagents). For example, the widely used luciferase-based detection reagents (Cell TiterGlo™ or biochemical assay formats based on pro-luciferin substrates, e.g. P450 Glo™) from Promega Corporation contain the P. pennsylvanica luciferase variant Ultra-Glo. Our profiling work indicates that Ultra-Glo is less likely to be inhibited by compounds used in screening (Auld et al., 2009b). Additionally, although many of the same chemotypes were found to inhibit both luciferases, these compounds were significantly less potent against a formulation of Ultra-Glo (PubChem AID: 1379; Figure 6C). An example of this is illustrated by the activity of a series of quinoline compounds, which demonstrated inhibition in the FLuc qHTS but were only weakly active against purified Ultra-Glo luciferase (Figure 6D). The quinoline/quinazoline scaffold is also found among known ATP-competitive protein kinase inhibitors. However, consistent with the unique ATP binding fold characteristic of FLuc, these specific compounds have not been found to inhibit protein kinases (Auld et al., 2008a). Interestingly, a qHTS assay for the protein kinase Clk4 constructed to detect ATP depletion or ADP production performed against a kinase-focused (quinazoline/pyrimidine) compound library using a formulation of Ultra-Glo showed potent inhibition of Clk4 but no inhibitors were identified for luciferase (PubChem AID:1379) (Tanega et al., 2009).
Another important consideration when performing a counterscreen/profile for compounds that may inhibit FLuc is detection reagent formulation. If one wants to identify even weakly competitive inhibitors, it is necessary to determine the activity of FLuc in the presence of KM concentrations of substrates. Profiling FLuc activity in the presence of detection reagents, which contain excess FLuc substrates (in addition to coenzyme A), may not identify weakly competitive inhibitors. However, if one is concerned only with identifying false positives (FLuc inhibitors) in the specific assay, detection reagents identical to those utilized in the assay can be used, as long as the FLuc enzyme concentration in the assays is very similar. Additionally, diluting commercially available FLuc detection reagents to save cost can result in detecting more FLuc inhibitors (relative to undiluted detection reagents), as the concentration of FLuc substrates is less, thus allowing inhibition by less potent FLuc inhibitors.
Effects of luciferase inhibitors in cell-based assays
Activity of FLuc in cell-based assays can be confounded by compounds that inhibit the FLuc enzyme, causing apparent inhibition (Bakhtiarova et al., 2006), or counter to one’s intuition, an increase of luminescence. It has been found that the FLuc protein is sensitive to the phenomenon of ligand-based stabilization by inhibitory compounds, leading to protein stabilization and accumulation of the affected protein (Thompson et al., 1991). This phenomenon has been observed for many proteins and has recently been described as a method to identify protein targets of compounds (Lomenick et al., 2009). Thompson and colleagues (Thompson et al., 1991) were the first to note that treatment of FLuc-expressing mammalian cells with simple benzthiazole inhibitors led to increased reporter protein levels in a transcriptional- and translational-independent manner, resulting in a counter-intuitive increase in luciferase signal upon detection.
As it turns out, intracellular stabilization of FLuc by inhibitory compounds is both a general and prevalent phenomena in cell-based assays. Using the SAR derived from the FLuc profile of the MLSMR described above, we performed a retrospective analysis of HTS experiments available within PubChem of cell-based assays that used FLuc as the reporter (Auld et al., 2008b). As expected we found an enrichment of FLuc inhibitors among compounds that scored positive in cell-based assays aimed at measuring a decrease in FLuc activity (38±8% of the positives were FLuc inhibitors). However, we also found a significant enrichment of FLuc inhibitors in cell-based assays designed to measure an increase in FLuc activity (46±10% of the positives were FLuc inhibitors), a result indicative of the apparent signal activation characteristic of inhibitor-based stabilization of FLuc. Inhibitor-based stabilization was found to occur for all the major chemotypes identified in the original profile study (Figure 6A). This finding supported previous work which indicated that intracellular FLuc inhibition can lead to an apparent activation response in cell-based assays - a counter-intuitive result for an enzyme inhibitor - which can confound the identification of compounds with target activity.
It is not always obvious, in cell-based assays, to identify compounds that cause FLuc inhibitor-based stabilization. This is because the type of inhibition (reversible, irreversible, competitive or non-competitive), affinity of the inhibitor for FLuc, basal luciferase expression, and assay format and protocol all influence whether or not the compound appears to cause an increase or decrease in FLuc activity at the compound concentration tested. Additionally, concentration response curves (CRCs) generated for these compounds in the cell-based FLuc assay can be sigmoidal or bell-shaped (Figure 7A) (Auld et al., 2008a; Inglese et al., 2009; Peltz et al., 2009). Upon incubation of compound with cells, a stable E•I complex is formed that prevents or slows FLuc enzyme degradation, thus allowing enzyme levels to increase in cell culture over time relative to controls not treated with inhibitor compound. Further, the concentration of E will be modulated by the affinity of the inhibitor (KI) for the free enzyme. When the ratio of inhibitor to KI is high, the slower degrading E•I complex is maximized and degradation is minimized. This model predicts that at different concentrations of I the amount of active enzyme will follow a CRC with an apparent EC50 centered on the KI value (Figure 7A). However, detection of FLuc activity is possible for cells treated with competitive FLuc inhibitors because these compounds are competitively displaced by luciferase detection reagents that contain high concentrations of FLuc substrates (ATP, D-LH2, and CoASH). When these inhibitors are competed off FLuc by excess substrates, the enzyme is able to catalyze the luminescence reaction, resulting in an apparent increase in assay signal, due to the accumulated FLuc. To reiterate, in the case of inhibitor-based stabilization, a competitive FLuc inhibitor will cause an apparent decrease in reporter activity when substrate levels are ≤ KM (as FLuc remains inhibited), however, when substrate levels are high enough to compete with the inhibitory compound (as is the case for detection reagents), an increase in reporter activity is detected. Experimentally, a concentration-dependent composite of FLuc activation due to stabilization at mid-level concentrations of compound followed by inhibition of FLuc activity at higher inhibitor concentrations, leading to bell-shaped CRCs, is observed (Figure 7B). In some cases, the inhibitory compounds may be non-competitive with FLuc substrates - for example, covalent irreversible inactivation, or possibly inhibitors that destabilize the FLuc enzyme intracellularly - which would lead to an apparent inhibitory response upon detection (Figure 7A).
Figure 7. Post-translational inhibitor-based reporter-stabilization.
A. Level of apparent reporter activation depends on properties of the luciferase inhibitor and the assay detection protocol used. Shown is the theoretically observed activity of the reporter enzyme in the presence of a FLuc inhibitor as determined in a biochemical assay in which substrate concentrations are held near their KM’s (orange line). Inset graph, the theoretical increase in enzyme concentration in the cell-based assay occurring as a result of inhibitor-based stabilization. Theoretical reporter activity in the cell-based assay for a fully reversible FLuc inhibitor is shown (blue line). In this case addition of excess beetle luciferase substrates in detection reagents fully relieves inhibition by the competitive inhibitor, causing apparent activation of FLuc that shows compound concentration dependence. This type of CRC can also result when a reversible inhibitor is removed by washing cells prior to detection. The theoretical activity for an irreversible FLuc inhibitor is shown for both a biochemical and cell-based assay (red line). Here, only inhibition of the reporter activity is observed in the cell-based assay. B. Observed effect for a FLuc inhibitor (PTC124) using a no-wash protocol and Steady-Glo® detection reagent (Promega). The top graph shows FLuc activity obtained from HEK293 cells transfected with the construct pGL3 (Promega; E1741) after treatment with PTC124 for 72 hrs (orange). The pGL3 construct contains a wild-type version of FLuc under the control of a SV40 promoter. A bell-shaped CRC is observed in the cell based assay due to the persistent inhibition of FLuc at high concentrations of compound. Also shown is the activity of PTC124 against purified FLuc assayed using KM levels of substrate (black data; bottom graph). Data is from Auld et al., (2009a) C. Experimental results for the three regioisomers shown at right, for the FLucUGA cell-based assay (top) and the enzyme assay at KM levels of substrate (bottom). Data is from Auld et al., (2010). Apparent activation in the cell-based assay and potency in the biochemical assay correlate with the reactivity of the carboxylate regioisomers for MAI formation (colors correspond to the structures shown).
Inhibition of enzymatic reporters is not limited to FLuc, and RLuc inhibitors have also been identified. In fact, the well-used tool compound H-89, used to modulate the protein kinase PKA (Herbst et al., 2009), inhibits RLuc. In addition, post-translational ligand-based reporter stabilization is not specific to FLuc, but has also been observed for RLuc (Auld et al., 2009a). Although RLuc has not been broadly profiled against a large chemical library, a pilot screen of a library available from Tocris Inc. indicated that approximately 10% of the library inhibited RLuc (in an assay using KM levels of coelenterazine; unpublished observation). Thus, it seems that profiling for RLuc inhibitors in small molecule libraries would prove informative and useful.
Role of dark reactions in the formation of novel potent inhibitors
One of the most potent synthetic FLuc inhibitors identified to date is PTC124 (Ataluren™), a compound derived from a series of FLuc assays aimed at identifying nonsense codon suppression agents (Welch et al., 2007). PTC124 possesses a 3,5-diaryloxadiazole scaffold - one of the prominent inhibitor scaffolds identified in the FLuc profile of the MLSMR chemical library (Figure 6A, iv). To examine if inhibitor-based reporter stabilization could explain the apparent activation of cellular FLuc activity by PTC124, we constructed the primary cell-based assay used to discover PTC124 -the FLucUGA assay in which cells are either stably or transiently transfected with a construct that contains the FLuc cDNA with a UGA stop codon. Compounds that appear to increase FLuc activity, presumably due to enhanced readthrough of the UGA stop codon and production of functional FLuc protein, are considered active in the assay. We were able to reproduce the apparent activation by PTC124 and related analogs in this cell-based assay. We also found that this assay was highly sensitive to FLuc stabilization by inhibitor compounds, as extremely low, but detectable, levels of nonspecific read-through of the FLucUGA gene provided a pool of FLuc protein with which compound could associate, eventually leading to accumulation of FLuc protein inside the cell (Auld et al., 2009a; Peltz et al., 2009). We also observed that activation in the cell-based FLucUGA assay mirrored the inhibition by the compounds against the purified FLuc enzyme, lending support to the premise that activation in the cell-based assay was due to formation of a stable cellular E•I complex. Additionally, we found that PTC124 was not active in an analogous RLuc UGA cell-based assay. Correspondingly, RLuc enzymatic activity was found to be refractory to inhibition by PTC124 and other structurally related diaryl-oxadiazole analogs, demonstrating that PTC124 activation in the FLucUGA cell-based assay was reporter-specific and independent of transcriptional or translational mechanisms. This study demonstrated that the discovery of PTC124 was highly biased by the FLuc reporter (Auld et al., 2009a).
The X-ray crystal structure of the E•I complex was determined in an effort to understand the structural basis for high affinity binding and stabilization of FLuc by PTC124 (Auld et al., 2010). Remarkably, when FLuc was crystallized in the presence of both PTC124 and ATP, a PTC124-AMP adduct was revealed in the active site of FLuc (Auld et al., 2010). Subsequent experiments confirmed that FLuc catalyzes the formation of this adenylate with PTC124 through nucleophilic displacement of the pyrophosphate from ATP by the m-carboxylate of PTC124 (Auld et al., 2010). In addition, PTC124-AMP was found to bind with high affinity to FLuc with a KD ~ 120 pM (Auld et al., 2010) (Figure 3C).
The formation of PTC124-AMP by FLuc exemplifies a dark reaction catalyzed by FLuc analogous to the naturally occurring side-reaction that results in L-AMP formation from LH2-AMP formation (see section above and Figure 3Aii, C). In fact, it appears that PTC124-AMP represents a structural mimic of the LH2-AMP intermediate. Indeed, overlay of the FLuc:PTC-AMP structure with the LcrLuc:DLSA structure indicated essentially identical binding modes and conformations of critical invariant residues in the FLuc/LcrLuc active sites with the respective MAI bound (Figure 5B) (Auld et al., 2010). Further, the precise positioning of the PTC124 carboxylate in the active site of the enzyme is required for efficient enzymatic formation of the MAI, which would explain why simple aromatic acids have not been observed as FLuc inhibitors. The m-carboxylate of PTC124 is optimal for formation of the MAI, as demonstrated by the reduced or eliminated ability of para- or ortho-carboxylate analogs of PTC124, respectively, to form MAIs. We reasoned that this is due to kinetic and energetic barriers which impair the formation of the proper near-attack conformers (NACs) required for the nucleophilic displacement reaction. Our studies indicate that the o-carboxylate cannot achieve the required NAC, as this would require rotation of the rigid oxadiazole core outside the luciferin pocket, and that MAI formation by the p-carboxylate analog is kinetically unfavorable (Auld et al., 2010).
Not surprisingly, formation of the PTC124-AMP MAI correlated with stabilization of the FLuc protein. Testing PTC124 and analogs, in addition to the PTC124-AMP adduct, in a thermal denaturation assay, showed PTC124-AMP to produce the greatest change in TM (ΔTM ~30°C), and that thermal stabilization of FLuc correlated with the ability the PTC124 carboxylate regioisomers to form the MAI (Auld et al., 2010). In addition, we found that the potency in both the purified FLuc enzyme assay and FLucUGA assay correlated with MAI formation - following the reactivity of the carboxylate with meta > para ≫ortho (Figure 7C; see also (Auld et al., 2010).
While these findings describe how the FLuc reporter catalyzes the production of a potent, stabilizing MAI (PTC124-AMP), less clear is how FLuc activity could be measured during the detection step of a cell-based FLuc reporter gene assay in the presence of this potent inhibitor. On the contrary, a decrease in luminescence would be expected due to the extremely high affinity of PTC124 for FLuc. Apparent activation of FLuc in cell-based assays upon PTC124 treatment can be explained by the presence of CoASH in detection reagents. As mentioned previously, FLuc catalyzes thiolysis of the potent L-AMP inhibitor with CoASH (Figure 3A ii). We also found that the potency of PTC124 is reduced by approximately 35-fold in the presence of 500μM CoASH, a concentration likely present in luciferase detection reagents. Formation of PTC124-CoA through a similarly FLuc-catalyzed thiolysis reaction would thus explain why PTC124 appears to increase, as oppose to inhibit, FLuc activity in an endpoint assay: PTC124-CoA (Figure 3C), which is expected to be a less potent inhibitor, could more readily dissociate and be competed by excess D-LH2 present in detection reagents.
It is routinely observed that the potency of a compound against its molecular target appears weaker in cellular settings due to the presence of competitive co-factors or substrates. In this case, intracellular reactivity or competition of the MAI by free CoASH or ATP, respectively could explain why PTC124 potency is nanomolar instead of picomolar in the cell-based assay. Thus, although activation in the cell-based FLucUGA assay has been used to suggest that PTC124 is a potent nonsense codon suppression agent (Peltz et al., 2009; Welch et al., 2007), a biologically plausible mechanism for these results is that FLuc is the molecular target of PTC124 in FLuc-cell-based assays.
Interpretation of FLuc assay results
The manifestation of the interaction of PTC124 with the FLuc reporter in biochemical and cell-based assays – as an inhibitor or activator of FLuc depending on the assay format - illustrates how manipulation of biology to accommodate a reporter output can introduce unexpected new complexities. For compounds identified as active in FLuc assays, it is essential to determine if the compound has activity attributable to a direct interaction with FLuc. A simple way to do this is to measure the potency of the compound(s) against purified FLuc using KM concentrations of substrates. In addition, PubChem is a publically available resource that contains HTS data from many different types of assays, and can be used to determine if certain structural classes of compounds display activity in other FLuc assays. While these datasets have been useful for defining common FLuc inhibitor chemotypes, it should be noted that they are not the optimal counter-screen for cell-based assay results. This is because the majority of this data is based on assay formats that involve FLuc detection reagents which often contain excess substrates, which may mask the inhibitory activity of substrate-competitive inhibitors. This is a particularly important consideration as not all competitive inhibitors of FLuc are obviously structurally related to the D-LH2 substrate, a good example being the 3,5-diaryloxadiazoles. To have greater certainty that a given compound doesn’t inhibit FLuc, we recommend an enzyme assay conducted with KM levels of ATP and D-LH2 substrates (Auld et al., 2009a; Auld et al., 2008b), where it is likely that the measured IC50 will approximate the KI (Cheng and Prusoff, 1973).
Ideally, it is always preferable to pursue compounds from FLuc reporter assays that have no activity against FLuc. However, if it is necessary to pursue compound(s) that potentially have dual activity – that is, activity against FLuc and the target of interest - a rigorous orthogonal assay should always be performed to confirm activity at the desired target. An orthogonal assay is directed against the same target in essentially the identical assay system, but utilizes a different mode of detection (Thorne et al., 2010). For FLuc-based assays, reporters such as RLuc, β-lactamase, or GFP can be used to test for reporter-specific effects. An orthogonal assay is ideally performed in advance of secondary assays to confirm that the activity of the compound(s) is reporter independent. Secondary assays are used to test the activity of the compounds found active in the primary (and orthogonal) assay (Thorne et al., 2010). It has been our experience that when more complex secondary assays (e.g. qRT-PCR, immunohistochemistry, Western blots) are used to define compound activity without first establishing reporter-independent activity, a common outcome is inconclusive or ambiguous results that may be misinterpreted.
Certain cell-based FLuc assays are more susceptible to FLuc inhibitor-based reporter stabilization than others. These cell-based assays tend to generate an initially low basal level of FLuc protein and thus FLuc signal, and active compounds are readily identified as those that cause an apparent increase in FLuc activity, an effect that is augmented with longer-compound incubation times. Therefore, if separate FLuc cell-based assays are intended as a counter-screen to eliminate compounds that may be FLuc inhibitors, it is important that the following variables be similar between the two assays, as each can affect apparent activation: basal level of luciferase signal, compound exposure time, and detection protocol. One research group developed a FLuc reporter with attenuated expression (e.g. an SV40 promoter) to mimic low basal levels of FLuc for the purpose of using the resulting data as a counter-screen (Lyssiotis et al., 2009).
Given all the complexities inherent to FLuc, the question that comes to mind is whether this enzymatic reporter is a good choice for the design of cell-based assays aimed at compound discovery efforts. It is our experience that the sensitivity inherent to FLuc as a reporter is currently required for many assays, and with its ease of use, it provides a robust assay design for use with HTS. Additionally, it is important to note that the artifacts associated with luciferases can be understood in relation to the enzymology of the reporter, making their activity relatively predictable. In contrast, artifacts associated with other assay formats, like fluorescence, can be less predictable (Simeonov et al., 2008). Further research and development of luciferases that demonstrate improved resistance to small molecule inhibition and reporter stabilization would be valuable.
Conclusion
As described in this review, the interaction of small molecules with luciferases can significantly interfere with HTS results. Oftentimes, this interaction is not recognized by researchers because of the insidious nature of the effect – for example, apparent activation of luciferase by compounds that inhibit the enzyme in cell-based assays. However, deceptive interactions of small molecules with assay components is not limited to the reporter FLuc. Recently, the activity of compounds thought to enhance the affinity of the human deacetylase SIRT1 for an acetylated p53 peptide (Howitz et al., 2003; Milne et al., 2007) have been shown to be confounded by the fluorescent-tag on the peptide used in the primary assays (Kaeberlein et al., 2005; Pacholec et al., 2010). For these reasons, we are trying to implement the policy of confirming compound activity in orthogonal assays, that is, an assay that uses a different reporter or assay format, to establish reporter-independent activity prior to experiments that inherently have more variables. Widespread use of FLuc reporter assays in chemical biology arose from their initial use as a reporter in molecular biology applications to study, for example, regulation of gene expression. However, adaptation of these reporter assays for HTS of large chemical libraries creates an underappreciated complexity that must be considered upon interpretation of results derived from these assays – how compounds interact and affect reporter enzymology and cellular half-life. Studies aimed at acquiring a better understanding of compound-reporter interactions, and the factors that can subvert meaningful results, should help the identification of improved leads for drug discovery and chemical biology research.
Supplementary Material
Acknowledgments
We thank Dr. Ryan MacArthur for performing the analysis on the distribution of assay types in PubChem and Dr. Min Shen for providing figures of the X-ray structures. The NIH Chemical Genomics Center is supported by funding from the Molecular Libraries Initiative of the NIH Roadmap for Medical Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almond B, Hawkins E, Stecha P, Garvin D, Paguio A, Butler B, Beck M, Wood M, Wood K. Introducing Chroma-Luc technology. Promega Notes. 2003;85:11–14. [Google Scholar]
- Auld DS, Lovell S, Thorne N, Lea WA, Maloney DJ, Shen M, Rai G, Battaile KP, Thomas CJ, Simeonov A, et al. Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124. Proc Natl Acad Sci U S A. 2010;107:4878–4883. doi: 10.1073/pnas.0909141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld DS, Southall NT, Jadhav A, Johnson RL, Diller DJ, Simeonov A, Austin CP, Inglese J. Characterization of chemical libraries for luciferase inhibitory activity. J Med Chem. 2008a;51:2372–2386. doi: 10.1021/jm701302v. [DOI] [PubMed] [Google Scholar]
- Auld DS, Thorne N, Maguire WF, Inglese J. Mechanism of PTC124 activity in cell-based luciferase assays of nonsense codon suppression. Proc Natl Acad Sci U S A. 2009a;106:3585–3590. doi: 10.1073/pnas.0813345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld DS, Thorne N, Nguyen DT, Inglese J. A specific mechanism for nonspecific activation in reporter-gene assays. ACS Chem Biol. 2008b;3:463–470. doi: 10.1021/cb8000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld DS, Zhang YQ, Southall NT, Rai G, Landsman M, Maclure J, Langevin D, Thomas CJ, Austin CP, Inglese J. A Basis for Reduced Chemical Library Inhibition of Firefly Luciferase Obtained from Directed Evolution. J Med Chem. 2009b;52:1450–1458. doi: 10.1021/jm8014525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhtiarova A, Taslimi P, Elliman SJ, Kosinski PA, Hubbard B, Kavana M, Kemp DM. Resveratrol inhibits firefly luciferase. Biochem Biophys Res Commun. 2006;351:481–484. doi: 10.1016/j.bbrc.2006.10.057. [DOI] [PubMed] [Google Scholar]
- Baldwin TO. Firefly luciferase: the structure is known, but the mystery remains. Structure. 1996;4:223–228. doi: 10.1016/s0969-2126(96)00026-3. [DOI] [PubMed] [Google Scholar]
- Binkowski B, Fan F, Wood K. Engineered luciferases for molecular sensing in living cells. Curr Opin Biotechnol. 2009;20:14–18. doi: 10.1016/j.copbio.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Cali JJ, Ma D, Sobol M, Simpson DJ, Frackman S, Good TD, Daily WJ, Liu D. Luminogenic cytochrome P450 assays. Expert Opin Drug Metab Toxicol. 2006;2:629–645. doi: 10.1517/17425255.2.4.629. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996;4:287–298. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein engineering. 1999;12:1035–1040. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- Davis RE, Zhang YQ, Southall N, Staudt LM, Austin CP, Inglese J, Auld DS. A Cell-Based Assay for IkappaBalpha Stabilization Using A Two-Color Dual Luciferase-Based Sensor. Assay Drug Dev Technol. 2007;5:85–104. doi: 10.1089/adt.2006.048. [DOI] [PubMed] [Google Scholar]
- de Wet JR, Wood KV, DeLuca M, Helinski DR, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wet JR, Wood KV, Helinski DR, DeLuca M. Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc Natl Acad Sci U S A. 1985;82:7870–7873. doi: 10.1073/pnas.82.23.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer BW, Ferrer FA, Klinedinst DK, Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal Biochem. 2000;282:158–161. doi: 10.1006/abio.2000.4605. [DOI] [PubMed] [Google Scholar]
- Fan F, Binkowski BF, Butler BL, Stecha PF, Lewis MK, Wood KV. Novel genetically encoded biosensors using firefly luciferase. ACS Chem Biol. 2008;3:346–351. doi: 10.1021/cb8000414. [DOI] [PubMed] [Google Scholar]
- Fan F, Wood KV. Bioluminescent assays for high-throughput screening. Assay Drug Dev Technol. 2007;5:127–136. doi: 10.1089/adt.2006.053. [DOI] [PubMed] [Google Scholar]
- Fontes R, Dukhovich A, Sillero A, Sillero MA. Synthesis of dehydroluciferin by firefly luciferase: effect of dehydroluciferin, coenzyme A and nucleoside triphosphates on the luminescent reaction. Biochem Biophys Res Commun. 1997;237:445–450. doi: 10.1006/bbrc.1997.7161. [DOI] [PubMed] [Google Scholar]
- Fontes R, Fernandes D, Peralta F, Fraga H, Maio I, Esteves da Silva JC. Pyrophosphate and tripolyphosphate affect firefly luciferase luminescence because they act as substrates and not as allosteric effectors. FEBS J. 2008;275:1500–1509. doi: 10.1111/j.1742-4658.2008.06309.x. [DOI] [PubMed] [Google Scholar]
- Fraga H, Esteves da Silva JC, Fontes R. Identification of luciferyl adenylate and luciferyl coenzyme a synthesized by firefly luciferase. Chembiochem. 2004;5:110–115. doi: 10.1002/cbic.200300735. [DOI] [PubMed] [Google Scholar]
- Fraga H, Fernandes D, Fontes R, Esteves da Silva JC. Coenzyme A affects firefly luciferase luminescence because it acts as a substrate and not as an allosteric effector. FEBS J. 2005;272:5206–5216. doi: 10.1111/j.1742-4658.2005.04895.x. [DOI] [PubMed] [Google Scholar]
- Fraga H, Fernandes D, Novotny J, Fontes R, Esteves da Silva JC. Firefly luciferase produces hydrogen peroxide as a coproduct in dehydroluciferyl adenylate formation. Chembiochem. 2006;7:929–935. doi: 10.1002/cbic.200500443. [DOI] [PubMed] [Google Scholar]
- Ghiradella H, Schmidt JT. Fireflies at One Hundred Plus: A New Look at Flash Control. INTEGR COMP BIOL. 2004;44:203–212. doi: 10.1093/icb/44.3.203. [DOI] [PubMed] [Google Scholar]
- Greer LF, 3rd, Szalay AA. Imaging of light emission from the expression of luciferases in living cells and organisms: a review. Luminescence. 2002;17:43–74. doi: 10.1002/bio.676. [DOI] [PubMed] [Google Scholar]
- Gulick AM. Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem Biol. 2009;4:811–827. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah RR, Jennens-Clough ML, Wood KV. Rapid luciferase reporter assay systems for high throughput studies. Promega Notes. 1998;65:9–14. [Google Scholar]
- Hart RC, Matthews JC, Hori K, Cormier MJ. Renilla reniformis bioluminescence: luciferase-catalyzed production of nonradiating excited states from luciferin analogues and elucidation of the excited state species involved in energy transfer to Renilla green fluorescent protein. Biochemistry. 1979;18:2204–2210. doi: 10.1021/bi00578a011. [DOI] [PubMed] [Google Scholar]
- Hart RC, Stempel KE, Boyer PD, Cormier MJ. Mechanism of the enzyme-catalyzed bioluminescent oxidation of coelenterate-type luciferin. Biochem Biophys Res Commun. 1978;81:980–986. doi: 10.1016/0006-291x(78)91447-x. [DOI] [PubMed] [Google Scholar]
- Herbst KJ, Allen MD, Zhang J. The cAMP-dependent protein kinase inhibitor H-89 attenuates the bioluminescence signal produced by Renilla Luciferase. PLoS One. 2009;4:e5642. doi: 10.1371/journal.pone.0005642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino H. Current advanced bioluminescence technology in drug discovery. Expert Opinion on Drug Discovery. 2009;9:374–389. doi: 10.1517/17460440902804372. [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- Inglese J, Thorne N, Auld DS. Reply to Peltz et al: Post-translational stabilization of the firefly luciferase reporter by PTC124 (Ataluren) PNAS. 2009;106:E65. [Google Scholar]
- Inouye S. Firefly luciferase: an adenylate-forming enzyme for multicatalytic functions. Cell Mol Life Sci. 2010;67:387–404. doi: 10.1007/s00018-009-0170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, et al. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- Loening AM, Fenn TD, Gambhir SS. Crystal structures of the luciferase and green fluorescent protein from Renilla reniformis. J Mol Biol. 2007;374:1017–1028. doi: 10.1016/j.jmb.2007.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S, Wang J, Wu RP, Gomez F, Loo JA, et al. Target identification using drug affinity responsive target stability (DARTS) Proc Natl Acad Sci U S A. 2009;106:21984–21989. doi: 10.1073/pnas.0910040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz WW, McCann RO, Longiaru M, Cormier MJ. Isolation and expression of a cDNA encoding Renilla reniformis luciferase. Proc Natl Acad Sci U S A. 1991;88:4438–4442. doi: 10.1073/pnas.88.10.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssiotis CA, Foreman RK, Staerk J, Garcia M, Mathur D, Markoulaki S, Hanna J, Lairson LL, Charette BD, Bouchez LC, et al. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc Natl Acad Sci U S A. 2009;106:8912–8917. doi: 10.1073/pnas.0903860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire CA, Deliolanis NC, Pike L, Niers JM, Tjon-Kon-Fat LA, Sena-Esteves M, Tannous BA. Gaussia Luciferase Variant for High-Throughput Functional Screening Applications. Anal Chem. 2009;81:7102–7106. doi: 10.1021/ac901234r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques SM, Esteves da Silva JC. Firefly bioluminescence: a mechanistic approach of luciferase catalyzed reactions. IUBMB Life. 2009;61:6–17. doi: 10.1002/iub.134. [DOI] [PubMed] [Google Scholar]
- Matthews JC, Hori K, Cormier MJ. Substrate and substrate analogue binding properties of Renilla luciferase. Biochemistry. 1977;16:5217–5220. doi: 10.1021/bi00643a009. [DOI] [PubMed] [Google Scholar]
- McElroy WD, DeLuca M, Travis J. Molecular uniformity in biological catalyses. The enzymes concerned with firefly luciferin, amino acid, and fatty acid utilization are compared. Science. 1967;157:150–160. doi: 10.1126/science.157.3785.150. [DOI] [PubMed] [Google Scholar]
- Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y, Kobayashi K, Yamagishi K, Enomoto T, Ohmiya Y. cDNA cloning and characterization of a secreted luciferase from the luminous Japanese ostracod, Cypridina noctiluca. Biosci Biotechnol Biochem. 2004;68:565–570. doi: 10.1271/bbb.68.565. [DOI] [PubMed] [Google Scholar]
- Nakatsu T, Ichiyama S, Hiratake J, Saldanha A, Kobashi N, Sakata K, Kato H. Structural basis for the spectral difference in luciferase bioluminescence. Nature. 2006;440:372–376. doi: 10.1038/nature04542. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Morange M, Bensaude O. Firefly luciferase luminescence assays using scintillation counters for quantitation in transfected mammalian cells. Anal Biochem. 1988;171:404–408. doi: 10.1016/0003-2697(88)90505-2. [DOI] [PubMed] [Google Scholar]
- Oba Y, Iida K, Inouye S. Functional conversion of fatty acyl-CoA synthetase to firefly luciferase by site-directed mutagenesis: a key substitution responsible for luminescence activity. FEBS Lett. 2009;583:2004–2008. doi: 10.1016/j.febslet.2009.05.018. [DOI] [PubMed] [Google Scholar]
- Oba Y, Ojika M, Inouye S. Firefly luciferase is a bifunctional enzyme: ATP-dependent monooxygenase and a long chain fatty acyl-CoA synthetase. FEBS Lett. 2003;540:251–254. doi: 10.1016/s0014-5793(03)00272-2. [DOI] [PubMed] [Google Scholar]
- Ow DW, JR, DEW, Helinski DR, Howell SH, Wood KV, Deluca M. Transient and Stable Expression of the Firefly Luciferase Gene in Plant Cells and Transgenic Plants. Science. 1986;234:856–859. doi: 10.1126/science.234.4778.856. [DOI] [PubMed] [Google Scholar]
- Pacholec M, Chrunyk BA, Cunningham D, Flynn D, Griffith DA, Griffor M, Loulakis P, Pabst B, Qiu X, Stockman B, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz SW, Welch EM, Jacobson A, Trotta CR, Naryshkin N, Sweeney HL, Bedwel DM. Nonsense suppression activity of PTC124 (ataluren) Proc Natl Acad Sci U S A. 2009;106:E64. doi: 10.1073/pnas.0901936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes WC, McElroy WD. The synthesis and function of luciferyl-adenylate and oxyluciferyl-adenylate. J Biol Chem. 1958;233:1528–1537. [PubMed] [Google Scholar]
- Ribeiro C, Esteves da Silva JCG. Kinetics of inhibition of firefly luciferase by oxyluciferin and dehydroluciferyl-adenylate. Photochem Photobiol Sci. 2008;7:1085 – 1090. doi: 10.1039/b809935a. [DOI] [PubMed] [Google Scholar]
- Roda A, Guardigli M, Michelini E, Mirasoli M. Bioluminescence in analytical chemistry and in vivo imaging. Trends in Analytical Chemistry. 2009;28:307–322. [Google Scholar]
- Simeonov A, Jadhav A, Thomas CJ, Wang Y, Huang R, Southall NT, Shinn P, Smith J, Austin CP, Auld DS, et al. Fluorescence spectroscopic profiling of compound libraries. J Med Chem. 2008;51:2363–2371. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- Singh P, Harden BJ, Lillywhite BJ, Broad PM. Identification of kinase inhibitors by an ATP depletion method. Assay Drug Dev Technol. 2004;2:161–169. doi: 10.1089/154065804323056503. [DOI] [PubMed] [Google Scholar]
- Stables J, Scott S, Brown S, Roelant C, Burns D, Lee MG, Rees S. Development of a dual glow-signal firefly and Renilla luciferase assay reagent for the analysis of G-protein coupled receptor signalling. J Recept Signal Transduct Res. 1999;19:395–410. doi: 10.3109/10799899909036660. [DOI] [PubMed] [Google Scholar]
- Tanega C, Shen M, Mott BT, Thomas CJ, MacArthur R, Inglese J, Auld DS. Comparison of bioluminescent kinase assays using substrate depletion and product formation. Assay Drug Dev Technol. 2009;7:606–614. doi: 10.1089/adt.2009.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Thompson JF, Hayes LS, Lloyd DB. Modulation of firefly luciferase stability and impact on studies of gene regulation. Gene. 1991;103:171–177. doi: 10.1016/0378-1119(91)90270-l. [DOI] [PubMed] [Google Scholar]
- Thorne N, Auld DS, Inglese J. Apparent Activity in High-Throughput Screening: Origins of Compound-Dependent Assay Interference. Curr Opin Chem Biol. 2010;14:315–324. doi: 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaegent M, Christopoulos TK. Recombinant Gaussia luciferase. Overexpression, purification, and analytical application of a bioluminescent reporter for DNA hybridization. Anal Chem. 2002;74:4378–4385. doi: 10.1021/ac025742k. [DOI] [PubMed] [Google Scholar]
- Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- Wood KV. The chemical mechanism and evolutionary development of beetle bioluminescence. Photochemistry and Photobiology. 1995;62:662–673. [Google Scholar]
- Wood KV. The chemistry of bioluminescent reporter assays. Promega Notes. 1998;65:14–20. [Google Scholar]
- Wood KV, Lam YA, McElroy WD, Seliger HH. Bioluminescent click beetles revisited. J Biolumin Chemilumin. 1989;4:31–39. doi: 10.1002/bio.1170040110. [DOI] [PubMed] [Google Scholar]
- Wurdinger T, Badr C, Pike L, de Kleine R, Weissleder R, Breakefield XO, Tannous BA. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat Methods. 2008;5:171–173. doi: 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.