Unstructured peptides merely garnished with moieties used for recognition rarely, if ever, possess function. By contrast, presentation of the same moieties by a globular protein can result in a high affinity ligand. Recently we described a strategy for the design of miniature functional proteins in which DNA-binding residues from the bZIP protein GCN4 were grafted on to the solvent exposed α-helix of avian pancreatic polypeptide (aPP).1,2,3 aPP contains a close-packed hydrophobic core comprised of six residues from the α–helix (L17, F20, L24, Y27, L28, V30 and V31) and four residues from a type II polyproline (PPII) helix (G1, P2, Q4, P5, and Y7) (Figure 1).4 In our first grafted peptide (PPBR4), Y27, L28 and V30 were changed to arginine to preserve DNA affinity and V31 was changed to alanine to improve helical propensity.1 These mutations destroyed the aPP core, and PPBR4 exhibited only nascent helicity at 4 °C and no DNA binding at ambient temperature. Mutations at positions 27, 28, 30, and 31 failed to produce molecules that were both helical and bound DNA.5
Figure 1.
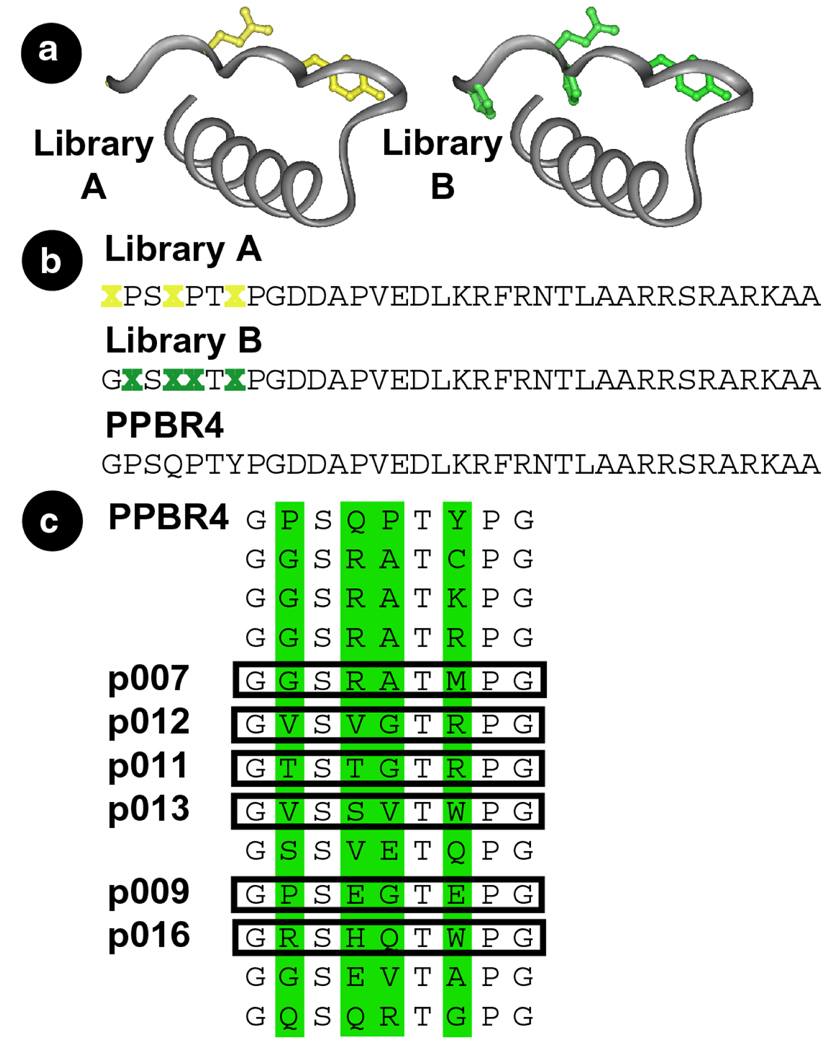
(a) Residues of PPBR4 targeted for variation mapped onto the crystal structure of aPP.4 Side chains varied in library A are in yellow, those varied in library B are in green. (b) Sequences of PPBR4 and the two libraries. Residues varied are indicated by an X. Each position was randomized at the DNA level using the NNS codon scheme.6 (c) Sequences of the N-terminal amino acids deduced from the DNA sequences of library B clones after three selection rounds. Peptides containing the boxed sequences followed by the remaining residues of PPBR4 were synthesized and their properties investigated in vitro.
We reasoned that some combinations of amino acids at positions 1, 2, 4, 5, and 7 of PPBR4 might rebuild the damaged hydrophobic core and pre-organize the α–helical residues into a conformation closer to that required for DNA binding. If so, peptides containing such sequences should bind specific DNA with high affinity. We conceived to locate appropriately pre-organized miniature proteins by affinity purification of a library of PPBR4 analogs containing mutations in the N-terminal PPII helix.
Two phage libraries were created to identify appropriately folded PPBR4 analogs (Figure 1). The members of libraries A and B differ from PPBR41 at 3 (library A) or 4 (library B) positions on the PPII helix. The proline residues retained at positions 2 and 5 of library A are highly conserved among PP-fold proteins.8 We anticipated that retention of these two prolines would effectively constrain the conformational space available to library A members9 and that most would contain N–terminal PPII helices. Such conformational constraints are absent in library B, acknowledging that there may be many ways to stabilize DNA-bound α–helices.10 Since the amino acids at positions 2 and 5 of library B are not restricted to proline, we anticipated that this library would sample a larger fraction of available phi-psi space.
Phage were sorted for three rounds on the basis of their ability to bind an oligonucleotide duplex containing the sequence ATGAC (hsCRE). To favor identification of sequences that bound hsCRE with high affinity at ambient temperature, two rounds of selection at 4 °C were followed by a single round at room temperature. By the final round, library A phage were retained at a level only comparable to PPBR4 phage and were not considered further. Library B phage were retained at a level comparable to PPBR4 phage after the first round, but at levels 15–16 times better than PPBR4 phage after the subsequent two rounds. Twelve library B clones were sequenced (Figure 1c) after round 3. Six sequences (p007, p009, p011, p012, p013, and p016) were synthesized and the DNA-binding properties of four analyzed in detail.
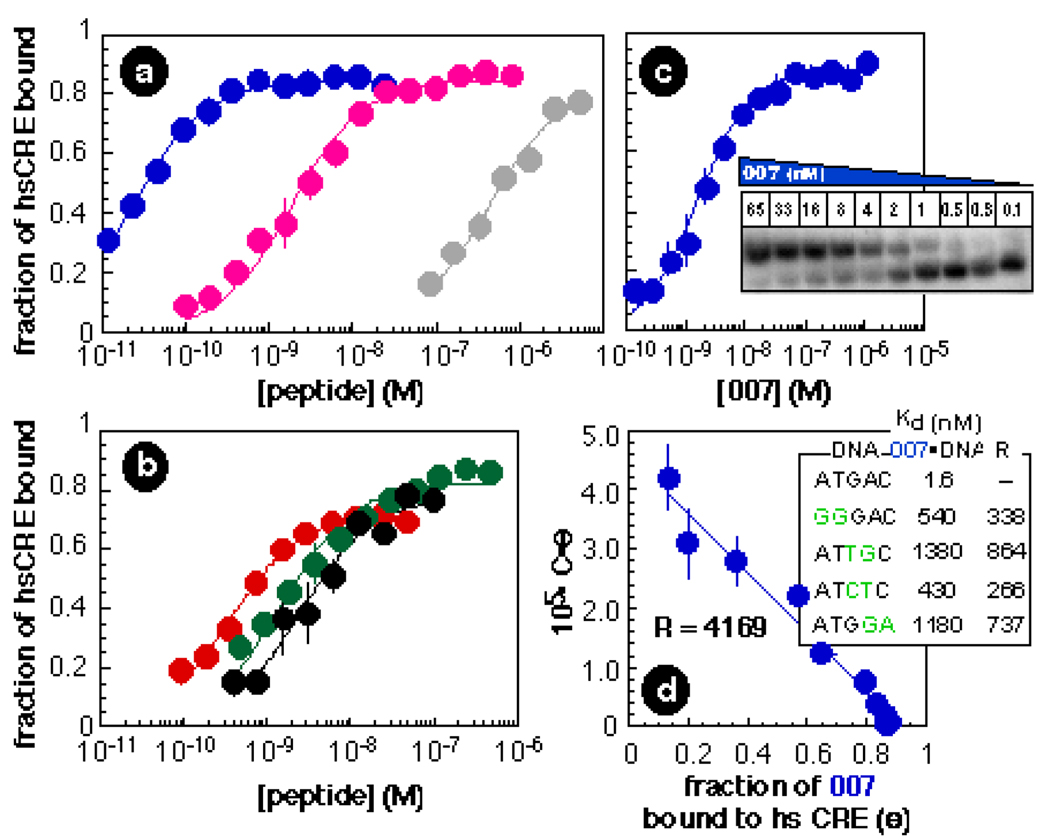
Quantitative electrophoretic mobility shift experiments were performed to assess the DNA affinities of p007, p011, p012, and p016 (Figure 2). All peptides tested bound hsCRE as well or better than did PPBR4 or G27 (the isolated basic region of GCN4). At 4 °C, p011 and p012 bound hsCRE with affinities of 1.5 ± 0.2 nM and 2.5 ± 0.5 nM, whereas p016 bound hsCRE with an affinity of 300 ± 60 pM. Of particular interest is p007, which bound hsCRE to form an exceptionally stable complex with a dissociation constant of 23 ± 1.2 pM (Figure 2a). This peptide bound specific DNA approximately 100 times better than did PPBR4 (Kd = 1.9 ± 0.2 nM) and approximately 20,000 times better than did G27 (Kd = 410 ± 53 nM).1 Moreover, at 25 °C p007 bound hsCRE with an affinity of 1.6 ± 0.1 nM (Figure 2c). Neither PPBR4 nor G27 showed evidence of DNA binding at this temperature. P007 binds specific DNA considerably more tightly than two fingers from the Tramtrack zinc finger protein, which binds 5 bp of DNA with an affinity of 400 nM.11
Figure 2.
(a & b) Plots illustrating the fraction of hsCRE bound (Θ) as a function of p011 ( ), p012 (●), p016 (
), p012 (●), p016 ( ), p007 (
), p007 ( ), PPBR4 (
), PPBR4 ( ), and G27 (
), and G27 ( ) concentration at 4 °C. Each point represents the average of at least three trials. Error bars show the standard error. (c) Autoradiogram and plot illustrating the fraction of hsCRE bound (Θ) by p007 at 25 °C. (d) Plot illustrating the relative affinity of p007 for γ-[32P] hsCRE versus calf thymus DNA. The molar concentration of competing binding sites on calf thymus DNA (C) was estimated from the concentration of DNA in mg/mL and the molecular weight of a base pair assuming that every base on either strand of DNA represents the start of a competitor site. The inset shows the affinity of p007 for four mismatched duplexes that differ from hsCRE at 2 of 5 base pairs. Binding reactions were performed and data analyzed as described.1,7
) concentration at 4 °C. Each point represents the average of at least three trials. Error bars show the standard error. (c) Autoradiogram and plot illustrating the fraction of hsCRE bound (Θ) by p007 at 25 °C. (d) Plot illustrating the relative affinity of p007 for γ-[32P] hsCRE versus calf thymus DNA. The molar concentration of competing binding sites on calf thymus DNA (C) was estimated from the concentration of DNA in mg/mL and the molecular weight of a base pair assuming that every base on either strand of DNA represents the start of a competitor site. The inset shows the affinity of p007 for four mismatched duplexes that differ from hsCRE at 2 of 5 base pairs. Binding reactions were performed and data analyzed as described.1,7
The specificity of DNA binding was investigated by determining the affinity of p007 for several duplex oligonucleotides containing 2 bp changes within the 5 bp hsCRE sequence. P007 was extremely discriminating, exhibiting specificity ratios R (defined as the ratio of the dissociation constants of specific and mutated complexes) between 200 and 800 (ΔΔG = −3.3 to 4.0 kcal•mol−1). This high level of discrimination was observed across the entire 5 bp hsCRE sequence (Figure 2d, inset), suggesting that no single interaction dominated the free energy of the p007•hsCRE complex and that the binding energy is partitioned across the entire protein–DNA interface. By contrast, at 4 °C PPBR4 discriminates poorly (ΔΔG = −1.7 kcal•mol−1) against sequences possessing mutations at the 5’ terminus of hsCRE (data not shown). We suggest the exceptional specificity of p007 results from structural imprinting of the correctly folded peptide in complex with hsCRE by functional selection.
To investigate the possibility that DNA sequences other than these four might bind p007 tightly, we measured the affinity of p007 for calf thymus DNA (CT DNA) which possesses a potential binding site in every register on either DNA strand. The specificity ratio for recognition of hsCRE in preference to any site in CT DNA was 4169 (Figure 2d). This ratio is considerably greater than the number of potential competitor sites (45=1024).7 Whereas the triple zinc finger construct Zif268 and variants thereof selected by phage display fail to uniquely specify 1–2 base pairs of their 9 bp binding sites,7 p007 completely specifies all 5 base pairs of its target sequence. In fact, even if each possible 5 base pair competitor site were present at equal molarity to the target site, 80% of the p007 molecules would be bound to hsCRE, despite the effects of mass action.
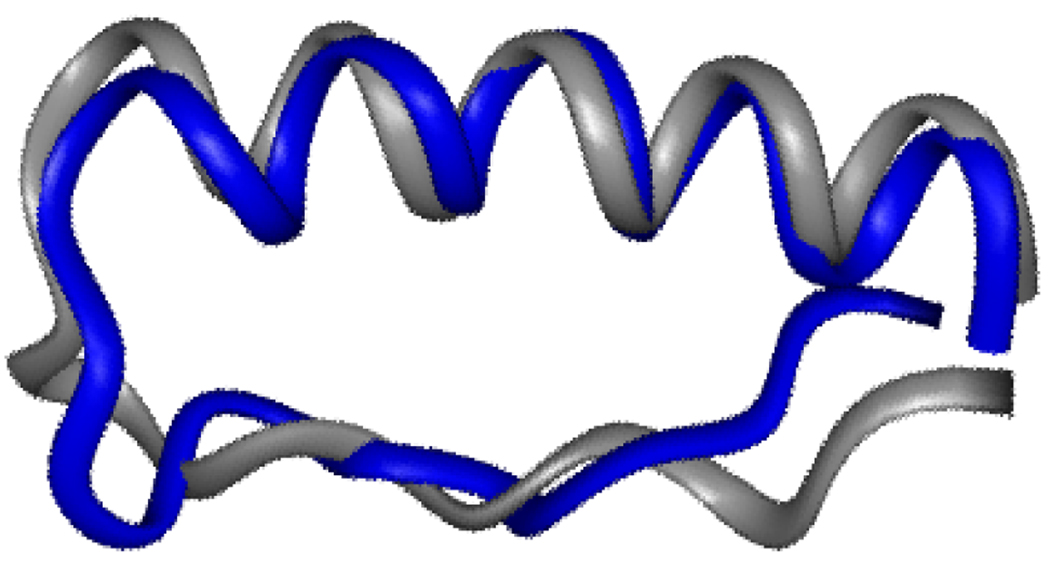
Multidimensional NMR experiments allowed us to characterize the structure of p007 in greater detail. The backbone and side-chain connectivities in p007 were assigned on the basis of reasonably disperse NOESY spectra. The presence of amide-amide cross peaks between residues at positions i and i+3 and i and i+4 defined an α–helical conformation for residues 14–30. Eleven long range NOEs between residues 8 and 17, 8 and 20, 7 and 20, 5 and 20, 4 and 27, 2 and 29, and 2 and 30 specify a folded structure that superimposes on residues 5–8 and 15–28 of aPP with an rmsd of 1.6 Å. Thus, the main chain folds of p007 and aPP are remarkably similar, with residues 5, 7, and 8 proximal to residue 20 and residues 1 and 2 proximal to residue 30. As found in earlier studies of pancreatic fold polypeptides,4 the PPII helix proposed for residues 1–8 of p007 is under-defined by the NMR data. However, in light of the similarity between the aPP and p007 folds, we suggest that p007 contains a structure similar to a PPII helix. It is notable that the PPII-like structure in p007 was selected from a pool of molecules that possessed many backbone conformations. Perhaps because of their extended yet regular structure, PPII helices may represent a general solution to the problem of α–helix recognition.
We have shown that protein grafting,1 in combination with combinatorial mutation and selection, provides rapid access to folded, functional, miniature proteins.2,3,12 Protein grafting involves installing on a miniature scaffold those residues – the functional epitope – responsible for recognition in their native context. As a result, refolding the grafted protein is largely independent of factors that stabilize the native epitope. This independence suggests that grafting will provide ready access to miniature versions of other proteins that use an α–helix to recognize macromolecules in a manner that is irrespective of the tertiary complexity of these proteins. Further, the successful selection of a folded, active miniature protein supports the hypothesis that functional selection of an organized active site may have driven evolution of folded protein receptors, alleviating the need for an otherwise exhaustive search of sequence space.13
Supplementary Material
Figure 3.
Residues 1–31 of the minimized mean structure of p007 superimposed on the corresponding residues of aPP.
Acknowledgment
We are grateful to the Fulbright Commission and the Arthur Wayland Dox Fund for fellowships to J.W.C. and to Dr. Brian Linton for assistance acquiring NMR spectra. This work was supported by the National Institutes of Health.
References
- 1. Zondlo NJ, Schepartz A. J. Am. Chem. Soc. 1999;121:6938. For earlier, successful efforts to design miniature, functional proteins, see references 2 and 3.
- 2.Cunningham BC, Wells JA. Curr. Op. Str. Biol. 1997;7:457. doi: 10.1016/s0959-440x(97)80107-8. [DOI] [PubMed] [Google Scholar]; Braisted AC, Wells JA. Proc. Natl. Acad. Sci., USA. 1996;93:5688. doi: 10.1073/pnas.93.12.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vita C, Roumestand C, Toma F, Menez A. Proc. Natl. Acad. USA. 1995;92:6404. doi: 10.1073/pnas.92.14.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lombardi A, Bryson JW, Ghirlanda G, DeGrado WF. J. Am. Chem. Soc. 1997;119:12378. [Google Scholar]; Vita C, et al. Proc. Natl. Acad. USA. 1999;96:13091. doi: 10.1073/pnas.96.23.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]; McColl DJ, Honchell CD, Frankel AD. Proc. Natl. Acad. USA. 1999;96:9521. doi: 10.1073/pnas.96.17.9521. [DOI] [PMC free article] [PubMed] [Google Scholar]; Domingues H, Cregut D, Sebald W, Oschkinat H, Serrano L. Nat. Str. Biol. 1999;6:652. doi: 10.1038/10706. [DOI] [PubMed] [Google Scholar]; Segal DJ, Barbas CF., III Curr. Op. Chem. Biol. 2000;4:34. doi: 10.1016/s1367-5931(99)00048-4. [DOI] [PubMed] [Google Scholar]
- 4.Blundell TL, Pitts JE, Tickle IC, Wood SP, Wu C-W. Proc. Natl. Acad. Sci. USA. 1981;78:4175. doi: 10.1073/pnas.78.7.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zondlo N. Ph.D. thesis. Yale University; 1999. [Google Scholar]
- 6.Kay BK, Winter J, McCafferty J. San Diego: Academic Press; 1995. [Google Scholar]
- 7.Greisman HA, Pabo CO. Science. 1997;275:657. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Sutcliffe MJ, Schwartz TW, Dobson CM. Biochemistry. 1992;31:1245. doi: 10.1021/bi00119a038. [DOI] [PubMed] [Google Scholar]
- 9.Adzubei AA, Sternberg MJE. Prot. Sci. 1994;3:2395. [Google Scholar]
- 10.Baranger AM. Curr. Opin. Chem. Biol. 1998;2:18. doi: 10.1016/s1367-5931(98)80031-8. [DOI] [PubMed] [Google Scholar]
- 11.Fairall L, Schwabe JWR, Chapman L, Finch JT, Rhodes D. Nature. 1993;366:483. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]; Fairall L, Harrison SD, Travers AA, Rhodes D. J. Mol. Biol. 1992;226:349. doi: 10.1016/0022-2836(92)90952-g. [DOI] [PubMed] [Google Scholar]
- 12.Nygren P, Uhlén M. Curr. Op. Str. Biol. 1997;7:463. doi: 10.1016/s0959-440x(97)80108-x. [DOI] [PubMed] [Google Scholar]; Shogren-Knaak MA, Imperiali B. Bioorg. Med. Chem. 1999;7:1993. doi: 10.1016/s0968-0896(99)00112-1. [DOI] [PubMed] [Google Scholar]
- 13.Yomo T, Saito S, Sasai M. Nat. Str. Biol. 1999;6:743. doi: 10.1038/11512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.