Abstract
The copper-zinc superoxide dismutase-1 (SOD1) is a highly structured protein and, a priori, one of the least likely proteins to be involved in a misfolding disease. However, more than 140, mostly missense, mutations in the SOD1 gene cause aggregation of the affected protein in familial forms of amyotrophic lateral sclerosis (ALS). The remarkable diversity of the effects of these mutations on SOD1 properties has suggested that they promote aggregation by a variety of mechanisms. Experimental assessment of surface hydrophobicity using a sensitive fluorescent-based assay, revealed that diverse ALS-causing mutations provoke SOD1 aggregation by increasing their propensity to expose hydrophobic surfaces. These findings could not be anticipated from analysis of the amino acid sequence. Our results uncover the biochemical nature of the misfolded aggregation-prone intermediate and reconcile the seemingly diverse effects of ALS-causing mutations into a unifying mechanism. Furthermore, the method we describe here will be useful for investigating and interfering with aggregation of various proteins and thereby provide insight into the molecular mechanisms underlying many neurodegenerative diseases.
Keywords: superoxide dismutase, amyotrophic lateral sclerosis, aggregation, neurodegeneration
Introduction
Aggregation of proteins of unrelated sequence is a major hallmark of neurodegenerative diseases. Amyotrophic lateral sclerosis (ALS) is the most common motor neuron disease. It has an adult onset and is rapidly progressive. The majority of ALS cases are sporadic but a large group of dominantly inherited forms of the disease are caused by mutations in the gene encoding the abundantly expressed cytosolic superoxide dismutase-1 (SOD1).1,2 How SOD1 mutants provoke motor neuron degeneration remains to be elucidated, but it is now well established that SOD1 mutations cause familial ALS (fALS) by a gain of toxic properties and not by a loss of enzymatic activity. Mice lacking SOD1 do not develop motor neuron disease.3 In contrast, transgenic mice expressing the ALS-causing SOD1 mutants, in addition to their endogenous SOD1, develop symptoms reminiscent of the human disease.1,4 Similar to other neurodegenerative diseases, a major hallmark of ALS is the presence of proteinaceous inclusions in affected neurons. In fALS patients with SOD1 mutations, aggregated SOD1 is the major component of the motor neuron inclusions5,6 and ALS-causing SOD1 mutants also form inclusions in transgenic mice.1,7 Thus, ALS-causing mutations confer aggregation propensity as well as some toxic properties to SOD1. Understanding how SOD1 mutations cause aggregation of the protein is of major importance, as it is one of the earliest events in the ALS pathogenesis.
SOD1 is an abundant and ubiquitously expressed 32 kDa homodimeric enzyme that catalyzes the dismutation of superoxide radicals. Each monomer folds as an eight-stranded Greek key β-barrel,2 binds one copper and one zinc ion, and contains a disulfide bond. The native protein is extremely stable and thus, a priori, one of the least likely proteins to be involved in a neurodegenerative disease. However, more than 140, mostly missense mutations in SOD1 cause fALS.2 SOD1 mutations are scattered through the entire sequence of the protein and have very diverse effects on the properties of the protein. SOD1 mutants partition in two groups, on the basis of their metal-binding properties and biological activities. One group is characterized by mutations in the metal-binding region (MBR) that perturb metal binding. The other fALS mutants generally retain wild-type metal content and biological activity and are referred to as wild-type like mutants (WTL).2 How such diverse ALS-causing SOD1 mutations all cause aggregation of the protein is a great puzzle. Most mutations in SOD1 only marginally reduce the stability of the native protein8 and a variety of different mechanisms have been proposed to explain the increased aggregation propensity of the diverse SOD1 mutants. Aggregation can arise as a consequence of perturbation of the structural integrity due to loss of metal binding,8–12 reduction of the intra-monomer disulfide bond13–16, oxidation,17 destabilization of the dimer18,19 or alteration of post-translational modifications.20,21 Some biochemical properties of the mutant proteins such as reduction of the repulsive charge of the protein,22 aberrant hydrophobicity of the apo-protein,23 perturbed folding24,25 or increased unfolding rates24,26 could account for the increased aggregation of SOD1 mutants, but it has not been established whether any of these alterations directly cause aggregation. Because of the diversity of the alterations provoked by SOD1 mutations, specific rules have been proposed to govern aggregation of distinct subsets of ALS-causing mutants.
Some structural alteration of the native fold ought to be required to elicit SOD1 misfolding. However, because SOD1 is such a stable protein, aggregation is likely to be initiated by local and possibly subtle unfolding of the native state, rather than global unfolding. Therefore, our ability to detect early conformational changes on the misfolding pathway depends on the availability of a sensitive assay. While using the conformation-sensitive dye Sypro Orange, which fluoresces in a hydrophobic environment,27 as a sensitive method to detect conformational transition, we have uncovered the biochemical nature of the aggregation-prone conformer. We found that ALS-causing mutations provoke aggregation by increasing the propensity of diverse SOD1 mutants to expose hydrophobic surfaces, a common feature that could not be anticipated from bioinformatic analysis of the biochemical alterations provoked by the mutations.28
Results
Increased Sypro Orange fluorescence is a common feature of ALS-causing SOD1 mutants
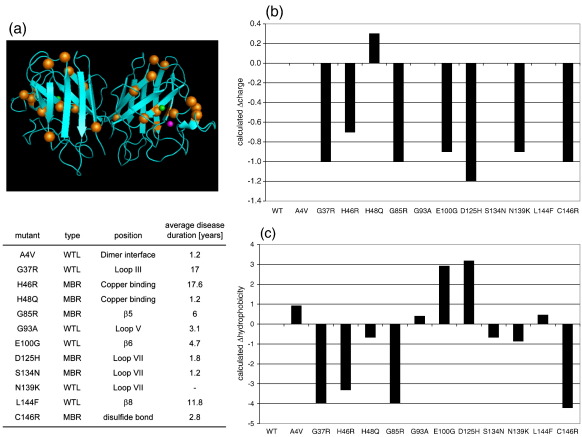
We selected a set of structurally diverse SOD1 mutants, including mutants that account for most of the fALS cases with SOD1 mutations.29 The selected mutants display the broad variety of biophysical and biochemical properties that characterize fALS-causing SOD1 mutations and different clinical characteristics (Fig. 1a). This comprises six WTL and six MBR mutants selected because 1) they all cause fALS, 2) their positions are scattered throughout the sequence of the protein, 3) they have been previously characterized and exhibit different enzymatic, physicochemical and structural properties, 4) the disease duration varies greatly for patients with these different SOD1 mutations (Fig. 1a). The selected mutants include the A4V mutant, which is both the most common and the most severe fALS-causing SOD1 mutant and retains WTL properties;30 the common H46R mutant with compromised copper binding and disease duration of about 17 years;31 and the N139K and S134N mutants, which were previously described as nearly indistinguishable from the wild-type protein.2,10 A bioinformatic analysis has previously revealed that SOD1 mutations have an overall tendency to decrease the net negative charge of SOD1.22 We also find this trend in the representative set of SOD1 mutants selected in this study, since seven out of the twelve mutants selected exhibited a decrease in their repulsive charge (Fig. 1b). In addition, we examined the change of hydrophobicity introduced by the mutations. Four mutations dramatically decreased and two increased the hydrophobicity, while the other mutations only caused subtle changes (Fig. 1c). These analyses confirmed that the selected SOD1 mutants cover the broad range of properties characteristic of SOD1 mutants.
Fig. 1.
Diverse properties of fALS-causing SOD1 mutants. (a) Schematic representation of SOD1 structure (PDB entry 1SPD).53 Mutants analyzed in this study are highlighted in orange, copper in green and zinc in magenta. Table listing key features of the mutants analyzed in this study. (b) Calculated change of charge (Δcharge) resulting from the mutations. Calculations were performed with PROTEIN CALCULATOR v3.3. (c) Calculated change of hydrophobicity (Δhydrophobicity) resulting from the mutations. Hydrophobicity values were calculated according to,42 with hydrophobicity values taken form.54
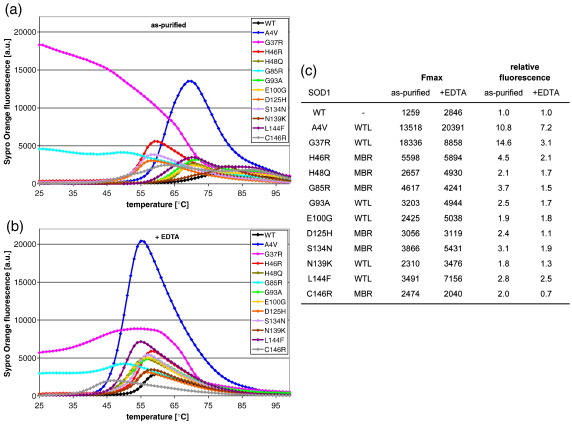
Aiming to set up an assay to detect the conformational rearrangements on the misfolding pathway, we analyzed the behaviour of SOD1 mutants in the presence of a conformation sensitive dye. Proteins were expressed in insect cells and purified as described9 (and Supplementary Fig. 1). We carried out thermal denaturation of SOD1 proteins in the presence of Sypro Orange (Fig. 2a, b and Supplementary Fig. 2). At room temperature, SOD1G37R and SOD1G85R already exhibited a significant fluorescence in the presence of the conformation sensitive dye, in contrast to all other mutants and wild-type protein (Fig. 2a). This confirmed that under native conditions, SOD1WT and most mutants did not expose hydrophobic surfaces as expected for tightly folded proteins. Heat denaturation of all mutants other than SOD1G37R and SOD1G85R, in the presence of Sypro Orange, resulted in a marked increase in fluorescence. Notably, the assay used here revealed differences between SOD1WT and the N139K and S134N mutants that have escaped previous analyses. The fluorescence maxima of the as-purified SOD1 mutants ranged from about 2-fold, to more than a 14-fold increase relative to the wild-type protein (Fig. 2a and c). We defined relative surface hydrophobicity values as the increase in fluorescence maxima relative to wild-type SOD1 (Fig. 2c). This revealed that the structurally diverse SOD1 mutants exposed more hydrophobicity than the wild-type protein.
Fig. 2.
Increased surface hydrophobicity upon thermal denaturation is a generic feature of ALS-causing SOD1 mutants. (a) Sypro Orange fluorescence during thermal unfolding of as-purified SOD1 without or (b) with 20 mM EDTA. Data are means of 3 independent experiments. s.d. are presented in Supplementary Fig. 2. (c) Hydrophobicity values measured in (a and b), as the mean Sypro Orange-derived fluorescence maxima (Fmax) or normalized to the Fmax of SOD1WT, using either as-purified or EDTA-treated proteins.
SOD1 variants expressed and purified in similar conditions as used here were found to exhibit various metal contents9 and previous studies have revealed that metal deficiency increases hydrophobicity.23,32 We next set up to determine the metallation status of most of the SOD1 variants used in this study, in order to determine whether the differences of hydrophobicity of the diverse SOD1 variants reflected differences in metal content. In agreement with a previous study, all the purified SOD1 variants analyzed had substochiometric copper content and the MBR mutants H46R, S134N and D125H were both copper- and zinc-deficient (Supplementary Fig. 3a and9). In the other mutants analyzed, the zinc site was fully occupied (Supplementary Fig. 3a). We failed to detect a correlation between low metal content and high amount of exposed hydrophobic surfaces (Supplementary Fig. 3b). However, it has long been suspected that metal deficiency could be at the origin of the pathogenicity of SOD1 mutants.8,9 We next exposed the purified SOD1 variants to 20 mM EDTA and monitored the effects of such treatment. As expected, the thermal denaturation profiles, in the presence of Sypro Orange, of the metal deficient mutants H46R, S134N and D125H, were very similar with or without 20 mM EDTA (Fig. 2a-c). The same was observed for the MBR mutant C146R. In contrast, the fluorescence maxima of the other mutants were higher when heat denatured in the presence of EDTA (Fig. 2a-c). Thus, EDTA treatment significantly reduced the metal content of the SOD1 variants and this reduction further enhanced exposure of hydrophobic surfaces of destabilized SOD1 mutants. As observed for the as-purified proteins, most EDTA-treated SOD1 mutants also exhibited fluorescence maxima that were higher than that of the wild-type protein (Fig. 2b and c). However, EDTA treatment attenuated the differences in surface hydrophobicity between the wild-type protein and the mutants (Fig. 2a-c). This is likely due to the finding that demetallation increased exposure of hydrophobic surfaces on SOD1WT (Fig. 2a-c).
Environmentally sensitive fluorescent dyes have been used previously to monitor thermal unfolding.34 The transition midpoint of the denaturation curves indicates the unfolding temperature (Tm). The apparent Tm values of the EDTA treated proteins were extracted from the Sypro Orange analyses (Supplementary Fig. 3c). We found that the apparent Tm values of our EDTA-treated mutants were nearly identical to the ones published for the same mutants in the apo-state (Supplementary Fig. 3c and10,33). This reveals that our EDTA-treated proteins are similar to apo-proteins prepared in other studies. As expected, EDTA treatment dramatically reduced the Tm of the biologically metallated proteins but had a minor effect on the metal deficient proteins H46R, S134N and D125H. Interestingly, except for SOD1N139K, all as-purified SOD1 mutants had a lower apparent Tm than the wild-type protein (Supplementary Fig. 3c). We then analyzed whether the extent of exposure of hydrophobicity directly correlated with Tm. While thermal unfolding of most SOD1 mutants is required to expose hydrophobic surfaces, the amount of exposed hydrophobic surfaces did not correlate directly with the apparent Tm of the different mutants (Supplementary Fig. 3d). This revealed that the knowledge of Tm can't predict the amount of surface hydrophobicity exposed by each mutant.
Together, these results reveal that exposure of hydrophobic surfaces is a common feature of diverse SOD1 mutants with different metal content and stability, when subjected to denaturation.
Increased surface hydrophobicity correlates with aggregation of SOD1A4V
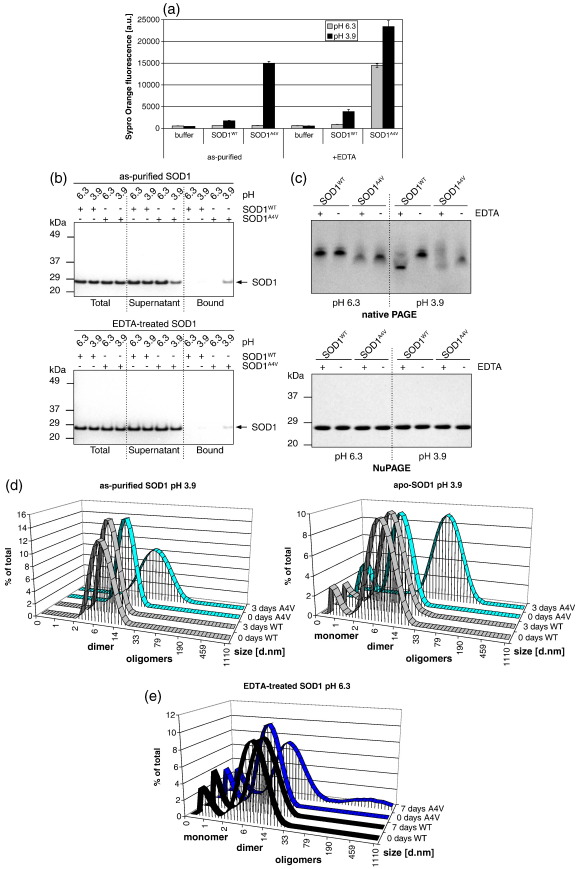
Having found an unprecedented common feature for these diverse SOD1 mutants we then asked whether increased exposure of hydrophobic surfaces caused aggregation. We used conditions previously established to elicit mutant SOD1 aggregation to address this question, such as acidic pH in the presence or absence of a chelator. In buffer alone, or in the presence of as-purified SOD1WT or SOD1A4V, Sypro Orange fluorescence was very low, indicating that hydrophobic residues are buried in both SOD1WT and SOD1A4V, as expected for native proteins (Fig. 3a). At physiological temperature, Sypro Orange fluorescence increased dramatically only in the presence of EDTA-treated SOD1A4V, indicating that EDTA-treated SOD1A4V exhibited greater surface hydrophobicity than the SOD1WT exposed to the same condition, or the untreated proteins (Fig. 3a), as previously observed.13
Fig. 3.
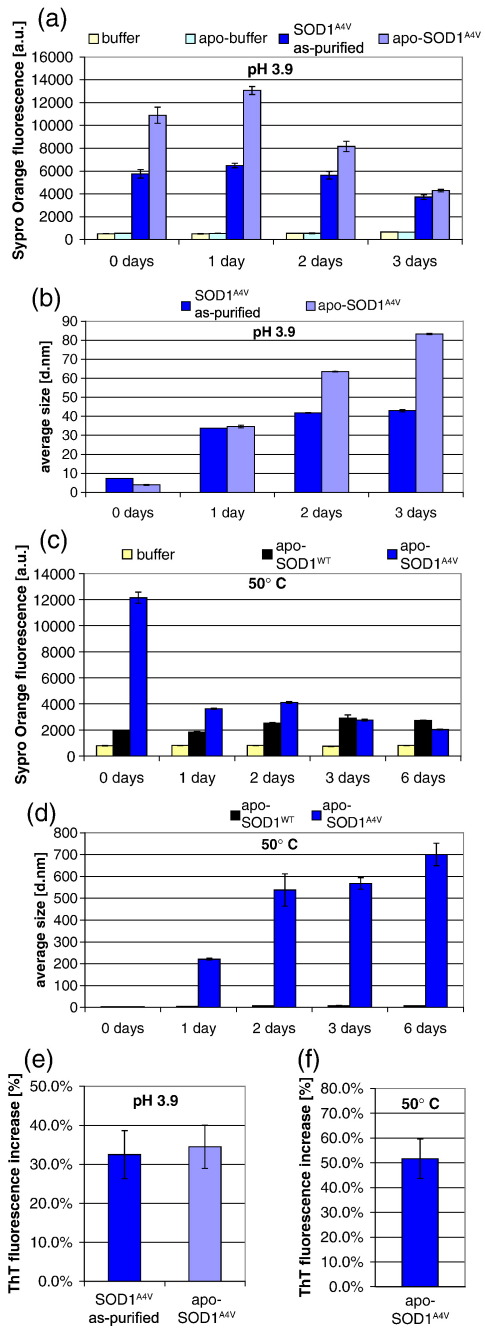
Destabilization of as-purified SOD1A4V by EDTA or low pH exposes hydrophobic surfaces and triggers aggregation. (a) Assessment of surface hydrophobicity by Sypro Orange fluorescence. Means and s.d. of replicate experiments (n = 3) are shown. (b) NuPAGE analysis of SOD1 binding to hydrophobic beads. (c) Electrophoretic mobilities of the SOD1 variants on native PAGE (1.2 μg) or denaturing NuPAGE (0.75 μg). Proteins were treated as indicated 30 min prior electrophoresis. (d, e) Aggregation of SOD1A4V monitored by DLS. d.nm: Diameter in nm.
Proteins were next exposed to low pH, a condition that elicits SOD1A4V aggregation, with or without EDTA.35 At pH 3.9, both EDTA-treated and as-purified SOD1A4V became strongly fluorescent in the presence of Sypro Orange, unlike SOD1WT exposed to the same conditions (Fig. 3a), revealing that EDTA or low pH exposed hydrophobic surfaces on SOD1A4V but not on the wild-type protein. To confirm these findings, SOD1WT and SOD1A4V were next incubated on Phenyl Sepharose. After extensive washing, a fraction of EDTA-treated and as-purified SOD1A4V exposed to low pH was selectively retained on the hydrophobic resin (Fig. 3b). These results confirm that the fluorescence-based assay is a highly sensitive method for detecting surface hydrophobicity.
To assess whether exposure to acidic pH in the presence of EDTA efficiently demetallated the proteins, SOD1WT and SOD1A4V were analyzed on native PAGE, since this method was shown to resolve differentially metallated species.9 The mobility of as-purified SOD1A4V was faster than as-purified SOD1WT (Fig. 3c), in good agreement with the finding that this mutant had reduced copper content, compared to the wild-type protein (Supplementary Fig. 3a). At pH 6.3, EDTA-treated SOD1A4V exhibited the same mobility as the untreated mutant on native PAGE (Fig. 3c), while Sypro Orange fluorescence increased in the presence of EDTA-treated SOD1A4V, but not the untreated mutant (Fig. 3a). This reveals that Sypro Orange fluorescence detected changes that were not detectable by native PAGE, suggesting that the Sypro Orange detects conformational changes that precede metal loss. At acidic pH, EDTA treatment increased mobility of both SOD1WT and SOD1A4V on native PAGE, indicating that such treatments efficiently depleted metal and produced apo-proteins (Fig. 3c). As previously observed,36 we also find that demetallation destabilized SOD1 dimers (Fig. 3d and e).
We next carried out analyses of SOD1 aggregation, using as-purified and apo-proteins at a 10 μM concentration.33 Aggregation was monitored by dynamic light scattering (DLS), which revealed that after 3 days at pH 3.9, both apo- and as-purified SOD1A4V were aggregated (Fig. 3d). As previously observed for wild-type SOD1WT, we found that low pH destabilized both SOD1WT and SOD1A4V dimers,37 but only apo-SOD1A4V aggregated. Under the same conditions, apo-SOD1A4V dramatically increased Sypro Orange fluorescence (Fig. 3a). In good agreement with the high fluorescence of demetallated SOD1A4V at low pH, compared to the as-purified protein (Fig. 3a), we observed that apo-SOD1A4V formed larger particles than as-purified SOD1A4V (Fig. 3d). After 7 days, EDTA-treated SOD1A4V also aggregated at pH 6.3 (Fig. 3e). In contrast, the wild-type protein remained soluble in each condition tested (Fig. 3d, e, Supplementary Fig. 4 and data not shown). All together, these results show that increased exposure of hydrophobic surfaces correlates with aggregation of SOD1A4V.
TFE exposes hydrophobic surfaces and provokes aggregation of SOD1H46R
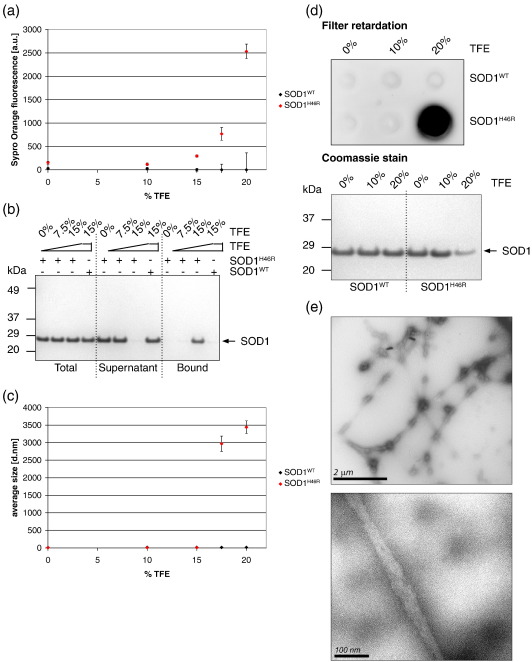
We next examined whether similar properties underlie aggregation of a mutant unrelated to the WTL mutant SOD1A4V. As-purified SOD1WT and the MBR mutant SOD1H46R were exposed to increasing trifluoroethanol (TFE) concentrations, a condition previously shown to trigger aggregation of SOD1 mutants,33 as well as other globular proteins.38 TFE increased Sypro Orange fluorescence (data not shown). However, when the fluorescence of the dye in buffer was subtracted, we found that the fluorescence of SOD1H46R, but not SOD1WT, increased dramatically in Sypro Orange, with increasing TFE concentrations (Fig. 4a). This result indicates that in the presence of TFE, SOD1H46R but not SOD1WT exposed hydrophobic surfaces, as confirmed by the specific retention of SOD1H46R, in the presence of TFE, on hydrophobic resin (Fig. 4b). In parallel, aggregation was monitored by DLS and revealed that TFE concentrations higher than 15% triggered aggregation of SOD1H46R, but not SOD1WT (Fig. 4c).
Fig. 4.
TFE treatment exposes hydrophobic surfaces on as-purified SOD1H46R and provokes fibrillogenesis. (a) Assessment of SOD1H46R surface hydrophobicity by Sypro Orange fluorescence in the presence of increasing TFE concentrations. (b) NuPAGE analysis of SOD1 binding to hydrophobic beads. (c) Aggregation of SOD1H46R monitored by DLS. (d) Aggregation of SOD1H46R revealed by filter retardation assay and NuPAGE analysis of aliquots of the samples used in the filter retardation assay. (e) Electron micrographs of negatively stained SOD1H46R fibrils formed in 20% TFE acquired at magnifications of 7100 (upper panel) and 52000 (lower panel). Means and s.d. of 3 independent experiments are shown in (a and c).
To confirm that the large particles observed upon incubation of SOD1H46R with TFE were aggregates, SOD1WT and SOD1H46R, in non-denaturing buffer or in the presence of increasing concentrations of TFE, were filtered through a cellulose acetate membrane and revealed with SOD1 antibodies, as described.39,40 While unaggregated SOD1 filtered through the membrane, aggregated SOD1H46R was retained when the mutant was exposed to 20% TFE (Fig. 4d), indicating that the large particles observed by DSL were SOD1H46R aggregates (Fig. 4c). Equal amounts of proteins were loaded on the filter retardation assay and on a denaturing NuPAGE gel (Fig. 4d, lower panel). However, the amount of SOD1H46R in 20% TFE that was resolved on the denaturing gel was consistently lower than that in absence of TFE. This indicated that aggregates were not resolved on the gel, suggesting that they might be resistant to boiling in reducing buffer. We noticed that SOD1H46R required lower TFE concentrations to trigger Sypro Orange fluorescence than to aggregate (Fig. 4a, c and d). This indicates that exposure of hydrophobic surfaces precedes aggregation.
We next examined the morphology of mutant SOD1 aggregates to determine whether they were amorphous or ordered. Electron microscopy revealed that SOD1H46R aggregates exhibited both fibrillar and granular components, reminiscent of the granule-coated fibrils found in affected neurons of ALS patients (Fig. 4e and Ref. 41). Together, these results demonstrate that aggregation of two distinct SOD1 mutants, elicited under different conditions, directly correlates with exposure of hydrophobic surfaces.
Exposure of hydrophobic surfaces precedes aggregation
Aggregation of SOD1H46R occurred within minutes after TFE addition, while a longer time was required to detect aggregation of SOD1A4V at low pH (Figs. 3, 4 and data not shown). We took advantage of the slow aggregation kinetics of both as-purified and apo-SOD1A4V at acidic pH to analyze the aggregation process in greater detail. As-purified and apo-SOD1A4V were adjusted to low pH (day 0) and aliquots taken every day to analyze both Sypro Orange-derived fluorescence and aggregation. Immediately after exposure to low pH, both as-purified and apo-SOD1A4V increased the fluorescence of the conformation-sensitive dye (Fig. 5a, day 0). The increase in fluorescence preceded the onset of aggregation (Fig. 5a and b, day 0). Fluorescence of Sypro Orange then reached a maximum after 1 day and gradually decreased after 2 days (Fig. 5a). Concomitantly, aggregates gradually grew (Fig. 5b and Supplementary Fig. 5a). Note that the soluble proteins were barely detectable after 2 days (Supplementary Fig. 5a). Together, these analyses reveal that low pH exposes hydrophobic regions on the surface of SOD1A4V and such regions are buried in aggregates.
Fig. 5.
Aggregation of as-purified SOD1 mutants follows exposure of hydrophobic surfaces. (a) Time course of measurements of exposed hydrophobicity monitored by Sypro Orange fluorescence and (b) aggregation of SOD1A4V at acidic pH monitored by DLS. Note that demetallation first provoked a decrease in the size of particles measured, indicating that metal loss provoked monomerization of the protein. (c) Time course of measurements of exposed hydrophobicity and (d) aggregation of apo-SOD1A4V exposed to 50 °C. (e, f) aggregation of SOD1A4V monitored by ThT after 3 days of incubation at acidic pH (e) or 50 °C (f). Data are means and s.d. values of replicate experiments (n = 3).
To determine whether these findings were restricted to the experimental conditions used to induce mutant SOD1 aggregation or were inherent to the aggregation mechanism, we used high temperature, since this condition was previously found to trigger SOD1A4V aggregation.33 Similar to what we observed upon destabilization of SOD1A4V at low pH, heating SOD1A4V to 50 °C increased the fluorescence of Sypro Orange (Fig. 5c). The fluorescence of the conformation-sensitive dye decreased over time, while aggregates grew (Fig. 5c and d). Note that no soluble proteins were detectable after 1 day (Supplementary Fig. 5c). In contrast, heat did not provoke fluorescence of SOD1WT or aggregation (Fig. 5c and d). Like the ALS deposits, 3 days-old aggregates were amyloid-like since they increased the fluorescence of Thioflavin T (ThT), an amyloid specific probe (Fig. 5e and f). In contrast, SOD1WT didn't increase ThT fluorescence (data not shown). The ThT increase for SOD1 aggregates was modest, in agreement with previous studies.33,35 Congo Red binding experiments also suggested that SOD1A4V aggregates, in contrast to the wild-type protein, contain an amyloid component, since they produced a small but reproducible shift in the absorbance spectra of Congo Red (Supplementary Fig. 5b, d), as previously reported.33,35 Taken together, these results indicate that denaturation of SOD1 mutants by diverse treatments exposes hydrophobic surfaces and that such regions engage non-native interactions to form amyloid-like aggregates. Thus, ordered assembly of SOD1 aggregates is likely to be driven by hydrophobic interactions. These experiments also reveal that Sypro Orange-derived fluorescence is a marker of one of the earliest conformational rearrangement in the aggregation pathway, rather than amyloids per se.
Increased propensity of diverse ALS-causing SOD1 mutants to expose hydrophobic surfaces causes aggregation
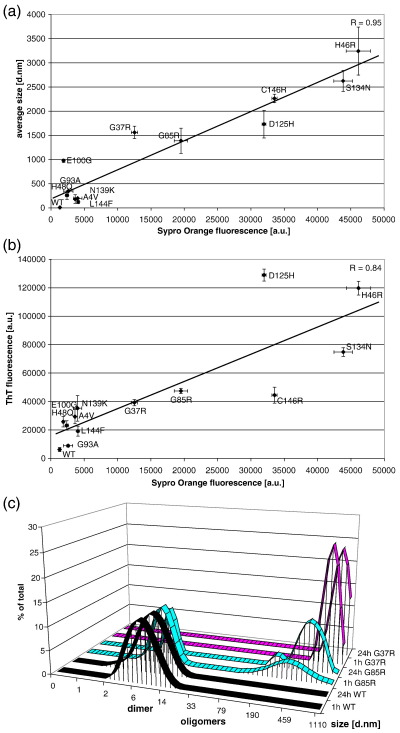
In all conditions examined using two specific SOD1 mutants, we observed a correlation between exposure of hydrophobic surfaces and aggregation. To determine whether increased exposure of hydrophobic surfaces correlates with aggregation of diverse SOD1 mutants, we monitored aggregation and exposure of hydrophobic surfaces of the representative set of 12 mutants. As-purified SOD1WT and mutant proteins were exposed to 20% TFE, a condition that was found to elicit aggregation of the diverse SOD1 mutants, but not the wild-type protein. This treatment converted the soluble MBR mutants into aggregates in 20 minutes and was also efficient to provoke aggregation of the WTL mutants (Supplementary Fig. 6a). Exposure of surface hydrophobicity was monitored by Sypro Orange fluorescence and aggregation monitored both by DLS and ThT fluorescence (Fig. 6a and b). ThT fluorescence indicated that aggregates have an amyloid component. Analysis of Congo Red binding confirmed this (Supplementary Fig. 6b). We found exposure of hydrophobicity and ordered assembly of SOD1 aggregates, monitored by these two independent methods, were strongly correlated. The strength of the correlation between exposure of hydrophobic surfaces and aggregation is attested by the very high linear correlation coefficient (Fig. 6a, R = 0.95 and b, R = 0.84). This revealed that exposure of hydrophobicity is a common feature that provokes aggregation of diverse SOD1 mutants.
Fig. 6.
Aggregation of as-purified ALS-causing SOD1 mutants is caused by their increased propensity to expose hydrophobic surfaces. (a, b) Correlation between surface hydrophobicity measured by Sypro Orange fluorescence and aggregation measured by DLS (a) and ThT fluorescence (b). Data are means and s.e.m. from 3 independent experiments. Measurements were made 20 minutes after TFE treatment. The high linear correlation coefficient R, denotes the strength of the correlation. Note that the MBR mutants are more susceptible to TFE-induced aggregation than the WTL mutants. (c) Aggregation of SOD1G85R and SOD1G37R at room temperature, revealed by DLS.
To further demonstrate that the exposure of hydrophobic surfaces predicts aggregation, we focused on SOD1G37R and SOD1G85R. These two mutants produced high Sypro Orange fluorescence at room temperature (Fig. 2a), suggesting that they might aggregate at room temperature. DLS analyses confirmed this hypothesis (Fig. 6c).
Discussion
A common feature of diverse neurodegenerative diseases is the deposition of aggregated proteins in affected neurons. Mutations of natively unstructured proteins associated with familial forms of these diseases cause aggregation of the proteins by altering simple intrinsic physicochemical properties such as hydrophobicity, secondary structure and charge.42,43 However, the rules governing aggregation propensity of highly structured proteins, such as SOD1, remained unknown. The results presented here reveal that increased surface hydrophobicity is a generic feature of structurally diverse ALS-causing SOD1 mutants that could not be anticipated by calculating the changes in hydrophobicity caused by the mutation or by using algorithms designed to predict protein aggregation.28 This common property is intrinsic to fALS mutants but not necessarily constitutive. The destabilizing mutations G85R and G37R constitutively expose hydrophobic surfaces. For the other SOD1 mutations, destabilization of the proteins exposes otherwise buried hydrophobic surfaces and causes aggregation. Previous studies have shown that SOD1 mutations increase unfolding rates24,26,33 and we found here that such unfolded intermediates expose aggregation-prone surfaces. While it is expected that partly unfolded proteins expose some hydrophobic surfaces, and such hydrophobic regions exist in SOD1,44 it could not be anticipated that diverse ALS-causing SOD1 point mutations, causing a variety of alterations on the properties of the proteins, all increase the propensity to expose hydrophobic surfaces and that this common feature is the molecular determinant underlying the formation of the ordered aggregates. Furthermore, the effects of the mutations analyzed here on the predicted hydrophobicity of the protein are completely random: Some mutations increase and some decrease hydrophobicity.
It has been previously reported that decreased stability and metal deficiency are important factors that influence SOD1 aggregation.8,9 We did not observe a direct correlation between metal content and exposure of hydrophobicity upon thermal denaturation of the as-purified proteins (Supplementary Fig. 3). However, treatment with a chelator further increased exposure of hydrophobic surfaces of the SOD1 variants that where loaded with metal. This indicates that demetallation increases exposure of hydrophobicity and thereby increases aggregation propensity. The MBR mutants were also more susceptible to TFE-induced aggregation than the WTL (Fig. 6), suggesting that these mutants are more vulnerable to destabilization. Similar to what we observed with metal contents, we found no direct correlation between Tm and exposed hydrophobicity (Supplementary Fig. 3). Yet, destabilization of the natively stable mutants is required to expose the otherwise buried hydrophobic surfaces. These observations are consistent with the following interpretation: metal deficiency, stability, susceptibility to unfolding are likely to be interconnected. Destabilization of metallated proteins is likely to provoke metal loss; conversely metal deficiency destabilizes the proteins. We found here that destabilizing diverse SOD1 mutants by various treatments systematically increased exposure of hydrophobic surfaces on the mutant but not the wild-type protein.
Taking advantage of the slow aggregation kinetics of SOD1 mutants at low pH and high temperature, we found that exposure of hydrophobic surfaces precedes aggregation (Fig. 5). This shows that aggregation of diverse pathogenic SOD1 mutants is driven by intermolecular hydrophobic interactions either between constitutively hydrophobic mutants or aggregation intermediates exposing hydrophobic surfaces. Our results are consistent with the structures of SOD1 amyloid-like filaments that have revealed that these amyloids are formed after local unfolding of the zinc and electrostatic loops that creates a hydrophobic interface between two proteins.45 In two previous studies, a subset of SOD1 mutants from mice tissues have been captured on hydrophobic resins.23,46 Our findings reveal that exposure of hydrophobic surfaces is a generic feature of diverse SOD1 mutants and this property governs aggregation propensity. The increased surface hydrophobicity of SOD1 mutants may also account for several of their properties: Their perturbed folding, both in vitro and in cells,24,25 their selective association with the cytoplasmic face of mitochondria,47 their recognition by chaperones48 as well as their decreased half-life,49 since hydrophobic surfaces, once recognized by chaperones may target the protein to the degradation machinery.50
Whether fALS with SOD1 mutations is a typical amyloidosis is not clear. Mutant SOD1 inclusions in human tissues are composed of 15- 25 nm granules-coated fibrils41 but they are not revealed by amyloid-specific dyes. However, inclusions are Thioflavin-S-positive in mice expressing SOD1 mutants.39 In vitro, we found that SOD1 aggregates increased ThT fluorescence, shifted Congo Red absorbance towards 541 nm and that they exhibited both fibrillar and granular components (Fig. 4e). Previous studies have also reported that SOD1 aggregates contain an amyloid component.11,15,33,35 Since minute amounts of misfolded SOD1 cause ALS,51 one hypothesis that can reconcile these seemingly conflicting observations is as follows: SOD1 aggregates contain an amyloid component but its low abundance precludes its detection by amyloid dyes in human tissues.
Aggregation-prone hydrophobic surfaces are only transiently exposed on the aggregation intermediate and later buried inside the aggregate. These findings may provide a molecular basis for the hypothesis of the toxic aggregation intermediate. Exposure of hydrophobic surfaces on the aggregation intermediate may be detrimental to the cell, while the mature fibrillar aggregate, burying these surfaces, is predicted to be less harmful. In aggregation reactions with slow kinetics, we found that Sypro Orange fluorescence precedes aggregation and ThT fluorescence (Fig. 5) indicating that Sypro Orange is a marker of an early species in the aggregation pathway rather than another amyloid marker. Sypro Orange reveals a soluble, yet aggregation-prone conformer that builds up aggregates. When aggregation is elicited with TFE, Sypro Orange remains high in the presence of TFE-treated SOD1 mutants after the onset of aggregation (data not shown). Since aggregation is observed rapidly after TFE addition, it is likely that TFE is a more potent destabilizing agent than pH or temperature. This suggests that under harsh conditions, some hydrophobic surfaces remained exposed on the aggregates, in contrast to the aggregation elicited in milder conditions such as low pH or high temperature (Fig. 5). Whether the aggregation pathways are different under different aggregation conditions is unclear. Regardless of the aggregation pathway, we found here that destabilization of SOD1 mutants, by several methods, exposes hydrophobic surfaces on the mutant proteins but not the wild-type. This reveals that, the initiation of aggregation is conserved. We propose that, any condition, yet to be identified, that will destabilize the protein in motor neurons, will expose hydrophobic surfaces and trigger the ordered assembly of fALS-causing SOD1 mutants.
This study identifies the common nature of the early aggregation-prone conformer of diverse ALS-causing SOD1 mutants and thereby provides essential information to elucidate the mechanism by which it provokes motor neuron death and to interfere with this process.
The method used here is broadly applicable, quantitative and high-throughput. The fluorescent-based assay can be used to screen for compounds that prevent the formation of the aggregation-prone conformer, one of the earliest events in the disease process. In addition, this method will be useful for investigating the aggregation propensity of a wide variety of proteins and thereby provide insight into the molecular mechanisms underlying many neurodegenerative diseases.
Materials and methods
Protein purification
cDNAs encoding human SOD1 mutants were generated by polymerase chain reactions. Wild-type and mutant cDNAs were cloned in pFastBac1 (Invitrogen) and expressed in Sf9 cells in the presence of 150 μM CuCl2 and ZnCl2. SOD1 proteins were purified as described.9 Assays were performed in the commonly used 10 mM MES buffer pH 6.38, with or without 20 mM EDTA to generate apo-SOD1. In Figs. 3 and 5 (a and b), 50 mM MES buffer pH 6.3 or sodium acetate buffer pH 3.9, were used. Protein concentrations were determined by measuring OD at 280 nm and by quantitative amino acid analyses.
Fluorescence measurements
Equal amounts of protein (3 μg in Figs. 2, 3, 5 and 6; 1 μg in Fig. 4) were mixed with an excess of Sypro Orange (10 × concentration of S5692 Sigma-Aldrich) in 20 μl in a 96 well plate and fluorescence was measured in the 7900HT Fast Real-Time PCR System from Applied Biosystems and expressed as arbitrary units (a.u.). Thermal denaturations were conducted with a heating rate of 1 °C/1.5 min. Note that Sypro Orange fluorescence measurements of as-purified SOD1 proteins were very similar in MES buffer pH 6.3 and in 50 mM Tris-HCl, pH 7.5, 150 mM NaCl and 10% glycerol (data not shown). Well to well variation was assessed and s.d. was quantified to be less than 10% in the different wells of a 96 well plate (data not shown).
Hydrophobic binding
Equal amounts of protein (10 μg) were incubated on 20 μl Phenyl Sepharose 6 Fast Flow (high sub, GE Healthcare) in 200 μl of the indicated buffer overnight at room temperature and washed in binding buffer. Total, bound and unbound (supernatant) proteins were resolved on NuPAGE 4-12% Bis-Tris gels (Invitrogen) and stained with Coomassie Brilliant Blue G-250.
Electrophoresis
For native PAGE, samples were equilibrated in Laemmli loading buffer lacking SDS and reducing agents, analyzed on 12% Tris-HCl gels and run in Laemmli buffer without SDS. NuPAGE 4-12% Bis-Tris gels (Invitrogen) were run in MES buffer and stained with Coomassie Brilliant Blue G-250.
Aggregation analyses by DLS, ThT fluorescence, electron microscopy and filter retardation assays
Wild-type or mutant SOD1 (10 μM) were incubated at temperatures between 25 to 50° C, in the indicated buffers, in the presence of 0 to 20% of 2-2,2-trifluoroethanol where indicated (Fluka). After 4 to 5 hours (day 0) up to 7 days, the sizes of the particles were measured using the Zetasizer Nano S (Malvern) and presented either as the distribution of particle size or the average size (Z average). To monitor amyloid content, reactions were diluted into ThT solutions with a final concentration of 10 μM ThT and 30 mM glycine, pH 8.5. Fluorescence emission between 480 nm and 500 nm was measured immediately after dilution, using an excitation wavelength of 446 nm in a Tecan Safire II. Electron micrographs were acquired by using either a Philips EM208 or a Tecnai T12 transmission electron microscope. A sample of 2.5 μl of a 10 μM protein solution was negatively stained with equal volume of a saturated solution of uranyl acetate. Filter retardation assays were performed as described in52 and revealed with SOD-100E antibody (Stressgen).
Note that Sypro Orange is not present during any aggregation reactions.
Acknowledgements
We are very grateful to Nigel Unwin for his help with preparing samples for electron microscopy and comments on the manuscript and to Dr Jason Day for the ICP-MS analyses. We thank Benjamin Dehay for cloning wild-type and G85R human SOD1, Lori Passmore and Graham Fraser for help with electron microscopes, David Owen for quantitative amino acid analyses, Sarath C. Janga and Madan Babu for help with data processing and Michel Goedert for discussions and comments on the manuscript. The Medical Research Council and the EMBO Young Investigator Programme supported this research.
Edited by S. Radford
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2010.04.019
Appendix A.
Supplementary Fig. 1. Characterization of purified SOD1. (a) NuPAGE analysis of 1.2 μg of purified SOD1WT and its mutants. Note that some mutants migrate differently from SOD1WT, in agreement with previous studies.1 (b) Circular dichroism analyses of SOD1 proteins showing the characteristic spectra with the typical shoulder at 230 nm for the metallated SOD1 proteins.2 Solid lines: WTL and WT. Dashed lines: MBR mutants. Supplementary Fig. 2. Increased surface hydrophobicity upon thermal denaturation of ALS-causing SOD1 mutants. (a) Sypro Orange fluorescence during thermal unfolding of as-purified SOD1 and (b) EDTA-treated SOD1. Data are means and s.d. of 3 independent experiments. Supplementary Fig. 3. Metal contents of as-purified SOD1 variants and comparison of metal contents, hydrophobicity and apparent Tm, derived from Sypro Orange analysis. (a) Copper (Cu) and Zinc (Zn) contents assessed by ICP-MS expressed as metal equivalents per dimer, considering that fully metallated SOD1 would contain 2 Cu and 2 Zn equivalents per dimer. (b) Hydrophobicity of mutant SOD1 relative to wild-type (Δhydrophobicity), measured by thermal unfolding in the presence of Sypro Orange (Fig. 2) and metal content. (c) apparent Tm values of SOD1 variants obtained from thermal denaturation in the presence of Sypro Orange. (d) Hydrophobicity of mutant SOD1 relative to wild-type (Δhydrophobicity) and melting temperature (ΔTm), measured by thermal unfolding in the presence of Sypro Orange, between SOD1 mutants and SOD1WT. Supplementary Fig. 4. As-purified SOD1WT and SOD1A4V do not aggregate at pH 6.3. Analyses of SOD1 particle size by DLS. Supplementary Fig. 5. Aggregation of SOD1 mutants monitored by DLS (a, c) and Congo Red binding (b, d). SOD1A4V were aggregated at pH 3.9 for 3 days or at 50°C for 5 days. A shift in the absorbance maxima suggests the presence of amyloid structures. Supplementary Fig. 6. Aggregation of as-purified ALS-causing SOD1 mutants is caused by their increased propensity to expose hydrophobic surfaces. (a) Correlation between surface hydrophobicity measured by Sypro Orange fluorescence and aggregation measured by DLS. Aggregation is expressed as the percentage of particles with a diameter larger than 12.5 nm. (b) Congo red binding to aggregated SOD1 proteins. Proteins were exposed to TFE for 1 day. A shift in the absorbance maxima suggests the presence of amyloid structures.
References
- 1.Bruijn L.I., Miller T.M., Cleveland D.W. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu. Rev. Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 2.Valentine J.S., Doucette P.A., Zittin Potter S. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu. Rev. Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 3.Reaume A.G., Elliott J.L., Hoffman E.K., Kowall N.W., Ferrante R.J., Siwek D.F. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat. Genet. 1996;13:43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 4.Gurney M.E., Pu H., Chiu A.Y., Dal Canto M.C., Polchow C.Y., Alexander D.D. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 5.Shibata N., Hirano A., Kobayashi M., Siddique T., Deng H.X., Hung W.Y. Intense superoxide dismutase-1 immunoreactivity in intracytoplasmic hyaline inclusions of familial amyotrophic lateral sclerosis with posterior column involvement. J. Neuropathol. Exp. Neurol. 1996;55:481–490. doi: 10.1097/00005072-199604000-00011. [DOI] [PubMed] [Google Scholar]
- 6.Kato S., Nakashima K., Horiuchi S., Nagai R., Cleveland D.W., Liu J. Formation of advanced glycation end-product-modified superoxide dismutase-1 (SOD1) is one of the mechanisms responsible for inclusions common to familial amyotrophic lateral sclerosis patients with SOD1 gene mutation, and transgenic mice expressing human SOD1 gene mutation. Neuropathology. 2001;21:67–81. doi: 10.1046/j.1440-1789.2001.00359.x. [DOI] [PubMed] [Google Scholar]
- 7.Bruijn L.I., Houseweart M.K., Kato S., Anderson K.L., Anderson S.D., Ohama E. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–1854. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- 8.Lindberg M.J., Tibell L., Oliveberg M. Common denominator of Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis: decreased stability of the apo state. Proc. Natl Acad. Sci. USA. 2002;99:16607–16612. doi: 10.1073/pnas.262527099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayward L.J., Rodriguez J.A., Kim J.W., Tiwari A., Goto J.J., Cabelli D.E. Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. J. Biol. Chem. 2002;277:15923–15931. doi: 10.1074/jbc.M112087200. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez J.A., Shaw B.F., Durazo A., Sohn S.H., Doucette P.A., Nersissian A.M. Destabilization of apoprotein is insufficient to explain Cu,Zn-superoxide dismutase-linked ALS pathogenesis. Proc. Natl Acad. Sci. USA. 2005;102:10516–10521. doi: 10.1073/pnas.0502515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banci L., Bertini I., Durazo A., Girotto S., Gralla E.B., Martinelli M. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: a possible general mechanism for familial ALS. Proc. Natl Acad. Sci. USA. 2007;104:11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durazo A., Shaw B.F., Chattopadhyay M., Faull K.F., Nersissian A.M., Valentine J.S., Whitelegge J.P. Metal-free superoxide dismutase-1 and three different amyotrophic lateral sclerosis variants share a similar partially unfolded beta-barrel at physiological temperature. J. Biol. Chem. 2009;284:34382–34389. doi: 10.1074/jbc.M109.052076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiwari A., Hayward L.J. Familial amyotrophic lateral sclerosis mutants of copper/zinc superoxide dismutase are susceptible to disulfide reduction. J. Biol. Chem. 2003;278:5984–5992. doi: 10.1074/jbc.M210419200. [DOI] [PubMed] [Google Scholar]
- 14.Lindberg M.J., Normark J., Holmgren A., Oliveberg M. Folding of human superoxide dismutase: disulfide reduction prevents dimerization and produces marginally stable monomers. Proc. Natl Acad. Sci. USA. 2004;101:15893–15898. doi: 10.1073/pnas.0403979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chattopadhyay M., Durazo A., Sohn S.H., Strong C.D., Gralla E.B., Whitelegge J.P., Valentine J.S. Initiation and elongation in fibrillation of ALS-linked superoxide dismutase. Proc. Natl Acad. Sci. USA. 2008;105:18663–18668. doi: 10.1073/pnas.0807058105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karch C.M., Prudencio M., Winkler D.D., Hart P.J., Borchelt D.R. Role of mutant SOD1 disulfide oxidation and aggregation in the pathogenesis of familial ALS. Proc. Natl Acad. Sci. USA. 2009;106:7774–7779. doi: 10.1073/pnas.0902505106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rakhit R., Cunningham P., Furtos-Matei A., Dahan S., Qi X.F., Crow J.P. Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J. Biol. Chem. 2002;277:47551–47556. doi: 10.1074/jbc.M207356200. [DOI] [PubMed] [Google Scholar]
- 18.Hough M.A., Grossmann J.G., Antonyuk S.V., Strange R.W., Doucette P.A., Rodriguez J.A. Dimer destabilization in superoxide dismutase may result in disease-causing properties: structures of motor neuron disease mutants. Proc. Natl Acad. Sci. USA. 2004;101:5976–5981. doi: 10.1073/pnas.0305143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Caruano-Yzermans A., Rodriguez A., Scheurmann J.P., Slunt H.H., Cao X. Disease-associated mutations at copper ligand histidine residues of superoxide dismutase 1 diminish the binding of copper and compromise dimer stability. J. Biol. Chem. 2007;282:345–352. doi: 10.1074/jbc.M604503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furukawa Y., Kaneko K., Yamanaka K., O'Halloran T.V., Nukina N. Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem. 2008;283:24167–24176. doi: 10.1074/jbc.M802083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilcox K.C., Zhou L., Jordon J.K., Huang Y., Yu Y., Redler R.L. Modifications of superoxide dismutase (SOD1) in human erythrocytes: a possible role in amyotrophic lateral sclerosis. J. Biol. Chem. 2009;284:13940–13947. doi: 10.1074/jbc.M809687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandelin E., Nordlund A., Andersen P.M., Marklund S.S., Oliveberg M. Amyotrophic lateral sclerosis-associated copper/zinc superoxide dismutase mutations preferentially reduce the repulsive charge of the proteins. J. Biol. Chem. 2007;282:21230–21236. doi: 10.1074/jbc.M700765200. [DOI] [PubMed] [Google Scholar]
- 23.Tiwari A., Xu Z., Hayward L.J. Aberrantly increased hydrophobicity shared by mutants of Cu,Zn-superoxide dismutase in familial amyotrophic lateral sclerosis. J. Biol. Chem. 2005;280:29771–29779. doi: 10.1074/jbc.M504039200. [DOI] [PubMed] [Google Scholar]
- 24.Lindberg M.J., Bystrom R., Boknas N., Andersen P.M., Oliveberg M. Systematically perturbed folding patterns of amyotrophic lateral sclerosis (ALS)-associated SOD1 mutants. Proc. Natl Acad. Sci. USA. 2005;102:9754–9759. doi: 10.1073/pnas.0501957102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruns C.K., Kopito R.R. Impaired post-translational folding of familial ALS-linked Cu, Zn superoxide dismutase mutants. EMBO J. 2007;26:855–866. doi: 10.1038/sj.emboj.7601528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rumfeldt J.A., Lepock J.R., Meiering E.M. Unfolding and folding kinetics of amyotrophic lateral sclerosis-associated mutant Cu,Zn superoxide dismutases. J. Mol. Biol. 2009;385:278–298. doi: 10.1016/j.jmb.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Hawe A., Sutter M., Jiskoot W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm. Res. 2008;25:1487–1499. doi: 10.1007/s11095-007-9516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nordlund A., Oliveberg M. SOD1-associated ALS: a promising system for elucidating the origin of protein-misfolding disease. HFSP J. 2008;2:354–364. doi: 10.2976/1.2995726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q., Johnson J.L., Agar N.Y., Agar J.N. Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS Biol. 2008;6:e170. doi: 10.1371/journal.pbio.0060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratovitski T., Corson L.B., Strain J., Wong P., Cleveland D.W., Culotta V.C., Borchelt D.R. Variation in the biochemical/biophysical properties of mutant superoxide dismutase 1 enzymes and the rate of disease progression in familial amyotrophic lateral sclerosis kindreds. Hum. Mol. Genet. 1999;8:1451–1460. doi: 10.1093/hmg/8.8.1451. [DOI] [PubMed] [Google Scholar]
- 31.Ohi T., Nabeshima K., Kato S., Yazawa S., Takechi S. Familial amyotrophic lateral sclerosis with His46Arg mutation in Cu/Zn superoxide dismutase presenting characteristic clinical features and Lewy body-like hyaline inclusions. J. Neurol. Sci. 2004;225:19–25. doi: 10.1016/j.jns.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Tiwari A., Liba A., Sohn S.H., Seetharaman S.V., Bilsel O., Matthews C.R. Metal deficiency increases aberrant hydrophobicity of mutant superoxide dismutases that cause amyotrophic lateral sclerosis. J. Biol. Chem. 2009;58:2565–2573. doi: 10.1074/jbc.M109.043729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stathopulos P.B., Rumfeldt J.A., Scholz G.A., Irani R.A., Frey H.E., Hallewell R.A. Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis show enhanced formation of aggregates in vitro. Proc. Natl Acad. Sci. USA. 2003;100:7021–7026. doi: 10.1073/pnas.1237797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo M.C., Aulabaugh A., Jin G., Cowling R., Bard J., Malamas M., Ellestad G. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal. Biochem. 2004;332:153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 35.DiDonato M., Craig L., Huff M.E., Thayer M.M., Cardoso R.M., Kassmann C.J. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization. J. Mol. Biol. 2003;332:601–615. doi: 10.1016/s0022-2836(03)00889-1. [DOI] [PubMed] [Google Scholar]
- 36.Arnesano F., Banci L., Bertini I., Martinelli M., Furukawa Y., O'Halloran T.V. The unusually stable quaternary structure of human Cu,Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. J. Biol. Chem. 2004;279:47998–48003. doi: 10.1074/jbc.M406021200. [DOI] [PubMed] [Google Scholar]
- 37.Khare S.D., Caplow M., Dokholyan N.V. The rate and equilibrium constants for a multistep reaction sequence for the aggregation of superoxide dismutase in amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. USA. 2004;101:15094–15099. doi: 10.1073/pnas.0406650101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiti F., Webster P., Taddei N., Clark A., Stefani M., Ramponi G., Dobson C.M. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc. Natl Acad. Sci. USA. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., Xu G., Gonzales V., Coonfield M., Fromholt D., Copeland N.G. Fibrillar inclusions and motor neuron degeneration in transgenic mice expressing superoxide dismutase 1 with a disrupted copper-binding site. Neurobiol. Dis. 2002;10:128–138. doi: 10.1006/nbdi.2002.0498. [DOI] [PubMed] [Google Scholar]
- 40.Rousseau E., Dehay B., Ben-Haiem L., Trottier Y., Morange M., Bertolotti A. Targeting expression of expanded polyglutamine proteins to the endoplasmic reticulum or mitochondria prevents their aggregation. Proc. Natl Acad. Sci. USA. 2004;101:9648–9653. doi: 10.1073/pnas.0403015101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato S., Hayashi H., Nakashima K., Nanba E., Kato M., Hirano A. Pathological characterization of astrocytic hyaline inclusions in familial amyotrophic lateral sclerosis. Am. J. Pathol. 1997;151:611–620. [PMC free article] [PubMed] [Google Scholar]
- 42.Chiti F., Stefani M., Taddei N., Ramponi G., Dobson C.M. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 43.Zibaee S., Jakes R., Fraser G., Serpell L.C., Crowther R.A., Goedert M. Sequence determinants for amyloid fibrillogenesis of human alpha-synuclein. J. Mol. Biol. 2007;374:454–464. doi: 10.1016/j.jmb.2007.09.039. [DOI] [PubMed] [Google Scholar]
- 44.Khare S.D., Wilcox K.C., Gong P., Dokholyan N.V. Sequence and structural determinants of Cu, Zn superoxide dismutase aggregation. Proteins. 2005;61:617–632. doi: 10.1002/prot.20629. [DOI] [PubMed] [Google Scholar]
- 45.Elam J.S., Taylor A.B., Strange R., Antonyuk S., Doucette P.A., Rodriguez J.A. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat. Struct. Biol. 2003;10:461–467. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- 46.Zetterstrom P., Stewart H.G., Bergemalm D., Jonsson P.A., Graffmo K.S., Andersen P.M. Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc. Natl Acad. Sci. USA. 2007;104:14157–14162. doi: 10.1073/pnas.0700477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vande Velde C., Miller T.M., Cashman N.R., Cleveland D.W. Selective association of misfolded ALS-linked mutant SOD1 with the cytoplasmic face of mitochondria. Proc. Natl Acad. Sci. USA. 2008;105:4022–4027. doi: 10.1073/pnas.0712209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J., Farr G.W., Zeiss C.J., Rodriguez-Gil D.J., Wilson J.H., Furtak K. Progressive aggregation despite chaperone associations of a mutant SOD1-YFP in transgenic mice that develop ALS. Proc. Natl Acad. Sci. USA. 2009;106:1392–1397. doi: 10.1073/pnas.0813045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borchelt D.R., Guarnieri M., Wong P.C., Lee M.K., Slunt H.S., Xu Z.S. Superoxide dismutase 1 subunits with mutations linked to familial amyotrophic lateral sclerosis do not affect wild-type subunit function. J. Biol. Chem. 1995;270:3234–3238. doi: 10.1074/jbc.270.7.3234. [DOI] [PubMed] [Google Scholar]
- 50.Arndt V., Rogon C., Hohfeld J. To be, or not to be–molecular chaperones in protein degradation. Cell Mol. Life Sci. 2007;64:2525–2541. doi: 10.1007/s00018-007-7188-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jonsson P.A., Ernhill K., Andersen P.M., Bergemalm D., Brannstrom T., Gredal O. Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain. 2004;127:73–88. doi: 10.1093/brain/awh005. [DOI] [PubMed] [Google Scholar]
- 52.Rousseau E., Kojima R., Hoffner G., Djian P., Bertolotti A. Misfolding of proteins with a polyglutamine expansion is facilitated by proteasomal chaperones. J. Biol. Chem. 2009;284:1917–1929. doi: 10.1074/jbc.M806256200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng H.X., Hentati A., Tainer J.A., Iqbal Z., Cayabyab A., Hung W.Y. Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase. Science. 1993;261:1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 54.Roseman M.A. Hydrophobicity of the peptide C = O.H-N hydrogen-bonded group. J. Mol. Biol. 1988;201:621–623. doi: 10.1016/0022-2836(88)90642-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Characterization of purified SOD1. (a) NuPAGE analysis of 1.2 μg of purified SOD1WT and its mutants. Note that some mutants migrate differently from SOD1WT, in agreement with previous studies.1 (b) Circular dichroism analyses of SOD1 proteins showing the characteristic spectra with the typical shoulder at 230 nm for the metallated SOD1 proteins.2 Solid lines: WTL and WT. Dashed lines: MBR mutants. Supplementary Fig. 2. Increased surface hydrophobicity upon thermal denaturation of ALS-causing SOD1 mutants. (a) Sypro Orange fluorescence during thermal unfolding of as-purified SOD1 and (b) EDTA-treated SOD1. Data are means and s.d. of 3 independent experiments. Supplementary Fig. 3. Metal contents of as-purified SOD1 variants and comparison of metal contents, hydrophobicity and apparent Tm, derived from Sypro Orange analysis. (a) Copper (Cu) and Zinc (Zn) contents assessed by ICP-MS expressed as metal equivalents per dimer, considering that fully metallated SOD1 would contain 2 Cu and 2 Zn equivalents per dimer. (b) Hydrophobicity of mutant SOD1 relative to wild-type (Δhydrophobicity), measured by thermal unfolding in the presence of Sypro Orange (Fig. 2) and metal content. (c) apparent Tm values of SOD1 variants obtained from thermal denaturation in the presence of Sypro Orange. (d) Hydrophobicity of mutant SOD1 relative to wild-type (Δhydrophobicity) and melting temperature (ΔTm), measured by thermal unfolding in the presence of Sypro Orange, between SOD1 mutants and SOD1WT. Supplementary Fig. 4. As-purified SOD1WT and SOD1A4V do not aggregate at pH 6.3. Analyses of SOD1 particle size by DLS. Supplementary Fig. 5. Aggregation of SOD1 mutants monitored by DLS (a, c) and Congo Red binding (b, d). SOD1A4V were aggregated at pH 3.9 for 3 days or at 50°C for 5 days. A shift in the absorbance maxima suggests the presence of amyloid structures. Supplementary Fig. 6. Aggregation of as-purified ALS-causing SOD1 mutants is caused by their increased propensity to expose hydrophobic surfaces. (a) Correlation between surface hydrophobicity measured by Sypro Orange fluorescence and aggregation measured by DLS. Aggregation is expressed as the percentage of particles with a diameter larger than 12.5 nm. (b) Congo red binding to aggregated SOD1 proteins. Proteins were exposed to TFE for 1 day. A shift in the absorbance maxima suggests the presence of amyloid structures.