This work explores how organogenesis of the shoot apical meristem (SAM)-less shoots evolved from that of the typical SAM-mediated shoots in the aquatic eudicot Podostemaceae. Based on the expression patterns of STM, WUS, and ARP orthologs, the leaf in SAM-less shoot species is initiated as a SAM and differentiates into a single apical leaf, resulting in the evolution of novel shoot-leaf mixed organs in Podostemaceae.
Abstract
Podostemaceae (the river weeds) are ecologically and morphologically unusual angiosperms. The subfamily Tristichoideae has typical shoot apical meristems (SAMs) that produce leaves, but Podostemoideae is devoid of SAMs and new leaves arise below the base of older leaves. To reveal the genetic basis for the evolution of novel shoot organogenesis in Podostemaceae, we examined the expression patterns of key regulatory genes for shoot development (i.e., SHOOT MERISTEMLESS (STM), WUSCHEL (WUS), and ASYMMETRIC LEAVES1/ROUGH SHEATH2/PHANTASTICA (ARP) orthologs) in Tristichoideae and Podostemoideae. In the SAM-mediated shoots of Tristichoideae, like in model plants, STM and WUS orthologs were expressed in the SAM. In the SAM-less shoots of Podostemoideae, STM and WUS orthologs were expressed in the initiating leaf/bract primordium. In older leaf/bract primordia, WUS expression disappeared and STM expression became restricted to the basal part, whereas ARP was expressed in the distal part in a complementary pattern to STM expression. In the reproductive shoots of Podostemoideae with a normal mode of flower development, STM and WUS were expressed in the floral meristem, but not in the floral organs, similar to the pattern in model plants. These results suggest that the leaf/bract of Podostemoideae is initiated as a SAM and differentiates into a single apical leaf/bract, resulting in the evolution of novel shoot-leaf mixed organs in Podostemaceae.
INTRODUCTION
One of the important questions in biology is how morphological novelty originated during evolution. Angiosperms have in common aerial-shoot and underground-root systems. The aerial system is derived from the shoot apical meristem (SAM), which generates indeterminate vegetative growth of the stem and produces determinate lateral leaves with shoot branches in their axils, whereas the underground system is derived from the root apical meristem (Esau, 1965). However, some plants, such as Podostemaceae (the river weeds), Streptocarpus (Gesneriaceae; the cape primroses), and Lemnaceae (the duckweeds), have extraordinary body plans (Bell, 2008). These plants show unusual shoot organogenesis involving modification of the SAM, which may reflect adaptation to new habitats and may yield lineages that can then diversify in a new adaptive zone.
Morphological novelty requires changes in development. Recent evolutionary developmental studies suggest that morphological innovations are associated with spatial and temporal changes in the expression of key regulatory genes, as well as mutations of their coding regions (Carroll, 2008). In plants, the spatial and temporal alteration of expression of key regulators is suggested to be involved in the evolution of novel organs, for example, the Chinese lantern (inflated calyx) of Physalis (He and Saedler, 2005), the Streptocarpus phyllomorph (Harrison et al., 2005), and compound leaves in diverse angiosperms (Bharathan et al., 2002).
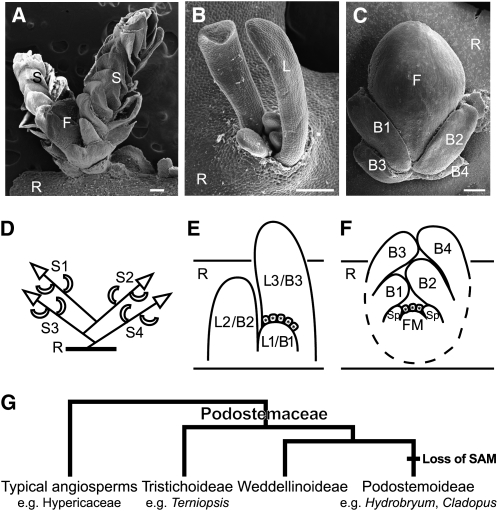
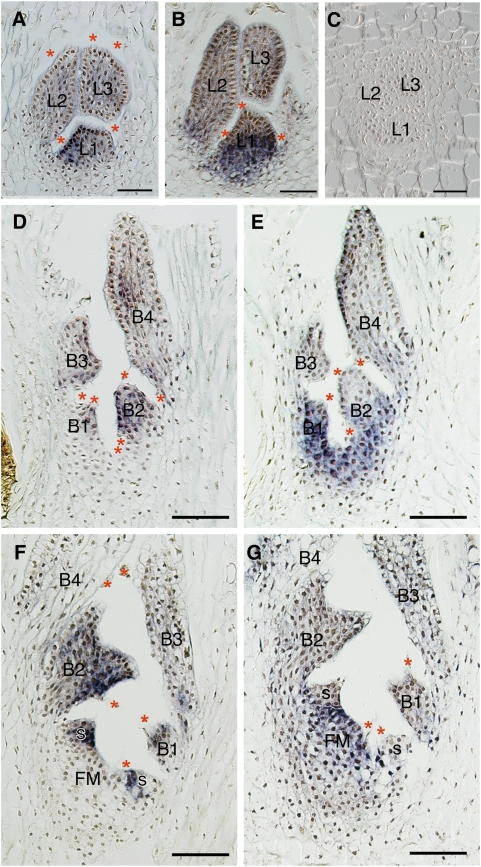
Aquatic eudicot Podostemaceae, the river weeds, exhibit unusual morphological features. They grow on rocks in rapids and waterfalls in the tropics and subtropics of the world. Most species have flattened roots that creep on rock surfaces, with adventitious shoots produced from the root (Figures 1A to 1C; see Supplemental Figure 1 online; Rutishauser, 1997). This is in contrast with most angiosperms, which have distinct aerial-shoot and underground-root systems. The three subfamilies of Podostemaceae exhibit different patterns of shoot development: SAM-mediated shoot development in Tristichoideae and Weddellinoideae and SAM-less shoot development in Podostemoideae. The subfamily Tristichoideae (e.g., Terniopsis) and Weddellinoideae show typical shoot organogenesis with tunica-corpus SAMs producing scaly leaves on their flanks and sympodial branching in which a new lateral shoot arises from the base of a parental shoot (Figures 1A and 1D; Imaichi et al., 1999; Koi and Kato, 2007; Fujinami and Imaichi, 2009). By contrast, in the subfamily Podostemoideae (e.g., Hydrobryum and Cladopus), the leaves form from an apparent shoot that lacks SAMs (Figures 1B and 1E). In well-examined Cladopus queenslandicus and Zeylanidium subulatum, each new leaf primordium arises below the base of an existing (the second youngest) leaf, without any structure recognizable as a SAM. The new leaf is histologically connected to its parental leaf, and the two leaves only become separated by an abscission-like process, in which vacuolated cells between the two leaves detach from the surrounding tissue (endogenous origin; Imaichi et al., 2005; Koi et al., 2005). This developmental process is also observed in reproductive shoots in Hydrobryum japonicum (Figure 1C; Katayama et al., 2008). The new bracts arise below the base of the second youngest bract in the same manner as vegetative leaf development (Figure 1E). Likewise, the floral meristem arises between and under the two youngest bracts endogenously and involves vacuolated cell detachment (Figure 1F). However, a cupular envelope (spathella: a floral organ unique to Podostemoideae) and other floral organs (tepals, stamens, and pistil) develop in an ordinary (exogenous) manner from a conventional floral meristem (Figures 1C and 1F; Katayama et al., 2008; Sehgal et al., 2009).
Figure 1.
Morphology, Development, and Phylogeny of Podostemaceae.
(A) to (C) Scanning electron microscopy images of T. minor (A), C. doianus (B), and H. japonicum (C). Bars = 200 μm.
(A) Shoots arising on the flank of a subcylindrical root of T. minor. A shoot complex consists of two determinate shoots (S) with scaly leaves and a floral bud (F).
(B) Vegetative shoot comprising a tuft of filliform leaves (L) arising from a root in C. doianus. Note that H. japonicum has a similar vegetative shoot (see Supplemental Figure 1 online).
(C) Reproductive shoot arising from the dorsal surface of a crustose root of H. japonicum. A floral bud forms between distichously arranged bracts and is enclosed by a spathella.
(D) to (F) Schematic illustrations showing the shoot development of Tristichoideae and Podostemoideae.
(D) Sympodial shoot branching of the Terniopsis shoot complex. Each determinate shoot arises at the base of an immediately older shoot.
(E) Longitudinal section of a vegetative shoot of Cladopus. A new leaf (L1) arises endogenously at the base of an existing leaf primordium (L3), involving detachment of vacuolated cells (boxes with asterisks). Note that H. japonicum has the same manner of leaf and bract development.
(F) Longitudinal section of a reproductive shoot of H. japonicum. A floral meristem (FM) arises between and under two youngest bracts (B1 and B2), involving vacuolated cell detachment (boxes with asterisks), and forms a spathella (Sp) on the flank. Tepals, stamens, and a pistil will arise later.
(G) Phylogenetic relationship of the subfamilies of Podostemaceae (Kita and Kato, 2001). The typical SAM disappeared in the early evolution of Podostemoideae.
B1 to B4, youngest to oldest bracts; F, floral bud; FM, floral meristem; L1 to L4, youngest to oldest leaves; R, root; S1 to S4, youngest to oldest shoots; Sp, spathella.
Molecular phylogenetic analyses suggest that the SAM-less shoot organogenesis of Podostemoideae is a derived character state, compared with the typical shoot organogenesis of Tristichoideae and Weddellinoideae (Figure 1G; Kita and Kato, 2001). The family is the most closely related to Hypericaceae (e.g., St. John’s wort) and Clusiaceae (e.g., mangosteen) in the eudicot Malpighiales (Wurdack and Davis, 2009), which have typical shoot development. Assuming that Tristichoideae and Weddellinoideae have retained the ancestral body plan, a major issue to be solved is how the organogenesis of the SAM-less shoots evolved from that of the typical SAM-mediated shoots of Tristichoideae and Weddellinoideae.
The genetic mechanisms underlying shoot development have been revealed in several model plants. The transcription factors homologous to SHOOT MERISTEMLESS (STM) and WUSCHEL (WUS) are involved in SAM initiation and maintenance. In Arabidopsis thaliana, STM, a class I KNOTTED-like homeobox (KNOX) gene, is expressed in the apical meristems of vegetative shoots, inflorescences, and flowers, where it maintains their cells in an undifferentiated state; STM is, by contrast, downregulated at the initiation site of lateral vegetative leaves and floral organs (Long et al., 1996; Long and Barton, 2000). WUS, a member of the WOX (for WUS HOMEOBOX) gene family, encodes a homeodomain transcription factor (Mayer et al., 1998). In Arabidopsis, WUS is expressed in the organizing center beneath a stem cell population within the SAM and establishes a feedback loop with CLAVATA3 to maintain a population size of stem cells (Brand et al., 2000; Schoof et al., 2000). Leaf development is regulated by an antagonistic expression between KNOX genes, which tend to suppress leaf development, and ASYMMETRIC LEAVES1/ROUGH SHEATH2/PHANTASTICA (ARP) genes, which tend to promote leaf identity. The ARP genes encode MYB-like transcription factors and are expressed at the site of leaf initiation and in young leaf primordia (Waites et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000). In Arabidopsis, downregulation of STM allows expression of AS1 at leaf initiation sites, while AS1 interacts with ASYMMETRIC LEAVES2 (AS2) to repress the expression of other class I KNOX genes in leaf primordia, resulting in leaf differentiation (Ori et al., 2000; Guo et al., 2008). Similar mutually exclusive expression patterns between KNOX1 and ARP genes are known in other model plants, and extensive conservation is suggested in the specification of the leaf initiation program in both monocots and eudicots (Waites et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000).
In this study, we isolated the orthologous STM, WUS, and ARP genes from three species of Podostemaceae (Terniopsis minor, H. japonicum, and Cladopus doianus) (see Supplemental Figures 2 to 4 online) and examined their expression patterns in the shoot of T. minor (Tristichoideae) and the SAM-less shoots of H. japonicum and C. doianus (Podostemoideae). The two genera of Podostemoideae to which these two species are assigned, along with closely related genera, form a derived monophyletic clade in the subfamily (Figure 1G; see Supplemental Figure 1 online; Kita and Kato, 2001). To elucidate whether cryptic SAMs exist in the SAM-less shoots of Podostemoideae, we investigated whether STM, WUS, and ARP genes are expressed in the SAM-less Podostemoideae shoots or not; and, if expressed, the patterns of the gene expressions in the SAM-less shoot development. Comparative analysis of gene expression patterns will provide insight into the evolutionary model underlying shoot novelty in the Podostemaceae.
RESULTS
Isolation and Phylogenetic Analyses of STM, WUS, and ARP Orthologs
We isolated STM, WUS, and ARP homologs from cDNA of T. minor, H. japonicum, and C. doianus (Tm STM, Tm WUS, and Tm PHAN from T. minor; Hj STM, Hj WUS, and Hj PHAN from H. japonicum; Cd STM, Cd WUS, and Cd PHAN from C. doianus). The inferred amino acid sequences were compared with those from Arabidopsis STM, WUS, and AS1, and phylogenetic analyses were performed along with other members of each gene family to examine orthology. The Podostemaceae STM genes had three conserved domains typical of class I KNOX genes (i.e., the MEINOX domain, ELK domain, and homeodomain). About 88% of the amino acids of the three domains were identical among the Podostemaceae STMs, and ~84% were shared between the Podostemaceae STMs and Arabidopsis STM (see Supplemental Figure 2A online). Phylogenetic analysis robustly supported a clade composed of Podostemaceae STM genes that included Ws STM from Weddellina squamulosa and Ps STM from Polypleurum stylosum and other STM orthologs (see Supplemental Figures 2B and 2C and Supplemental Data Set 1 online).
Tm WUS, Hj WUS, and Cd WUS also had the conserved regions encoding a WUSCHEL-type homeodomain, a WUS box, and an EAR-like domain (see Supplemental Figure 3A online). The inferred amino acid sequences of these domains were identical among the Podostemaceae WUS genes and showed 88% similarity to Arabidopsis WUS. Phylogenetic analysis showed that WUS orthologs, including Podostemaceae WUS genes, formed a clade in a tree of WOX family members, supported by a high (94% in neighbor-joining analysis and 82% in maximum parsimony analysis) bootstrap value (see Supplemental Figures 3B and 3C and Supplemental Data Set 2 online). Although this clade appears to be more closely related to asterid sequences than to the rosid sequence (WUS from Arabidopsis), it is likely an artifact from analysis of so few rosid WUS genes.
Podostemaceae ARP genes, Tm PHAN, Hj PHAN, and Cd PHAN, had the conserved MYB DNA binding domain at the N terminus (see Supplemental Figure 4A online), including two imperfect repeats (Jin and Martin, 1999). The inferred amino acid sequences of the MYB and CTD domains showed ~85% similarity among the Podostemaceae ARP genes and ~76% similarity among Podostemaceae ARPs and Arabidopsis AS1. Phylogenetic analysis placed the Podostemaceae ARP orthologs in a clade consisting of angiosperm ARP orthologs (see Supplemental Figures 4B and 4C and Supplemental Data Set 3 online). The relationship between the podostemad ARP genes and other ARP genes is unresolved, but the data are compatible with the podostemad ARP genes being embedded in a clade composed of other ARP-like genes. Therefore, we conclude that the isolated Podostemaceae STM, WUS, and ARP genes are orthologous to the STM, WUS, and ARP genes, respectively.
Expression of STM and WUS Orthologs in the SAM-Mediated Shoots of T. minor (Tristichoideae)
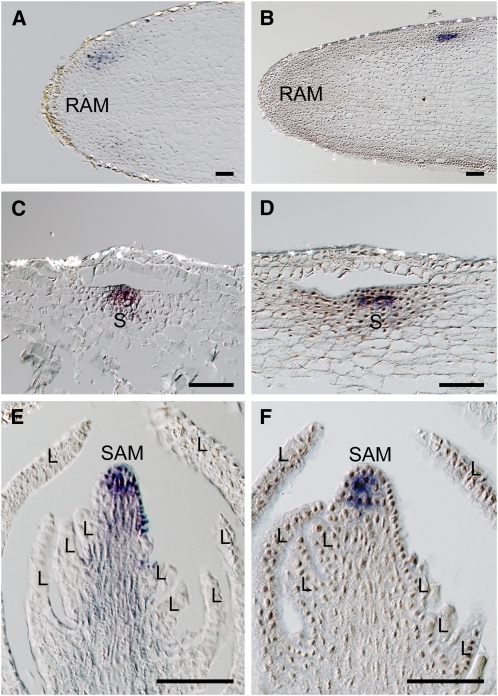
To assess whether the shoot development of T. minor has a typical genetic basis, we performed RNA in situ hybridization (ISH) using STM and WUS orthologs. Tm STM was expressed in the root in a site proximal to the root apical meristem, where a new shoot will arise (Figure 2A). The shoot primordium was initiated below the epidermal and subdermal layers of the root and involved vacuolated cell detachment in the subdermal layer, so that the third cell layer of the root became the epidermal layer of the arising shoot. Tm STM expression was detected in the new epidermal layer and inner tissue of the incipient shoot primordium (Figure 2C). In a mature shoot, Tm STM was expressed in the apex, including the epidermal layer, while it was not detected in the leaf primordium arising on its flank (Figure 2E). Tm WUS expression was observed in the inner tissue of the shoot primordium when the shoot arose within the root (Figures 2B and 2D). In a mature shoot, Tm WUS was expressed in all inner cells of the SAM, but neither in its epidermal layer nor in the leaf primordium (Figure 2F). Thus, the expression patterns of STM and WUS in T. minor are consistent with those of typical angiosperms.
Figure 2.
Expression of Tm STM and Tm WUS in T. minor Roots and Shoots.
(A) to (D) Frontal longitudinal sections of roots.
(A) Tm STM expression was detected close to the root apical meristem.
(B) Tm WUS expression was detected close to the root apical meristem.
(C) Tm STM was expressed in a shoot primordium below the epidermal and vacuolated subdermal layers of the root.
(D) Tm WUS was expressed in inner tissue of a shoot primordium at a developmental stage equivalent to (C).
(E) and (F) Longitudinal sections of a shoot apex.
(E) Tm STM was expressed in the whole apex. Note Tm STM was not detected in leaf primordia.
(F) Tm WUS was expressed in the inner tissue of the apex.
L, leaf; RAM, root apical meristem; S, shoot. Bars = 50 μm.
Expression of STM, ARP, and WUS Orthologs in the SAM-Less Shoots of H. japonicum (Podostemoideae)
To compare the gene expression patterns of the SAM-less shoot of this species with those of the T. minor shoot, we analyzed the STM and WUS orthologs and also the ARP ortholog. Results of RT-PCR showed that Hj STM, Hj WUS, and Hj PHAN expression occurs in young vegetative shoots, young reproductive shoots, flower buds, and root tips (see Supplemental Figure 5A online). Because no expression of Hj STM, Hj WUS, and Hj PHAN was detected in the root tip in ISH analyses (see below), all three signals in the root tip detected in the RT-PCR analyses are likely caused by the presence of young leaf primordia arising in close proximity to the root marginal meristem (see Supplemental Table 1 online). In addition, weaker Hj WUS expression was detected in mature flowers, and Hj PHAN expression was detected in mature leaves and mature flowers and other shoots that contained leaf-like organs (see Supplemental Figure 5A and Supplemental Table 1 online).
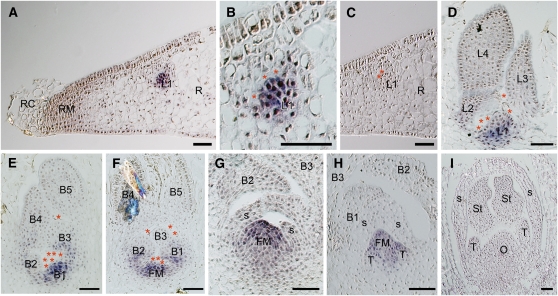
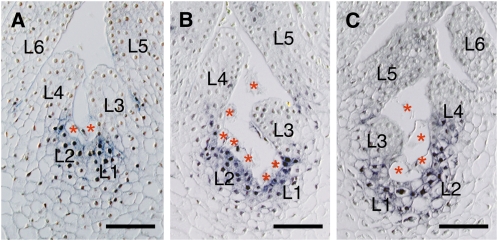
In H. japonicum, the first primordium arising at the proximal site to the root meristem develops into a first leaf, while the primordium from the root develops into a shoot in T. minor (Ota et al., 2001). Results of ISH analyses showed that Hj STM was expressed in the entire first leaf primordium (L1) (Figures 3A to 3C). No signal was detected in the root tip itself (Figure 3A). As the leaf primordium developed and new leaf primordia formed, accompanied by vacuolated cell detachment, Hj STM expression disappeared in the distal part of the youngest leaf primordium (L1) (i.e., was restricted to its basal part and eventually disappeared entirely from the older primordia [L3 and L4]) (Figure 3D). In the reproductive shoot, which contains distichous bracts, Hj STM was expressed in the entire youngest bract primordium (B1) and in the basal part of the second and third youngest bract primordia (B2 and B3) (Figure 3E), as in the vegetative leaf development. The floral meristem arises endogenously from the tissue between and under the two youngest bracts (B1 and B2), involving vacuolated cell detachment among the bracts and the emerging meristem, and Hj STM expression was detected in the incipient floral meristem (Figure 3F). As the dome-shaped floral meristem developed and formed a spathella (a cupular envelope) exogenously on its flank, Hj STM was expressed in the entire floral meristem (Figure 3G). Hj STM was not detected in the incipient spathella (S in Figure 3G). Furthermore, Hj STM expression was also not detected in the tepal primordia (T in Figure 3H), which arose exogenously from the floral meristem following the spathella. Subsequently, as the floral organs matured, Hj STM expression disappeared, and no Hj STM expression was detected in any mature tissues (Figure 3I; see Supplemental Figure 5A online).
Figure 3.
Expression of Hj STM in H. japonicum Shoots.
(A) to (D) Longitudinal sections of vegetative shoots at different stages of development.
(A) First leaf primordium (L1) arising from root tissue near the root margin.
(B) Magnification of (A). Hj STM was detectable in the entire L1.
(C) Negative control using Hj STM sense probe in the same leaf as (A).
(D) Developing vegetative shoot comprising of four young leaves. Hj STM was expressed in the basal part of L1.
(E) to (I) Longitudinal sections of reproductive shoots at different stages.
(E) Hj STM was expressed in the entire youngest bract primordium (B1) and the basal part of the second and third youngest bract primordia (B2 and B3).
(F) Hj STM was detectable in the emerging floral meristem (FM) and the basal part of B1 and B2.
(G) Expression of Hj STM was restricted to FM.
(H) No expression of Hj STM was detected in tepal primordia (T), but it was detectable in FM.
(I) Hj STM expression was undetectable in mature flower.
Asterisks indicate vacuolated cells. B1 to B5, youngest to oldest bracts; FM, floral meristem; L1 to L4, youngest to oldest leaves; O, ovary; R, root; RC, root cap; RM, root marginal meristem; S, spathella; St, stamen; T, tepal. Bars = 50 μm.
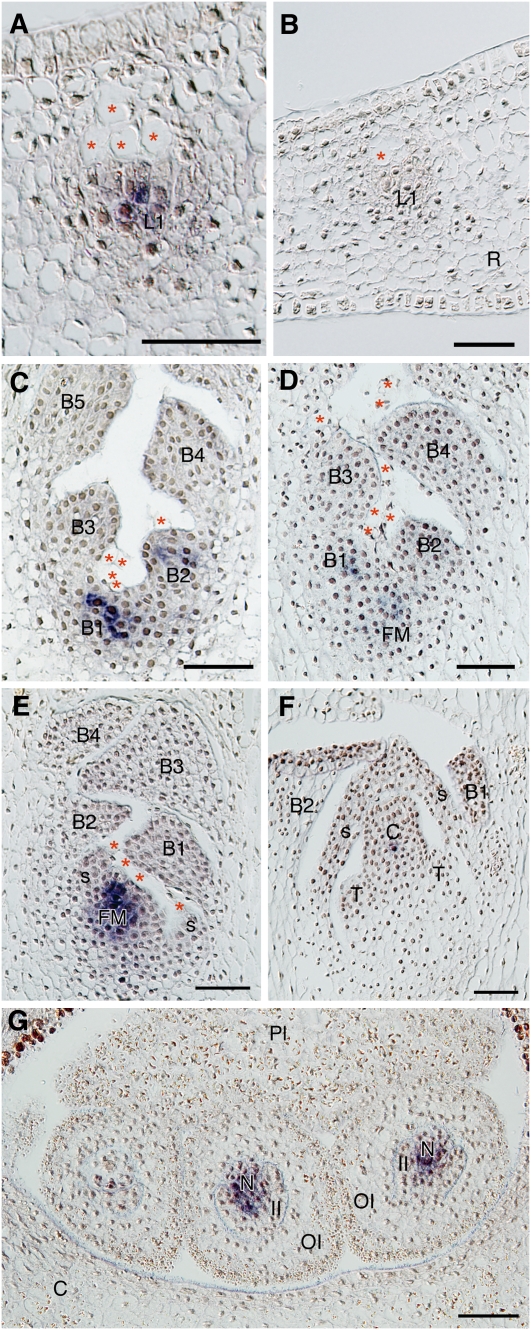
Hj WUS was expressed in the center of the first leaf primordium (L1) from the root but not in the epidermal layer (Figures 4A and 4B; at the same stage as in Figure 3A). Similar to the vegetative leaves, Hj WUS was expressed in the center of the two youngest bract primordia (B1 and B2 in Figure 4C), whereas it was not detected in older primordia. During floral meristem initiation, Hj WUS was detected in the tissue between and under B1 and B2 and in the center of the youngest bract primodium (B1) (Figure 4D). As the dome-shaped floral meristem emerged, Hj WUS was expressed in its central area (Figure 4E). However, Hj WUS expression was not detected in the developed floral organs (Figure 4F) but was present in the nucellus of the developing ovule (N in Figure 4G).
Figure 4.
Expression Pattern of Hj WUS in H. japonicum Shoots.
(A) and (B) Longitudinal sections of an initiating leaf.
(A) Hj WUS was detectable within the first leaf primordium (L1) arising from the inner root tissue near the root margin.
(B) Negative control using Hj WUS sense probe.
(C) to (G) Longitudinal sections of reproductive shoots at successive stages.
(C) Hj WUS was expressed within the youngest and second youngest bract primordia (B1 and B2) in a young shoot without flowers.
(D) Hj WUS was detected in B1 and at the site where the floral meristem would arise (FM).
(E) Expression of Hj WUS was restricted to within the emerging FM, which is flanked by spathella primordia.
(F) Weak restricted expression of Hj WUS in the center of a carpel (C).
(G) Hj WUS expression was detected in nucelli (N).
Asterisks indicate vacuolated cells. B1 to B5, youngest to oldest bracts; C, carpel; FM, floral meristem; II, inner integument; L1, youngest leaf; N, nucellus; OI, outer integument; Pl, placenta; R, root; S, spathella; T, tepal. Bars = 50 μm.
To compare the expression patterns between Hj STM and Hj PHAN, we conducted ISH analyses using serial sections of the same tissue. Hj PHAN was expressed in the distal part of the developing vegetative leaf primordia and complemented Hj STM expression (Figures 5A to 5C). Hj PHAN expression was also detected in the distal part of the developing bract primordia, showing a complementary pattern to Hj STM expression (Figures 5D and 5E). In flower development, Hj PHAN was expressed in the young spathella, where Hj STM expression was not detected (Figures 5F and 5G), consistent with typical flower development in other angiosperms.
Figure 5.
Comparison of Hj PHAN and Hj STM Expression in H. japonicum Shoots.
(A) to (C) Longitudinal sections of the same vegetative shoot.
(A) Hj PHAN was expressed in the distal part of the youngest leaf primordium (L1).
(B) Expression of Hj STM was restricted to the basal part of L1.
(C) Negative control using the Hj PHAN sense probe.
(D) and (E) Longitudinal sections of the same reproductive shoot.
(D) Hj PHAN was expressed in the distal part of the youngest bract primordia (B1 and B2).
(E) Expression of Hj STM was restricted to the basal part of B1 and B2.
(F) and (G) Longitudinal sections of the same reproductive shoot older than ([D] and [E]).
(F) Hj PHAN was expressed in a spathella (S) and distal parts of B1 and B2.
(G) Hj STM was expressed in the entire floral meristem (FM) and not in the spathella (S).
Asterisks indicate vacuolated cells. B1 to B4, youngest to oldest leaves; FM, floral meristem; L1 to L3, youngest to oldest leaves; S, spathella. Bars = 50 μm.
Expression of STM Ortholog in the SAM-Less Shoot of C. doianus (Podostemoideae)
To determine whether the disposition of STM in young leaves is general in Podostemoideae, STM expression in developing vegetative leaves was examined in C. doianus, which is related to H. japonicum. In RT-PCR analyses, Cd STM was detected only in young vegetative shoots (see Supplemental Figure 5B online). Results of ISH analysis of the vegetative shoot in successive developmental stages showed that Cd STM expression was first detected in the site below the vacuolated cells at the base of developing leaf primordia (L3 and L4), where new leaf primordia arise (L1 and L2) (Figure 6A). As the leaf primordium bulged, Cd STM was expressed in the entire body (L2 in Figure 6B). As the leaf primordium developed further, Cd STM expression disappeared in L3 (Figure 6C). The expression of Cd STM remained at the base of L3, where a new leaf primordium (L1) was arising (Figure 6C).
Figure 6.
Expression of Cd STM in C. doianus Shoots.
(A) to (C) Longitudinal sections of vegetative shoots at successive developmental stages.
(A) Cd STM was detected at the sites where new leaf primordia would arise (L1 and L2).
(B) Cd STM was detected in a slightly bulged leaf primordium (L2) and a site where a leaf primordium would arise (L1).
(C) Cd STM was not detected in a more developed leaf primordium (L3) while the expression remained at the bases of L3. A new leaf primordium would arise at the site where was Cd STM expressed (L1 and L2).
Asterisks indicate vacuolated cells. L1 to L6, youngest to oldest leaves. Bars = 50 μm.
DISCUSSION
Genetic Basis of the SAM-Less Shoot in Podostemoideae
Comparison of the amino acid sequences of the STM, WUS, and ARP orthologs shows that the three Podostemaceae species examined have the homeodomains and other functional domains. Furthermore, the expression patterns of the STM and WUS orthologs in the vegetative SAM of T. minor and those of the STM, WUS, and ARP orthologs in the floral meristem of H. japonicum correspond to those of STM, WUS, and AS1 in Arabidopsis (Figure 7A) and to their orthologs in other angiosperms (Piazza et al., 2005; see references cited in this review). They imply that the orthologs of the Podostemaceae preserve biochemical and developmental features identical or similar to those of other angiosperms, allowing one to use these genes as markers for cell identity.
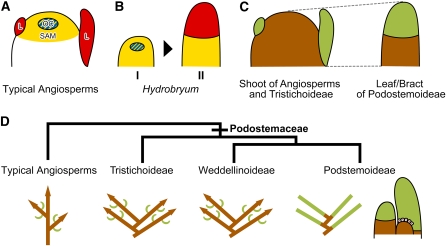
Figure 7.
Schematic Illustration of Gene Expression Patterns, Comparison of Organ Identity, and Podostemaceae Shoot Evolution.
(A) and (B) Expression patterns of STM (yellow), WUS (blue), and ARP (red) genes in the shoot apex of ordinary angiosperms (A) and in the leaf/bract of H. japonicum (B).
(A) The STM gene is expressed in the SAM but not in leaf primordia (L). The WUS gene is expressed in the organization center (OC). The ARP gene is expressed in leaf primordia (L), complementary to the STM gene.
(B) At an early stage of leaf/bract development (I), Hj STM is expressed in the entire leaf/bract primordium and Hj WUS is expressed in the center. At a later stage (II) (not at maturity), Hj STM expression is restricted in the basal part of the leaf/bract primordium, and Hj PHAN is expressed in the distal part.
(C) The same colors indicate comparable organs that have the same genetic identity. The leaf/bract of Podostemoideae (H. japonicum and C. doianus) is comparable to the shoot of Tristichoideae (T. minor) and typical angiosperms. The proportion of green and brown areas is not to scale.
(D) The sympodium model (modified from Koi and Kato, 2007). Illustrations below the phylogenetic tree (Kita and Kato, 2001) show branching patterns in which brown and green bars indicate shoot and leaf, respectively. Arrowheads indicate the presence of SAMs. A chain of leaves in Podostemoideae is derived from the sympodial shoots of Tristichoideae and Weddellinoideae. On the extreme right side is the development of a SAM-less shoot complex. A new SSM arises from the base of an older SSM and differentiates into an apical leaf.
In leaf/bract development of the SAM-less shoot of H. japonicum, Hj STM is expressed in the entire leaf/bract primordium, and Hj WUS is expressed in an inner subset of the STM-expressing cells (I in Figure 7B). This expression is similar to that in the floral meristem of H. japonicum and the SAMs of other angiosperms (Figure 7A). On the other hand, Hj PHAN is expressed in the distal part of the developing leaf/bract primordium, concomitantly with restriction of Hj STM expression to the basal part and loss of Hj WUS expression (II in Figure 7B). This complementary expression pattern of Hj STM and Hj PHAN is similar to the process of shoot versus leaf development in Arabidopsis and other angiosperms, in which STM genes are downregulated in the initiating leaf primordium while ARP genes are expressed (Figure 7A; Waites et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000). Taking the phylogeny of Podostemaceae into account, it is likely that the changes in expression pattern of genes involved in SAM maintenance and leaf initiation cause the change in the position of leaf initiation sites from the periphery of the SAM to its apex. Therefore, we interpret that the single leaf/bract of the Podostemoideae shoot is initiated as a SAM and later differentiates into an apical leaf/bract, and from a proximal shoot domain, a new leaf/bract primordium arises. Such a leaf/bract is not a leaf and bract in a strict sense, but comparable to a sympodial unit composed of a meristematic shoot zone and a single apical leaf and bract (Figure 7C). By contrast, the expression patterns of the three genes support the interpretation that the spathella, like the tepal and stamen, is a typical floral organ of leaf homology, while the flower as a whole is a typical shoot homolog, as suggested from morphological evidence (Katayama et al., 2008).
In Arabidopsis, WUS expression in the organizing center keeps stem cells in an undifferentiated state (Mayer et al., 1998). The SAM with a stem cell population continues indeterminate growth, forming lateral organs on the flank. The wus-1 mutant loses SAMs after several leaves have formed because the stem cell population has not been maintained (Laux et al., 1996). In H. japonicum, WUS expression is lost soon after shoot initiation. The loss, along with the changes of STM and ARP expression pattern in the shoot apex, might be involved in the determinate growth of the shoot, which eventually is differentiated to an apical leaf.
The expression of both KNOX and ARP genes in leaves has also been shown in many compound leaves. In such compound leaves, leaflet initiation appears to be regulated by the same mechanisms that are responsible for the initiation of leaves in the shoot apex (Kim et al., 2003; Hay and Tsiantis, 2006; Blein et al., 2008). KNOX expression is downregulated at the initiation of leaf primordia but is reestablished during the development of compound leaves. We suggest that compound leaves and Podostemoideae leaves/bracts are similarly formed via modifications in the expression of SAM- and leaf-specific genes, although the genetic regulation and resulting leaf morphology differ substantially between the two.
In Podostemaceae, adventitious shoots are produced near the root meristem. In this study, we found that the STM and WUS orthologs are expressed near the root apex where adventitious shoots arise in T. minor and H. japonicum. In Arabidopsis, the ectopic expression of WUS in the root induces adventitious shoots in close proximity to the root apical meristem (Gallois et al., 2004). This similarity suggests that such ectopic expression of shoot characteristic genes in roots may be involved in the development of the root-borne shoot in Podostemaceae.
Evolution of Novel Shoot Organogenesis of Podostemoideae
Based on the results of this study and previous developmental anatomical studies (Imaichi et al., 2005; Koi et al., 2005; Koi and Kato, 2007; Fujinami and Imaichi, 2009), we put forward a model of the evolution of novel shoot organogenesis in Podostemoideae (Figure 7D). We hypothesize that there are histologically invisible SAMs in Podostemoideae, which we denote sympodial shoot meristem (SSM). Repetitive SSM formation occurs by proliferation on the adaxial side of the existing SSM, and subsequently each SSM differentiates into a determinate organ (i.e., an apical leaf/bract), resulting in a chain of conventionally interpreted leaves of mixed shoot-leaf identity. Under this interpretation, plants in Tristichoideae and Weddellinoideae represent a transitional step toward the developmental pattern seen in Podostemoideae: each new SAM arises in a place proximal to a previously formed SAM, resulting in sympodial shoot branching (Koi and Kato, 2007; Fujinami and Imaichi, 2009). We suggest that the repetitive SSM formation of Podostemoideae is derived from the sympodial shoot branching of Tristichoideae and Weddellinoideae (the sympodium model; Figure 7D). Thus, the apparent absence of a SAM in Podostemoideae is not due to its loss but to the transformation of the shoot to a leaf-stem fuzzy organ.
The derivation of the Podostemoideae leaf from the more conventional shoot (stem + leaves) of Tristichoideae and Weddellinoideae presumably involves changes in the spatial and temporal expression of a genetic regulatory module comprising of STM, WUS, and ARP. It is known that some other angiosperms exhibit both stem (SAM) and leaf features in the same structures (e.g., leaf-shoot continuum) (Arber, 1950; Sinha, 1999). For example, the unifoliate plants of Streptocarpus (Gesneriaceae) are devoid of typical SAMs and have a single, inflorescence-bearing, indeterminate cotyledon (phyllomorph) (Jong and Burtt, 1975). The monocot Ruscus (Asparagaceae) has leaf-like, determinate stems (phylloclades) in the axil of scaly leaves (Arber, 1924). Molecular genetic studies in these two cases have suggested that the key regulatory genes for both SAM and leaf development are expressed in the same organ (Harrison et al., 2005; Hirayama et al., 2007), implying that similar network co-option of genes may be involved in the evolution of those shoot-leaf mixed organs.
Most Podostemaceae have a horizontal leading axis comprising of roots, which creep on rocks and produce adventitious determinate shoots. Our study suggests that the changes in expression patterns of shoot-characteristic genes may be involved in the evolution of leaf-like, root-borne shoots. The acquisition of novel shoot organogenesis resulted in the transformation of body plan from the typical aerial-shoot and underground-root system to the horizontal root and reduced shoot system of Podostemaceae, which may permit this family to invade submerged rock surfaces under high current pressure.
METHODS
Scanning Electron Microscopy Observation
The materials were fixed with FAA (formalin:acetic acid:50% ethanol = 5:5:90), dehydrated in an ethanol series, critical point dried, and coated with platinum-palladium. Observation was done using a JMS-6390LV SEM (Jeol) at 15 kV.
Isolation of Genes
Plants of Terniopsis minor, Hydrobryum japonicum, and Cladopus doianus were collected in Thailand and Japan (see Supplemental Table 2 online). They were immediately soaked in RNA Later (Ambion) and stored at 4°C for preservation of RNA. Total RNA was extracted using PureLink Plant RNA reagent (Invitrogen) and RNeasy mini columns from the RNeasy mini kit (Qiagen) and treated with DNase I (TaKaRa). First-strand cDNA was synthesized from the extracted RNA using SuperScript III reverse transcriptase (Invitrogen) and oligo(dT)-containing adaptor primer. Orthologs of STM, WUS, and PHAN were amplified using degenerate primers (see Supplemental Table 7 online) and TaKaRa Ex Taq under appropriate PCR conditions. The sequences of the 5′-ends of the genes were determined with the 5′-RACE system for rapid amplification of cDNA ends (Invitrogen) or the GeneRacer kit (Invitrogen). The amplified products were gel purified and cloned into the pGEM-T plasmid (Promega) following Shindo et al. (1999). Sequences were deposited in GenBank, and their accession numbers are given in Supplemental Table 3 online.
Phylogenetic Analysis
The deduced amino acid sequences of homologous genes were gathered from the National Center for Biotechnology Information DNA database (see Supplemental Tables 4 to 6 online). The amino acid sequences of the obtained genes of T. minor, H. japonicum, and C. doianus and the retrieved genes were aligned using ClustalX version 2.0.7 (Larkin et al., 2007) based on the Gonnet series matrices. Phylogenetic analyses were performed with PAUP* 4.0b10 (Swofford, 2002) based on the only conserved domains of the amino acid sequences in neighbor-joining and maximum parsimony analysis because alignment of nonconserved regions, which show high variability, was difficult in the whole gene family (see Supplemental Figures 2 to 4 online). For the neighbor-joining analyses, the distance metric was generated using mean character difference. Bootstrap values were calculated with 1000 replicates. For the maximum parsimony analyses, we performed heuristic searches. Trees were started using stepwise addition, followed by 100 random addition sequence replicates with the tree-bisection-reconnection branch-swapping algorithm. Bootstrap values were calculated with 100 replicates for STM and 1000 replicates for ARP and WUS. BELL1 and H1 of STM homologs, WOX10, WOX13, and WOX14 of WUS homologs, and At MYB97, At MYB124, Hv GAMYB, Lt GAMYB, and Os GAMYB of PHAN homologs were treated as outgroups.
RT-PCR
Collected samples of entire plants were soaked in RNA Later (Ambion), and several kinds of organs were dissected (young vegetative shoots, mature leaves, young reproductive shoots, flower buds, mature flowers, roots, and root tips in H. japonicum; young vegetative shoots, mature leaves, roots, and root tips in C. doianus) under a binocular microscope (WILD M10; Leica). Total RNA was extracted from each organ. The samples and contained organ type(s) are summarized in Supplemental Table 1 online. First-strand cDNAs were synthesized for each sample by the methods described above and used as a template for RT-PCR. We designed gene-specific primers (see Supplemental Table 7 online). RT-PCR reactions were performed according to standard procedures except for annealing temperature (50°C for Cd WUS, 52°C for HJ PHAN, 55°C for Hj GAPDH, Hj WUS, Cd GAPDH, and Cd STM, and 57°C for HJ STM) and cycle number (32 cycles for H. japonicum genes and 40 cycles for C. doianus), which were optimized for each gene to set up amplification below the saturation point. PCR products were subjected to gel electrophoresis and detected by ethidium bromide staining.
RNA ISH
Pieces of the shoots and roots were dissected in the field. Samples were immediately soaked in 4% paraformaldehyde (and 0.25% glutaraldehyde for the Hj WUS probe) in 0.1 M sodium phosphate buffer, pH 7.2, and fixed for 12 h at 4°C. The fixed materials were dehydrated in an ethyl and tertiary butyl alcohol series and mounted in Paraplast Plus (Oxford Labware). Microtome sections (6 to 8 μm thick) were mounted on MAS-coated glass slides (Matsunami). In preparing templates for all probes, nearly the full length of the coding regions of Hj STM, Hj WUS, Hj ARP, and Cd STM and the partial sequences of Tm STM and Tm WUS were amplified from the cDNA by PCR using gene-specific primers (see Supplemental Table 7 online) and TaKaRa Ex Taq followed by ligation into the pGEM-T vector (Promega) and amplified using the following primer sets: M13F and M13R, M13F and Sp6, or T7 and M13R. For the Tm STM probe template, the less conserved 5′-region (amplified with GeneRacer 5′ nested primer and Pod-STM-Pn in the 5′-RACE experiment) was used. To generate antisense or sense probes labeled with digoxigenin, RNAs were transcribed with T7 or SP6 RNA polymerase from the above templates with the DIG RNA labeling kit (Roche). The synthesized RNA probes were partially hydrolyzed to ~300 bp with alkaline solution (60 mM Na2CO3 and 40 mM NaHCO3, pH 10.2) at 60°C for an appropriate time. ISH was performed following the methods of Kouchi and Hata (1993). Signals were detected with the DIG nucleic acid detection kit (Roche) and observed under a light microscope (BX-50; Olympus).
Voucher specimens and seeds of T. minor, H. japonicum, and C. doianus are deposited in the Herbarium of the National Museum of Nature and Science (TNS) in Japan.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB512740 to AB512746, AB512748, and AB512750 to AB512754 (see Supplemental Table 3 online). Other sequence data are provided in Supplemental Tables 4 to 7 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Morphology of Podostemaceae.
Supplemental Figure 2. Structure and Phylogeny of Podostemaceae STM Genes.
Supplemental Figure 3. Structure and Phylogeny of Podostemaceae WUS Genes.
Supplemental Figure 4. Structure and Phylogeny of Podostemaceae ARP Genes.
Supplemental Figure 5. Results of RT-PCR Analyses of Gene Expression in Different Organs of Hydrobryum japonicum and Cladopus doianus.
Supplemental Table 1. Organs Contained in Samples Used in RT-PCR Analysis.
Supplemental Table 2. Sample Collection Sites.
Supplemental Table 3. GenBank Accession Numbers for Genes Isolated in This Study.
Supplemental Table 4. Gene Accession Numbers for KNOX Phylogeny.
Supplemental Table 5. Gene Accession Numbers for WOX Phylogeny.
Supplemental Table 6. Gene Accession Numbers for ARP Phylogeny.
Supplemental Table 7. Primers Used in This Study.
Supplemental Data Set 1. The Sequences Used to Generate the Phylogenies of the KNOX Family.
Supplemental Data Set 2. The Sequences Used to Generate the Phylogenies of the WOX Family.
Supplemental Data Set 3. The Sequences Used to Generate the Phylogenies of the ARP Family.
Supplementary Material
Acknowledgments
We thank H. Hirano and T. Suzaki for their technical assistance with the ISH experiments. We also thank T. Wongprasert for his help during the collection trips in Thailand, N. Yagi for phylogenetic analyses, and T. Yamada and R. Kofuji for reading the manuscript and providing helpful comments. This study was supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science.
References
- Arber A. (1924). Danae, Ruscus and Semele: A morphological study. Ann. Bot. (Lond.) 38: 229–260 [Google Scholar]
- Arber A. (1950). The Natural Philosophy of Plant Form. (Cambridge, UK: Cambridge University Press; ). [Google Scholar]
- Bell A.D. (2008). Plant Form: An Illustrated Guide to Flowering Plant Morphology. (Portland, OR: Timber Press; ). [Google Scholar]
- Bharathan G., Goliber T.E., Moore C., Kessler S., Pham T., Shinha N.R. (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860 [DOI] [PubMed] [Google Scholar]
- Blein T., Paulido A., Vialette-Guiraud A., Nikovics K., Morin H., Hay A., Johansen I.E., Tsiantis M., Laufs P. (2008). A conserved molecular framework for compound leaf development. Science 322: 1835–1839 [DOI] [PubMed] [Google Scholar]
- Brand U., Fletcher J.C., Hobe M., Meyerowitz E.M., Simon R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Byrne M.E., Barley R., Curtis M., Arroyo J.M., Dunham M., Hudson A., Martienssen R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Carroll S.B. (2008). Evo-Devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134: 25–36 [DOI] [PubMed] [Google Scholar]
- Esau K. (1965). Plant Anatomy, 2nd ed. (New York: Wiley; ). [Google Scholar]
- Fujinami R., Imaichi R. (2009). Developmental anatomy of Terniopsis malayana (Podostemaceae, subfamily Tristichoideae), with implications for body plan evolution. J. Plant Res. 122: 551–558 [DOI] [PubMed] [Google Scholar]
- Gallois J.L., Nora F.R., Mizukami Y., Sablowski R. (2004). WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 18: 375–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Thomas J., Collins G., Timmermans M.C.P. (2008). Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20: 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J., Möller M., Langdale J., Cronk Q., Hudson A. (2005). The role of KNOX genes in the evolution of morphological novelty in Streptocarpus. Plant Cell 17: 430–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsutra. Nat. Genet. 38: 942–947 [DOI] [PubMed] [Google Scholar]
- He C., Saedler H. (2005). Heterotopic expression of MPF2 is the key to the evolution of the Chinese lantern of Physalis, a morphological novelty in Solanaceae. Proc. Natl. Acad. Sci. USA 102: 5779–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama Y., Yamada Y., Oya Y., Ito M., Kato M., Imaichi R. (2007). Expression pattern of class I KNOX and YABBY genes in Ruscus aculeatus (Asparagaceae) with implications for phylloclade homology. Dev. Genes Evol. 217: 363–372 [DOI] [PubMed] [Google Scholar]
- Imaichi R., Hiyama Y., Kato M. (2005). Leaf development in the absence of a shoot apical meristem in Zeylanidium subulatum (Podostemaceae). Ann. Bot. (Lond.) 96: 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaichi R., Ichiba T., Kato M. (1999). Developmental morphology and anatomy of the vegetative organs in Malaccotristicha malayana (Podostemaceae). Int. J. Plant Sci. 160: 253–259 [Google Scholar]
- Jin H.L., Martin C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41: 577–585 [DOI] [PubMed] [Google Scholar]
- Jong K., Burtt B.L. (1975). The evolution of morphological novelty exemplified in the growth patterns of some Gesneriaceae. New Phytol. 75: 297–311 [Google Scholar]
- Katayama N., Koi S., Kato M. (2008). Developmental anatomy of the reproductive shoot in Hydrobryum japonicum (Podostemaceae). J. Plant Res. 121: 417–424 [DOI] [PubMed] [Google Scholar]
- Kim M., McCormimick S., Timmermans M., Sinha N. (2003). The expression domain of PHANTASTICA determines leaflet placement in compound leaves. Nature 424: 438–443 [DOI] [PubMed] [Google Scholar]
- Kita Y., Kato M. (2001). Infrafamilial phylogeny of the aquatic angiosperm Podostemaceae inferred from the nucleotide sequence of the matK gene. Plant Biol. 3: 156–163 [Google Scholar]
- Koi S., Imaichi R., Kato M. (2005). Endogenous leaf initiation in the apical-meristemless shoot of Cladopus queenslandicus (Podostemaceae) and implications for evolution of shoot morphology. Int. J. Plant Sci. 166: 199–206 [Google Scholar]
- Koi S., Kato M. (2007). Developmental morphology of the shoot in Weddellina squamulosa and implications for shoot evolution in the Podostemaceae. Ann. Bot. (Lond.) 99: 1121–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H., Hata S. (1993). Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 238: 106–119 [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Laux T., Mayer K.F.X., Berger J., Jürgens G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Long J.A., Barton M.K. (2000). Initiation of axillary and floral meristems in Arabidopsis. Dev. Biol. 218: 341–353 [DOI] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Mayer K.F., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998). Role of WUSHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Ori N., Eshed Y., Chuck G., Bowman J.L., Hake S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532 [DOI] [PubMed] [Google Scholar]
- Ota M., Imaichi R., Kato M. (2001). Developmental morphology of the thalloid Hydrobryum japonicum (Podostemaceae). Am. J. Bot. 88: 382–390 [PubMed] [Google Scholar]
- Piazza P., Jasinski S., Tsiantis M. (2005). Evolution of leaf developmental mechanisms. New Phytol. 167: 693–710 [DOI] [PubMed] [Google Scholar]
- Rutishauser R. (1997). Structural and developmental diversity in Podostemaceae (river-weeds). Aquat. Bot. 57: 29–70 [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F.X., Jürgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL gene. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Sehgal A., Sethi M., Mohan Ram H.Y. (2009). Development of the floral shoot and pre-anthesis cleistogamy in Hydrobryopsis sessilis (Podostemaceae). Bot. J. Linn. Soc. 159: 222–236 [Google Scholar]
- Shindo S., Ito M., Ueda K., Kato M., Hasebe M. (1999). Characterization of MADS genes in the gymnosperm Gnetum parvifolium and its implication on the evolution of reproductive organs in seed plants. Evol. Dev. 1: 180–190 [DOI] [PubMed] [Google Scholar]
- Sinha N. (1999). Leaf development in angiosperms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 419–446 [DOI] [PubMed] [Google Scholar]
- Swofford D.L. (2002). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. (Sunderland, MA: Sinauer Associates; ). [Google Scholar]
- Timmermans M.C., Hudson A., Becraft P.W., Nelson T. (1999). ROUGH SHEATH2: An Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284: 151–153 [DOI] [PubMed] [Google Scholar]
- Tsiantis M., Schneeberger R., Golz J.F., Freeling M., Langdale J.A. (1999). The maize rough sheathe2 gene and leaf development programs in monocot and dicot plants. Science 284: 154–156 [DOI] [PubMed] [Google Scholar]
- Waites R., Selvadurai H.R., Oliver I.R., Hudson A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93: 779–789 [DOI] [PubMed] [Google Scholar]
- Wurdack K.J., Davis C. (2009). Malpighiales phylogenetics: Gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am. J. Bot. 96: 1551–1570 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.