Abstract
A20 is a ubiquitin modifying enzyme that restricts NF-κB signals and protects cells against tumor necrosis factor (TNF) induced programmed cell death. Given recent data linking A20 (TNFAIP3) with human B cell lymphomas and systemic lupus erythematosus (SLE), we have generated mice bearing a floxed allele of Tnfaip3 to interrogate A20’s roles in regulating B cell functions. A20-deficient B cells are hyper-responsive to multiple stimuli and display exaggerated NF-κB responses to CD40 induced signals. Mice expressing absent or hypomorphic amounts of A20 in B cells possess elevated numbers of germinal center B cells, autoantibodies, and glomerular immunoglobulin deposits. A20 deficient B cells are resistant to Fas mediated cell death, likely due to increased expression of NF-κB-dependent anti-apoptotic proteins such as Bcl-x. These findings show that A20 can restrict B cell survival, while A20 protects other cells from TNF induced cell death. Our studies demonstrate how reduced A20 expression predisposes to autoimmunity.
Keywords: A20, ubiquitin, haploinsufficiency, NFkB, B cell selection, autoimmunity, programmed cell death, SLE
Introduction
Maintenance of B cell homeostasis requires proper intracellular integration of signals delivered from multiple surface receptors such as the B cell antigen receptor, Toll-like receptors (TLRs), B cell activating factor (BAFF) receptor, and CD40, as well as intracellular cues. Failure to integrate pathways such as NF-κB signaling can lead to B cell deficiency, aberrant B cell activity, or even lymphoma. Aberrant B cell tolerance and selection can cause production of autoantibodies, formation of immune complexes (IC), and ultimately tissue damage and autoimmune disease (Fairhurst et al., 2006).
Tnfaip3 encodes the A20 protein, a ubiquitin-modifying enzyme (Wertz et al., 2004; Boone et al., 2004). A20 was initially identified as a TNF-induced molecule that restricts TNF induced signaling (Opipari et al., 1990). Targeting of Tnfaip3 in mice revealed A20’s critical anti-inflammatory functions, as A20-deficient (Tnfaip3−/−) mice exhibit severe spontaneous multi-organ inflammation, cachexia, and perinatal death (Lee et al., 2000). Epistasis experiments revealed that A20 restricts TLR and nucleosome-binding oligomerization domain (NOD) triggered NF-κB signaling, in addition to TNF induced NF-κB and programmed cell death (PCD) signaling (Lee et al., 2000; Boone et al., 2004; Hitotsumatsu et al., 2008). Thus, A20 restricts a number of innate immune signaling pathways in macrophages and fibroblasts. The severe systemic inflammation and cachexia caused by A20 deficiency is ameliorated in mice that also lack the TLR adapter protein MyD88 (Turer et al., 2008). Radiation chimeras bearing Tnfaip3−/− hematopoietic cells also develop spontaneous systemic inflammation, which is alleviated by depletion of commensal intestinal bacteria with antibiotics (Turer et al., 2008). Thus, A20 maintains immune homeostasis and restricts the potentially pro-inflammatory nature of basal MyD88-dependent signals.
In addition to the innate immune functions described above in macrophages and fibroblasts, A20 is also expressed in T and B cells (Sarma et al., 1995; Lee et al., 2000). During T cell activation, A20 is recruited to the MALT-1-Bcl-10 scaffold complex, and is cleaved by the paracaspase MALT-1 (Coornaert et al., 2008). A20 has also been reported to de-ubiquitinate MALT-1 to restrict TCR signals (Duwel et al., 2009). A20 cleavage is also observed in B lymphoma cell lines in response to BCR stimulation (Coornaert et al., 2008). Other clues that A20 may play important roles in adaptive lymphocytes derives from human genetic studies that implicate A20 (or TNFAIP3) as a susceptibility gene for systemic lupus erythematosus (SLE)--an autoimmune disease associated with aberrant B cell function--as well as studies showing that A20 is a tumor suppressor in B cell lymphomas (Graham et al., 2008; Musone et al., 2008; Compagno et al, 2009; Kato et al, 2009; Novak et al, 2009; Schmitz et al, 2009). Nevertheless, the physiological roles of A20 in T and B cells are largely undefined.
B cells are regulated by BCR, TLR, BAFF and CD40 signals. These signaling cascades share some of the same ubiquitin-dependent signaling molecules utilized by TNF and TLR ligands (e.g., TRAF2, TRAF6, IKKγ) (Hayden and Ghosh, 2008). Given A20’s role in preventing inflammation, its genetic linkage to human B cell lymphomas and SLE, and the central role B cells play in SLE pathogenesis, we hypothesized that A20 may regulate B cell homeostasis and prevent autoimmunity. To determine the cell intrinsic functions of A20 in regulating B cells, we have generated mice lacking A20 specifically in these cells.
Results
B-lineage deletion of A20 perturbs lymphoid homeostasis
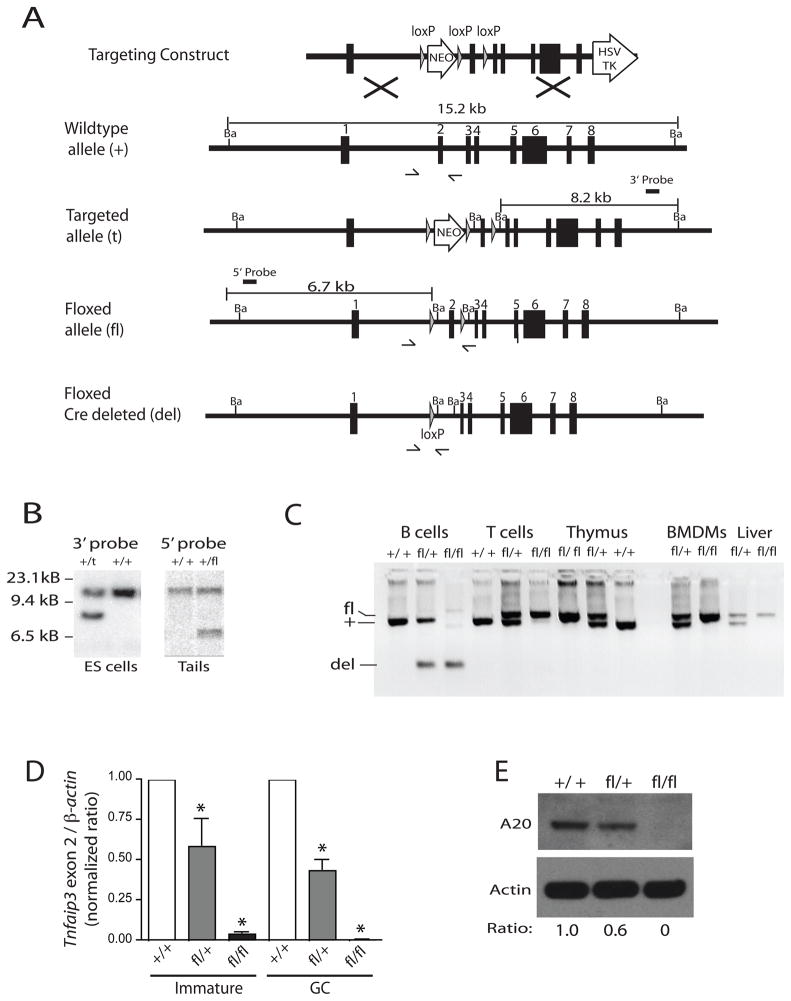
In order to analyze the cell intrinsic functions of A20 in B cells, we generated a targeting construct in which exon 2 of the Tnfaip3 gene was flanked by loxP sites, a “floxed” allele. The targeting construct was transfected into C57BL/6 ES cells and neomycin resistant clones were screened for the targeted allele (Figures 1A and B). Transient transfection of Cre recombinase resulted in removal of the neomycin cassette to obtain the floxed Tnfaip3 allele (Figures 1A and B). ES clones were injected into albino C57BL/6 blastocysts, and the resultant chimeras were bred with albino C57BL/6 mice. Non-albino C57BL/6 progeny were screened for the presence of the floxed allele, Tnfaip3fl (Figure 1B).
Figure 1. Gene targeting strategy to generate mice lacking tnfaip3 in B cells.
(A) Schematic representation of the gene targeting construct and screening strategy for obtaining the tnfaip3 floxed (fl) allele. Half arrows indicate locations of PCR primers for distinguishing wild type (+), floxed (fl) and deleted (del) alleles. (B) Southern blots of BamHI digested genomic DNA from ES cells showing the targeted allele (left blot) and from tails from mice with germline inheritance of the fl allele (right blot). (C) Genomic DNA PCR analysis of wild type (+), floxed (fl) and deleted (del) alleles of tnfaip3 exon 2 in the indicated cell types from mice of the indicated genotypes using PCR primers shown in (A). BMDMs are bone marrow derived macrophages. All mice are CD19-Cre+/−. PCR products for floxed (fl), wild type (WT) and deleted (Del) alleles are indicated. Data are representative of 5 mice per genotype. (D) Quantitative genomic DNA PCR analysis of tnfaip3 exon 2 in flow cytometry-sorted populations of immature (CD19+ CD93[AA4.1]+) and GC (CD19+ GL7+ CD95+) B cells from mice of the indicated genotypes. PCR primers described in Methods. Error bars show S.E.M. of 3 mice per genotype. (E) Immunoblot analysis of A20 expression in B cells from the indicated genotypes of mice. The A20 to actin protein ratio relative to Tnfaip3+/+ CD19-Cre cells is shown below the blots.
Mice carrying the Tnfaip3 fl allele were bred with Cd19-Cre knock-in mice to generate Tnfaip3+/+, Tnfaip3fl/+ and Tnfaip3fl/fl mice bearing a single copy of the CD19-Cre allele (Rickert et al., 1997). All mice described in this study were heterozygous for the CD19-Cre targeted allele (CD19-Cre +/−) to control for potential nonspecific effects of Cre expression while maintaining CD19 expression. For simplicity, CD19-Cre +/− mice will subsequently be referred to as CD19-Cre mice. As has been found for other “floxed” alleles, Tnfaip3fl/fl CD19-Cre mice had efficient and B cell specific deletion of Tnfaip3 exon 2, as assessed by genomic polymerase chain reaction (PCR) and Southern blot (Figure 1C and data not shown). Flow cytometry sorted immature and germinal center (GC) B cells, subsets represented in smaller proportions, were also nearly 100% deleted as measured by quantitative genomic PCR (Figure 1D). A20 protein is constitutively expressed in B cells and T cells (Figure 1E). Deletion of tnfaip3 exon 2 on both alleles (Tnfaip3fl/fl CD19-Cre) led to complete loss of A20 protein in splenic B cells (Figure 1E). Note that deletion of one allele of tnfaip3 in Tnfaip3fl/+ CD19-Cre mice causes hypomorphic (~50%) expression of A20 protein in B cells (Figure 1E).
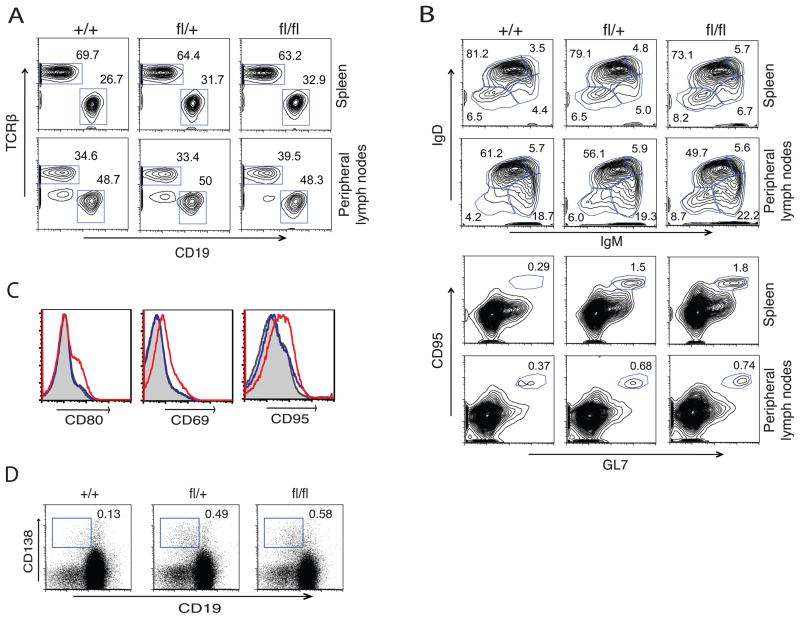
Tnfaip3fl/fl CD19-Cre mice were obtained in Mendelian numbers and developed normally. Hence, these mice differed dramatically from mice lacking A20 in all cells or in all hematopoietic cells, both of which develop severe spontaneous inflammation and early lethality (Lee et al., 2000; Boone et al., 2004; Turer et al., 2008). To begin to assess the roles of A20 in regulating B cells, we quantitated lymphoid populations from 5–7 week old Tnfaip3fl/fl CD19-Cre, Tnfaip3fl/+ CD19-Cre and Tnfaip3+/+ CD19-Cre littermates by flow cytometry (Table 1, top panel). Tnfaip3fl/fl CD19-Cre mice contained moderately increased numbers of B cells (CD19+), particularly immature B cells (CD19+IgMhi) and germinal center (GC) B cells, when compared to Tnfaip3+/+ CD19-Cre control mice (Table 1, Figure 2A, B, C). Although the percentage of B1a (IgM+, CD5+) cells in the peritoneal cavity of Tnfaip3fl/fl CD19-Cre mice was lower than Tnfaip3+/+ CD19-Cre and Tnfaip3fl/+ CD19-Cre mice, the absolute number was not significantly different (Figure 2C, S1A, Table 1). Although A20 deletion in Tnfaip3fl/fl CD19-Cre mice occurs in B cells and not T cells (Figure 1C), both B cells (CD19+) and T cells (TCRβ+) were modestly expanded in Tnfaip3fl/fl CD19-Cre mice (Figure 2A and Table 1). The relative percentages of T cell subpopulations (CD4+, CD8+, and Tregulatory) were normal (data not shown). Taken together, these findings suggest that A20 restricts the numbers of B cells, particularly immature and GC B cells.
Table 1. Cellulariry of B lymphocytes populations in Tnfaip3+/+, Tnfaip3fl/+ and Tnfaip3fl/flCD19-Cre mice.
Cellularity of lymphoid organs and respective subpopulations in Tnfaip3fl CD19-Cre mice.
Quantitation of lymphoid populations in the indicated tissues from the indicated genotypes of 5–7 week old mice. The total cellularity of lymphoid organs is shown in the top panel. Subpopulations were identified by flow cytometry using the indicated markers: B cells (CD19+); T-cells (TCRβ+); myeloid cells (Mac-1+); mature B-cells (CD19+, IgMLo, IgD+); immature B-cells (CD19+ IgMhi); marginal zone B-cells (CD21/35Hi CD23Lo); GC B cells (CD19+, GL7+, CD95+); B1a (IgM+, CD5+).
| Total cellularity(×106) | +/+ | fl/+ | fl/fl |
|---|---|---|---|
| Lymph Nodes | 14.3 ± 0.7 | 15.8 ± 1.2 | 25.7 ± 3.7* |
| Spleen | 44.4 ± 11 | 55.4 ± 9.0 | 74.2 ± 11 |
| Bone Marrow | 31.4 ± 3.2 | 30.7 ± 5.6 | 31.2 ± 4.1 |
| Peritoneum | 4.7 ± 0.7 | 3.8 ± 1.1 | 8.1 ± 1.6 |
| Lymph Nodes (×106) | |||
| B-cells | 3.9 ± 0.2 | 4.7 ± 0.5 | 8.8 ± 1.1* |
| T-cell | 8.9 ± 0.7 | 8.4 ± 0.8 | 12.9 ± 1.6* |
| Myeloid Cells | 0.15 ± 0.03 | 0.15 ± 0.03 | 0.24 ± 0.03 |
| Mature B-cells | 2.5 ± 0.1 | 2.9 ± 0.3 | 3.3 ± 0.5 |
| Immature B-cells | 1.2 ± 0.1 | 1.1 ± 0.2 | 3.9 ± 0.3* |
| Germinal Center B-cells | 0.01 ± 0.003 | 0.08 ± 0.02 * | 0.05 ± 0.02* |
| Spleen (×106) | |||
| B-cells | 23.7 ± 6.0 | 26.5 ± 4.5 | 37.8 ± 6.4 |
| T-cell | 16.5 ± 0.8 | 16.1 ± 1.7 | 24.7 ± 2.5* |
| Myeloid Cells | 0.7 ± 0.03 | 1.0 ± 0.2 | 1.4 ± 0.3 |
| Mature B-cells | 11.2 ± 1.4 | 15.1 ± 3.6 | 17.9 ± 3.08 |
| Immature B-cells | 9.8 ± 0.6 | 9.2 ± 0.7 | 14.8 ± 2.6* |
| Germinal Center B-cells | 0.17 ± 0.03 | 0.6 ± 0.15 * | 0.45 ± 0.08* |
| Marginal Zone B-cells | 1.6 ± 0.08 | 1.1 ± 0.3 | 0.9 ± 0.2 |
| Peritoneum (×106) | |||
| B-cells | 1.6 ± 0.34 | 2.7± 0.25 | 4.1 ± 0.37 |
| B1a | 0.45± 0.04 | 0.69 ± 0.16 | 0.6 ± 0.24 |
| B2 | 0.55± 0.16 | 0.93 ± 0.11 | 1.6 ± 0.12 |
indicates significant difference relative to +/+ (p < 0.05, using one-way Anova).
For GC B-cells, means are from 5 mice per genotype; for all other subpopulations, means of 5 Tnfaip3+/+ CD19-Cre, 11 Tnfaip3fl/+ CD19-Cre and 11 Tnfaip3fl/fl CD19-Cre mice are shown.
Figure 2. Flow cytometric analyses of B lymphocyte populations in Tnfaip3fl/fl CD19-Cre mice.
Flow cytometric analyses of lymphoid tissues. (A) Analyses of lymphoid populations in spleens and peripheral lymph nodes from 5 – 7 week-old mice. (B) Analysis of CD19+ gated cells showing B-cell maturation (IgM, IgD) from 5 – 7 week-old mice.. (C) Analysis of CD19+ gated cells showing GC B cells (GL7+ CD95+) from 5 – 7 week-old mice. (D) Plasma cells (CD19+ low CD138+) within TCRβ− CD19+ gated splenic B cells from 6 month old mice are shown. Percentages of cells within the indicated gates are shown on plots. E) Histograms comparing expression of B cell activation markers (CD80, CD69, and CD95) on CD19+ gated cells from Tnfaip3+/+ cells (shaded histogram) Tnfaip3fl/+ cells (blue line) and Tnfaip3fl/fl B cells (red line). All data compare littermates of the indicated genotypes and are representative of 3–5 mice per genotype.
Heterozygous Tnfaip3fl/+ CD19-Cre mice possess largely normal numbers of lymphoid populations, even though Tnfaip3fl/+ CD19-Cre B cells express half the amount of A20 protein as wild type Tnfaip3+/+ CD19-Cre B cells (Figure 1E). A notable exception is that the numbers of germinal center (GC) (CD95+GL7+) B cells in Tnfaip3fl/+ CD19-Cre mice approximates the number present in Tnfaip3fl/fl CD19-Cre mice (Figure 2C and Table 1). Thus, proper regulation of GC B cell homeostasis requires more A20 protein than other B cell populations.
Bone marrow from Tnfaip3fl/fl CD19-Cre and Tnfaip3fl/+ CD19-Cre mice contained normal numbers of B lineage cells, with normal proportions of pro-B (CD43+, IgM−) and pre-B (CD43−, IgM−) cells (Figure S1B). There was a small decrease in the percentage of IgM+ B cells in Tnfaip3fl/fl CD19-Cre bone marrow, which reflected reductions in mature or recirculating (IgM+, IgD+) B cells (Figure S1B). As CD19 is expressed throughout B cell development, these results suggest that A20 is not required for early B cell differentiation.
The differences in peripheral lymphocyte populations described above persisted but were not further exaggerated in 6 month old mice (Figure S1C and data not shown). In addition, six month old Tnfaip3fl/+ CD19-Cre and Tnfaip3fl/fl CD19-Cre mice contained increased percentages of splenic plasma cells when compared to Tnfaip3+/+ CD19-Cre mice (Figure 2D). While markers of B cell activation were expressed normally in 5–7 week old mice, spontaneous B cell activation became apparent in 6 month old Tnfaip3fl/fl CD19-Cre but not Tnfaip3fl/+ CD19-Cre mice (Figure 2E, S1D). Spontaneous T cell activation was not observed (data not shown). These findings suggest that A20 expression in B cells prevents spontaneous activation and differentiation of B cells over time.
Overall, elevated numbers of T and B cells and spontaneous B cell activation were observed in Tnfaip3fl/fl CD19-Cre mice by six months of age. In addition, elevated numbers of GC B cells were observed in both heterozygous Tnfaip3fl/+ CD19-Cre mice and homozygous Tnfaip3fl/fl CD19-Cre mice.
Exaggerated responses of Tnfaip3fl/fl CD19-Cre B cells in vitro
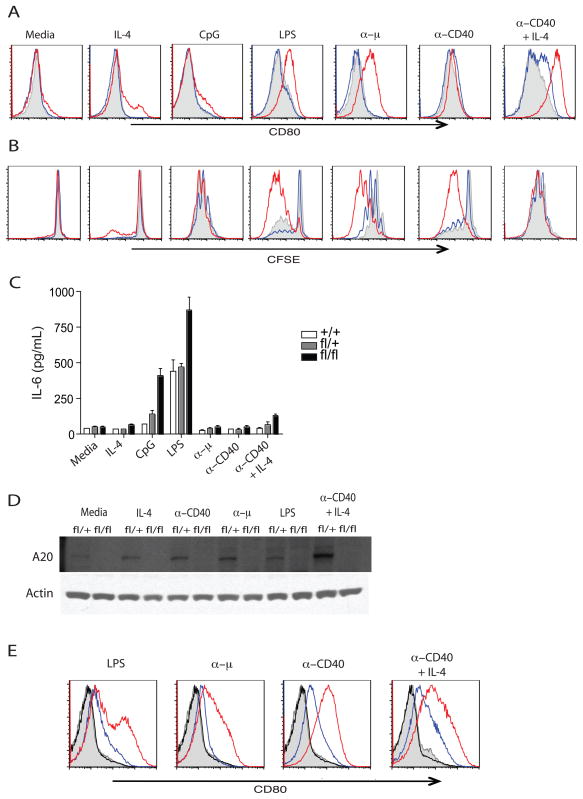
Our prior work indicated that A20 restricts TNF, TLR, and NOD induced NF-κB signals as well as TNF induced PCD in fibroblasts and macrophages (Lee et al., 2000; Boone et al., 2004; Hitotsumatsu et al., 2008; Turer et al., 2008). B cells receive activation, proliferation and survival signals from BCR, CD40 receptor, and other receptors (Skaug et al., 2009). To test if A20 directly regulates B cell responses, we assayed responses of Tnfaip3fl/fl CD19-Cre, Tnfaip3fl/+ CD19-Cre and Tnfaip3+/+ CD19-Cre splenic B cells to LPS, cytosine phosphoguanine oligodeoxynucleotide (CpG) and to agonist anti-IgM and anti-CD40. As noted above, unstimulated splenic B cells from young (5–7 week old) Tnfaip3fl/fl CD19-Cre mice resembled B cells from Tnfaip3fl/+ CD19-Cre and Tnfaip3+/+ CD19-Cre mice (Supplementary Figure 1). Tnfaip3fl/fl CD19-Cre B cells expressed higher amounts of CD80 (B7.1) and CD69 after stimulation with several B cell agonists (Figure 4A and data not shown). Tnfaip3fl/fl CD19-Cre B cells also proliferated to a greater extent than control cells (Figure 4B). Thirdly, Tnfaip3fl/fl CD19-Cre B cells produced more IL-6 than Tnfaip3fl/+ CD19-Cre and Tnfaip3+/+ CD19-Cre cells after treatment with LPS and CpG (Figure 4C).
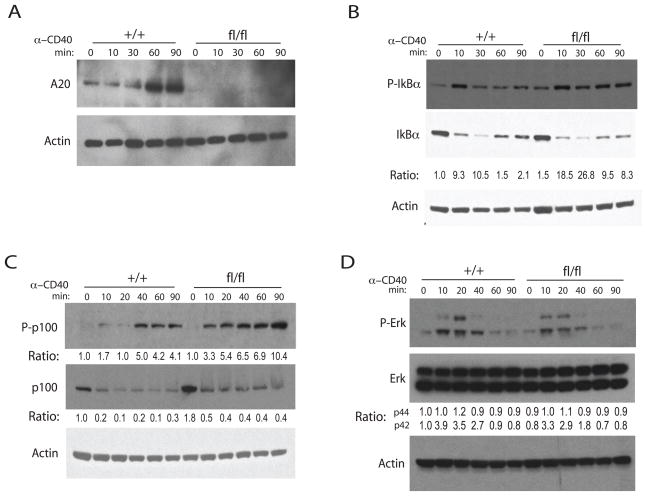
Figure 4. A20 restricts NF-κB signaling downstream of CD40 signals.
(A) Immunoblot analysis of A20 protein induction by agonist anti-CD40. (B) Immunoblot analyses of phospho-IκBα and IκBα after CD40 stimulation. Ratios of pIkBα/IkBα were normalized to time 0 of Tnfaip3+/+ CD19-Cre cells and are shown below. (C) Immunoblot analyses of NF-κB P-p100 (upper panel) and p100 (middle panel) protein amounts in response to agonist anti-CD40 antibody. Ratios of P-p100/actin, normalized to time 0 sample in Tnfaip3+/+ CD19-Cre B cells, are shown below P-p100 immunoblot. Ratios of p100/actin, normalized to time 0 sample in Tnfaip3+/+ CD19-Cre B cells, are shown below p100 immunoblot. (D) Immunoblot analysis of Erk signaling. Ratios of pErk/Erk were normalized to time 0 of Tnfaip3+/+ CD19-Cre cells and are shown below. Actin protein amounts shown below all panels as loading controls. Data are representative of 3 independent experiments.
To control for potential differences in B cell populations and to avoid potential caveats associated with developmental abnormalities, we sought to eliminate A20 expression in mature B cells after B cell development. Accordingly, we interbred Tnfaip3fl mice with estrogen receptor (ER)-Cre [Gt(ROSA)26ER-Cre] mice to obtain mice in which A20 (Tnfaip3) deletion would not occur until cells were exposed to 4-hydroxytamoxifen (4-OH-T). Splenic B cells enriched from Tnfaip3fl/fl ER-Cre mice and treated with 4-OH-T in vitro effectively ablated A20 protein expression (Figure 4D). We then tested responses of these B cells. Consistent with our findings with Tnfaip3fl/fl CD19-Cre B cells, mature splenic Tnfaip3fl/fl ER-Cre B cells rendered acutely A20 deficient with 4-OH-T exhibited exaggerated responses to all receptor stimuli when compared to control Tnfaip3fl/+ ER-Cre cells (Figure 4E). Thus, A20 expression in mature B cells restricts B cell responses independently of any potential roles of A20 in regulating B cell development.
A20 restricts NF-κB activation signals downstream of CD40
A20 restricts cellular responses to TNF, TLR, and NOD2 ligands by restricting NF-κB signaling (Lee et al., 2000; Boone et al., 2004; Hitotsumatsu et al., 2008; Turer et al., 2008). CD40 is a tumor necrosis factor receptor (TNFR) family member that triggers NF-κB signals and supports B cell activation and survival (Elgueta et al., 2009). To test whether A20 directly restricts CD40 induced NF-κB signals, splenic B cells from Tnfaip3+/+ CD19-Cre and Tnfaip3fl/fl CD19-Cre mice were stimulated with agonist anti-CD40 antibody. A20 protein was dramatically induced by CD40 engagement in Tnfaip3+/+ CD19-Cre B cells, whereas no A20 expression was observed in Tnfaip3fl/fl CD19-Cre B cells (Figure 5A). Tnfaip3fl/fl CD19-Cre B cells displayed increased and prolonged canonical NF-κB signaling as measured by IκBα phosphorylation in response to anti-CD40 (Figure 5B). Agonist anti-CD40 also induced greater amounts of phospho-p100, an indicator of non-canonical NF-κB signaling, in Tnfaip3fl/fl CD19-Cre B cells when compared to Tnfaip3+/+ CD19-Cre B cells (Figure 5C). This increased non-canonical NF-κB signaling correlates with increased basal p100 protein amounts in Tnfaip3fl/fl CD19-Cre B cells (Figure 5C). By contrast, Erk phosphorylation was similarly induced in Tnfaip3fl/fl CD19-Cre and control B cells (Figure 5D).
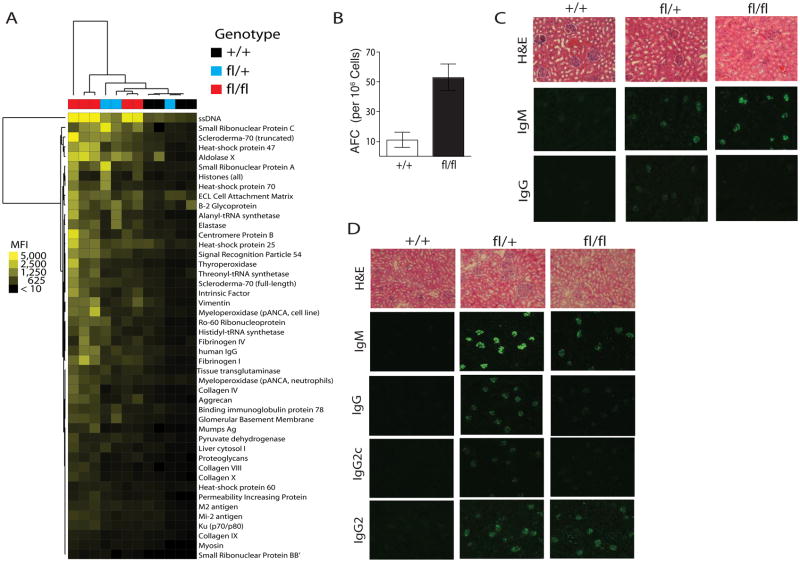
Figure 5. Spontaneous autoantibody production in Tnfaip3fl/+ CD19-Cre and Tnfaip3fl/fl CD19-Cre mice.
(A) Protein array analyses of autoantibodies in sera from 3 month old Tnfaip3fl/fl CD19-Cre (n=5), Tnfaip3fl/+ CD19-Cre (n=3), and Tnfaip3+/+ CD19-Cre (n=4) mice. Heat map shows relative reactivity to the respective antigens on the arrays, hierarchically clustered in both axes by Euclidean Distance. The reactivity intensities (MFI) are depicted on a relative color scale. Statistically different antigens were identified using 2-class Significant Analysis of Microarrays (SAM) with an unpaired t-test. (B) Antibody Forming Cells (AFC) measured on an Elispot for anti-dsDNA Ig producing B-cells. Counts were plotted as the mean of triplicate wells and S.D. is shown. Data are representative of 3 independent experiments. (C) Immunofluorescent analyses of glomerular Ig deposition in 6 month old mice of indicated genotypes. Analyses of IgM and IgG deposits shown in upper and middle panels, respectively. Data are representative of at 3 mice per genotype. Sections stained with hematoxylin and eosin are shown above. (D) Immunofluorescent analyses of glomerular deposition of Igs of the indicated isotypes after CpG treatment. 8–10 week old mice (n=4) of the indicated genotypes were treated with 40 μg of CpG intraperitoneally every other day for 2 weeks. Mice were analyzed 6 weeks after start of treatment. H&E sections shown above. All sections 100× magnification.
Normal antigen specific B cell responses in Tnfaip3fl/fl CD19-Cre mice
A20’s capacity to restrict B cell activation in vitro suggests that Tnfaip3fl/fl CD19-Cre mice might exhibit exaggerated antigen specific B cell responses in vivo. We thus tested antigen specific B cell responses to NP-KLH in these mice. At baseline, Tnfaip3fl/fl CD19-Cre mice have more IgM and modestly elevated amounts of IgGs (Figure S2A). Tnfaip3fl/fl CD19-Cre mice produced proportionately higher amount of anti-NP IgM antibodies before and after immunization with NP-KLH (Figure S2B). All genotypes of mice generated similar amounts of anti-NP IgG (Figure S2C). Thus, T cell dependent B cell responses occur largely normally in Tnfaip3fl/fl CD19-Cre mice. Anti-NP-Ficoll IgM responses paralleled T cell dependent NP-KLH responses in Tnfaip3fl/fl CD19-Cre mice, indicating that T cell independent B cell responses also occur normally in these mice (data not shown).
Tnfaip3fl/fl CD19-Cre and Tnfaip3fl/+ CD19-Cre mice develop autoimmunity
Increased numbers of immature and germinal center B cells suggest that auto-reactive B cells may accumulate and produce autoantibodies in Tnfaip3fl/fl CD19-Cre mice. In addition, several SNPs near the human A20 (TNFAIP3) gene are independently associated with susceptibility to SLE (Graham et al., 2008; Musone et al., 2008). To characterize the autoantibody profile of Tnfaip3fl/+ CD19-Cre mice and Tnfaip3fl/fl CD19-Cre mice as compared to Tnfaip3+/+ CD19-Cre mice, we used large-scale 1152-feature protein and peptide microarrays to detect autoantibodies directed against over 140 antigens (Robinson et al., 2002). Antibodies to over 46 self antigens were detected, including antibodies to nuclear antigens (e.g., single stranded DNA, small ribonuclear proteins A and C, Ku protein), glomerular antigens (e.g., vimentin, collagen X, proteogylcan, and aggrecan), and heat shock proteins (Figure 5A). Importantly, the serum autoantibody profiles from Tnfaip3fl/fl CD19-Cre and Tnfaip3fl/+ CD19-Cre mice clustered well with each other and away from Tnfaip3+/+ CD19-Cre mice (Figure 5A). These autoantibodies were observed in both male and female mice and were observed in C57BL/6 inbred mice, a strain that is relatively resistant to SLE-like disease. Elispot analysis for anti-DNA indicate both an increase in the number and size of anti-DNA producing B cells in Tnfaip3fl/fl CD19-Cre mice (Figure 5B and data not shown). These findings indicate that A20 expression in B cells prevents spontaneous production of autoantibodies.
Autoantibodies can be deposited in glomeruli of kidneys of SLE patients and ultimately cause glomerulonephritis. To determine whether serum autoantibodies in Tnfaip3fl/fl CD19-Cre and Tnfaip3fl/+ CD19-Cre mice lead to glomerular Ig deposits, we examined kidneys from 6 month old mice by histology and immunofluorescence. Although kidney sections appeared histologically normal, IgM deposits were observed in the kidneys of both Tnfaip3fl/fl CD19-Cre and Tnfaip3fl/+ CD19-Cre mice (Figure 5C). IgG deposits were more prominent in Tnfaip3fl/+ CD19-Cre mice (Figure 5C).
In addition to autoantibody producing B cells “escaping” from negative selection, activation of innate immune cells and type I interferon (IFN I) secretion may be key factors contributing to autoantibody production and the pathogenesis of SLE (Fairhurst et al., 2006; Shlomchik, 2008). CpG triggers B cell activation and production of type I interferons (IFN), and increases class switching to pathogenic autoantibody isotypes (Ehlers et al., 2006). We thus asked whether Tnfaip3fl/+ CD19-Cre and Tnfaip3fl/fl CD19-Cre mice develop more autoimmune disease after stimulation with CpG. CpG treatment of intact mice enhanced production of IgG anti-dsDNA antibodies in serum as well as pathogenic deposition of IgG in renal glomeruli of both Tnfaip3fl/fl CD19-Cre and Tnfaip3fl/+ CD19-Cre mice but not Tnfaip3+/+ CD19-Cre mice (Figure 5D and data not shown). Taken together, these findings indicate that A20 expression in B cells prevents autoimmunity.
A20 restricts B cell survival to Fas-mediated PCD
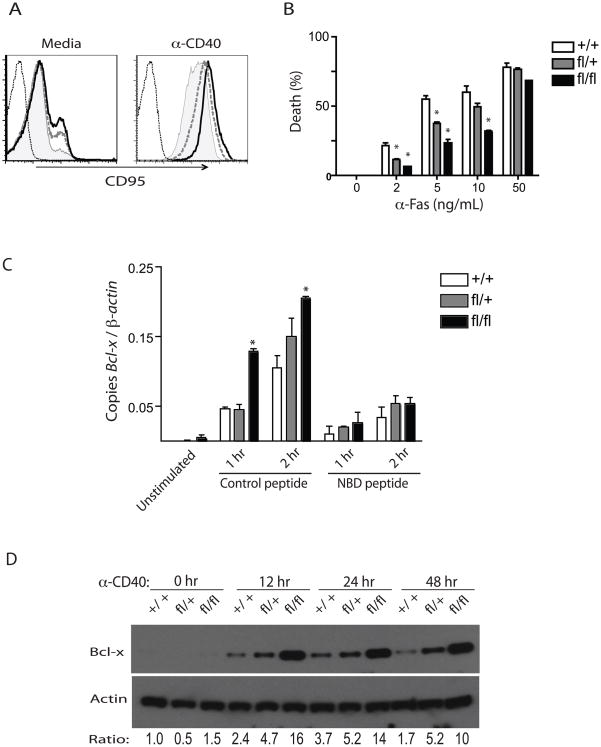
Tnfaip3fl/+ CD19-Cre mice share two key features with Tnfaip3fl/fl CD19-Cre mice: increased numbers of GC B cells and susceptibility to autoimmunity. These observations suggest that the amount of A20 expression regulates B cell selection in germinal centers. One possible mechanism for negative selection of autoreactive B cells in GCs is Fas-induced PCD (Hao et al., 2008). We thus tested the susceptibility of Tnfaip3+/+ CD19-Cre, Tnfaip3fl/+ CD19-Cre, and Tnfaip3fl/fl CD19-Cre B cells to Fas-induced PCD. Splenic B cells were activated with agonist anti-CD40 to induce Fas sensitivity. As expected, activation of Tnfaip3+/+ CD19-Cre B cells caused increased expression of Fas and dose dependent sensitivity to Fas mediated PCD (Figure 6A, B). Tnfaip3fl/fl CD19-Cre B cells expressed higher amounts of surface Fas than Tnfaip3+/+ CD19-Cre B cells after CD40 stimulation, while Tnfaip3fl/+ CD19-Cre B cells expressed intermediate amounts (Figure 6A). Remarkably, Tnfaip3fl/fl CD19-Cre and Tnfaip3fl/+ CD19-Cre B cells were resistant to Fas-mediated PCD when compared to control Tnfaip3+/+ CD19-Cre cells--even though Tnfaip3fl/fl CD19-Cre and Tnfaip3fl/+ CD19-Cre B cells expressed greater amounts of Fas (Figure 6B). This finding is particularly surprising given our prior observation that A20 deficient fibroblasts are more susceptible to TNF induced PCD than normal cells (Lee et al, 2000). Thus, A20 supports Fas mediated PCD in B cells while inhibiting TNFR induced PCD in fibroblasts.
Figure 6. A20 deficient and hypomorphic B-cells are resistant to programmed cell death.
(A) Flow cytometric analysis of CD95 (Fas) expression in enriched B cells after agonist anti-CD40 stimulation. Overlays of CD95 histograms on gated CD19+ cells from Tnfaip3+/+ CD19-Cre (shaded), Tnfaip3fl/+ CD19-Cre (dashed line), Tnfaip3fl/fl CD19-Cre (black line), and unstained control (dotted line) are shown. (B) Enriched B-cells stimulated for 48 hours with agonist anti-CD40 and treated with the indicated concentrations of agonist anti-CD95 for 12 hours were analyzed for survival by measuring the percentage of dead (Annexin-V+ DAPI+) cells by flow cytometry. Percent death was calculated as [% Fas induced dead - % control dead/100% - % control dead]. Data are plotted as mean ± S.D of triplicate wells. * indicates p < 0.001 using two way Anova. (C) Bcl-x mRNA amounts measured by real time quantitative (RT qPCR) in enriched B-cells. Cells of the indicated genotypes were stimulated with agonist anti-CD40 for the indicated times in the presence of an inhibitor or a control peptide, or left unstimulated. (D) Immunoblot analysis for the expression of Bcl-x protein in B-cells of the indicated genotypes stimulated with agonist anti-CD40 for the indicated times. Actin protein amounts are shown below as control. Ratios of Bcl-x to actin protein, normalized to the Tnfaip3+/+ B cells at time 0, are shown below each sample. All data are representative of 3 independent experiments.
To understand why A20 deficient B cells are resistant to Fas mediated PCD, we hypothesized that increased NF-κB signaling in these cells might lead to increased expression of anti-apoptotic proteins such as Bcl-x. After stimulation with agonist anti-CD40 in vitro, Bcl-x mRNA expression increased within one hour, and rose to a higher amount in Tnfaip3fl/fl CD19-Cre B cells compared with Tnfaip3fl/+ CD19-Cre or Tnfaip3+/+ CD19-Cre B cells (Figure 6C). CD40 triggered induction of Bcl-x mRNA was blocked by the NF-κB inhibitor, NEMO-binding domain (NBD) peptide, but not by control peptide (Figure 6C). Hence, NF-κB signaling appears directly required for induced Bcl-x mRNA transcription. Consistent with enhanced Bcl-x mRNA expression, markedly higher quantities of Bcl-x protein appear in Tnfaip3fl/fl CD19-Cre B cells compared with Tnfaip3fl/+ CD19-Cre or Tnfaip3+/+ CD19-Cre B cells (Figure 6D). Taken together, these findings indicate that A20 restricts the survival of activated B cells by limiting the NF-κB dependent transcription of Bcl-x mRNA and the subsequent production of Bcl-x protein. They also provide a potential mechanism by which A20 deficient B cells are resistant to negative selection in germinal centers.
Discussion
The generation and characterization of mice lacking A20 specifically in B cells, Tnfaip3fl/fl CD19-Cre mice, has allowed us to unveil several novel functions for A20. We have discovered that B cell specific expression of A20 restricts CD40 and BCR responses, terminates CD40 triggered NF-κB signals, restricts B cell survival, and prevents autoimmunity. These studies provide unique molecular insights into B cell homeostasis, human SLE and B cell lymphomas.
Tnfaip3fl/fl CD19-Cre mice are largely healthy, in marked contrast to mice lacking A20 expression in all cells or in all hematopoietic cells. This observation is consistent with our prior suggestion that the cachexia, myeloid dysregulation and perinatal lethality observed in globally A20-deficient mice is largely due to myeloid cell dysfunction. While Tnfaip3fl/fl CD19-Cre mice contained mild expansion of T cell numbers, these perturbations are probably not due to aberrant A20 deletion as judged by our molecular analyses and by the published literature for this Cre strain (Rickert et al., 1997; Schmidt-Supprian and Rajewsky, 2007). Rather, these mice express increased quantities of splenic IL-4 mRNA (data not shown), and Tnfaip3fl/fl CD19-Cre B cells produced more IL-6 and expressed higher amounts of co-stimulatory molecules upon stimulation. Thus, T cell expansion may be due to antigen-independent bystander effects induced by B cell derived cytokines and/or co-stimulatory molecules, as has been observed in other settings of B cell hyper-responsiveness (Homig-Holzel et al., 2008; Hao et al., 2008).
Our findings demonstrate that A20 performs important functions in adaptive immune cells in addition to previously described functions in innate immune cells (Lee et al, 2000; Boone et al, 2004; Hitotsumatsu et al, 2008). A20’s roles in restricting CD40 and BCR triggered NF-κB signals add to the spectrum of signaling cascades regulated by this ubiquitin modifying enzyme. NF-κB signaling is important for regulating B cell homeostasis (Sen 2006; Siebenlist et al, 2005). The phenotypes of our mice lacking A20 expression in B cells reveal the importance of tightly regulating basal NF-κB signals in these cells.
B cell specific loss of A20 expression leads to increased numbers of autoantibody producing cells. B cells undergo several stages of negative selection to eliminate autoreactive cells both in the bone marrow and in peripheral lymphoid organs (Jacobi and Diamond, 2005; Shlomchik, 2008; Yurasov and Nussenzweig, 2007; von Boehmer and Melchers, 2010). In the bone marrow, immature B cells are selected as a consequence of BCR and BAFF signals, and intracellular cell survival factors. In the periphery, selection occurs during GC maturation. Germinal centers are sites where B cells undergo expansion, immunoglobulin class switching, somatic hypermutation, and affinity maturation (Klein and Dalla-Favera, 2008). B cells with low affinity for antigen or reactivity for self antigens are negatively selected within GCs (Shlomchik, 2008). Deletion of autoreactive B cells helps prevent autoimmunity, and defective GC selection of autoreactive B cells has been observed in human SLE patients (Cappione et al., 2005). Fas (CD95) is highly expressed on GC B cells (Watanabe et al, 1995). While the role of Fas mediated PCD in GC selection has been controversial (Smith et al., 1995; Takahashi et al., 2001; Mizuno et al., 2003; Hoa et al., 2008), Fas mediated PCD likely plays an important role in eliminating autoreactive B cells (Rathmell et al., 1995; Fukuyama et alk., 2002; William et al., 2002). Thus, the accumulation of GC B cells in Tnfaip3fl/+ CD19-Cre and Tnfaip3fl/fl CD19-Cre mice may be due to the increased resistance of A20 deficient B cells to physiological PCD, leading to the escape of autoreactive B cells.
How might A20 deficiency in B cells render them resistant to PCD? One possibility stems from the observation that NF-κB dependent proteins protect B cells against PCD. Indeed, both canonical and non-canonical NF-κB signaling downstream of BCR, CD40, BAFF, and TLR receptors are thought to promote B cell survival as well as proliferation and activation (Siebenlist et al., 2005; Sen, 2006; Homig-Holzel et al., 2008). NF-κB has been suggested to be necessary for mediating BCR induced resistance to Fas mediated PCD (Mizuno and Rothstein, 2003; Schram and Rothstein, 2003). Our studies indicate that A20 directly restricts canonical NF-κB signals and suggest that these signals may lead to elevated non-canonical NF-κB signals. These increased NF-κB signals lead to increased expression of anti-apoptotic proteins such as Bcl-2 and Bcl-x. Deregulated expression of these proteins has been shown to cause altered GC B cell selection (Grillot et al., 1996; Takahashi et al., 1999). Hence, increased expression of Bcl-x and/or other NF-κB dependent proteins may provide a molecular underpinning for increased numbers of GC B cells in Tnfaip3fl/fl CD19-Cre mice.
Heterozygous Tnfaip3fl/+ CD19-Cre mice contain similarly increased numbers of GC B cells and autoantibodies as homozygous Tnfaip3fl/fl CD19-Cre mice at young ages (i.e., 5–7 weeks old), suggesting that a high threshold of A20 expression must be maintained for properly selecting (or deleting) these cells. Reduced A20 expression in other cell types leads to increased production of NF-κB dependent gene products, so endogenous amounts of A20 protein appear to be limiting (O. Hitotsumatsu, S. Oshima, G. Hammer, unpublished data). Reduced (rather than absent) quantities of A20 expression or hypomorphic A20 proteins may also link A20 (TNFAIP3) susceptibility SNPs with SLE in human patients (Musone et al, 2008). Thus, mice expressing reduced amounts of A20 may prove to be highly relevant models of human autoimmune diseases.
While reduced A20 expression in B cells leads to accumulation of GC B cells and IgG autoantibodies, absent A20 expression also causes accumulation of immature B cells and IgM in Tnfaip3fl/fl CD19-Cre mice and progressive activation of B cells with age. Hence, a lower amount of A20 is necessary to preserve selection of immature B cells and to restrict spontaneous B cell activation than the amount required for proper GC selection. As IgM autoantibodies may be protective against IgG mediated autoimmune disease, higher IgM amounts in homozygous Tnfaip3fl/fl CD19-Cre mice may reduce the degree of autoimmune disease observed in these mice (Witte, 2008). A20 amounts are dynamically regulated, largely in response to NF-κB signals (Krikos et al, 1992). Thus, A20 expression amounts appear to be finely tuned to regulate NF-κB signaling and survival of distinct subsets of B cells.
Tnfaip3fl/fl CD19-Cre mice exhibit largely normal antigen specific B cell responses in vivo, despite the fact that mature Tnfaip3fl/fl CD19-Cre B cells exhibit increased responses to BCR, CD40, and TLR ligands in vitro. These findings suggest that B cell independent factors such as T cells and myeloid cells can properly restrict antigen specific B cell responses, even if they allow progressive accumulation of autoreactive B cells in Tnfaip3fl/fl CD19-Cre mice.
Our experiments indicate that A20 expression in B cells regulates GC B cell selection as well as B cell activation, thereby regulating key aspects of B cell tolerance. It is remarkable that B cell specific deletion of A20 alone is sufficient for autoimmunity in C57BL/6 mice. The appearance of IgG deposits suggests that abnormal B cells are sufficient for at least the initial stages of the autoimmunity and are part of the continuum to full blown disease. Hence, lupus prone Tnfaip3fl/fl CD19-Cre and Tnfaip3fl/+ CD19-Cre mice as well as genetic derivatives of these mice should be useful models for understanding human SLE. Heterozygous Tnfaip3fl/+ CD19-Cre mice may be particularly relevant, as reduced, rather than absent A20 expression may characterize this human condition.
Recent studies have shown that somatic loss of A20 in B cells causes several types of Hodgkin, non-Hodgkin, and marginal zone B cell lymphomas in humans (Kato et al., 2009; Compagno et al., 2009; Schmitz et al., 2009; Novak et al., 2009). Our findings that A20 deficient B cells express high amounts of Bcl-x and are resistant to Fas mediated PCD provide molecular insights into how A20 functions as a tumor suppressor in B cells. Remarkably, A20 is a pro-apoptotic protein in B cells even though it restricts TNF induced apoptosis in fibroblasts and hepatocytes (Lee et al., 2000; Arvelo et al., 2002). Hence, it is critical to analyze A20’s physiological functions in cell type-specific contexts. Future studies testing the potential of A20 deficiency to collaborate with other B cell oncogenes may reveal the spectrum of A20’s tumor suppressor functions in B cells.
In conclusion, we have demonstrated functions for A20 in regulating B cell responses, including the restriction of CD40 induced NF-κB signals. These cell autonomous functions are critical for B cell homeostasis and the prevention of autoreactive B cells and autoimmunity. In addition to unveiling new molecular mechanisms of B cell homeostasis, these studies provide critical insights into the pathogenesis of human SLE and B cell lymphomas.
Methods
Generation of A20 conditionally targeted (Tnfaip3fl/fl) mice
Recombineering was used to generate a gene targeting construct from a bacterial artificial chromosome (BAC) from the C57BL/6J strain containing the tnfaip3 gene. C57BL/6 ES cells (PRX-B6T, Primogenix) were transfected with this construct, and successfully targeted ES cells were identified by Southern blot analysis. Correctly targeted clones were then transfected with recombinant Cre and screened for removal of the neomycin cassette to obtain the tnfaip3 “floxed” allele. Blastocyst injections of targeted ES cells were performed by the UCSF Transgenic Core. Chimeras were bred to albino C57BL/6J mice and non-albino progeny were screened for the presence of the floxed allele (Tnfaip3fl/+). Mice bearing the targeted allele in the germline were interbred with CD19-Cre mice on the C57BL/6 background (B6.129P2(C)-CD19tm1(cre)Cgn/J) (Rickert et al, 1997). Tnfaip3fl/fl or Tnfaip3 fl/+ mice homozygous for CD19-Cre were bred with Tnfaip3fl/fl or Tnfaip3 fl/+ mice without the CD19-Cre allele to generate experimental mice of various Tnfaip3 genotypes (Tnfaip3+/+, Tnfaip3fl/+ and Tnfaip3fl/fl) that have one copy of Cre and are heterozygous for CD19. Genotypes were initially confirmed by Southern blot analysis and subsequently identified by PCR using the following primers: 5′-AACTTTACAGTCCCCAGCAATGG-3′ (sense); and 5′ GAGGAGGTTGGAAGACATAGAATCG-3′ (antisense).
Cell preparation and analyses
Single cell suspensions were prepared and incubated with the designated conjugated antibodies (all from BD Biosciences, except anti-CD5 and anti-CD93 (AA4.1), eBioscience), and live cells (DAPI−) were analyzed by flow cytometry (LSRII, BD Biosciences) using FlowJo software (Tree Star Inc.). BMDMs were prepared as previously described (Boone et al., 2004). For in vitro assays, B-cells were isolated by negative depletion with TCRβ, Mac-1, NK1.1 and Ter119 biotinylated antibodies (BD Biosciences) bound to streptavidin coated magnetic beads (M-280 Dynabeads, Invitrogen). Cells were stimulated with anti-CD40 (HM40-3, BD Biosciences) at 1 μg/ml, anti-μ chain (Jackson Immunoresearch) at 2 μg/ml, IL-4 (Peprotech) at 10 ng/ml and LPS (Sigma) at 1μg/ml. For in vitro deletion of tnfaip3 exon 2 from GT-Rosa Cre B cells, cells were treated with 4-OH-T (2.5 nM) for the first 12 hours of stimulation. For NF-κB inhibition, control peptide or NBD Peptide (Calbiochem) were added at 0.5 μM one hour prior to stimulation with agonist anti-CD40 antibody.
Mouse immunizations
For in vivo antigen responses, 6–8 week old mice were injected intraperitoneally (ip) and bled on the indicated days. Mice were injected with 50 μg NP-KLH (Biosearch Technologies) mixed 1:1 with Imject Alum (Thermo Scientific). For CpG treatment, 8 week old mice were injected ip, every other day for 2 weeks, with 40 μg CpG ODN 2395 (tcgtcgttttcggcgcgcgccg) with phosphorothioate bases (Invitrogen). Serum was collected before treatment, and 6 weeks post-treatment. All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of California, San Francisco.
Immunoglobulin and AFC determination
Quantities of serum and supernatant immunoglobulins (total Ig, IgM, IgG1, IgG2c, IgG2b, IgG3 and IgA) were determined by isotype specific ELISA (Southern Biotechnology, Birmingham, Alabama). NP (4-Hydroxy-3-nitrophenylacetyl)-specific antibody titers were determined by ELISA with plates coated with NP23-BSA (Biosearch Technologies). dsDNA-specific antibody titers were determined by ELISA with plates coated with Hind III (New England Biolabs) digested pUC19 in 0.1M Tris, over-night at room temperature. Cytokines amounts were measured by ELISA as recommended by the manufacturer (BD Biosciences). Elispot was performed by incubating enriched B cells on plates coated with dsDNA as above. After overnight incubation, plates were washed and incubated with alkaline phosphatase (AP) conjugated anti-IgM (Southern Biotech), and subsequently developed with BCIP substrate (Sigma) dissolved into alkaline phosphatase buffer (0.1M Tris, 0.1 M NaCl, 5 mM MgCl2) and 0.6 % LMP Agarose (Sigma).
Autoantigen arrays were printed and processed as previously described. Arrays were probed with goat antibody specific for mouse Ig (Jackson Immunoresearch). Detailed protocols and lists of antigens have been published and are available online at http://utzlab.stanford.edu/protocols (Robinson et al., 2002). Significance Analysis of Microarrays (SAM) was applied to the data to identify antigens with statistically significant differences in array reactivity between mutant and wild type mice (Tusher et al., 2001).
Histology and immunohistochemistry
Kidneys and spleens were fixed in 10% formalin. Sections and H&E stain were performed by the UCSF VAMC Pathology Core. For immunohistochemistry, kidneys were embedded in Tissue-Tek OCT™ compound and snap frozen in Methyl-butane with dry ice. Tissue sections were then stained with IgM-FITC or IgG-FITC (Jackson Immunoresearch), IgG2c-FITC or IgG2b-FITC (Bethyl Laborotories Inc.).
Cell signaling assays
Enriched B cells were stimulated as indicated in the figures, lysed in 0.1% NP-40 (Calbiochem) lysis buffer, and nuclei spun down to yield cytoplasmic lysates. Lysates were cleared by centrifugation at 14,000 g for 20 min at 4 ° C, supernatants were removed, heated in Laemmli buffer and run on SDS-PAGE (Novex System, Invitrogen). Immunobots were probed for A20 (Boone et al, 2004), actin (JLA20, Calbiochem), phospho-IκBα, IκBα, phospho-Erk, Erk, phospho-NF-κB2 p100 and NF-κB2 p100/p52 (Cell Signaling), Bcl-x (Transduction Biotechnologies).
Fas-induced cell death assay
Fas-induced cell death assays were performed as described (Watanabe et al, 1995; Wang et al., 1996). Briefly, enriched B-cells were plated with agonist anti-CD40 (1 μg/ml) for 60 hours. Agonist anti-CD95 (Fas) was added to the cultures at the indicated doses for the last 12 hours. Cells were harvested and stained for flow cytometry with Annexin-V (BD Biosciences) and DAPI.
Real-time PCR assays
For quantitation of genomic Tnfaip3 exon 2, DNA was prepared using DNeasy Kit (Qiagen), after which qPCR was performed using SYBR Green (Qiagen). Primers for exon 2 of Tnfaip3 were the following: 5′-CTGACCTGGTCCTGAGGAAG-3′ (sense); and 5′-GCAAAGTCCTGTTTCCACAA-3′ (anti-sense). This qPCR assay was shown to detect less than 1% of exon 2 DNA in titrated mixtures of Tnfaip3−/− and Tnfaip3+/+ cells (data not shown). For quantitation of Bcl-x mRNA expression, mRNA was prepared using RNeasy Kit (Qiagen) and cDNA was obtained with Quantitect Reverse Transcription Kit (Qiagen). Primers for Bcl-x cDNA were the following: 5′-GCAGACCCAGTAAGTGAGCA-3′ (sense) and 5′-AGAAAGTCGACCACCAGCTC-3′ (antisense). In both types of assays, β-actin primers used as a reference were: 5′-AAGTGTGACGTTGACATCCGTAA-3′ (sense) and 5′-TGCCTGGGTACATGGTGG TA-3′ (antisense). Assays were performed using an ABI 7300 real-time PCR machine (Applied Biosystems).
Supplementary Material
Figure 3. Hyper-responsiveness of A20-deficient B cells.
In vitro responses of purified B cells after stimulation with the indicated stimuli for 72h. Flow cytometric analyses of (A) surface expression of the activation marker CD80 (B7.1) and (B) dilution of CFSE-labeled cells. Tnfaip3fl/fl CD19-Cre cells are shown in red lines; Tnfaip3fl/+ CD19-Cre cells shown in blue lines; and Tnfaip3+/+ CD19-Cre cells shown in shaded gray histograms. (C) ELISA determination of IL-6 secretion in the supernatants of 72 hr B cell cultures. Means and standard deviations of triplicate wells are shown. (D) Immunoblots of A20 and actin protein expression in purified Rosa-ER-Cre+ B cells of the indicated genotypes. Lysates were isolated after 72 hr stimulation with the indicated ligands in the presence of 4-OH-T. (E) Expression of CD80 in purified B cells after treatment with 4-OH-T and the indicated ligands for 72hr. Tnfaip3fl/fl GT-Rosa-Cre+ B cells treated with IL-4 or media (black lines) or the indicated ligands (red lines); Tnfaip3fl/+ GT-Rosa-Cre+ B cells treated with IL-4 or media (gray shaded histograms) or the indicated ligands (blue lines). All data are representative of 3 independent experiments.
Acknowledgments
This work was supported by the National Institutes of Health and the Fundação para a Ciência e Tecnologia (to R. T.), UCSF-VAMC Pathology Core, the Cancer Center Pathology Core, UCSF Liver Center Immunology and Pathology Cores, and the UCSF Transgenic and Targeted Mutagenesis Core Facility. We thank the skillful technical assistance of Ivy Hsieh (UCSF-VAMC Pathology) and Jean Publicover (UCSF Liver Center), and helpful advice from João Pedro Pereira and Lee Reinhardt. We thank Tony DeFranco and Jason Cyster for critically reading the manuscript.
Footnotes
Author Contributions
R.M.T. planned and performed the experiments, analyzed the data, and assisted in writing the manuscript. E.E.T. made the conditional gene targeting construct, performed gene targeting in ES cells, and characterized the original Tnfaip3fl germline mice. R.A. performed mice breeding and genotyping, and assisted with mouse injections and bleeding. C.L.L. performed the antibody array experiments and performed statistical analysis, under the supervision of P.J.U. C.A.L. and P.J.U. edited the manuscript. P.S. assisted in the analysis of the autoantibody data and mouse histology and immunohistochemistry, under the supervision of C.A.L. L.R. performed some of the initial experiments with R.M.T. J.B. provided excellent technical assistance. B.A.M. and A.M. helped plan and supervise the experiments and data analysis, and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arvelo MB, Arvelo MB, Cooper JT, Longo C, Daniel S, Grey ST, Mahiou J, Czismadia E, Abu-Jawdeh G, Ferran C. A20 protects mice from D-galactosamine/lipopolysaccharide acute toxic lethal hepatitis. Hepatology. 2002;35:535–543. doi: 10.1053/jhep.2002.31309. [DOI] [PubMed] [Google Scholar]
- Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- Cappione A, Anolik JH, Pugh-Bernard A, Barnard J, Dutcher P, Silverman G, Sanz I. Germinal center exclusion of autoreactive B cells is defective in human systemic lupus erythematosus. J Clin Invest. 2005;115:3205–3216. doi: 10.1172/JCI24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J, Sun L, Chen ZJ, Marynen P, Beyaert R. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat Immunol. 2008;9:263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düwel M, Welteke V, Oeckinghaus A, Baens M, Kloo B, Ferch U, Darnay BG, Ruland J, Marynen P, Krappmann D. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J Immunol. 2009;182:7718–7728. doi: 10.4049/jimmunol.0803313. [DOI] [PubMed] [Google Scholar]
- Ehlers M, Fukuyama H, McGaha TL, Aderem A, Ravetch JV. TLR9/MyD88 signaling is required for class switching to pathogenic IgG2a and 2b autoantibodies in SLE. J Exp Med. 2006;203:553–561. doi: 10.1084/jem.20052438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. 2009;229:152–172. doi: 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhurst A-M, Wandstrat AE, Wakeland EK. Systemic lupus erythematosus: multiple immunological phenotypes in a complex genetic disease. Adv Immunol. 2006;92:1–69. doi: 10.1016/S0065-2776(06)92001-X. [DOI] [PubMed] [Google Scholar]
- Fukuyama H, Adachi M, Suematsu S, Miwa K, Suda T, Yoshida N, Nagata S. Requirement of Fas expression in B cells for tolerance induction. Eur J Immunol. 2002;32:223. doi: 10.1002/1521-4141(200201)32:1<223::AID-IMMU223>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, Burtt NP, Guiducci C, Parkin M, Gates C, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillot DA, Merino R, Pena JC, Fanslow WC, Finkelman FD, Thompson CB, Nunez G. Bcl-x exhibits regulated expression during B cell development and activation and modulates lymphocyte survival in transgenic mice. J Exp Med. 1996;183:381–391. doi: 10.1084/jem.183.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z, Duncan GS, Seagal J, Su Y-W, Hong C, Haight J, Chen N-J, Elia A, Wakeham A, Li WY, et al. Fas receptor expression in germinal center B cells is essential for T and B lymphocyte homeostasis. Immunity. 2008;29:615–627. doi: 10.1016/j.immuni.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Hitotsumatsu O, Ahmad RC, Tavares R, Wang M, Philpott D, Turer EE, Lee BL, Shiffin N, Advincula R, Malynn BA, Wertz C, Ma A. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain-containing 2 triggered signals. Immunity. 2008;28:381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homig-Holzel C, Hojer C, Rastelli J, Casola S, Strobl L, Muller W, Quintanilla-Martinez L, Gewies A, Ruland J, Rajewsky K, Zimber-Strobl U. Constitutive CD40 signaling in B cells selectively activates the noncanonical NF-κB pathway and promotes lymphomagenesis. J Exp Med. 2008;205:1317–1329. doi: 10.1084/jem.20080238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi AM, Diamond B. Balancing diversity and tolerance: lessons from patients with systemic lupus erythematosus. J Exp Med. 2005;202:341–344. doi: 10.1084/jem.20050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, Niwa A, Chen Y, Nakazaki K, Nomoto J, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- Krikos A, Laherty CD, Dixit VM. Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by kappa B elements. J Biol Chem. 1992;267:17971–6. [PubMed] [Google Scholar]
- Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodoce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Rothstein TL. Fas-induced apoptosis in B cells. Apoptosis. 2003;8:451–60. doi: 10.1023/a:1025534223168. [DOI] [PubMed] [Google Scholar]
- Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, Shifrin N, Petri MA, Kamboh MI, Manzi S, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak U, Rinaldi A, Kwee I, Nandula SV, Rancoita PM, Compagno M, Cerri M, Rossi D, Murty VV, Zucca E, et al. The NF-kappaB negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone lymphomas. Blood. 2009;113:4918–4921. doi: 10.1182/blood-2008-08-174110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opipari AW, Jr, Boguski MS, Dixit VM. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J Biol Chem. 1990;265:14705–14708. [PubMed] [Google Scholar]
- Rathmell JC, Cooke MP, Ho WY, Grein J, Townsend SE, Davis MM, Goodnow CC. CD95 (Fas)-dependent elimination of self-reactive B cells upon interaction with CD4+ T cells. Nature. 1995;376:181–184. doi: 10.1038/376181a0. [DOI] [PubMed] [Google Scholar]
- Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WH, DiGennaro C, Hueber W, Haab BB, Kamachi M, Dean EJ, Fournel S, Fong D, Genovese MC, de Vegvar HE, et al. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nature Med. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- Sarma V, Lin Z, Clark L, Rust BM, Tewari M, Noelle RJ, Dixit VM. Activation of the B-cell surface receptor CD40 induces A20, a novel zinc finger protein that inhibits apotosis. J Biol Chem. 1995;270:12343–12346. doi: 10.1074/jbc.270.21.12343. [DOI] [PubMed] [Google Scholar]
- Schmidt-Supprian M, Rajewsky K. Vagaries of conditional gene targeting. Nat Immunol. 2007;8:665–668. doi: 10.1038/ni0707-665. [DOI] [PubMed] [Google Scholar]
- Schmitz R, Hansmann ML, Bohle V, Martin-Subero JI, Hartmann S, Mechtersheimer G, Klapper W, Vater I, Giefing M, Gesk S, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med. 2009;206:981–989. doi: 10.1084/jem.20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schram BR, Rothstein T. NF-kappa B is required for surface Ig-induced Fas resistance in B cells. J Immunol. 2003;170:3118–24. doi: 10.4049/jimmunol.170.6.3118. [DOI] [PubMed] [Google Scholar]
- Sen R. Control of B lymphocyte apoptosis by the transcription factor NF-kappaB. Immunity. 2006;25:871–883. doi: 10.1016/j.immuni.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Shlomchik MJ. Sites and stages of autoreactive B cell activation and regulation. Immunity. 2008;28:18–28. doi: 10.1016/j.immuni.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-κB regulatory pathways. Ann Rev Biochem. 2009;78:769–96. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- Siebenlist U, Brown K, Claudio E. Control of lymphocyte development by nuclear factor-κB. Nat Rev Immunol. 2005;5:435–445. doi: 10.1038/nri1629. [DOI] [PubMed] [Google Scholar]
- Smith KGC, Nossal GJV, Tarlinton DM. FAS is highly expressed in the germinal center but is not required for regulation of the B cell response antigen. Proc Natl Acad Sci USA. 1995;92:11628–11632. doi: 10.1073/pnas.92.25.11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Cerasoli DM, Dal Porto JM, Shimoda M, Freund R, Fang W, Telander DG, Malvey EN, Mueller DL, Behrens TW, Kelsoe G. Relaxed negative selection in germinal centers and impaired affinity maturation in Bcl-xL transgenic mice. J Exp Med. 1999;190:399–410. doi: 10.1084/jem.190.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Ohta H, Takemori T. Fas is required for clonal selection in germinal centers and the subsequent establishment of the memory B cell repertoire. Immunity. 2001;14:181–192. doi: 10.1016/s1074-7613(01)00100-5. [DOI] [PubMed] [Google Scholar]
- Turer EE, Tavares RM, Mortier E, Hitotsumatsu O, Advincula R, Lee B, Shifrin N, Malynn BA, Ma A. Homeostatic MyD88-dependent signals cause lethal inflammation in the absence of A20. J Exp Med. 2008;205:451–64. doi: 10.1084/jem.20071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci, USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Boehmer H, Melchers F. Checkpoints in lymphocyte develoment and autoimmune disease. Nat Immunol. 2010;11:14–20. doi: 10.1038/ni.1794. [DOI] [PubMed] [Google Scholar]
- Wang J, Taniuchi I, Maekawa Y, Howard M, Cooper MD, Watanabe T. Expression and function of Fas antigen on activated murine B cells. Eur J Immunol. 1996;26:92–96. doi: 10.1002/eji.1830260114. [DOI] [PubMed] [Google Scholar]
- Watanabe D, Suda T, Nagata S. Expression of Fas in B cells of the mouse germinal center and Fas-dependent killing of activated B cells. Int Immunol. 1995;7:1949–1956. doi: 10.1093/intimm/7.12.1949. [DOI] [PubMed] [Google Scholar]
- Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. Deubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- William J, Euler C, Christensen S, Schlomchik MJ. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science. 2002;297:2066–70. doi: 10.1126/science.1073924. [DOI] [PubMed] [Google Scholar]
- Witte T. IgM antibodies against dsDNA in SLE. Clinic Rev Allerg Immunol. 2008;34:345–47. doi: 10.1007/s12016-007-8046-x. [DOI] [PubMed] [Google Scholar]
- Yurasov S, Nussenzweig MC. Regulation of autoreactive antibodies. Curr Opin Rheumatol. 2007;19:421–426. doi: 10.1097/BOR.0b013e328277ef3b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.