Abstract
The mucolipin (TRPML) subfamily of transient receptor potential (TRP) cation channels consists of three members that play various roles in the regulation of membrane and protein sorting along endo-lysosomal pathways. Loss-of-function mutations in TRPML1 cause the neurodegenerative lysosomal storage disorder, mucolipidosis type IV (MLIV), whereas a gain-of-function mutation in TRPML3 is principally implicated in the hearing-impaired and abnormally pigmented varitint-waddler mouse. Currently, TRPML2 is not implicated in any pathological disorder, but we have recently shown that it is a functional cation channel that physically interacts with TRPML1 and TRPML3 to potentially regulate lysosomal integrity. Here, we show that mutant TRPMLs heteromultimerize with other mutant and wild-type TRPMLs to regulate cell viability and starvation-induced autophagy, a process that mediates macromolecular and organellar turnover under cell starvation conditions. Heteromultimerization of dominant-negative TRPMLs with constitutively active TRPMLs rescues cells from the cytotoxic effects of TRPML constitutive activity. Moreover, dominant-negative TRPML1 channels, including a mutant channel directly implicated in MLIV pathology, also inhibit starvation-induced autophagy by interacting with and affecting native TRPML channel function. Collectively, our results indicate that heteromultimerization of TRPML channels plays a role in various TRPML-regulated mechanisms.
Keywords: Mucolipin, TRPML, Ion channel heteromultimerization, TRP channels, Starvation-induced autophagy
Introduction
The mucolipins (TRPML) are a subfamily of channel proteins within the transient receptor potential (TRP) superfamily of cation channels (Venkatachalam and Montell, 2007) and the mucolipins 1, 2 and 3 (TRPML1, TRPML2 and TRPML3, respectively) each feature predicted cation channel pores between their transmembrane segments 5 (TM5) and 6 (TM6) (Puertollano and Kiselyov, 2009). Loss-of-function mutations in TRPML1 are implicated in mucolipidosis type IV (MLIV; MIM# 252650) pathogenesis, whereas a spontaneous gain-of-function mutation in TRPML3 is principally implicated in the varitint-waddler mouse mutant (Cuajungco and Samie, 2008; Zeevi et al., 2007). The varitint-waddler mouse features hearing loss, vestibular dysfunction, and abnormal pigmentation (Di Palma et al., 2002). On the cellular level, the A419P gain-of-function mutation in TRPML3 causes Ca2+ overload, which leads to cell death in mouse inner ear cells and melanosomes (Grimm et al., 2007; Grimm et al., 2009; Xu et al., 2007). MLIV is an autosomal recessive neurodegenerative lysosomal storage disorder characterized by mental and motor retardation, hypotonia, and opthalmological abnormalities (Bach, 2005). Loss-of-function mutations in TRPML1 lead to heterogeneous lysosomal storage of hydrophobic and hydrophilic macromolecules in cells of MLIV patients and other animal models (Bach, 2001; Venkatachalam et al., 2008; Venugopal et al., 2007). At present, TRPML2 is not implicated in any pathological disorder; however, we have recently described lysosomal inclusions in HEK 293 cells featuring significantly reduced native TRPML2 expression (Zeevi et al., 2009).
TRPML channels have been implicated in various membrane and protein sorting mechanisms along endo-lysosomal pathways (Puertollano and Kiselyov, 2009). MLIV pathogenesis is directly linked to TRPML1 loss-of-function within the late endosomal/lysosomal compartments where the protein is generally found (Manzoni et al., 2004; Pryor et al., 2006; Vergarajauregui and Puertollano, 2006) and, hence, several lysosomal regulatory roles have been proposed for TRPML1 in these compartments (Dong et al., 2008; Dong et al., 2009; LaPlante et al., 2006; Miedel et al., 2008; Piper and Luzio, 2004; Pryor et al., 2006; Soyombo et al., 2006; Treusch et al., 2004; Venugopal et al., 2009; Vergarajauregui et al., 2008). TRPML2 features cell surface and recycling endosomal localization and, as a result, has been implicated in recycling of glycosylphosphatidylinositol (GPI)-anchored proteins from recycling endosomes to the cell surface along the Arf6-regulated pathway (Karacsonyi et al., 2007). TRPML3 appears to play roles in the regulation of endocytosis and autophagy that correspond with localization of the protein to the cell surface and early endosomes under basal conditions, and autophagosome localization under stress conditions (Kim et al., 2009; Martina et al., 2009). Taken together, the differing intracellular distributions and proposed protein functions of each respective TRPML might suggest that paralogous TRPMLs are unlikely to co-regulate particular celluar tasks. However, recent evidence argues otherwise.
Although TRPMLs have shown to be distinct from each other in their primary intracellular distributions (Karacsonyi et al., 2007; Kim et al., 2009; Martina et al., 2009; Pryor et al., 2006; Vergarajauregui and Puertollano, 2006), all three channels do also colocalize with each other, in dual combinations, in a subset of intracellular vesicles (Zeevi et al., 2009). Furthermore, recombinant as well as native TRPML channels also physically interact with each other in homo- and heteromultimeric combinations (Curcio-Morelli et al., 2010; Venkatachalam et al., 2006; Zeevi et al., 2009). In addition, TRPML2 and TRPML3 traffic, in part, to lysosomes (Karacsonyi et al., 2007; Kim et al., 2009; Martina et al., 2009; Zeevi et al., 2009) where, like TRPML1, they also appear to regulate lysosomal function, in an as-yet-unknown manner. Indeed, gene-specific knockdown of TRPML2 or TRPML3 leads to lysosomal inclusions reminiscent of those found in MLIV patient TRPML1-null cells (Zeevi et al., 2009), suggesting that TRPMLs might dynamically interact with each other to sustain lysosomal integrity. Other discoveries in the TRPML field also point towards commonalities in TRPML function. Acidic pH appears to regulate the ion current properties of all three TRPMLs (Cantiello et al., 2005; Dong et al., 2008; Grimm et al., 2007; Kim et al., 2008; Lev et al., 2010; Raychowdhury et al., 2004; Xu et al., 2007), and paralogous missense mutations, within TRPML putative ion channel pores, regulate the activity of each TRPML channel in the same manner (Dong et al., 2008; Grimm et al., 2007; Lev et al., 2010; Xu et al., 2007). Recently, TRPML1 was also shown to regulate the gene expression of TRPML2 (Samie et al., 2009), and recombinant TRPML1 was shown to reduce the surface expression of recombinant TRPML3 (Grimm et al., 2010). Together, these findings suggest a more concerted role for TRPMLs in cell regulatory events.
Although, at present, the precise mechanism by which TRPMLs regulate each other's function to affect cell biological processes is unclear, recent evidence suggests that TRPMLs do so by heteromultimerizing to form distinct cation channel assemblies (Curcio-Morelli et al., 2010). Functional TRPML1–TRPML2 and TRPML1–TRPML3 channel assemblies have been characterized in a synthetic lipid-bilayer system by single-channel analysis (Curcio-Morelli et al., 2010), but a more comprehensive physiological interrogation of these and other TRPML channel assemblies has still to be carried out. Here, we assayed TRPML channel assemblies, in every possible homo- and heteromeric combination, in the physiological context of native TRPML-expressing tissue-culture cells. Furthermore, in order to demonstrate the direct application of these channel assemblies upon cell biology and homeostasis, we examined the affect of heterologous–heterologous and heterologous–native TRPML subfamily interactions upon cellular function. We show that dominant-negative loss-of-function TRPML mutants physically interact with constitutively active TRPMLs to form functional complexes. These homo- and heteromultimeric channel complexes feature reduced activity, which affects cell viability. Finally, we show that heteromultimerization of TRPML channels can play a role in starvation-induced autophagy, which suggests that this process is regulated, at least in part, by a concerted effort of more than just one TRPML channel.
Results
All TRPMLs feature similar ion current regulatory elements that affect cell viability in a similar manner
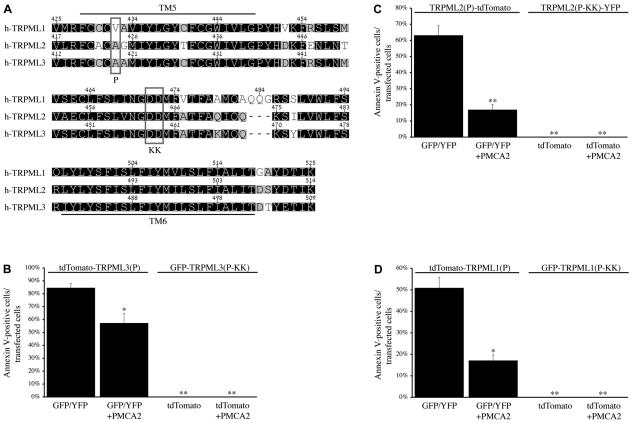
When comparing the amino acid sequence of the putative channel pore region (ranging from TM5 to TM6) of each respective TRPML channel with that of its other subfamily members, a striking similarity is observed (Fig. 1A). The amino acid identity and similarity of these channels in this region is ~82% and ~89%, respectively. This suggests that all three TRPMLs feature a highly similar ion channel pore, at least with regard to structure, implying that missense mutations within this region might give rise to similar effects on the channel activity of each TRPML. Indeed, recent studies have shown that paralogous mutations in this region have similar effects on TRPML channel function, regardless of which particular TRPML is mutated. The A419P mutation in TRPML3, which is implicated in varitint-waddler pathogenesis (Di Palma et al., 2002), is a gain-of-function mutation that leads to robust, inwardly rectifying currents through Ca2+-permeable and constitutively active TRPML3 channels (Grimm et al., 2007; Kim et al., 2007; Lev et al., 2010; Nagata et al., 2008; Xu et al., 2007). Likewise, we and others have recently shown that paralogous mutations in TRPML2 and TRPML1 lead to a similar effect on the channel function of the respective proteins (Dong et al., 2008; Lev et al., 2010; Samie et al., 2009; Xu et al., 2007). On the flip-side, the DD458/459KK mutation in TRPML3 is a loss-of-function mutation that renders the channel inactive (Kim et al., 2009; Xu et al., 2007), whereas the paralogous mutation in TRPML2 also inactivates the channel (Lev et al., 2010). To date, the electrophysiological properties of the paralogous TRPML1 mutant have not been characterized; however, a cell-based assay with this mutant protein suggests that it lacks channel function as well (Pryor et al., 2006). Thus, all three TRPMLs can be activated and inactivated by the same paralogous mutations, which lends support to the notion that all three channels feature similar current regulatory elements.
Fig. 1.
Paralogous current-modulating mutations in all three TRPML channels affect cell viability in a similar manner. (A) Amino acid sequence alignment of the TM5–TM6 regions of TRPML1, TRPML2, and TRPML3. Residues are shaded by similarity from low (white) to high (black). The locations of the P and KK mutations (see Table 1) are indicated. (B) HeLa cells were co-transfected with tdTomato-TRPML3(P) + GFP/YFP or GFP-TRPML3(P-KK) + tdTomato, with or without PMCA2. Alexa-Fluor-647–AnnexinV staining was performed in order to detect early apoptotic cells. The histogram indicates the mean percentage of cells exhibiting both GFP/YFP and tdTomato fluorescence, under each condition, that also stained positively for Alexa-Fluor-647–AnnexinV. (C) Same as B, with TRPML2(P)-tdTomato or TRPML2(P-KK)-YFP co-transfected cells. (D) Same as B, with tdTomato-TRPML1(P) or GFP-TRPML1(P-KK) co-transfected cells. For B–D, n=3 independent experiments of at least 42 cells per assay. Indicated P values are relative to the left-most bar in each histogram: *P<0.05, **P<0.01.
Importantly, the TRPML activating and inactivating mutations described above have also been shown to play a role in cellular function and homeostasis. The inwardly rectifying, Ca2+-permeable, and constitutively active TRPML3 A419P [TRPML3(P); see Table 1] mutant channel causes intracellular Ca2+ overload, which ultimately leads to cell death in TRPML3(P)-expressing cells (Grimm et al., 2007; Kim et al., 2007; Nagata et al., 2008; Xu et al., 2007). This Ca2+-mediated cytotoxic effect of TRPML3(P) mutant channels is reduced, in part, by coexpression with the calcium extrusion pump, plasma membrane calcium ATPase type 2 (PMCA2) (Grimm et al., 2009) (Fig. 1B). Likewise, we have recently demonstrated a similar effect upon cell viability when expressing the paralogous TRPML2 A424P [TRPML2(P); see Table 1] mutant in the presence and absence of PMCA2 (Lev et al., 2010) (Fig. 1C). Here, we show that the paralogous TRPML1 V432P [TRPML1(P); see Table 1] mutant presents with cytotoxic effects that can also be partially rescued when the channel is coexpressed with PMCA2 (Fig. 1D). This indicates, for the first time, that cell death in TRPML1(P)-expressing cells is also caused by intracellular Ca2+ overload. Interestingly, the TRPML channel-inactivating mutations, described above, have shown to be even more effective at neutralizing the cytotoxic effects of TRPML(P) mutants than coexpression with PMCA2 (Fig. 1B–D). A previous study demonstrated that introduction of the TRPML3 channel-inactivating DD458/459KK mutation in cis with the activating A419P mutation [TRPML3(P-KK); see Table 1] leads to a fully inactive channel with reduced cytotoxic effects in melan-a2 cells (Xu et al., 2007). Consistent with this study, we show that, under our assay conditions in HeLa cells, the TRPML3(P-KK) mutant presents with virtually nonexistent cytotoxic effects (Fig. 1B). Moreover, the same dominant-negative effect of the KK mutation over the P mutation is also observed with paralogous TRPML2(P-KK) and TRPML1(P-KK) mutants (Fig. 1C,D; Table 1). Therefore, we conclude that paralogous current-modulating mutations in all three TRPMLs affect cell viability in a similar manner.
Table 1.
Abbreviated notation of TRPML mutants described in this study
TRPMLs with mutations in their putative ion channel pores still physically interact with each other in homo- and heteromultimeric combinations
Having established the cytotoxic effects of TRPML(P) expression and the non-cytotoxic effects of TRPML(P-KK) expression, we utilized our constructs as tools by which to assay the functionality of homo- and heteromultimeric TRPML subfamily channel assemblies. Recent studies have demonstrated that the KK mutations in TRPML2 and TRPML3 exert a dominant-negative inhibitory effect on the function of their respective native channels. This effect was attributed to the interactions of TRPML2-KK and TRPML3-KK with and neutralization of the activity of their corresponding native channel (Karacsonyi et al., 2007; Kim et al., 2009). In a similar manner, we hypothesized that if the TRPML(P) and TRPML(P-KK) mutants were to form channel assemblies with each other, regardless of channel identity, then the channel-inactive TRPML(P-KK) mutants should exert a dominant-negative effect on constitutively active TRPML(P) mutants by rendering the complex inactive.
Because this hypothesis is predicated upon the ability of all TRPML(P) channels to physically interact with each TRPML(P-KK) channel in functional complexes, we performed co-immunoprecipitation (co-IP) assays to ensure that the channel pore mutations (P and KK) do not interfere with previously described TRPML subfamily protein–protein interactions (Venkatachalam et al., 2006; Zeevi et al., 2009). In supplementary material Fig. S1, we show the physical interactions of TRPML mutants from the same protein of origin [for example, TRPML1(P) vs. TRPML1(P-KK)] (supplementary material Fig. S1A). Indeed, the introduction of P and KK mutations into all three TRPML channels bears no effect upon such homomeric physical interactions. In supplementary material Fig. S2, we show the physical interactions of TRPML mutants from differing proteins of origin. Indeed, heteromeric interactions between TRPML(P) and TRPML(P-KK) mutants from differing proteins of origin [for example, TRPML3(P) and TRPML2(P-KK)] are also unaffected by the P and KK mutations (supplementary material Fig. S2A). These heteromeric TRPML channel interactions appear to be TRPML subfamily-specific because none of the TRPML channels co-immunoprecipitated with TRPM8, a related channel from a different TRP channel subfamily (supplementary material Fig. S2B). Thus, we demonstrate that TRPMLs, selectively, homo- and heteromultimerize with each other even when possessing mutations in their putative ion channel pores.
Dominant-negative TRPMLs attenuate the cytotoxic effects of constitutively active TRPML channels
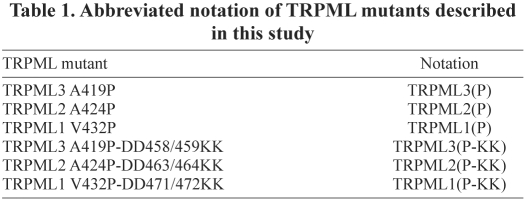
Having established that TRPML(P) and TRPML(P-KK) channels physically interact with each other in homo- and heteromultimeric combinations, we proceeded to assay the effect of these interactions upon cellular function. In particular, we sought to clarify the importance of heteromultimeric TRPML subfamily interactions. Thus, based on the cytotoxic effects of TRPML(P) expression and the non-cytotoxic effects of TRPML(P-KK) expression, as described above (Fig. 1B–D), we established cell viability as the indicator in our experiments. If the non-toxic TRPML(P-KK) mutants do not exert a dominant-negative effect on the toxic TRPML(P) mutants, then when the proteins heteromultimerize, we would not expect to observe a significant rescue of TRPML(P) and TRPML(P-KK) coexpressing cells from TRPML(P)-induced apoptosis. However, if TRPML(P-KK)s do exert a dominant-negative effect over TRPML(P)s, then we would expect to observe the opposite when the proteins heteromultimerize.
Coexpression of TRPML3(P) with TRPML2(P-KK) significantly reduced cell death, with respect to TRPML3(P) and GFP/YFP coexpressing control cells, from 84.7±3.2 to 26.8±2.5% AnnexinV-positive (early apoptotic) cells (Fig. 2A). This observation suggests that TRPML2(P-KK) channels exert a dominant-negative effect over TRPML3(P) channels. In addition, we found that TRPML3(P)-induced apoptosis is further reduced when coexpressing the channel with the dominant-negative TRPML2(P-KK) channel and the calcium extrusion pump, PMCA2 (Fig. 2A), indicating that the dominant-negative effect of TRPML2(P-KK) stems from reducing the cytotoxicity of TRPML3(P)-induced Ca2+ overload. Moreover, we also observed similar cytotoxic rescue in cells coexpressing TRPML3(P) with TRPML1(P-KK) and in cells coexpressing TRPML3(P) with TRPML3(P-KK) (Fig. 2A), which suggests that any TRPML(P-KK) mutant is capable of rescuing cells from TRPML3(P)-induced apoptosis. Importantly, this TRPML(P-KK)-mediated rescue was not limited to TRPML3(P)-expressing cells only, because similar neutralizing effects of dominant-negative TRPML(P-KK) channels on the cytotoxic effects of TRPML2(P) (Fig. 2B) and TRPML1(P) (Fig. 2C) expression were also observed in the presence and absence of PMCA2 (Fig. 2B,C). Therefore, these data combined with the findings of the co-IP assays described above (supplementary material Figs S1, S2), suggest that dominant-negative TRPML(P-KK)s attenuate the cytotoxic effects of constitutively active TRPML(P) channels vis-à-vis homo- and heteromeric physical interactions.
Fig. 2.
Dominant-negative TRPML(P-KK)s attenuate the cytotoxic effects of constitutively active TRPML(P) channels. (A) HeLa cells were co-transfected with tdTomato-TRPML3(P) in combination with PMCA2 (+), empty vector (−), and GFP/YFP-tagged expression constructs, as indicated. Alexa-Fluor-647–AnnexinV staining was performed and the results of staining are summarized as in Fig. 1B. (B) Same as A, with TRPML2(P)-tdTomato co-transfected cells. (C) Same as A, with tdTomato-TRPML1(P) co-transfected cells. For A–C, n=3 independent experiments of at least 45 cells per assay. Indicated P values are relative to the left-most bar in each histogram: *P<0.05, **P<0.01.
Dominant-negative TRPML2(P-KK) channels significantly attenuate the currents of TRPML3(P) channels in heteromeric complexes
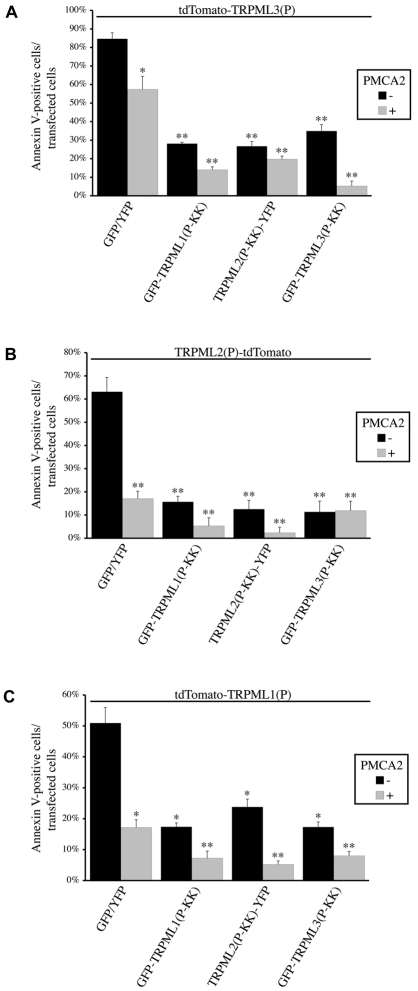
As suggested by the cell viability experiments, it is likely that inactive TRPML(P-KK) channel subunits exert their dominant-negative effect over constitutively active TRPML(P) channel subunits by functionally interacting with and affecting the current of TRPML(P) channels. To support this notion, we looked at whether expression of TRPML3(P) together with TRPML2(P-KK) reduces the robust, TRPML3(P)-associated currents that lead to cell Ca2+ overload and death. In HEK cells, where our current recordings were performed, we observed a clear difference in overall cell morphology between cells expressing TRPML3(P) only and those expressing TRPML2(P-KK) + TRPML3(P). Consistent with the results in Fig. 2A, we found that when TRPML3(P) was expressed alone, most TRPML3(P)-positive cells featured membrane blebbing and loss of morphology, which was reduced when the channel was coexpressed with TRPML2(P-KK) (Fig. 3A).
Fig. 3.
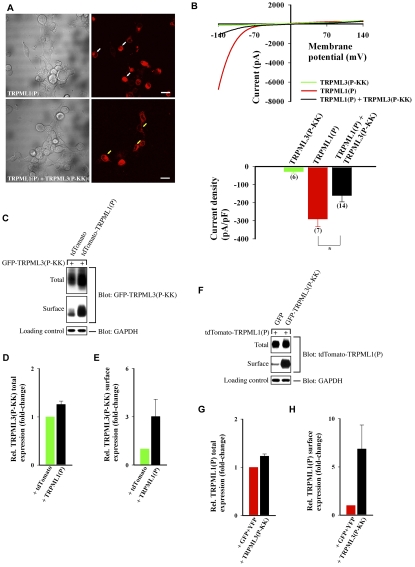
TRPML3(P) current density is significantly reduced when the channel is coexpressed with dominant-negative TRPML2(P-KK). (A) Wide-field confocal images showing representative cell morphology of HEK cells transfected with the indicated constructs. tdTomato-TRPML3(P) fluorescence (right panels) is shown next to DIC images (left panels). White arrows indicate cells with abnormal morphology. Yellow arrows indicate cells with normal morphology. Scale bars: 20 μm. (B) Top: Representative I-V curves of whole-cell currents measured from HEK cells coexpressing the indicated channels together with PMCA2. Bottom: Histogram of the average current density (at −120 mV) exhibited in HEK cells co-transfected with the indicated channels and PMCA2; n is number of cells analyzed; *P<0.05. (C) HEK cells were co-transfected with PMCA2 and the indicated expression constructs (above top panel). Surface proteins were biotinylated and subsequently isolated from normalized total cell lysates. Pictured are immunoblots of total lysate (Total) and surface protein (Surface) fractions that were probed for GFP+YFP and TRPML2(P-KK)-YFP. The endogenous GAPDH protein was probed in total lysates as a loading control. Transfection of unconjugated GFP and YFP fluorescent proteins served as a negative control for the assays. (D,E) Mean total and surface expression of TRPML2(P-KK) in the presence of TRPML3(P) (black), relative to its expression in the absence of TRPML3(P) (green), as determined from the experiments depicted in C. Experiments involving TRPML2(P-KK) alone were assigned a value of 1; n=3. (F) Same as C, with tdTomato-tagged proteins being probed for total and surface expression. Transfection of the unconjugated tdTomato fluorescent protein served as a negative control for the assays. (G,H) Mean total and surface expression of TRPML3(P) in the presence of TRPML2(P-KK) (black), relative to its expression in the absence of TRPML2(P-KK) (red), as determined from the experiments depicted in F. Experiments involving TRPML3(P) alone were assigned a value of 1; n=3.
In order to clarify the role of the TRPML3(P) current in the experiments depicted in Fig. 3A, we recorded whole-cell currents from HEK cells expressing TRPML3(P) and TRPML2(P-KK) in the presence of PMCA2. The coexpression of PMCA2 was used to help maintain cell integrity, which facilitates whole-cell recordings. Cells expressing TRPML3(P) in the absence of TRPML2(P-KK) feature robust, inward rectifying currents (Fig. 3B, red), consistent with previous reports describing the constitutive channel activity of TRPML3(P) (Grimm et al., 2007; Grimm et al., 2009; Kim et al., 2007; Lev et al., 2010; Nagata et al., 2008; Xu et al., 2007). By contrast, cells expressing TRPML2(P-KK) in the absence of TRPML3(P) do not exhibit any significant currents (Fig. 3B, green), implying that TRPML2(P-KK) is an inactive channel. Importantly, significantly reduced current density was observed in cells coexpressing both channels (Fig. 3B, black). Because this effect might be attributed to reduced surface expression of TRPML3(P) in the presence of TRPML2(P-KK), we assayed the total and surface expression of both channels when expressed alone and in combination. The total and surface expression of TRPML2(P-KK) are modestly enhanced by the presence of TRPML3(P) (Fig. 3C–E) and the total expression of TRPML3(P) is modestly enhanced by the presence of TRPML2(P-KK)(Fig. 3F,G). However, the surface expression of TRPML3(P) is greatly enhanced by the presence of TRPML2(P-KK)(Fig. 3F,H). This suggests that TRPML3(P)-mediated current is reduced in the presence of TRPML2(P-KK), despite enhanced overall surface expression of the constitutively active channel.
These findings (Fig. 3), together with the co-IP (supplementary material Fig. S2A) and cell viability (Fig. 2A) assays described above, indicate that TRPML2(P-KK) rescues cells from TRPML3(P)-induced apoptosis by physically interacting with TRPML3(P), in dominant-negative fashion, to form surface-expressed inactive heteromeric channel complexes. Previous studies have shown that extracellular Ca2+ is the primary driving force in causing intracellular Ca2+ overload and cell death in the presence of the surface-expressed, constitutively active, inwardly rectifying, Ca2+-permeable TRPML3(P) channel (Grimm et al., 2007; Kim et al., 2007). To confirm that functional physical interactions underlie the effects we have described in Fig. 3, we analyzed the TRPM8 current when the channel was coexpressed with TRPML2(P-KK). TRPM8, as we have shown, does not physically interact with TRPML2(P-KK) or any other TRPML(P-KK) dominant-negative mutant (supplementary material Fig. S2B). Consistent with this finding, coexpression of TRPML2(P-KK) with TRPM8 did not reduce the outwardly rectifying and menthol-facilitated TRPM8-derived (Varnai et al., 2006) whole-cell currents (supplementary material Fig. S3). This control further supports the notion that the reduction of TRPML3(P) channel activity that occurs in the presence of TRPML2(P-KK) (Fig. 3B, black) can be explained by specific physical interactions between the two channels (supplementary material Fig. S2A) that lead to the formation of heteromultimeric channel assemblies with reduced channel activity. Conversely, where there is no interaction of TRPML2(P-KK) with another TRP channel (such as TRPM8), there is no dominant-negative inhibitory effect on the current density of that channel.
Dominant-negative TRPML3(P-KK) channels significantly attenuate the currents of TRPML1(P) channels in heteromeric complexes
Like recombinant TRPML3(P), recombinant TRPML1(P) is also a surface-expressed, constitutively active, inwardly rectifying, Ca2+-permeable channel (Dong et al., 2008; Dong et al., 2009; Samie et al., 2009; Xu et al., 2007) that is implicated in causing cell death (Fig. 1D). In HEK cells, TRPML1(P) causes cell degeneration that is attenuated when the channel is coexpressed with the inactive TRPML3(P-KK) channel (Fig. 4A). Consistent with this finding, TRPML1(P)-derived whole-cell currents (Fig. 4B, red) are significantly reduced in the presence of TRPML3(P-KK) (Fig. 4B, black). However, given that native TRPML1 is strictly intracellular (Kim et al., 2009; Zeevi et al., 2009), it is possible that coexpression with TRPML3(P-KK) reduces TRPML1(P)-derived whole-cell currents by also reducing the surface expression of TRPML1(P) in favor of the natural TRPML1 intracellular localization. Fig. 4C–E shows that the overall surface expression of TRPML3(P-KK) is enhanced in the presence of TRPML1(P), and Fig. 4F–H shows that the overall surface expression of TRPML1(P) is enhanced even more so in the presence of TRPML3(P-KK). Thus, TRPML1(P)-mediated current is reduced in the presence of TRPML3(P-KK) despite its enhanced overall surface expression. Together, these results indicate that TRPML3(P-KK) reduces TRPML1(P)-induced apoptosis (Fig. 2C) by physically interacting with TRPML1(P), in dominant-negative fashion, to form surface-expressed inactive heteromeric channel complexes.
Fig. 4.
TRPML1(P) current density is significantly reduced when the channel is coexpressed with dominant-negative TRPML3(P-KK). (A) Same experimental procedures as in Fig. 3A with the indicated constructs. tdTomato-TRPML1(P) fluorescence (right panels) is shown next to DIC images (left panels); Scale bars: 20 μm. (B) Same experimental procedures as in Fig. 3B; *P<0.05. (C) HEK cells were co-transfected with PMCA2, GFP-TRPML3(P-KK), and either tdTomato or tdTomato-TRPML1(P). Total and surface expression of GFP-TRPML3(P-KK) was analyzed as described in Fig. 3C. (D,E) Summary of experiments in C as in Fig. 3D,E; n=3. (F) HEK cells were co-transfected with PMCA2, tdTomato-TRPML1(P), and either GFP or GFP-TRPML3(P-KK). Total and surface expression of tdTomato-TRPML1(P) was analyzed as described in Fig. 3F. (G,H) Summary of experiments in F as in Fig. 3G,H; n=3.
Dominant-negative TRPML1(P-KK) channels significantly attenuate the currents of TRPML2(P) channels in heteromeric complexes
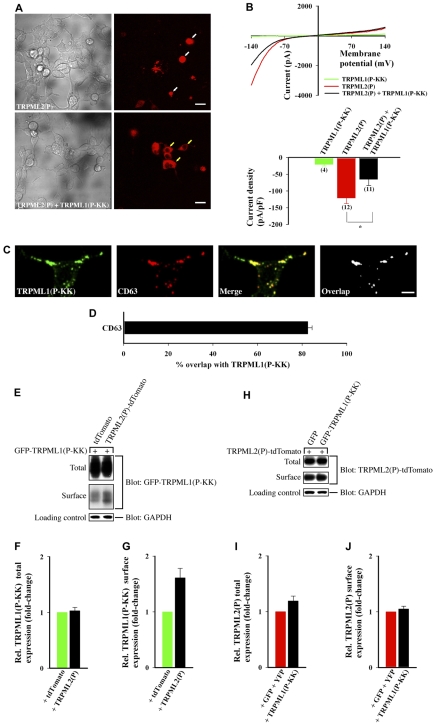
Like recombinant TRPML3(P) and TRPML1(P), recombinant TRPML2(P) is also a surface-expressed, constitutively active, inwardly rectifying, Ca2+-permeable channel (Dong et al., 2008; Lev et al., 2010; Samie et al., 2009) that is implicated in causing cell death (Fig. 1C). In HEK cells, TRPML2(P) causes cell degeneration that is attenuated when the channel is coexpressed with the inactive TRPML1(P-KK) channel (Fig. 5A). Consistent with this finding, TRPML2(P)-derived whole-cell currents (Fig. 5B, red) are significantly reduced in the presence of TRPML1(P-KK) (Fig. 5B, black).
Fig. 5.
TRPML2(P) current density is significantly reduced when the channel is coexpressed with dominant-negative TRPML1(P-KK). (A) Same experimental procedures as in Fig. 3A with the indicated constructs. TRPML2(P)-tdTomato fluorescence (right panels) is shown next to DIC images (left panels). Scale bars: 20 μm. (B) Same experimental procedures as in Fig. 3B; *P<0.05. (C) HeLa cells were transfected with GFP-TRPML1(P-KK) and immunostained for CD63. Cells were then imaged by confocal microscopy. Scale bar: 10 μm. (D) Images similar to those in C, from at least 15 cells in three independent experiments, were used to determine the mean percentage of GFP-TRPML1(P-KK) that overlapped with CD63. (E–J) Same experimental procedures as in Fig. 4C–H with the indicated constructs; n=3 independent experiments per assay.
We have found that the majority of recombinant TRPML1(P-KK) is expressed intracellularly and colocalizes, for the most part, with the late endosomal/lysosomal marker CD63 (Fig. 5C,D), like wild-type TRPML1 (Vergarajauregui and Puertollano, 2006). Still, a small fraction of TRPML1(P-KK) also reaches the cell surface (Fig. 5E). Interestingly, a recent report has shown that wild-type (WT)-TRPML3 surface expression and agonist-induced whole-cell currents are reduced when the channel is coexpressed with the intracellular WT-TRPML1 (Grimm et al., 2010). However, the same report also suggested that coexpression of WT-TRPML1 with TRPML3(P) did not bear any effect upon TRPML3(P) whole-cell currents. Thus, we asked whether TRPML1(P-KK) exerts its dominant-negative effect over TRPML2(P) by interacting with the channel intracellularly and preventing it from reaching the cell surface, or by interacting with the channel on the cell surface and attenuating its current.
Fig. 5E–G shows that a small fraction of overexpressed TRPML1(P-KK) reaches the cell surface. Interestingly, this fraction is enhanced when the channel is coexpressed with TRPML2(P) (Fig. 5E–G). Fig. 5H–J shows that TRPML2(P) total and surface expression are modestly enhanced in the presence of TRPML1(P-KK). This suggests that TRPML2(P) remains surface-expressed even in the presence of the primarily intracellular, but partially surface-expressed TRPML1(P-KK). Thus, our data indicate that TRPML1(P-KK) exerts its dominant-negative effect on TRPML2(P) currents in surface-expressed heteromeric inactive channel assemblies that prevent TRPML2(P)-mediated apoptosis (Fig. 2B).
Together, our findings from co-IP, cell viability assays, whole-cell current recordings, and cell surface expression assays indicate that TRPML mutant regulation of cell viability is probably mediated by cell-surface-localized heteromultimeric protein–protein interactions that constitute functional or non-functional heteromultimeric TRPML channel assemblies.
Dominant-negative TRPML1 channels inhibit starvation-induced autophagy
Having established that gain-of-function and dominant-negative, loss-of-function TRPML mutants form heteromultimeric TRPML channel assemblies that affect cell viability, we next asked whether heteromultimeric TRPML channel assemblies composed of dominant-negative and WT-TRPMLs affect starvation-induced autophagy (SIA). The process of SIA is not directly regulated by TRPML1 (Vergarajauregui et al., 2008), but is directly regulated by TRPML3 (Kim et al., 2009; Martina et al., 2009) and possibly TRPML2 (whose role in SIA has not yet been determined).
Autophagy is a process whereby macromolecules and organelles are processed for degradation in lysosomes via specialized intracellular vesicles in response to various cell stressors including, but not limited to, starvation conditions (Cuervo, 2004b; Yorimitsu and Klionsky, 2005). The autophagic process begins with the formation of double-membrane autophagosomes and can be detected by examining the distribution of autophagosomal light chain three (LC3) in cells because this protein is recruited into autophagosomes upon induction of autophagy (Klionsky et al., 2007). LC3 displays a soluble cytosolic distribution under basal conditions, which transforms into a punctate vesicular distribution as LC3 becomes lipidated during autophagy. Thus, autophagy induction, under starvation conditions, can be quantified by counting LC3-positive puncta within RFP-LC3-transfected cells.
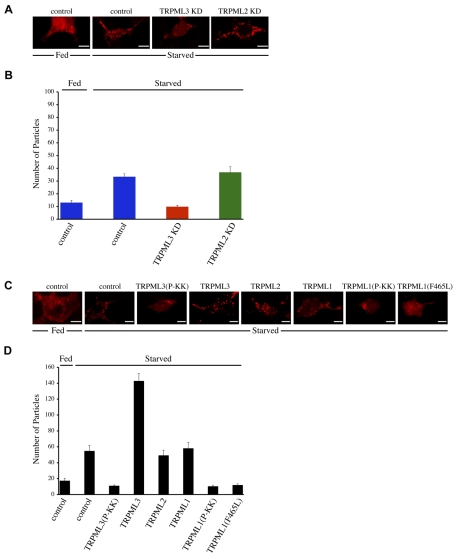
The potential role of TRPML2 in the regulation of starvation-induced autophagy has not been elucidated. Thus, we assayed RFP-LC3 distribution in cells stably transfected with microRNA-adapted short hairpin RNAs (shRNAmirs) designed to knockdown native TRPML2 or TRPML3 expression (Zeevi et al., 2009). As seen in Fig. 6A and the summary in Fig. 6B, a primarily diffuse cytosolic RFP-LC3 distribution in fed control cells (transfected with non-silencing shRNAmir) is transformed into a punctate distribution in starved control cells, where the autophagic machinery is expected to be upregulated. In addition, consistent with a previous report describing the active role of TRPML3 in the regulation of SIA (Kim et al., 2009), we confirm that knockdown of native TRPML3 expression reduces autophagosome formation in starved TRPML3 knockdown cells, with respect to starved control cells (Fig. 6A,B). Importantly, in starved TRPML2 knockdown cells, we did not detect any discrepancy in autophagic levels with respect to control (Fig. 6A,B). Moreover, whereas overexpression of recombinant WT-TRPML3 markedly enhances autophagosome formation in starved transfected cells (Kim et al., 2009; Martina et al., 2009) with respect to starved control cells; overexpression of WT-TRPML2 does not affect typical autophagic levels under the same conditions (Fig. 6C,D). Together, these results indicate for the first time that, unlike TRPML3, TRPML2 does not appear to play an active role in SIA.
Fig. 6.
Dominant-negative TRPML1 mutants inhibit starvation-induced autophagy. (A,B) Knockdown of native TRPML2 expression does not affect autophagic response in starved cells. (A) HEK cells, stably transfected with non-silencing (control), TRPML3-depleting (TRPML3 KD), or TRPML2-depleting (TRPML2 KD) shRNAmirs, were transfected with RFP-LC3 followed by incubation in feeding (Fed) or starvation (Starved) media for 2 hours. Representative confocal images of RFP-LC3 are presented for each experiment. Scale bars: 10 μm. (B) Histogram of the average number of RFP-LC3 puncta in A, as determined from at least 13 cells in three independent experiments. TRPML3-specific and TRPML2-specific knockdown was verified in the appropriate cell lines, as previously described (Zeevi et al., 2009), in parallel with each experiment. (C,D) TRPML1(P-KK) and TRPML1(F465L) mutants inhibit starvation-induced autophagy. (C) HeLa cells were co-transfected with RFP-LC3 and the indicated GFP/YFP-tagged expression constructs, followed by treatment as in A. Representative confocal images of RFP-LC3 are presented. Scale bars: 10 μm. (D) Histogram of the average number of RFP-LC3 puncta in C, as determined from at least 10 cells in three independent experiments.
Overexpression of a dominant-negative TRPML3-KK construct inhibits starvation-induced autophagy by, presumably, interacting with native TRPML3 and reducing its channel activity (Kim et al., 2009). Here, we show that the TRPML3(P-KK) dominant-negative mutant also inhibits autophagosome formation in starved cells (Fig. 6C,D). Strikingly, we also found that overexpression of the TRPML1(P-KK) dominant-negative mutant unexpectedly inhibited SIA (Fig. 6C,D; see below).
The TRPML1(P-KK) mutant physically interacts with WT-TRPML1 and WT-TRPML2 (supplementary material Fig. S4), suggesting that endogenous TRPML1 and TRPML2 function can be compromised when these native channels complex with the TRPML1(P-KK) dominant-negative channel. However, previously described experiments indicate that SIA is not regulated by endogenous TRPML1 function because neither the absence (Vergarajauregui et al., 2008) nor overexpressed presence of the TRPML1 channel (confirmed in Fig. 6C,D) leads to any discernible effect upon SIA (Vergarajauregui et al., 2008). Our data also demonstrate a similar argument regarding endogenous TRPML2 function in SIA regulation (Fig. 6A–D). Thus, the inhibition of SIA by TRPML1(P-KK) cannot be attributed to physical interactions with, and channel inactivation of, native TRPML1 or native TRPML2. Rather, a more likely explanation for this effect implicates heteromultimeric interactions between TRPML1(P-KK) and native TRPML3, a channel that plays a definitive SIA-stimulatory role (Kim et al., 2009; Martina et al., 2009). Indeed, supplementary material Fig. S4 demonstrates that, in addition to interacting with WT-TRPML1 and WT-TRPML2, TRPML1(P-KK) also physically interacts with WT-TRPML3, which suggests that endogenous TRPML3 function can also be compromised when the native channel complexes with the TRPML1(P-KK) dominant-negative channel. Therefore, we propose that inhibition of SIA, resulting from TRPML1(P-KK) overexpression, is caused by formation of inactive heteromultimeric channel assemblies comprised of TRPML1(P-KK) and native TRPML3. This is a striking demonstration of how a single TRPML1 mutant can affect a physiological function mediated by another TRPML channel (i.e. TRPML3).
In view of our findings in Figs 1, 2 and 5, it is likely that TRPML1(P-KK) is an inactive channel. We also propose that the dominant-negative nature of TRPML1(P-KK) correlates with its ability to physically interact with wild-type or constitutively active TRPML channels to form inactive channel assemblies (supplementary material Figs S1, S2, S4). Accordingly, we tested SIA in cells expressing the known channel-impaired TRPML1(F465L) mutant (Dong et al., 2008; Kiselyov et al., 2005). The F465L missense mutation in TRPML1 was first described in compound heterozygosity with a TRPML1-null allele in an MLIV patient (Bargal et al., 2001; Kiselyov et al., 2005). In addition, this mutation does not interfere with the ability of TRPML1 to physically interact with both WT-TRPMLs (supplementary material Fig. S5A) and constitutively active TRPML(P) mutants (data not shown). Therefore, we reasoned that if impaired channel activity of one component of a TRPML channel assembly leads to a dominant-negative effect, then heteromultimeric channel assemblies composed of native TRPML3 and the channel-impaired TRPML1(F465L) mutant should result in functionally reduced activity of these assemblies. This in turn might affect SIA, if SIA is regulated by TRPML heteromultimerization. As seen in Fig. 6C,D, this is precisely what occurs. Inhibition of SIA by TRPML1(F465L) parallels the inhibition found in TRPML3(P-KK)- and TRPML1(P-KK)-expressing cells. This suggests that significantly reduced channel activity of one, naturally occurring, mutated TRPML channel results in dominant-negative inhibition of native TRPML3 protein function at the cellular level by rendering the heteromultimeric complex less active.
Dominant-negative properties of TRPML1(F465L)
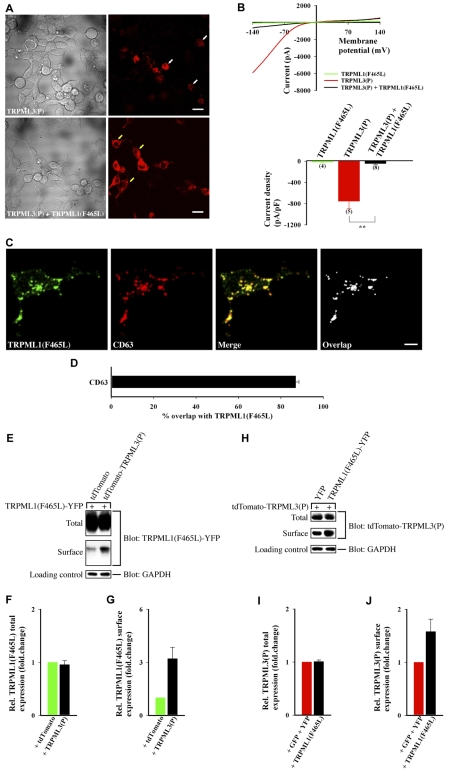
The TRPML1(F465L) mutation displays autosomal recessive inheritance in humans (Bargal et al., 2001), meaning that the dominant-negative behavior, demonstrated in our assay of SIA, should not necessarily be expected even when the channel is overexpressed in cells. As a result, we sought to further characterize the dominant-negative properties of TRPML1(F465L) in our cell viability assays (described in Fig. 2). As a negative control for the assay, we compared the cell viability of TRPML(P) + TRPML1(F465L) co-transfected cells with that of TRPML(P) + WT-TRPML1 co-transfected cells in order to preclude the possibility that WT-TRPML1 channels [which physically interact with TRPML(P) mutants (data not shown)] might also rescue TRPML(P) expressing cells. Our results indicate that neither WT-TRPML1 nor TRPML1(F465L) channels cause apoptosis (supplementary material Fig. S5B). However, only coexpression of TRPML(P) mutants with TRPML1(F465L), as opposed to WT-TRPML1, significantly rescued cells from apoptosis (supplementary material Fig. S5B). This finding is corroborated in HEK cells where TRPML1(F465L) rescues TRPML3(P)-expressing cells from cell degeneration (Fig. 7A).
Fig. 7.
TRPML3(P) current density is significantly reduced when the channel is coexpressed with dominant-negative TRPML1(F465L). (A) Same experimental procedures as in Fig. 3A with the indicated constructs. tdTomato-TRPML3(P) fluorescence (right panels) is shown next to DIC images (left panels). Scale bars: 20 μm. (B) Same experimental procedures as in Fig. 3B; **P<0.01. (C,D) Same experimental procedure as in Fig. 5C,D with the TRPML1(F465L)-YFP construct. n=3 independent experiments of at least 15 cells. (E–J) Same experimental procedures as in Fig. 4C–H with the indicated constructs. n=3 independent experiments per assay.
TRPML1(F465L) displays impaired channel activity (Fig. 7B, green), but its inhibition of the TRPML3(P) whole-cell current density is noteworthy (Fig. 7B, black) because most of the overexpressed channel is expressed intracellularly and colocalizes, for the most part, with CD63 (Fig. 7C,D). Nevertheless, Fig. 7E–G demonstrates that despite its predominant intracellular localization, a small fraction of overexpressed TRPML1(F465L) reaches the cell surface. Moreover, this surface fraction is tripled when the channel is coexpressed with TRPML3(P) (Fig. 7E–G). In addition, Fig. 7H–I shows that although total TRPML3(P) expression levels are unaffected by TRPML1(F465L), its surface expression levels are modestly enhanced (Fig. 7J). This suggests that TRPML3(P) remains surface-expressed even in the presence of the primarily intracellular, but partially surface-expressed TRPML1(F465L). Thus, our results indicate that TRPML1(F465L) can function as a dominant-negative channel by complexing with surface-expressed TRPML3(P) to form heteromeric inactive channel assemblies that prevent TRPML3(P)-mediated apoptosis (supplementary material Fig. S5B).
Together, our data suggest that TRPML channels with reduced activity, such as the naturally occurring TRPML1(F465L) mutant, are capable of affecting the function of homo- and heteromultimeric TRPML channel assemblies in dominant-negative fashion.
Discussion
In this report, we demonstrated the ability of all three TRPMLs to interact with each other, in every possible dual combination, to form homo- and heteromultimeric channel assemblies. Moreover, we also found that TRPML channel assemblies form, even when various missense mutations are located within the putative channel pore region of each subunit. This property of TRPML channels became useful when we assayed the effects of heteromultimeric TRPML channel assemblies, comprised of mutant and wild-type channel subunits, upon cellular function. Indeed, our observation that TRPML1(P-KK) inhibits starvation-induced autophagy suggests a nascent function for TRPML1 in the regulation of SIA. This arises from the ability of TRPML1 to complex with, and affect the functionality of, TRPML3 channels even when possessing dominant-negative mutations in its channel pore. Furthermore, our findings that the MLIV-associated channel-impaired TRPML1(F465L) mutant inhibits SIA, and also behaves in dominant-negative fashion in multiple assays, implies that channel functionality is key to establishing some TRPML-based dominant-negative phenotypes. Indeed, this report is the first to demonstrate dominant-negative properties in an MLIV-associated missense mutant protein.
Although a thorough clinical and phenotypic assessment has not been performed with carriers of the F465L mutation in TRPML1, the mutation originally presented with apparent autosomal recessive inheritance in the family where the mutation was identified (Bargal et al., 2001). On the other hand, we have seen that this mutation accrues dominant-negative properties to the TRPML1 channel, and it should also be noted that heterozygote carriers of MLIV-associated mutations, in general, exhibit slightly more lysosomal inclusions than normal in electron micrographs (our unpublished observations). Hence, if the F465L mutation is truly dominant-negative, why are the carriers of this mutation presumably asymptomatic for MLIV? We offer three possible explanations. First, it could be that the F465L mutant transcript is degraded in F465L/wild-type heterozygotes, thereby eliminating the negative effects of TRPML1(F465L) expression. Second, assuming that F465L expression levels might parallel wild-type transcript expression levels, it is possible that functional TRPML1 channel complexes (comprised of wild-type-only channels), and non-functional F465L-containing TRPML1 channel complexes, coexist within cells. If proper lysosomal function requires only a minimal amount of functional TRPML1 channels, then this might also explain why F465L carriers are asymptomatic for MLIV. Third, it is possible that F465L channels are only selectively dominant, which means that they are dominant in regulating certain cellular functions but not necessarily in other more crucial cellular functions. Due to inaccessibility of cultured cells from the family of the F465L-bearing MLIV patient and those of the patient himself, we cannot explore the possible explanations for the recessive inheritance of the F465L mutation in TRPML1. Nonetheless, the notion that an MLIV-associated mutation exhibits a dominant-negative effect on physiological mechanisms offers new possibilities in developing therapeutically relevant strategies for treating TRPML-related pathologies.
Recombinant TRPML1(F465L) traffics primarily to late endosomal/lysosomal intracellular compartments, as does native TRPML1 and a fraction of native TRPML3 (Kim et al., 2009; Zeevi et al., 2009). Autophagy is an intracellular process (Cuervo, 2004a) that is also regulated by TRPML3 (Kim et al., 2009). This suggests that TRPML1(F465L) is likely to inhibit SIA by complexing with native TRPML3 in intracellular compartments. However, because some recombinant TRPML1(F465L) reaches the cell surface (Kiselyov et al., 2005), we cannot preclude the possibility that the channel also complexes with surface-localized TRPML3 to inhibit SIA. However, this latter possibility is unlikely because autophagy is an intracellular process (Cuervo, 2004a). Nevertheless, the partial surface expression of TRPML1(F465L) and other recombinant TRPML1 mutants did become useful in this study in describing functional surface-expressed heteromeric channel complexes. Future studies should investigate these physiological interactions in a native system in order to verify the implications of heteromeric channel assemblies.
The notion that heteromeric interactions among TRPMLs can potentially affect cellular function and homeostasis has significant ramifications when investigating TRPML function. Our study demonstrates the importance of investigating the effect of a single missense TRPML mutant on the functions and related mechanisms of not one but all three native TRPML channels. Indeed, recombinant dominant-negative missense TRPML mutants afford investigators a valuable tool for potentially downregulating all three native TRPMLs at once. This might facilitate comparisons between three-TRPML-coding human and murine TRPML models and Caenorhabditis elegans and Drosophila models that code for a single TRPML ortholog. Such comparisons could ultimately serve to translate discoveries from TRPML-based animal models into therapies for TRPML-associated disorders such as MLIV.
In addition, future studies should also take into account the potential residual effects of the absence or abundance of a single TRPML upon the functions of remaining or other TRPML paralogs. For instance, it could be that MLIV pathogenesis results, at least in part, from the downregulation of native TRPML2 and TRPML3 that results from the absence of TRPML1 in MLIV cells. For example, high levels of basal (as opposed to starvation-induced) autophagy have been detected in MLIV patient TRPML1-null cells (Vergarajauregui et al., 2008), whereas overexpression of TRPML3 leads to upregulation of basal autophagic levels (Kim et al., 2009; Martina et al., 2009). This, taken together with our observation that native TRPML1 dynamically interacts with native TRPML3 (Zeevi et al., 2009), suggests that there might be interplay between TRPML1 and TRPML3 in the regulation of basal autophagy. When TRPML1 is present, the ability of TRPML3 to stimulate the formation of autophagosomes is attenuated accordingly; but when TRPML1 is absent, as is mostly the case in MLIV cells (Bargal et al., 2001; Sun et al., 2000), stimulation of autophagy by TRPML3 is left unregulated, allowing for the increased autophagosome formation that is found in MLIV patient cells. Future studies should investigate this potential TRPML1–TRPML3 interplay in light of our findings here.
Materials and Methods
Expression constructs
The TRPML1-YFP expression construct, used in this study, was described previously (Zeevi et al., 2007). The EGFP ORF, minus a stop codon, and the complete EYFP ORF were derived from the pEGFP-C2 and pEYFP-N1 vectors (Clontech), respectively, and cloned into a slightly modified version of the pcDNA3 vector (Invitrogen). tdTomato nucleotide sequences, with and without a stop codon, were derived from the pRSET-B tdTomato bacterial expression construct, kindly provided by Roger Tsien (UCSD, San Diego, CA), and cloned into the same pcDNA3 vector. Subsequently, the human TRPML1 and TRPML3 ORFs were cloned downstream to and in-frame with the GFP and tdTomato sequences on the pcDNA3-GFP and pcDNA3-tdTomato vectors, respectively. Similarly, the human TRPML2 ORF was cloned upstream to and in-frame with EYFP and tdTomato sequences on the appropriate pcDNA3-EYFP and pcDNA3-tdTomato vectors, respectively. The resulting TRPML fusion-protein constructs were then used to derive TRPML mutants by site-directed mutagenesis according to a previously described method (Ho et al., 1989). Insert orientation and polymerase fidelity were verified by restriction enzyme mapping and sequencing for each construct. The TRPM8, pmRFP-LC3, pEGFP-TRPML3, and PMCA2z/a constructs, used in this study, were kindly provided by Sharona E. Gordon (University of Washington, Seattle, WA), Tamotsu Yoshimori (Osaka University, Osaka, Japan), Shmuel Muallem (University of Texas Southwestern Medical Center, Dallas, TX), and Peter G. Gillespie (Oregon Hearing Research Center, Portland, OR), respectively.
Cell culture
HeLa and HEK cells were grown at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% Pen-Strep (Biological Industries). Transfections were performed with the Fugene 6 (Roche) or TransIt (Mirus) transfection reagents with equal amounts of cDNA. Wherever not specified, transfections were supplemented with GFP, YFP, tdTomato, or empty vector cDNA in order to equalize DNA concentrations in all transfections. For immunoprecipitation and electrophysiology experiments, PMCA2 was added to all transfections unless the experiment included WT-TRPML constructs. HEK cells, stably transfected with non-silencing, TRPML3-depleting, or TRPML2-depleting shRNAmirs were described previously (Zeevi et al., 2009).
AnnexinV staining
Hela cells were grown on coverslips and transfected with fluorescent protein constructs and PMCA2 or empty vector. At 18 hours post-transfection, cells were stained with Alexa-Fluor-647–AnnexinV (BioLegend) and analyzed on a confocal microscope (Olympus Fluoview 300 IX70) as previously described (Lev et al., 2010).
Antibodies
Monoclonal anti-GFP (which recognizes both GFP and YFP) and polyclonal anti-DsRed (which recognizes tdTomato) antibodies were purchased from Clontech. Polyclonal anti-GFP (598; which recognizes both GFP and YFP), polyclonal anti-TRPM8 (including a TRPM8-blocking peptide), monoclonal anti-GAPDH (6C5), and monoclonal anti-CD63 were from MBL (Woburn, MA), Alomone Labs (Jerusalem, Israel), Santa Cruz Biotechnology and BD Biosciences, respectively. All secondary antibodies, except for Alexa Fluor 647 goat anti-mouse IgG (Invitrogen), were purchased from Jackson ImmunoResearch.
Immunoprecipitation and immunoblotting
HEK cells were solubilized on ice in lysis buffer containing 1% Triton X-100, 25 mM Tris–HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, and complete TM protease inhibitor cocktail (Santa Cruz Biotechnology). After centrifugation to clear debris, cell lysates were then divided into separate tubes for immunoprecipitation with two different antibodies in the presence of Protein A/G PLUS-Agarose (Santa Cruz Biotechnology) at 4°C overnight, with shaking. Unless indicated otherwise, immunoprecipitation was performed with the GFP/YFP-recognizing polyclonal anti-GFP antibody, or with the tdTomato-recognizing polyclonal anti-DsRed antibody. Following overnight incubation, all immunoprecipitates were washed five times with lysis buffer followed by a final wash in 50 mM Tris–HCl (pH 8). Precipitated proteins were eluted with 2× LDS sample buffer containing 2× NuPAGE Reducing Agent (Invitrogen) and heated to 60°C for 10 minutes prior to separation on 4–12% NuPAGE Novex Bis-Tris gels (Invitrogen), and then immunoblotted according to standard procedures. Immunoprecipitates were blotted for GFP/YFP with the monoclonal anti-GFP antibody; for tdTomato, with the polyclonal anti-DsRed antibody; or for TRPM8, with the polyclonal anti-TRPM8 antibody. Secondary detection was performed with HRP-conjugated goat anti-mouse or mouse anti-rabbit IgG light-chain-specific antibodies, where appropriate.
Surface biotinylation assay
Transfected HEK cell surface proteins were biotinylated and isolated from normalized cell lysates, as previously described (Lev et al., 2010). Total lysate and cell surface tdTomato-tagged and GFP/YFP-tagged proteins were probed by immunoblot using either the polyclonal anti-DsRed or the polyclonal anti-GFP antibodies, respectively. The mouse monoclonal anti-GAPDH antibody was used as loading control. Immunoblots were quantitated by densitometry analysis using Photoshop software (Adobe Systems).
Electrophysiology
HEK cells were seeded onto polylysine-coated plates at 25% confluence. At 20–24 hours before the experiment, cells were transfected to induce expression of channels. Whole-cell currents were recorded at room temperature using borosilicate patch pipettes of 3–5 MΩ and an Axopatch 200B (Molecular Devices) voltage-clamp amplifier. Voltage-clamp pulses in the whole-cell configuration, using voltage ramps from −140 to 140 mV in 1 second, were generated and the data captured using a Digidata 1440A interfaced to a computer running the pClamp 10 software (Molecular Devices). Currents were filtered using the 8-pole low pass Bessel filter of the patch-clamp amplifier at 5 kHz and sampled at 50 kHz. To measure I–V curves with minimal distortions, only cells with low (<10 MΩ) series resistance were used and the series resistance was compensated by 80%.
Solutions
The extracellular Tyrode solution was prepared as a Ca2+ nominal based solution to which either 0.5 mM EGTA was added for TRPM8 experiments, or 1.5 mM CaCl2 for all other experiments. The solution was titrated to pH 7.4 and otherwise contained 145 mM NaCl, 5 mM KCl, 1 mM MgCl2, 15 mM HEPES, and 10 mM glucose. The intracellular solution was titrated to pH 7.2 and contained 130 mM CsCl, 1 mM MgCl2, 10 mM EGTA, 10 mM HEPES, 2 mM Na2-ATP and 4.1 mM CaCl2. Cells were perfused via a BPS-4 valve control system (Scientific Instruments) at a rate of ~30 chamber volumes per minute. Menthol was added to the extracellular solution at a final concentration of 200 μM.
Confocal microscopy
For immunofluorescence, HeLa cells were grown on coverslips and transfected with either GFP-TRPML1(P-KK) or TRPML1(F465L)-YFP. Cells were then fixed and immunostained according to a previously described method (Zeevi et al., 2009). Immunostaining for CD63 was performed with monoclonal anti-CD63 and Alexa Fluor 647 goat anti-mouse antibodies. Confocal images were acquired on an Olympus Fluoview 300 IX70 microscope, with a 60× oil immersion objective, and processed off-line with ImageJ software.
For assaying autophagy, HeLa cells were co-transfected with pmRFP-LC3 and another GFP/YFP-tagged protein construct. At 18 hours post-transfection, the growth medium was replaced with either fresh growth or starvation medium (Earle's balanced salt solution; Biological Industries) for 2 hours at 37°C. Following treatment, cells were fixed in 4% formaldehyde for 30 minutes at room temperature, washed three times with 200 mM NH4Cl in phosphate-buffered saline, and subsequently mounted onto slides with Antifade Solution (Vysis). Confocal images of cells exhibiting both RFP and GFP/YFP emission were then acquired and processed as described above. Confocal images of transfected HEK cells were acquired as previously described (Lev et al., 2010).
Data analysis
Data were analyzed and plotted using pClamp 10 (Molecular Devices) and Sigma Plot 8.02 (Systat software) software. Confocal images were imported as tiff single images into Photoshop (Adobe Systems), where they were subsequently cropped and resized.
Statistical analysis
Student's t-test was used for statistical analysis. All error bars show s.e.m.
Supplementary Material
Acknowledgments
This work was supported by the Rita Altura foundation (to G.B.), the Moscona and Minerva Foundations (to B.M.), and by Grants from the National Institute of Health (RO1 EY 03529 to B.M.), the Israel Science Foundation (ISF, to G.B. and B.M.), the US-Israel Binational Science Foundation (BSF to B.M.), and the German-Israel Foundation (GIF to B.M.). Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/18/3112/DC1
References
- Bach G. (2001). Mucolipidosis type IV. Mol. Genet. Metab. 73, 197-203 [DOI] [PubMed] [Google Scholar]
- Bach G. (2005). Mucolipin 1, endocytosis and cation channel-a review. Pflugers Arch. 451, 313-317 [DOI] [PubMed] [Google Scholar]
- Bargal R., Avidan N., Olender T., Ben Asher E., Zeigler M., Raas-Rothschild A., Frumkin A., Ben-Yoseph O., Friedlender Y., Lancet D., et al. (2001). Mucolipidosis type IV: Novel MCOLN1 mutations in Jewish and non-Jewish patients and the frequency of the disease in the Ashkenazi Jewish population. Hum. Mutat. 17, 397-402 [DOI] [PubMed] [Google Scholar]
- Cantiello H. F., Montalbetti N., Goldmann W. H., Raychowdhury M. K., Gonzalez-Perrett S., Timpanaro G. A., Chasan B. (2005). Cation channel activity of mucolipin-1: the effect of calcium. Pflugers Arch. 451, 304-312 [DOI] [PubMed] [Google Scholar]
- Cuajungco M. P., Samie M. A. (2008). The varitint-waddler mouse phenotypes and the TRPML3 ion channel mutation: cause and consequence. Pflugers Arch. Eur. J. Physiol. 457, 463-473 [DOI] [PubMed] [Google Scholar]
- Cuervo A. M. (2004a). Autophagy: in sickness and in health. Trends Cell Biol. 14, 70-77 [DOI] [PubMed] [Google Scholar]
- Cuervo A. M. (2004b). Autophagy: many paths to the same end. Mol. Cell. Biochem. 263, 55-72 [DOI] [PubMed] [Google Scholar]
- Curcio-Morelli C., Zhang P., Venugopal B., Charles F. A., Browning M. F., Cantiello H. F., Slaugenhaupt S. A. (2010). Functional multimerization of mucolipin channel proteins. J. Cell Physiol. 222, 328-335 [DOI] [PubMed] [Google Scholar]
- Di Palma F., Belyantseva I. A., Kim H. J., Vogt T. F., Kachar B., Noben-Trauth K. (2002). Mutations in Mcoln3 associated with deafness and pigmentation defects in varitint-waddler (Va) mice. Proc. Natl. Acad. Sci. USA 99, 14994-14999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. P., Cheng X., Mills E., Delling M., Wang F., Kurz T., Xu H. (2008). The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature 455, 992-996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. P., Wang X., Shen D., Chen S., Liu M., Wang Y., Mills E., Cheng X., Delling M., Xu H. (2009). Activating mutations of the TRPML1 channel revealed by proline-scanning mutagenesis. J. Biol. Chem. 284, 32040-32052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C., Cuajungco M. P., van Aken A. F., Schnee M., Jors S., Kros C. J., Ricci A. J., Heller S. (2007). A helix-breaking mutation in TRPML3 leads to constitutive activity underlying deafness in the varitint-waddler mouse. Proc. Natl. Acad. Sci. USA 104, 19583-19588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C., Jors S., Heller S. (2009). Life and death of sensory hair cells expressing constitutively active TRPML3. J. Biol. Chem. 284, 13823-13831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C., Jors S., Saldanha S. A., Obukhov A. G., Pan B., Oshima K., Cuajungco M. P., Chase P., Hodder P., Heller S. (2010). Small molecule activators of TRPML3. Chem. Biol. 17, 135-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51-59 [DOI] [PubMed] [Google Scholar]
- Karacsonyi C., Miguel A. S., Puertollano R. (2007). Mucolipin-2 localizes to the Arf6-associated pathway and regulates recycling of GPI-APs. Traffic 8, 1404-1414 [DOI] [PubMed] [Google Scholar]
- Kim H. J., Li Q., Tjon-Kon-Sang S., So I., Kiselyov K., Muallem S. (2007). Gain-of-function mutation in TRPML3 causes the mouse Varitint-Waddler phenotype. J. Biol. Chem. 282, 36138-36142 [DOI] [PubMed] [Google Scholar]
- Kim H. J., Li Q., Tjon-Kon-Sang S., So I., Kiselyov K., Soyombo A. A., Muallem S. (2008). A novel mode of TRPML3 regulation by extracytosolic pH absent in the varitint-waddler phenotype. EMBO J. 27, 1197-1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Soyombo A. A., Tjon-Kon-Sang S., So I., Muallem S. (2009). The Ca(2+) channel TRPML3 regulates membrane trafficking and autophagy. Traffic 10, 1157-1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselyov K., Chen J., Rbaibi Y., Oberdick D., Tjon-Kon-Sang S., Shcheynikov N., Muallem S., Soyombo A. (2005). TRP-ML1 is a lysosomal monovalent cation channel that undergoes proteolytic cleavage. J. Biol. Chem. 280, 43218-43223 [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Cuervo A. M., Seglen P. O. (2007). Methods for monitoring autophagy from yeast to human. Autophagy 3, 181-206 [DOI] [PubMed] [Google Scholar]
- LaPlante J. M., Sun M., Falardeau J., Dai D., Brown E. M., Slaugenhaupt S. A., Vassilev P. M. (2006). Lysosomal exocytosis is impaired in mucolipidosis type IV. Mol. Genet. Metab. 89, 339-348 [DOI] [PubMed] [Google Scholar]
- Lev S., Zeevi D. A., Frumkin A., Offen-Glasner V., Bach G., Minke B. (2010). Constitutive activity of the human TRPML2 channel induces cell degeneration. J. Biol. Chem. 285, 2771-2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoni M., Monti E., Bresciani R., Bozzato A., Barlati S., Bassi M. T., Borsani G. (2004). Overexpression of wild-type and mutant mucolipin proteins in mammalian cells: effects on the late endocytic compartment organization. FEBS Lett. 567, 219-224 [DOI] [PubMed] [Google Scholar]
- Martina J. A., Lelouvier B., Puertollano R. (2009). The calcium channel mucolipin-3 is a novel regulator of trafficking along the endosomal pathway. Traffic 10, 1143-1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedel M. T., Rbaibi Y., Guerriero C. J., Colletti G., Weixel K. M., Weisz O. A., Kiselyov K. (2008). Membrane traffic and turnover in TRP-ML1-deficient cells: a revised model for mucolipidosis type IV pathogenesis. J. Exp. Med. 205, 1477-1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K., Zheng L., Madathany T., Castiglioni A. J., Bartles J. R., Garcia-Anoveros J. (2008). The varitint-waddler (Va) deafness mutation in TRPML3 generates constitutive, inward rectifying currents and causes cell degeneration. Proc. Natl. Acad. Sci. USA 105, 353-358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper R. C., Luzio J. P. (2004). CUPpling calcium to lysosomal biogenesis. Trends Cell Biol. 14, 471-473 [DOI] [PubMed] [Google Scholar]
- Pryor P. R., Reimann F., Gribble F. M., Luzio J. P. (2006). Mucolipin-1 is a lysosomal membrane protein required for intracellular lactosylceramide traffic. Traffic 7, 1388-1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R., Kiselyov K. (2009). TRPMLs: in sickness and in health. Am. J. Physiol. Renal. Physiol. 296, F1245-F1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychowdhury M. K., Gonzalez-Perrett S., Montalbetti N., Timpanaro G. A., Chasan B., Goldmann W. H., Stahl S., Cooney A., Goldin E., Cantiello H. F. (2004). Molecular pathophysiology of mucolipidosis type IV: pH dysregulation of the mucolipin-1 cation channel. Hum. Mol. Genet. 13, 617-627 [DOI] [PubMed] [Google Scholar]
- Samie M. A., Grimm C., Evans J. A., Curcio-Morelli C., Heller S., Slaugenhaupt S. A., Cuajungco M. P. (2009). The tissue-specific expression of TRPML2 (MCOLN-2) gene is influenced by the presence of TRPML1. Pflugers Arch. 459, 79-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyombo A. A., Tjon-Kon-Sang S., Rbaibi Y., Bashllari E., Bisceglia J., Muallem S., Kiselyov K. (2006). TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. J. Biol. Chem. 281, 7294-7301 [DOI] [PubMed] [Google Scholar]
- Sun M., Goldin E., Stahl S., Falardeau J. L., Kennedy J. C., Acierno J. S., Jr, Bove C., Kaneski C. R., Nagle J., Bromley M. C., et al. (2000). Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet. 9, 2471-2478 [DOI] [PubMed] [Google Scholar]
- Treusch S., Knuth S., Slaugenhaupt S. A., Goldin E., Grant B. D., Fares H. (2004). Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc. Natl. Acad. Sci. USA 101, 4483-4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P., Thyagarajan B., Rohacs T., Balla T. (2006). Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J. Cell Biol. 175, 377-382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam K., Montell C. (2007). TRP channels. Annu. Rev. Biochem. 76, 387-417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam K., Hofmann T., Montell C. (2006). Lysosomal localization of TRPML3 depends on TRPML2 and the mucolipidosis-associated protein TRPML1. J. Biol. Chem. 281, 17517-17527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam K., Long A. A., Elsaesser R., Nikolaeva D., Broadie K., Montell C. (2008). Motor deficit in a drosophila model of mucolipidosis type IV due to defective clearance of apoptotic cells. Cell 135, 838-851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal B., Browning M. F., Curcio-Morelli C., Varro A., Michaud N., Nanthakumar N., Walkley S. U., Pickel J., Slaugenhaupt S. A. (2007). Neurologic, gastric, and opthalmologic pathologies in a murine model of Mucolipidosis type IV. Am. J. Hum. Genet. 81, 1070-1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal B., Mesires N. T., Kennedy J. C., Curcio-Morelli C., LaPlante J. M., Dice J. F., Slaugenhaupt S. A. (2009). Chaperone-mediated autophagy is defective in mucolipidosis Type IV. J. Cell. Physiol. 219, 344-353 [DOI] [PubMed] [Google Scholar]
- Vergarajauregui S., Puertollano R. (2006). Two di-leucine motifs regulate trafficking of mucolipin-1 to lysosomes. Traffic 7, 337-353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergarajauregui S., Connelly P. S., Daniels M. P., Puertollano R. (2008). Autophagic dysfunction in mucolipidosis type IV patients. Hum. Mol. Genet. 17, 2723-2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Delling M., Li L., Dong X., Clapham D. E. (2007). Activating mutation in a mucolipin transient receptor potential channel leads to melanocyte loss in varitint-waddler mice. Proc. Natl. Acad. Sci. USA 104, 18321-18326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J. (2005). Autophagy: molecular machinery for self-eating. Cell Death Differ. 12, 1542-1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi D. A., Frumkin A., Bach G. (2007). TRPML and lysosomal function. Biochim. Biophys. Acta 1772, 851-858 [DOI] [PubMed] [Google Scholar]
- Zeevi D. A., Frumkin A., Offen-Glasner V., Kogot-Levin A., Bach G. (2009). A potentially dynamic lysosomal role for the endogenous TRPML proteins. J. Pathol. 219, 153-162 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.