Abstract
We have identified and characterized an Arabidopsis thaliana rad50 mutant plant containing a T-DNA insertion in the AtRAD50 gene and showing both meiotic and DNA repair defects. We report here that rad50/rad50 mutant cells show a progressive shortening of telomeric DNA relative to heterozygous rad50/RAD50 controls and that the mutant cell population rapidly enters a crisis, with the majority of the cells dying. Surviving rad50 mutant cells have longer telomeres than wild-type cells, indicating the existence in plants of an alternative RAD50-independent mechanism for telomere maintenance. These results report the role of a protein essential for double-strand break repair in telomere maintenance in higher eukaryotes.
Keywords: double-strand break repair, recombination, senescence
Telomeres are specialized structures at the ends of chromosomes essential for the avoidance of fusion, recombination, and degradation of these ends. They are composed of short G-rich, tandemly repeated sequences, conforming to the consensus sequence Tx(A)Gy. The total length of these telomeric repeat tracts can vary in different cells or tissues of an organism (1–3), with the subject of this report, Arabidopsis thaliana, having 2–4-kb-long repeats of the sequence (TTTAGGG)n (4). Telomeric repeats are elongated by telomerase, a ribonucleoprotein polymerase (5, 6), the RNA component of which provides the template for the synthesis of de novo telomeric repeats on chromosome ends in vitro and in vivo. The catalytic component of telomerase is a reverse transcriptase, named TERT (telomerase reverse transcriptase) (7, 8).
The expression of telomerase is developmentally regulated in mammals, being largely restricted to the germ line with undetectable or very low levels of expression in most somatic cells (9). Normal human somatic cells cultured in vitro show shortening of telomeres as they divide, and these cells senesce after a number of divisions (10–13). However, most tumor or virus-transformed cell lines with unlimited proliferation capacity present high telomerase activity and stable telomere length (14, 15). Even so, about 10% of human tumors lack telomerase activity and possess long telomeres, suggesting the activation of telomerase-independent mechanisms of telomere-length maintenance (16–19). Telomerase activity is also developmentally regulated in plant cells. Telomerase activity is not detectable in vegetative tissues but is highly expressed in reproductive tissues, as well as in plant tissue culture cells (3, 20–25).
In addition to telomerase, several additional proteins have been identified in yeast as being required for telomere replication, such as the Cdc13 protein that binds the single-stranded portion of telomere DNA (26), as well as proteins involved in DNA double-strand break (DSB) repair (27, 28). DNA ends are present in cells as result of breaks induced by irradiation or other types of DNA-damaging agents or by replication/recombination processes. Cells have developed pathways to assure maintenance of the genome by sensing and repairing double-strand breaks. Two main pathways operate in yeast and higher eukaryotic cells to repair double-strand breaks: homologous recombination and nonhomologous end joining, with the latter constituting the primary DNA double-strand break repair pathway in mammals and plants. Two protein complexes, the Ku heterodimer and the Rad50-Mre11-Xrs2 complex, are known to be required for nonhomologous end joining (29–36). In yeast cells, mutations in KU, RAD50, MRE11, or XRS2 genes give rise to shorter telomeres (37–41).
These and other genes have been classified genetically into three epistasis groups for telomere maintenance: the telomerase, Cdc13, and Ku groups. Cdc13 protects the telomeres against strand-specific degradation as loss of Cdc13 function results in rapid removal of the C-rich strand of the telomere. Telomere replication and end-protection appear to be distinct activities, given that a telomerase-CDC13 double mutant presents an exacerbated growth defect as compared with either single mutant strain (42). In a genetic screen designed to identify genes that act in parallel to CDC13 and/or telomerase, Nugent et al. identified KU80 and RAD50 (39). However, the double Ku and cdc13 mutation resulted in more strongly impaired growth than either single mutant, indicating that Cdc13 and Ku are two independent activities both required for full telomere function. The same genetic screen has shown that mutations in RAD50 or MRE11 do not exacerbate a telomerase defect but do increase the growth defect observed in Ku and cdc13 mutated strains. These genetic data indicate that the Rad50-Mre11-Xrs2 complex is needed for telomere replication, and the authors propose that the Rad50-Mre11-Xrs2 complex is required to prepare or present DNA ends to telomerase for further replication. This epistatic relationship between telomerase and Rad50 is not perfect, however, as yeast double tlc1 rad50 mutants have delayed senescence relative to tcl1 mutants (43), as do double mutants of tel1 (believed to act in telomere maintenance via its action on the Rad50-Mre11-Xrs2 complex) and tlc1 (44).
The question of whether Ku and the Rad50-Mre11-Xrs2 complexes are essential for telomere maintenance in higher eukaryotes remains open. The mammalian Ku protein has been shown in vivo and in vitro to be associated with telomeres, suggesting a role of Ku in mammalian cells as a telomere end factor (45, 46). However, no information concerning the roles of other proteins involved in nonhomologous end joining in telomere maintenance in higher eukaryotes is available.
We have isolated and characterized the Arabidopsis thaliana homologue of the RAD50 gene and a rad50 mutant plant (47). The mutant plant is sterile, showing a clear meiotic defect (unpublished observations), and in vitro callus cultures derived from rad50/rad50 plants show a clear sensitivity to the DNA-damaging agent methylmethane sulfonate, suggesting a role in DNA repair of RAD50 in plant cells (47).
Here, we have addressed the possible role of Rad50 in the maintenance of telomeres in plant cells. Homozygous rad50 mutant cells exhibit shortening of telomeres and cell senescence. These cells express telomerase activity as shown in vitro by the telomere repeat amplification protocol (TRAP) assay, suggesting that Rad50 is not needed for expression of the telomerase activity itself but rather for its in vivo action on chromosome ends. A fraction of cells survive long periods of culture and these present longer telomeres, suggesting the presence in plant cells of an alternative, Rad50-independent mechanism for telomere maintenance, perhaps analogous to that seen in yeast telomerase mutants.
Materials and Methods
Plant Growth and Callus Induction.
A. thaliana plants (ws ecotype) were grown in a greenhouse under standard conditions. Seedlings carrying the rad50 insertional mutation were selected on Murashige and Skoog medium containing 50 mg/liter kanamycin (48). Homozygous and heterozygous plants for the RAD50 allele were distinguished by phenotypic and PCR analysis.
In vitro callus cultures were derived from leaves of young homozygous and heterozygous plants as follows (J. Lucht, personal communication). Leaves, surface sterilized in 0.5% sodium hypochlorite, were placed on callus induction medium (CIM) agar [Gamborg's B-5 (Sigma), 30 g/liter sucrose/1 mg/liter 2,4-dichlorophenoxyacetic acid/0.2 mg/ml kinetin/Bacto-agar (Difco)], 0.8% wt/vol, for 1 week at 22°C and 16 h light/8 h dark. They were then transferred to shoot induction medium (SIM) agar (Gamborg's B-5, 30 g/liter sucrose/0.1 mg/liter naphthalene acetic acid/1 mg/liter 6-benzyl amino purine), and after 2–3 weeks, green callus was transferred to fresh SIM (solid or liquid) and maintained in this medium by regular subculture.
DNA Isolation and Southern Analysis.
DNA was prepared from plant tissues or callus as follows. After grinding in liquid nitrogen, 1 g of powdered frozen tissue or cells was resuspended in 2.5 ml of extraction buffer (0.3 M NaCl/50 mM Tris, pH 7.5/20 mM EDTA/2% sarkosyl/0.5% SDS/5 M urea/5% phenol), mixed with 2.5 ml of phenol/chloroform (1:1). The aqueous phase was recovered after centrifugation at 5,000 × g for 15 min at 20°C. DNA was precipitated with 0.7 volume isopropanol, and the resulting pellet was washed with ethanol and resuspended in 10 mM Tris-Cl/2.5 mM EDTA, pH 8 (TE), plus RNaseA (10 mg/liter) incubated 25 min at 37°C, phenol extracted, ethanol precipitated, and dissolved in TE. DNA (1 μg) was digested with 20 units of restriction enzyme in a final volume of 100 μl for 16 h following the manufacturer's recommendations. Digested DNA samples were phenol/chloroform extracted, ethanol precipitated, resuspended in TE, and electrophoresed in 0.8% agarose/TBE gels. Gels were blotted into a positively charged nylon membrane (Hybond N+, Amersham Pharmacia), which were hybridized in 0.5 M phosphate buffer/7% (wt/vol) SDS/1% (wt/vol) BSA at 62°C (49). Blots were washed with 0.5% SSC/0.1% SDS at 55°C. Subtelomeric probes were labeled with [α-32P]dCTP by using the Prime-it II kit (Stratagene) according to the manufacturer's instructions. Telomeric repeat probe [5′(TTTAGGG)6] was 5′ end labeled by using T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol, Amersham Pharmacia).
The subtelomeric probe was isolated by PCR using two oligonucleotide primers designed to amplify a putatively coding, genomic DNA region in the subtelomeric DNA sequence of Arabidopsis chromosome II (50). This chromosome end was chosen because of the availability of DNA sequence and the fact that this sequence clearly runs into the telomeric repeat DNA. The primers used had the following sequences: 5′-CTAAACTAGTTGTGTTCCCGTCTCTACT and 5′-GGTGGGCGACCTTGTGCTTGCCAAAGTC.
Preparation of Plant Extracts and Telomerase Assay Telomere Repeat Amplification Protocol.
Total protein extracts were prepared from plant callus tissue as described (20). Telomerase was detected by the TRAP assay (20). In this assay, a forward primer is mixed with the plant extracts in the presence of dNTPs. This primer is recognized by telomerase, and telomeric repeats (TTTAGGG) are synthesized onto its 3′ end by the enzyme's reverse transcriptase activity by using its RNA component as template. PCR is performed to increase the sensitivity of the assay by using as reverse primer the complement to the consensus plant telomere sequence, which ensures that amplification will occur only if an activity in the plant extract added TTTAGGG repeats. The oligos used were 5′-CACTATCGACTACGCGATCGG as substrate for telomerase activity and 5′-CCCTAAACCCTAAAACCCTAAA added for PCR amplification. Addition of T4 gene 32 single-stranded DNA-binding protein was not necessary.
Results
The rad50 Mutation Leads to Telomere Shortening in Plant Cells.
We have identified and characterized an A. thaliana rad50 mutant plant, caused by the insertion of a T-DNA into the AtRAD50 gene. Plants homozygous for the T-DNA insertion present a sterility phenotype, and callus cultures derived from these plants are methylmethane sulfonate hypersensitive, suggesting a role in plant cells for this protein in meiosis and DNA DSB repair (47). In Saccharomyces cerevisiae, in addition to its roles in meiosis and DSB repair, Rad50 protein is needed for normal telomere maintenance. Thus, yeast rad50 mutants present telomere shortening (31). Given the conserved Rad50 function in meiosis and DSB repair between Saccharomyces and plants, we have analyzed whether mutation of the A. thaliana RAD50 gene affects telomere length. Telomere length was measured by Southern analysis, with the telomere repeat probe, of genomic DNA prepared from flowers of wild-type, rad50/RAD50 heterozygous, and rad50/rad50 homozygous plants. HinfI or MboI digested DNA prepared from wild-type and mutant plants showed the expected telomeric smear present in Arabidopsis between 3 and 6 kb (Fig. 1). The smaller bands correspond to previously described centromeric degenerate telomere motifs in the Arabidopsis genome (51). Telomeric DNA tracts of the same length were observed in rosette leaves (data not shown) and floral buds, whether the DNA was prepared from wild-type or rad50 mutant plants. These results agree with previous published data demonstrating the absence of telomere shortening with aging in Arabidopsis plants (52).
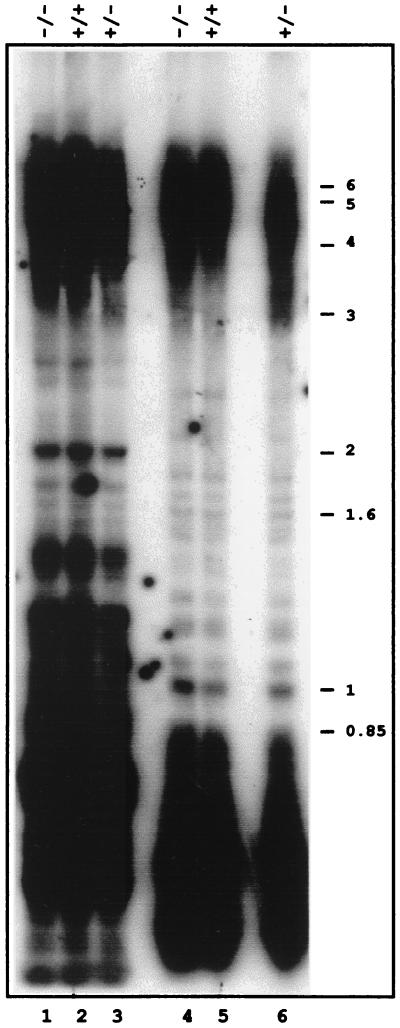
Figure 1.
Comparison of telomeres lengths in DNA prepared from flowers of wild-type and rad50 mutant plants. Southern analysis of flower DNA from wild-type (+/+), heterozygous (+/−), and homozygous (−/−) rad50 mutant plants digested with either MboI (lanes 1–3) or HinfI (lanes 4–6). Positions of molecular weight size markers (in kb) are indicated to the right.
It has been shown recently that an Arabidopsis telomerase mutant presents telomere shortening at a rate of approximately 500 bp per generation (52). We have not been able to observe this, as the Arabidopsis rad50 mutant is sterile and must be maintained in the heterozygous state (47). Thus, each mutant plant is a first mutant generation. The results presented in Fig. 1 correspond to the third heterozygous generation of plants carrying the rad50 mutation, showing that as for the meiosis, the telomere maintenance defect of the rad50 mutation is recessive at least up to three generations. Thus, to examine the requirement of Arabidopsis telomeres for the Rad50 protein, we generated actively dividing in vitro callus cultures from leaf tissue of rad50/rad50 homozygous and RAD50/rad50 heterozygous plants.
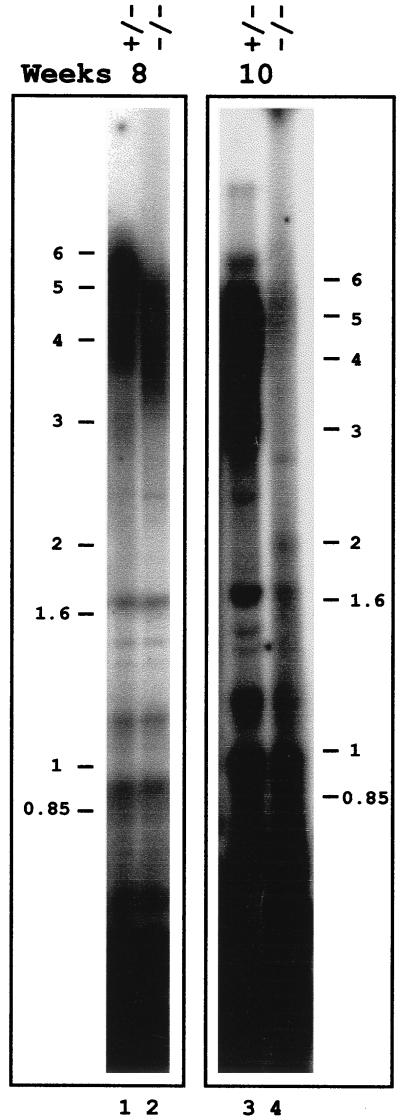
Callus cells induced on solid medium were transferred to and grown in liquid medium by regular subculture. DNA was prepared from rad50 heterozygous and homozygous mutant cells 8 weeks old. Southern analysis of MboI-digested genomic DNA by using the telomeric probe showed shortening of the telomeres in the rad50/rad50 cells (Fig. 2, lane 2) as compared with the heterozygous cells (Fig. 2, lane 1). Further reduction in telomere size was observed with DNA prepared from rad50/rad50 cells after 10 weeks of growth (Fig. 2, lane 4). In fact, for an equivalent amount of DNA (as defined from the centromeric hybridization of the probe corresponding to the bottom part of the gel), very little telomeric DNA is detected in DNA from the homozygous cells (lane 4) as compared with the heterozygous cells (lane 3). These results demonstrate that the RAD50 gene is needed for telomere maintenance in plant cells.
Figure 2.
Telomere shortening in homozygous rad50/rad50 mutant cells. DNA was prepared from calli generated from leaves heterozygous (+/−) or homozygous (−/−) for the mutant rad50 allele. DNA was prepared from cells grown in liquid culture for 8 (lanes 1 and 2) or 10 (lanes 3 and 4) weeks. MboI-digested DNA was analyzed by Southern analysis by using the telomeric repeat probe. Positions of molecular weight size markers (in kb) are indicated at the side of each panel (note that the two panels are from two different gels).
rad50 Mutant Cells Display a Senescent Phenotype and Survivors Present Longer Telomeres as Compared with Wild-Type Cells.
With prolonged growth we observed an increase in the frequency of cell death in cultures of the rad50/rad50 homozygous as compared with the heterozygous cells. This phenomenon is most easily observed after growth of the rad50 mutant cells on solid medium (Fig. 3), and we attribute it to the dramatic loss of telomeric DNA observed in the homozygous mutant cells. Sectors of growing cells appeared on the dying rad50 mutant callus (Fig. 3, arrows).
Figure 3.
Mutant rad50/rad50 cells present a senescent phenotype. rad50 homozygous (Upper) and rad50 heterozygous (Lower) cells of the same age grown on nutrient agar. The rad50 homozygous cells are dying, whereas the heterozygous cells are alive. Sectors of surviving homozygous cells are shown with arrows.
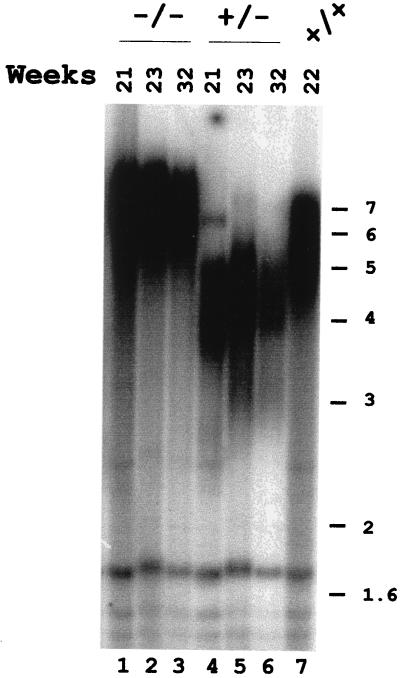
To further investigate the relationship between the decline in cell viability and shortening of the telomeres, we assayed samples of MboI-digested genomic DNA from surviving rad50 homozygous and heterozygous cells after 21, 23, and 32 weeks of growth by Southern analysis. Homozygous rad50 cells grown for 21 weeks present longer telomeres (Fig. 4, lane 1) as compared with the wild-type and heterozygous controls (Fig. 4, lanes 4–7). These longer telomeres are maintained in mutant cells that have been growing up to at least 32 weeks (Fig. 4, lanes 2 and 3). The appearance of longer telomeres in rad50 mutant cells surviving long periods of growth implies the existence in plant cells of a RAD50-independent mechanism for telomere maintenance.
Figure 4.
Surviving rad50/rad50 mutant cells present longer telomeres. Southern analysis of MboI-digested DNA using the telomeric repeat probe. DNA was prepared from rad50 heterozygous (+/−) and homozygous (−/−) mutant cells as well as wild-type (+/+) cells after different periods in culture. Positions of molecular weight size markers (in kb) are indicated to the right.
Interestingly, after longer periods of growth in culture, the heterozygote cells also show attrition of telomeric DNA (Fig. 4, lanes 4–6) compared with wild-type cells in culture (Fig. 4, lane 7). Although this telomere shortening is significantly less severe than that observed in the homozygous cells, these results indicate that the rad50 mutation is not fully recessive for telomere maintenance in Arabidopsis cells.
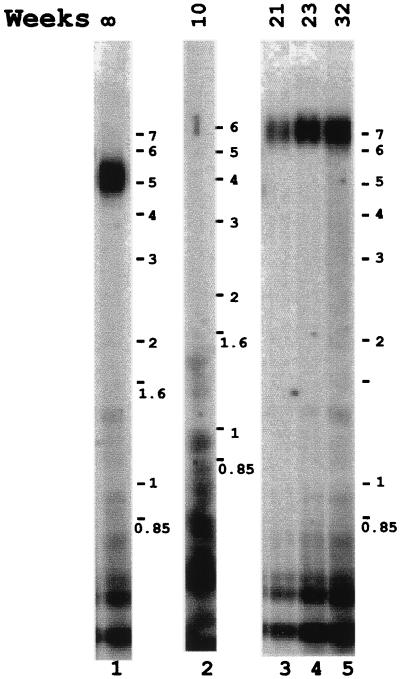
One possible mechanism to explain the appearance of longer telomeres in the surviving cells is that the telomerase becomes able to add new telomere sequences to the existing (shortened) telomeric repeats. In this case, loss of chromosome end sequence would not extend beyond the telomeric repeats. To test this hypothesis, we prepared a subtelomeric probe. This probe is a DNA fragment present 2.5 kb proximal to the start of the telomeric repeats on the long arm of chromosome II. MboI-digested DNA from rad50/rad50 homozygous cells 8, 10, 21, 23, and 32 weeks old was analyzed by Southern analysis by using this subtelomeric probe. DNA from 8-week-old cell culture presents a broad band that corresponds in size to the telomeric length (Fig. 5, lane 1). However, as described above for the telomeric repeat probe, no band was detected with DNA prepared from 10-week-old cells (Fig. 5, lane 2). That this absence of subtelomeric DNA is not because of a generalized chromosomal fragmentation and degradation is shown by the continued presence of the centromeric telomere DNA repeats in this DNA (Fig. 2, lane 4) and has been confirmed by reprobing this blot with an internal chromosome II probe 6.1 Mb from the nearest end (data not shown). These results indicate that after 10 weeks of growth, chromosome II has been degraded at least 2.5 kb proximal of the telomeric repeat sequences in the majority of mutant cells. However, as shown in Fig. 5 (lanes 3–5), the chromosome II present in cells grown for longer periods (at least up to 32 weeks) has retained its subtelomeric sequences. Thus, the longer telomeres seen after extended growth in culture of rad50 mutant cells represent the repopulation of the senescent culture with cells that had never fully degraded their telomeres, at least in the case of the chromosome II arm examined here. These surviving cells thus initially represent a sufficiently small fraction of the population so as not to have been visible in the earlier (10 week) time point. This surviving subpopulation is also seen as green, growing sectors on the senescent rad50 mutant callus show in Fig. 3 (arrow).
Figure 5.
Dynamics of the chromosome II subtelomeric region of rad50/rad50 mutant cells with culture time. MboI-digested DNA was prepared from rad50/rad50 mutant cells grown for 8, 10, 21, 23, and 32 weeks after initiation of cell culture. The Southern blot was probed with the subtelomeric region present in chromosome II. Position of marker sizes are shown to the right of each panel (note that lanes 1, 3, 4, and 5 are from the same gel and that lane 2 is from a reprobing of the filter shown in Fig. 2, lane 4).
Telomerase Is Expressed in rad50 Mutant Cells.
By analogy with the situation in yeast, we suppose that Rad50 protein is needed for normal telomerase function in Arabidopsis. To test whether the telomere loss described above was caused by an absence of telomerase activity in the mutant, we used the TRAP assay to measure telomerase activity in vitro in total protein extracts of mutant and heterozygous cells. Results of the TRAP assay are presented in Fig. 6. Telomere elongation is undetected in the absence of plant protein extracts (Lanes 1, 4, and 7), nor in the presence of RNaseA-treated extracts (lanes 5 and 6). The expected pattern of bands with seven-nucleotide periodicity is observed in extracts prepared from both homozygous and heterozygous cells (lanes 2, 3, 8, and 9). RNase treatment after the telomere extension step has no effect on the PCR reaction (lanes 8 and 9). Thus, extracts from both types of cells contain similar levels of telomerase activity, indicating that Rad50 protein is not needed for telomerase activation associated with cellular dedifferentiation on the induction of growth in cell culture.
Figure 6.
Telomerase activity in rad50 mutant cells. Protein extracts were prepared from heterozygous (+/−) and homozygous (−/−) rad50 mutant cells. Telomerase activity was detected by the TRAP assay as described in Materials and Methods. Products were separated on a 10% sequencing gel to reveal the periodic band profile. The TRAP assay was realized with extracts prepared from heterozygous (lanes, 3, 6, and 9) and homozygous (lanes 2, 5, and 8) rad50 mutant cells as indicated at the top of the figure. Lanes 1, 4, and 7 are the no-extract controls. Lanes 1–3 are from the TRAP assay under standard conditions. For, lanes 4–6, cell extracts were treated with RNaseA before the telomerase step. For lanes 7–9, samples were treated with RNaseA after the telomerase step and before the PCR step.
Discussion
The Rad50-Mre11-Xrs2 complex has been shown in yeast cells to be essential to the induction and processing of double-strand breaks in meiosis. This complex also plays an important role in DNA double-strand break repair and recombination. Finally, these proteins have been shown to be essential for telomere maintenance (28). We have recently cloned the Arabidopsis RAD50 gene and characterized a rad50 mutant plant. rad50 mutant plants are sterile and callus derived from them is hypersensitive to methyl methanesulfonate, suggesting a role of this protein in meiosis and DSB repair in plant cells (47). These observations indicate a strong conservation of RAD50 function between plants and yeast. Here, we show that Rad50 is involved in telomere maintenance in plants. As mammalian rad50 mutants are inviable at the cellular level, the Rad50 protein has thus been very little studied in vivo in metazoans (see ref. 53). We thus report the involvement of Rad50 in telomere maintenance in a multicellular organism.
We have analyzed whether the observed role of Rad50 in telomere maintenance in yeast is conserved in higher eukaryotes. As expected, Southern analysis showed the Arabidopsis telomeres as a smear of DNA between 3 and 6 kb, representing variations in telomere length between different chromosomes (4). We did not detect any difference in telomere sizes between leaves and flowers or after callus induction from plants heterozygous for the rad50 mutation. These results are in agreement with observations in Nicotiana tabacum (25) and Melandrium album (23), where no differences in telomere dynamics were observed on induction of callus cultures and contrast with the telomere lengthening observed in barley calli (24). In contrast, callus from plants homozygous for the rad50 mutation present progressive shortening of the telomeres and the cells senesce more rapidly than the controls. Shortening of telomeres is also seen in S. cerevisiae rad50 null mutants (37–41, 54). Furthermore, yeast rad50 mutants grow slowly (55, 56) and show reduced plating efficiencies (41, 54). The actively growing Arabidopsis liquid suspension cultures described here are diluted 5-fold at each weekly subculture, which would give approximately two to three cell population doublings per week. However, this estimation of the number of divisions preceding the detection of shortened telomeres in the Arabidopsis cell culture is made difficult and extremely imprecise by the “clumpy” nature of these cultures and the fact that as the population becomes senescent, the proportion of actively dividing cells in the population is changing. For this reason, we limit ourselves to a comparative analysis of the behavior of these mutant cell cultures with respect to the control, non-mutant cells.
The telomere shortening observed in yeast and Arabidopsis rad50 mutants could be the result of an indirect effect associated with the failure of these mutants to repair damaged DNA. However, the demonstration that mre11 and rad50 yeast mutants lie in the same epistasis group as the telomerase mutant for telomere length (39) argues against this hypothesis. At present, no evidence of direct interaction of Rad50 with the telomeres and/or the telomerase is available. We argue that if RAD50 function is required in Arabidopsis cells (e.g., preparing chromosome DNA ends for telomerase action) and not for the telomerase function in itself, we should be able to detect telomerase activity in rad50− protein extracts in vitro. Results presented in Fig. 6 support this hypothesis, as we detected similar levels of in vitro telomerase activity in extracts prepared from rad50 heterozygous or homozygous cells. These results also indicate that although Rad50 protein is essential for normal telomere maintenance, it is not necessary for the telomerase up-regulation accompanying cellular dedifferentiation upon culture. Analysis of the phenotype of a rad50-telomerase double mutant in Arabidopsis will be needed to confirm the direct role of RAD50 in telomere maintenance in Arabidopsis.
We observed that the rad50 mutant cell population passes through a crisis from which only a fraction of cells survive. These survivors present longer telomeres as compared with wild-type cells. This is the first observation of the generation of longer telomeres in rad50 survivors, implying the existence in plant cells of a Rad50-independent mechanism for telomere maintenance. This is reminiscent of the longer telomeres seen in yeast telomerase mutant survivors (57) and of the exceptionally long telomeres observed in human tumors (58) or cultured human cells that lack telomerase (17). Interestingly, yeast rad50 mutants have stable, short telomeres in contrast to the Arabidopsis rad50 mutant described here.
Two patterns of telomeric DNA are detected in yeast telomerase mutant survivors. The more common type I survivors correspond to tandem amplification of the subtelomeric Y′ region followed by short telomeric tracts of C1–3A/TG1–3 DNA. In type II survivors, chromosomes were shown to end in very long telomeric tracts of C1–3A/TG1–3 DNA, heterogeneous in length. Upon further growth, most type I survivors convert to type II, which are then maintained for at least 250 cell divisions (59). These survivors are dependent on RAD52-mediated yeast recombination (57, 60). Interestingly, telomeres of type II survivors returned to wild-type lengths when telomerase was reintroduced (59). Whether the longer telomeres generated in mammalian cells lacking telomerase activity or in rad50 mutant plants are generated by recombination remains an open question. Analysis of individual chromosome ends by using specific subtelomeric sequences should address whether the longer telomeres detected in Arabidopsis rad50 mutant survivors contain longer repetitions of the telomeric tract. We observed that most rad50/rad50 mutant cells lose the subtelomeric sequence present on (at least) chromosome II (Fig. 5, lane 2). Survivors presenting longer telomeres have retained the subtelomeric sequence in chromosome II (Fig. 5, lanes 3–5), arguing for the growing out of a small subpopulation of cells that had not lost all of their telomeric DNA before developing long telomeres. Two mechanisms could be envisaged to explain the appearance of longer telomeres: (i) reduction of telomere length beyond a critical size permits the telomerase to function in the absence of the Rad50 protein, and (ii) as has been suggested for the yeast telomerase mutant, survivals result from recombination between telomeric sequences. Discrimination between these two hypotheses must await the generation and analysis of rad50-telomerase double-mutant plants.
Abbreviations
- DSB
double-strand break
- TRAP
telomere repeat amplification protocol
References
- 1.Zakian V A. Annu Rev Genet. 1996;30:141–172. doi: 10.1146/annurev.genet.30.1.141. [DOI] [PubMed] [Google Scholar]
- 2.Greider C W. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 3.McKnight T D, Fitzgerald M S, Shippen D E. Biochemistry. 1997;62:1224–1231. [PubMed] [Google Scholar]
- 4.Richards E J, Ausubel F M. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- 5.Greider C W, Blackburn E H. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 6.Greider C W, Blackburn E H. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 8.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 9.Wright W E, Piatyszek M A, Rainey W E, Byrd W, Shay J W. Dev Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 10.Harley C B, Futcher A B, Greider C W. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 11.Hastie N D, Dempster M, Dunlop M G, Thompson A M, Green D K, Allshire R C. Nature (London) 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 12.de Lange T, Shiue L, Myers R M, Cox D R, Naylor S L, Killery A M, Varmus H E. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindsey J, McGill N I, Lindsey L A, Green D K, Cooke H J. Mutat Res. 1991;256:45–48. doi: 10.1016/0921-8734(91)90032-7. [DOI] [PubMed] [Google Scholar]
- 14.Counter C M, Avilion A A, LeFeuvre C E, Stewart N G, Greider C W, Harley C B, Bacchetti S. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Autexier C, Greider C W. Trends Biochem Sci. 1996;21:387–391. [PubMed] [Google Scholar]
- 16.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L, Coviello G M, Wright W E, Weinrich S L, Shay J W. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 17.Bryan T M, Englezou A, Gupta J, Bacchetti S, Reddel R R. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murnane J P, Sabatier L, Marder B A, Morgan W F. EMBO J. 1994;13:4953–4962. doi: 10.1002/j.1460-2075.1994.tb06822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strahl C, Blackburn E H. Mol Cell Biol. 1996;16:53–65. doi: 10.1128/mcb.16.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzgerald M S, McKnight T D, Shippen D E. Proc Natl Acad Sci USA. 1996;93:14422–14427. doi: 10.1073/pnas.93.25.14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heller K, Kilian A, Piatyszek M A, Kleinhofs A. Mol Gen Genet. 1996;252:342–345. doi: 10.1007/BF02173780. [DOI] [PubMed] [Google Scholar]
- 22.Fajkus J, Kovarik A, Kralovics R. FEBS Lett. 1996;391:307–309. doi: 10.1016/0014-5793(96)00757-0. [DOI] [PubMed] [Google Scholar]
- 23.Riha K, Fajkus J, Siroky J, Vyskot B. Plant Cell. 1998;10:1691–1698. doi: 10.1105/tpc.10.10.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilian A, Stiff C, Kleinhofs A. Proc Natl Acad Sci USA. 1995;92:9555–9559. doi: 10.1073/pnas.92.21.9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fajkus J, Fulneckova J, Hulanova M, Berkova K, Riha K, Matyasek R. Mol Gen Genet. 1998;260:470–474. doi: 10.1007/s004380050918. [DOI] [PubMed] [Google Scholar]
- 26.Lin J J, Zakian V A. Proc Natl Acad Sci USA. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shore D. Science. 1998;281:1818–1819. doi: 10.1126/science.281.5384.1818. [DOI] [PubMed] [Google Scholar]
- 28.Bertuch A, Lundblad V. Trends Cell Biol. 1998;8:339–342. doi: 10.1016/s0962-8924(98)01331-2. [DOI] [PubMed] [Google Scholar]
- 29.Fox M E, Smith G R. Prog Nucleic Acid Res. 1998;61:345–378. doi: 10.1016/s0079-6603(08)60831-4. [DOI] [PubMed] [Google Scholar]
- 30.Jeggo P A. Adv Genet. 1998;38:185–218. doi: 10.1016/s0065-2660(08)60144-3. [DOI] [PubMed] [Google Scholar]
- 31.Paques F, Haber J E. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrini J H, Bressan D A, Yao M S. Semin Immunol. 1997;9:181–188. doi: 10.1006/smim.1997.0067. [DOI] [PubMed] [Google Scholar]
- 33.Roeder G S. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 34.Smith G R. DNA Damage and Repair. I. Totowa, NJ: Humana; 1998. pp. 135–162. [Google Scholar]
- 35.Smith K N, Nicolas A. Curr Opin Genet Dev. 1998;8:200–211. doi: 10.1016/s0959-437x(98)80142-1. [DOI] [PubMed] [Google Scholar]
- 36.Tsukamoto Y, Ikeda H. Genes Cells. 1998;3:135–144. doi: 10.1046/j.1365-2443.1998.00180.x. [DOI] [PubMed] [Google Scholar]
- 37.Boulton S J, Jackson S P. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porter S E, Greenwell P W, Ritchie K B, Petes T D. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nugent C I, Bosco G, Ross L O, Evans S K, Salinger A P, Moore J K, Haber J E, Lundblad V. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- 40.Boulton S J, Jackson S P. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kironmai K M, Muniyappa K. Genes Cells. 1997;2:443–455. doi: 10.1046/j.1365-2443.1997.1330331.x. [DOI] [PubMed] [Google Scholar]
- 42.Nugent C I, Hughes T R, Lue N F, Lundblad V. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 43.Le S, Moore J K, Haber J E, Greider C W. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritchie K B, Mallory J C, Petes T D. Mol Cell Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bianchi A, de Lange T. J Biol Chem. 1999;274:21223–21227. doi: 10.1074/jbc.274.30.21223. [DOI] [PubMed] [Google Scholar]
- 46.Hsu H L, Gilley D, Blackburn E H, Chen D J. Proc Natl Acad Sci USA. 1999;96:12454–12458. doi: 10.1073/pnas.96.22.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gallego M E, Jeanneau M, Granier F, Bouchez D, Bechtold N, White C I. Plant J. 2001;25:1–13. doi: 10.1046/j.1365-313x.2001.00928.x. [DOI] [PubMed] [Google Scholar]
- 48.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 49.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin X, Kaul S, Rounsley S, Shea T P, Benito M I, Town C D, Fujii C Y, Mason T, Bowman C L, Barnstead M, et al. Nature (London) 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 51.Richards E J, Goodman H M, Ausubel F M. Nucleic Acids Res. 1991;19:3351–3357. doi: 10.1093/nar/19.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fitzgerald M S, Riha K, Gao F, Ren S, McKnight T D, Shippen D E. Proc Natl Acad Sci USA. 1999;96:14813–14818. doi: 10.1073/pnas.96.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo G B, Yao M S, Bender C F, Mills M, Bladl A R, Bradley A, Petrini J H J. Proc Natl Acad Sci USA. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchie K B, Petes T D. Genetics. 2000;155:475–479. doi: 10.1093/genetics/155.1.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alani E, Padmore R, Kleckner K. Cell. 1990;63:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 56.Haber J E. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 57.Lundblad V, Blackburn E H. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 58.Bryan T M, Englezou A, Dalla-Pozza L, Dunham M A, Reddel R R. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 59.Teng S C, Zakian V A. Mol Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kass-Eisler A, Greider C W. Trends Biochem Sci. 2000;25:200–204. doi: 10.1016/s0968-0004(00)01557-7. [DOI] [PubMed] [Google Scholar]