Abstract
Chromatin plays a central role in eukaryotic gene regulation. We have performed genome-wide mapping of epigenetically-marked nucleosomes to determine their position both near transcription start sites and at distal regulatory elements including enhancers. In prostate cancer cells where androgen receptor (AR) binds primarily to enhancers, we found that androgen treatment dismisses a central nucleosome present over AR binding sites that is flanked by a pair of marked nucleosomes. A novel quantitative model built on the behavior of such nucleosome pairs correctly identified regions bound by the regulators of the immediate androgen response including AR and FoxA1. More importantly this model also correctly predicted novel binding sites for other transcription factors present following prolonged androgen stimulation including Oct1 and NKX3.1. Thus quantitative modeling of enhancer structure provides a powerful predictive method to infer the identity of transcription factors involved in cellular responses to specific stimuli.
Transcription in eukaryotes is regulated by transcription factors that associate with the genome in a cell-type and condition-specific manner. Chromatin organization forms part of the basis for this cell-type specificity by allowing or denying transcription factor (TF) access to DNA. The basic units of chromatin structure are the nucleosomes, which are known to restrict the in vivo access of certain classes of transcription factors 1. Intensive work has been done to reveal the correlation between nucleosome position, histone modification and gene expression 2–4. Genome-wide nucleosome occupancy maps have been generated in S. cerevisiae 5–7, Drosophila 8 and C. elegans 9, but high quality human nucleosome occupancy data is more difficult to acquire due to the large size of the human genome 10,11. While the nucleosome positioning patterns are well established at transcription start sites (TSS), they are less well known at enhancers. Functional enhancers are cis-regulatory DNA elements that are orientation and position independent and can act at variable distances from the TSS of the genes they regulate 12,13. Monomethylated H3K4 (H3K4me) has been shown to be associated with transcription factor binding at enhancers, trimethylated H3K4 (H3K4me3) with the TSS, and dimethylated H3K4 (H3K4me2) with both the TSS and enhancers 14,15. To characterize the pattern of nucleosome positioning at enhancers, we used nucleosome-resolution ChIP-seq of H3K4me, H3K4me2 and H3k4me3 in the prostate cancer cell line LNCaP in response to a time-course of stimulation by the AR agonist 5α-dihydrotestoterone (DHT).
By comparing regions of enriched histone modification (Supplementary Table 1) to AR and FoxA1 binding sites (4h DHT) and the promoter regions of RefSeq genes we found H3K4me2 ChIP-seq to be the most efficient in identifying both promoters and putative enhancers (Supplementary Fig. 1). To differentiate intergenic TF binding sites from promoters, we removed H3K4me2 regions with strong H3K4me3 ChIP-seq signals from subsequent analyses.
We next examined whether H3K4me2 shows positional trends relative to the known AR and FoxA1 binding sites. H3K4me2 signal (based on average tag count) is highest near AR binding sites in the absence of DHT (Fig. 1a). Upon DHT stimulation and concomitant AR binding, H3K4me2 signal decreases at the binding sites and increases in the flanking regions (Fig. 1b). The same analysis relative to the binding sites of FoxA1 shows a bimodal tag count distribution both before (Fig. 1c) and after (Fig. 1d) DHT treatment, consistent with the role of FoxA1 as a pioneer factor to facilitate AR binding16.
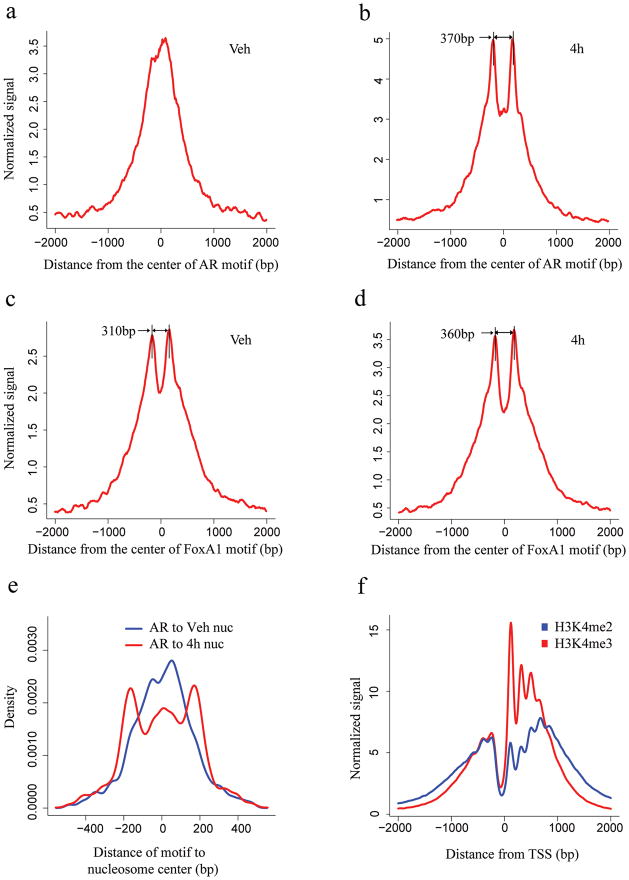
Figure 1. Signal distribution and nucleosome position analysis in the AR and FoxA1 binding regions identified by ChIP-chip experiment and the TSS.
H3K4me2 signal distribution relative to the center of the AR motif (a, b) and FoxA1 motif (c, d) in the binding regions. The x-axis represents the distance to the center of the best AR or FoxA1 motif match in a given binding site. The y-axis represents normalized ChIP-Seq tag count numbers. “Veh” represents the unstimulated condition; “4h” represents stimulated conditions with treatment of DHT for 4 hours. (e) Distance from the AR motif to the center of the nearest nucleosome in the AR binding sites under vehicle (red) and 4 hours after DHT stimulation (blue). (f) H3K4m2 and H3K4me3 signal distribution relative to the TSS.
To investigate whether nucleosome positioning could explain the pattern observed in the previous analysis, we determined the likely positions of H3K4me2 marked nucleosomes using the NPS algorithm (Supplementary Table 1) 17. The distance from the AR motif in the binding site to the nearest detected nucleosome increases to ~200bp upon DHT stimulation (Fig. 1e, Supplementary Fig. 2a), indicating that nucleosomes tend to be less occupied (destabilized) at the binding site itself and more occupied (stabilized) at adjacent nucleosomes. Interestingly, the locations of the most positioned nucleosomes are concordant between DHT and vehicle (Supplementary Fig. 2b). This suggests that prior to AR activation, AR binding loci are already marked with two well-positioned H3K4me2-containing nucleosomes, 250–450 bp apart, flanking the precise binding sites, along with a well-positioned nucleosome occluding the binding site itself. Upon AR activation, H3K4me2 modified nucleosomes are destabilized at the AR binding sites and are better positioned at the two flanking loci. While the chromatin structure relative to TSS is characterized by a nucleosome free region immediately upstream and a series of well positioned nucleosomes downstream (Fig. 1f), we found that in general only two well-positioned nucleosomes are evident at TF bound enhancers (Fig. 1b, c and d).
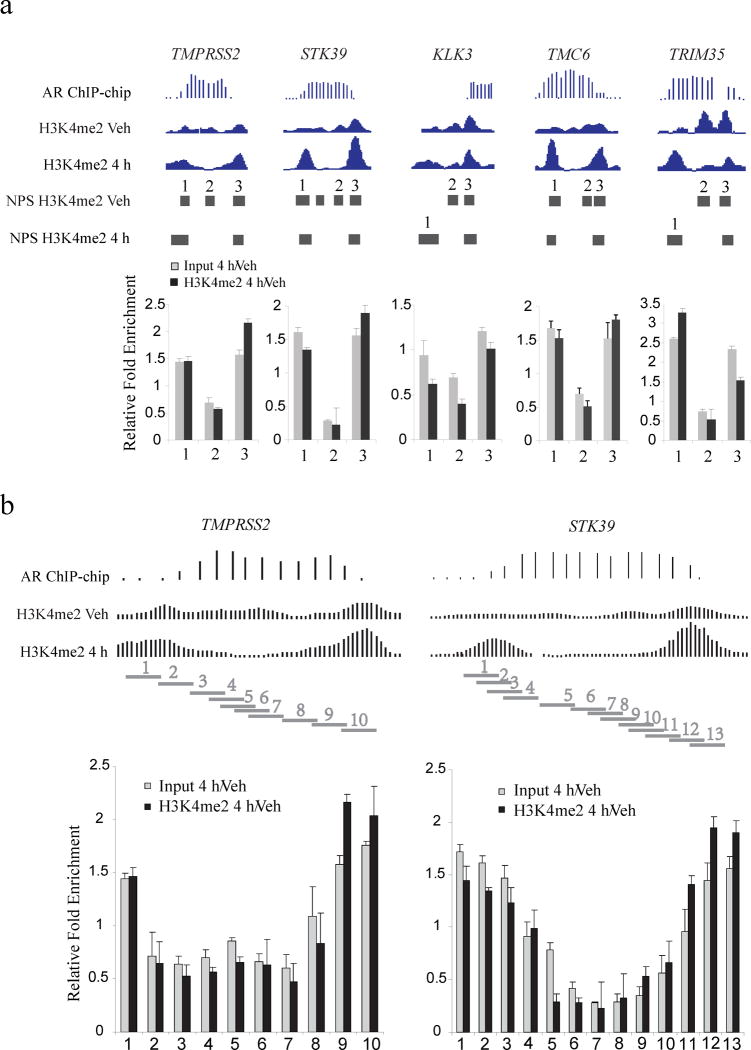
To validate our observations and rule out the possibility of ChIP-seq artifacts or loss of the H3K4me2 mark rather than the nucleosome, we conducted H3K4me2 ChIP-qPCR and Input DNA-qPCR on five AR binding sites that exhibited these H3K4me2 patterns near the genes TMPRSS2, STK39, KLK3, TMC6 and TRIM35. In all cases, the Input DNA-qPCR results show that overall nucleosome density decreases over the transcription factor binding sites and increases in the flanking regions to the same extent as the H3K4me2 marked nucleosomes (Fig. 2a). We examined two of these cases, TMPRSS2 and STK39, in greater detail using primers that tile regions of interest at a finer resolution (Fig. 2b), and obtained similar results.
Figure 2. qPCR validation of the nucleosomes stabilized-destabilized around AR binding sites.
(a) Five AR binding sites near the genes TMPRSS2, STK39, KLK3, TMC6 and TRIM35. “AR ChIP-chip” represents the AR ChIP-chip signals; “H3K4me2 Veh” and “H3K4me2 4h” represent H3K4me2 ChIP-Seq signals before and after 4 hours of DHT treatment. “Input 4h/Veh” represents the qPCR assay of nucleosome fold change for DHT treatment relative to vehicle; “H3K4me2 4h/Veh” represents the qPCR assay of fold change for H3K4me2 signal for DHT treatment relative to vehicle, standard deviation is shown. Each horizontal bar represents a NPS peak region. (b) Detailed qPCR analysis of the AR binding sites near the genes TMPRSS2 and TMC6. Each horizontal bar represents a qPCR amplification region.
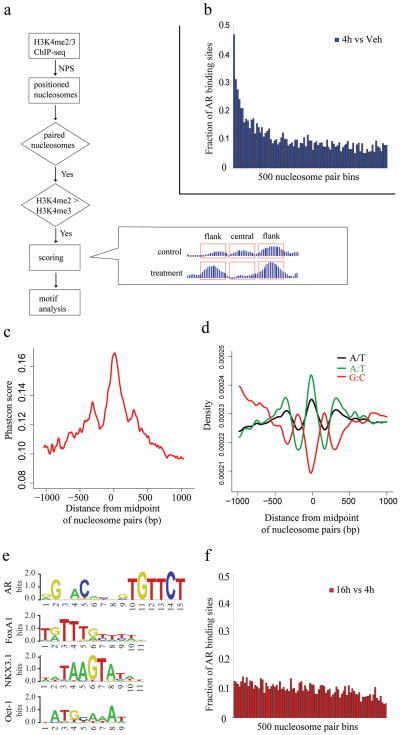
In order to determine whether the pattern of nucleosome positioning we observed at AR binding sites could be applied more broadly to other TFs, we used the change in H3K4me2 marked nucleosomes at AR binding sites following acute androgen stimulation to develop a general model of nucleosome positions at enhancers. We identified ~65,000 well-positioned nucleosome pairs (4h DHT) separated by the characteristic distance of 250–450bp in which promoter proximal pairs were removed on the basis of having H3K4me3 > H3K4me2 (Fig. 3a). From the analysis of the AR binding sites we developed a quantitative model based on the changes in the H3K4me2 signal in the flanking nucleosomes and in the region between them (Fig. 3a). Running the model results in a “nucleosome stabilization – destabilization” (NSD) score S (as defined in Fig. 3a and having a distribution as in Supplementary Fig. 3) for each pair of appropriately spaced nucleosomes. Indeed, when we ranked all the nucleosome pairs by NSD score and grouped them into bins of 500, we found that the top scoring bins show the highest enrichment in AR binding sites (Fig. 3b).
Figure 3. Motif analysis in the paired nucleosome regions.
(a) Flowchart of the prediction model. The formula for the NSD score is described in “Methods” section. “treatment” and “control” refer to treatment and vehicle control conditions, respectively. “flank” refers to the 200bp of sequence centered on each flanking nucleosome, and “central” refers to the sequence between these regions. (b) The fraction of AR binding sites in score ranked paired nucleosome bins with decreasing score (4h vs. Veh). Paired nucleosome regions are ranked by scores representing the differences in H3K4me2 tag counts before and after DHT treatment. These ranked regions are grouped into bins of 500. Represented here is the number of regions in each bin that overlap with AR ChIP-chip enriched regions. (c) Evolutionary conservation in the vicinity of the 5000 highest scoring nucleosome pairs. Mean PhastCons scores representing DNA sequence conservation over 17 species is plotted as a function of the distance from the midpoint between paired nucleosomes. (d) DNA sequence content associated with nucleosome positioning. The 5000 highest scoring paired nucleosome regions, aligned at the midpoint, were analyzed for simple DNA sequence features: the distribution of A/T mononucleotides (black), G:C dinucleotides (red) or A:T dinucleotides (green). (e) Logos of AR, FoxA1, NKX3.1 and Oct1 motifs from TRANSFAC library. (f) The fraction of AR binding sites in score ranked paired nucleosome bins with decreasing score (16h vs. 4h).
To further test the functional relevance of the regions identified by the model, we examined the evolutionary conservation across the 5,000 highest-scoring paired nucleosomes. We see three PhastCons conservation peaks, one major peak at the nucleosome depleted regions between the paired nucleosomes, and one flanking each of these nucleosomes (Fig. 3c), this suggests evolutionary pressure not only on the TF binding sites between the paired nucleosomes but also on the regions immediately outside the paired nucleosomes.
To investigate the nature of nucleosome depletion in the regions between the paired nucleosomes, we studied the DNA sequence features in these regions. We observe that, consistent with previous models 18,19, simple A/T content and AA/TT/TA/AT dinucleotides are depleted in nucleosome-enriched regions and enriched in nucleosome-depleted regions, while G:C dinucleotides show the opposite trend (Fig. 3d). In addition, the stabilization of nucleosomes flanking the TF binding sites supports a model in which binding of non-nucleosomal proteins such as transcription factors forms boundaries that direct the in vivo positioning of nearby nucleosomes 20. A recent study also suggests that H3.3/H2A.Z-containing nucleosomes are intrinsically labile, which facilitate TFs access at regulatory sites in vivo 21. We performed H2A.Z ChIP-qPCR at 5 representative AR binding sites (Supplementary Fig. 4). These results show that H2A.Z is indeed enriched at the central nucleosome compared with the two flanking ones for all the five sites tested suggesting that it may be more labile. Significantly, the mean positions of the paired nucleosomes at AR binding loci appear to be the same in both bound (DHT) and unbound (Vehicle) conditions (Fig. 2, Supplementary Fig. 2b) suggesting the existence of a common enhancer architecture that enables access of transcription factors to DNA.
To determine whether the model could be used to impute the identity of TFs that bind between nucleosome pairs, we searched for motifs in the 1000 top NSD scoring nucleosome paired regions. The top motifs identified are from the forkhead and steroid receptor families (Supplementary Table 2 and Fig. 3e, Supplementary Fig. 5a, 5b), previously shown to be responsible for the androgen response in prostate cancer 16. Interestingly, this approach predicted AR binding at several sites that were not previously identified by ChIP-chip, and all of a representative sample of these were validated by ChIP-qPCR (Fig. 4a, Supplementary Fig. 6a).
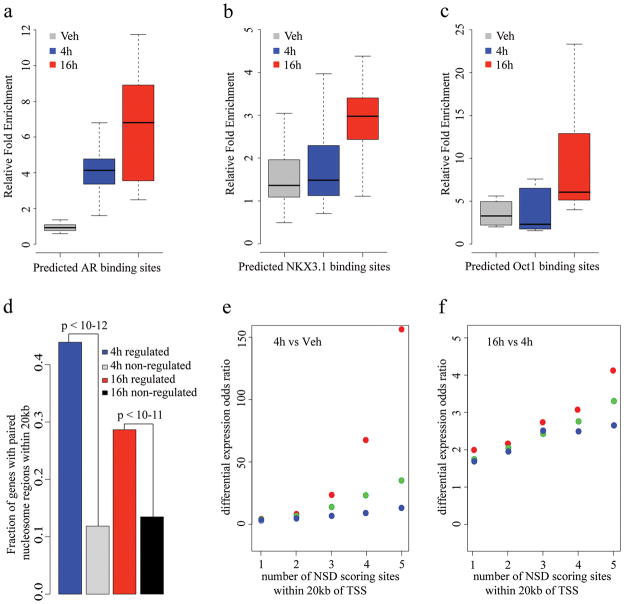
Figure 4. ChIP-qPCR and gene expression analysis of NSD scoring sites.
ChIP-qPCR validation of predicted (a)AR, (b)NKX3.1 and (c)Oct1 binding sites. Box plots were generated from ChIP-qPCR data obtained from three independent experiment testing 10 sites for AR, 22 sites for NKX3.1 and 9 sites forOct1. The individual ChIP-qPCR assays are shown in Supplementary Fig. 5. (d) Correlation of paired nucleosome regions with gene expression. The fraction of differentially regulated genes with paired nucleosome regions within 20 kb is shown. The top 5000 paired nucleosome regions were selected under the conditions of DHT 4 hours vs. vehicle and DHT 16 hours vs. DHT 4 hours. Differentially regulated genes were identified as described in the “Methods” section. “4hregulated” represents the fraction of DHT 4 hours vs. vehicle differentially regulated genes with at least one DHT 4 hour vs. vehicle paired nucleosome region within 20kb of the transcription start site. “4h non-regulated” represents the fraction of non-regulated genes under the same condition. “16h regulated” and “16h non-regulated” represent the fractions under the condition of DHT 16 hours vs. DHT 4 hours. Correlation of score and number of NSD scoring sites and up-regulated gene expression, (e) 4h vs Veh, (f) 16h vs 4h. X axis represents the lower bound n of the number of sites within 20kb of the TSS of a gene, Y axis represents the odds ratio calculated by the formula (up-regulated genes with at least n sites/non-regulated genes with at least n sites)/(all up-regulated genes/all non-regulated genes). Red, green and blue dots represent the top 5000, 10,000 and 20,000 NSD score sites, respectively.
If the model is valid more generally, it should identify key transcription factors regulating the response of a cell population to a stimulus using the H3K4me2/3 nucleosome-resolution ChIP-seq data alone. As the LNCaP response to 4 hours of androgen exposure was dominated by AR and FoxA1, we generated nucleosome-resolution H3K4me2 ChIP-seq data at 16 hours after DHT treatment, when we predicted secondary transcriptional responses would dominate. Applying our prediction model we found NKX3.1 and Oct1 motifs to be very significantly associated with high NSD scoring regions (Supplementary Table 2, p-value < 1e-7, Supplementary Fig. 5c, Fig. 5d), while the AR motif is not (Supplementary Table 2, Fig. 3f). NKX3.1 is a homeobox gene involved in normal prostate development that marks the prostate luminal epithelial stem cell and is a putative tumor suppressor gene in the prostate 22,23. Oct1 has been shown previously to collaborate with AR at a subset of AR binding loci in LNCaP cells 24. While Oct1 is constitutively expressed in LNCaP cells, NKX3.1 was induced 4-fold by androgens both at the 4h and 16h time point 25.
To test whether these factors were truly differentially bound, we selected sites with high NSD scores (16h vs. 4h) and central sequences closely resembling the TRANSFAC-defined NKX3.1 or Oct1 motifs (Fig. 3e). ChIP-qPCR was performed to compare binding under vehicle conditions to that after DHT stimulation (4h and 16h). DHT-dependent NKX3.1 recruitment was validated on 18 out of 22 selected regions, to a degree comparable with that of two known 26 NKX3.1 binding sites (Fig. 4b, Supplementary Fig. 6b). While previously identified Oct1 binding sites have been located in close proximity to AR binding sites 24, the model predicted a set of putative DHT responsive Oct1 binding sites that are independent of AR binding. ChIP-qPCR of these sites shows a strong response to DHT stimulation, where all nine sites have greater enrichment of Oct1 binding at 16 hours compared with 4 hours (Fig. 4c, Supplementary Fig. 6c).
We investigated whether the genomic regions identified with this model might be of regulatory importance. We defined “4 hour” sites and “16 hour” sites as the 5,000 regions with highest NSD scores after 4h DHT treatment versus vehicle, and 16h versus 4h DHT treatment, respectively. We then examined gene expression microarray assays at the vehicle, 4h, and 16h time points, and compared imputed differential binding with differential gene expression. At both 4h and 16h, the differentially expressed genes are more highly associated with imputed binding sites than non-differentially expressed genes (Fig. 4d). Further analysis shows that the likelihood of a gene being up-regulated increases with the number and score of paired nucleosome sites in the vicinity of the TSS (Fig. 4e and 4f). In contrast, when we examined the relationship between number and score of paired nucleosome sites for down-regulated genes, we found no correlation (Supplementary Fig. 7). These results suggest that the high NSD scoring sites are functional enhancers that play a functional role in gene regulation.
By performing ChIP-seq for H3K4me2 and H3K4me3, we profiled at high resolution the changes in nucleosome occupancy that occur at enhancers in a human prostate cancer cell line in response to androgen stimulation. Analysis of nucleosome occupancy near AR and FoxA1 binding sites revealed a striking pattern of nucleosome stabilization in a pair of nucleosomes flanking the binding site and elimination of a nucleosome at the site itself. We found several intrinsic characteristics of the middle nucleosome that distinguish it from the flanking ones, its sequence is more evolutionarily conserved and has a higher A/T content, while its histone octamer is more likely to contain the H2A.Z histone variant. This suggests that it may be intrinsically less stable than the flanking nucleosomes. Thus the apparent differences in nucleosome stability may be the result of the combination of DNA sequence, histone octamer composition and transcription factor binding. We developed a novel quantitative model and scoring function, the NSD score, that correctly identified not only the sites of AR binding but also allowed the prediction of the binding of other factors including NKX3.1 and Oct1 that mediate secondary transcriptional responses. Thus this model defines the characteristic pattern of nucleosome occupancy changes associated with enhancers and can be used to infer dynamic TF binding events that occur when a cell population transitions between states.
METHODS
ChIP-seq and ChIP-qPCR
The prostate cancer cell line LNCaP was obtained from the American Type Culture Collection. ChIP and ChIP-seq libraries construction for histone marks H3K4me1/2/3 were performed as previously described 14, the libraries were sequenced to 35bp with the Illumina Genome Analyzer. ChIP experiments for AR, NKX3.1 and Oct-1 as well as ChIP-qPCR were performed as previously described 27.
Peak calling
Significantly enriched regions were detected using the MACS software 28 using default parameters. Mononucleosomes were detected using the NPS analysis of nucleosome positions with default parameters 17.
AR and FoxA1 binding sites definition
Data sets of AR (FDR of 5%) and FoxA1 (FDR of 1%) binding sites were from our previous works 16,25.
Model for identifying differential transcription factor binding locations
NPS was used to identify nucleosome positions based on treatment condition data. Nucleosome intervals were defined as the 200bp centered on the center position of the NPS identified nucleosomes. Nucleosome pairs with center positions lying in a range between 250 and 450 bp were identified for further analysis. For each nucleosome pair the number of tags in the nucleosomes and in the inter-nucleosomal region was counted. A tag was considered to belong to a genomic interval if, when shifted 73 bp in a strand directed direction, the entire tag fell within that interval. These pairs are then given a NSD score (S) by the formula
That takes into account changes in H3K4me2 ChIP-seq tag counts falling on the flanking nucleosomes as well as the region lying between them. Tag counts were scaled in proportion to the overall counts in the treatment and control samples. In this formula, n is the tag count, superscript “treat” and “control” refer to DHT and vehicle conditions, respectively. Subscript “flank” refers to the 200bp of sequence centered on each flanking nucleosome, and “central” refers to the sequence between these regions (Fig. 3a).
Motif statistics
Known DNA motifs that are enriched relative to the center of model predicted TF binding sites were identified using the following statistic. Motif analysis was conducted on 600bp DNA segments, each segment representing one nucleosome pair. Each segment was derived from a 1kb sequence centered at the midpoint between a nucleosome pair from which 200bp centered on each of the nucleosomes was excluded. All subsequences within these sequences were scored by a TRANSFAC motif 29 and the genomic background sequence composition to identify hits above a probability cutoff. Let xi, be a value between 0 and 1, which denotes the relative location of motif hit i out of N total motif hits (where 0 and 1 represent the respective center and edge of the sequence). We define a z score, z=Σi=1 to N (xi − 0.5)/√(N/12) that represents the positional bias of a motif toward the centers of the regions. Different integer cutoffs were tested for each motif, and the cutoff resulting in the highest z was selected. This statistic is based on the assumptions that insignificant DNA motifs will be uniformly distributed across the sequences and the null distribution of Σi = 1 to N xi can be estimated as the N-fold convolution of uniform density functions.
Gene expression analysis
Affymetrix U133 Plus 2.0 microarray data were analyzed using the RMA algorithm 30 using acustom CDF probe (v11) mapping to RefSeq genes 31. Differentially expressed genes were identified using the SAM algorithm at a local FDR of 10% 32. The nearest putative TF binding sites were associated with each non-redundant RefSeq gene transcription start site. The statistical significance association between putative TF binding sites was calculated using Fisher’s exact test where genes were categorized according to whether they were differentially expressed, and whether they had at least one putative TF binding site within 20kb of the transcription start site.
Supplementary Material
Acknowledgments
This work was supported by grants from National Institutes of Health (1R01 HG004069-02 to X.S.L., and 2P50 CA090381-06 to X.S.L. and M.B.), the Department of Defense (W81XWH-07-1-0037 to X.S.L.) and the Prostate Cancer Foundation (to M.B.).
Footnotes
AUTHOR CONTRIBUTIONS H.H.H., C.A.M., K.Z., J.D.L., X.S.L. and M.B. designed experiments. H.H.H., S.T.B., G.W., Q.W., K.X., M.N., M.L. and P.M. performed experiments. C.A.M., H.H.H., H.S., and Y.Z. performed data analysis. C.A.M., H.H.H., X.S.L., M.B., J.D.L. and M.L. wrote the manuscript.
Accession codes. NCBI Short Read Archive: raw sequence tags and processed peak files have been deposited with accession code GSE20042.
References
- 1.Beato M, Eisfeld K. Transcription factor access to chromatin. Nucleic Acids Res. 1997;25:3559–3563. doi: 10.1093/nar/25.18.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Narlikar L, Gordan R, Hartemink AJ. A nucleosome-guided map of transcription factor binding sites in yeast. PLoS Comput Biol. 2007;3:e215. doi: 10.1371/journal.pcbi.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozsolak F, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan GC, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 6.Lee W, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 7.Mavrich TN, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mavrich TN, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valouev A, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 11.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blackwood EM, Kadonaga JT. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 13.Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 14.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 16.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Shin H, Song JS, Lei Y, Liu XS. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics. 2008;9:537. doi: 10.1186/1471-2164-9-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peckham HE, et al. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan GC, Liu JS. Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput Biol. 2008;4(1):e13. doi: 10.1371/journal.pcbi.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornberg RD, Stryer L. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 1988;16:6677–6690. doi: 10.1093/nar/16.14.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin C, et al. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korkmaz CG, et al. Analysis of androgen regulated homeobox gene NKX3.1 during prostate carcinogenesis. JUrol. 2004;172:1134–1139. doi: 10.1097/01.ju.0000136526.78535.b8. [DOI] [PubMed] [Google Scholar]
- 23.Asatiani E, et al. Deletion, methylation, and expression of the NKX3.1 suppressor gene in primary human prostate cancer. Cancer Res. 2005;65:1164–1173. doi: 10.1158/0008-5472.CAN-04-2688. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W, et al. Characterization of two functional NKX3.1 binding sites upstream of the PCAN1 gene that are involved in the positive regulation of PCAN1 gene transcription. BMC Mol Biol. 2008;9:45. doi: 10.1186/1471-2199-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 31.Dai M, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.