Abstract
Hsp104, a hexameric AAA+ ATPase found in yeast, transduces energy from cycles of ATP binding and hydrolysis to resolve disordered protein aggregates and cross-β amyloid conformers. These disaggregation activities are often co-ordinated by the Hsp70 chaperone system and confer considerable selective advantages. First, renaturation of aggregated conformers by Hsp104 is critical for yeast survival after various environmental stresses. Second, amyloid remodeling by Hsp104 enables yeast to exploit multifarious prions as a reservoir of beneficial and heritable phenotypic variation. Curiously, although highly conserved in plants, fungi and bacteria, Hsp104 orthologues are absent from metazoa. Indeed, metazoan proteostasis seems devoid of a system that couples protein disaggregation to renaturation. Here, we review recent endeavors to enhance metazoan proteostasis by applying Hsp104 to the specific protein-misfolding events that underpin two deadly neurodegenerative amyloidoses. Hsp104 potently inhibits Aβ42 amyloidogenesis, which is connected with Alzheimer's disease, but appears unable to disaggregate preformed Aβ42 fibers. By contrast, Hsp104 inhibits and reverses the formation of α-synuclein oligomers and fibers, which are connected to Parkinson's disease. Importantly, Hsp104 antagonizes the degeneration of dopaminergic neurons induced by α-synuclein misfolding in the rat substantia nigra. These studies raise hopes for developing Hsp104 as a therapeutic agent.
Keywords: AAA+ protein, Hsp104, Prion, Amyloid, Oligomer
The protein-misfolding problem
The successful functioning of all cells is dependent on proper protein folding. Functional native structure is encoded by primary sequence (Anfinsen 1973; Englander et al. 2007). However, native structures are dynamic systems constructed by complex networks of mutually supportive, weak interactions that cannot be readily established simultaneously during folding (Bartlett and Radford 2009; Englander et al. 2007). Consequently, folding energy landscapes can be rugged and pose challenges to successful folding (Bartlett and Radford 2009). Polypeptides can become trapped in non-native intermediate states or go astray into off-pathway conformations. Even once folding is completed, co-operative structural units of functional native structure unfold and refold iteratively (Englander et al. 2007). Additionally, mutations or decreased transcriptional or translational fidelity can yield primary sequences less able to achieve functional native structure (Dobson 2003; Lee et al. 2006). Thermal, oxidative, or chemical stress can also profoundly subvert protein folding (Parsell and Lindquist 1993). Thus, proteins may fail to fold or fail to remain correctly folded, making them prone to aggregation. The highly crowded macromolecular environment that cells must maintain further exacerbates this risk (Dobson 2003; Ellis and Minton 2006). Therefore, sophisticated protein homeostasis (proteostasis) networks have evolved to ensure that protein biogenesis is successful and that polypeptides effectively acquire, maintain, and (if necessary) reacquire their functional native structure (Balch et al. 2008; Powers et al. 2009).
Even before translation is complete, molecular chaperones associate with the growing polypeptide chain to ensure a folding-competent state (Kramer et al. 2009). Following translation, molecular chaperones help prevent misfolding and aggregation and assist polypeptides in acquiring their native state (Young et al. 2004). For example, proteins that enter the yeast secretory pathway are triaged through a series of sequential checkpoints that monitor folding status and determine conformational fitness for transport to subsequent destinations (Vashist and Ng 2004). Any terminally mis-folded proteins are targeted for degradation by the ubiquitin–proteasome pathway (Varshavsky 2005; Vembar and Brodsky 2008). However, aggregated proteins are more resistant to proteasomal degradation (Bence et al. 2001), but can be catabolized by autophagy (Cuervo 2008). Finally, sophisticated disaggregases reverse protein aggregation. Disaggregation can be coupled to protein degradation (Cohen et al. 2006) or renaturation (Doyle and Wickner 2009; Glover and Lum 2009; Shorter 2008; Weibezahn et al. 2005).
Once individuals reach post-reproductive age, these proteostatic safeguards invariably decline and errors in protein folding can arise with devastating effects (Cohen et al. 2006; Cuervo 2008; Morimoto 2006; Skovronsky et al. 2006). One pernicious and recurring problem is that the functional native structure is not always the lowest free-energy form (Englander et al. 2007). Rather, many proteins, regardless of primary sequence, can spontaneously access a generic, β-sheet-rich polymeric form of even lower free energy, termed amyloid (Dobson 2003).
Amyloid: pathogenic, protective, or beneficial
Amyloidogenesis of specific proteins is connected with several debilitating disorders, including Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease (HD), type II diabetes, prion diseases, and various cardiovascular and systemic amyloidoses (Caughey and Lansbury 2003; Kholová and Niessen 2005; Skovronsky et al. 2006; Taylor et al. 2002). There are no effective treatments for any of these disorders. Moreover, a major risk factor for these specific diseases is aging. As life spans increase through improvements in medicine and public health, these disorders will inevitably become among the most prevalent and recalcitrant impediments to living longer, more fulfilling lives.
All amyloids adopt a “cross-β” conformation in which the strands of the b-sheets run orthogonal to the fiber axis (Nelson and Eisenberg 2006; Sunde and Blake 1997; Sunde et al. 1997). The extreme stability of this fold, which resists disruption by proteases, detergents, chaotropes, and high temperatures, makes amyloid extremely difficult to clear (Eisenberg et al. 2006; Knowles et al. 2007; Smith et al. 2006). Moreover, the ends of amyloid fibers recruit other copies of the same protein and rapidly induce them to form a complementary cross-β structure. Once initiated, this self-templating process can rapidly convert entire populations of a given protein to the amyloid fold (Lansbury and Caughey 1995; Nelson and Eisenberg 2006). Steric effects often cause proteins to lose functionality once sequestered in the amyloid state (Baxa et al. 2002). This loss of function can contribute to pathogenesis in some disorders (Forman et al. 2004). Furthermore, amyloids can deplete other cellular components that co-precipitate (Chen et al. 2005). In systemic amyloidoses, amyloid deposition can be so extensive that tissue architecture becomes mechanically disrupted (Merlini and Westermark 2004).
In other diseases, the quantity of amyloid deposits can be minimal and inclusion formation can correlate with cell survival (Arrasate et al. 2004; Dobson 2003). This has led to proposals that despite being a space-occupying lesion, the amyloid form may be relatively benign and reflect a defense mechanism to sequester more toxic soluble species (Bucciantini et al. 2002; Kayed et al. 2003). Indeed, soluble oligomers that form during the characteristic lag phase of amyloidogenesis are often highly toxic and share a generic conformation, distinct from fibers, irrespective of primary sequence (Bucciantini et al. 2002; Haass and Selkoe 2007; Kayed et al. 2003; Lashuel et al. 2002; Lesné et al. 2006). These shared facets of amyloidogenesis suggest that effective therapeutics might have widespread applicability (Skovronsky et al. 2006). Despite these similarities, a major unsolved issue is how amyloidogenesis of specific proteins determines the selective cell death that differentiates various neurodegenerative amyloidoses (Skovronsky et al. 2006).
Accumulating evidence, however, suggests that amyloid is not invariably benign. In a mouse model of AD, β-amyloid plaques can appear extremely rapidly and are critical mediators of pathology (Meyer-Luehmann et al. 2008). Another danger is that amyloid might slowly release toxic misfolded species. Indeed, natural lipids can destabilize amyloid fibers, leading to liberation of toxic oligomers (Martins et al. 2008). Another issue originates from the tendency of amyloidogenic proteins to fold into multiple structurally distinct amyloid forms, or strains, which confer distinct phenotypes (Legname et al. 2006; Safar et al. 1998; Tanaka et al. 2006). Beyond sharing the cross-β amyloid conformation, little is known about the underlying atomic structures of these distinct strains or how structural polymorphism enciphers distinct phenotypes or disease states. However, it is clear that depending on the environmental conditions (e.g., pH, temperature) distinct strains can form, which are distinguished by distinct intermolecular contacts between fiber protomers and different lengths of primary sequence sequestered in cross-β structure (Krishnan and Lindquist 2005; Tessier and Lindquist 2007; Toyama et al. 2007). Importantly, different strains of Aβ40 fibers, which are connected with AD, and polyglutamine (polyQ), which are connected to HD, confer different levels of toxicity (Nekooki-Machida et al. 2009; Petkova et al. 2005). Some strains are relatively benign, whereas others are highly toxic (Nekooki-Machida et al. 2009; Petkova et al. 2005). Intriguingly, the more toxic polyQ strains harbor more flexible loops and turns, and expose a higher fraction of glutamine residues (Nekooki-Machida et al. 2009). Moreover, in a mouse model of HD, the most affected brain regions contained higher levels of the cytotoxic strain, whereas in less affected regions more benign strains predominated (Nekooki-Machida et al. 2009). Local microenvironmental differences including expression levels or proteostatic buffers might generate strain biases. It will be critical to determine how strain variation correlates with affected brain regions in other neurodegenerative amyloidoses. Identifying which species are toxic and which are benign will be key to the development of potential targeted therapies.
Benign amyloid conformers have been harnessed during evolution for adaptive purposes (Fowler et al. 2007; Shorter and Lindquist 2005b). For example, Pmel17 amyloids are critical for melanosome biogenesis (Berson et al. 2003; Fowler et al. 2006) and many peptide hormones in pituitary secretory granules are packaged as amyloids that slowly release active monomers upon secretion (Maji et al. 2009). The self-templating nature of CPEB amyloids might function in long-term memory formation (Si et al. 2003). In yeast, multiple proteins can form infectious amyloids, termed prions, which confer heritable phenotypes and selective advantages under diverse environmental conditions (Alberti et al. 2009; Griswold et al. 2009; King and Masel 2007; Shorter and Lindquist 2005b; True and Lindquist 2000; Tyedmers et al. 2008). In these cases, the proteostasis network ensures that benign amyloid conformers assemble at the expense of toxic intermediates (Douglas et al. 2008; Shorter and Lindquist 2004; Treusch et al. 2009). An accurate understanding of how amyloids have been exploited for beneficial purposes may yield key insights into how to safely eliminate toxic amyloid fibers and preamyloid oligomers.
To date, there are no cures or effective treatments for any of the neurodegenerative amyloidoses that afflict human-kind. Currently prescribed therapies remain palliative in nature, and do not antagonize the underlying causative continuum of amyloid forms or cytotoxic oligomers. A key therapeutic advance will be the ability to enhance proteostasis, such that entire spectra of toxic amyloid strains and preamyloid oligomers are eliminated, while beneficial amyloids are left intact. Here, we entertain the possibility of enhancing mammalian proteostasis with an AAA+ protein from yeast, Hsp104, which can rapidly dissolve amyloid conformers and preamyloid oligomers (Shorter 2008). First, however, we consider the mechanistic basis of Hsp104 activity.
Hsp104: a protein disaggregase
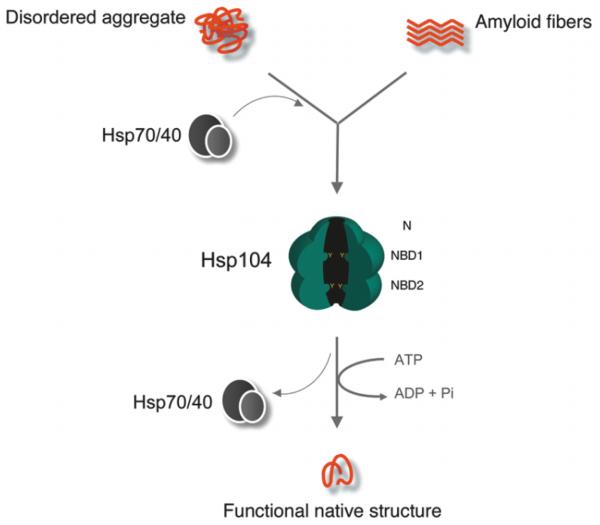
Hsp104 enhances yeast survival ~10,000-fold after a variety of environmental stresses that induce protein misfolding and subsequent aggregation (Sanchez and Lindquist 1990; Sanchez et al. 1992). Orthologues in plants (Hsp101) and bacteria (ClpB) confer similar advantages (Queitsch et al. 2000; Squires et al. 1991). These advantages stem from the ability of Hsp104 (and orthologues) to synergize with Hsp70 and Hsp40 to rapidly resolve chemically or thermally denatured protein aggregates and return proteins to native structure and function (Fig. 1) (Cashikar et al. 2005; Glover and Lindquist 1998; Goloubinoff et al. 1999; Mogk et al. 1999; Parsell et al. 1994b; Parsell et al. 1993; Weibezahn et al. 2004). Hsp70 and Hsp40 help present aggregated substrates to Hsp104, and likely assist substrate refolding after release from the aggregate (Glover and Lindquist 1998; Tessarz et al. 2008). This rapid renaturation of proteins obviates the large energetic cost of protein degradation and de novo synthesis that would be required to eliminate and replace the aggregated protein.
Fig. 1.
Protein disaggregation by Hsp104. Hsp104 couples energy from ATP binding and hydrolysis to dissolve denatured protein aggregates and amyloid fibers. Hsp70 and Hsp40 co-ordinate the disaggregation of denatured protein aggregates, whereas they are less essential for the dissolution of amyloid structure. Note the 3-tiered structure of Hsp104, and the substrate-binding Tyr motifs that project into the central channel (denoted by small Y-shapes).
Hsp104 is a modular protein comprising an N-terminal domain, a first AAA+ nucleotide-binding domain (NBD1), a distinctive coiled-coil middle domain, a second AAA+ nucleotide-binding domain (NBD2), and a short C-terminal domain (Doyle and Wickner 2009). Like many AAA+ proteins, Hsp104 is only active once it forms hexamers, which requires ADP or ATP binding to NBD2 (Parsell et al. 1994a; Schirmer et al. 1998; Schirmer et al. 2001). Cryo-electron microscopy and single particle reconstruction demonstrate that Hsp104 hexamers possess a 3-tiered hexameric ring structure that envelops a large central channel (Wendler et al. 2007; Wendler et al. 2009). Homology models of Hsp104 based on the atomic structure of the T. thermophilus orthologue, tClpB protomer (Lee et al. 2003), have been fitted into the cryo-EM reconstructions. A small ring of N-terminal domains form the top tier, whereas expanded rings of NBD1 and NBD2 form the middle and lower tiers, respectively (Fig. 1) (Wendler et al. 2007; Wendler et al. 2009). The prominent middle domain, which consists of 2 antiparallel coiled-coil motifs reminiscent of a 2-bladed propeller (Lee et al. 2003), interposes in subunit packing by intercalating between NBD1 and NBD2 (Wendler et al. 2007; Wendler et al. 2009). This hexameric model of Hsp104 differs significantly from a hexameric model proposed for tClpB, where the coiled-coil domains are proposed to protrude laterally from the surface of the hexameric ring (Lee et al. 2007; Lee et al. 2003). Potential reasons for these differences are discussed in detail elsewhere (Wendler and Saibil 2010).
Several lines of evidence suggest that Hsp104 and orthologues disaggregate substrates by coupling ATP binding and hydrolysis to the translocation of polypeptides from the aggregate surface across the central channel to solution (Lum et al. 2008; Lum et al. 2004; Schlieker et al. 2004; Weibezahn et al. 2004; reviewed in Shorter and Lindquist, 2005a). However, we are only at the inception of understanding the mechanistic and structural basis of this activity. Hsp104 initially engages misfolded substrates when NBD1 is in an ATP-bound conformation (Bösl et al. 2005; Schaupp et al. 2007). Simultaneous elimination of ATPase activity at both NBDs abolishes disaggregase activity (Doyle et al. 2007b; Shorter and Lindquist 2004). Both NBDs catalyze ATP hydrolysis co-operatively and allosteric communication occurs within and between NBD1 and NBD2 (Cashikar et al. 2002; Doyle et al. 2007b; Hattendorf and Lindquist 2002; Schaupp et al. 2007; Schirmer et al. 2001). In the absence of substrate, NBD1 makes the major contribution to ATPase activity (kcat ~ 76 min−1, KM ~ 170 μmol·L−1, nh = 2.3,) but has a lower affinity for nucleotide compared with NBD2 (kcat ~ 0.27 min−1, KM ~ 4.7 μmol·L−1, nh = 1.6) (Hattendorf and Lindquist 2002). Yet, it seems likely that these patterns need to be subtly altered to elicit disaggregation. Deceleration of ATPase activity at NBD1 but not at NBD2, or vice versa, facilitates disaggregation (Doyle et al. 2007a; Doyle et al. 2007b; Schaupp et al. 2007). Why the hydrolysis pattern must be decelerated asymmetrically to elicit disaggregation remains an open question requiring further study. However, it seems likely that this alteration has allosteric sequelae that enable Hsp104 to simultaneously bind, unfold, and translocate substrates. Without this deceleration, substrate binding is likely too transient to be productive (Fig. 2a).
Fig. 2.
Co-ordination of Hsp104 ATPase activity is required for protein disaggregation. (a) In the presence of ATP, Hsp104 engages denatured aggregates too transiently to initiate disaggregation. (b) In the presence of specific mixtures of ATP and ATPγS (e.g., 3 ATP : 1 ATPγS), Hsp104 is able to productively couple substrate binding to translocation and disaggregation. Only specific ratios of ATP and ATPγS are effective, and pure ATPγS inhibits activity (Doyle et al. 2007b). (c) In the presence of ATP, specific Hsp104 mutants with reduced ATPase activity at NBD2, such as N728A or K620T, can productively couple substrate binding to translocation and disaggregation (Doyle et al. 2007b). Despite this innate ability to disaggregate some disordered aggregates, these mutants fail to disaggregate amyloid and are unable to synergize with the Hsp70 chaperone system and are defective in vivo (Doyle et al. 2007b; Hattendorf and Lindquist 2002; Shorter and Lindquist 2004). (d) In the presence of ATP, Hsp70 and Hsp40 ensure productive substrate binding by Hsp104, which leads to translocation and disaggregation. (e) In the presence of ATP, Hsp104 is able to bind and disaggregate amyloid substrates.
In the absence of Hsp70 and Hsp40, dissolution of denatured aggregates can be triggered by artificially choreographing the requisite mode of ATPase cycling with specific mixtures of ATP and ATPγS, a slowly hydrolyzable ATP analogue (Fig. 2b) (Doyle et al. 2007a; Doyle et al. 2007b). Alternatively, mutation of conserved AAA+ motifs (Walker A, Walker B, or sensor-1) at one NBD to slow ATP hydrolysis at that site can also elicit disaggregase or substrate unfolding activity in the absence of Hsp70 and Hsp40 (Fig. 2c) (Doyle et al. 2007a; Doyle et al. 2007b; Schaupp et al. 2007). However, Hsp70 and Hsp40 are usually required to present denatured aggregated substrates to Hsp104 (Glover and Lindquist 1998; Tessarz et al. 2008; Weibezahn et al. 2004), and likely play some role in co-ordinating the requisite mode of Hsp104 ATPase cycling (Fig. 2d) (Doyle et al. 2007a; Doyle et al. 2007b). In some cases, the substrate itself (e.g., amyloid) can impose the requisite changes (Fig. 2e) (Doyle et al. 2007b; Shorter and Lindquist 2004). Collectively, these data suggest that a cooperative division of labor among the 12 AAA+ domains drives protein disaggregation. One subset slowly hydrolyzes ATP to facilitate substrate binding, whereas another subset rapidly hydrolyzes ATP to promote substrate unfolding and translocation.
How does this co-operative division of labor facilitate substrate translocation across the central channel? The N-and C-terminal domains may help bind substrates and cofactors (Barnett et al. 2005; Cashikar et al. 2002; Mackay et al. 2008). However, critical substrate interactions are mediated by an α-helical insertion in NBD1 and a β-hairpin in NBD2, located before helix α2 in the αβ subdomain in both NBDs (Lum et al. 2008; Lum et al. 2004; Schlieker et al. 2004; Tessarz et al. 2008; Weibezahn et al. 2004). Short, highly conserved loops, KYKG in NBD1 and GYVG in NBD2, project into the channel (Fig. 1) (Wendler et al. 2009). Of particular importance is the tyrosine residue in these loops, as mutation of this residue to alanine disrupts substrate interactions and disaggregation activity in vitro and in vivo (Lum et al. 2008; Lum et al. 2004; Tessarz et al. 2008). Mutation of the NBD2 loop tyrosine confers the most drastic effects in vivo and phenocopies deletion of Hsp104 (Lum et al. 2004). More conservative substitutions of the NBD2 loop tyrosine, such as phenylalanine and tryptophan, maintain partial functionality (Hung and Masison 2006). Dynamic rearrangements of channel loop tyrosines, which are proposed to grip substrate, synchronized with ATPase cycling likely provide a series of motions that translocate substrates across the channel.
Recent cryo-EM reconstructions of Hsp104 hexamers in the presence of ATPγS, ATP, and ADP have provided structural insight into the conformational changes that facilitate substrate translocation (Wendler et al. 2009). This study employed the NBD2 sensor-1 mutant, Hsp104N728A, which is able to disaggregate denatured aggregates without Hsp70 or Hsp40 (Doyle et al. 2007b), but is defective in prion disaggregation and provides only limited thermotolerance in vivo (Hattendorf and Lindquist 2002; Shorter and Lindquist 2004). Reconstructions with imposed 6-fold symmetry reveal that ATP binding and hydrolysis induce large domain movements in NBD1 that impart a peristaltic mechanism for substrate translocation. Upon ATP binding, the NBD1 substrate-binding KYKG motifs move up toward the N-terminal end of the channel and are poised to receive substrate. Upon ATP hydrolysis, NBD1 generates a large motion that displaces the KYKG motif from the N-terminal end to the center of the channel. Simultaneously, the NBD2 substrate-binding GYVG motifs rotate into the center of the channel to receive substrate translocated by NBD1. Subsequent ATP binding to NBD1 then moves the NBD1 KYKG motifs back up toward the N-terminal entrance, while simultaneously moving the NBD2 GYVG motifs down toward the C-terminal end of the channel. Thus, the NBD2 GYVG motif is able to exert a pulling force without ATP hydrolysis by NBD2. The ADP state of the hexamer suggests that ATP hydrolysis at NBD2 might induce a dramatic rotation of this domain that would eject substrate. These interdependent motions of NBD1 and NBD2 ensure continuous substrate handling during disaggregation (Wendler et al. 2009).
Single particle reconstructions without imposed symmetry reveal that Hsp104 hexamers are strongly asymmetric in the presence of ATPγS or ATP (Wendler et al. 2009). Thus, the NBD1 domain movements are highly likely to proceed in a sequential rather than concerted or probabilistic manner. Substrate handover appears to be most compatible with a clockwise order of ATP hydrolysis in NBD1. The extremely large size of the channel compared to other AAA+ proteins might enable the translocation of exposed loops or more than one polypeptide, rather than having to search for exposed N- or C-termini of individual polypeptides (Haslberger et al. 2008; Wendler et al. 2007). Moreover, Hsp104 hexamers are likely to be highly dynamic and rapidly exchange monomers like ClpB hexamers (Haslberger et al. 2008; Werbeck et al. 2008), which might enable recycling of Hsp104 monomers should disaggregation stall or fail. Throughout the Hsp104 ATPase cycle the coiled-coil middle domain, which distinguishes Hsp104 and orthologues from all other AAA+ proteins, appears to play a critical structural role that facilitates the dramatic rotations of NBD1 and NBD2 that forcibly drive substrate translocation (Wendler and Saibil 2010; Wendler et al. 2007; Wendler et al. 2009).
What role the coiled-coil domain might play in substrate translocation is much less clear in the hexameric model of tClpB, where the coiled-coil domains radiate laterally from the surface of the hexamer (Lee et al. 2007; Lee et al. 2003). Functional evidence suggests that helix 3 of the coiled-coil collaborates with the Hsp70 chaperone system to shuffle aggregated substrate into the ClpB channel (Haslberger et al. 2007). However, this region is in a similar position in the hexameric models of both tClpB and Hsp104 (Haslberger et al. 2007; Wendler and Saibil 2010; Wendler et al. 2007; Wendler et al. 2009). Further biochemical and structural studies will be required to evaluate the various predictions of these models.
Hsp104: powerful amyloid-remodeling activity
In addition to the ability to resolve denatured aggregates, Hsp104 possesses an unusually powerful amyloid-remodeling activity and couples ATP hydrolysis to the rapid deconstruction of amyloid fibers composed of yeast prion proteins Sup35 and Ure2 (Narayanan et al. 2006; Savistchenko et al. 2008; Shorter and Lindquist 2004; Shorter and Lindquist 2006). Importantly, even brief overexpression of Hsp104 is sufficient to eliminate Sup35 prions (Chernoff et al. 1995). Dissolution of amyloid structure does not require Hsp70 and Hsp40 (Fig. 1, 2e), although their presence can improve Hsp104 activity against various amyloids in vitro (Lo Bianco et al. 2008; Shorter and Lindquist 2008; Sweeny and Shorter 2008) and in vivo (Higurashi et al. 2008; Tipton et al. 2008). Hsp104 is also able to resolve preamyloid oligomers of Sup35 (Shorter and Lindquist 2004; Shorter and Lindquist 2006), which adopt a generic conformation shared by many disease-associated amyloidogenic proteins (Kayed et al. 2003).
Perplexingly, no clear metazoan homologue or analogue of Hsp104 has been identified. Moreover, no activity that couples protein disaggregation to renaturation has been identified in metazoa. Initial attempts to isolate an analogous disaggregase by biochemical fractionation of mammalian cytosol have been unsuccessful (Mosser et al. 2004). Crude whole-animal extracts from Caenorhabditis elegans are able to slowly disaggregate and degrade Aβ42 fibers (Cohen et al. 2006). However, the factor(s) involved remain unidentified (Cohen et al. 2006). The search for functional equivalents of Hsp104 has led researchers to test other conserved AAA+ proteins. One highly conserved candidate, p97 (Meyer and Popp 2008), appears to collaborate with Hsp70 and Hsp40 to refold soluble misfolded conformers (Thoms 2002), but no convincing demonstration of coupled disaggregation and renaturation has been forthcoming using solely pure proteins (Nishikori et al. 2008). p97 appears to assist in the recovery of luciferase acivity from the insoluble fraction of HeLa cells after mild heat stress; however, whether this activity is direct or extends to other substrates remains unclear (Kobayashi et al. 2007). Despite this curious hint, it appears that metazoan proteostasis is more centered on clearing aggregated proteins by autophagy and other proteolytic systems rather than by disaggregation and renaturation (Cohen et al. 2006; Cuervo 2008).
The ability of Hsp104 to rapidly deconstruct the generic cross-β structures of yeast prions as well as the shared generic form of preamyloid oligomers raises the possibility of applying Hsp104 to metazoan systems to prevent or reverse various amyloidoses. The ability to reverse amyloid formation would counter several intractable issues that likely synergize to various extents in the etiology of various amyloid disorders: (i) the toxic gain of function of amyloid or preamyloid conformers; (ii) the loss of function of the aggregated protein; and (iii) the loss of other essential proteins that co-aggregate with the disease-associated protein.
Remarkably, despite being a yeast protein, Hsp104 is very well tolerated in metazoan systems and displays no overt toxicity. In fact, expression of Hsp104 in various mammalian cell lines confers increased stress tolerance (Dandoy-Dron et al. 2006; Mosser et al. 2004). Moreover, Hsp104 can synergize with the mammalian Hsp70 chaperone system to promote the renaturation of aggregated conformers (Glover and Lindquist 1998; Mosser et al. 2004; Schaupp et al. 2007). Hsp104 protects against other proteotoxic insults in a tissue culture setting, including polyQ misfolding (Carmichael et al. 2000; Perrin et al. 2007) and poly(A)-binding protein 2 misfolding associated with oculopharyngeal muscular dystrophy (Bao et al. 2002). Furthermore, Hsp104 has been safely expressed in C. elegans to counter polyQ toxicity (Satyal et al. 2000). Transgenic mice expressing Hsp104 have been successfully generated and appear grossly normal, indicating that Hsp104 does not interfere with mammalian development (Dandoy-Dron et al. 2006; Vacher et al. 2005). Moreover, Hsp104 prolonged lifespan of an HD mouse model by ~20% and reduced polyQ aggregation in this setting (Vacher et al. 2005). These studies provide clear precedent for the utility of Hsp104 in enhancing metazoan proteostasis to counter protein aggregation and amyloidogenesis. In the remainder of this review, we consider two recent applications of Hsp104 to the amyloidogenic events that distinguish AD and PD.
Hsp104 and AD
Alzheimer's disease (AD) is the most common fatal neurodegenerative disorder, afflicting ~27 million people worldwide and is projected to increase ~4-fold by 2050 (Brookmeyer et al. 2007). Beyond minor symptomatic relief, there are no effective treatments (Roberson and Mucke 2006). AD is characterized by gross diffuse atrophy of the brain and neurodegeneration in the cerebral cortex and certain subcortical regions (Wenk 2003). The defining pathological lesions are extracellular neuritic plaques composed primarily of amyloid forms of the β-amyloid (Aβ) peptides Aβ42 and Aβ40 (Glenner and Wong 1984; Iwatsubo et al. 1994; Masters et al. 1985), and intracellular neurofibrillary tangles composed of amyloid forms of the microtubule-binding protein tau (Skovronsky et al. 2006). Several treatment strategies are in clinical trials (Roberson and Mucke 2006) and several small molecules have been described that can inhibit (Gestwicki et al. 2004) and sometimes even reverse Aβ42 fibrillization (Wang et al. 2008). Thus far, only the effects of Hsp104 on Aβ42 have been reported (Arimon et al. 2008; Schirmer and Lindquist 1997).
Schirmer and Lindquist first discovered that Hsp104 could interact with Aβ42. Aβ42 very potently inhibited Hsp104 ATPase activity (Schirmer and Lindquist 1997). A more recent study found that Aβ42 stimulates Hsp104 ATPase activity (Arimon et al. 2008). Importantly, Hsp104 very potently inhibited de novo Aβ42 fibrillization, even when Aβ42 was in excess by 1000-fold (Arimon et al. 2008). Such substoichiometric inhibition suggests that Hsp104 specifically antagonizes an obligate intermediate that is ‘on pathway’ for fiber assembly. Indeed, Hsp104 also antagonized the conversion of Aβ42 oligomers into fibers, and interacted directly with Aβ42 monomers and oligomers (Arimon et al. 2008). Additionally, Hsp104 potently inhibited Aβ42 fibrillization assembly seeded by preformed fibers (Arimon et al. 2008). Consistent with these data, Hsp104 inhibited Aβ42 fibrillization when added during lag phase or assembly phase (Arimon et al. 2008). Curiously, however, Hsp104 was unable to disassemble Aβ42 fibers or oligomers (Arimon et al. 2008). This deficiency might reflect a need for Hsp70 and Hsp40, which can assist Hsp104 in disassembly of amyloid (Higurashi et al. 2008; Lo Bianco et al. 2008; Shorter and Lindquist 2008; Sweeny and Shorter 2008; Tipton et al. 2008). Alternatively, the particular strain of Aβ42 fibers that formed under these conditions may be refractory to Hsp104, or all strains of Aβ42 fibers may be refractory. Nonetheless, the ability of Hsp104 to bind Aβ42 monomers and inhibit seeded assembly, coupled to the fact that amyloids very slowly exchange monomers via a soluble pool (Carulla et al. 2005), might enable Hsp104 to slowly shift the equilibrium away from the assembled fibrous state. Thus, Hsp104 might slowly resolve Aβ42 fibers over a time frame longer than those thus far explored (Arimon et al. 2008).
These findings are promising; however, all experiments presented were performed in vitro and no complementary in vivo approaches were adopted. Extension to cell culture and animal models is needed for validation. Neuroblastoma cell lines have been widely used to assess the toxicity of Aβ fibers and oligomers (Kayed et al. 2003; Petkova et al. 2005), and this system might be easily adapted to test whether the Hsp104-Aβ42 interactions buffer toxicity to cultured neurons. Another issue is that the majority of Aβ42 fibers are extracellular in AD, which may make them challenging targets for Hsp104. However, intraneuronal Aβ42 is also found in AD and may contribute to disease progression (Grundke-Iqbal et al. 1989; LaFerla et al. 2007; Wertkin et al. 1993). Hsp104 might be efficacious against intraneuronal pools of misfolded Aβ42.
Hsp104 and PD
There are no efficacious treatments for Parkinson's disease (PD), the most common neurodegenerative movement disorder, which afflicts several million people worldwide (Dorsey et al. 2007). PD is due to a severe and selective devastation of dopaminergic neurons from the substantia nigra pars compacta, although neuropathology extends into other regions of the brain (Braak et al. 2003). Intracellular inclusions termed Lewy bodies and Lewy neurites, composed of amyloid forms of the small pre-synaptic protein α-synuclein (α-syn), are the signature lesion of PD (Spillantini et al. 1997). Although PD is most frequently a sporadic disorder, mutations in α-syn (e.g., A30P, A53T, E46K) and duplication or triplication of the wild-type gene are linked with early-onset PD in rare familial forms of the disease (Moore et al. 2005). α-Syn function is uncertain, but may play a key regulatory role in dopamine release from synaptic vesicle pools (Gitler and Shorter 2007; Larsen et al. 2006). Pure α-syn readily accesses amyloid forms in vitro that bear remarkable similarities to α-syn fibers isolated from synucleinopathy patients (Crowther et al. 2000; Spillantini et al. 1998).
Hsp104 potently inhibited fibrillization of α-syn and PD-linked variants (A30P, A53T, and E46K) in vitro (Lo Bianco et al. 2008). Moreover, Hsp104 coupled ATP hydrolysis to the disassembly of toxic oligomers composed of α-syn A30P (Lo Bianco et al. 2008). Hsp104 also coupled ATP hydrolysis to the disassembly of α-syn fibers (Lo Bianco et al. 2008). Disassembly was enhanced by the mammalian Hsp70 chaperone system, and in particular by the specific combination of Hsc70 and Hdj2 (Lo Bianco et al. 2008). All α-syn variant fibers were effectively disassembled, except for the E46K PD-linked mutant (Lo Bianco et al. 2008). This might indicate that α-syn E46K forms a different strain of amyloid. Indeed, α-syn E46K fibers tend to form compact bundles and meshwork arrays not observed with wild-type α-syn (Choi et al. 2004; Greenbaum et al. 2005). Nonetheless, this battery of remodeling activities suggested that Hsp104 might effectively buffer α-syn toxicity in vivo.
The development of PD therapies has been hindered by a lack of animal models that successfully recreate the progressive and selective degeneration of dopaminergic neurons and formation of phosphorylated α-syn inclusions. However, a rat PD model based on the lentiviral-mediated expression of human α-syn A30P in the substantia nigra is able to recapitulate these features (Lo Bianco et al. 2002; Lo Bianco et al. 2004). Thus, Hsp104 and α-syn A30P were expressed simultaneously in the rat substantia nigra using the lentiviral delivery system. Remarkably, Hsp104 reduced formation of phosphorylated α-syn A30P inclusions and prevented nigrostriatal dopaminergic neurodegeneration (Lo Bianco et al. 2008). Thus, Hsp104 is able to buffer α-syn A30P toxicity in the physiological arena of the mammalian substantia nigra.
Although these results are promising, several questions remain. First, it has not yet been possible to express Hsp104 after α-syn has already aggregated, a situation that might mimic more closely a potential treatment. Thus, whether Hsp104 can reverse α-syn aggregation in the setting of the rat substantia nigra remains unclear. Another issue concerns releasing a large pulse of soluble α-syn from Lewy Bodies in surviving neurons. Such a pulse might be detrimental since high levels of soluble α-syn can inhibit synaptic vesicle release and perturb other membrane trafficking events (Gitler et al. 2008; Gitler and Shorter 2007; Larsen et al. 2006). However, this situation is likely to be preferable to the persistence of toxic α-syn conformers. Finally, further study is needed to assess any dangers of long-term Hsp104 expression in the mammalian brain.
Concluding remarks
The foregoing sections raise hopes that Hsp104 might hold therapeutic potential for antagonizing amyloidogenic events associated with AD and PD. Furthermore, the ability of Hsp104 to prevent or reverse the formation of non-amyloid, disease-associated aggregates should also be considered. For example, two devastating neurodegenerative disorders, amyotrophic lateral sclerosis and frontotemporal lobar degeneration with ubiquitin-positive inclusions, are connected with the formation of non-amyloid, aggregated species of a conserved hnRNP, TDP-43 (Johnson et al. 2009; Neumann et al. 2006). Nevertheless, a plethora of issues must be overcome if Hsp104 is to be developed as a therapeutic agent. Not least is the issue that gene therapy would seem necessary to apply Hsp104 as a therapeutic agent. Gene therapy has produced encouraging preclinical outcomes for a number of disorders (Bainbridge et al. 2008; Hacein-Bey-Abina et al. 2002; Maguire et al. 2008), yet technical and safety issues have restricted translation to the clinic. Indeed, gene therapy approaches to treat neurodegenerative amyloidoses remain in early developmental stages and considerable caution is needed at this time. However, initial studies suggest that gene therapy in the adult brain might be safe for various neurodegenerative disorders, including PD (Feigin et al. 2007; Kaplitt et al. 2007; Stoessl 2007). Thus, even though we await several key advances in gene therapy before any chaperone treatment becomes feasible, it remains important to develop solutions to amyloid problems and to test these solutions both in vitro and in animal models.
Existing Hsp104 specificity or activity is unlikely to be optimal against substrates that it never ordinarily encounters, such as α-syn or Aβ42. Indeed, disassembly of α-syn fibers requires considerably larger amounts of Hsp104 than disassembly of Sup35 or Ure2, two natural amyloid substrates (Lo Bianco et al. 2008; Shorter and Lindquist 2006). Even for Sup35 and Ure2, high concentrations of Hsp104 are required to reverse amyloid formation (Shorter and Lindquist 2006). Acting at lower concentrations, Hsp104 fragments Sup35 and Ure2 prions, which generates more fiber ends that can convert soluble copies of the protein to the prion form (Shorter and Lindquist 2006). Thus, an important therapeutic consideration is to express Hsp104 above a certain threshold that reduces and does not exacerbate the amyloid burden.
Hsp104 is likely to be a generalist, since it must catalyze the disaggregation of large portions of the yeast proteome after environmental stress. Regarding amyloid conformers, it seems likely that Hsp104 might be adapted to remodel cross-b structures comprised of the uncharged polar residues that distinguish the prion domains of many proteins in yeast (Alberti et al. 2009). Promiscuous disaggregation activity might also be undesirable in a therapeutic setting. Ideally, a therapeutic disaggregase would selectively eliminate toxic strains and misfolded species, and not eradicate benign strains or even beneficial amyloids such as CPEB prions, which might encode long-term memory (Si et al. 2003; reviewed in Shorter and Lindquist 2005b). Thus, an important goal is to engineer or evolve Hsp104 variants with enhanced and selective ability to eradicate specific amyloid or aggregated conformers. Ultimately, designer disaggregases might be developed to annihilate purely toxic conformers unique to each particular disease.
Acknowledgements
We thank M. Noelle Knight for expert graphical assistance and Petra Wendler for critical reading of the manuscript. Our work is supported by an NIH Director's New Innovator Award (DP2OD002177); an Ellison New Scholar in Aging Award (AG-NS-0611–09); an American Heart Association Scientist Development Award; and University of Pennsylvania Institute on Aging, Alzheimer's Disease Core Center and Diabetes and Endocrinology Research Center pilots (J.S.).
Footnotes
This paper is one of a selection of papers published in this special issue entitled 8th International Conference on AAA Proteins and has undergone the Journal's usual peer review process.
References
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137(1):146–158. doi: 10.1016/j.cell.2009.02.044. doi:10.1016/j.cell.2009.02.044. PMID:19345193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181(96):223–230. doi: 10.1126/science.181.4096.223. doi:10.1126/science.181.4096.223. PMID:4124164. [DOI] [PubMed] [Google Scholar]
- Arimon M, Grimminger V, Sanz F, Lashuel HA. Hsp104 targets multiple intermediates on the amyloid pathway and suppresses the seeding capacity of Abeta fibrils and protofibrils. J. Mol. Biol. 2008;384(5):1157–1173. doi: 10.1016/j.jmb.2008.09.063. doi:10.1016/j.jmb.2008.09.063. PMID:18851977. [DOI] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431(7010):805–810. doi: 10.1038/nature02998. doi:10.1038/nature02998. PMID:15483602. [DOI] [PubMed] [Google Scholar]
- Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, et al. Effect of gene therapy on visual function in Leber's congenital amaurosis. N. Engl. J. Med. 2008;358(21):2231–2239. doi: 10.1056/NEJMoa0802268. doi:10.1056/NEJMoa0802268. PMID:18441371. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. doi: 10.1126/science.1141448. doi:10.1126/science.1141448. PMID:18276881. [DOI] [PubMed] [Google Scholar]
- Bao YP, Cook LJ, O'Donovan D, Uyama E, Rubinsztein DC. Mammalian, yeast, bacterial, and chemical chaperones reduce aggregate formation and death in a cell model of oculopharyngeal muscular dystrophy. J. Biol. Chem. 2002;277(14):12263–12269. doi: 10.1074/jbc.M109633200. doi:10.1074/jbc.M109633200. PMID:11796717. [DOI] [PubMed] [Google Scholar]
- Barnett ME, Nagy M, Kedzierska S, Zolkiewski M. The amino-terminal domain of ClpB supports binding to strongly aggregated proteins. J. Biol. Chem. 2005;280(41):34940–34945. doi: 10.1074/jbc.M505653200. doi:10.1074/jbc.M505653200. PMID:16076845. [DOI] [PubMed] [Google Scholar]
- Bartlett AI, Radford SE. An expanding arsenal of experimental methods yields an explosion of insights into protein folding mechanisms. Nat. Struct. Mol. Biol. 2009;16(6):582–588. doi: 10.1038/nsmb.1592. doi:10.1038/nsmb.1592. PMID:19491935. [DOI] [PubMed] [Google Scholar]
- Baxa U, Speransky V, Steven AC, Wickner RB. Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc. Natl. Acad. Sci. U.S.A. 2002;99(8):5253–5260. doi: 10.1073/pnas.082097899. doi:10.1073/pnas.082097899. PMID:11959975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292(5521):1552–1555. doi: 10.1126/science.292.5521.1552. doi:10.1126/science.292.5521.1552. PMID:11375494. [DOI] [PubMed] [Google Scholar]
- Berson JF, Theos AC, Harper DC, Tenza D, Raposo G, Marks MS. Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J. Cell Biol. 2003;161(3):521–533. doi: 10.1083/jcb.200302072. doi:10.1083/jcb.200302072. PMID:12732614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bösl B, Grimminger V, Walter S. Substrate binding to the molecular chaperone Hsp104 and its regulation by nucleotides. J. Biol. Chem. 2005;280(46):38170–38176. doi: 10.1074/jbc.M506149200. doi:10.1074/jbc.M506149200. PMID:16135516. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K, Rüb U, de Vos RA, Jansen Steur EN, Braak E. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol. Aging. 2003;24(2):197–211. doi: 10.1016/s0197-4580(02)00065-9. doi:10.1016/S0197-4580(02)00065-9. PMID:12498954. [DOI] [PubMed] [Google Scholar]
- Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer's disease. Alzheimers Dement. 2007;3(3):186–191. doi: 10.1016/j.jalz.2007.04.381. doi:10.1016/j.jalz.2007.04.381. PMID:19595937. [DOI] [PubMed] [Google Scholar]
- Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416(6880):507–511. doi: 10.1038/416507a. doi:10.1038/416507a. PMID:11932737. [DOI] [PubMed] [Google Scholar]
- Carmichael J, Chatellier J, Woolfson A, Milstein C, Fersht AR, Rubinsztein DC. Bacterial and yeast chaperones reduce both aggregate formation and cell death in mammalian cell models of Huntington's disease. Proc. Natl. Acad. Sci. U.S.A. 2000;97(17):9701–9705. doi: 10.1073/pnas.170280697. doi:10.1073/pnas.170280697. PMID:10920207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carulla N, Caddy GL, Hall DR, Zurdo J, Gairí M, Feliz M, et al. Molecular recycling within amyloid fibrils. Nature. 2005;436(7050):554–558. doi: 10.1038/nature03986. doi:10.1038/nature03986. PMID:16049488. [DOI] [PubMed] [Google Scholar]
- Cashikar AG, Schirmer EC, Hattendorf DA, Glover JR, Ramakrishnan MS, Ware DM, Lindquist SL. Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol. Cell. 2002;9(4):751–760. doi: 10.1016/s1097-2765(02)00499-9. doi:10.1016/S1097-2765(02)00499-9. PMID:11983167. [DOI] [PubMed] [Google Scholar]
- Cashikar AG, Duennwald M, Lindquist SL. A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J. Biol. Chem. 2005;280(25):23869–23875. doi: 10.1074/jbc.M502854200. doi:10.1074/jbc.M502854200. PMID:15845535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 2003;26(1):267–298. doi: 10.1146/annurev.neuro.26.010302.081142. doi:10.1146/annurev.neuro.26.010302.081142. PMID:12704221. [DOI] [PubMed] [Google Scholar]
- Chen Q, Liu JB, Horak KM, Zheng H, Kumarapeli AR, Li J, et al. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ. Res. 2005;97(10):1018–1026. doi: 10.1161/01.RES.0000189262.92896.0b. doi:10.1161/01.RES.0000189262.92896.0b. PMID:16210548. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268(5212):880–884. doi: 10.1126/science.7754373. doi:10.1126/science.7754373. PMID:7754373. [DOI] [PubMed] [Google Scholar]
- Choi W, Zibaee S, Jakes R, Serpell LC, Davletov B, Crowther RA, et al. Mutation E46K increases phospholipid binding and assembly into filaments of human alpha-synuclein. FEBS Lett. 2004;576(3):363–368. doi: 10.1016/j.febslet.2004.09.038. doi:10.1016/j.febslet.2004.09.038. PMID:15498564. [DOI] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313(5793):1604–1610. doi: 10.1126/science.1124646. doi:10.1126/science.1124646. PMID:16902091. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Daniel SE, Goedert M. Characterisation of isolated alpha-synuclein filaments from substantia nigra of Parkinson's disease brain. Neurosci. Lett. 2000;292(2):128–130. doi: 10.1016/s0304-3940(00)01440-3. doi:10.1016/S0304-3940(00)01440-3. PMID:10998565. [DOI] [PubMed] [Google Scholar]
- Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24(12):604–612. doi: 10.1016/j.tig.2008.10.002. doi:10.1016/j.tig.2008.10.002. PMID:18992957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandoy-Dron F, Bogdanova A, Beringue V, Bailly Y, Tovey MG, Laude H, et al. Infection by ME7 prion is not modified in transgenic mice expressing the yeast chaperone Hsp104 in neurons. Neurosci. Lett. 2006;405(3):181–185. doi: 10.1016/j.neulet.2006.05.066. doi:10.1016/j.neulet.2006.05.066. PMID:16884849. [DOI] [PubMed] [Google Scholar]
- Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–890. doi: 10.1038/nature02261. doi:10.1038/nature02261. PMID:14685248. [DOI] [PubMed] [Google Scholar]
- Dorsey ER, Constantinescu R, Thompson JP, Biglan KM, Holloway RG, Kieburtz K, et al. Projected number of people with Parkinson disease in the most populous nations, 2005 through 2030. Neurology. 2007;68(5):384–386. doi: 10.1212/01.wnl.0000247740.47667.03. doi:10.1212/01.wnl.0000247740.47667.03. PMID:17082464. [DOI] [PubMed] [Google Scholar]
- Douglas PM, Treusch S, Ren HY, Halfmann R, Duennwald ML, Lindquist S, Cyr DM. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc. Natl. Acad. Sci. U.S.A. 2008;105(20):7206–7211. doi: 10.1073/pnas.0802593105. doi:10.1073/pnas.0802593105. PMID:18480252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Wickner S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem. Sci. 2009;34(1):40–48. doi: 10.1016/j.tibs.2008.09.010. doi:10.1016/j.tibs.2008.09.010. PMID:19008106. [DOI] [PubMed] [Google Scholar]
- Doyle SM, Hoskins JR, Wickner S. Collaboration between the ClpB AAA+ remodeling protein and the DnaK chaperone system. Proc. Natl. Acad. Sci. U.S.A. 2007a;104(27):11138–11144. doi: 10.1073/pnas.0703980104. doi:10.1073/pnas.0703980104. PMID:17545305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Shorter J, Zolkiewski M, Hoskins JR, Lindquist S, Wickner S. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat. Struct. Mol. Biol. 2007b;14(2):114–122. doi: 10.1038/nsmb1198. doi:10.1038/nsmb1198. PMID:17259993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Nelson R, Sawaya MR, Balbirnie M, Sambashivan S, Ivanova MI, et al. The structural biology of protein aggregation diseases: Fundamental questions and some answers. Acc. Chem. Res. 2006;39(9):568–575. doi: 10.1021/ar0500618. doi:10.1021/ar0500618. PMID:16981672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ, Minton AP. Protein aggregation in crowded environments. Biol. Chem. 2006;387(5):485–497. doi: 10.1515/BC.2006.064. doi:10.1515/BC.2006.064. PMID:16740119. [DOI] [PubMed] [Google Scholar]
- Englander SW, Mayne L, Krishna MM. Protein folding and misfolding: mechanism and principles. Q. Rev. Biophys. 2007;40(4):287–326. doi: 10.1017/S0033583508004654. PMID:18405419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigin A, Kaplitt MG, Tang C, Lin T, Mattis P, Dhawan V, et al. Modulation of metabolic brain networks after subthalamic gene therapy for Parkinson's disease. Proc. Natl. Acad. Sci. U.S.A. 2007;104(49):19559–19564. doi: 10.1073/pnas.0706006104. doi:10.1073/pnas.0706006104. PMID:18042721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman MS, Trojanowski JQ, Lee VM. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat. Med. 2004;10(10):1055–1063. doi: 10.1038/nm1113. doi:10.1038/nm1113. PMID:15459709. [DOI] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4(1) doi: 10.1371/journal.pbio.0040006. e6.doi:10.1371/journal.pbio.0040006. PMID:16300414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid – from bacteria to humans. Trends Biochem. Sci. 2007;32(5):217–224. doi: 10.1016/j.tibs.2007.03.003. doi:10.1016/j.tibs.2007.03.003. PMID:17412596. [DOI] [PubMed] [Google Scholar]
- Gestwicki JE, Crabtree GR, Graef IA. Harnessing chaperones to generate small-molecule inhibitors of amyloid beta aggregation. Science. 2004;306(5697):865–869. doi: 10.1126/science.1101262. doi:10.1126/science.1101262. PMID:15514157. [DOI] [PubMed] [Google Scholar]
- Gitler AD, Shorter J. Prime time for alpha-synuclein. J. Neurosci. 2007;27(10):2433–2434. doi: 10.1523/JNEUROSCI.0094-07.2007. doi:10.1523/JNEUROSCI.0094-07.2007. PMID:17344380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler AD, Bevis BJ, Shorter J, Strathearn KE, Hamamichi S, Su LJ, et al. The Parkinson's disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc. Natl. Acad. Sci. U.S.A. 2008;105(1):145–150. doi: 10.1073/pnas.0710685105. doi:10.1073/pnas.0710685105. PMID:18162536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenner GG, Wong CW. Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 1984;120(3):885–890. doi: 10.1016/s0006-291x(84)80190-4. doi:10.1016/S0006-291X(84)80190-4. PMID:6375662. [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94(1):73–82. doi: 10.1016/s0092-8674(00)81223-4. doi:10.1016/S0092-8674(00)81223-4. PMID:9674429. [DOI] [PubMed] [Google Scholar]
- Glover JR, Lum R. Remodeling of protein aggregates by Hsp104. Protein Pept. Lett. 2009;16(6):587–597. doi: 10.2174/092986609788490087. doi:10.2174/092986609788490087. PMID:19519516. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P, Mogk A, Zvi AP, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. U.S.A. 1999;96(24):13732–13737. doi: 10.1073/pnas.96.24.13732. doi:10.1073/pnas.96.24.13732. PMID:10570141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum EA, Graves CL, Mishizen-Eberz AJ, Lupoli MA, Lynch DR, Englander SW, et al. The E46K mutation in alpha-synuclein increases amyloid fibril formation. J. Biol. Chem. 2005;280(9):7800–7807. doi: 10.1074/jbc.M411638200. doi:10.1074/jbc.M411638200. PMID:15632170. [DOI] [PubMed] [Google Scholar]
- Griswold CK, Masel J, Úbeda F. Complex adaptations can drive the evolution of the capacitor [PSI], even with realistic rates of yeast sex. PLoS Genet. 2009;5(6) doi: 10.1371/journal.pgen.1000517. e1000517.doi:10.1371/journal.pgen.1000517. PMID:19521499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundke-Iqbal I, Iqbal K, George L, Tung YC, Kim KS, Wisniewski HM. Amyloid protein and neurofibrillary tangles coexist in the same neuron in Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 1989;86(8):2853–2857. doi: 10.1073/pnas.86.8.2853. doi:10.1073/pnas.86.8.2853. PMID:2649895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007;8(2):101–112. doi: 10.1038/nrm2101. doi:10.1038/nrm2101. PMID:17245412. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Le Deist F, Carlier F, Bouneaud C, Hue C, De Villartay JP, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N. Engl. J. Med. 2002;346(16):1185–1193. doi: 10.1056/NEJMoa012616. doi:10.1056/NEJMoa012616. PMID:11961146. [DOI] [PubMed] [Google Scholar]
- Haslberger T, Weibezahn J, Zahn R, Lee S, Tsai FT, Bukau B, Mogk A. M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol. Cell. 2007;25(2):247–260. doi: 10.1016/j.molcel.2006.11.008. doi:10.1016/j.molcel.2006.11.008. PMID:17244532. [DOI] [PubMed] [Google Scholar]
- Haslberger T, Zdanowicz A, Brand I, Kirstein J, Turgay K, Mogk A, Bukau B. Protein disaggregation by the AAA+ chaperone ClpB involves partial threading of looped polypeptide segments. Nat. Struct. Mol. Biol. 2008;15(6):641–650. doi: 10.1038/nsmb.1425. doi:10.1038/nsmb.1425. PMID:18488042. [DOI] [PubMed] [Google Scholar]
- Hattendorf DA, Lindquist SL. Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J. 2002;21(1-2):12–21. doi: 10.1093/emboj/21.1.12. doi:10.1093/emboj/21.1.12. PMID:11782421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc. Natl. Acad. Sci. U.S.A. 2008;105(43):16596–16601. doi: 10.1073/pnas.0808934105. doi:10.1073/pnas.0808934105. PMID:18955697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung GC, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics. 2006;173(2):611–620. doi: 10.1534/genetics.106.056820. doi:10.1534/genetics.106.056820. PMID:16582428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, Nukina N, Ihara Y. Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: evidence that an initially deposited species is A beta 42(43) Neuron. 1994;13(1):45–53. doi: 10.1016/0896-6273(94)90458-8. doi:10.1016/0896-6273(94)90458-8. PMID:8043280. [DOI] [PubMed] [Google Scholar]
- Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 2009;284(30):20329–20339. doi: 10.1074/jbc.M109.010264. doi:10.1074/jbc.M109.010264. PMID:19465477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplitt MG, Feigin A, Tang C, Fitzsimons HL, Mattis P, Lawlor PA, et al. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: an open label, phase I trial. Lancet. 2007;369(9579):2097–2105. doi: 10.1016/S0140-6736(07)60982-9. doi:10.1016/S0140-6736(07)60982-9. PMID:17586305. [DOI] [PubMed] [Google Scholar]
- Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300(5618):486–489. doi: 10.1126/science.1079469. doi:10.1126/science.1079469. PMID:12702875. [DOI] [PubMed] [Google Scholar]
- Kholová I, Niessen HW. Amyloid in the cardiovascular system: a review. J. Clin. Pathol. 2005;58(2):125–133. doi: 10.1136/jcp.2004.017293. doi:10.1136/jcp.2004.017293. PMID:15677530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, Masel J. The evolution of bet-hedging adaptations to rare scenarios. Theor. Popul. Biol. 2007;72(4):560–575. doi: 10.1016/j.tpb.2007.08.006. doi:10.1016/j.tpb.2007.08.006. PMID:17915273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles TP, Fitzpatrick AW, Meehan S, Mott HR, Vendruscolo M, Dobson CM, Welland ME. Role of intermolecular forces in defining material properties of protein nanofibrils. Science. 2007;318(5858):1900–1903. doi: 10.1126/science.1150057. doi:10.1126/science.1150057. PMID:18096801. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Manno A, Kakizuka A. Involvement of valosin-containing protein (VCP)/p97 in the formation and clearance of abnormal protein aggregates. Genes Cells. 2007;12(7):889–901. doi: 10.1111/j.1365-2443.2007.01099.x. doi:10.1111/j.1365-2443.2007.01099.x. PMID:17584300. [DOI] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 2009;16(6):589–597. doi: 10.1038/nsmb.1614. doi:10.1038/nsmb.1614. PMID:19491936. [DOI] [PubMed] [Google Scholar]
- Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435(7043):765–772. doi: 10.1038/nature03679. doi:10.1038/nature03679. PMID:15944694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFerla FM, Green KN, Oddo S. Intracellular amyloid-beta in Alzheimer's disease. Nat. Rev. Neurosci. 2007;8(7):499–509. doi: 10.1038/nrn2168. doi:10.1038/nrn2168. PMID:17551515. [DOI] [PubMed] [Google Scholar]
- Lansbury PT, Jr., Caughey B. The chemistry of scrapie infection: implications of the ‘ice 9’ metaphor. Chem. Biol. 1995;2(1):1–5. doi: 10.1016/1074-5521(95)90074-8. doi:10.1016/1074-5521(95)90074-8. PMID:9383397. [DOI] [PubMed] [Google Scholar]
- Larsen KE, Schmitz Y, Troyer MD, Mosharov E, Dietrich P, Quazi AZ, et al. Alpha-synuclein overexpression in PC12 and chromaffin cells impairs catecholamine release by interfering with a late step in exocytosis. J. Neurosci. 2006;26(46):11915–11922. doi: 10.1523/JNEUROSCI.3821-06.2006. doi:10.1523/JNEUROSCI.3821-06.2006. PMID:17108165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr. Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature. 2002;418(6895):291. doi: 10.1038/418291a. doi:10.1038/418291a. PMID:12124613. [DOI] [PubMed] [Google Scholar]
- Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, Yoshida M, Tsai FT. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115(2):229–240. doi: 10.1016/s0092-8674(03)00807-9. doi:10.1016/S0092-8674(03)00807-9. PMID:14567920. [DOI] [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443(7107):50–55. doi: 10.1038/nature05096. doi:10.1038/nature05096. PMID:16906134. [DOI] [PubMed] [Google Scholar]
- Lee S, Choi JM, Tsai FT. Visualizing the ATPase cycle in a protein disaggregating machine: structural basis for substrate binding by ClpB. Mol. Cell. 2007;25(2):261–271. doi: 10.1016/j.molcel.2007.01.002. doi:10.1016/j.molcel.2007.01.002. PMID:17244533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legname G, Nguyen HO, Peretz D, Cohen FE, DeArmond SJ, Prusiner SB. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc. Natl. Acad. Sci. U.S.A. 2006;103(50):19105–19110. doi: 10.1073/pnas.0608970103. doi:10.1073/pnas.0608970103. PMID:17142317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesné S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, et al. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440(7082):352–357. doi: 10.1038/nature04533. doi:10.1038/nature04533. PMID:16541076. [DOI] [PubMed] [Google Scholar]
- Lo Bianco C, Ridet JL, Schneider BL, Deglon N, Aebischer P. alpha -Synucleinopathy and selective dopaminergic neuron loss in a rat lentiviral-based model of Parkinson's disease. Proc. Natl. Acad. Sci. U.S.A. 2002;99(16):10813–10818. doi: 10.1073/pnas.152339799. doi:10.1073/pnas.152339799. PMID:12122208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Bianco C, Schneider BL, Bauer M, Sajadi A, Brice A, Iwatsubo T, Aebischer P. Lentiviral vector delivery of parkin prevents dopaminergic degeneration in an alpha-synuclein rat model of Parkinson's disease. Proc. Natl. Acad. Sci. U.S.A. 2004;101(50):17510–17515. doi: 10.1073/pnas.0405313101. doi:10.1073/pnas.0405313101. PMID:15576511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Bianco C, Shorter J, Régulier E, Lashuel H, Iwatsubo T, Lindquist S, Aebischer P. Hsp104 antagonizes alpha-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. J. Clin. Invest. 2008;118(9):3087–3097. doi: 10.1172/JCI35781. doi:10.1172/JCI35781. PMID:18704197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J. Biol. Chem. 2004;279(28):29139–29146. doi: 10.1074/jbc.M403777200. doi:10.1074/jbc.M403777200. PMID:15128736. [DOI] [PubMed] [Google Scholar]
- Lum R, Niggemann M, Glover JR. Peptide and protein binding in the axial channel of Hsp104. Insights into the mechanism of protein unfolding. J. Biol. Chem. 2008;283(44):30139–30150. doi: 10.1074/jbc.M804849200. doi:10.1074/jbc.M804849200. PMID:18755692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay RG, Helsen CW, Tkach JM, Glover JR. The C-terminal extension of Saccharomyces cerevisiae Hsp104 plays a role in oligomer assembly. Biochemistry. 2008;47(7):1918–1927. doi: 10.1021/bi701714s. doi:10.1021/bi701714s. PMID:18197703. [DOI] [PubMed] [Google Scholar]
- Maguire AM, Simonelli F, Pierce EA, Pugh EN, Jr., Mingozzi F, Bennicelli J, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N. Engl. J. Med. 2008;358(21):2240–2248. doi: 10.1056/NEJMoa0802315. doi:10.1056/NEJMoa0802315. PMID:18441370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325(5938):328–332. doi: 10.1126/science.1173155. doi:10.1126/science.1173155. PMID:19541956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins IC, Kuperstein I, Wilkinson H, Maes E, Vanbrabant M, Jonckheere W, et al. Lipids revert inert Abeta amyloid fibrils to neurotoxic protofibrils that affect learning in mice. EMBO J. 2008;27(1):224–233. doi: 10.1038/sj.emboj.7601953. doi:10.1038/sj.emboj.7601953. PMID:18059472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. U.S.A. 1985;82(12):4245–4249. doi: 10.1073/pnas.82.12.4245. doi:10.1073/pnas.82.12.4245. PMID:3159021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini G, Westermark P. The systemic amyloidoses: clearer understanding of the molecular mechanisms offers hope for more effective therapies. J. Intern. Med. 2004;255(2):159–178. doi: 10.1046/j.1365-2796.2003.01262.x. doi:10.1046/j.1365-2796.2003.01262.x. PMID:14746554. [DOI] [PubMed] [Google Scholar]
- Meyer H, Popp O. Role(s) of Cdc48/p97 in mitosis. Biochem. Soc. Trans. 2008;36(Pt 1):126–130. doi: 10.1042/BST0360126. doi:10.1042/BST0360126. PMID:18208399. [DOI] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Spires-Jones TL, Prada C, Garcia-Alloza M, de Calignon A, Rozkalne A, et al. Rapid appearance and local toxicity of amyloid-beta plaques in a mouse model of Alzheimer's disease. Nature. 2008;451(7179):720–724. doi: 10.1038/nature06616. doi:10.1038/nature06616. PMID:18256671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Tomoyasu T, Goloubinoff P, Rüdiger S, Röder D, Langen H, Bukau B. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18(24):6934–6949. doi: 10.1093/emboj/18.24.6934. doi:10.1093/emboj/18.24.6934. PMID:10601016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DJ, West AB, Dawson VL, Dawson TM. Molecular pathophysiology of Parkinson's disease. Annu. Rev. Neurosci. 2005;28(1):57–87. doi: 10.1146/annurev.neuro.28.061604.135718. doi:10.1146/annurev.neuro.28.061604.135718. PMID:16022590. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Stress, aging, and neurodegenerative disease. N. Engl. J. Med. 2006;355(21):2254–2255. doi: 10.1056/NEJMcibr065573. doi:10.1056/NEJMcibr065573. PMID:17124027. [DOI] [PubMed] [Google Scholar]
- Mosser DD, Ho S, Glover JR. Saccharomyces cerevisiae Hsp104 enhances the chaperone capacity of human cells and inhibits heat stress-induced proapoptotic signaling. Biochemistry. 2004;43(25):8107–8115. doi: 10.1021/bi0493766. doi:10.1021/bi0493766. PMID:15209506. [DOI] [PubMed] [Google Scholar]
- Narayanan S, Walter S, Reif B. Yeast prion-protein, sup35, fibril formation proceeds by addition and substraction of oligomers. ChemBioChem. 2006;7(5):757–765. doi: 10.1002/cbic.200500382. doi:10.1002/cbic.200500382. PMID:16570324. [DOI] [PubMed] [Google Scholar]
- Nekooki-Machida Y, Kurosawa M, Nukina N, Ito K, Oda T, Tanaka M. Distinct conformations of in vitro and in vivo amyloids of huntingtin-exon1 show different cytotoxicity. Proc. Natl. Acad. Sci. U.S.A. 2009;106(24):9679–9684. doi: 10.1073/pnas.0812083106. doi:10.1073/pnas.0812083106. PMID:19487684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R, Eisenberg D. Structural models of amyloid-like fibrils. Adv. Protein Chem. 2006;73:235–282. doi: 10.1016/S0065-3233(06)73008-X. doi:10.1016/S0065-3233(06)73008-X. PMID:17190616. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. doi:10.1126/science.1134108. PMID:17023659. [DOI] [PubMed] [Google Scholar]
- Nishikori S, Yamanaka K, Sakurai T, Esaki M, Ogura T. p97 Homologs from Caenorhabditis elegans, CDC-48.1 and CDC-48.2, suppress the aggregate formation of huntingtin exon1 containing expanded polyQ repeat. Genes Cells. 2008;13(8):827–838. doi: 10.1111/j.1365-2443.2008.01214.x. doi:10.1111/j.1365-2443.2008.01214.x. PMID:18782221. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993;27(1):437–496. doi: 10.1146/annurev.ge.27.120193.002253. doi:10.1146/annurev.ge.27.120193.002253. PMID:8122909. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Taulien J, Lindquist S, Viitanen P, Jaenicke R, Horwich A, et al. The role of heat-shock proteins in thermotolerance. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1993;339(1289):279–285. doi: 10.1098/rstb.1993.0026. discussion 285-286. doi:10.1098/rstb.1993.0026. PMID:8098532. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Lindquist S. Saccharomyces cerevisiae Hsp104 protein. Purification and characterization of ATP-induced structural changes. J. Biol. Chem. 1994a;269(6):4480–4487. PMID:8308017. [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994b;372(6505):475–478. doi: 10.1038/372475a0. doi:10.1038/372475a0. PMID:7984243. [DOI] [PubMed] [Google Scholar]
- Perrin V, Régulier E, Abbas-Terki T, Hassig R, Brouillet E, Aebischer P, et al. Neuroprotection by Hsp104 and Hsp27 in lentiviral-based rat models of Huntington's disease. Mol. Ther. 2007;15(5):903–911. doi: 10.1038/mt.sj.6300141. doi:10.1038/mt.sj.6300141. PMID:17375066. [DOI] [PubMed] [Google Scholar]
- Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005;307(5707):262–265. doi: 10.1126/science.1105850. doi:10.1126/science.1105850. PMID:15653506. [DOI] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 2009;78(1):959–991. doi: 10.1146/annurev.biochem.052308.114844. doi:10.1146/annurev.biochem.052308.114844. PMID:19298183. [DOI] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2000;12(4):479–492. doi: 10.1105/tpc.12.4.479. doi:10.1105/tpc.12.4.479. PMID:10760238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED, Mucke L. 100 years and counting: prospects for defeating Alzheimer's disease. Science. 2006;314(5800):781–784. doi: 10.1126/science.1132813. doi:10.1126/science.1132813. PMID:17082448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, et al. Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 1998;4(10):1157–1165. doi: 10.1038/2654. doi:10.1038/2654. PMID:9771749. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248(4959):1112–1115. doi: 10.1126/science.2188365. doi:10.1126/science.2188365. PMID:2188365. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Taulien J, Borkovich KA, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11(6):2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. PMID:1600951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, Morimoto RI. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 2000;97(11):5750–5755. doi: 10.1073/pnas.100107297. doi:10.1073/pnas.100107297. PMID:10811890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savistchenko J, Krzewska J, Fay N, Melki R. Molecular chaperones and the assembly of the prion Ure2p in vitro. J. Biol. Chem. 2008;283(23):15732–15739. doi: 10.1074/jbc.M800728200. doi:10.1074/jbc.M800728200. PMID:18400756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaupp A, Marcinowski M, Grimminger V, Bösl B, Walter S. Processing of proteins by the molecular chaperone Hsp104. J. Mol. Biol. 2007;370(4):674–686. doi: 10.1016/j.jmb.2007.04.070. doi:10.1016/j.jmb.2007.04.070. PMID:17543332. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Lindquist S. Interactions of the chaperone Hsp104 with yeast Sup35 and mammalian PrP. Proc. Natl. Acad. Sci. U.S.A. 1997;94(25):13932–13937. doi: 10.1073/pnas.94.25.13932. doi:10.1073/pnas.94.25.13932. PMID:9391130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer EC, Queitsch C, Kowal AS, Parsell DA, Lindquist S. The ATPase activity of Hsp104, effects of environmental conditions and mutations. J. Biol. Chem. 1998;273(25):15546–15552. doi: 10.1074/jbc.273.25.15546. doi:10.1074/jbc.273.25.15546. PMID:9624144. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Ware DM, Queitsch C, Kowal AS, Lindquist SL. Subunit interactions influence the biochemical and biological properties of Hsp104. Proc. Natl. Acad. Sci. U.S.A. 2001;98(3):914–919. doi: 10.1073/pnas.031568098. doi:10.1073/pnas.031568098. PMID:11158570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C, Weibezahn J, Patzelt H, Tessarz P, Strub C, Zeth K, et al. Substrate recognition by the AAA+ chaperone ClpB. Nat. Struct. Mol. Biol. 2004;11(7):607–615. doi: 10.1038/nsmb787. doi:10.1038/nsmb787. PMID:15208691. [DOI] [PubMed] [Google Scholar]
- Shorter J. Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals. 2008;16(1):63–74. doi: 10.1159/000109760. doi:10.1159/000109760. PMID:18097161. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304(5678):1793–1797. doi: 10.1126/science.1098007. doi:10.1126/science.1098007. PMID:15155912. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Navigating the ClpB channel to solution. Nat. Struct. Mol. Biol. 2005a;12(1):4–6. doi: 10.1038/nsmb0105-4. doi:10.1038/nsmb0105-4. PMID:15689967. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 2005b;6(6):435–450. doi: 10.1038/nrg1616. doi:10.1038/nrg1616. PMID:15931169. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol. Cell. 2006;23(3):425–438. doi: 10.1016/j.molcel.2006.05.042. doi:10.1016/j.molcel.2006.05.042. PMID:16885031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27(20):2712–2724. doi: 10.1038/emboj.2008.194. doi:10.1038/emboj.2008.194. PMID:18833196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115(7):879–891. doi: 10.1016/s0092-8674(03)01020-1. doi:10.1016/S0092-8674(03)01020-1. PMID:14697205. [DOI] [PubMed] [Google Scholar]
- Skovronsky DM, Lee VM, Trojanowski JQ. Neurodegenerative diseases: new concepts of pathogenesis and their therapeutic implications. Annu. Rev. Pathol. 2006;1(1):151–170. doi: 10.1146/annurev.pathol.1.110304.100113. doi:10.1146/annurev.pathol.1.110304.100113. PMID:18039111. [DOI] [PubMed] [Google Scholar]
- Smith JF, Knowles TP, Dobson CM, Macphee CE, Welland ME. Characterization of the nanoscale properties of individual amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 2006;103(43):15806–15811. doi: 10.1073/pnas.0604035103. doi:10.1073/pnas.0604035103. PMID:17038504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388(6645):839–840. doi: 10.1038/42166. doi:10.1038/42166. PMID:9278044. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Crowther RA, Jakes R, Cairns NJ, Lantos PL, Goedert M. Filamentous alpha-synuclein inclusions link multiple system atrophy with Parkinson's disease and dementia with Lewy bodies. Neurosci. Lett. 1998;251(3):205–208. doi: 10.1016/s0304-3940(98)00504-7. doi:10.1016/S0304-3940(98)00504-7. PMID:9726379. [DOI] [PubMed] [Google Scholar]
- Squires CL, Pedersen S, Ross BM, Squires C. ClpB is the Escherichia coli heat shock protein F84.1. J. Bacteriol. 1991;173(14):4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. PMID:2066329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoessl AJ. Gene therapy for Parkinson's disease: early data. Lancet. 2007;369(9579):2056–2058. doi: 10.1016/S0140-6736(07)60957-X. doi:10.1016/S0140-6736(07)60957-X. PMID:17586286. [DOI] [PubMed] [Google Scholar]
- Sunde M, Blake C. The structure of amyloid fibrils by electron microscopy and X-ray diffraction. Adv. Protein Chem. 1997;50:123–159. doi: 10.1016/s0065-3233(08)60320-4. doi:10.1016/S0065-3233(08)60320-4. PMID:9338080. [DOI] [PubMed] [Google Scholar]
- Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CC. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 1997;273(3):729–739. doi: 10.1006/jmbi.1997.1348. doi:10.1006/jmbi.1997.1348. PMID:9356260. [DOI] [PubMed] [Google Scholar]
- Sweeny EA, Shorter J. Prion proteostasis: Hsp104 meets its supporting cast. Prion. 2008;2(4):135–140. doi: 10.4161/pri.2.4.7952. PMID:19242125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442(7102):585–589. doi: 10.1038/nature04922. doi:10.1038/nature04922. PMID:16810177. [DOI] [PubMed] [Google Scholar]
- Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296(5575):1991–1995. doi: 10.1126/science.1067122. doi:10.1126/science.1067122. PMID:12065827. [DOI] [PubMed] [Google Scholar]
- Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol. Microbiol. 2008;68(1):87–97. doi: 10.1111/j.1365-2958.2008.06135.x. doi:10.1111/j.1365-2958.2008.06135.x. PMID:18312264. [DOI] [PubMed] [Google Scholar]
- Tessier PM, Lindquist S. Prion recognition elements govern nucleation, strain specificity and species barriers. Nature. 2007;447(7144):556–561. doi: 10.1038/nature05848. doi:10.1038/nature05848. PMID:17495929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms S. Cdc48 can distinguish between native and non-native proteins in the absence of cofactors. FEBS Lett. 2002;520(1-3):107–110. doi: 10.1016/s0014-5793(02)02777-1. doi:10.1016/S0014-5793(02)02777-1. PMID:12044880. [DOI] [PubMed] [Google Scholar]
- Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol. Cell. 2008;32(4):584–591. doi: 10.1016/j.molcel.2008.11.003. doi:10.1016/j.molcel.2008.11.003. PMID:19026788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449(7159):233–237. doi: 10.1038/nature06108. doi:10.1038/nature06108. PMID:17767153. [DOI] [PubMed] [Google Scholar]
- Treusch S, Cyr DM, Lindquist S. Amyloid deposits: protection against toxic protein species? Cell Cycle. 2009;8(11):1668–1674. doi: 10.4161/cc.8.11.8503. PMID:19411847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407(6803):477–483. doi: 10.1038/35035005. doi:10.1038/35035005. PMID:11028992. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Madariaga ML, Lindquist S, Weissman J. Prion switching in response to environmental stress. PLoS Biol. 2008;6(11):e294. doi: 10.1371/journal.pbio.0060294. doi:10.1371/journal.pbio.0060294. PMID:19067491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher C, Garcia-Oroz L, Rubinsztein DC. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington's disease. Hum. Mol. Genet. 2005;14(22):3425–3433. doi: 10.1093/hmg/ddi372. doi:10.1093/hmg/ddi372. PMID:16204350. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. Regulated protein degradation. Trends Biochem. Sci. 2005;30(6):283–286. doi: 10.1016/j.tibs.2005.04.005. doi:10.1016/j.tibs.2005.04.005. PMID:15950869. [DOI] [PubMed] [Google Scholar]
- Vashist S, Ng DT. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 2004;165(1):41–52. doi: 10.1083/jcb.200309132. doi:10.1083/jcb.200309132. PMID:15078901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9(12):944–957. doi: 10.1038/nrm2546. doi:10.1038/nrm2546. PMID:19002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Duennwald ML, Roberts BE, Rozeboom LM, Zhang YL, Steele AD, et al. Direct and selective elimination of specific prions and amyloids by 4,5-dianilinophthalimide and analogs. Proc. Natl. Acad. Sci. U.S.A. 2008;105(20):7159–7164. doi: 10.1073/pnas.0801934105. doi:10.1073/pnas.0801934105. PMID:18480256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119(5):653–665. doi: 10.1016/j.cell.2004.11.027. doi:10.1016/j.cell.2004.11.027. PMID:15550247. [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Schlieker C, Tessarz P, Mogk A, Bukau B. Novel insights into the mechanism of chaperone-assisted protein disaggregation. Biol. Chem. 2005;386(8):739–744. doi: 10.1515/BC.2005.086. doi:10.1515/BC.2005.086. PMID:16201868. [DOI] [PubMed] [Google Scholar]
- Wendler P, Saibil HR. Cryo-electron microscopy structures of Hsp100 proteins-crowbars in or out. Biochem. Cell Biol. 2010;88 doi: 10.1139/o09-164. this issue. [DOI] [PubMed] [Google Scholar]
- Wendler P, Shorter J, Plisson C, Cashikar AG, Lindquist S, Saibil HR. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell. 2007;131(7):1366–1377. doi: 10.1016/j.cell.2007.10.047. doi:10.1016/j.cell.2007.10.047. PMID:18160044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler P, Shorter J, Snead D, Plisson C, Clare DK, Lindquist S, Saibil HR. Motor mechanism for protein threading through Hsp104. Mol. Cell. 2009;34(1):81–92. doi: 10.1016/j.molcel.2009.02.026. doi:10.1016/j.molcel.2009.02.026. PMID:19362537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk GL. Neuropathologic changes in Alzheimer's disease. J. Clin. Psychiatry. 2003;64(Suppl 9):7–10. PMID:12934968. [PubMed] [Google Scholar]
- Werbeck ND, Schlee S, Reinstein J. Coupling and dynamics of subunits in the hexameric AAA+ chaperone ClpB. J. Mol. Biol. 2008;378(1):178–190. doi: 10.1016/j.jmb.2008.02.026. doi:10.1016/j.jmb.2008.02.026. PMID:18343405. [DOI] [PubMed] [Google Scholar]
- Wertkin AM, Turner RS, Pleasure SJ, Golde TE, Younkin SG, Trojanowski JQ, Lee VM. Human neurons derived from a teratocarcinoma cell line express solely the 695-amino acid amyloid precursor protein and produce intracellular beta-amyloid or A4 peptides. Proc. Natl. Acad. Sci. U.S.A. 1993;90(20):9513–9517. doi: 10.1073/pnas.90.20.9513. doi:10.1073/pnas.90.20.9513. PMID:8415732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 2004;5(10):781–791. doi: 10.1038/nrm1492. doi:10.1038/nrm1492. PMID:15459659. [DOI] [PubMed] [Google Scholar]