Abstract
The O-antigen polymerase of Gram-negative bacteria has been difficult to characterize. Herein we report the biochemical and functional characterization of the protein product (Wzy) of the gene annotated as the putative O-antigen polymerase, which is located in the O-antigen biosynthetic locus of Francisella tularensis. In silico analysis (homology searching, hydropathy plotting, and codon usage assessment) strongly suggested that Wzy is an O-antigen polymerase whose function is to catalyze the addition of newly synthesized O-antigen repeating units to a glycolipid consisting of lipid A, inner core polysaccharide, and one repeating unit of the O-polysaccharide (O-PS). To characterize the function of the Wzy protein, a non-polar deletion mutant of wzy was generated by allelic replacement, and the banding pattern of O-PS was observed by immunoblot analysis of whole-cell lysates obtained by SDS-PAGE and stained with an O-PS-specific monoclonal antibody. These immunoblot analyses showed that O-PS of the wzy mutant expresses only one repeating unit of O-antigen. Further biochemical characterization of the subcellular fractions of the wzy mutant demonstrated that (as is characteristic of O-antigen polymerase mutants) the low molecular weight O-antigen accumulates in the periplasm of the mutant. Site-directed mutagenesis based on protein homology and topology, which was carried out to locate a catalytic residue of the protein, showed that modification of specific residues (Gly176, Asp177, Gly323, and Tyr324) leads to a loss of O-PS polymerization. Topology models indicate that these amino acids most likely lie in close proximity on the bacterial surface.
Keywords: Fusion Protein, Lipopolysaccharide (LPS), Membrane Proteins, Site-directed Mutagenesis, Subcellular Fractionation, Francisella tularensis, O-antigen Polymerase, Characterization, wzy
Introduction
Francisella tularensis is a highly infectious Gram-negative facultative intracellular pathogen that causes tularemia, an often fatal zoonotic disease (1–4). Considered a potential biological weapon (5), F. tularensis is classified as a CDC category A select agent because of its capacity to disseminate by aerosol and to induce severe morbidity and high mortality. F. tularensis encompasses four closely related subspecies: tularensis, holarctica, mediasiatica, and novicida (6–8). F. tularensis subsp. tularensis (type A) is highly virulent to many mammalian species, including humans, and is found predominantly in North America. F. tularensis subsp. holarctica (type B), which is isolated primarily in Europe and northern Asia, causes a milder infection in humans but is still highly virulent in mice. A live attenuated vaccine strain, LVS (live vaccine strain), was derived from F. tularensis subsp. holarctica (5), but its use has not been licensed in the United States because of untoward reactions and an incomplete understanding of the precise mechanism of attenuation.
In Gram-negative bacteria, an important component of the outer membrane is the lipopolysaccharide (LPS) or endotoxin. LPS comprises three chemically linked components: lipid A, a hydrophobic glycolipid that is integral to the membrane; core polysaccharide, a nonrepeating oligosaccharide located at the membrane surface that is ketosidically linked to an N-acetylglucosamine disaccharide residue of lipid A; and O-antigen, a polysaccharide made up of repeating units of sugars extending from the membrane surface into the environment (9). The O-antigen structure varies with the bacterial species, but in most Gram-negative bacteria, each repeating unit is composed of up to eight sugars linked glycosidically to each other and to the core polysaccharide. O-antigens are responsible for many of the properties of Gram-negative bacteria, including serologic specificity, interactions with the innate immune system, and bacteriophage attachment. The structure of LPS from F. tularensis has been well described (10–14). The O-antigen of F. tularensis strains LVS and SchuS4 contains the rare sugars 2-acetamido-2,6-dideoxy-d-glucose (QuiNAc)2 and 4,6-dideoxy-4-formamido-d-glucose (Qui4NFm) as well as 2 mol of 2-acetoamido-2-deoxy-d-galacturonamide (GalNAcAN); the resulting repeat structure is 4-α-GalNAcAN-1,4-α-GalNAcAN-1,3-β-QuiNAc-1,2-β-Qui4NFm.
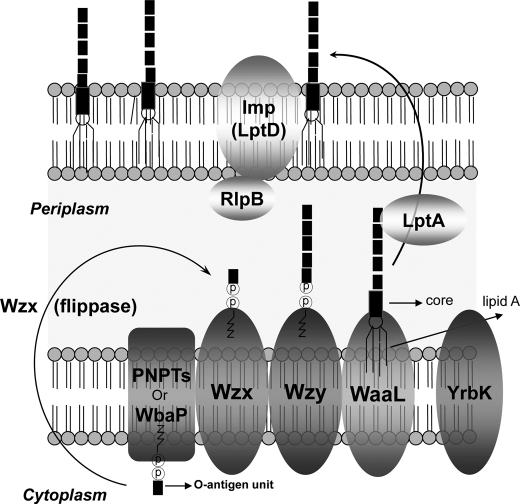
The three known modes of O-antigen assembly (Wzy-dependent, ABC transporter-dependent, and synthase-dependent) rely on export mechanisms and the presence of specific enzymes in a given pathway (15). In the Wzy-dependent pathway (Fig. 1), O-repeating unit synthesis is initiated from two different classes of integral membrane proteins: N-acetylhexosamine-1-phosphate transferase (PNPT) and polyisoprenyl-phosphate hexose-1-phosphate transferase. These proteins catalyze the transfer of sugar nucleotides to undecaprenol phosphate (16–18). The O-antigen subunits are then assembled from undecaprenol phosphate-sugar by serial actions of glycosyltransferases at the cytoplasmic face of the inner membrane (19) and are transported to the periplasmic face by the protein Wzx (flippase). Assembly and polymerization of the subunits by Wzy (O-antigen polymerase) and subsequent ligation to the lipid A core by WaaL occur at the periplasmic portion of the inner membrane (20). Finally, the entire structure is translocated to the outer membrane (21, 22) with the help of several essential proteins, including LtpA and Imp (23–25). Two genes in the F. tularensis LVS O-antigen gene cluster encode proteins whose high degree of sequence similarity to Wzy and Wzx suggests that O-antigen is transported and polymerized via the wzy-dependent pathway.
FIGURE 1.
The assembly of O-PS. The biosynthesis of O-antigen is independent of core-lipid A synthesis. O-PS biosynthesis starts on the cytoplasmic side of the cytoplasmic membrane with assembly of the repeating unit of O-unit by sequential transfer of sugars onto a lipid carrier, undecaprenol pyrophosphate. The repeating units are transferred to the periplasm and polymerized from the reducing end by the O-antigen polymerase (Wzy). The O-antigen chains are then ligated to the core-lipid A by O-antigen ligase (WaaL), and the complete LPS is translocated to the outer membrane of the bacterial cell by several essential proteins, such as LptA and Imp. WbaP is a prototype of polyisoprenyl-phosphate hexose-1-phosphate transferase. PNPT, N-acetylhexosamine-1-phosphate transferase.
The O-antigen gene cluster encoding the enzymes for the biosynthesis of nucleotide sugars and the glycosyltransferases for O-unit assembly have been found in many bacteria (16, 26–28). The F. tularensis putative O-antigen biosynthetic gene cluster has been identified (12); it is estimated to be ∼17 kb in length and is predicted to contain 15 genes involved in O-antigen biosynthesis (12). Deletion of any single gene within the locus might result in significant attenuation of the organism's virulence (29–31) because an incomplete LPS would probably be synthesized. Because most of the genes within the cluster have been assigned putative functions on the basis of sequence similarity with genes in O-antigen biosynthetic clusters from other bacteria (26, 32), additional studies are required to elucidate the actual role of each protein.
The O-antigen polymerase (Wzy) is responsible for adding oligosaccharide repeating units of the O-antigen to the LPS core region. Although Wzy is essential for O-polysaccharide synthesis, amino acid residues that are critical to the function of this protein have not been identified. The Wzy proteins from several bacterial species have been identified, and all are predicted to be integral membrane proteins with 11–13 transmembrane domains. Even at the amino acid level, these proteins show little similarity to each other in terms of primary sequence (27, 33). The absence of conserved regions has complicated the identification of catalytic and binding residues in Wzy; consequently, the mechanism of its activity has not been unraveled. Crystallographic resolution of the structure of Wzy has not been reported, perhaps because of the difficulty of defining structure in proteins that are so highly integrated into membranes. This study describes the functional and biochemical characterization of the putative Wzy in F. tularensis LVS and the identification of specific amino acid residues that play an important role in maintaining this protein's catalytic function.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
Bacterial strains and plasmids used in this study are summarized in supplemental Table 1. F. tularensis LVS (kindly provided by Dr. Karen Elkins, United States Food and Drug Administration, Bethesda, MD) was grown either in modified Muller-Hinton broth (Difco) supplemented with ferric pyrophosphate and IsoVitaleX (BD Biosciences) or in cysteine heart agar supplemented with 1% hemoglobin (CHAH) for 72 h at 37 °C in 5% CO2. Escherichia coli strain DH5α was used for cloning experiments, and Shigella flexneri 2α (kindly provided by Dr. Marcia Goldberg, Massachusetts General Hospital, Boston, MA) was grown either in Luria-Bertani (LB) broth (Difco) or on LB agar plates. Ampicillin was added to a final concentration of 100 μg/ml and kanamycin to a final concentration of 50 μg/ml in LB medium and 10 μg/ml in CHAH if antibiotic selection was necessary.
DNA Methods
Plasmids were introduced into F. tularensis LVS by electroporation (34). Miniplasmids were prepared for E. coli DH5α and F. tularensis LVS with a QIAprep® spin miniprep kit (Qiagen, Hilde, Germany). Restriction enzymes and DNA-modifying enzymes were purchased from New England Biolabs (Beverly, MA) and were used as recommended by the manufacturer. Standard molecular cloning and transformation procedures were employed (35).
Construction of the Putative wzy Mutant of F. tularensis LVS
Construction of the wzy mutant of F. tularensis LVS was based on allelic exchange of the target gene by homologous recombination (36, 37). To construct a suicide plasmid for the recombination event, two fragments (fragments 1 and 2, each consisting of ∼1,000 bp of up- and downstream regions of the putative wzy gene) were amplified by PCR with two pairs of primers. For fragment 1, the forward primer was wzYm_F1 (5′-ACCCCCGGGATAAATCTGAGTTTGAAAAAG-3′; XmaI site underlined), and the reverse primer was wzYm_R1 (5′-TGTGGTACCTTAATTAATTAAATACCACTC-3′; KpnI site underlined). For fragment 2, the forward primer was wzYm_F2 (5′-ACCGGTACCATTTGAAAAAGGTTTGTACAT-3′; KpnI site underlined), and the reverse primer was wzYm_R2 (5′-TGTGAATTCATAAGTGATATAACCGTAATA-3′; EcoRI site underlined). The two amplified fragments were subsequently ligated into the XmaI and KpnI sites (fragment 1) and into the XmaI and EcoRI sites (fragment 2) of pEX18.Kan to yield pTH29. The sacB gene, which was used as a counterselection marker, was amplified from pPV (38) and inserted into the PstI site of pTH29 to compensate for the low expression of sacB in pEX18.Kan; the final result was pTH30. For the first homologous recombination, suicide plasmid pTH30 was introduced into the target bacteria by electroporation, as described previously (34). After electroporation, the transformed cells were resuspended in 1 ml of tryptic soy broth supplemented with cysteine, incubated at 37 °C without antibiotics on a rotary shaker until an A620 of ∼0.6 was reached, and then plated onto CHAH supplemented with 10% sucrose to induce a second recombination. Individual sucrose-resistant, kanamycin-sensitive colonies were selected as the mutant candidates. PCR was used to screen and select mutants, and the selected mutants were confirmed by genomic sequencing as described previously (39).
Construction and Expression of the Wzy-GFP Fusion Protein in F. tularensis LVS
Site-directed mutant wzy genes in each plasmid were amplified by PCR with forward primer wzy_ERI_F (5′-ACCGAATTCATGTACATAAAAAAAGTG-3′; EcoRI site underlined) and reverse primer wzy_ERI_R (5′-TGTGAATTCCCAAATCACACTCCTAGT-3′; EcoRI site underlined), and ligated into the EcoRI site of pTH32, which yielded F. tularensis LVS ompA promoter-controlled Wzy-GFP fusion proteins (PompA-wzy-GFP). The constructs were introduced into F. tularensis LVS, and each transformant was subcultured and resuspended with MH broth to normalize the A value (A620 = 1.0). The intensity of GFP fluorescence was measured with a fluorometer (Fluoroskan Ascent, Theomo Electronic Corp., Waltham, MA) at wavelengths of 485 nm (excitation) and 520 nm (emission).
Immunoblot Analysis
Whole-cell lysates of wild-type and mutant F. tularensis LVS were suspended in 1× Laemmli sample buffer (Bio-Rad), heated at 100 °C for 10 min, and subjected to SDS-PAGE on a 4–20% gradient gel. For immunoblot analysis, the bands were transferred overnight onto an Immobilon-P transfer membrane (Millipore, Billerica, MA) in Towbin buffer (25 mm Tris-HCl, 192 mm glycine, 20% methanol). After transfer, the membranes were blocked in 5% alkali-soluble casein (Novagen, Madison, WI) and probed with primary antibodies at a dilution of 1:3,000 in 1% casein (Novagen) at room temperature for 1 h. Monoclonal antibody (mAb) ab2033 was purchased from Abcam Inc. (Cambridge, MA). The rabbit polyclonal antiserum to F. tularensis (α-F. tularensis LVS serum) used in this study was generated with the Classic-Line protocol of Lampire Biological Laboratories (Pipersville, PA). The appropriate secondary antibodies conjugated to alkaline phosphatase were used at a dilution of 1:3,000 in 1% casein and incubated for 1 h at room temperature. The blots were developed with an alkaline phosphatase detection kit (Novagen) according to the manufacturer's protocol.
Complementation Analysis
To express the putative wzy of F. tularensis LVS under the influence of the groEL promoter (PgroEL), the 238-bp PgroEL was amplified by PCR with the following primer pairs: PgroELRBS_F (5′-ACCGGTACCTATACCCTTCAAGCTTTG-3′; KpnI site underlined) and PgroELRBS_R (5′-TGTGAATTCGCGTTAACAATCTTACTCCTTTG-3′; EcoRI site underlined). The amplified groEL promoter fragment was ligated into the KpnI and EcoRI sites of broad host range shuttle plasmid pFNLTP6 to yield pTH17. Subsequent amplification of 1,230 bp of the putative wzy gene was carried out with primers wzyC_F (5′-ACCGAATTCATGTACATAAAAAAAGTG-3′; EcoRI site underlined) and wzyC_R (5′-TGTGGATCCTCAAATCACACTCCTAGTTGT-3′; BamHI site underlined). The amplified putative wzy fragment was then ligated into the EcoRI and BamHI sites of pTH17 to obtain pTH21. For complementation studies, recombinant plasmid pTH21 was introduced into the mutant strain under standard electroporation conditions (34), and transformants were selected on CHAH supplemented with 2% bovine hemoglobin and kanamycin (10 μg/ml) after incubation at 37 °C as described above. The clones harboring pTH21 were confirmed by DNA sequencing and were subsequently used for immunoblot analysis to confirm complementation. The other complementation study used wzy of S. flexneri 2α and was conducted according to the same scheme. The 1,149-bp S. flexneri 2α wzy gene was amplified with primers Shi_wzy_F (5′-ACCGAATTCATGAATATAAATAAAATT-3′; EcoRI site underlined) and Shi_wzy_R (5′-TGTGGATCCTTATTTTGCTCCAGAAGTGAG-3′; BamHI site underlined), and the amplified fragment was subsequently ligated into pTH17 to yield pTH33. Plasmid pTH33 was also electroporated into the F. tularensis LVS wzy mutant strain for further immunoblot analysis.
Immunoelectron Microscopy
Negative contrast immune electron microscopy was performed as previously described (40). Bacterial cells grown on CHAH plates were labeled with primary antibody (mAb ab2033) and rabbit anti-mouse bridging antibody at final dilutions of 1:20 and 1:100, respectively. Secondary antibody tagged with 15 nm of protein A gold (Cell Microscopy Center, University Medical Center, Utrecht, The Netherlands) was used at a final dilution of 1:70. All images were recorded at a primary magnification of ×18,500.
LPS Purification
LPS was purified from the F. tularensis LVS wzy mutant strain by a modification of the hot phenol-water method (41). First, cells were inoculated from CHAH plates into five 250-ml vented cap baffled flasks containing 100 ml of tryptic soy broth with 0.1% cysteine, 0.025% ferric pyrophosphate, and 0.1% antifoam 204 (all from Sigma-Aldrich). After incubation overnight at 37 °C and 200 rpm, these “seed” cells were inoculated into five 3-liter vented cap baffled flasks containing 2 liters of the broth with additives just described, one 250-ml flask per 3-liter flask, and this preparation was incubated at 37 °C and 200 rpm for 72 h. Cells were harvested by centrifugation and were frozen at −80 °C until used. For LPS purification, a hot solution of fresh 50% phenol was added to thawed cells (ratio, 1 g of cells to 5 ml of phenol solution). This preparation was mixed first for 2 h at 68 °C with sterile glass beads and an overhead mixer and then on a stir plate overnight at 4 °C. Cell debris and phenol were removed by centrifugation (6000 × g for 20 min at 4 °C) in Teflon FEP centrifuge bottles (Nalgene, Rochester, NY). The top aqueous phase was separated and diluted with one volume of water and two volumes of ether to remove additional phenol. The solution was mixed vigorously for 10 min in a separatory funnel and then allowed to separate overnight at room temperature. The bottom aqueous phase containing LPS was decanted. Residual ether was removed from the collected aqueous phase using a rotary evaporator, and the volume was reduced by lyophilization. The sample was then thawed and dialyzed against water before enzyme treatment. Nucleic acid and protein were degraded by sequential treatment with DNase, RNase, and Pronase (Worthington). The solution was collected and clarified by low speed centrifugation (5000 × g overnight at 4 °C). The supernatant was lyophilized to dryness and resuspended in PBS before further purification on a Sephacryl 300-HR 26/100 column (GE Healthcare). For LPS detection, refractive index-positive samples were screened for LPS in a competitive inhibition ELISA with wild-type LPS as the coating antigen and mAb ab2033. Fractions containing LPS were pooled and dialyzed against water. A portion of the solution was lyophilized to dryness, and the LPS concentration was determined.
Preparation of Membrane Fraction
The method used for preparation of the periplasmic portion of wild-type F. tularensis LVS and the wzy mutant was modified on the basis of previous descriptions (42, 43). The wild-type and mutant strains were cultured in 2-liter baffled flasks until an A620 of ∼1.7 (stationary phase) was reached. Cultures were subjected to centrifugation at 8,000 × g for 30 min at 10 °C for collection of cells, and the resulting cell pellets were washed with phosphate-buffered saline. Cell pellets were suspended in 35 ml of 0.75 m sucrose (in 5 mm Tris, pH 7.5) and were transferred to a sterile flask with a stir bar. A 70-ml volume of 10 mm EDTA (in 5 mm Tris, pH 7.8) was slowly added. After incubation for 30 min at room temperature, lysozyme was added slowly to a final concentration of 200 μg/ml. After further incubation for 30 min at room temperature, the suspensions were centrifuged at 7,500 × g for 30 min at 10 °C. The supernatant was dialyzed for 2 days in sterile deionized water to remove sucrose. After dialysis, the supernatant fractions that contained periplasm and outer membrane fractions were centrifuged at 210,000 × g (∼40,000 rpm in an OptimaTM LE80K, Beckman Coulter, Fullerton, CA) for 2 h at 4 °C for separation of the periplasmic (supernatant) and the outer membrane (pellet) fractions. The fractionated periplasmic portions were concentrated by lyophilization (Lyo-center, VirTis, Gardiner, NY) for further study.
Site-directed Mutagenesis
In vitro site-directed mutagenesis was performed with the QuikChange® site-directed mutagenesis kit (Stratagene, La Jolla, CA), which was used according to the manufacturer's instructions. The plasmid pTH21 was used as a template for mutant strand synthesis, and newly synthesized mutated plasmids were introduced into E. coli XL-10-Gold competent cells (Stratagene) for nick repair. Mutagenesis was confirmed by sequence analysis. After sequence verification, the plasmids harboring mutated Wzy residues were introduced into the F. tularensis LVS wzy mutant strain, and complementation of O-PS expression was assessed.
RESULTS
Analysis of F. tularensis Putative O-antigen Polymerase (Wzy)
The putative O-antigen biosynthetic gene cluster of F. tularensis is estimated to be ∼17 kb in length and has been predicted to contain 15 genes involved in O-antigen biosynthesis (12). All of the genes within the cluster were assigned putative functions on the basis of sequence similarity with genes from O-antigen biosynthetic clusters in other Gram-negative bacteria. The putative O-antigen polymerase, located at the 7th position in the cluster, showed 53.5% similarity with that of Pseudomonas aeruginosa PA 103 (accession number AAD45264), 50.8% similarity with that of Salmonella enterica subsp. enterica serovar Typhi strain CT18 (accession number NP_456180), and 48.7% similarity with that of Shigella dysenteriae Sd197 (accession number YP_403785) by ClustalW alignment (45).
The O-antigen polymerases of other bacterial species are generally extremely hydrophobic and have ≥11 putative membrane-spanning domains (27, 28, 33). The deduced number of membrane-spanning helices in the putative O-antigen polymerase of F. tularensis LVS was 11. Hydropathy plotting (46) of the O-antigen polymerase of F. tularensis LVS showed a high degree of hydrophobicity similar to that of other bacterial polymerases (data not shown).
It has been reported that the G + C content of the coding region of O-antigen polymerase is generally lower than that of the bulk DNA of the bacterium (27, 47). Indeed, the deduced G + C content of the putative O-antigen polymerase in F. tularensis LVS was 23%, a value lower than that of the bacterium itself (33.2%) (12).
Analysis of codon usage in the putative wzy gene showed a higher rate of use of “rare” or modulator codons as defined by Grosjean and Fiers (48) than in other genes (27). The rare codons are CUA (leucine), AUA (isoleucine), AGA/AGG/CGA/CGG (arginine), and GGA/GGG (glycine), and the rate of the rare codon use for the putative wzy gene in F. tularensis LVS was 10.7%; this value represents high frequency rare codon use similar to that documented for other bacterial wzy genes (e.g. S. dysenteriae, 9.2%; S. enterica M40, 10.9%; S. enterica M32, 12.5%).
Construction and Characterization of wzy Mutant
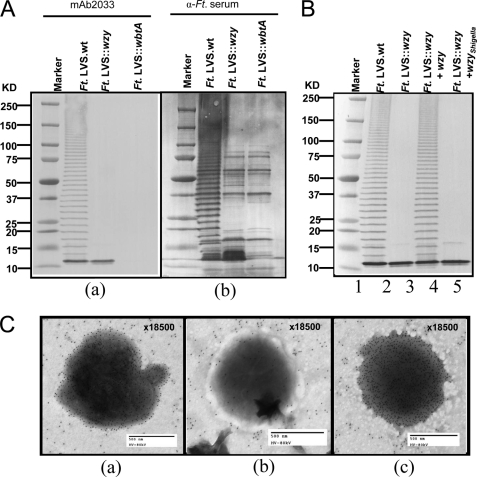
F. tularensis LVS wzy mutation was generated as described previously (36, 37) (see “Experimental Procedures”). The non-polar nature of the wzy mutant was confirmed by PCR and genomic DNA sequencing (data not shown). The expression of outer membrane O-PS in the wild-type and wzy mutant strains was investigated by immunoblot analysis. Whole-cell lysates were prepared from wild-type F. tularensis LVS, a previously described wbtA mutant (31), and the wzy mutant. WbtA is a dehydratase that is presumably necessary for the synthesis of α-d-QuiNAc, a sugar required for O-antigen subunit assembly. The wbtA mutant produces lipid A and core polysaccharide but cannot produce O-PS. Lysates of the LVS strain and its wbtA and wzy mutants were compared by immunoblot analysis with monoclonal antibody specific for the O-antigen of F. tularensis LVS (ab2033) (Fig. 2A, a); as a control to ensure that the quantities of lysate present were sufficient for visualization of O-PS should it exist, we also stained the wzy mutant and the wbtA mutant with polyclonal antiserum to LVS (Fig. 2A, b). A characteristic laddering pattern of O-PS was seen when wild-type F. tularensis LVS was examined (Fig. 2A, a); the laddering bands were dense enough to obscure the visualization of any protein in the wild-type preparation. In contrast, an accumulation of only low molecular size O-antigen was observed in the whole-cell lysate from the wzy mutant strain (Fig. 2A, a), and, as has been previously reported (31), no O-antigen was seen with the wbtA mutant (Fig. 2A, a). Probing with antiserum to whole F. tularensis LVS revealed distinct protein bands in the wzy mutant lane that were similar to those observed with the wbtA mutant (Fig. 2A, b).
FIGURE 2.
Analysis of O-PS expression. A, O-PS polymerization in F. tularensis (Ft.) LVS and F. tularensis LVS wzy mutant strain. Immunoblot analysis of whole cell lysates from F. tularensis LVS and F. tularensis LVS wzy mutant on 4–20% gradient gels (10 μl/lane), probed with either mouse monoclonal Ab ab2033 to F. tularensis O-antigen (A, a) or α-F. tularensis serum (rabbit polyclonal antibody to F. tularensis LVS) (A, b). B, complementation of wzy mutant strain by wzy of F. tularensis or S. flexneri 2α. All lanes show bacterial lysates from the indicated strain. C, the result of negative contrast immunoelectron microscopy of F. tularensis LVS (a), wbtA mutant (b), and wzy mutant (c). Immunogold-labeled LPSs are shown as black dots (see arrowheads).
Complementation of the wzy Mutant
To demonstrate that the abrogation of O-antigen polymerization in the wzy mutant strain was due solely to inactivation of the wzy gene, we complemented the phenotype by introducing an intact copy of the wzy gene in trans. Immunoblot analysis of the complemented wzy mutant demonstrated the typical ladder pattern of LPS, with complete rescue of O-antigen (Fig. 2B, lane 4). This result indicated that Wzy participates in the multienzyme complex biosynthetic pathway for O-antigen polysaccharide in F. tularensis LVS. Complementation of the wzy mutant F. tularensis strain with wzy of S. flexneri 2α was unsuccessful (Fig. 2B, lane 5), probably because of the low degree of sequence identity between the wzy genes of these two strains (16.1%).
Purification of LPS in the wzy Mutant
Although the wzy mutant cannot polymerize the O-antigen repeating unit, we hypothesized that the presence of an intact waaL gene (ligase), would result in an LPS mutant having lipid A, the core polysaccharide plus a single, non-polymerized oligosaccharide as the O-antigen unit. To examine the degree of O-PS polymerization in the wzy mutant, we purified LPS from both wild type and wzy mutant and subjected them to immunoblot analysis with mAb ab2033 specific for the O-antigen of F. tularensis. As we expected, LPS of wild-type F. tularensis LVS showed the typical laddering pattern of O-PS expression; however, the mutant strain's LPS had only one repeating unit of O-antigen (data not shown).
Outer Surface Expression of O-PS in the wzy Mutant
The surface expression of O-PS was compared for the wild-type, wzy mutant, and wbtA mutant strains of F. tularensis by negative contrast immune electron microscopy with mAb ab2033 specific for the O-antigen. O-PS was seen on the surface of the wzy mutant strain (Fig. 2C, c) and on the wild-type LVS strain (Fig. 2C, a). However, as shown previously (31), the wbtA mutant (Fig. 2C, b) had no O-PS on its surface. These are consistent with the hypothesis that the wzy mutant synthesizes and attaches only one repeating unit of O-PS to the core.
Membrane Fractionation of Putative O-polymerase Wzy and Analysis of the O-antigen Polysaccharide Expression Pattern
In the wzy-dependent O-PS biosynthetic pathway, polymerization of O-antigen subunits by Wzy occurs at the periplasmic side of the inner membrane after newly synthesized O-antigen subunits are translocated from the cytoplasm into the periplasmic region by Wzx (Fig. 1). To further examine the O-PS expression pattern of both the wzy mutant and wild-type strain at the subcellular level, periplasmic and outer membrane fractions of both the wzy mutant and the wild-type strain were isolated, and the expression of O-PS in each fraction was examined by immunoblot analyses using O-antigen-specific mAb ab2033 (Fig. 3). The purity of the subcellular fractions was confirmed by mass spectrometry (Taplin Mass Spectrometry Facility, Harvard Medical School, Boston, MA). Briefly, the isolated fractions that were analyzed by SDS-PAGE and the protein bands (including the bands that have the same molecular weight but were isolated from the different fractions) that were analyzed by MS from each fraction were determined to identify the proteins in there and to predict their subcellular location (see supplemental Figs. 1 and 2). Isolated fractions were subjected to SDS-PAGE, and the separated protein bands were analyzed by mass spectrometry to confirm the expected location of subcellular marker proteins identified in the fraction. Immunoblots of the periplasmic fractions (Fig. 3, lanes 4 and 5) showed polymerization of wild-type LPS and the presence of one repeating unit of O-antigen in the wzy mutant. Thus, the wzy mutant fails to polymerize the O-PS in the periplasm of the cell, a finding consistent with the characterization of this gene as an O-antigen polymerase. By comparing the density of the Western blots with purified known concentration standards of the LPS from LVS and from the LVS wzy mutant, it was determined that there is ∼2.6-fold more O-antigen in the outer membrane fraction of LVS (∼80 pg/2-liter culture) than the wzy mutant strain (∼30 pg/2-liter culture) and ∼2.4-fold more O-PS in the periplasmic fraction of wzy mutant (∼60 pg/2-liter culture) than parental LVS strain (∼25 pg/2-liter culture) (data not shown; the amount of LPS in each fraction had been normalized to A600 = 1.7, 2-liter culture).
FIGURE 3.
Immunoblot analysis of subcellular fractions of F. tularensis (Ft.). Patterns of O-PS polymerization were analyzed by immunoblot analysis in both wild-type and wzy mutant subcellular fractions with mAb ab2033 outer membrane fraction (lanes 2 and 3) and periplasm (lanes 4 and 5). The subcellular localization and purity of each fraction was confirmed by mass spectrometry analysis of the proteins in each sample.
Identification of Putative Catalytic Residues That Maintain the Function of Wzy
To identify catalytic residues that might be responsible for the polymerase function of Wzy, amino acid sequences of the O-antigen polymerases in various bacteria, including F. tularensis LVS, were aligned by ClustalW (45), and topology maps were made. All of the topology maps were based on the TMHMM server V.2.0 (available on the World Wide Web), the previously published topology map of Shigella (33), and other prediction programs (SOSUI, TopPred 2, and TMpred, available on the World Wide Web).
Alignment of the protein sequences from other bacteria showed that there were no obvious conserved regions of O-antigen polymerase. Therefore, we compared the Wzy protein sequences from bacteria originating from the same phylogenetic tree (for F. tularensis LVS, the γ subdivision of Proteobacteria) (Fig. 4A). Multiple alignment of eight other evolutionarily related O-antigen polymerases revealed that a Gly323 is the only absolutely conserved residue among bacterial sequences examined (Fig. 4A). On the basis of the alignment results, we chose 11 candidates for the generation of site-specific mutations by substitution of amino acid residues. Plasmids with single site-specific amino acid substitutions in F. tularensis LVS wzy were generated (see “Experimental Procedures”), and their ability to restore the expression of O-antigen in the wzy mutant was examined. The failure of the plasmid harboring the site-specific amino acid substitution to restore the expression of the O-antigen would suggest a critical role for the amino acid. Among the 11 site-specific mutations created, we observed that the substitution of Gly323 with glutamic acid, serine, and arginine (Fig. 4C, b) resulted in loss of the protein's polymerization function. We also observed the same results in substitutions with proline, tyrosine, leucine, glutamine, and aspartic acid (data not shown). The TMHMM server and the previously published topology map of Shigella (33) indicated that Gly323 is located in the periplasm. Comparison of the topology of the O-antigen polymerase in various Gram-negative bacteria predicted the presence of 4 or 5 peptide loops in the periplasm (Fig. 5). The second (or third) and fourth (or fifth) loops are generally the largest, and the fourth (or fifth) is usually larger than the second (or third). The fifth loop in F. tularensis (amino acids 268–327) is composed of >50 amino acids and has abundant glycine residues (Fig. 4B). The presence of glycine-abundant loops in the periplasm of Wzy is interesting because glycine residues have been found to contribute to the flexibility of some loops associated with enzyme catalysis (50). Topology indicated that Gly323 is located on the fifth loop, which is both the largest and most glycine-abundant.
FIGURE 4.
Putative catalytic residues of Wzy in F. tularensis LVS. A, ClustalW analysis based on the bacteria present in the same phylogeny. C.S, Chromohalobacter salexigens DSM 3043; H.H, Halorhodospira halophila SL1; M.sp, Marinobacter sp. ELB17; N.M, Nitrococcus mobilis Nb-231; P. AR, Psychrobacter arcticus 273-4; P. AT, Pseudoalteromonas atlantica T6c; S.D, Saccharophagus degradans 2–40; T.C, Thiomicrospira crunogena XCL-2; F.T, Francisella tularensis LVS. B, topology of the O-antigen polymerase in F. tularensis LVS based on TMHMM-2.0 and the previously published topology map of Shigella (33). C, immunoblot of wzy mutants harboring plasmids containing site-specific amino acid substitutions in Wzy. Alanine substitutions on the charged amino acids in the periplasmic loop of Wzy (a), mutation of the Gly323, Asp177 and their adjacent residues (b).
FIGURE 5.
Comparison of topology of O-antigen polymerase in several Gram-negative bacteria. Theoretical topology map of the O-antigen polymerases in E. coli O148 (A), Salmonella typhimurium LT2 (B), Bacteroides fragilis YCH46 (C), Pseudoalteromonas atlantica T6c (D), Marinobacter sp. ELB17 (E), and Thiomicrospira crunogena XCL-2 (F). Topology maps were predicted by TMHMM-2.0 and the previously published topology map of Shigella (33). Each circle represents an amino acid residue, and closed circles represent glycine residues. L1–L6, numbering of periplasmic loop (L).
Many active site residues are composed of amino acids with side chain charged groups, such as aspartic acid, glutamic acid, lysine, arginine, and histidine. Because polymerization is thought to occur in the periplasm, we focused our next group of experimental substitutions on the charged amino acids in the periplasmic loops of the protein. Seventeen randomly chosen candidate amino acids were mutated to alanine in an assay for O-PS expression complementation in the wzy mutant (Fig. 4C, a). Substitution of Asp177 with other amino acids resulted in a functional defect of the Wzy protein (Fig. 4C, b). We found that Asp177 is located on the third loop (amino acids 142–178), which is the second largest loop in the periplasm and also contains abundant glycine residues. Mutation of residues adjacent to Gly323 and Asp177 (Gly176, Gly178, and Tyr324) revealed a contribution of Gly176 to the catalytic performance of the protein (Fig. 4C, b). Similarly, we found that Tyr324 (adjacent to Gly323) plays a role in maintaining the function of the protein (Fig. 4C, b).
To ensure that the functional results described above were not the result of interference with Wzy expression, we determined the relative expression of Wzy in each mutant clone. Each Wzy clone was fused with a GFP reporter gene and introduced into F. tularensis LVS, and the expression level was measured by fluorometry (Fig. 6). F. tularensis LVS expressing only GFP without the Wzy (P-GFP) showed a ∼125-fold increase in fluorescence compared with that of the promoterless negative control (wzy-GFP) or the parental bacteria, which does not harbor the plasmid (LVS). The intensity of the fluorescence in bacteria expressing the intact Wzy (P-wzy-GFP) or mutagenized but still able to complement the O-PS ladder pattern (P-wzy(K152A)-GFP) was increased ∼13-fold higher than the negative controls. The expression level of Wzy in the mutant clones, which were not able to complement the O-PS ladder pattern (P-wzy(G176E)-GFP, P-wzy(D177A)-GFP, P-wzy(G323E)-GFP, and P-wzy(Y324E)-GFP) were increased 10–13-fold more than the negative control, indicating that Wzy in each mutant clone was expressed at nearly the same level as that of the intact Wzy.
FIGURE 6.
Expression of Wzy in site-directed mutagenized clones. Each mutagenized Wzy was fused with GFP reporter and expressed in F. tularensis LVS. The relative quantity of expressed Wzy was measured by fluorometry. Bacterial strains were as follows: 1) wzy-GFP, bacteria harboring the plasmid containing Wzy-GFP fusion without promoter; 2) P-GFP, bacteria harboring the plasmid containing ompA promoter-GFP fusion without Wzy; 3) P-wzy-GFP, bacteria harboring the plasmid containing ompA promoter-GFP fusion with intact Wzy; 4) P-wzy(G176E)-GFP, bacteria harboring the plasmid containing ompA promoter-Wzy (G176E)-GFP; 5) P-wzy(D177A)-GFP, bacteria harboring the plasmid containing ompA promoter-Wzy (D177A)-GFP; 6) P-wzy(G323E)-GFP, bacteria harboring the plasmid containing ompA promoter-Wzy (G323E)-GFP; 7) P-wzy(Y324E)-GFP, bacteria harboring the plasmid containing ompA promoter-Wzy (Y324E)-GFP; 8) P-wzy(K152E)-GFP, bacteria harboring the plasmid containing ompA promoter-Wzy (K352E)-GFP. All bacterial strains were normalized by A620 = 1.0.
DISCUSSION
The O-antigen polymerase (Wzy), an integral membrane protein with 11–13 transmembrane domains, is responsible for the polymerization of O-antigens (43) as well as capsules (51) and other cell surface polysaccharides (52). Studies of O-antigen polymerases of many bacteria are hindered by the absence of a conserved region within and between species, an inability to recombinantly express in vitro the protein because of its hydrophobicity, the higher rate of use of rare codons, and a weak ribosomal binding site (47). Differences in primary amino acid sequences are probably necessary because Wzy recognizes cognate O-units that usually differ significantly in structure. Although Wzy apparently mediates the formation of a typical glycosidic linkage between O-units, it displays no homology to known glycosyltransferases.
Some studies have partially characterized the O-antigen polymerase (27, 47); however, this protein has received relatively little attention because most investigations have focused on the entire O-antigen biosynthetic locus, of which Wzy is only one component (12, 33, 53). The synthesis of O-antigens involves two distinct polymerization models that are distinguished by the direction of chain elongation (54). One model entails growth at the reducing terminus and follows a wzz- and wzy-dependent polymerization pathway; the other entails growth at the nonreducing terminus and follows a wzz- and wzy-independent pathway. In the model of growth at the reducing terminus, polymerization involves the addition of nascent O-polymer from one undecaprenyl pyrophosphate carrier to the nonreducing terminus of a newly synthesized single O-unit attached to a second undecaprenyl pyrophosphate carrier (55). The presence of the putative Wzy and the complex structure of the O-antigen repeating unit in F. tularensis indicate that polymerization of this organism's O-antigen would follow the model of growth at the reducing terminus.
Feldman et al. (56) demonstrated that only a single undecaprenol phosphate-linked sugar is required for Wzx-mediated translocation from the cytoplasm to the periplasm (i.e. that the complete O-antigen subunit is not required for the activity of Wzx). Likewise, our electron microscopy study with immunogold-labeled LPS (Fig. 2C, c) showed that a single O-antigen unit also can be translocated from the periplasm to the outer membrane surface. This observation suggests that the protein involved in translocation from the periplasm to the outer surface does not require the complete set of polymerized O-antigen subunits.
It is interesting that we did not observe the typical modal distribution of O-PS molecular size in wild-type F. tularensis LVS. The modal size distribution of O-PS is thought to be dependent on the Wzz protein (22, 32, 57), the chain length determinant molecule. The absence of a Wzz homolog in F. tularensis LVS may explain why the O-PS of this strain does not follow the modal size distribution pattern of the LPS of many other bacteria expressing Wzz. Loss of modal size distribution of LPS has been reported in other organisms lacking Wzz (58).
It has previously been shown that O-antigen polymerization by wzy occurs in the periplasm (20) and that the polymerized O-antigen chain is translocated into the outer membrane. We have demonstrated that, after subcellular fractionation, O-antigens were present in the periplasmic region of both the wild-type and the mutant strain. However, in the wzy mutant, the O-PS was of very small molecular size and did not appear to be polymerized. In contrast to that for O-PS, the mechanism for translocation of synthesized capsular polysaccharide from the periplasm to the outer membrane has been well described (44, 59, 60).
As noted previously, difficulties in expressing these highly membrane-integrated proteins in vitro and the absence of a conserved motif among other O-antigen polymerases have made it difficult to elucidate the exact functional and biochemical mechanism of polymerization. On the basis of multiple alignments of hydrophobicity plots, Morona et al. (27) hypothesized that the periplasmic domain of the O-antigen polymerase is involved in bonding the O-antigen repeat units and in polymerizing them into the O-antigen chain. Moreover, McGrath and Osborn (49) reported that O-antigen polymerase is active on the periplasmic side of the cytoplasmic membrane. Studying the predicted topology of O-antigen polymerases, we observed that usually two relatively large peptide loops exist in the periplasm and that these loops have a tendency to include more glycine residues than do other loops (Fig. 5). We have now shown that some amino acids in these loops are important in maintaining the function of Wzy.
Overall, our site-directed mutagenesis study and the comparison of alignment data for bacterial species of the same phylogeny, combined with topology comparison of the proteins, revealed that the third (amino acids 142–178) and fifth (amino acids 268–327) periplasmic loops of the Wzy in F. tularensis LVS are functionally important and that a single amino acid substitution (Gly176, Asp177, Gly323, or Tyr324) leads to a loss of Wzy function. Among these residues, Asp177 may be particularly important, given that even a highly conserved substitution of Asp to Ala resulted in a functional defect in polymerization.
The studies reported herein do not differentiate mutations at a putative catalytic site from folding mutants. Loss of function, even with conservative mutations, could result from conformational changes of the periplasmic loop of the protein associated with mutation adjacent to a C-terminal membrane reinsertion point. Our findings indicate that these two glycine-abundant loops could, with the help of glycine residues, change the conformation of the Wzy protein and ultimately provide a substrate binding site for polymerization. The residues that we have found to be involved in polymerase function are all predicted to be located in adjacent loops in the periplasm near the periplasmic surface of the cytoplasmic membrane. Although it is conceivable that these residues compose the catalytic site, further studies are required to elucidate the exact mechanism by which these residues fulfill the polymerization function. However, the predicted localization of these amino acids makes it possible that the catalytic residues in the O-antigen polymerase are found therein. In summary, we have characterized the putative O-antigen polymerase in F. tularensis LVS by biochemical and functional approaches, and we have used protein alignment and topology to identify a site necessary for this protein's function.
Supplementary Material
Acknowledgments
We greatly appreciate the gifts of F. tularensis LVS from Dr. Karen Elkins and S. flexneri 2α from Dr. Marcia Goldberg. We thank Barbara G. Reinap for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health New England Regional Center of Excellence Grant R01-AI47484.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
- QuiNAc
- 2-acetamido-2,6-dideoxy-d-glucose
- Qui4NFm
- 4,6-dideoxy-4-formamido-d-glucose
- GalNAcAN
- 2-acetoamido-2-deoxy-d-galacturonamide
- O-PS
- O-polysaccharide.
REFERENCES
- 1.Ellis J., Oyston P. C., Green M., Titball R. W. (2002) Clin. Microbiol. Rev. 15, 631–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Francis E. (1925) JAMA 84, 1243–1250 [Google Scholar]
- 3.Mörner T. (1992) Rev. Sci. Tech. 11, 1123–1130 [PubMed] [Google Scholar]
- 4.Sjöstedt A. (2007) Ann. N.Y. Acad. Sci. 1105, 1–29 [DOI] [PubMed] [Google Scholar]
- 5.Dennis D. T., Inglesby T. V., Henderson D. A., Bartlett J. G., Ascher M. S., Eitzen E., Fine A. D., Friedlander A. M., Hauer J., Layton M., Lillibridge S. R., McDade J. E., Osterholm M. T., O'Toole T., Parker G., Perl T. M., Russell P. K., Tonat K. (2001) JAMA 285, 2763–2773 [DOI] [PubMed] [Google Scholar]
- 6.Forsman M., Sandström G., Jaurin B. (1990) Appl. Environ. Microbiol. 56, 949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forsman M., Sandström G., Sjöstedt A. (1994) Int. J. Syst. Bacteriol. 44, 38–46 [DOI] [PubMed] [Google Scholar]
- 8.Keim P., Johansson A., Wagner D. M. (2007) Ann. N.Y. Acad. Sci. 1105, 30–66 [DOI] [PubMed] [Google Scholar]
- 9.Jann K., Goldemann G., Weisgerber C., Wolf-Ullisch C., Kanegasaki S. (1982) Eur. J. Biochem. 127, 157–164 [DOI] [PubMed] [Google Scholar]
- 10.Dreisbach V. C., Cowley S., Elkins K. L. (2000) Infect. Immun. 68, 1988–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunn J. S., Ernst R. K. (2007) Ann. N.Y. Acad. Sci. 1105, 202–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prior J. L., Prior R. G., Hitchen P. G., Diaper H., Griffin K. F., Morris H. R., Dell A., Titball R. W. (2003) J. Med. Microbiol. 52, 845–851 [DOI] [PubMed] [Google Scholar]
- 13.Vinogradov E. V., Shashkov A. S., Knirel Y. A., Kochetkov N. K., Tochtamysheva N. V., Averin S. F., Goncharova O. V., Khlebnikov V. S. (1991) Carbohydr. Res. 214, 289–297 [DOI] [PubMed] [Google Scholar]
- 14.Vinogradov E., Perry M. B., Conlan J. W. (2002) Eur. J. Biochem. 269, 6112–6118 [DOI] [PubMed] [Google Scholar]
- 15.Raetz C. R., Whitfield C. (2002) Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexander D. C., Valvano M. A. (1994) J. Bacteriol. 176, 7079–7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keenleyside W. J., Whitfield C. (1996) J. Biol. Chem. 271, 28581–28592 [DOI] [PubMed] [Google Scholar]
- 18.Lehrer J., Vigeant K. A., Tatar L. D., Valvano M. A. (2007) J. Bacteriol. 189, 2618–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osborn M. J., Gander J. E., Parisi E. M. (1972) J. Biol. Chem. 247, 3973–3986 [PubMed] [Google Scholar]
- 20.Mulford C. A., Osborn M. J. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 1159–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Batchelor R. A., Alifano P., Biffali E., Hull S. I., Hull R. A. (1992) J Bacteriol 174, 5228–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burrows L. L., Charter D. F., Lam J. S. (1996) Mol Microbiol 22, 481–495 [DOI] [PubMed] [Google Scholar]
- 23.Bos M. P., Tefsen B., Geurtsen J., Tommassen J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9417–9422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genevrois S., Steeghs L., Roholl P., Letesson J. J., van der Ley P. (2003) EMBO J. 22, 1780–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tran A. X., Trent M. S., Whitfield C. (2008) J. Biol. Chem. 283, 20342–20349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knirel Y. A., Vinogradov E. V., Shashkov A. S., Dmitriev B. A., Kochetkov N. K., Stanislavsky E. S., Mashilova G. M. (1985) Eur. J. Biochem. 150, 541–550 [DOI] [PubMed] [Google Scholar]
- 27.Morona R., Mavris M., Fallarino A., Manning P. A. (1994) J. Bacteriol. 176, 733–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao Z., Valvano M. A. (1994) J. Bacteriol. 176, 4133–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J., Ryder C., Mandal M., Ahmed F., Azadi P., Snyder D. S., Pechous R. D., Zahrt T., Inzana T. J. (2007) Microbiology 153, 3141–3153 [DOI] [PubMed] [Google Scholar]
- 30.Raynaud C., Meibom K. L., Lety M. A., Dubail I., Candela T., Frapy E., Charbit A. (2007) Infect. Immun. 75, 536–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sebastian S., Dillon S. T., Lynch J. G., Blalock L. T., Balon E., Lee K. T., Comstock L. E., Conlan J. W., Rubin E. J., Tzianabos A. O., Kasper D. L. (2007) Infect. Immun. 75, 2591–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bélanger M., Burrows L. L., Lam J. S. (1999) Microbiology 145, 3505–3521 [DOI] [PubMed] [Google Scholar]
- 33.Daniels C., Vindurampulle C., Morona R. (1998) Mol. Microbiol. 28, 1211–1222 [DOI] [PubMed] [Google Scholar]
- 34.Baron G. S., Myltseva S. V., Nano F. E. (1995) Methods Mol. Biol. 47, 149–154 [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 36.Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998) Gene 212, 77–86 [DOI] [PubMed] [Google Scholar]
- 37.Lauriano C. M., Barker J. R., Nano F. E., Arulanandam B. P., Klose K. E. (2003) FEMS Microbiol. Lett. 229, 195–202 [DOI] [PubMed] [Google Scholar]
- 38.Golovliov I., Sjöstedt A., Mokrievich A., Pavlov V. (2003) FEMS Microbiol. Lett. 222, 273–280 [DOI] [PubMed] [Google Scholar]
- 39.Kawula T. H., Hall J. D., Fuller J. R., Craven R. R. (2004) Appl. Environ. Microbiol. 70, 6901–6904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huebner J., Wang Y., Krueger W. A., Madoff L. C., Martirosian G., Boisot S., Goldmann D. A., Kasper D. L., Tzianabos A. O., Pier G. B. (1999) Infect. Immun. 67, 1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westphal O., Jann K. (1965) Methods in Carbohydrate Chemistry, Volume V, pp. 83–91, Academic Press, Maryland Heights, MO [Google Scholar]
- 42.Brandon L. D., Goldberg M. B. (2001) J. Bacteriol. 183, 951–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huntley J. F., Conley P. G., Hagman K. E., Norgard M. V. (2007) J. Bacteriol. 189, 561–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drummelsmith J., Whitfield C. (2000) EMBO J. 19, 57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kyte J., Doolittle R. F. (1982) J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 47.Wong D. K., Morris C., Lam T. L., Wong W. K., Hackett J. (1999) Microbiology 145, 2443–2451 [DOI] [PubMed] [Google Scholar]
- 48.Grosjean H., Fiers W. (1982) Gene 18, 199–209 [DOI] [PubMed] [Google Scholar]
- 49.McGrath B. C., Osborn M. J. (1991) J. Bacteriol. 173, 649–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milne J. J., Malthouse J. P. G. (1995) Biochem. J. 311, 1015–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amor P. A., Whitfield C. (1997) Mol. Microbiol. 26, 145–161 [DOI] [PubMed] [Google Scholar]
- 52.Stevenson G., Andrianopoulos K., Hobbs M., Reeves P. R. (1996) J. Bacteriol. 178, 4885–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bengoechea J. A., Pinta E., Salminen T., Oertelt C., Holst O., Radziejewska-Lebrecht J., Piotrowska-Seget Z., Venho R., Skurnik M. (2002) J. Bacteriol. 184, 4277–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitfield C. (1995) Trends Microbiol. 3, 178–185 [DOI] [PubMed] [Google Scholar]
- 55.Robbins P. W., Bray D., Dankert B. M., Wright A. (1967) Science 158, 1536–1542 [DOI] [PubMed] [Google Scholar]
- 56.Feldman M. F., Marolda C. L., Monteiro M. A., Perry M. B., Parodi A. J., Valvano M. A. (1999) J. Biol. Chem. 274, 35129–35138 [DOI] [PubMed] [Google Scholar]
- 57.Van den Bosch L., Manning P. A., Morona R. (1997) Mol. Microbiol. 23, 765–775 [DOI] [PubMed] [Google Scholar]
- 58.Whitfield C., Amor P. A., Köplin R. (1997) Mol. Microbiol. 23, 629–638 [DOI] [PubMed] [Google Scholar]
- 59.Collins R. F., Beis K., Dong C., Botting C. H., McDonnell C., Ford R. C., Clarke B. R., Whitfield C., Naismith J. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2390–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong C., Beis K., Nesper J., Brunkan-LaMontague A. L., Clarke B. R., Whitfield C., Naismith J. H. (2006) Nature 444, 226–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.