Abstract
To study the structure, function, and interactions of proteins, a plethora of techniques is available. Many techniques sample such parameters in non-physiological environments (e.g. in air, ice, or vacuum). Atomic force microscopy (AFM), however, is a powerful biophysical technique that can probe these parameters under physiological buffer conditions. With the atomic force microscope operating under such conditions, it is possible to obtain images of biological structures without requiring labeling and to follow dynamic processes in real time. Furthermore, by operating in force spectroscopy mode, it can probe intramolecular interactions and binding strengths. In structural biology, it has proven its ability to image proteins and protein conformational changes at submolecular resolution, and in proteomics, it is developing as a tool to map surface proteomes and to study protein function by force spectroscopy methods. The power of AFM to combine studies of protein form and protein function enables bridging various research fields to come to a comprehensive, molecular level picture of biological processes. We review the use of AFM imaging and force spectroscopy techniques and discuss the major advances of these experiments in further understanding form and function of proteins at the nanoscale in physiologically relevant environments.
To understand biological processes at the molecular level it is essential to identify the involved proteins and proteinaceous assemblies, to characterize their structure and function, and to unravel their interplay with other proteins and molecules (1). Techniques like x-ray crystallography, electron microscopy, nuclear magnetic resonance spectroscopy, and mass spectrometry have contributed massively to elucidate such protein properties. These techniques can easily sample the properties of a large ensemble of proteins; however, they require subjecting the sample to harsh treatments such as drying, crystallizing, or vaporizing in vacuum, thereby limiting the range of measurable dynamical properties of the sample. One powerful method that permits the investigation of molecules in their native physiological buffer condition is atomic force microscopy (AFM)1 (2). An atomic force microscope is a microscope and force spectrometer at the same time. The imaging resolution of the atomic force microscope is comparable with that of electron microscopes, and it has the special capability to image samples in a variety of environments such as in vacuum, air, or liquid, which therefore enables studying biological specimens in their native environments (i.e. in buffer solutions) (3, 4). In addition, its ability to “touch” the sample gives it the advantage to manipulate single particles/molecules and probe their mechanical properties (5–8). However, AFM force spectroscopy is currently a technique with rather fast pulling and pushing speeds, thereby often operating out of equilibrium conditions. Improvements with ultrastable atomic force microscopes are underway to tackle this problem with promising results (9, 10). Furthermore, AFM is not well suited to apply and resolve forces at the single piconewton range due to large size tips and relatively stiff cantilevers. The issue of nonspecificity of the tip interaction with the sample is also of concern, especially in pulling experiments that require the capability to accurately recognize and select the appropriate molecule or point of interest. The current introduction of carbon nanotube tips can address the former issue (11, 12), whereas techniques in chemical functionalization can provide directed tip specificity and recognition capability (13–18), thereby further improving and widening the applicability of AFM in the future. In addition, the coupling of the atomic force microscope to fluorescence microscopes further enhances its versatility by adding (single molecule) fluorescence imaging to the AFM imaging capability (19–21), and the development of high speed systems makes it possible for AFM to probe fast dynamics of various biological processes (22–26).
The applicability of AFM in proteomics is diverse and includes the characterization of the cell surface proteome (for a recent review, see Ref. 27), label-free detection and counting of single proteins (28, 29), and force spectroscopy measurements of binding and unbinding events (30, 31). In structural biology, AFM has shown to be a powerful tool for high resolution imaging of proteins in near native conditions (3, 6) and structural studies of supramolecular assemblies like protein filaments and viruses by nanoindentation methods (32, 33). These experiments show the potential of AFM to study both “form” and “function” of proteins, thereby resolving questions in proteomics and structural biology quasi-simultaneously. In the following, we will explain the principles of atomic force microscopy and its different operation modes and finally discuss examples of imaging, nanoindentation, and protein (un)binding and unfolding studies using AFM.
PRINCIPLES OF AFM
The atomic force microscope is a member of the scanning probe microscopy techniques that utilize a probing tip that scans the surface of a sample. The measured interaction between the sample and the probe (e.g. current in scanning tunneling microscopy or force in AFM) renders a three-dimensional image of the sample surface.
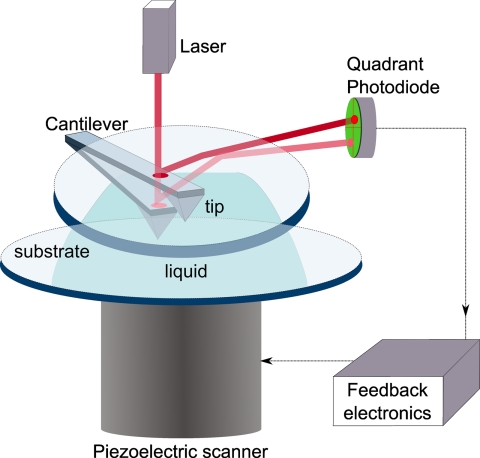
In AFM, the probe is a sharp tip with a typical radius of about 1–20 nm mounted on a cantilever. The interaction between the sample and the tip is recorded by the bending of the cantilever as shown in Fig. 1. The deflection of the cantilever is monitored by the change in direction of the reflected light (laser), which is recorded by a position-sensitive detector (quadrant photodiode). Soft cantilevers with spring constants of about 0.01–0.1 newton/m can sense forces as low as a few piconewtons, whereas modern piezoelectric scanners can translate the sample or tip in x-, y-, and z-directions with subnanometer resolutions. This combined functionality provides the atomic force microscope with the capability of rendering three-dimensional images of the sample with atomic resolution and manipulating them at single molecular level with nanoscale forces (34–37).
Fig. 1.
Schematic diagram of atomic force microscopy in liquid. The cantilever is mounted on glass to facilitate access of the laser while probing a sample in a liquid cell. Light is reflected from the cantilever and detected by the quadrant photodiode. The photodiode signal is used to instruct the piezoelectric scanner to translate the z-position of the sample through a feedback loop mechanism.
As a microscope, there are various modes available to construct an image of the sample. Choosing the appropriate imaging mode and parameters influences the amount of contact force between the sample and the probe tip (38–40). The different operation modes can be divided into static and dynamic categories (41–43). In static imaging mode, the sample is brought into physical contact with the tip until their interaction reaches a condition where it satisfies a predefined parameter setting, such as the specified z-position of the sample (constant height mode) or the amount of interaction force between the tip and sample (constant force mode). In constant height mode, the sample is moved in x-y direction while maintaining its z-position. The surface or height information of the sample is reconstructed from the deflection of the cantilever, thus the measured contact force. However, because the distance between the tip and sample is fixed, lateral dragging of the sample during imaging is quite common. In constant force mode, it is the imaging force that is set on a fixed value. The sample is brought in contact with the tip by moving the piezoelectric scanner in the z-direction until the contact force reaches the set value. The height of the sample at that position is then determined by how much the sample was moved in z. The tip is then moved to the next x-y position and using a feedback mechanism will retract or approach the sample until the set contact force is obtained. The constant force mode provides a careful and cautious way of handling the sample by setting the contact force to a minimum. However, the cantilever normally moves laterally in a continuous manner, and therefore the retraction or approach during feedback can still impart considerable dragging or damage during these lateral movements, especially in imaging soft biological samples. An alternative way of performing the constant force imaging mode is called jumping mode. This mode provides a safer scheme by retracting the tip to a defined height away from the sample before moving on to the next lateral x-y position (44). With this mode, there is less probability of dragging the sample laterally because each time it moves to another position it is first retracted away from the sample surface, which could be set such that it is above the highest structural feature. In addition, because it also operates in contact force mode, the tip-sample interaction force can be preset to the lowest possible value, thereby preventing damage to the sample.
In dynamic imaging mode, the tip is made to oscillate while it approaches the sample. The change in the oscillation amplitude or frequency during approach is used to construct its surface information. In this mode, the tip can be intermittently touching the sample (tapping mode) or not touching the sample at all (non-contact dynamic mode). In tapping mode, the contact between the tip and sample is minimized by oscillating the cantilever so that the tip bounces up and down and therefore only “taps” the surface as it moves around the sample (45). The cantilever is made to oscillate lower than the natural resonance frequency; but when it approaches the sample, the resonance frequency decreases because of damping effects. The result is an amplitude increase because the driving frequency is now closer to the new resonance. Finally, the oscillation amplitude decreases again when the tip hits the sample. The monitored change in the oscillation amplitude is used as the feedback signal for constructing the image of the sample surface. In non-contact dynamic mode, the tip is never in contact with the sample. To implement this, the cantilever is made to oscillate at a frequency higher than resonance. As it approaches the sample, its amplitude of oscillation decreases because of damping: the closer it is to the sample, the lower its amplitude of oscillation. The tip is prevented from coming too close to the sample by setting a limit on the lowest possible oscillation amplitude. As with tapping mode, this parameter is used as the feedback signal for image construction. The dynamic modes provide the option of little physical contact with the sample during imaging.
In force spectroscopy, the atomic force microscope is operating in the force-distance measurement mode (46, 47). In this mode, the force is now specifically monitored and recorded as the tip is brought into contact (e.g. pushing/indentation experiments) or out of contact from the sample (e.g. unbinding and stretching experiments) while simultaneously recording the amount of distance of approach or retraction of the tip. In this mode, no lateral movement is performed during the approach and retraction cycles. Typical applications of the spectroscopy mode in protein studies are in pushing or nanoindentation of viruses and microtubules to probe their mechanical properties (32, 33, 48) and in pulling or stretching experiments to measure the (un)binding strength of two protein molecules (49–52) or to measure the interactions of the different structural elements comprising large molecules such as muscle protein titin (53, 54), ubiquitin (55, 56), and membrane protein bacteriorhodopsin (57–59). In these experiments, the plot of the measured force against the deformation of the sample (force-distance curve) provides information on the material properties such as the spring constants, Young moduli, and the binding and unbinding, stretching and breaking forces of the probed structures.
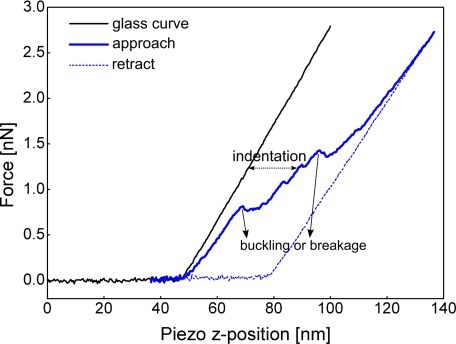
A typical force-distance curve generated by nanoindenting a virus particle is depicted in Fig. 2. In the beginning, the tip is far from the sample; thus, the measured interaction force between them is zero. At the instance the tip touches the sample surface, the cantilever starts to bend, and the force-distance curve starts to rise as the tip pushes further on the sample. When the tip is retracted, the retraction curve may simply trace back the curve during approach (in the case of a hard surface), or it retraces a completely different curve depending on the properties of the sample (37, 47, 60, 61). As illustrated in Fig. 2, the virus particle has exceeded its elastic limits during approach at about 0.8 nanonewton and thereafter suffered irreversible deformation, maybe breakage. During retraction, the tip did not interact anymore with an intact virus particle, and the retraction curve resembled that of the glass curve.
Fig. 2.
Example of force-distance curve. The plot shows a force-distance curve when the AFM tip is pushed onto a hard surface (e.g. the glass substrate) and pushing on a supramolecular proteinaceous structure. During approach, as the tip is still far from the sample, the measured force on the cantilever is zero. As the tip pushes on the sample, the force increases with the amount of distance pushed. In pushing hard samples like glass, this relationship is linear, showing the elastic property of the cantilever (glass curve). In this case, the retraction curve retraces the approach curve. For deformable protein samples, in this example a viral capsid in buffer solution, the curve may not be completely linear, and sudden drops of the force can occur. This can be interpreted as buckling or breakage in the sample. In this example, the retraction curve is very different and implies that the indented particle was irreversibly deformed during approach (for at least the time scale of the experiment and likely permanently). nN, nanonewtons.
To ensure that the measured parameters are accurate and precise, the piezoelectric scanner, spring constant of the cantilever, and photodiode signal must be calibrated. The scanner can be calibrated by imaging surfaces or microgrids with known dimensions. For the cantilever calibration, there are three different methods in measuring its spring constant: dimensional, static, and dynamic methods. Dimensional methods calculate the spring constant based on the physical dimensions and material properties of the cantilever (62, 63), and the static method is based on the deflection of the cantilever in response to a known force (64, 65), whereas the dynamic method measures the resonance frequency and Q-factor by oscillating the cantilever (66–68). Most atomic force microscopes use the beam bounce method to measure the cantilever deflection using a photodiode. Thus, an a priori calibration procedure is important to convert the photodiode signal into the actual force applied by the tip. Force calibration is usually implemented by pushing the tip on a hard surface such as a mica or glass substrate. This curve measures the deflection due to the cantilever alone, referred to as the glass curve in Fig. 2. When pushing or pulling a sample, its actual indentation or stretching can be measured by subtracting the deflection of the cantilever from the recorded force-distance curve as demonstrated by the line with double arrow in Fig. 2.
APPLICATIONS OF AFM TO PROTEIN RESEARCH
Since its development as an imaging technique, AFM has become a widespread tool in academic and industry settings for solid state and materials research (2, 39). Because it is a contact microscope, it does not suffer from diffraction limits as optical microscopes do, thereby providing it with the capability to reach atomic resolutions. Furthermore, specific tip-sample interactions such as electrostatic and magnetic forces were further exploited to characterize material properties of solid state and polymer samples. The application of AFM has since then encompassed other areas of research. In this section, we discuss the advances in imaging and force spectroscopy in the study of proteins and other biological samples.
High Resolution Imaging
AFM can render images of samples in different ways based on the measurement parameters used. The most common imaging is topographic; in this case, the image of the sample surface is reconstructed from the z-movement of the piezoelectric scanner. An intuitive image can be also be derived from the error signal, i.e. the input to the feedback loop system, which can be interpreted as the first-order derivative of the topography image. This image enhances the fine details of the sample at the expense of the coarse features. The side-to-side difference signal of the photodiode, which measures the frictional force experienced by the tip as it scans the sample (69–71), and the phase information in dynamic mode AFM (42, 72, 73) again render different aspects of the sample. These images reveal material characteristics of the sample such as viscoelastic and electrostatic properties. However, this information is quite complex and hard to quantify; thus, these images should be carefully analyzed and in most cases only serve as intuitive supplements.
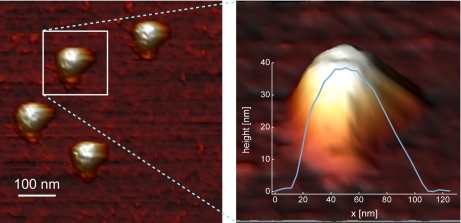
The capability of AFM to image in liquids caught the attention of the biology community and has since then been added to the palette of available techniques to image biological samples (74–78). A myriad of examples ranging from imaging cells, bacteria, and viruses to the imaging of isolated molecules such as nucleic acids (DNA and RNA) and individual proteins shows the flexibility of the atomic force microscope to image and manipulate (fragile) samples in their native state without destroying them. One of the first biological specimens that was imaged in aqueous solutions is the human blood protein fibrinogen, which was followed in real time during its clotting process (79). AFM images of the fibrinogen before and after the addition of thrombin clearly show how it transforms the protein to fibrin monomers and catalyzes the spontaneous polymerization of the monomers to chains and the subsequent cross-linking of these chains to form intricate fibrin nets. These fibrin nets play key roles in processes such as wound healing and the occurrence of blood clots that cause heart attacks and strokes. The first AFM images of isolated viruses and phages were recorded in air and were used for tip calibration and reference purposes (7, 80). Around the same time, cell-associated viruses in liquid were also imaged (81). Since then a series of experiments have been performed to improve the quality of AFM images of viruses and viral subunits (7, 82) (an example is shown in Fig. 3). Nowadays it is possible to image specific substructures like bacteriophage connectors and tails (72, 87, 88), individual capsomeres of plant and human viruses (82, 89), new viruses budding from cells (7, 71, 81, 90, 91), and expelled viral genomes (92–94) under liquid conditions.
Fig. 3.
Sample AFM image of noroviruses in liquid. On the left is a top view image showing several viral particles. In this view, structural details of the particles (cuplike depressions) can already be recognized. On the right is a zoomed-in side view image on one of the particles shown in a three-dimensional representation. The cross-sectional profile shows the apparent overestimation of the lateral dimensions of the sphere-like particle, which results from the tip convolution effect inherent to AFM imaging (80, 83). However, the diameter can be accurately determined by measuring the height. The z-profile indicates a maximum height of about 38 nm, which is in close agreement to the diameter determined by electron microscopy and x-ray crystallography (84, 85). The images were recorded using jumping mode AFM (see Ref. 86 for full experimental details with the sole difference that these images were taken in sodium phosphate buffer).
Visualization of fragile proteins in their native state such as mitochondrial and photosynthetic membrane proteins (e.g. bacteriorhodopsin) (95, 96) was also performed during the early developments of AFM. The surface topography of bacteriorhodopsin was resolved down to 1 nm (97), and since then vertical resolution of up to ∼1 Å was attained as demonstrated by the imaging of the photosynthetic core complex in native Rhodopseudomonas viridis membranes (98). The images of the latter experiment show a single reaction center encircled by an ellipsoid of 16 light-harvesting subunits. Furthermore, by applying loading forces to the tip, different protein complexes were nanodissected out of the photosynthetic core, and by imaging afterward, the structural changes and rearrangement of the protein assembly were observed (98). AFM images were also used to measure and probe material properties such as persistence lengths of DNA (99–102) and the structure and defects of lipid films (103). Aside from visualization of biological objects, topographic AFM images were also used for surface roughness analyses to study the biocompatibility of scaffolds by correlating surface characteristics with (stem) cell proliferation (104, 105) or the adsorbed proteome repertoire (106). Furthermore, time-lapsed AFM imaging enabled tracking the motion of proteins at subnanometer resolutions (107, 108), real time probing of protein-protein interactions such as the binding and dissociation of chaperonin protein GroES to individual GroEL proteins (77, 109, 110), and observing the disassembly dynamics of microtubules (111) and diffusion and transcription of Escherichia coli RNA polymerase (112, 113).
Single Particle/Molecule Manipulation and Force Measurements
Aside from topographic imaging, AFM is also widely used for single molecule manipulation and for the application of forces in force-distance measurement mode. In these measurements, which can be categorized into pushing or pulling experiments, the application of force by the tip is well controlled while the indentation or stretching of the sample is precisely monitored. The capability of AFM to physically sense interactions allows the study of inter- and intramolecular forces within the sample (114). In biological applications, experiments range from determining the mechanics and structure of cells, supramolecular assemblies, and single molecules as exemplified by (i) nanoindentation of cells (115–118), microtubules (48, 119, 120), viruses (32, 121), and globular proteins (122, 123) to probing molecular bond strengths as demonstrated by (ii) unbinding, stretching, and unfolding of receptor/ligands (124–126), proteins (51, 52, 127), and nucleic acids (37, 101, 128–130). In addition, inherent tip-sample interaction forces, which include adhesion, capillary, van der Waals, and electrostatic forces, can also be used to reveal material and chemical properties of the sample.
Pushing and Deformation Experiments
Nanoindentation experiments are performed by using the tip to push on the sample and recording the force-distance curve. In this case, the recorded deflection of the cantilever depends on the viscoelastic property of the sample (33, 60, 131). When the measured stress (force) and strain (deformation) are linearly related, the material is said to be linearly elastic, and it will normally regain its original form after relaxation. Nanoindentation experiments can measure local or global elasticity and can be expressed in terms of the Young (or elastic) modulus. Furthermore, by applying higher forces, the atomic force microscope can also probe the structural strength of the sample from the force at which it starts to irreversibly deform or break. For instance, in a subfield of the emerging research domain of physical virology (i.e. studies of material properties of viruses, forces involved in viral DNA packaging, genome ejection mechanisms, etc. (132, 133)), AFM is used to analyze the mechanical structure of viral particles. These nanoindentation experiments have shown that the Young moduli between viruses can differ significantly (32) but that one can correlate those differences with the density of the protein packing in the capsid (134). Viruses with a loose protein packing like cowpea chlorotic mottle virus (CCMV) and hepatitis B virus (HBV) have a Young modulus below 0.5 gigapascal (135, 136), whereas viruses with a capsid structure that exhibits more extensive protein-protein contacts like the bacteriophages ϕ29 and λ and the minute virus of mice (MVM) possess Young moduli of at least 1.0 gigapascal (121, 137, 138). Remarkably the differences in Young moduli also seem to reflect the packaging mode for genome encapsidation. Viruses that self-assemble around their genome (CCMV and HBV) do not need a rigid shell as it is unlikely that high pressures will be exerted onto their inner walls by the genome. However, when a packaging motor is used to encapsidate the genome into preformed capsids, such as phage ϕ29, phage λ, MVM (see also Ref. 32), and herpes simplex virus type 1 (89, 139), higher internal forces will generally be exerted onto the capsid walls.
The combination of AFM nanoindentation measurements with theoretical and modeling approaches has provided additional information on the studied viral capsids like the Young modulus of the particle and the deformation behavior under high forces (for a review on modeling approaches of viral capsids, see Ref. 140). Finite element simulations and continuum elastic theory have been applied to understand the experimental results of, for instance, phage ϕ29 (121, 140, 141), CCMV (136, 140–143), MVM (138), HBV (144), murine leukemia virus (145), human immunodeficiency virus (146), and tobacco mosaic virus (147). As these modeling approaches are generally completely reversible, they cannot explain in detail irreversible processes like plastic deformation or capsid breakage at large deformations. To do this, one needs to resort to other modeling approaches like, for instance, molecular dynamics (MD) simulations (134). Using such models permits interpretation of the experimental results at the protein level, a major step forward in our understanding of the mechanics of protein shells that is not possible with experimental techniques alone.
AFM nanoindentation experiments on other regular supramolecular assemblies like microtubules have been performed to study the material properties of these self-assembling protein tubes (48, 119, 120, 148, 149). Noteworthy is a nanoindentation study of the decoration of microtubules with the microtubule-associated protein tau (149). In this study, it was shown how tau binds in a 1-nm-thick layer around the microtubules, presumably along the protofilament tops. The radial spring constant of the microtubules was unchanged upon tau binding; however, the resistance against failure increased slightly. This experiment shows how form and function of protein assemblies can be studied using AFM. In another example of AFM studies of microtubules, not only the Young modulus but also the shear modulus of microtubules was determined. These measurements were performed by indenting microtubules that were lying over holes in the supporting substrate (120, 150). This approach was further developed for mechanical studies of a different cytoskeletal protein polymer, vimentin intermediate filaments (151). A length-dependent bending modulus was inferred from the data, suggesting axial sliding between the subunits of intermediate filaments. In a similar approach, the mechanical properties of elastic fibers and collagen fibrils were probed by freely suspending them over microchannels in a solid support (152–154). The 2 orders of magnitude difference between the Young modulus and shear modulus of type I collagen fibrils that was determined confirmed their mechanical anisotropy. As with viral capsids, the studies of protein filaments have also benefited from modeling approaches like finite element and MD simulations to determine, for instance, accurate values of the Young modulus (150, 155, 156).
The mechanical properties of selected globular proteins were also studied by AFM nanoindentation as discussed previously (33, 123). Examples include force curves on lysozyme molecules (122) and on bovine carbonic anhydrase II, a zinc metalloprotein (157). The latter experiments describe the change in mechanics of this zinc metalloprotein under increasing denaturing conditions, revealing a significant softening of the molecules at the onset of denaturation. In another experiment by the same group, the compression of surface-adsorbed green fluorescent proteins was studied. They showed a reversible quenching of the fluorescence around a force of 5 nanonewtons (158, 159). Whereas AFM nanoindentation experiments on supramolecular assemblies have provided insight into the mechanics and structure of a variety of biological complexes, such experiments on single proteins are still very scarce. This is probably because of the difficulty to interpret single molecule compression results, but this situation might change with further developments in modeling approaches (33). Contrasting with the small number of “pushing” experiments on single molecules, there has been a wealth of “pulling” experiments as we will discuss below.
Pulling Experiments
The interactions between isolated pairs of molecules, such as protein-protein or receptor-ligand bonding (17, 30, 52, 61, 160, 161), have been probed by AFM force spectroscopy as well as the distribution and interaction forces of proteins on cell surfaces (17, 27). These experiments generally require functionalized AFM tips and substrates and immobilization of the partners either on the tip or on the surface while taking force-distance curves. From these experiments, binding strengths of structural and chemical bonds and their breaking forces were measured. The binding strength between biotin and (strept)avidin for fixed loading rates (the ratio of pulling force or piezo movement over time) was probed by various groups, and averaged interaction forces of tens to hundreds of piconewtons were found (52, 124, 126, 162). To study the energy landscape of bond strengths, however, one should probe the interaction force for different loading rates (163–165). Doing so for the biotin-streptavidin bond shows that bond strengths increase with increasing loading rates (125). Next to receptor-ligand interactions, antibody-antigen interactions were also probed by AFM force spectroscopy. The unbinding force between human serum albumin and a polyclonal antibody to this molecule was found to be ∼250 piconewtons (15). The interaction of intercellular adhesion molecule-1 with a specific antibody was probed in a combined force spectroscopy and adhesion imaging approach (166). Here it was shown that topography information was correlated with specific molecular recognition. This is a nice example of how form and function of proteins can be simultaneously studied with one and the same technique. Interactions between other proteins have also been probed, for instance the binding strength between the motor protein myosin and its molecular track actin (167) and the interaction force between E. coli chaperonin GroEL and two different substrates (60). In the latter study, it was found that the binding strengths are lower in the presence of ATP and that denatured proteins bind more strongly to GroEL than proteins in a native-like state. Finally, intermolecular forces in nucleic acids were probed by AFM force spectroscopy in an approach to pull apart two single strands of DNA using complementary oligonucleotides consisting of 20 bases (129).
In AFM stretching and unfolding experiments of proteins, single or multidomain proteins are functionally attached from one end to an AFM tip and from the other end to the surface. Then the tip is pulled away from the surface, stretching and elongating each domain of the protein. In this scheme, at certain force ranges, a molecular bond or a structural element unfolds, resulting in a sudden increase of the contour length and drop of the force. Depending on the protein, this could be considered an elastic extension or an irreversible rupture, which depends on the pulling speed and temperature and solvent conditions among other factors (30, 114). The first publication among these experiments was the repeated stretching of the giant muscle protein titin, exhibiting a reversible unfolding of the individual immunoglobulin domains (127). Furthermore, it was shown that one can mechanically induce the binding of ATP to titin kinase, which means that titin kinase acts as a biological force sensor (168). Stretching experiments on enhanced yellow fluorescent protein (169) and green fluorescent protein (170) showed a complex unfolding energy landscape of the latter. In a recent high resolution force spectroscopy approach, the equilibrium fluctuations of calmodulin, a calcium-dependent signal transducer, were studied (10). It was shown that target peptides have a stabilizing function on folded calmodulin domains and that the folding kinetics of these domains is influenced by calcium ions. The atomic force microscope was also used to unzip the neck coiled coil domains of the motor protein kinesin-1 (171). As with nanoindentation experiments on microtubules and viruses and force spectroscopy of unbinding experiments, for unfolding measurements, the comparison of experimental data with simulations also have yielded additional insights into the exact molecular processes that take place. Examples include MD simulations on unfolding of specific titin domains (172, 173), titin kinase (168, 174), and green fluorescent protein (175).
CONCLUSION
In this review, we have shown the advances in the study of protein form and function using atomic force microscopy. The strength of the atomic force microscope in comparison with other biophysical techniques lies in its capability to image biological samples in their native environment and its versatility to do force spectroscopy. Hence, AFM can be considered as a worthy addition to the standard methods and techniques in protein studies as exemplified by the experiments discussed in this review. With continuing development and advancement of AFM, it is expected to become an indispensable tool in the study of single proteins and supramolecular protein assemblies to elucidate the relationships between their form (structure) and function.
Acknowledgments
We thank M. Estes, B. Prasad, and S. Crawford (Baylor College of Medicine, Houston TX) and A. Heck, G. Shoemaker, and C. Uetrecht (Universiteit Utrecht, Utrecht, The Netherlands) for providing and collaborating on the noroviruses.
* This work was supported through a Stichting voor de Technische Wetenschappen administered NanoSci-E+ grant and by the Nederlandse Organisatie voor Wetenschappelijk onderzoek through a CW-ECHO grant (both to G. J. L. W.).
1 The abbreviations used are:
- AFM
- atomic force microscopy
- CCMV
- cowpea chlorotic mottle virus
- HBV
- hepatitis B virus
- MVM
- minute virus of mice
- MD
- molecular dynamics.
REFERENCES
- 1.Heck A. J. (2008) Native mass spectrometry: a bridge between interactomics and structural biology. Nat. Methods 5, 927–933 [DOI] [PubMed] [Google Scholar]
- 2.Binnig G., Quate C. F., Gerber C. (1986) Atomic force microscope. Phys. Rev. Lett. 56, 930–933 [DOI] [PubMed] [Google Scholar]
- 3.Fechner P., Boudier T., Mangenot S., Jaroslawski S., Sturgis J. N., Scheuring S. (2009) Structural information, resolution, and noise in high-resolution atomic force microscopy topographs. Biophys. J. 96, 3822–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheuring S., Sturgis J. N. (2005) Chromatic adaptation of photosynthetic membranes. Science 309, 484–487 [DOI] [PubMed] [Google Scholar]
- 5.Henderson E. (1992) Imaging and nanodissection of individual supercoiled plasmids by atomic force microscopy. Nucleic Acids Res. 20, 445–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engel A., Müller D. J. (2000) Observing single biomolecules at work with the atomic force microscope. Nat. Struct. Biol. 7, 715–718 [DOI] [PubMed] [Google Scholar]
- 7.Baclayon M., Wuite G. J., Roos W. H. (2010) Imaging and manipulation of single viruses by atomic force microscopy. Soft Matter, doi:10.1039/b923992h [Google Scholar]
- 8.Scheuring S., Fotiadis D., Moller C., Muller S. A., Engel A., Muller D. J. (2001) Single proteins observed by atomic force microscopy. Single Mol. 2, 59–67 [Google Scholar]
- 9.King G. M., Carter A. R., Churnside A. B., Eberle L. S., Perkins T. T. (2009) Ultrastable atomic force microscopy: atomic-scale stability and registration in ambient conditions. Nano Lett. 9, 1451–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Junker J. P., Ziegler F., Rief M. (2009) Ligand-dependent equilibrium fluctuations of single calmodulin molecules. Science 323, 633–637 [DOI] [PubMed] [Google Scholar]
- 11.Hafner J. H., Cheung C. L., Woolley A. T., Lieber C. M. (2001) Structural and functional imaging with carbon nanotube AFM probes. Prog. Biophys. Mol. Biol. 77, 73–110 [DOI] [PubMed] [Google Scholar]
- 12.Cheung C. L., Hafner J. H., Lieber C. M. (2000) Carbon nanotube atomic force microscopy tips: direct growth by chemical vapor deposition and application to high-resolution imaging. Proc. Natl. Acad. Sci. U.S.A. 97, 3809–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebner A., Wildling L., Zhu R., Rankl C., Haselgrubler T., Hinterdorfer P., Gruber H. J. (2008) Functionalization of probe tips and supports for single-molecule recognition force microscopy, in STM and AFM Studies On (Bio)Molecular Systems: Unravelling the Nanoworld, pp. 29–76, Springer-Verlag, Berlin: [DOI] [PubMed] [Google Scholar]
- 14.Ebner A., Hinterdorfer P., Gruber H. J. (2007) Comparison of different aminofunctionalization strategies for attachment of single antibodies to AFM cantilevers. Ultramicroscopy 107, 922–927 [DOI] [PubMed] [Google Scholar]
- 15.Hinterdorfer P., Baumgartner W., Gruber H. J., Schilcher K., Schindler H. (1996) Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. U.S.A. 93, 3477–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong S. S., Joselevich E., Woolley A. T., Cheung C. L., Lieber C. M. (1998) Covalently functionalized nanotubes as nanometre-sized probes in chemistry and biology. Nature 394, 52–55 [DOI] [PubMed] [Google Scholar]
- 17.Hinterdorfer P., Dufrêne Y. F. (2006) Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods 3, 347–355 [DOI] [PubMed] [Google Scholar]
- 18.Ebner A., Wildling L., Kamruzzahan A. S., Rankl C., Wruss J., Hahn C. D., Hölzl M., Zhu R., Kienberger F., Blaas D., Hinterdorfer P., Gruber H. J. (2007) A new, simple method for linking of antibodies to atomic force microscopy tips. Bioconjug. Chem. 18, 1176–1184 [DOI] [PubMed] [Google Scholar]
- 19.Hards A., Zhou C., Seitz M., Bräuchle C., Zumbusch A. (2005) Simultaneous AFM manipulation and fluorescence imaging of single DNA strands. ChemPhysChem 6, 534–540 [DOI] [PubMed] [Google Scholar]
- 20.Kufer S. K., Puchner E. M., Gumpp H., Liedl T., Gaub H. E. (2008) Single-molecule cut-and-paste surface assembly. Science 319, 594–596 [DOI] [PubMed] [Google Scholar]
- 21.Kassies R., van der Werf K. O., Lenferink A., Hunter C. N., Olsen J. D., Subramaniam V., Otto C. (2005) Combined AFM and confocal fluorescence microscope for applications in bio-nanotechnology. J. Microsc. 217, 109–116 [DOI] [PubMed] [Google Scholar]
- 22.Ando T., Kodera N., Takai E., Maruyama D., Saito K., Toda A. (2001) A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl. Acad. Sci. U.S.A. 98, 12468–12472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sulchek T., Hsieh R., Adams J. D., Minne S. C., Quate C. F., Adderton D. M. (2000) High-speed atomic force microscopy in liquid. Rev. Sci. Instrum. 71, 2097–2099 [Google Scholar]
- 24.Casuso I., Kodera N., Le Grimellec C., Ando T., Scheuring S. (2009) Contact-mode high-resolution high-speed atomic force microscopy movies of the purple membrane. Biophys. J. 97, 1354–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fantner G. E., Barbero R. J., Gray D. S., Belcher A. M. (2010) Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nat. Nanotechnol. 5, 280–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansma P. K., Schitter G., Fantner G. E., Prater C. (2006) Applied physics—high-speed atomic force microscopy. Science 314, 601–602 [DOI] [PubMed] [Google Scholar]
- 27.Dufrêne Y. F. (2009) Atomic force microscopy: a powerful molecular toolkit in nanoproteomics. Proteomics 9, 5400–5405 [DOI] [PubMed] [Google Scholar]
- 28.Yu X., Xu D., Cheng Q. (2006) Label-free detection methods for protein microarrays. Proteomics 6, 5493–5503 [DOI] [PubMed] [Google Scholar]
- 29.Archakov A. I., Ivanov Y. D., Lisitsa A. V., Zgoda V. G. (2007) AFM fishing nanotechnology is the way to reverse the Avogadro number in proteomics. Proteomics 7, 4–9 [DOI] [PubMed] [Google Scholar]
- 30.Rief M., Grubmüller H. (2002) Force spectroscopy of single biomolecules. ChemPhysChem 3, 255–261 [DOI] [PubMed] [Google Scholar]
- 31.Zeng G., Chen J., Zhong L., Wang R., Jiang L., Cai J., Yan L., Huang D., Chen C. Y., Chen Z. W. (2009) NSOM- and AFM-based nanotechnology elucidates nano-structural and atomic-force features of a Y-pestis V immunogen-containing particle vaccine capable of eliciting robust response. Proteomics 9, 1538–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roos W. H., Wuite G. L. (2009) Nanoindentation studies reveal material properties of viruses. Adv. Mater. 21, 1187–1192 [Google Scholar]
- 33.Kasas S., Dietler G. (2008) Probing nanomechanical properties from biomolecules to living cells. Pflugers Arch. 456, 13–27 [DOI] [PubMed] [Google Scholar]
- 34.Binnig G., Gerber C., Stoll E., Albrecht T. R., Quate C. F. (1987) Atomic resolution with atomic force microscope. Europhys. Lett. 3, 1281–1286 [Google Scholar]
- 35.Giessibl F. J., Binnig G. (1992) Investigation of the (001) cleavage plane of potassium-bromide with an atomic force microscope at 4.2-k in ultra-high vacuum. Ultramicroscopy 42, 281–289 [Google Scholar]
- 36.Parot P., Dufrêne Y. F., Hinterdorfer P., Le Grimellec C., Navajas D., Pellequer J. L., Scheuring S. (2007) Past, present and future of atomic force microscopy in life sciences and medicine. J. Mol. Recognit. 20, 418–431 [DOI] [PubMed] [Google Scholar]
- 37.Zlatanova J., Leuba S. H. (2002) Stretching and imaging single DNA molecules and chromatin. J. Muscle Res. Cell Motil. 23, 377–395 [DOI] [PubMed] [Google Scholar]
- 38.Hansma H. G., Hoh J. H. (1994) Biomolecular imaging with the atomic-force microscope. Annu. Rev. Biophys. Biomol. Struct. 23, 115–139 [DOI] [PubMed] [Google Scholar]
- 39.Giessibl F. J. (2003) Advances in atomic force microscopy. Rev. Mod. Phys. 75, 949–983 [Google Scholar]
- 40.Jalili N., Laxminarayana K. (2004) A review of atomic force microscopy imaging systems: application to molecular metrology and biological sciences. Mechatronics 14, 907–945 [Google Scholar]
- 41.Morris V. J., Kirby A. R., Gunning A. P. (2010) Atomic Force Microscopy for Biologists, 2nd Ed., Chapter 3, Imperial College Press, London [Google Scholar]
- 42.Garcia R., Perez R. (2002) Dynamic atomic force microscopy methods. Surf. Sci. Rep. 47, 197–301 [Google Scholar]
- 43.Braga P. C., Ricci D. (2004) Atomic Force Microscopy: Biomedical Methods and Applications, Humana Press, Totowa, NJ [Google Scholar]
- 44.de Pablo P. J., Colchero J., Gomez-Herrero J., Baro A. M. (1998) Jumping mode scanning force microscopy. Appl. Phys. Lett. 73, 3300–3302 [Google Scholar]
- 45.Zhong Q., Inniss D., Kjoller K., Elings V. B. (1993) Fractured polymer silica fiber surface studied by tapping mode atomic force microscopy. Surf. Sci. 290, L688–L692 [Google Scholar]
- 46.Cappella B., Dietler G. (1999) Force-distance curves by atomic force microscopy. Surf. Sci. Rep. 34, 1–104 [Google Scholar]
- 47.Butt H. J., Cappella B., Kappl M. (2005) Force measurements with the atomic force microscope: technique, interpretation and applications. Surf. Sci. Rep. 59, 1–152 [Google Scholar]
- 48.de Pablo P. J., Schaap I. A., MacKintosh F. C., Schmidt C. F. (2003) Deformation and collapse of microtubules on the nanometer scale. Phys. Rev. Lett. 91, 098101 [DOI] [PubMed] [Google Scholar]
- 49.Chilkoti A., Boland T., Ratner B. D., Stayton P. S. (1995) The relationship between ligand-binding thermodynamics and protein-ligand interaction forces measured by atomic force microscopy. Biophys. J. 69, 2125–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takano H., Kenseth J. R., Wong S. S., O'Brien J. C., Porter M. D. (1999) Chemical and biochemical analysis using scanning force microscopy. Chem. Rev. 99, 2845–2890 [DOI] [PubMed] [Google Scholar]
- 51.Leckband D. (2000) Measuring the forces that control protein interactions. Annu. Rev. Biophys. Biomol. Struct. 29, 1–26 [DOI] [PubMed] [Google Scholar]
- 52.Zlatanova J., Lindsay S. M., Leuba S. H. (2000) Single molecule force spectroscopy in biology using the atomic force microscope. Prog. Biophys. Mol. Biol. 74, 37–61 [DOI] [PubMed] [Google Scholar]
- 53.Oberhauser A. F., Hansma P. K., Carrion-Vazquez M., Fernandez J. M. (2001) Stepwise unfolding of titin under force-clamp atomic force microscopy. Proc. Natl. Acad. Sci. U.S.A. 98, 468–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Linke W. A., Grützner A. (2008) Pulling single molecules of titin by AFM—recent advances and physiological implications. Pflugers Arch. 456, 101–115 [DOI] [PubMed] [Google Scholar]
- 55.Fernandez J. M., Li H. B. (2004) Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science 303, 1674–1678 [DOI] [PubMed] [Google Scholar]
- 56.Schlierf M., Li H., Fernandez J. M. (2004) The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc. Natl. Acad. Sci. U.S.A. 101, 7299–7304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oesterhelt F., Oesterhelt D., Pfeiffer M., Engel A., Gaub H. E., Müller D. J. (2000) Unfolding pathways of individual bacteriorhodopsins. Science 288, 143–146 [DOI] [PubMed] [Google Scholar]
- 58.Kedrov A., Janovjak H., Sapra K. T., Müller D. J. (2007) Deciphering molecular interactions of native membrane proteins by single-molecule force spectroscopy. Annu. Rev. Biophys. Biomol. Struct. 36, 233–260 [DOI] [PubMed] [Google Scholar]
- 59.Fisher T. E., Marszalek P. E., Fernandez J. M. (2000) Stretching single molecules into novel conformations using the atomic force microscope. Nat. Struct. Biol. 7, 719–724 [DOI] [PubMed] [Google Scholar]
- 60.Vinckier A., Gervasoni P., Zaugg F., Ziegler U., Lindner P., Groscurth P., Plückthun A., Semenza G. (1998) Atomic force microscopy detects changes in the interaction forces between GroEL and substrate proteins. Biophys. J. 74, 3256–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willemsen O. H., Snel M. M., Cambi A., Greve J., De Grooth B. G., Figdor C. G. (2000) Biomolecular interactions measured by atomic force microscopy. Biophys. J. 79, 3267–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Senden T. J., Ducker W. A. (1994) Experimental determination of spring constants in atomic force microscopy. Langmuir 10, 1003–1004 [Google Scholar]
- 63.Clifford C. A., Seah M. P. (2005) The determination of atomic force microscope cantilever spring constants via dimensional methods for nanomechanical analysis. Nanotechnology 16, 1666–1680 [Google Scholar]
- 64.Chen G. Y., Warmack R. J., Thundat T., Allison D. P., Huang A. (1994) Resonance response of scanning force microscopy cantilevers. Rev. Sci. Instrum. 65, 2532–2537 [Google Scholar]
- 65.Albrecht T. R., Akamine S., Carver T. E., Quate C. F. (1990) Microfabrication of cantilever styli for the atomic force microscope. J. Vac. Sci. Technol. A 8, 3386–3396 [Google Scholar]
- 66.Hutter J. L., Bechhoefer J. (1993) Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 64, 1868–1873 [Google Scholar]
- 67.Sader J. E., Larson I., Mulvaney P., White L. R. (1995) Method for the calibration of atomic force microscope cantilevers. Rev. Sci. Instrum. 66, 3789–3798 [Google Scholar]
- 68.Sader J. E., Chon J. W., Mulvaney P. (1999) Calibration of rectangular atomic force microscope cantilevers. Rev. Sci. Instrum. 70, 3967–3969 [Google Scholar]
- 69.Ogletree D. F., Carpick R. W., Salmeron M. (1996) Calibration of frictional forces in atomic force microscopy. Rev. Sci. Instrum. 67, 3298–3306 [Google Scholar]
- 70.Staii C., Wood D. W., Scoles G. (2008) Verification of biochemical activity for proteins nanografted on gold surfaces. J. Am. Chem. Soc. 130, 640–646 [DOI] [PubMed] [Google Scholar]
- 71.Gladnikoff M., Shimoni E., Gov N. S., Rousso I. (2009) Retroviral Assembly and Budding Occur through an Actin-Driven Mechanism. Biophys. J. 97, 2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Melcher J., Carrasco C., Xu X., Carrascosa J. L., Gómez-Herrero J., José de Pablo P., Raman A. (2009) Origins of phase contrast in the atomic force microscope in liquids. Proc. Natl. Acad. Sci. U.S.A. 106, 13655–13660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Magonov S. N., Elings V., Whangbo M. H. (1997) Phase imaging and stiffness in tapping-mode atomic force microscopy. Surf. Sci. 375, L385–L391 [Google Scholar]
- 74.Worcester D. L., Miller R. G., Bryant P. J. (1988) Atomic force microscopy of purple membranes. J. Microsc. 152, 817–821 [DOI] [PubMed] [Google Scholar]
- 75.Lyubchenko Y. L., Gall A. A., Shlyakhtenko L. S., Harrington R. E., Jacobs B. L., Oden P. I., Lindsay S. M. (1992) Atomic force microscopy imaging of double-stranded DNA and RNA. J. Biomol. Struct. Dyn. 10, 589–606 [DOI] [PubMed] [Google Scholar]
- 76.Hansma H. G., Bezanilla M., Zenhausern F., Adrian M., Sinsheimer R. L. (1993) Atomic force microscopy of DNA in aqueous solutions. Nucleic Acids Res. 21, 505–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Müller D. J., Janovjak H., Lehto T., Kuerschner L., Anderson K. (2002) Observing structure, function and assembly of single proteins by AFM. Prog. Biophys. Mol. Biol. 79, 1–43 [DOI] [PubMed] [Google Scholar]
- 78.Kasas S., Thomson N. H., Smith B. L., Hansma P. K., Miklossy J., Hansma H. G. (1997) Biological applications of the AFM: from single molecules to organs. Int. J. Imaging Syst. Technol. 8, 151–161 [Google Scholar]
- 79.Drake B., Prater C. B., Weisenhorn A. L., Gould S. A., Albrecht T. R., Quate C. F., Cannell D. S., Hansma H. G., Hansma P. K. (1989) Imaging crystals, polymers, and processes in water with the atomic force microscope. Science 243, 1586–1589 [DOI] [PubMed] [Google Scholar]
- 80.Thundat T., Zheng X. Y., Sharp S. L., Allison D. P., Warmack R. J., Joy D. C., Ferrell T. L. (1992) Calibration of atomic force microscope tips using biomolecules. Scanning Microsc. 6, 903–910 [PubMed] [Google Scholar]
- 81.Häberle W., Hörber J. K., Ohnesorge F., Smith D. P., Binnig G. (1992) In situ investigations of single living cells infected by viruses. Ultramicroscopy 42, 1161–1167 [DOI] [PubMed] [Google Scholar]
- 82.Kuznetsov Y. G., Malkin A. J., Lucas R. W., Plomp M., McPherson A. (2001) Imaging of viruses by atomic force microscopy. J. Gen. Virol. 82, 2025–2034 [DOI] [PubMed] [Google Scholar]
- 83.Allen M. J., Hud N. V., Balooch M., Tench R. J., Siekhaus W. J., Balhorn R. (1992) Tip-radius-induced artifacts in AFM images of protamine-complexed DNA fibers. Ultramicroscopy 42, 1095–1100 [DOI] [PubMed] [Google Scholar]
- 84.Prasad B. V., Rothnagel R., Jiang X., Estes M. K. (1994) 3-Dimensional structure of Baculovirus-expressed Norwalk virus capsids. J. Virol. 68, 5117–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prasad B. V., Hardy M. E., Dokland T., Bella J., Rossmann M. G., Estes M. K. (1999) X-ray crystallographic structure of the Norwalk virus capsid. Science 286, 287–290 [DOI] [PubMed] [Google Scholar]
- 86.Shoemaker G. K., van Duijn E., Crawford S. E., Uetrecht C., Baclayon M., Roos W. H., Wuite G. J., Estes M. K., Prasad B. V., Heck A. J. (April22, 2010) Norwalk virus assembly and stability monitored by mass spectrometry. Mol. Cell. Proteomics 10.1074/mcp.M900620-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Valle M., Valpuesta J. M., Carrascosa J. L., Tamayo J., Garcia R. (1996) The interaction of DNA with bacteriophage phi 29 connector: a study by AFM and TEM. J. Struct. Biol. 116, 390–398 [DOI] [PubMed] [Google Scholar]
- 88.Müller D. J., Engel A., Carrascosa J. L., Vélez M. (1997) The bacteriophage phi 29 head-tail connector imaged at high resolution with the atomic force microscope in buffer solution. EMBO J. 16, 2547–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roos W. H., Radtke K., Kniesmeijer E., Geertsema H., Sodeik B., Wuite G. J. L. (2009) Scaffold expulsion and genome packaging trigger stabilization of herpes simplex virus capsids. Proc. Natl. Acad. Sci. U.S.A. 106, 9673–9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gladnikoff M., Rousso I. (2008) Directly monitoring individual retrovirus budding events using atomic force microscopy. Biophys. J. 94, 320–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ohnesorge F. M., Hörber J. K., Häberle W., Czerny C. P., Smith D. P., Binnig G. (1997) AFM review study on pox viruses and living cells. Biophys. J. 73, 2183–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kolbe W. F., Ogletree D. F., Salmeron M. B. (1992) Atomic force microscopy imaging of T4 bacteriophages on silicon substrates. Ultramicroscopy 42, 1113–1117 [DOI] [PubMed] [Google Scholar]
- 93.Kuznetsov Y. G., Daijogo S., Zhou J., Semler B. L., McPherson A. (2005) Atomic force microscopy analysis of icosahedral virus RNA. J. Mol. Biol. 347, 41–52 [DOI] [PubMed] [Google Scholar]
- 94.Kienberger F., Zhu R., Moser R., Blaas D., Hinterdorfer P. (2004) Monitoring RNA release from human rhinovirus by dynamic force microscopy. J. Virol. 78, 3203–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Müller D. J., Sapra K. T., Scheuring S., Kedrov A., Frederix P. L., Fotiadis D., Engel A. (2006) Single-molecule studies of membrane proteins. Curr. Opin. Struct. Biol. 16, 489–495 [DOI] [PubMed] [Google Scholar]
- 96.Engel A., Gaub H. E. (2008) Structure and mechanics of membrane proteins. Annu. Rev. Biochem. 77, 127–148 [DOI] [PubMed] [Google Scholar]
- 97.Butt H. J., Downing K. H., Hansma P. K. (1990) Imaging the membrane-protein bacteriorhodopsin with the atomic force microscope. Biophys. J. 58, 1473–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scheuring S., Seguin J., Marco S., Lévy D., Robert B., Rigaud J. L. (2003) Nanodissection and high-resolution imaging of the Rhodopseudomonas viridis photosynthetic core complex in native membranes by AFM. Proc. Natl. Acad. Sci. U.S.A. 100, 1690–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moukhtar J., Fontaine E., Faivre-Moskalenko C., Arneodo A. (2007) Probing persistence in DNA curvature properties with atomic force microscopy. Phys. Rev. Lett. 98, 178101. [DOI] [PubMed] [Google Scholar]
- 100.Lyubchenko Y. L., Shlyakhtenko L. S. (1997) Visualization of supercoiled DNA with atomic force microscopy in situ. Proc. Natl. Acad. Sci. U.S.A. 94, 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hansma H. G., Kim K. J., Laney D. E., Garcia R. A., Argaman M., Allen M. J., Parsons S. M. (1997) Properties of biomolecules measured from atomic force microscope images: a review. J. Struct. Biol. 119, 99–108 [DOI] [PubMed] [Google Scholar]
- 102.Hansma H. G., Vesenka J., Siegerist C., Kelderman G., Morrett H., Sinsheimer R. L., Elings V., Bustamante C., Hansma P. K. (1992) Reproducible imaging and dissection of plasmid DNA under liquid with the atomic force microscope. Science 256, 1180–1184 [DOI] [PubMed] [Google Scholar]
- 103.Dufrêne Y. F., Lee G. U. (2000) Advances in the characterization of supported lipid films with the atomic force microscope. Biochim. Biophys. Acta 1509, 14–41 [DOI] [PubMed] [Google Scholar]
- 104.Domke J., Dannöhl S., Parak W. J., Müller O., Aicher W. K., Radmacher M. (2000) Substrate dependent differences in morphology and elasticity of living osteoblasts investigated by atomic force microscopy. Colloids Surf. B Biointerfaces 19, 367–379 [DOI] [PubMed] [Google Scholar]
- 105.Kroeze R. J., Helder M. N., Roos W. H., Wuite G. J., Bank R. A., Smit T. H. (2010) The effect of ethylene oxide, glow discharge and electron beam on the surface characteristics of poly(L-lactide-co-caprolactone) and the corresponding cellular response of adipose stem cells. Acta Biomater. 6, 2060–2065 [DOI] [PubMed] [Google Scholar]
- 106.Bonomini M., Pavone B., Sirolli V., Del Buono F., Di Cesare M., Del Boccio P., Amoroso L., Di Ilio C., Sacchetta P., Federici G., Urbani A. (2006) Proteomics characterization of protein adsorption onto hemodialysis membranes. J. Proteome Res. 5, 2666–2674 [DOI] [PubMed] [Google Scholar]
- 107.Thomson N. H., Fritz M., Radmacher M., Cleveland J. P., Schmidt C. F., Hansma P. K. (1996) Protein tracking and detection of protein motion using atomic force microscopy. Biophys. J. 70, 2421–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Müller D. J., Engel A., Matthey U., Meier T., Dimroth P., Suda K. (2003) Observing membrane protein diffusion at subnanometer resolution. J. Mol. Biol. 327, 925–930 [DOI] [PubMed] [Google Scholar]
- 109.Viani M. B., Pietrasanta L. I., Thompson J. B., Chand A., Gebeshuber I. C., Kindt J. H., Richter M., Hansma H. G., Hansma P. K. (2000) Probing protein-protein interactions in real time. Nat. Struct. Biol. 7, 644–647 [DOI] [PubMed] [Google Scholar]
- 110.Yokokawa M., Wada C., Ando T., Sakai N., Yagi A., Yoshimura S. H., Takeyasu K. (2006) Fast-scanning atomic force microscopy reveals the ATP/ADP-dependent conformational changes of GroEL. EMBO J. 25, 4567–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thomson N. H., Kasas S., Riederer B. M., Catsicas S., Dietler G., Kulik A. J., Forró L. (2003) Large fluctuations in the disassembly rate of microtubules revealed by atomic force microscopy. Ultramicroscopy 97, 239–247 [DOI] [PubMed] [Google Scholar]
- 112.Guthold M., Zhu X., Rivetti C., Yang G., Thomson N. H., Kasas S., Hansma H. G., Smith B., Hansma P. K., Bustamante C. (1999) Direct observation of one-dimensional diffusion and transcription by Escherichia coli RNA polymerase. Biophys. J. 77, 2284–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kasas S., Thomson N. H., Smith B. L., Hansma H. G., Zhu X., Guthold M., Bustamante C., Kool E. T., Kashlev M., Hansma P. K. (1997) Escherichia coli RNA polymerase activity observed using atomic force microscopy. Biochemistry 36, 461–468 [DOI] [PubMed] [Google Scholar]
- 114.Puchner E. M., Gaub H. E. (2009) Force and function: probing proteins with AFM-based force spectroscopy. Curr. Opin. Struct. Biol. 19, 605–614 [DOI] [PubMed] [Google Scholar]
- 115.Rotsch C., Jacobson K., Radmacher M. (1999) Dimensional and mechanical dynamics of active and stable edges in motile fibroblasts investigated by using atomic force microscopy. Proc. Natl. Acad. Sci. U.S.A. 96, 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mahaffy R. E., Shih C. K., MacKintosh F. C., Käs J. (2000) Scanning probe-based frequency-dependent microrheology of polymer gels and biological cells. Phys. Rev. Lett. 85, 880–883 [DOI] [PubMed] [Google Scholar]
- 117.Li Q. S., Lee G. Y., Ong C. N., Lim C. T. (2008) AFM indentation study of breast cancer cells. Biochem. Biophys. Res. Commun. 374, 609–613 [DOI] [PubMed] [Google Scholar]
- 118.Radmacher M., Fritz M., Kacher C. M., Cleveland J. P., Hansma P. K. (1996) Measuring the viscoelastic properties of human platelets with the atomic force microscope. Biophys. J. 70, 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vinckier A., Dumortier C., Engelborghs Y., Hellemans L. (1996) Dynamical and mechanical study of immobilized microtubules with atomic force microscopy. J. Vac. Sci. Technol. B Microelectron. Nanometer Struct. Process. Meas. Phenom. 14, 1427–1431 [Google Scholar]
- 120.Kis A., Kasas S., Babiæ B., Kulik A. J., Benoît W., Briggs G. A., Schönenberger C., Catsicas S., Forró L. (2002) Nanomechanics of microtubules. Phys. Rev. Lett. 89, 248101. [DOI] [PubMed] [Google Scholar]
- 121.Ivanovska I. L., de Pablo P. J., Ibarra B., Sgalari G., MacKintosh F. C., Carrascosa J. L., Schmidt C. F., Wuite G. J. L. (2004) Bacteriophage capsids: tough nanoshells with complex elastic properties. Proc. Natl. Acad. Sci. U.S.A. 101, 7600–7605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Radmacher M., Fritz M., Cleveland J. P., Walters D. A., Hansma P. K. (1994) Imaging adhesion forces and elasticity of lysozyme adsorbed on mica with the atomic-force microscope. Langmuir 10, 3809–3814 [Google Scholar]
- 123.Ikai A., Afrin R., Sekiguchi H. (2007) Pulling and pushing protein molecules by AFM. Curr. Nanosci. 3, 17–29 [Google Scholar]
- 124.Florin E. L., Moy V. T., Gaub H. E. (1994) Adhesion forces between individual ligand-receptor pairs. Science 264, 415–417 [DOI] [PubMed] [Google Scholar]
- 125.Merkel R., Nassoy P., Leung A., Ritchie K., Evans E. (1999) Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature 397, 50–53 [DOI] [PubMed] [Google Scholar]
- 126.Lee G. U., Kidwell D. A., Colton R. J. (1994) Sensing discrete streptavidin biotin interactions with atomic-force microscopy. Langmuir 10, 354–357 [Google Scholar]
- 127.Rief M., Gautel M., Oesterhelt F., Fernandez J. M., Gaub H. E. (1997) Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 276, 1109–1112 [DOI] [PubMed] [Google Scholar]
- 128.Fotiadis D., Scheuring S., Müller S. A., Engel A., Müller D. J. (2002) Imaging and manipulation of biological structures with the AFM. Micron 33, 385–397 [DOI] [PubMed] [Google Scholar]
- 129.Lee G. U., Chrisey L. A., Colton R. J. (1994) Direct measurement of the forces between complementary strands of DNA. Science 266, 771–773 [DOI] [PubMed] [Google Scholar]
- 130.Colton R. J. (2004) Nanoscale measurements and manipulation. J. Vac. Sci. Technol. B Microelectron. Nanometer Struct. Process. Meas. Phenom. 22, 1609–1635 [Google Scholar]
- 131.Costa K. D., Yin F. C. (1999) Analysis of indentation: implications for measuring mechanical properties with atomic force microscopy. J. Biomech. Eng. 121, 462–471 [DOI] [PubMed] [Google Scholar]
- 132.Roos W. H., Ivanovska I. L., Evilevitch A., Wuite G. J. L. (2007) Viral capsids: mechanical characteristics, genome packaging and delivery mechanisms. Cell. Mol. Life Sci. 64, 1484–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gelbart W. M., Knobler C. M. (2009) Pressurized viruses. Science 323, 1682–1683 [DOI] [PubMed] [Google Scholar]
- 134.Arkhipov A., Roos W. H., Wuite G. J., Schulten K. (2009) Elucidating the mechanism behind irreversible deformation of viral capsids. Biophys. J. 97, 2061–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Uetrecht C., Versluis C., Watts N. R., Roos W. H., Wuite G. J., Wingfield P. T., Steven A. C., Heck A. J. R. (2008) High-resolution mass spectrometry of viral assemblies: molecular composition and stability of dimorphic hepatitis B virus capsids. Proc. Natl. Acad. Sci. U.S.A. 105, 9216–9220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Michel J. P., Ivanovska I. L., Gibbons M. M., Klug W. S., Knobler C. M., Wuite G. J., Schmidt C. F. (2006) Nanoindentation studies of full and empty viral capsids and the effects of capsid protein mutations on elasticity and strength. Proc. Natl. Acad. Sci. U.S.A. 103, 6184–6189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ivanovska I., Wuite G., Jönsson B., Evilevitch A. (2007) Internal DNA pressure modifies stability of WT phage. Proc. Natl. Acad. Sci. U.S.A. 104, 9603–9608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Carrasco C., Carreira A., Schaap I. A., Serena P. A., Gómez-Herrero J., Mateu M. G., de Pablo P. J. (2006) DNA-mediated anisotropic mechanical reinforcement of a virus. Proc. Natl. Acad. Sci. U.S.A. 103, 13706–13711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liashkovich I., Hafezi W., Kühn J. E., Oberleithner H., Kramer A., Shahin V. (2008) Exceptional mechanical and structural stability of HSV-1 unveiled with fluid atomic force microscopy. J. Cell Sci. 121, 2287–2292 [DOI] [PubMed] [Google Scholar]
- 140.Gibbons M. M., Klug W. S. (2007) Mechanical modeling of viral capsids. J. Mater. Sci. 42, 8995–9004 [Google Scholar]
- 141.Buenemann M., Lenz P. (2007) Mechanical limits of viral capsids. Proc. Natl. Acad. Sci. U.S.A. 104, 9925–9930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Klug W. S., Bruinsma R. F., Michel J. P., Knobler C. M., Ivanovska I. L., Schmidt C. F., Wuite G. J. (2006) Failure of viral shells. Phys. Rev. Lett. 97, 228101. [DOI] [PubMed] [Google Scholar]
- 143.Gibbons M. M., Klug W. S. (2008) Influence of nonuniform geometry on nanoindentation of viral capsids. Biophys. J. 95, 3640–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Roos W. H., Gibbons M. M., Arkhipov A., Uetrecht C., Watts N. R., Wingfield P. T., Steven A. C., Heck A. J., Schulten K., Klug W. S., Wuite G. J. (2010) Squeezing protein shells: how continuum elastic models, molecular dynamics simulations and experiments coalesce at the nanoscale. Biophys. J., doi: 10.1016/j.bpj.2010.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kol N., Gladnikoff M., Barlam D., Shneck R. Z., Rein A., Rousso I. (2006) Mechanical properties of murine leukemia virus particles: effect of maturation. Biophys. J. 91, 767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kol N., Shi Y., Tsvitov M., Barlam D., Shneck R. Z., Kay M. S., Rousso I. (2007) A stiffness switch in human immunodeficiency virus. Biophys. J. 92, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhao Y., Ge Z., Fang J. (2008) Elastic modulus of viral nanotubes. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 78, 031914 [DOI] [PubMed] [Google Scholar]
- 148.Munson K. M., Mulugeta P. G., Donhauser Z. J. (2007) Enhanced mechanical stability of microtubules polymerized with a slowly hydrolyzable nucleotide analogue. J. Phys. Chem. B 111, 5053–5057 [DOI] [PubMed] [Google Scholar]
- 149.Schaap I. A., Hoffmann B., Carrasco C., Merkel R., Schmidt C. F. (2007) Tau protein binding forms a 1 nm thick layer along protofilaments without affecting the radial elasticity of microtubules. J. Struct. Biol. 158, 282–292 [DOI] [PubMed] [Google Scholar]
- 150.Kasas S., Kis A., Riederer B. M., Forró L., Dietler G., Catsicas S. (2004) Mechanical properties of microtubules explored using the finite elements method. ChemPhysChem 5, 252–257 [DOI] [PubMed] [Google Scholar]
- 151.Guzmán C., Jeney S., Kreplak L., Kasas S., Kulik A. J., Aebi U., Forró L. (2006) Exploring the mechanical properties of single vimentin intermediate filaments by atomic force microscopy. J. Mol. Biol. 360, 623–630 [DOI] [PubMed] [Google Scholar]
- 152.Koenders M. M., Yang L., Wismans R. G., van der Werf K. O., Reinhardt D. P., Daamen W., Bennink M. L., Dijkstra P. J., van Kuppevelt T. H., Feijen J. (2009) Microscale mechanical properties of single elastic fibers: the role of fibrillin-microfibrils. Biomaterials 30, 2425–2432 [DOI] [PubMed] [Google Scholar]
- 153.Yang L., van der Werf K. O., Koopman B. F., Subramaniam V., Bennink M. L., Dijkstra P. J., Feijen J. (2007) Micromechanical bending of single collagen fibrils using atomic force microscopy. J. Biomed. Mater. Res. A 82, 160–168 [DOI] [PubMed] [Google Scholar]
- 154.Yang L., van der Werf K. O., Fitié C. F., Bennink M. L., Dijkstra P. J., Feijen J. (2008) Mechanical properties of native and cross-linked type I collagen fibrils. Biophys. J. 94, 2204–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schaap I. A., Carrasco C., de Pablo P. J., MacKintosh F. C., Schmidt C. F. (2006) Elastic response, buckling, and instability of microtubules under radial indentation. Biophys. J. 91, 1521–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sept D., MacKintosh F. C. (2010) Microtubule elasticity: connecting all-atom simulations with continuum mechanics. Phys. Rev. Lett. 104, 018101 [DOI] [PubMed] [Google Scholar]
- 157.Afrin R., Alam M. T., Ikai A. (2005) Pretransition and progressive softening of bovine carbonic anhydrase II as probed by single molecule atomic force microscopy. Protein Sci. 14, 1447–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kodama T., Ohtani H., Arakawa H., Ikai A. (2005) Mechanical perturbation-induced fluorescence change of green fluorescent protein. Appl. Phys. Lett. 86, 043901 [Google Scholar]
- 159.Kodama T., Ohtani H., Arakawa H., Ikai A. (2005) Observation of the destruction of biomolecules under compression force. Ultramicroscopy 105, 189–195 [Google Scholar]
- 160.Lin S., Chen J. L., Huang L. S., Lin H. W. (2005) Measurements of the forces in protein interactions with atomic force microscopy. Curr. Proteomics 2, 55–81 [Google Scholar]
- 161.Browning-Kelley M. E., Wadu-Mesthrige K., Hari V., Liu G. Y. (1997) Atomic force microscopic study of specific antigen/antibody binding. Langmuir 13, 343–350 [Google Scholar]
- 162.Moy V. T., Florin E. L., Gaub H. E. (1994) Intermolecular forces and energies between ligands and receptors. Science 266, 257–259 [DOI] [PubMed] [Google Scholar]
- 163.Grubmüller H., Heymann B., Tavan P. (1996) Ligand binding: molecular mechanics calculation of the streptavidin biotin rupture force. Science 271, 997–999 [DOI] [PubMed] [Google Scholar]
- 164.Izrailev S., Stepaniants S., Balsera M., Oono Y., Schulten K. (1997) Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys. J. 72, 1568–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Evans E., Ritchie K. (1997) Dynamic strength of molecular adhesion bonds. Biophys. J. 72, 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Willemsen O. H., Snel M. M., van der Werf K. O., de Grooth B. G., Greve J., Hinterdorfer P., Gruber H. J., Schindler H., van Kooyk Y., Figdor C. G. (1998) Simultaneous height and adhesion imaging of antibody-antigen interactions by atomic force microscopy. Biophys. J. 75, 2220–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nakajima H., Kunioka Y., Nakano K., Shimizu K., Seto M., Ando T. (1997) Scanning force microscopy of the interaction events between a single molecule of heavy meromyosin and actin. Biochem. Biophys. Res. Commun. 234, 178–182 [DOI] [PubMed] [Google Scholar]
- 168.Puchner E. M., Alexandrovich A., Kho A. L., Hensen U., Schäfer L. V., Brandmeier B., Gräter F., Grubmüller H., Gaub H. E., Gautel M. (2008) Mechanoenzymatics of titin kinase. Proc. Natl. Acad. Sci. U.S.A. 105, 13385–13390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Perez-Jimenez R., Garcia-Manyes S., Ainavarapu S. R., Fernandez J. M. (2006) Mechanical unfolding pathways of the enhanced yellow fluorescent protein revealed by single molecule force spectroscopy. J. Biol. Chem. 281, 40010–40014 [DOI] [PubMed] [Google Scholar]
- 170.Dietz H., Rief M. (2004) Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. U.S.A. 101, 16192–16197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bornschlögl T., Woehlke G., Rief M. (2009) Single molecule mechanics of the kinesin neck. Proc. Natl. Acad. Sci. U.S.A. 106, 6992–6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lu H., Isralewitz B., Krammer A., Vogel V., Schulten K. (1998) Unfolding of titin immunoglobulin domains by steered molecular dynamics simulation. Biophys. J. 75, 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Marszalek P. E., Lu H., Li H., Carrion-Vazquez M., Oberhauser A. F., Schulten K., Fernandez J. M. (1999) Mechanical unfolding intermediates in titin modules. Nature 402, 100–103 [DOI] [PubMed] [Google Scholar]
- 174.Gräter F., Shen J., Jiang H., Gautel M., Grubmüller H. (2005) Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys. J. 88, 790–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hisatomi Y., Katagiri D., Neya S., Hara M., Hoshino T. (2008) Analysis of the unfolding process of green fluorescent protein by molecular dynamics simulation. J. Phys. Chem. B 112, 8672–8680 [DOI] [PubMed] [Google Scholar]