Abstract
The molecular conformation of the cardiac myosin motor is modulated by intermolecular interactions among the heavy chain, the light chains, myosin binding protein-C, and titin and is governed by post-translational modifications (PTMs). In-gel digestion followed by LC/MS/MS has classically been applied to identify cardiac sarcomeric PTMs; however, this approach is limited by protein size, pI, and difficulties in peptide extraction. We report a solution-based work flow for global separation of endogenous cardiac sarcomeric proteins with a focus on the regulatory light chain (RLC) in which specific sites of phosphorylation have been unclear. Subcellular fractionation followed by OFFGEL electrophoresis resulted in isolation of endogenous charge variants of sarcomeric proteins, including regulatory and essential light chains, myosin heavy chain, and myosin-binding protein-C of the thick filament. Further purification of RLC using reverse-phase HPLC separation and UV detection enriched for RLC PTMs at the intact protein level and provided a stoichiometric and quantitative assessment of endogenous RLC charge variants. Digestion and subsequent LC/MS/MS unequivocally identified that the endogenous charge variants of cardiac RLC focused in unique OFFGEL electrophoresis fractions were unphosphorylated (78.8%), singly phosphorylated (18.1%), and doubly phosphorylated (3.1%) RLC. The novel aspects of this study are that 1) milligram amounts of endogenous cardiac sarcomeric subproteome were focused with resolution comparable with two-dimensional electrophoresis, 2) separation and quantification of post-translationally modified variants were achieved at the intact protein level, 3) separation of intact high molecular weight thick filament proteins was achieved in solution, and 4) endogenous charge variants of RLC were separated; a novel doubly phosphorylated form was identified in mouse, and singly phosphorylated, singly deamidated, and deamidated/phosphorylated forms were identified and quantified in human non-failing and failing heart samples, thus demonstrating the clinical utility of the method.
Modulation of sarcomeric protein function ensures that the heart ejects blood against a systemic resistance to supply peripheral tissues with oxygen and nutrients and to remove carbon dioxide and wastes. Muscle contraction occurs by myosin motors of the thick filament reacting with actin and propelling thin filaments toward the center of the sarcomere. In cardiac muscle, contraction is activated by a release of sarcoplasmic reticular Ca2+ that binds to troponin C, which is positioned on the actin thin filament. It was long assumed that regulation of Ca2+ levels and cycling were the sole avenue by which contraction was modulated. It is now evident, however, that post-translational modifications (PTMs)1 are important in regulating the function of ejecting ventricles as mechanisms downstream of Ca2+ fluxes at the level of the sarcomere appear to dominate ejection and sustain ventricular elastance during systole (1). Intra- and intermolecular interactions of sarcomeric thick and thin filament proteins are modifiable by PTMs, and it has been demonstrated that the intensity and dynamics of contraction and relaxation can be finely tuned via PTMs, particularly those of thin filament proteins (2, 3). Mechanisms by which PTMs tune molecular interactions of the thick filament are less well understood. Extrapolating from molecular mechanisms to the cardiac disorder requires an understanding of the endogenous charge state of proteins that can be governed by phosphorylation, acetylation, oxidation, and other post-translational modifications.
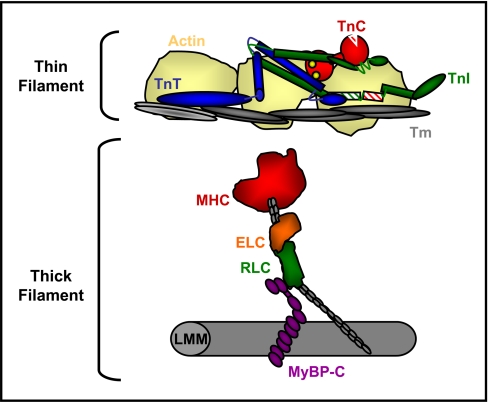
The thick filament is composed mainly of myosin, a motor protein that interacts with neighboring proteins, including the essential (ELC) and regulatory light chains (RLC), and myosin-binding protein-C (MyBP-C) (Fig. 1). RLC binds the S1-S2 lever arm of the myosin motor and is optimally positioned at the fulcrum to modulate interactions between the globular myosin heavy chain (MHC) head, the coiled coil light meromyosin thick filament backbone, and the neighboring MyBP-C (Fig. 1). Cardiac RLC is phosphorylated in large part by myosin light chain kinase (4), which induces an increase in tension at submaximally activating levels of Ca2+ (5–7) in skinned cardiac fibers bathed in exogenous myosin light chain kinase. Furthermore, mice expressing a transgenic cardiac RLC with alanine residues in place of N-terminal serines have significantly impaired systolic kinetics in vivo (8). Separation of endogenous RLC by two-dimensional electrophoresis (2DE) clearly illustrates multiple phosphorylated spots of RLC (8). The most abundant phosphorylated spot of RLC was found to represent phosphorylation at Ser-15 (9). The second, more acidic phosphorylated spot is 3–5 times lower in abundance and has been difficult to capture using in-gel digestion and mass spectrometry likely due to protein loss during in-gel digestion combined with poor ionization potential of the phosphopeptide. A possible approach to overcome this limitation is the use of a gel-free (in-solution) technique for separating charged species of RLC that is designed to minimize loss and increase sample load.
Fig. 1.
Schematic illustrating cardiac sarcomeric proteins. The myofibrillar lattice is composed mainly of actin thin filaments and myosin thick filaments, each bound to regulatory proteins. Activation of cardiac contraction during systole proceeds by calcium binding to troponin C (TnC), which induces conformational changes and altered interactions among troponin I (TnI), troponin T (TnT), and tropomyosin (Tm), resulting in the removal of steric inhibition over the myosin binding site on actin. An activated thin filament allows the binding of myosin heads, which then propel the actin filaments toward the center of the sarcomere. The thick filament is composed mainly of MHC, which binds two light chains, ELC and RLC, and associates with MyBP-C in the hinge and light meromyosin (LMM) regions.
Biochemically, proteins comprising the thick filament present unique purification challenges in part due to their large size (MHC, 223 kDa; MyBP-C, 150 kDa), producing difficulties in the isolation of intact proteins (10–12). In contrast, non-covalently bound ELC and RLC (i.e. the light chains of myosin) are more amenable to separation using standard biochemical methods due in part to their moderate sizes (22.4 and 18.9 kDa, respectively); however, because purification methods for either MHC or ELC/RLC are not time- and cost-effective, novel strategies for preparing/enriching these proteins in a single step are warranted.
When the research objective is to identify all proteins in a sample, a general, unbiased method is appropriate as two peptide ions with high quality, comprehensive MS/MS spectra are sufficient to reliably identify a protein. However, in targeted proteomics approaches where the objective is to distinguish functionally relevant charge variants (isoforms or post-translationally modified proteins) in a smaller subset of proteins, cleaving proteins into constituent peptides results in a loss of a significant amount of information and produces protein inference problems when assigning identities to modified versions of the same protein (13). A reliable and advantageous strategy to combat the issue of protein inference would be to discriminate charged variants at the intact protein level. Reverse-phase (RP) HPLC and OFFGEL electrophoresis (OGE) are two solution-based separation methods that exploit hydrophobicity and isoelectric point, respectively. Both techniques have been optimized for discriminatory separation at the peptide level (14, 15) but in the past have been underutilized in proteomics work flows for separation of intact proteins.
We report here a rapid solution-based method for purifying endogenous sarcomeric proteins, allowing for the enrichment and identification of the low abundance phosphospecies of cardiac RLC. The approach uses a tandem OGE/HPLC work flow that discriminately separates RLC at the intact protein level in a quantifiable manner. The novelty and strengths of this method are that 1) milligram quantities of sarcomeric subproteome are focused with resolution qualitatively similar to that of 2DE, 2) quantifiable separation of post-translationally modified variants is achieved at the whole protein level, 3) high sequence coverage of the protein under study, RLC, is achieved due to the substantial enrichment of proteins, and 4) separation of high molecular weight cardiac thick filament proteins can be achieved while maintaining proteins in solution in their intact forms. This work flow was used to identify a novel doubly charged phosphospecies of mouse RLC phosphorylated at adjacent Ser-14 and Ser-15. Additionally, we successfully used our method for identifying and quantifying post-translational modifications of RLC occurring in human heart failure, thus demonstrating clinical utility for future studies.
EXPERIMENTAL PROCEDURES
Animal Subjects
All experimental procedures performed using mice and mouse tissue procurement were approved by the Animal Care and Use Committee at the University of Illinois at Chicago (Animal Care Committee number 05-240).
Human Subjects
All procedures and informed consent involved in the procurement of human tissue were approved by the Institutional Review Board (Colorado Multiple Institutional Review Board number 00-242) at the University of Colorado, and all clinical materials were deidentified according to Health Insurance Portability and Accountability Act policy. Human tissue samples used were either from explanted hearts with dilated, non-ischemic cardiomyopathy (n = 2) or control hearts (n = 2) considered for cardiac transplantation. Samples were from the University of Colorado cardiology biobank, which includes a large collection of explanted diseased hearts as well as control heart specimens considered for transplantation. Tissue was collected not as part of a formal study design but rather to develop the biobank, which could be made available to investigators with an interest in the biology of human heart failure. Table I provides clinical details of the four samples used in the current study. A similar procurement strategy was used for all specimens: at the time of explant and following cardioplegia, a small portion of the left ventricle (<0.5 g) was excised, promptly placed in liquid N2 in the operating room, and then subsequently stored at −70 °C for less than 6 months. Samples were shipped on dry ice from Denver to Chicago and placed immediately at −70 °C until the time of analysis.
Table I. Definition of diseased state of patients.
The human specimens used in this study included cardiac muscle from two non-failing (NF) and two end stage DCM patients. The detailed descriptions of each are given. NYHA, New York Heart Association classification; EFx, ejection fraction; n/a, not applicable; F, female; M, male.
| Patient | Gender | Age | Ethnicity | Type | Subtype | Stage | EFx | Comments |
|---|---|---|---|---|---|---|---|---|
| % | ||||||||
| NF 1 | F | 57 | Caucasian | NF | n/a | n/a | 65 | Intracranial hemorrhage |
| NF 2 | M | 37 | Hispanic | NF | n/a | n/a | 60 | Intraventricular hemorrhage |
| DCM 3 | M | 20 | Caucasian | DCM | Non-ischemic | NYHA IV | 10 | None |
| DCM 4 | M | 47 | Hispanic | DCM | Non-ischemic | NYHA IV | <20 | None |
Myofilament Enrichment and Sample Preparation
Subcellular fractionation proceeded according to methods described previously (16) with modifications. Hearts from 4-month-old FVB male and female mice or from human biopsies were excised and immediately placed on ice in 75 mm KCl, 10 mm imidazole (pH 7.2), 2 mm MgCl2, 2 mm EDTA, 1 mm NaN3 with 1% Triton X-100, phosphatase inhibitor mixture I (Calbiochem), and protease inhibitor mixture (Sigma-Aldrich). Tissue was homogenized and centrifuged (18,000 × g for 10 min at 4 °C), and the supernatant fraction was removed. The pellets following two extractions were washed in the absence of Triton X-100, then extracted in IEF buffer (8 m urea, 2 m thiourea, 4% CHAPS), and clarified by centrifuging (18,000 × g for 5 min).
OFFGEL Electrophoresis
A myofilament-enriched fraction was diluted in OFFGEL electrophoresis buffer (7 m urea, 2 m thiourea, 6% glycerol, 65 mm DTT, 1% ampholytes 4–7), and the sample was partitioned among 24 1-cm wells superimposed on a pH 4–7, 24-cm OFFGEL strip (Agilent Technologies, Santa Clara, CA). Proteins were separated using the following conditions: 8000-V maximum voltage, 50-μA maximum current, and 200-milliwatt maximum power for 64,000 V-h. Individual fractions were aspirated and stored at −80 °C for no longer than 1 week.
Reverse-phase HPLC Separation and Digestion
OFFGEL fractions of interest were acidified with TFA to pH 2, filtered with 0.45-μm PVDF spin filters (Millipore, Billerica, MA), and separated using C4 reverse-phase chromatography (4.6 × 150 mm, 300-Å pore size; Grace Vydac) at a flow rate of 1.0 ml/min at 37 °C on a U3000 analytical HPLC instrument (Dionex, Bannockburn, IL). Proteins were eluted from 100% buffer A (95% H2O, 5% ACN, 0.1% TFA) to 90% buffer B (95% ACN, 5% H2O, 0.1% TFA) with the following protocol (time post-injection/buffer B): 0 min/0%, 5 min/0%, 20 min/20%, 80 min/65%, 85 min/90%, 90 min/90%, 91 min/0%, 101 min/0%. Fractions were collected every 1 min (1 ml); speed vacuumed to reduce volume; resolubilized in 100 mm NH4HCO3 (pH 8.4), 1 mm DTT; and digested with Lys-C (1:50 enzyme:protein; Sigma-Aldrich) for 16 h at 37 °C. Digests were acidified, lyophilized, and resuspended in 3% ACN, 0.1% formic acid with vigorous vortexing.
LC/MS/MS of Lys-C Peptides for Identification of Endogenous PTMs
LC/MS/MS of peptides was performed using a Q-TOF 6510 instrument (Agilent Technologies) equipped with a nano-LC-chip cube. The HPLC system consisted of a nanoflow analytical pump and a capillary loading pump (Agilent 1200 series). HPLC grade water and ACN used for HPLC mobile phases were obtained from Burdick and Jackson (Morristown, NJ). Formic acid was obtained from Sigma-Aldrich. Parameters for the analytical (nano) pump were as follows. Buffer A was HPLC grade water, 0.1% formic acid; buffer B was 90% ACN, 10% HPLC water, 0.1% formic acid. The flow rate for the nanopump was 550 nl/min, and the elution gradient was as follows: 3% buffer B at 0 min, 3% buffer B at 1 min, 40% buffer B at 12 min, 80% buffer B at 13 min, 80% buffer B at 16 min, 3% buffer B at 16.1 min, and 3 min post-time. Parameters for loading the capillary pump were as follows: 3% ACN, 97% HPLC grade water, 0.1% formic acid. The loading pump was operated in isocratic mode. The Q-TOF instrument was externally calibrated. Peptides were enriched and separated via nano-LC (0.075 × 43 mm, packed with Zorbax 300SB-C18, 5-μm material, 300-Å pore size) integrated in the HPLC Chip (G4240-62001). A full scan was acquired over a range of m/z 290–3000 at eight spectra per second, and an MS/MS scan was acquired over m/z 100–2400 at three spectra per second with a maximum of five precursors per cycle. Fragmentation energy was applied at a slope of 3.0 V/100 Da with a 2.0 offset. Spectra were deconvoluted and analyzed using Spectrum Mill, version A.03.03.080 SR1 (Agilent Technologies). Peak picking was performed with the following parameters: signal to noise was set at 25:1, a maximum charge state of 4 was allowed (z ≤ 4), and the program was directed to attempt to find a precursor charge state. Spectra were searched with the following search criteria: database, International Protein Index (IPI) mouse version 3.32 (49,427 total proteins) or IPI human version 3.68 (87,061 total proteins) (17); digest, Lys-C; maximum number of missed cleavages, 2; protein pI, from 3.0 to 10.0; variable modifications: oxidation of Met, phosphorylation of Ser, Thr, Tyr, and deamidation of Asn, Gln; instrument, Agilent ESI Q-TOF; precursor mass tolerance, 20 ppm; product mass tolerance, 20 ppm; maximum ambiguous precursor charge, 5. All other criteria were set to software default settings. MS/MS spectra were first autovalidated with Spectrum Mill software followed by manual inspection for confirmation of PTM identification.
Targeted Analysis and Quantification of RLC Variants in Dilated Cardiomyopathy (DCM) Versus Control by LC/MS/MS
LC/MS/MS of peptides was performed using a QQQ 6410 instrument (Agilent Technologies) equipped with an electrospray source. The HPLC system consisted of a binary analytical pump and a quaternary loading pump (Agilent 1200 series). The flow rate for the binary pump was 200 μl/min, and the gradient was as follows: 20% buffer B from 0–1 min, 20–40% buffer B from 1–12 min. The gradient and flow rates for the quaternary pump were as follows: 3% buffer B at 0.2 ml/min for 1 min and 100% buffer B at 1 ml/min from 1.01 to 12 min. The column switching valve was switched from enrichment to analysis at 1.0 min and then back to enrichment at 9 min. Peptides were trapped on a Zorbax 17 × 1.0-mm 5-μm SB-C18 enrichment column and then separated using a Zorbax 150 × 1.0-mm 3.5-μm 300SB-C18 300-Å-pore size analytical column. The electrospray ion source parameters were 325 °C for the gas temperature, 10 liters/min for the gas flow, 30 p.s.i. for the nebulizer, 4000 V for the capillary voltage, and 175 V for the fragmentor voltage. The collision energies for each transition were obtained from the Q-TOF data. The following transitions, which correspond to the resulting b9 product ion, were monitored for the peptide (K↓RAGGANSNVFSMFEQTQIQEFK↓E) and used to distinguish isoforms: unmodified (14/16NS15), 830.4 → 827.4; deamidated (14/16nS15), 830.70 → 828.4; phosphorylated (14/16Ns15), 857.1 → 907.4; and deamidated and phosphorylated (14/16ns15), 857.4 → 908.4. MassHunter software was used to integrate peaks and determine area. Although isotopic overlap resulted in apparent chromatographic peak overlap between deamidated and non-deamidated isoforms, adequate chromatographic separation of the isoforms allowed for unadulterated integration of individual transition peaks. Individual fractions were analyzed in triplicate, and the area was determined for each transition. Limited sample resulted in low signal to noise and poor reproducibility for four transitions from a total of 18 experiments (total of 72 transitions), and these values were discarded. The total area for each transition was combined between fractions for simplicity. Three experiments were conducted, and the average and S.D. were calculated. t tests were used to determine whether significant differences existed between isoforms from control and heart disease samples.
2DE
The mouse myofilament subproteome was prepared according to subfractionation methods described above. Proteins were solubilized in IEF buffer, clarified with centrifugation (18,000 × g for 5 min at 25 °C), and labeled with Cy3. Proteins were separated over a non-linear, broad pI range of 3–11 on 18-cm IPG strips according to methods described previously (8). Strips were overlaid on 12% SDS-polyacrylamide second dimension gels (18) for separation by molecular weight. Gels were rinsed in water and imaged using a Typhoon 9410 (GE Healthcare).
RESULTS
Preparation of Sarcomeric Subproteome
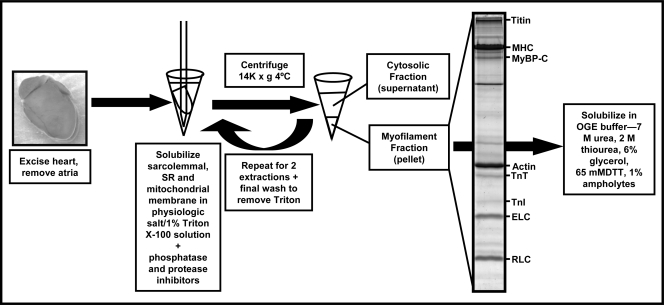
Fig. 2 shows the myofilament enrichment using the Triton X-100 extraction method, originally described by Solaro et al. (16), resulting in myofibrils that are essentially free of sarcolemmal, sarcoplasmic reticular, and mitochondrial contaminants. This method was adapted to expedite fractionation by omitting the initial series of washes in 0.3 m sucrose, 10 mm imidazole (pH 7.0) and 60 mm KCl, 30 mm imidazole (pH 7.0) (16) and homogenizing heart tissue directly in physiologic salt solution containing 1% Triton X-100 with high concentrations of protease and phosphatase inhibitor mixtures using glass-on-glass homogenizers on ice. This rapid protocol resulted in a clean and reproducible enriched sarcomeric subproteome, which was solubilized in 8 m urea, 2 m thiourea, 4% CHAPS to minimize further alterations in PTMs or degradation.
Fig. 2.
Protocol for preparation of sarcomeric subproteome using Triton X-100. This protocol was originally described by Solaro et al. (16). This method uses the non-ionic detergent Triton X-100 to solubilize the sarcolemmal, sarcoplasmic reticular (SR), and mitochondrial membranes, allowing the release of soluble proteins into solution and pelleting of the insoluble myofibrillar lattice in a rapid and efficient manner. All fractionation is performed in the presence of phosphatase and protease inhibitors to preserve the endogenous phosphostate and integrity of sarcomeric proteins. TnI, troponin I; TnT, troponin T.
In-solution Separation of Intact Cardiac Sarcomeric Proteins with OGE
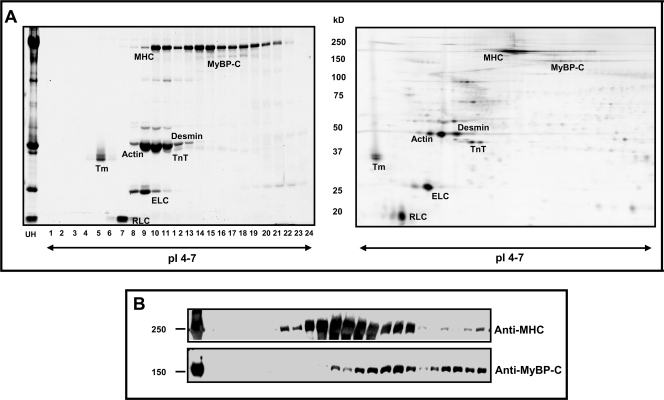
The resolving capability of OGE at the intact protein level is illustrated in Fig. 3. Separation of 3.5 mg of sarcomeric subproteome was achieved over a narrow pH range of 4–7 and 24-cm focusing distance. Shown in Fig. 3A (left panel) is a one-dimensional SDS-PAGE gel, with 2 μl from each OGE fraction loaded, illustrating composite proteins of each fraction. OGE wells contained a volume of 100–200 μl of buffer at cessation of focusing depending on the protein concentration per well. Thus, Coomassie-stained bands shown in Fig. 3 represent of the actual protein content harvested per OGE well. To compare the focusing pattern of OGE with traditional 2DE, 40 μg of sarcomeric subproteome was separated using a pH 3–11 non-linear, 18-cm IPG strip, and the area containing pH 4–7 was cropped and run on a 12% second dimension SDS-polyacrylamide gel (Fig. 3A, right panel). Comparison of the OGE fractions with 2DE demonstrated similar protein profiles despite the variation in focusing distance. Importantly, ∼100-fold more protein was resolved using OGE with resolution qualitatively similar to that of 2DE. Using this method, intact charge variants of thick filament proteins MHC, MyBP-C, ELC, and RLC were successfully separated in solution. To our knowledge, this is the first in-solution separation of endogenous charged variants demonstrated for full-length intact MHC and My-BPC (Fig. 3B). The reproducibility of OGE separation of sarcomeric proteins is illustrated in supplemental Fig. S1. Total myofibrillar extract from one mouse heart was divided into two samples (∼2 mg each) and separated in successive OGE runs to obtain technical replicates. SDS-PAGE profiles of the fractions illustrate significant reproducibility of OGE separation (supplemental Fig. S1), demonstrating the applicability of this technique to studies undergoing comparative analysis.
Fig. 3.
Comparison of OGE and 2DE focusing patterns. The cardiac sarcomeric subproteome (load, 3.5 mg) was separated in solution by OGE over a narrow pI range of 4–7 and 24-cm distance. A, left panel, approximately the volume (2 μl) of each OGE fraction was separated by 12% SDS-PAGE to illustrate constituents of OGE fractions (labeled 1–24, x axis) and the unfractionated homogenate (UH). Detection is via Coomassie G-250. Right panel, 2DE of the same sarcomeric subproteome with a 40-μg protein load demonstrates resolution of sarcomeric proteins comparable with OGE. The method of detection is Cy3 fluorescence. Importantly, note that 3.5 mg of protein separates in solution using OGE with resolution qualitatively similar to 40 μg in gel. B, OGE fractions probed with anti-MHC and anti-MyBP-C antibodies illustrates the first in-solution separation of full-length endogenous high molecular weight thick filament proteins. TnT, troponin T; Tm, tropomyosin.
RP-HPLC Separation Purifies Co-focusing Sarcomeric Proteins in OGE
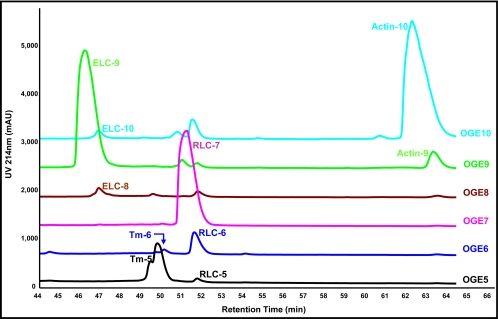
To purify RLC charged species in each OGE fraction from co-migrating proteins, OGE fractions of interest were sequentially separated using RP-HPLC. Fig. 4 shows overlaid chromatograms of OGE fractions 5–10. This method separated sarcomeric proteins with high resolution. Because the goal was to determine site-specific phosphorylation of RLC, to further purify RLC from contaminating proteins in OGE fractions, 1-min (1-ml) RP-HPLC fractions from OGE fractions 5–7 were collected and subjected to subsequent analyses. It was evident from Western blotting of each collected fraction for RLC (data not shown) and corresponding peak overlays that there were three isoelectric variants of RLC, denoted RLC-5, -6, and -7 (Fig. 4), at the intact protein level in OGE fractions 5–7, respectively, consistent with previous 2DE. From the integration of peaks detected with a UV detector (214 nm), we quantified RLC species as RLC-5 (Area = 75.2 mAU × min), RLC-6 (Area = 446.3 mAU × min), and RLC-7 (Area = 1943.5 mAU × min); thus, RLC exists endogenously in cardiac mouse myofibrils as three populations (3.1% RLC-5, 18.1% RLC-6, and 78.8% RLC-7). A significant strength of this approach was in the use of UV detection as a quantitative measure of endogenous charged species of RLC as UV detection at 214 nm offers unbiased detection of peptide bonds and is linear over several orders of magnitude.
Fig. 4.
Purification of sarcomeric proteins in OGE fractions using RP-HPLC. Shown are OGE fractions 5–10, each run separately on a 4.6 × 150-mm C4 reverse-phase column (TP5415, Grace Vydac) with UV detection at 214 nm, and the chromatograms were overlaid for comparison. Separation of endogenous charge variants of ELC, tropomyosin (Tm), RLC, and actin is demonstrated in the retention time range 44–66 min. Quantification of the three unique RLC species demonstrated a 3.1/18.1/78.8% distribution of endogenous RLC in fractions RLC-5, -6, and -7, respectively, which were collected for further analysis by LC/MS/MS.
LC/MS/MS of RLC Charged Species Reveals Two Novel Findings: Phosphorylation at Serine 14 and Presence of Doubly Phosphorylated Species
To identify site-specific differences in RLC charged variants separated by OFFGEL and purified by RP-HPLC, proteins were digested with Lys-C and subjected to nano-LC/MS/MS analysis on a Q-TOF mass spectrometer. Peptides were eluted over a 16-min linear gradient of ACN and scanned in a data-dependent manner with the five most abundant peaks of each scan subjected to fragmentation. Spectra were extracted and searched against the mouse IPI database (17) using Spectrum Mill (Agilent Technologies). Confident protein hits with two or more distinct peptides detected, spectral intensities, and amino acid sequence coverage from off-line LC fractions RLC-5, -6, and -7 were aligned for comparison and are listed in Table II. RLC showed a striking 95% sequence coverage. The region of RLC not covered was the N-terminal MAPKKAK sequence due to the high lysine content. This high coverage demonstrates the power of using prefractionation and/or enrichment techniques in cases where detecting the majority of peptides is critical as during PTM or isoform identification.
Table II. Protein-protein comparison analysis of mouse LC/MS/MS data.
Shown are constituents of HPLC fractions RLC-5, -6, and -7. Proteins detected by two or more distinct peptides are shown. RLC exhibited 95% sequence coverage. Database acc. no., IPI database accession number; AA cov., percent amino acid sequence coverage; dist. pep., distinct peptides.
| RLC-5 |
RLC-6 |
RLC-7 |
Database acc. no. | AA cov. | No. dist. pep. | Distinct summed MS/MS score | Protein name | |||
|---|---|---|---|---|---|---|---|---|---|---|
| No. spectra | Mean intensity | No. spectra | Mean intensity | No. spectra | Mean intensity | |||||
| 40 | 2.11e+06 | 69 | 4.09e+06 | 78 | 5.46e+06 | IPI00555015 | 95 | 24 | 467.94 | Myl2; myosin regulatory light chain 2, ventricular/cardiac muscle |
| 56 | 7.20e+05 | 30 | 3.47e+05 | 29 | 3.99e+05 | IPI00123316 | 71 | 33 | 578.58 | Tpm1; 33-kDa protein |
| 9 | 8.89e+04 | 13 | 2.10e+05 | 7 | 1.35e+05 | IPI00400016 | 7 | 10 | 179.67 | Lamc1; laminin subunit γ-1 precursor |
| 1 | 5.92e+04 | 2 | 1.14e+05 | 0 | 0.00e+00 | IPI00227299 | 8 | 3 | 43.46 | Vim; vimentin |
| 0 | 0.00e+00 | 2 | 4.88e+04 | 3 | 4.52e+04 | IPI00130102 | 7 | 3 | 38.97 | Des; desmin |
| 1 | 2.87e+04 | 2 | 8.35e+04 | 2 | 2.26e+05 | IPI00346834 | 2 | 2 | 31.94 | Similar to c |
| 0 | 0.00e+00 | 0 | 0.00e+00 | 2 | 2.91e+05 | IPI00120900 | 4 | 2 | 19.35 | Ovgp1; oviduct-specific glycoprotein precursor |
| 1 | 3.29e+05 | 0 | 0.00e+00 | 1 | 9.70e+05 | IPI00120455 | 4 | 2 | 18.53 | Snx13; sorting nexin-13 |
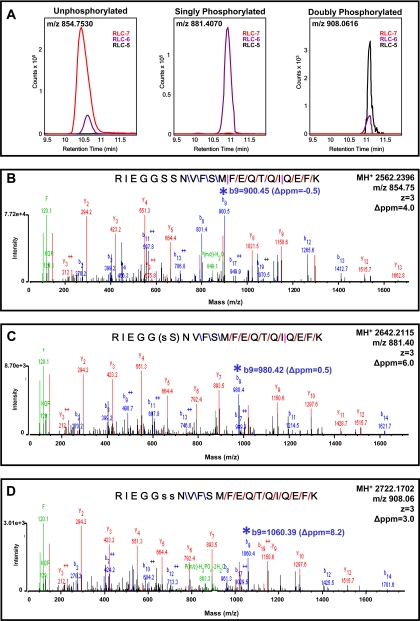
Fig. 5 shows extracted ion chromatograms, obtained using MassHunter qualitative analysis software (Agilent Technologies), of the triply charged N-terminal 9–30 peptide RIEGGSSNVFSMFEQTQIQEFK showing its abundance as unphosphorylated, singly phosphorylated, and doubly phosphorylated, corresponding to fractions RLC-7, -6, and -5, respectively. Fig. 5A, left panel, demonstrates that the majority of unphosphorylated m/z 854.7530 was present in RLC-7 with a small amount (5%) detected in RLC-6, suggesting minor amounts of overlap between fractions 6 and 7 during OGE focusing. The corresponding MS/MS spectra to demonstrate the identity of the unphosphorylated peptide as confirmed by MH+ 2562.2396 matching within 6 ppm is shown in Fig. 5B. Fig. 5A, middle panel, demonstrates that essentially all of the singly phosphorylated peptide m/z 881.4070 was in RLC-6, and the corresponding MS/MS spectra in Fig. 5C identified a singly phosphorylated peptide, MH+ 2642.2115 (mass error of 4.2 ppm), with the phosphorylation residing on Ser15 or Ser14. Our previous analysis of this singly phosphorylated species with LTQ-FT-ICR showed that when a successful Ser-Ser break was achieved with either CID or electron capture dissociation Ser14 and Ser15 were alternately phosphorylated, roughly 20 and 80% of the time, respectively (data not shown).2 This singly phosphorylated charged species of RLC migrating in fraction 6 of OGE thus likely represents a mixture of RLC-Ser14 and RLC-Ser15 phosphorylated molecules. The majority of doubly phosphorylated RLC m/z 908.0616 shown in Fig. 5A, right panel, was in RLC-5 with some overlap evident in RLC-6. MS/MS spectra of MH+ 2722.1702 (mass error of 3.0 ppm) in Fig. 5D unequivocally demonstrated that these two phosphorylations were on residues Ser14 and Ser15. This novel finding is the first report demonstrating a doubly phosphorylated endogenous species of cardiac RLC. We unequivocally identified three distinct charge species of RLC with high mass accuracy of both precursor and fragment ions inherent to the Q-TOF (see low mass errors in Fig. 5). Importantly, RP-HPLC chromatograms of intact RLC protein detected with a UV detector shown in Fig. 4 demonstrate doubly phosphorylated RLC as 3.1%, the singly phosphorylated as 18.1%, and the unphosphorylated as 78.8% of total sarcomeric RLC. It is also worth noting that MS/MS spectra of un-, singly, and doubly phosphorylated peptides shown in Fig. 5 have scored peak intensities (SPI) between 92 and 96% and Spectrum Mill peptide scores between 19 and 21. The SPI represents the percentage of the MS/MS peak-detected spectral ion current explained by the search interpretation, demonstrating the remarkably high confidence and fidelity of our MS/MS spectra. The peptide score reflects the information content in the MS/MS spectrum; a score of 13–25 is considered of excellent quality representing extensive peptide fragmentation, and when combined with an SPI of 70% or greater, the result is of high confidence and validity.
Fig. 5.
MS/MS spectra showing identification of phosphospecies in murine RLC. A, left panel, extracted ion chromatogram of unphosphorylated RLC peptide 9–30 (m/z 854.7530) showing the majority in fraction RLC-7 with minor carryover in RLC-6. Middle panel, extracted ion chromatogram of singly phosphorylated RLC peptide 9–30 (m/z 881.4070) showing the presence of this peptide exclusively in RLC-6. Right panel, extracted ion chromatogram of doubly phosphorylated RLC peptide 9–30 (m/z 908.0616) of RLC showing the majority of this peptide in fraction RLC-5 with minor carryover in RLC-6. B, MS/MS spectra (SPI of 96%; peptide score of 20.8) of unphosphorylated RLC peptide m/z 854.7530 (precursor mass error of 5.8 ppm) with peptide bond cleavage type denoted as follows: blue, b ion; red, y ion; and pink, both b and y ion formation. C, MS/MS spectra (SPI of 91%; peptide score of 20.8) of singly phosphorylated RLC peptide m/z 881.4070 (precursor mass error of 4.2 ppm). s, phosphorylated serine. D, MS/MS spectra (SPI of 96%; peptide score of 19.2) of doubly phosphorylated RLC peptide m/z 908.0616 (precursor mass error of 3.0 ppm). Note the mass shifts and mass accuracy of b9 fragments (*) going from B to D, illustrating a 79.966 shift in each as an additional HPO3 is added to the peptide.
In-solution Work Flow Applied to Clinical Samples Reveals Significant Changes in RLC PTMs in Human Heart Failure
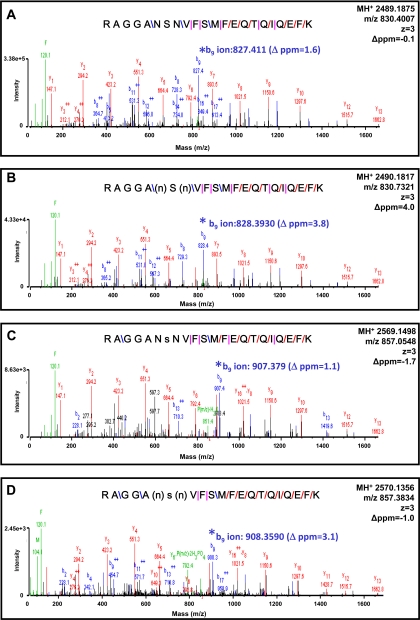
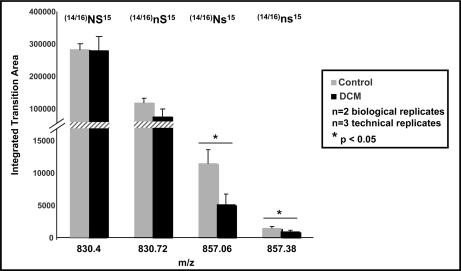
To examine whether differential modifications are present in human heart failure, non-failing and failing DCM samples were analyzed using the OGE-HPLC-MS/MS work flow. Human samples used were either “control” left ventricular tissue from donor hearts that were considered for heart transplantation or hearts from patients with dilated, non-ischemic cardiomyopathy explanted at the time of heart transplantation. Interestingly, human RLC was identified in OGE fractions 7, 8, and 9, whereas mouse RLC was identified in OGE fractions 5, 6, and 7. In silico alignment of human (Swiss-Prot accession number P10916) versus mouse (Swiss-Prot accession number P51667) RLC demonstrated slight variations in the primary sequence of RLC with an overall isoelectric point difference (excluding all variance due to PTMs) of 4.92 (human) to 4.86 (mouse). RLC in each fraction was subjected to MS/MS analysis on the Q-TOF instrument (search results shown in Table III), and three arrangements of modifications on the 9–30 N-terminal peptide of RLC were detected, including deamidation, phosphorylation, and simultaneous deamidation/phosphorylation (Fig. 6). Interestingly, the identified deamidation was the conversion of asparagine to aspartate at position 14 or 16, immediately adjacent of the solely identified phosphorylation site, serine 15. Note the sequence difference between the mouse (Fig. 5) 9–30 peptide and human (Fig. 6) 9–30 peptide. The position 14/15 serine/serine double phosphorylation in mouse is a position 14/15 or 15/16 deamidation/phosphorylation or phosphorylation/deamidation, respectively, in human; thus, the net charge on the N-terminal region of RLC in each species remains similar, albeit through different chemistries (Table IV). No doubly phosphorylated species was detected on other serine/threonine residues in the 9–30 peptide of human RLC as in mouse. This is the first site-specific identification of post-translational modifications in human RLC. To quantify differences in RLC PTMs in non-failing versus failing ventricular tissue, targeted multiple reaction monitoring was used. Fig. 7 illustrates the abundance of each RLC 9–30 peptide species as quantified from the b9 ion transitions and normalized to total RLC protein amount determined via UV detection in the off-line HPLC separation of intact RLC. Failing DCM samples demonstrated a significant decrease in the singly phosphorylated species (control, 11,411.2 ± 2230.3 versus DCM, 5051.0 ± 1703.5) and the combined deamidated/phosphorylated species (control, 1456.8 ± 265.7 versus DCM, 869.4 ± 270.5) relative to non-failing controls, suggesting that the negative charge in the N-terminal region of RLC is reduced in human heart failure.
Table III. Protein-protein comparison of human LC/MS/MS data.
Shown are characteristics of RLC-7, -8, and -9 in non-failing (NF) and DCM samples. Proteins detected by two or more distinct peptides are shown. Database acc. no., IPI database accession number; AA cov., percent amino acid sequence coverage; dist. pep., distinct peptides.
| NF RLC-7 |
NF RLC-8 |
NF RLC-9 |
DCM RLC-7 |
DCM RLC-8 |
DCM RLC-9 |
Database acc. no. | AA cov. | No. dist. pep. | Distinct summed MS/MS score | Protein name | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. spectra | Mean intensity | No. spectra | Mean intensity | No. spectra | Mean intensity | No. spectra | Mean intensity | No. spectra | Mean intensity | No. spectra | Mean intensity | |||||
| 49 | 1.87e+08 | 42 | 2.79e+08 | 44 | 5.21e+08 | 39 | 1.20e+08 | 50 | 6.87e+08 | 44 | 4.28e+08 | IPI00216798 | 89 | 14 | 306.85 | MYL2; myosin regulatory light chain 2, ventricular/cardiac muscle form |
| 30 | 4.63e+07 | 0 | 0.00e+00 | 0 | 0.00e+00 | 42 | 4.61e+07 | 1 | 4.26e+05 | 0 | 0.00e+00 | IPI00014581 | 54 | 15 | 287.95 | TPM1; isoform 1 of tropomyosin α-1 chain |
| 9 | 4.12e+05 | 0 | 0.00e+00 | 3 | 1.17e+06 | 5 | 1.66e+05 | 1 | 6.15e+04 | 1 | 3.63e+05 | IPI00025880 | 8 | 11 | 176.6 | MYH7; myosin-7 |
| 5 | 2.71e+06 | 5 | 4.37e+06 | 10 | 2.81e+07 | 6 | 2.90e+06 | 6 | 8.95e+06 | 6 | 6.97e+06 | IPI00418471 | 20 | 7 | 131.66 | VIM; vimentin |
| 2 | 4.69e+05 | 1 | 7.02e+05 | 2 | 1.47e+06 | 2 | 5.12e+05 | 3 | 3.60e+06 | 5 | 5.17e+06 | IPI00465084 | 16 | 5 | 85.47 | DES; desmin |
| 0 | 0.00e+00 | 0 | 0.00e+00 | 1 | 1.01e+05 | 0 | 0.00e+00 | 3 | 3.33e+05 | 3 | 5.98e+05 | IPI00025465 | 9 | 3 | 48.14 | OGN; osteoglycin |
| 2 | 2.01e+05 | 0 | 0.00e+00 | 0 | 0.00e+00 | 0 | 0.00e+00 | 0 | 0.00e+00 | 0 | 0.00e+00 | IPI00243742 | 16 | 2 | 37.39 | MYL3; myosin light chain 3 |
| 1 | 2.98e+04 | 3 | 1.00e+06 | 2 | 3.42e+05 | 0 | 0.00e+00 | 3 | 4.68e+05 | 0 | 0.00e+00 | IPI00021854 | 23 | 2 | 36.96 | APOA2; apolipoprotein A-II |
| 0 | 0.00e+00 | 2 | 3.38e+05 | 0 | 0.00e+00 | 0 | 0.00e+00 | 0 | 0.00e+00 | 0 | 0.00e+00 | IPI00783987 | 2 | 2 | 33.56 | C3; complement C3 (fragment) |
| 0 | 0.00e+00 | 0 | 0.00e+00 | 0 | 0.00e+00 | 2 | 7.03e+04 | 0 | 0.00e+00 | 0 | 0.00e+00 | IPI00019359 | 4 | 2 | 30.11 | KRT9; keratin, type I cytoskeletal 9 |
Fig. 6.
MS/MS spectra showing identification of phospho- and deamidated species in human RLC. A, MS/MS spectra (SPI of 100%; peptide score of 21) of unphosphorylated, non-deamidated RLC peptide m/z 830.4007 (precursor mass error of −0.1 ppm) with peptide bond cleavage type denoted as follows: blue, b ion; red, y ion; and pink, both b and y ion formation. B, MS/MS spectra (SPI of 93%; peptide score of 20.2) of singly deamidated RLC peptide m/z 830.7321 (precursor mass error of 4.0 ppm). n, deamidated asparagine. (n) denotes ambiguity in the assignment of the deamidation to the asparagine at position 14 or position 16. C, MS/MS spectra (SPI of 81%; peptide score of 18.4) of singly phosphorylated RLC peptide m/z 857.0548 (precursor mass error of −1.7 ppm). s, phosphorylated serine. D, MS/MS spectra (SPI of 89%; peptide score of 19.1) of singly deamidated and singly phosphorylated RLC peptide m/z 857.3834 (precursor mass error of −1.0 ppm). Note the mass shifts and mass accuracy of b9 fragments (*) going from A to D.
Table IV. Details of RLC N-terminal peptide 9–30 mouse and human PTMs measured via LC-MS/MS. Modified amino acid residues are shown in bold.
p, phosphorylation; d, deamidation; Spec Mill, Spectrum Mill; mods, modifications.
| Species | Peptide sequence | Precursor m/z | Precursor MH+ | Precursor charge | Δppm | SPI | Spec Mill score | No. detected mods |
|---|---|---|---|---|---|---|---|---|
| % | ||||||||
| Mouse | RIEGGSSNVFSMFEQTQIQEFK | 854.75 | 2562.2396 | 3 | 5.8 | 96 | 20.8 | 0 |
| Mouse | RIEGG(sS)NVFSMFEQTQIQEFK | 881.40 | 2642.2115 | 3 | 4.2 | 91 | 20.8 | 1 p |
| Mouse | RIEGGssNVFSMFEQTQIQEFK | 908.06 | 2722.1702 | 3 | 3.0 | 96 | 19.2 | 2 p |
| Human | RAGGANSNVFSMFEQTQIQEFK | 830.40 | 2489.1875 | 3 | −0.1 | 100 | 21.0 | 0 |
| Human | RAGGAnSNVFSMFEQTQIQEFK | 830.73 | 2490.1817 | 3 | 4.0 | 93 | 20.2 | 1 d |
| Human | RAGGANsNVFSMFEQTQIQEFK | 857.05 | 2569.1498 | 3 | −1.7 | 81 | 18.4 | 1 p |
| Human | RAGGAnsNVFSMFEQTQIQEFK | 857.38 | 2570.1356 | 3 | −1.0 | 89 | 19.1 | 1 d, 1 p |
Fig. 7.
Quantification of human RLC species in DCM versus control by multiple reaction monitoring reveals differences in phosphorylation and deamidation. Heart tissue from non-failing and DCM patients was extracted and fractionated using OGE. Fractions (OGE-7, OGE-8, and OGE-9) from two biological replicates were pooled, and RLC species were separated via RP-HPLC as described above. Fractions were digested using Lys-C and subjected to liquid chromatography tandem mass spectrometry using a triple quadrupole mass spectrometer. The following transitions were monitored for the peptide (K↓RAGGANSNVFSMFEQTQIQEFK↓E) and used to distinguish isoforms: unmodified ((14/16)NS15), 830.4 → 827.4; singly deamidated ((14/16)nS15), 830.70→ 828.4; singly phosphorylated ((14/16)Ns15), 857.1 → 907.4; and singly deamidated/singly phosphorylated ((14/16)ns15), 857.4 → 908.4. Each transition corresponds to the resulting b9 product ion. Individual fractions were analyzed, the area was determined for each transition, and the total area for each transition was combined between fractions for simplicity. Three experiments were conducted, and the averages are shown. Error bars represent the standard deviation. Transitions corresponding to the singly phosphorylated and the phosphorylated/deamidated forms are significantly different between heart disease and control, whereas singly deamidated and unphosphorylated showed no difference.
DISCUSSION
Effectiveness of Rapid, In-solution Method to Purify Endogenous Cardiac Proteins
Although 2DE is a powerful method for protein separation and resolution, when used in preparation for analysis by mass spectrometry, in-gel approaches can be less than ideal. An inherent attribute of in-gel approaches is that proteins are encased inside a matrix and must be efficiently eluted for further analysis. Although protein digestion into smaller peptides should in theory cleave proteins into easily diffusible pieces, the reality is that losses incurred by this approach due to unavoidable incomplete digestion are still greater than if kept in solution. Additional limitations of 2DE include difficulties encountered in analysis of membrane proteins; extremely basic proteins, which are resistant to focusing; and high molecular weight proteins that will not migrate into the first dimension IPG strip or from the IPG strip into the second dimension gel. We have taken advantage of a relatively new methodology, OGE (19), and a classic biochemical method, RP-HPLC, and applied these methods to cardiac proteins of the sarcomeric subproteome. With the era of “shotgun” proteomics steering the development of separation technologies, RP-HPLC of intact proteins has been widely underutilized despite the fact that analyzing proteins in their intact form seems an attractive strategy. Nevertheless, there have been a few cases where RP-HPLC has been utilized at the protein level as the second dimension in tandem for two-dimensional protein separation methods following ion exchange chromatography, chromatofocusing, electroelution, or size exclusion chromatography (20–24), although these tandem technologies have not been routinely applied to cardiac muscle proteins. However, separate from two-dimensional LC applications, RP-HPLC has been used alone to separate sarcomeric proteins from failing versus non-failing swine hearts by Neverova and Van Eyk (25). Following buffer optimization, they were able to enhance separation of myofilament proteins and identify alterations in MHC in heart failure. Our work flow therefore expanded on this work by incorporating OGE as a reproducible method of sample simplification prior to RP-HPLC, thus creating a two-dimensional, in-solution separation of the sarcomeric subproteome that partitioned enriched pools of post-translationally modified protein. For our molecule of interest, cardiac RLC, this approach separated RLC charge variants into OFFGEL fractions with minimal overlap, thus providing an effective preparatory step for downstream PTM identification with mass spectrometry. Prior to using this approach, we had used 2DE to separate RLC charge variants followed by in-gel digestion and mass spectrometry. Although this approach was successful in identifying the highly abundant, singly phosphorylated RLC species in rodent cardiac muscle (9), a doubly phosphorylated peptide was undetectable with the mass spectrometry. The current approach has proven to be more effective and will be of value for PTM identification of other sarcomeric proteins in future studies.
Advantages of OGE as Preparatory Purification Step for High Molecular Weight Sarcomeric Proteins
Myosin and MyBP-C are high molecular weight proteins of the thick filament for which the identification of isoforms and post-translational modifications remains ambiguous. Muscle MHC exists as a heterogenous population (12) expressed as two unique isoforms, α and β (26), with post-translational modifications (27–31). MHC generates 150 tryptic peptides when unmodified and fully cleaved; therefore the endogenous number of MHC charge variants induced by isoforms and PTMs may be greater than 104. Thus, identifying and quantifying endogenous charge variants of MHC within a non-simplified cell lysate becomes difficult because of ion suppression effects and incomplete sequence coverage common to complex sample analysis combined with ambiguities in protein identifications couched in the problem of protein inference (13). Previous in-solution methods at separating full-length MHC have been explored using hydroxyapatite HPLC (12) and hydrophobic interaction HPLC (32); however, these separation chemistries have demonstrated limited and incomplete resolution of individual charged variants of MHC and have been successful only with purified myosin sample following a lengthy, multiple step biochemical purification. Furthermore, MHC is not amenable to separation via RP-HPLC because of irreversible binding to free silanol groups or strong hydrophobic interactions between the column packing material and myosin protein (33). MyBP-C, a second large molecular weight protein, is digested into 86 fully cleaved/unmodified tryptic peptides. Previous two-dimensional in-gel analysis demonstrated MyBP-C to be highly modified in vivo with 10 charged variants evident in mouse (8), suggesting that MyBP-C, like MHC, possesses numerous endogenous charge variants. At least three sites of MyBP-C phosphorylation have been identified (34–36); however, these cannot account for all MyBP-C charge variants observed, and therefore further analysis is required. To our knowledge, no one has demonstrated an in-solution separation of full-length cardiac MyBP-C into unique charge variants. The techniques described in our study should be applicable to the separation and analysis of full-length MyBP-C and MHC.
Importance of Identifying PTMs in RLC
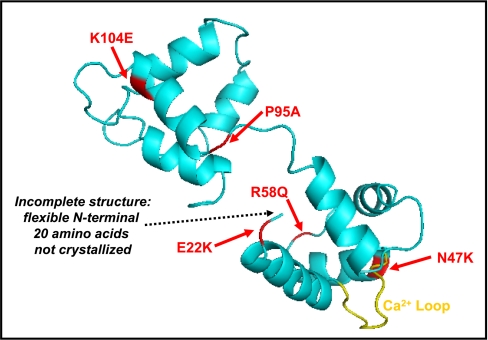
The identification of precise sites of RLC modifications, as has been accomplished in this study, leads to a more complete understanding of intra- and intermolecular interactions that occur within the myosin motor and contribute to disease mechanisms. It has been noted from familial hypertrophic cardiomyopathic (FHC) mutations occurring in all thick filament proteins (MHC, MyBP-C, ELC, and RLC) that a change in one amino acid side chain can have an enormous effect on cardiac morphology and function. The exchange of a Ser or Thr hydroxyl with a bulky, charged 80-dalton phosphoester is chemically more substantial than the physical or chemical changes induced by a number of FHC mutations. In the case of RLC, several FHC mutations have been documented that cause mid-left ventricular obstruction and papillary muscle hypertrophy, and not coincidentally, these residues tend to cluster around the Ca2+ binding domain and the N-terminal phosphorylatable region (37) (Fig. 8). The importance of identifying sites of endogenous phosphorylation is underscored by the observation that RLC Ca2+ binding, phosphorylation potential, and secondary structure were significantly altered in FHC mutants A13T, F18L, E22K, R58Q, P95A, and N47Q (37). Interestingly, phosphorylation of wild-type RLC by myosin light chain kinase significantly decreased the helical content of the molecule, suggesting that RLC undergoes a substantial conformational change induced by phosphorylation (37). In addition, neighboring thick filament proteins, including MyBP-C and titin, are phosphorylated (38, 39), and there is evidence to suggest that the phosphorylation of MyBP-C governs its interaction with the S2 hinge region of MHC (40), the precise location of RLC binding. In this context, our laboratory has observed dephosphorylation of MyBP-C in mice expressing a non-phosphorylatable RLC, suggesting that the phosphorylation of MyBP-C and RLC may be functionally linked (8). Thus, it is clear that the definition of sites of endogenous modifications in RLC (as well as other cardiac thick filament proteins) will lead to a molecular description of cardiac muscle function.
Fig. 8.
FHC mutations identified in RLC. Shown is a three-dimensional illustration of RLC structure (turquoise) adapted from Protein Data Bank code 1wdc. Identified FHC point mutations are shown in red, and the Ca2+ binding loop is shown in yellow.
RLC Modifications in Human Heart Failure
Previous studies demonstrated a decrease in RLC phosphorylation in human end stage heart failure (41–43). Our data show a similar trend in phosphorylation with the novel addition of alterations in deamidation in the N-terminal region of RLC in human heart failure. White et al. (44) previously identified deamidation and phosphorylation of N-terminal RLC in rabbit cardiac muscle; however, our study is the first to map these modifications in human RLC and to quantify the differences in human heart failure. The functional significance of the observed deamidation remains obscure as this can be either a biologically relevant signal or a chemical artifact. Deamidation is a relatively common modification in human proteins and has been generally thought of as a negative functional change via signaling proteins for degradation or replacement (45). It is surprising, therefore, that in our study the non-failing tissue showed significantly greater deamidation than the DCM samples. It is possible that cleavage of RLC at the residue 14/15 junction is occurring in DCM samples as has been observed in human DCM (46) and rabbit ischemia-reperfusion (44), thus lowering the effective concentration of 9–30 peptide present in these samples. How the differences in each variant of RLC may contribute to the contractile dysfunction observed in heart failure is uncertain. It is unlikely that the observed decreases in RLC phosphorylation and deamidation are causative for heart failure, but rather the important finding reported here is that RLC is modified (along with other proteins) in a diseased state, suggesting that this change may functionally contribute within the context of the pathogenesis of heart failure.
RLC in Therapeutic Interventions for Cardiac Failure
The myosin motor is a prime target for therapeutic intervention in clinical heart failure. Small molecules that specifically target myosin and produce a positive functional effect, termed “sarcomere activators,” are now in clinical trials (47–49). A sarcomere activator that increases the intensity of cardiac contraction per unit of activating Ca2+ bound to troponin C without altering the Ca2+ transient would be energetically favorable for the heart. The myosin light chains (ELC and RLC) bound between the active site and the lever arm fulcrum of MHC are ideally located to regulate enzymatic activity of the myosin head and are a plausible site of action for sarcomere activators. Endogenous MHC-ELC-RLC conformations are governed by local charge changes induced by phosphorylation (50) that subsequently affect function. Indeed, it has been demonstrated that in isolated skinned cardiac fibers treated with myosin light chain kinase there was an increase in MgATPase activity (51) and tension per unit of Ca2+ (5–7). Furthermore, our laboratory demonstrated that in mice expressing RLC with serines 14/15/19 mutated to alanine (TG-RLC(P−)) myofibrillar ATPase was significantly decreased (8). Okafor et al. (52) examined the effect of MCI-154, a myofilament Ca2+ sensitizer with unknown mechanism, on skinned fibers from non-failing, idiopathic, or ischemic cardiomyopathic patients and reported significant increases in myosin-specific ATPase activity independent of the thin filament regulatory proteins troponin and tropomyosin. Additionally, our laboratory demonstrated that TG-RLC(P−) mice had altered in vivo systolic kinetics measured by an increase in ejection time (8), and coincidentally, the sarcomere activator CK-1827452, now in clinical trials, increases left ventricular ejection time and stroke volume in healthy patients and those with heart failure (47–49). Although the detailed myosin-specific mechanism of action of both MCI-154 and CK-1827452 are unknown, it is highly likely that alterations in charge near the binding region of RLC, which is modulated by phosphorylation, play a role.
Concluding Remarks
We have demonstrated a novel application of current state-of-the-art in-solution separation technologies to cardiac sarcomeric proteins, allowing the novel identification and quantification of charge variants of cardiac RLC: a singly and doubly phosphorylated species in mouse and a singly phosphorylated, singly deamidated, and deamidated-phosphorylated species in human. Although it is understood that the phosphorylation of RLC is critical for cardiac function (8), future studies must address the significance of the multiple additional charge variants present in human cardiac RLC.
Supplementary Material
Acknowledgments
We thank Dr. Lori A. Walker from the University of Colorado at Denver for helpful discussions and Dr. Peipei Ping from the University of California at Los Angeles for generous support of the project. We acknowledge the Proteomics and Mass Spectrometry Facility at National Jewish Health and the University of Illinois at Chicago (UIC) Chicago Biomedical Consortium and Research Resources Center (CBC-RRC) for assistance. The CBC-RRC was established by a grant from The Searle Funds at the Chicago Community Trust to the Chicago Biomedical Consortium.
* This work was supported, in whole or in part, by National Institutes of Health Grants P01 HL62426 (to R. J. S. and P. M. B.), R01 HL22231 (to R. J. S.), T32 HL07962 (to S. B. S.), and P01 HL80111 (to P. Ping). This work was also supported by American Heart Association Grant 0710031Z (to S. B. S.).
 This article contains supplemental Fig. S1.
This article contains supplemental Fig. S1.
2 S. B. Scruggs, R. J. Solaro, and P. M. Buttrick, unpublished observations.
1 The abbreviations used are:
- PTM
- post-translational modification
- RP
- reverse-phase
- ELC
- essential light chain
- FHC
- familial hypertrophic cardiomyopathic
- MHC
- myosin heavy chain
- MyBP-C
- myosin-binding protein-C
- OGE
- OFFGEL electrophoresis
- RLC
- regulatory light chain
- SPI
- scored peak intensity
- 2DE
- two-dimensional electrophoresis
- IPI
- International Protein Index
- mAU
- milliabsorbance unit
- DCM
- dilated cardiomyopathy.
REFERENCES
- 1.Hinken A. C., Solaro R. J. (2007) A dominant role of cardiac molecular motors in the intrinsic regulation of ventricular ejection and relaxation. Physiology 22, 73–80 [DOI] [PubMed] [Google Scholar]
- 2.Solaro R. J. (2001) Heart Physiology and Pathophysiology, 4th Ed., Academic Press, New York: 519–526 [Google Scholar]
- 3.Solaro R. J., Rarick H. M. (1998) Troponin and tropomyosin: proteins that switch on and tune in the activity of cardiac myofilaments. Circ. Res. 83, 471–480 [DOI] [PubMed] [Google Scholar]
- 4.Chan J. Y., Takeda M., Briggs L. E., Graham M. L., Lu J. T., Horikoshi N., Weinberg E. O., Aoki H., Sato N., Chien K. R., Kasahara H. (2008) Identification of cardiac-specific myosin light chain kinase. Circ. Res. 102, 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sweeney H. L., Stull J. T. (1986) Phosphorylation of myosin in permeabilized mammalian cardiac and skeletal muscle cells. Am. J. Physiol. Cell Physiol. 250, C657–C660 [DOI] [PubMed] [Google Scholar]
- 6.Olsson M. C., Patel J. R., Fitzsimons D. P., Walker J. W., Moss R. L. (2004) Basal myosin light chain phosphorylation is a determinant of Ca2+ sensitivity of force and activation dependence of the kinetics of myocardial force development. Am. J. Physiol. Heart Circ. Physiol. 287, H2712–H2718 [DOI] [PubMed] [Google Scholar]
- 7.Morano I., Hofmann F., Zimmer M., Rüegg J. C. (1985) The influence of P-light chain phosphorylation by myosin light chain kinase on the calcium sensitivity of chemically skinned heart fibers. FEBS Lett. 189, 221–224 [DOI] [PubMed] [Google Scholar]
- 8.Scruggs S. B., Hinken A. C., Thawornkaiwong A., Robbins J., Walker L. A., de Tombe P. P., Geenen D. L., Buttrick P. M., Solaro R. J. (2009) Ablation of ventricular myosin regulatory light chain phosphorylation in mice causes cardiac dysfunction in situ and affects neighboring myofilament protein phosphorylation. J. Biol. Chem. 284, 5097–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan C., Sheng Q., Tang H., Li Y., Zeng R., Solaro R. J. (2008) Quantitative comparison of sarcomeric phosphoproteomes of neonatal and adult rat hearts. Am. J. Physiol. Heart Circ. Physiol. 295, H647–H656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warren C. M., Krzesinski P. R., Greaser M. L. (2003) Vertical agarose gel electrophoresis and electroblotting of high-molecular-weight proteins. Electrophoresis 24, 1695–1702 [DOI] [PubMed] [Google Scholar]
- 11.Libera L. D. (1988) A comparative study of chicken ventricular and slow skeletal myosin light chains. Cell Biol. Int. Rep. 12, 1089–1098 [DOI] [PubMed] [Google Scholar]
- 12.Kawasaki T., Takahashi S., Ikeda K. (1985) Hydroxyapatite high-performance liquid chromatography: column performance for proteins. Eur. J. Biochem. 152, 361–371 [DOI] [PubMed] [Google Scholar]
- 13.Nesvizhskii A. I., Aebersold R. (2005) Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics 4, 1419–1440 [DOI] [PubMed] [Google Scholar]
- 14.Hörth P., Miller C. A., Preckel T., Wenz C. (2006) Efficient fractionation and improved protein identification by peptide OFFGEL electrophoresis. Mol. Cell. Proteomics 5, 1968–1974 [DOI] [PubMed] [Google Scholar]
- 15.Ishihama Y. (2005) Proteomic LC-MS systems using nanoscale liquid chromatography with tandem mass spectrometry. J. Chromatogr. A 1067, 73–83 [DOI] [PubMed] [Google Scholar]
- 16.Solaro R. J., Pang D. C., Briggs F. N. (1971) The purification of cardiac myofibrils with Triton X-100. Biochim. Biophys. Acta 245, 259–262 [DOI] [PubMed] [Google Scholar]
- 17.Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. (2004) The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 [DOI] [PubMed] [Google Scholar]
- 18.Fritz J. D., Swartz D. R., Greaser M. L. (1989) Factors affecting polyacrylamide gel electrophoresis and electroblotting of high-molecular-weight myofibrillar proteins. Anal. Biochem. 180, 205–210 [DOI] [PubMed] [Google Scholar]
- 19.Ros A., Faupel M., Mees H., Oostrum J., Ferrigno R., Reymond F., Michel P., Rossier J. S., Girault H. H. (2002) Protein purification by Off-Gel electrophoresis. Proteomics 2, 151–156 [DOI] [PubMed] [Google Scholar]
- 20.Wagner K., Racaityte K., Unger K. K., Miliotis T., Edholm L. E., Bischoff R., Marko-Varga G. (2000) Protein mapping by two-dimensional high performance liquid chromatography. J. Chromatogr. A 893, 293–305 [DOI] [PubMed] [Google Scholar]
- 21.Isobe T., Uchida K., Taoka M., Shinkai F., Manabe T., Okuyama T. (1991) Automated two-dimensional liquid chromatographic system for mapping proteins in highly complex mixtures. J. Chromatogr. 588, 115–123 [DOI] [PubMed] [Google Scholar]
- 22.Chong B. E., Yan F., Lubman D. M., Miller F. R. (2001) Chromatofocusing nonporous reversed-phase high-performance liquid chromatography/electrospray ionization time-of-flight mass spectrometry of proteins from human breast cancer whole cell lysates: a novel two-dimensional liquid chromatography/mass spectrometry method. Rapid Commun. Mass Spectrom. 15, 291–296 [DOI] [PubMed] [Google Scholar]
- 23.Rose D. J., Opiteck G. J. (1994) Two-dimensional gel electrophoresis/liquid chromatography for the micropreparative isolation of proteins. Anal. Chem. 66, 2529–2536 [DOI] [PubMed] [Google Scholar]
- 24.Opiteck G. J., Ramirez S. M., Jorgenson J. W., Moseley M. A., 3rd. (1998) Comprehensive two-dimensional high-performance liquid chromatography for the isolation of overexpressed proteins and proteome mapping. Anal. Biochem. 258, 349–361 [DOI] [PubMed] [Google Scholar]
- 25.Neverova I., Van Eyk J. E. (2002) Application of reversed phase high performance liquid chromatography for subproteomic analysis of cardiac muscle. Proteomics 2, 22–31 [PubMed] [Google Scholar]
- 26.Palmer B. M. (2005) Thick filament proteins and performance in human heart failure. Heart Fail. Rev. 10, 187–197 [DOI] [PubMed] [Google Scholar]
- 27.Schilder R. J., Marden J. H. (2007) Parasites, proteomics and performance: effects of gregarine gut parasites on dragonfly flight muscle composition and function. J. Exp. Biol. 210, 4298–4306 [DOI] [PubMed] [Google Scholar]
- 28.Hedou J., Cieniewski-Bernard C., Leroy Y., Michalski J. C., Mounier Y., Bastide B. (2007) O-linked N-acetylglucosaminylation is involved in the Ca2+ activation properties of rat skeletal muscle. J. Biol. Chem. 282, 10360–10369 [DOI] [PubMed] [Google Scholar]
- 29.Ramamurthy B., Höök P., Larsson L. (1999) An overview of carbohydrate-protein interactions with specific reference to myosin and ageing. Acta Physiol. Scand. 167, 327–329 [DOI] [PubMed] [Google Scholar]
- 30.Cieniewski-Bernard C., Bastide B., Lefebvre T., Lemoine J., Mounier Y., Michalski J. C. (2004) Identification of O-linked N-acetylglucosamine proteins in rat skeletal muscle using two-dimensional gel electrophoresis and mass spectrometry. Mol. Cell. Proteomics 3, 577–585 [DOI] [PubMed] [Google Scholar]
- 31.Ramamurthy B., Höök P., Jones A. D., Larsson L. (2001) Changes in myosin structure and function in response to glycation. FASEB J. 15, 2415–2422 [DOI] [PubMed] [Google Scholar]
- 32.Malmqvist U. P., Aronshtam A., Lowey S. (2004) Cardiac myosin isoforms from different species have unique enzymatic and mechanical properties. Biochemistry 43, 15058–15065 [DOI] [PubMed] [Google Scholar]
- 33.Libera L. D. (2001) High performance liquid chromatography in the analysis and separation of contractile proteins. Basic Appl. Myol. 11, 115–118 [Google Scholar]
- 34.Yuan C., Guo Y., Ravi R., Przyklenk K., Shilkofski N., Diez R., Cole R. N., Murphy A. M. (2006) Myosin binding protein C is differentially phosphorylated upon myocardial stunning in canine and rat hearts—evidence for novel phosphorylation sites. Proteomics 6, 4176–4186 [DOI] [PubMed] [Google Scholar]
- 35.Mohamed A. S., Dignam J. D., Schlender K. K. (1998) Cardiac myosin-binding protein C (MyBP-C): identification of protein kinase A and protein kinase C phosphorylation sites. Arch. Biochem. Biophys 358, 313–319 [DOI] [PubMed] [Google Scholar]
- 36.Schlender K. K., Bean L. J. (1991) Phosphorylation of chicken cardiac C-protein by calcium/calmodulin-dependent protein kinase II. J. Biol. Chem. 266, 2811–2817 [PubMed] [Google Scholar]
- 37.Szczesna D., Ghosh D., Li Q., Gomes A. V., Guzman G., Arana C., Zhi G., Stull J. T., Potter J. D. (2001) Familial hypertrophic cardiomyopathy mutations in the regulatory light chains of myosin affect their structure, Ca2+ binding, and phosphorylation. J. Biol. Chem. 276, 7086–7092 [DOI] [PubMed] [Google Scholar]
- 38.Yamasaki R., Wu Y., McNabb M., Greaser M., Labeit S., Granzier H. (2002) Protein kinase A phosphorylates titin's cardiac-specific N2B domain and reduces passive tension in rat cardiac myocytes. Circ. Res. 90, 1181–1188 [DOI] [PubMed] [Google Scholar]
- 39.Hartzell H. C., Glass D. B. (1984) Phosphorylation of purified cardiac muscle C-protein by purified cAMP-dependent and endogenous Ca2+-calmodulin-dependent protein kinases. J. Biol. Chem. 259, 15587–15596 [PubMed] [Google Scholar]
- 40.Gruen M., Prinz H., Gautel M. (1999) cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on-off fashion. FEBS Lett. 453, 254–259 [DOI] [PubMed] [Google Scholar]
- 41.van der Velden J., Papp Z., Boontje N. M., Zaremba R., de Jong J. W., Janssen P. M., Hasenfuss G., Stienen G. J. (2003) The effect of myosin light chain 2 dephosphorylation on Ca2+ sensitivity of force is enhanced in failing human hearts. Cardiovasc. Res. 57, 505–514 [DOI] [PubMed] [Google Scholar]
- 42.van Der Velden J., Klein L. J., Zaremba R., Boontje N. M., Huybregts M. A., Stooker W., Eijsman L., de Jong J. W., Visser C. A., Visser F. C., Stienen G. J. (2001) Effects of calcium, inorganic phosphate, and pH on isometric force in single skinned cardiomyocytes from donor and failing human hearts. Circulation 104, 1140–1146 [DOI] [PubMed] [Google Scholar]
- 43.Morano I. (1992) Effects of different expression and posttranslational modifications of myosin light chains on contractility of skinned human cardiac fibers. Basic Res. Cardiol. 87, 129–141 [DOI] [PubMed] [Google Scholar]
- 44.White M. Y., Cordwell S. J., McCarron H. C., Tchen A. S., Hambly B. D., Jeremy R. W. (2003) Modifications of myosin-regulatory light chain correlate with function of stunned myocardium. J. Mol. Cell. Cardiol. 35, 833–840 [DOI] [PubMed] [Google Scholar]
- 45.Robinson N. E., Robinson A. B. (2001) Deamidation of human proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 12409–12413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holt J. C., Caulfield J. B., Norton P., Chantler P. D., Slayter H. S., Margossian S. S. (1995) Human cardiac myosin light chains: sequence comparisons between myosin LC1 and LC2 from normal and idiopathic dilated cardiomyopathic hearts. Mol. Cell. Biochem. 145, 89–96 [DOI] [PubMed] [Google Scholar]
- 47.Cleland J. G., Coletta A. P., Clark A. L. (2006) Clinical trials update from the Heart Failure Society of America meeting: FIX-CHF-4, selective cardiac myosin activator and OPT-CHF. Eur. J. Heart Fail. 8, 764–766 [DOI] [PubMed] [Google Scholar]
- 48.Coletta A. P., Cleland J. G., Cullington D., Clark A. L. (2008) Clinical trials update from Heart Rhythm 2008 and Heart Failure 2008: ATHENA, URGENT, INH study, HEART and CK-1827452. Eur. J. Heart Fail. 10, 917–920 [DOI] [PubMed] [Google Scholar]
- 49.Solaro R. J. (2009) CK-1827452, a sarcomere-directed cardiac myosin activator for acute and chronic heart disease. IDrugs 12, 243–251 [PubMed] [Google Scholar]
- 50.Levine R. J., Kensler R. W., Yang Z., Stull J. T., Sweeney H. L. (1996) Myosin light chain phosphorylation affects the structure of rabbit skeletal muscle thick filaments. Biophys. J. 71, 898–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clement O., Puceat M., Walsh M. P., Vassort G. (1992) Protein kinase C enhances myosin light chain kinase effects on force development and ATPase activity in rat single skinned cardiac cells. Biochem. J. 285, 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okafor C., Liao R., Perreault-Micale C., Li X., Ito T., Stepanek A., Doye A., de Tombe P., Gwathmey J. K. (2003) Mg-ATPase and Ca+ activated myosin ATPase activity in ventricular myofibrils from non-failing and diseased human hearts—effects of calcium sensitizing agents MCI-154, DPI 201–106, and caffeine. Mol. Cell. Biochem. 245, 77–89 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.