Abstract
The nucleus is a highly structured organelle and contains many functional compartments. Although the structural basis for this complex spatial organization of compartments is unknown, a major component of this organization is likely to be the non-chromatin scaffolding called nuclear matrix (NuMat). Experimental evidence over the past decades indicates that most of the nuclear functions are at least transiently associated with the NuMat, although the components of NuMat itself are poorly known. Here, we report NuMat proteome analysis from Drosophila melanogaster embryos and discuss its links with nuclear architecture and functions. In the NuMat proteome, we found structural proteins, chaperones, DNA/RNA-binding proteins, chromatin remodeling and transcription factors. This complexity of NuMat proteome is an indicator of its structural and functional significance. Comparison of the two-dimensional profile of NuMat proteome from different developmental stages of Drosophila embryos showed that less than half of the NuMat proteome is constant, and the rest of the proteins are stage-specific dynamic components. These NuMat dynamics suggest a possible functional link between NuMat and embryonic development. Finally, we also showed that a subset of NuMat proteins remains associated with the mitotic chromosomes, implicating their role in mitosis and possibly the epigenetic cellular memory. NuMat proteome analysis provides tools and opens up ways to understand nuclear organization and function.
The eukaryotic cell nucleus contains a number of structural features like heterochromatin, nucleolus, nuclear lamina, and nuclear territories and functional features like transcription foci, replication foci, Cajal bodies, stress-induced foci, etc. (1–7). Packaging of the genome on the other hand also results in non-random distribution of a variety of structural features like chromatin domains that help to regulate expression of genes (8). How these complex structural and functional domains are established and maintained is not known. It is likely that the non-chromatin scaffolding called nuclear matrix (9) plays an important role in this process. Nuclear matrix (NuMat)1 mainly consists of the nuclear lamina, the nucleolar remnants, and an internal nuclear meshwork of fibers of unknown constitution that can be seen by electron microscopy (10). These fibers have ultrastructural features reminiscent of intermediate filaments of the cytosol (11). The protein component of nucleoplasmic filaments, however, is largely unknown. Earlier studies on a few proteins isolated from nuclear matrix from different organisms have shown them to be DNA- and RNA-binding and structural components of the nuclear pore complex (12), enzymes (13), and nuclear membrane proteins (14). This diversity among the small number of identified NuMat constituents reflects the link of nuclear architecture with the variety of nuclear functions. The importance of NuMat constituents in nuclear processes has been apparent for a long time, and several studies have been carried out to identify the proteins of this complex structure in different organisms (15–18). Major advancements in the field of proteomics now provide further scope for extensive analysis of NuMat in this context.
Here, we present a comprehensive analysis of the NuMat proteome of Drosophila melanogaster. NuMat preparations from D. melanogaster embryos were separated by one-dimensional gel electrophoresis, and the proteins were identified by LC-MS/MS. We also report a remarkable variation in the NuMat proteome depending on developmental stages, indicating a link between embryonic development and nuclear architecture. Finally, immunostaining using antibody against NuMat components points to a link between the nuclear architecture and epigenetic cellular memory.
MATERIALS AND METHODS
Drosophila S2 Cell and Embryo Collections
S2 cells were grown on Schneider's medium (Himedia Laboratories) with 10% fetal bovine serum and penicillin and streptomycin antibiotics. Embryos were collected on plates containing solidified fly food base streaked with yeast paste in boxes containing Canton S flies. Embryos were collected after 16 h (0–16 h). For developmental stage-specific analysis, 0–2-, 6–8-, and 14–16-h embryos were collected.
NuMat Preparation from Drosophila Embryos
D. melanogaster embryos were collected after 16 h and dechorionated with 50% Chlorex (4–6% sodium hypochlorite) to remove chorion for 5 min. After washing thoroughly under running water, embryos were homogenized in 0.25 m sucrose in nuclear isolation buffer (15 mm Tris, pH 7.4, 40 mm KCl, 1 mm EDTA, 0.1 mm EGTA, 0.1 mm PMSF, 0.25 mm spermidine) and filtered through Miracloth (Calbiochem). The filtrate obtained was spun at 600 × g for 2 min to remove the debris. The supernatant was adjusted to 1.8 m sucrose and centrifuged at 6000 × g to isolate the nuclear pellet. The pellet was washed by resuspending in nuclear isolation buffer containing 0.25 m sucrose and spinning at 3000 × g for 10 min. The nuclear pellet was incubated in digestion buffer (20 mm Tris, pH 7.4, 20 mm KCl, 70 mm NaCl, 10 mm MgCl2, 0.125 mm spermidine, 0.1 mm PMSF, 0.5% Triton X-100, and 40 units/μl DNase I) at 4 °C for 1 h. Digestion was followed by extraction with 0.4 m NaCl for 5 min and after increasing the concentration to 2 m NaCl extraction for an additional 5 min in extraction buffer (10 mm Hepes, pH 7.5, 4 mm EDTA, 0.25 mm spermidine, 0.1 mm PMSF, 0.5% Triton X-100). The final pellet after extraction was washed twice with wash buffer (5 mm Tris, 20 mm KCl, 1 mm EDTA, 0.25 mm spermidine, 0.1 mm PMSF) 5 min each time. After washing, the pellet was stored at −70 °C or used for further analysis.
NuMat Preparation from S2 Cells on Microscopic Glass Slides
Approximately 104 cells were spun onto microscopic glass slides with a Cytospin at 900 rpm for 10 min. The cells attached to the glass slides were washed with PBS. Attached cells were extracted with 2 m NaCl extraction buffer to prepare halos. DNase I digestion was carried out on halos as described above to get NuMat. At all the stages, the samples were fixed with 4% formaldehyde and washed before proceeding for immunostaining and imaging.
Difference Gel Electrophoresis (DIGE)
Three samples of 25 μg of NuMat proteins, each from three different stage embryo collections, were labeled with CyDye DIGE Fluor (Amersham Biosciences) according to the manufacturer's specifications. For two-dimensional gel electrophoresis, NuMat proteins were solubilized in rehydration buffer (8 m urea, 2% CHAPS, 50 mm DTT). 11-cm IEF strips (3–10 pH range ReadyStrip™, Bio-Rad) were soaked in rehydration buffer containing proteins at 20 °C overnight. Isoelectric focusing was carried out at 8000 V until 45,000 V-h. Second dimension protein separation was done by SDS-PAGE. The gels were scanned using a Typhoon Trio scanner (Amersham Biosciences).
Gel Electrophoresis and In-gel Digestion for LC-MS/MS
For each LC-MS/MS experiment, 200 μg of the nuclear matrix preparation was fractionated by 12% SDS-PAGE. The gels were stained with Coomassie Brilliant Blue (R-250) for 1 h, destained, and washed with Milli-Q water several times. Each gel lane was sliced into several pieces and washed three times for 30 min each in 50% acetonitrile (ACN) with 25 mm ammonium bicarbonate (ABC), pH 8.0 to remove excess Coomassie stain. One final wash was done with 50% ACN, 10 mm ABC to remove excess salt. After the washings, the gel slices were soaked in 100% ACN for 5 min to dehydrate the gels. Excess ACN was removed, and the gel slices were vacuum-dried for 30 min. The dried gels were rehydrated and trypsinized with 30 μl of cold trypsin (Promega) solution (10 μg/ml in 25 mm ABC, pH 8.0) and incubated at 37 °C for 16 h. The tryptic peptides were extracted by soaking the gel slices in 50 μl of 50% ACN, 5% trifluoroacetic acid (TFA) for 60 min with gentle agitation at room temperature. The supernatant was collected and transferred to a second clean microcentrifuge tube. The gels were extracted again with another 50-μl aliquot of 50% ACN, 5% TFA for 60 min. The two extracts were pooled and vacuum-dried to complete dryness for about 1 h. Before loading, the samples were reconstituted in 12 μl of 5% ACN, 0.1% formic acid.
LC-MS/MS Analysis
All LC-MS/MS experiments were carried out on an ESI mass spectrometer with a linear ion trap mass analyzer (LTQ-IT, Thermo Fischer, Waltham, MA) equipped with a Finnigan Surveyor MS Pump Plus. 10 μl of the sample was loaded with a constant flow of 2 μl/min onto a reverse phase Micro LC column (Bio Basic C18, Thermo Fischer). Peptides were eluted on a gradient of 120 min for each gel slice starting with 100% water for the first 20 min in which 10 min were for retention of peptides in water. The acetonitrile gradient was set from 0 to 100% over the next 90 min followed by a 100% water wash for the last 10 min. Chromatographically separated peptides were sprayed through a 20-cm metal needle emitter, and the mass spectrometer was operated in the data-dependent mode to acquire MS and MS/MS spectra, switching automatically between MS and MS/MS modes. One full MS scan from 200 to 2000 m/z followed by seven data-dependent MS/MS scans was recorded. The electrospray voltage was set at 4.0 kV, and the capillary temperature was set at 200 °C. The peptides were fragmented using CID with a normalized collision energy of 35%. The top seven peptide precursor ions were selected for MS/MS analysis. The raw files acquired were compiled and subjected to bioinformatics analysis.
Bioinformatics Analysis
The mass spectra obtained were searched against the protein sequences of the D. melanogaster assembly 5.2 obtained from NCBI (20,513 proteins) using the SEQUEST algorithm incorporated in the BioWorks Browser (Version 3.2 EF2, Thermo Electron Corp.) (19). Enzyme specificity was set to full trypsin digestion with only one missed cleavage. Methionine oxidation was set as a variable modification. The other parameters were set as follows: precursor ion tolerance was 1 amu, and fragment ion tolerance was 0.35 amu.
Peptide identifications were accepted if they passed the following filter criteria: ΔCN value of 0.100; Rsp of 5; Xcorr versus charge values of 1.90 (1+ charge), 2.20 (2+ charge), and 3.30 (3+ charge); and protein probability of 0.001. An in-house program was used to remove redundant peptides and retrieve data from FlyBase (a database for Drosophila genetics and molecular biology). Protein identifications were accepted only if they contained at least two unique peptides Supplemental Table 1. We also found multiple isoforms of several proteins, but because no isoform-specific peptide was detected, we considered isoforms of one protein as a single entry in our count for NuMat proteome. Single peptide hits are listed in Supplemental Table 2 and the corresponding spectra are provided in Supplemental data files 1, 2, 3 & 4.
Antibodies and Immunostaining
Drosophila embryo NuMat (0–16 h) was solubilized in 6 m guanidinium chloride in PBS and serially dialyzed against 3, 1.5, 0.75, and 0.36 m guanidinium chloride, and the insoluble fractions were collected. Rats were immunized against these insoluble fractions of the NuMat. Antisera were used for immunostaining 0–2-h D. melanogaster embryos following a published protocol (20).
RESULTS
NuMat Preparation and Quality Control
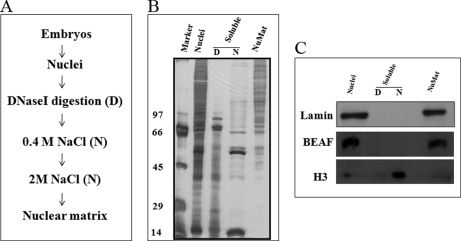
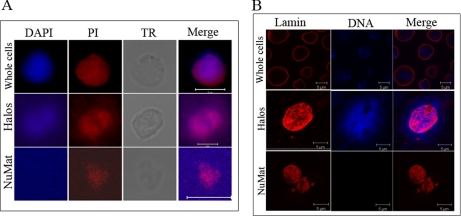
NuMat is a complex structure that contains proteins, RNA, and matrix-associated regions (MARs) of genomic DNA. We modified slightly the work flow of existing NuMat extraction protocols (Fig. 1A) and introduced several quality control checks to ensure a consistent NuMat preparation. As shown in Fig. 1B, NuMat preparations are enriched in high molecular weight proteins. The most reliable criterion for good NuMat preparation is the presence of known NuMat proteins and the absence of non-NuMat proteins in Western blot (Fig. 1C) analysis. The nuclear versus NuMat proportion of RNA, DNA, and proteins is yet another relatively convenient parameter for reliable preparation (21). Retention of RNA (∼45%), a small fraction of DNA (∼0.92%), and proteins (∼10%) in the NuMat preparation from Drosophila cells/embryos is considered acceptable (21). The quality of the NuMat preparation used for proteomics analysis was confirmed by PAGE (Fig. 1) and Western analysis. Known NuMat proteins like BEAF and lamin are positive controls, and histone (H3) is a negative control. Histones are eluted out in salt washes and are, therefore, not retained in matrix, whereas high molecular weight proteins get enriched (Fig. 1C). Finally, we used the cell biology criteria for a good NuMat preparation by looking for the shape of the nuclear structures, the presence of RNA as seen by propidium iodide (PI) stain, and an undetectable amount of DNA in the DAPI-stained preparations (Fig. 2A). Distribution of lamin was checked by immunostaining of S2 cell and NuMat preparations. In S2 whole cells, lamin showed nuclear rim staining, whereas in NuMat preparations it was seen forming a fibrous network (Fig. 2B). It has been shown earlier that an internal lamin network is visualized by immunostaining only after the removal of chromatin from the nucleus (22).
Fig. 1.
Isolation of NuMat. A, outline of NuMat preparation protocol from D. melanogaster embryos. B, extracts in each step on a silver-stained PAGE gel. High molecular weight proteins are retained in the NuMat, whereas histones are extracted by salt extraction. C, Western blot to show that BEAF and lamin, known NuMat proteins, are retained in NuMat, whereas histone H3, a non-NuMat protein, is extracted as a soluble fraction. In all panels, D and N represent the soluble fractions of the nucleus after DNase I digestion and NaCl extraction, respectively.
Fig. 2.
RNA and lamin are components of S2 cell NuMat. A, DAPI and PI are seen staining the entire nuclear volume in the whole cell, whereas DNA is seen around the nucleus after extraction in halos, and DAPI signal is lost in the NuMat, indicating complete chromatin digestion. PI staining, which stains both DNA and RNA, persists in NuMat, indicating the presence of RNA component. TR is the transmission microscopic image. B, lamin shows nuclear rim staining in the whole cell, whereas in halos and NuMat the internal meshwork of lamin is accessible for staining. The absence of DAPI signal in NuMat indicates efficient removal of chromatin, leaving clean NuMat in the preparation. Scale bars, 5 micron.
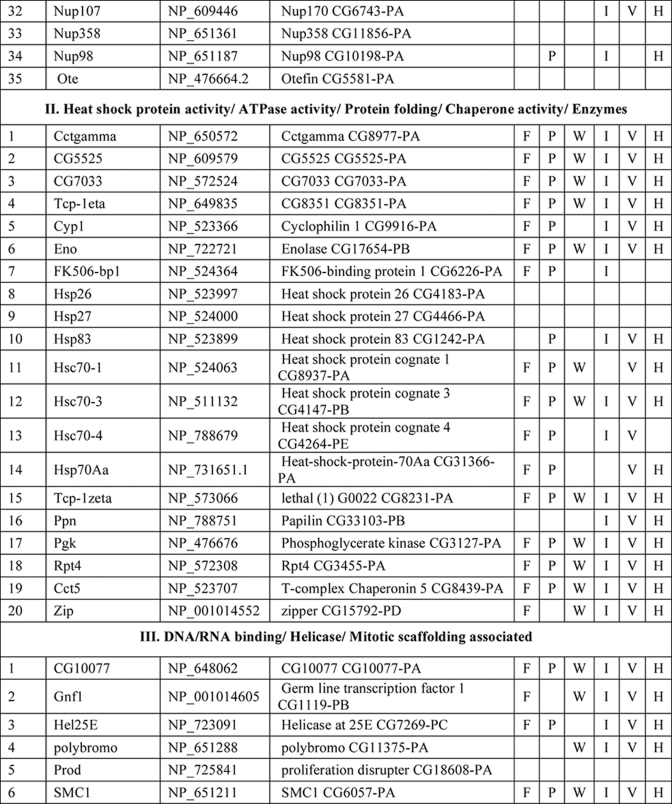
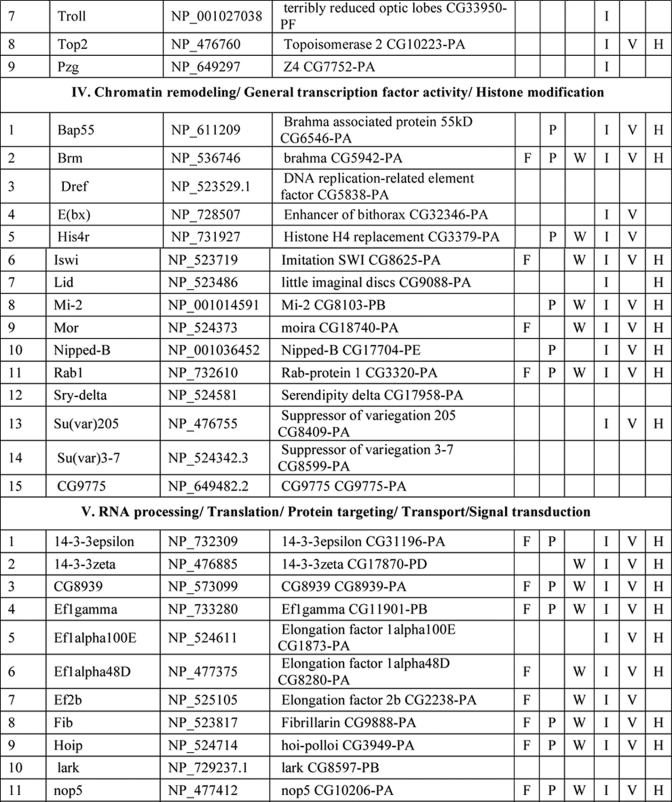
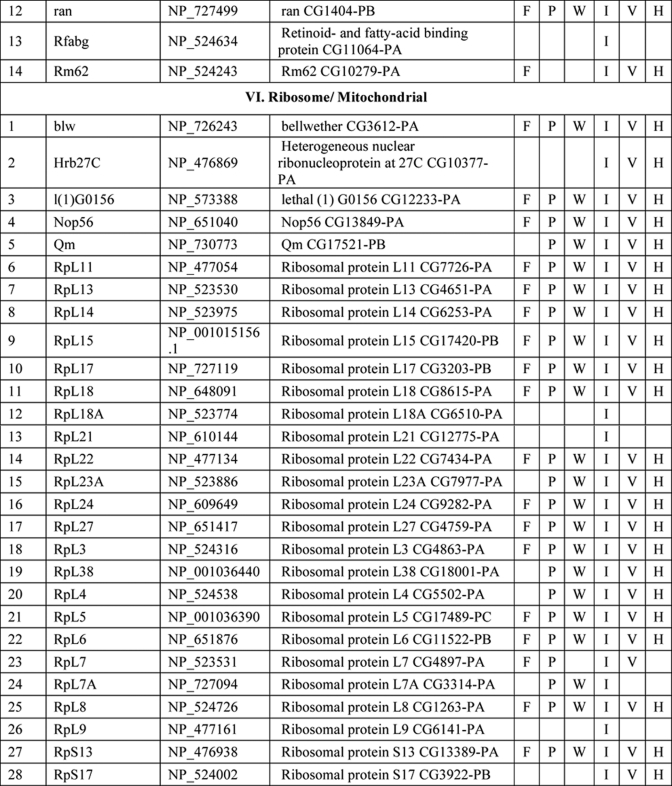
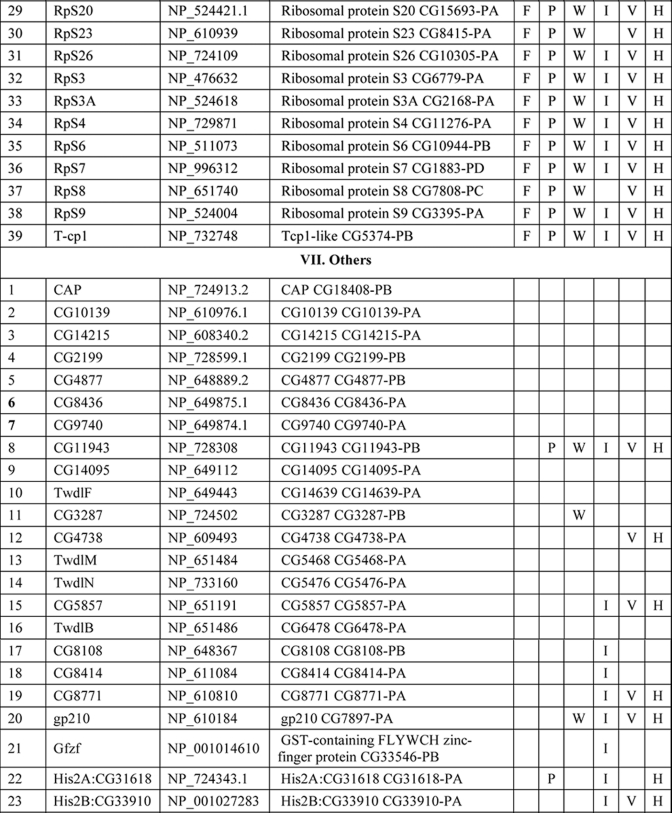
NuMat Proteome of D. melanogaster
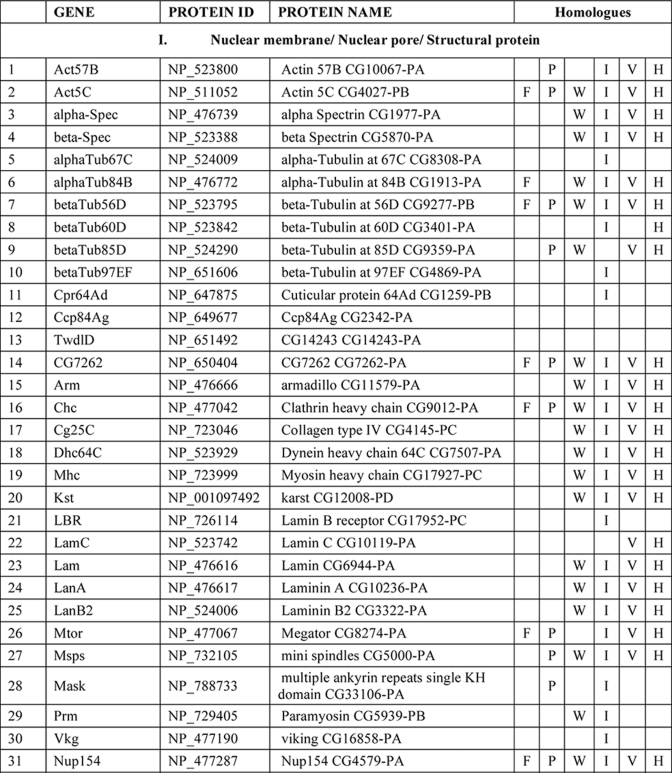
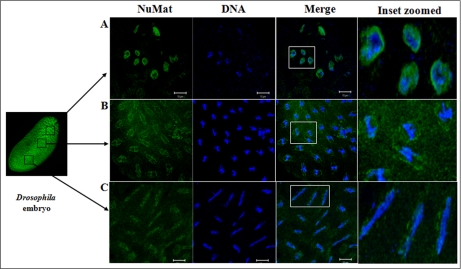
Several hundred spots were detected by two-dimensional gel electrophoresis of NuMat proteins, indicating the complexity of the NuMat proteome (Fig. 3). NuMat of 0–16-h embryos gave ∼200 spots when counted on a Coomassie Brilliant Blue-stained two-dimensional electrophoresis gel. By LC-MS/MS analysis, the nuclear matrix proteome profile of 0–16-h embryos was identified as described under “Materials and Methods.” An annotated MS/MS spectrum is shown as representative of these data (Fig. 4). Two biological replicates as well as technical replicates were used for the study, and it was found that 65% of the proteins were reproducibly detected in the experiments. However, the total number of proteins presented in this study (354 proteins) is inclusive of both lists (supplemental Tables 1 and 2). The molecular functions, biological processes, and cellular component for each protein were assigned based on gene ontology classification (23). The homologues of each protein were searched in the NCBI HomoloGene database (24). We found that of 354 proteins 167 were identified by two or more unique peptides, and these were classified according to their known or putative function based on sequence signatures (Table I and supplemental Table 1). Major classes that emerged from this analysis reflect the rich variety of factors associated with NuMat, indicating its involvement in diverse functions. Of the proteins identified in the NuMat preparation, 21% were structural proteins of the nucleus, 12% were heat shock proteins/chaperones and related proteins, whereas 5% were DNA/RNA-binding proteins, and 9% were chromatin remodeling complexes or transcription-related proteins. 8% of the proteome has proteins implicated in RNA processing, translation, signal transduction, and trafficking; 23% were ribosomal; and 2% were mitochondrial proteins, which may be cytoplasmic contaminants or may have hitherto unknown roles in the nucleus. The presence of actin and related proteins in our preparation reinforces their role in nuclear architecture (25). It is likely that many of the structural features of NuMat are dependent on these kinds of proteins. We also found a number of heat shock proteins and chaperones. Earlier studies have shown that HSC3 and HSC4 co-immunoprecipitate and genetically interact with Polycomb group proteins (26, 27), indicating their regulatory function mediated by chromatin modulation. The presence of these proteins in the nuclear matrix provides a possible structural basis for PcG protein compartmentalization and function.
Fig. 3.
Two-dimensional gel protein profile of NuMat prepared from 0–16-h embryos. About 200 protein spots can be detected by two-dimensional gel electrophoresis. The size of the proteins ranged from 100 to 14 kDa, and the pI of the spots ranged from 3 to 10.
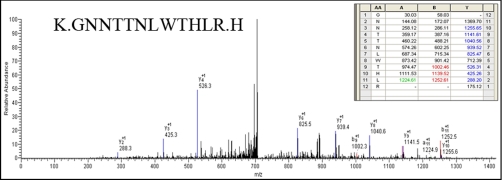
Fig. 4.
Annotated MS/MS spectrum of BEAF32B. The figure shows the peptide and fragment assignment table as an inset in the spectrum. The x axis and the y axis in the spectrum represent the m/z and the relative abundance, respectively. The matching A, B, and Y ions in the spectrum are highlighted in green, red, and blue, respectively, in the fragment assignment table.
Table I. List of NuMat proteins identified by LC-MS/MS.
F, fungi, including Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Dictyostelium discoideum; P, plants, including Oryza sativa and Arabidopsis thaliana; W, worms, including Caenorhabditis elegans and Caenorhabditis briggsae; I, insects, including Anopheles gambiae; V, vertebrates, including Danio rerio and Takifugu rubripes; H, Homo sapiens. KH domain, K Homology domain
The interaction of chromatin and NuMat is mediated by AT-rich regions of DNA known as MARs (28). The DNA-binding proteins in our analysis (Table I) are good candidates to specifically bind to MARs. Because RNA is an integral part of NuMat (29–31), the RNA-binding proteins identified in our analysis (Table I) similarly present potential factors that stabilize NuMat RNA in its nuclear structural framework. Many biological processes have been found to be associated with NuMat and MARs, including replication of DNA (32) and transcription (33). Chromatin remodeling and transcription factors that appear in the NuMat preparation may provide the structural link between the nuclear architecture and chromatin dynamics. Finally, targeting a protein/RNA to a specific compartment of the nucleus needs precise trafficking of these macromolecules. NuMat and its associated proteins are likely to be involved in the nuclear transport and may also play a vital role by providing a scaffold for the transport machinery. In addition, post-translational modifications appear to be important for NuMat proteins (21, 22). The presence of protein kinases in the NuMat may be relevant in this context.
As is clear from the two-dimensional profiles in Fig. 3, the NuMat proteome is much more complex than the 167 proteins analyzed here. Indeed, we indeed identified 187 additional proteins based on a single peptide hit (supplemental Table 2). Although we have not included single peptide hit proteins for extensive analysis with the proteins listed in Table I, these proteins are highly likely to be bona fide NuMat components as there are a number of circumstances in which a single peptide is all one can obtain (e.g. for small proteins or much less abundant proteins). We indeed identified one such protein, BEAF32B, by a single peptide in the present LC-MS/MS analysis (Fig. 4) that is a known NuMat component (20) as also shown by the Western blotting (Fig. 1C). Therefore, we think that the number of proteins in the NuMat may be around 350, including those from the single peptide list. Because the proteins from the single peptide hits could also fit within the classes of proteins in Table I (data not shown), the number of classes of proteins is not likely to increase from what is discussed here even with elucidation of the complete NuMat proteome.
Developmental and Cell Cycle Dynamics of NuMat Proteome
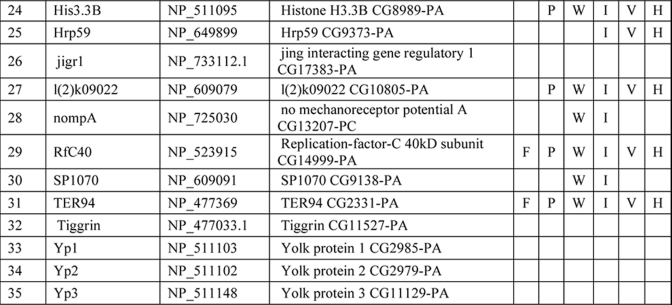
We compared the NuMat proteome from 0–2-, 6–8-, and 14–16-h-old Drosophila embryos. Several hundred spots were detected by DIGE (Fig. 5A). The experiment was performed three times, and each time the numbers of spots were reproducible. However, the average number rounded off to the nearest whole number was used for further analysis. About 40% of these proteins were present in all three stages of D. melanogaster embryos and may be considered as the minimal or core NuMat constituents (Fig. 5B). A significant proportion of spots showed stage-specific association with the NuMat (Fig. 5B). We observed a more complex NuMat proteome in early (2-h) stages of development compared with 8-h and late stage (16-h) NuMat proteomes. Although 17, 8, and 9% of protein spots were specific to the NuMat proteome of 2-, 8-, and 16-h embryos, respectively, there are a considerable number of proteins that are common to more than one stage. For instance, intersections of different developmental stages show that 11% of protein spots are present in very early stages, whereas they disappear at the 8-h stage and reappear in late stages of development. Further studies will be needed to molecularly and functionally characterize these classes of proteins to put them in the context of nuclear architecture and embryonic development.
Fig. 5.
Developmental dynamics of NuMat proteome. A, 2-, 8-, and 16-h-old Drosophila embryos were used for NuMat isolation and were separated by two-dimensional DIGE. The NuMat proteome is dynamic during the development of the embryo. B, Venn diagram to show the distribution of spots in various developmental stages of Drosophila embryos. Up to several hundred protein spots can be detected by DIGE, and there are more NuMat proteins in the early stages of development. Colors of the circles correspond to the spot colors in the gels.
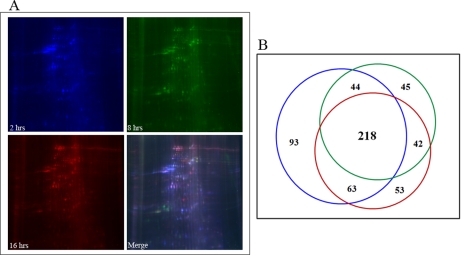
We followed the fate of NuMat during mitosis by immunostaining the early Drosophila embryos with NuMat mixture antibodies. The breakdown of the nuclear envelope and lamin structure shows that NuMat is dynamic during cell division. During interphase the NuMat antigens are present at the envelope and interior of the cell nuclei (Fig. 6A), whereas they form spindle-like structures during metaphase (Fig. 6B). Interestingly, at least some constituents of NuMat proteins seem to follow the DNA closely as the cells progress to anaphase (Fig. 6C). This association can be compared with page marking (34) and may have relevance to epigenetic marks being carried through mitosis by means of NuMat signatures along the chromosomes to unfurl the expression state in the imminent interphase. It is important to note that much of NuMat is not accessible to staining reagents during interphase and perhaps during other stages of the cell cycle. New protocols need to be developed to stain NuMat at different stages of the cell cycle to have a more realistic view of nuclear architecture during mitosis. Nevertheless, our studies provide insight into how the mitotic fidelity may be ensured with the help of continuity provided by a subset of NuMat proteins.
Fig. 6.
NuMat dynamic during mitosis. Immunostaining with a mixture of NuMat antibodies in early D. melanogaster embryos reveals the dynamics of NuMat in different mitotic stages. The distribution of NuMat proteins during mitosis is dynamic and follows specific patterns during interphase (A), metaphase (B), and anaphase (C). A subset of NuMat proteins stays associated with the chromosomes during mitosis (C) even when the nuclear envelope and much of the nuclear matrix are dissolved.
DISCUSSION
There is growing evidence indicating that genomic organization, function, and regulation are interlinked through nuclear architecture. For example, sites of replication (32), transcription (11, 33, 35), and post-transcriptional processing and other processes (8, 36) take place in specific nuclear compartments. Also, more recent studies indicate that localization of genes and regulatory elements to a specific compartment in a developmental stage-specific manner is a widespread mechanism for regulation of genes. The structural basis of these compartments, however, is not understood. We think that NuMat, the non-diffusible fibrous meshwork in the nucleus, is most likely a framework to provide a structural platform to various nuclear processes. The groups of NuMat constituents that we report here provide further structural and potentially mechanistic insights into the role of NuMat in a variety of nuclear processes (Table 1). One of the classes of proteins in this study includes nuclear membrane, nuclear pore, and structural proteins. Our results support earlier findings that suggested involvement of lamins, nuclear envelope proteins, actins, and myosin in the core nucleoskeleton (16). In addition to these core nucleoskeleton proteins, we identified a number of novel structural proteins involved in actin and microtubule binding as NuMat components. These include karst; spectrins; minispindles (microtubule binding), multiple ankyrin repeats single KH domain (structural constituent of the cytoskeleton); and paramyosin. The presence of viking, an extracellular matrix constituent, and some cuticular proteins in the NuMat proteome of Drosophila brings out new and interesting roles of these structural proteins.
The second general category of proteins in our study includes heat shock proteins/chaperones and enzymes. It has been shown in many earlier studies that heat shock proteins and other chaperones are associated with nuclear matrix (37, 38). It has also been shown in a recent study that molecular chaperone Hsp90 interacts with Trithorax to maintain active chromatin at sites of gene expression (39). In addition to a number of heat shock proteins such as Hsp26, -27, -83, and -70A, we identified few novel molecular chaperones that have ATPase activity and unfolded protein binding activity as a part of the nuclear matrix. These include lethal (1), Cctγ, and T-complex chaperonin 5. Interestingly, we also found certain enzymes to be associated with the nuclear matrix. These include enolase, phosphoglycerate kinase (glycolysis), FK506-binding protein 1 (peptidyl-prolyl cis-trans isomerase), papilin (metalloendopeptidase activity and serine type endopeptidase inhibitor activity), and Rpt4 (endopeptidase activity). Earlier studies have shown the association of several kinases with nuclear matrix, prominent among them being phosphatidylinositol 4-kinase, phosphatidylinositol-4-phosphate 5-kinase, diacylglycerol kinase, phospholipase C (40), and protein kinases C (PKC α and ζ), but the association of the enzymes reported in this study is the first of its kind to our knowledge. Thus, the association of these novel molecular chaperones and enzymes with the nuclear matrix in our study calls for further analysis on the roles played by these molecules in the context of chromatin organization, signal transduction, development, and differential gene expression.
The role of nuclear matrix in DNA replication, RNA transcription, hnRNA processing, DNA damage and repair, chromatin remodeling, nuclear transport, and translation has been well established in many elegant studies. We also identified a number of proteins associated with these nuclear processes (Table I and supplemental Table 1) to be part of the nuclear matrix. However, a few proteins in our study have not been reported earlier in the context of nuclear matrix and turned out to be novel proteins. These include Rm62 (mRNA binding and ATP-dependent RNA helicase activity), hoi-polloi (mRNA binding and nuclear mRNA splicing), little imaginal discs (histone H3-K4 demethylase activity), terribly reduced optic lobes (DNA methylation), and 14-3-3 ε and ζ.
Taken together, we have found more than 350 different proteins to be associated with the NuMat in our study, which fairly covers the complexity of the NuMat proteome (supplemental Tables 1 and 2). Our results also provide specific candidates that need further analysis by genetic, molecular, and cell biological approaches to understand the precise molecular interactions and mechanisms of nuclear functions in the context of the NuMat framework. Such studies would help us to understand whether a protein is transiently bound to NuMat or represents an essential NuMat constituent as a building block of the structure.
We also compared the nuclear matrix proteome of Drosophila with earlier studies on the nuclear matrix fractions of different cells and tissues of different species and found that several classes of functional proteins in the nuclear matrix are shared between kingdoms. A proteomics study of the Arabidopsis nuclear matrix shows the presence of nucleolar proteins such as IMP4, Nop56, Nop58, fibrillarins, and nucleolin; ribosomal proteins L7, L5, and L18; a number of α-tubulins and β-tubulins; homologues of translation elongation factor eEF-1; and the heat shock protein At-hsc70-3 (15). These proteins have also been identified in the nucleolus and nuclear matrix fractions of human cells. Our study also reports the presence of a large number of ribosomal proteins, Nop56, tubulins, heat shock proteins, and fibrillarin. When compared with the nuclear matrix fractions of human peripheral blood cells, the presence of molecular chaperones (Tcp-1-related protein and certain members of the heat shock protein family) and protein folding catalysts was found to be common with our study (38). The association of heat shock proteins with nuclear matrix fractions has also been shown in cancer and stressed cells (37, 41). The presence of topoisomerases in the nuclear matrix fractions shown by earlier studies is yet another feature consistent with our study (42). Thus, the presence of the homologues of Drosophila NuMat proteome constituents in human and other organisms underscores the evolutionary conservation of the NuMat organization.
Our studies also reveal two new aspects of NuMat in addition to its conservation and link to a variety of nuclear processes, namely the high degree of developmental dynamics of the NuMat composition and retention of a subset of the NuMat proteome with chromatin through mitosis. Our observations suggest that more than half of the NuMat proteome is dynamic and corresponds to a particular developmental stage. Although much of these differences may be quantitative, this nevertheless provides a new angle to study and understand the cellular differentiation process. Nuclear matrix is the architectural framework of the nucleus and therefore is involved in the higher order chromatin organization, which is closely linked to differential gene expression. In the complex process of embryonic development where dramatic changes occur in the embryo, including rearrangement in the organization of DNA and DNA-protein interactions, nuclear matrix may also undergo significant alterations in its composition. This is supported by many earlier studies that have shown that nuclear matrix is a dynamic structure whose morphology and composition vary with the functional stage of the nuclei (43). Previous studies have also shown that the protein composition of the nuclear matrix can be modified during cell differentiation (44–47). It has also been shown that the nuclear matrix proteome varies significantly between cancerous and healthy tissue, and a recent study regarding identification of proteins enriched in the nuclear matrix fraction and regulation in adenoma to carcinoma progression further strengthens the reason why developmental regulation of the NuMat proteome would be of importance (48).
The second new aspect of the NuMat proteome is the finding that a fraction of the NuMat proteome remains associated with chromosomes during mitosis when the nuclear envelope and most of the NuMat structures are thought to be dissolved. During mitosis, the unique NuMat framework that is paired with the cell type-specific epigenome is dismantled or drastically rearranged. With the onset of cell division, most of the transcriptional processes are stopped, and the genome enters a “mitotic blackout” as it is not clear how memory of the expression state is retained to re-establish the transcriptional state in the interphase of daughter cells. Association of some of the NuMat components with mitotic chromosomes may suggest that a set of NuMat constituents might function as cellular memory tools to replicate the chromatin structure in the following interphase. Identification of proteins associated with the nuclear matrix and mitotic chromosomes could provide vital clues to this problem (49–51). A genome-wide RNAi screening for genes that lead to spindle defect (52) identified a number of factors, and 13 of them are part of the NuMat proteome in our study. These studies not only implicate a role for NuMat in epigenetic cellular memory, but they also present a set of proteins that can be tested in vivo from this perspective to help understand the mechanism behind this process.
Our analysis of the NuMat proteome from D. melanogaster embryos that led to identification of more than 350 proteins reflects the structural and functional complexity of NuMat. We also found that NuMat composition varies significantly during development and that some of the NuMat constituents remain associated with chromosomes during mitosis, pointing to a role of NuMat in cellular memory. With the identification of a significant proportion of NuMat constituents, further studies that can lead to a better understanding of the structural basis and molecular mechanisms of these complex processes will be possible.
Supplementary Material
Acknowledgments
We thank Sreenath for embryo collection and Centre for Cellular and Molecular Biology proteomics facility-associated members K. S. Rakesh, P. Ravindra Varma, and Priyanka Bolan for help. We especially thank Ravi Sirdeshmukh, C. S. Sundaram, and M. V. Jagannadhan for help in proteomics experiments.
* This work was supported in part by a young investigator grant from the Human Frontier Science Program and a grant from the Indo-French Centre for the Promotion of Advanced Research.
 This article contains supplemental Tables 1 and 2.
This article contains supplemental Tables 1 and 2.
1 The abbreviations used are:
- NuMat
- nuclear matrix
- PI
- propidium iodide
- MAR
- matrix-associated region
- ABC
- ammonium bicarbonate
- BEAF
- boundary element associated factor; PRIDE database = Accession Numbers 12879–12896; Supplemental Table 1 is list of proteins with 2 or more unique peptide hits, Supplemental Table 2 is list of single peptide hits. Supplemental Data files 1, 2, 3 & 4 provide spectra of single peptide hits
REFERENCES
- 1.Cockell M., Gasser S. M. (1999) Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev. 9, 199–205 [DOI] [PubMed] [Google Scholar]
- 2.Cremer T., Kreth G., Koester H., Fink R. H., Heintzmann R., Cremer M., Solovei I., Zink D., Cremer C. (2000) Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture. Crit. Rev. Eukaryot. Gene Expr. 10, 179–212 [PubMed] [Google Scholar]
- 3.Cremer T., Küpper K., Dietzel S., Fakan S. (2004) Higher order chromatin architecture in the cell nucleus: on the way from structure to function. Biol. Cell 96, 555–567 [DOI] [PubMed] [Google Scholar]
- 4.Doucas V., Evans R. M. (1996) The PML nuclear compartment and cancer. Biochim. Biophys. Acta 1288, M25–M29 [DOI] [PubMed] [Google Scholar]
- 5.Fakan S., van Driel R. (2007) The perichromatin region: a functional compartment in the nucleus that determines large-scale chromatin folding. Semin. Cell Dev. Biol. 18, 676–681 [DOI] [PubMed] [Google Scholar]
- 6.Moen P. T., Jr., Smith K. P., Lawrence J. B. (1995) Compartmentalization of specific pre-mRNA metabolism: an emerging view. Hum. Mol. Genet. 4, 1779–1789 [DOI] [PubMed] [Google Scholar]
- 7.Ogg S. C., Lamond A. I. (2002) Cajal bodies and coilin—moving towards function. J. Cell Biol. 159, 17–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isogai Y., Tjian R. (2003) Targeting genes and transcription factors to segregated nuclear compartments. Curr. Opin. Cell Biol. 15, 296–303 [DOI] [PubMed] [Google Scholar]
- 9.Fawcett D. W. (1966) An Atlas of Fine Structure: the Cell, Its Organelles and Inclusions, W. B. Saunders Co., Philadelphia: p.49–62 [Google Scholar]
- 10.Fey E. G., Wan K. M., Penman S. (1984) Epithelial cytoskeletal framework and nuclear matrix-intermediate filament scaffold: three-dimensional organization and protein composition. J. Cell Biol. 98, 1973–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson D. A. (2005) The amazing complexity of transcription factories. Brief. Funct. Genomic. Proteomic. 4, 143–157 [DOI] [PubMed] [Google Scholar]
- 12.Miller B. R., Forbes D. J. (2000) Purification of the vertebrate nuclear pore complex by biochemical criteria. Traffic 1, 941–951 [PMC free article] [PubMed] [Google Scholar]
- 13.Tubo R. A., Berezney R. (1987) Identification of 100 and 150 S DNA polymerase alpha-primase megacomplexes solubilized from the nuclear matrix of regenerating rat liver. J. Biol. Chem. 262, 5857–5865 [PubMed] [Google Scholar]
- 14.Berezney R., Coffey D. S. (1977) Nuclear matrix. Isolation and characterization of a framework structure from rat liver nuclei. J. Cell Biol. 73, 616–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calikowski T. T., Meulia T., Meier I. (2003) A proteomic study of the Arabidopsis nuclear matrix. J. Cell. Biochem. 90, 361–378 [DOI] [PubMed] [Google Scholar]
- 16.Gerner C., Gotzmann J., Fröhwein U., Schamberger C., Ellinger A., Sauermann G. (2002) Proteome analysis of nuclear matrix proteins during apoptotic chromatin condensation. Cell Death Differ. 9, 671–681 [DOI] [PubMed] [Google Scholar]
- 17.He D., Zeng C., Brinkley B. R. (1995) Nuclear matrix proteins as structural and functional components of the mitotic apparatus. Int. Rev. Cytol. 162B, 1–74 [DOI] [PubMed] [Google Scholar]
- 18.Mika S., Rost B. (2005) NMPdb: Database of Nuclear Matrix Proteins. Nucleic Acids Res. 33, D160–D163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eng J. K., McCormack A. L., Yates J. R., 3rd (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 20.Mitchison T. J., Sedat J. (1983) Localization of antigenic determinants in whole Drosophila embryos. Dev. Biol. 99, 261–264 [DOI] [PubMed] [Google Scholar]
- 21.Pathak R. U., Rangaraj N., Kallappagoudar S., Mishra K., Mishra R. K. (2007) Boundary element-associated factor 32B connects chromatin domains to the nuclear matrix. Mol. Cell. Biol. 27, 4796–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muralikrishna B., Thanumalayan S., Jagatheesan G., Rangaraj N., Karande A. A., Parnaik V. K. (2004) Immunolocalization of detergent-susceptible nucleoplasmic lamin A/C foci by a novel monoclonal antibody. J. Cell. Biochem. 91, 730–739 [DOI] [PubMed] [Google Scholar]
- 23. (2010) The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 38, D331–D335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sayers E. W., Barrett T., Benson D. A., Bryant S. H., Canese K., Chetvernin V., Church D. M., DiCuccio M., Edgar R., Federhen S., Feolo M., Geer L. Y., Helmberg W., Kapustin Y., Landsman D., Lipman D. J., Madden T. L., Maglott D. R., Miller V., Mizrachi I., Ostell J., Pruitt K. D., Schuler G. D., Sequeira E., Sherry S. T., Shumway M., Sirotkin K., Souvorov A., Starchenko G., Tatusova T. A., Wagner L., Yaschenko E., Ye J. (2009) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 37, D5–D15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olave I. A., Reck-Peterson S. L., Crabtree G. R. (2002) Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 71, 755–781 [DOI] [PubMed] [Google Scholar]
- 26.Mollaaghababa R., Sipos L., Tiong S. Y., Papoulas O., Armstrong J. A., Tamkun J. W., Bender W. (2001) Mutations in Drosophila heat shock cognate 4 are enhancers of Polycomb. Proc. Natl. Acad. Sci. U.S.A. 98, 3958–3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saurin A. J., Shao Z., Erdjument-Bromage H., Tempst P., Kingston R. E. (2001) A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412, 655–660 [DOI] [PubMed] [Google Scholar]
- 28.Hakes D. J., Berezney R. (1991) DNA binding properties of the nuclear matrix and individual nuclear matrix proteins. Evidence for salt-resistant DNA binding sites. J. Biol. Chem. 266, 11131–11140 [PubMed] [Google Scholar]
- 29.He D. C., Martin T., Penman S. (1991) Localization of heterogeneous nuclear ribonucleoprotein in the interphase nuclear matrix core filaments and on perichromosomal filaments at mitosis. Proc. Natl. Acad. Sci. U.S.A. 88, 7469–7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long B. H., Huang C. Y., Pogo A. O. (1979) Isolation and characterization of the nuclear matrix in Friend erythroleukemia cells: chromatin and hnRNA interactions with the nuclear matrix. Cell 18, 1079–1090 [DOI] [PubMed] [Google Scholar]
- 31.Nickerson J. A., Krochmalnic G., Wan K. M., Penman S. (1989) Chromatin architecture and nuclear RNA. Proc. Natl. Acad. Sci. U.S.A. 86, 177–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anachkova B., Djeliova V., Russev G. (2005) Nuclear matrix support of DNA replication. J. Cell. Biochem. 96, 951–961 [DOI] [PubMed] [Google Scholar]
- 33.S'Iakste N. I., S'Iakste T. G. (2001) Transcription factors and the nuclear matrix. Mol. Biol. 35, 739–749 [PubMed] [Google Scholar]
- 34.Delcuve G. P., He S., Davie J. R. (2008) Mitotic partitioning of transcription factors. J. Cell. Biochem. 105, 1–8 [DOI] [PubMed] [Google Scholar]
- 35.Travis J. (1993) Looking for Cancer in Nuclear Matrix Proteins. Science 259, 1258. [DOI] [PubMed] [Google Scholar]
- 36.Thiry M., Lafontaine D. L. (2005) Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol. 15, 194–199 [DOI] [PubMed] [Google Scholar]
- 37.Willsie J. K., Clegg J. S. (2002) Small heat shock protein p26 associates with nuclear lamins and HSP70 in nuclei and nuclear matrix fractions from stressed cells. J. Cell. Biochem. 84, 601–614 [PubMed] [Google Scholar]
- 38.Gerner C., Holzmann K., Meissner M., Gotzmann J., Grimm R., Sauermann G. (1999) Reassembling proteins and chaperones in human nuclear matrix protein fractions. J. Cell. Biochem. 74, 145–151 [PubMed] [Google Scholar]
- 39.Tariq M., Nussbaumer U., Chen Y., Beisel C., Paro R. (2009) Trithorax requires Hsp90 for maintenance of active chromatin at sites of gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 1157–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Payrastre B., Nievers M., Boonstra J., Breton M., Verkleij A. J., Van Bergen en Henegouwen P. M. (1992) A differential location of phosphoinositide kinases, diacylglycerol kinase, and phospholipase C in the nuclear matrix. J. Biol. Chem. 267, 5078–5084 [PubMed] [Google Scholar]
- 41.Roti Roti J. L., Kampinga H. H., Malyapa R. S., Wright W. D., vanderWaal R. P., Xu M. (1998) Nuclear matrix as a target for hyperthermic killing of cancer cells. Cell Stress Chaperones 3, 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berrios M., Osheroff N., Fisher P. A. (1985) In situ localization of DNA topoisomerase II, a major polypeptide component of the Drosophila nuclear matrix fraction. Proc. Natl. Acad. Sci. U.S.A. 82, 4142–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bladon T., Brasch K., Brown D. L., Setterfield G. (1988) Changes in structure and protein composition of bovine lymphocyte nuclear matrix during concanavalin-A-induced mitogenesis. Biochem. Cell Biol. 66, 40–53 [DOI] [PubMed] [Google Scholar]
- 44.Fey E. G., Penman S. (1988) Nuclear matrix proteins reflect cell type of origin in cultured human cells. Proc. Natl. Acad. Sci. U.S.A. 85, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stuurman N., Van Driel R., De Jong L., Meijne A. M., Van Renswoude J. (1989) The protein composition of the nuclear matrix of murine P19 embryonal carcinoma cells is differentiation-stage dependent. Exp. Cell Res. 180, 460–466 [DOI] [PubMed] [Google Scholar]
- 46.Dworetzky S. I., Fey E. G., Penman S., Lian J. B., Stein J. L., Stein G. S. (1990) Progressive changes in the protein composition of the nuclear matrix during rat osteoblast differentiation. Proc. Natl. Acad. Sci. U.S.A. 87, 4605–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bidwell J. P., Van Wijnen A. J., Fey E. G., Dworetzky S., Penman S., Stein J. L., Lian J. B., Stein G. S. (1993) Osteocalcin gene promoter-binding factors are tissue-specific nuclear matrix components. Proc. Natl. Acad. Sci. U.S.A. 90, 3162–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albrethsen J., Knol J. C., Piersma S. R., Pham T. V., de Wit M., Mongera S., Carvalho B., Verheul H. M., Fijneman R. J., Meijer G. A., Jimenez C. R. (2010) Subnuclear proteomics in colorectal cancer. Identification of proteins enriched in the nuclear matrix fraction and regulation in adenoma to carcinoma progression. Mol. Cell. Proteomics 9, 988–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gassmann R., Henzing A. J., Earnshaw W. C. (2005) Novel components of human mitotic chromosomes identified by proteomic analysis of the chromosome scaffold fraction. Chromosoma 113, 385–397 [DOI] [PubMed] [Google Scholar]
- 50.Mancini M. A., He D., Ouspenski II, Brinkley B. R. (1996) Dynamic continuity of nuclear and mitotic matrix proteins in the cell cycle. J. Cell. Biochem. 62, 158–164 [DOI] [PubMed] [Google Scholar]
- 51.Morrison C., Henzing A. J., Jensen O. N., Osheroff N., Dodson H., Kandels-Lewis S. E., Adams R. R., Earnshaw W. C. (2002) Proteomic analysis of human metaphase chromosomes reveals topoisomerase II alpha as an Aurora B substrate. Nucleic Acids Res. 30, 5318–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goshima G., Wollman R., Goodwin S. S., Zhang N., Scholey J. M., Vale R. D., Stuurman N. (2007) Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316, 417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.