Abstract
Venezuelan equine encephalitis virus (VEEV) is a prototypical enveloped ssRNA virus of the family Togaviridae. To better understand alphavirus assembly, we analyzed newly formed nucleocapsid particles (termed pre-viral nucleocapsids) isolated from infected cells. These particles were intermediates along the virus assembly pathway, and ultimately bind membrane-associated viral glycoproteins to bud as mature infectious virus. Purified pre-viral nucleocapsids were spherical with a unimodal diameter distribution. The structure of one class of pre-viral nucleocapsids was determined with single particle reconstruction of cryo-electron microscopy images. These studies showed that pre-viral nucleocapsids assembled into an icosahedral structure with a capsid stoichiometry similar to the mature nucleocapsid. However, the individual capsomers were organized significantly differently within the pre-viral and mature nucleocapsids. The pre-viral nucleocapsid structure implies that nucleocapsids are highly plastic and undergo glycoprotein and/or lipid-driven rearrangements during virus self-assembly. This mechanism of self-assembly may be general for other enveloped viruses.
Keywords: alphavirus, virus assembly, cryo-electron microscopy, virus structure
Introduction
Venezuelan equine encephalitis virus (VEEV) is a single-stranded RNA enveloped virus (family: Togaviridae, genus: alphavirus) (Weaver et al., 2004) that causes highly debilitating flu-like symptoms following human infection. Encephalitis and severe neurological complications occur in ~10% of infections and correlate to infection of the central nervous system. Mortality rates are typically less than 1%. Reoccurring outbreaks and epidemics extend from South America (Rico-Hesse et al., 1995; Weaver et al., 1996; Weaver et al., 2004) to the southern portion of the United States (Weaver et al., 2004). Unfortunately, no VEEV antivirals or vaccines are approved for use by the general public.
Mature infectious VEEV is composed of E1 and E2 virus-encoded glycoproteins organized with icosahedral symmetry and anchored within a host-derived lipid bilayer (David, 1971; Hirschberg and Robbins, 1974; Paredes et al., 2001; Schlesinger and Schlesinger, 2001). The bilayer surrounds a compact nucleocapsid formed from the virus-encoded capsid protein and viral RNA (vRNA) (Cheng et al., 1994; Mancini et al., 2000; Vogel et al., 1986; Weaver et al., 2004). The mature VEEV nucleocapsid structure has capsid proteins organized into pentameric and hexameric capsomers with T=4 icosahedral symmetry that complements the envelope protein icosahedral organization (Paredes et al., 2001). Similar to other alphaviruses, the VEEV complementary icosahedral shells are separated by a lipid bilayer.
The structures of VEEV and other mature alphaviruses provide little insight into their assembly mechanisms. Studies of VEEV isolated nucleocapsids, produced by removing the lipid bilayer and envelope glycoproteins from whole virus, revealed that interactions between VEEV capsid proteins and vRNA were sufficient to maintain the nucleocapsid T=4 icosahedral structure, although these nucleocapsids underwent radial expansion and capsomer rearrangement relative to the mature virus (Paredes et al., 2003). In cell-free systems, purified alphavirus capsid proteins self-assembled with small synthetic nucleotide oligomers into icosahedral particles (Murphy and Fields, 1967). However, it remains unclear how capsid proteins and vRNA assemble within cells, and if viral nucleocapsids form a single assembly state or reorganize in response to glycoprotein and/or lipid interactions and the changing environments experienced during assembly and maturation (Bykovsky et al., 1969; Garoff et al., 2004; Haag et al., 2002; Schlesinger and Schlesinger, 2001; Wengler et al., 2003).
To better understand the process of alphavirus assembly within cells, we determined the three-dimensional (3D) structure of a VEEV nucleocapsid assembly intermediate isolated from infected cells prior to virus budding (termed a pre-viral nucleocapsid). Pre-viral nucleocapsids were purified, cryogenically preserved, and imaged with electron microscopy. Images were processed to produce 3D electron density maps using icosahedral reconstruction methods (Baker et al., 1999; Fuller et al., 1996). These structures clearly showed that pre-viral nucleocapsids, although plastic, assembled as icosahedral particles in the cytoplasm of infected cells. Interestingly, the capsid protein organization differed significantly between the pre-viral and mature nucleocapsid particles, although these particles had similar capsid protien stoichiometry (Paredes et al., 2001; Paredes et al., 2003). The pre-viral nucleocapsid structure suggests that VEEV capsid protein and vRNA are sufficient to self-assemble into an icosahedral (or pseudo-icosahedral) particle in the cytosol, that nucleocapsids are plastic structures, and that nucleocapsids likely undergo extensive glycoprotein- and/or lipid-driven rearrangements during virus assembly and budding. This work represents one of the few cryo-EM reconstructions of an in vivo nucleocapsid intermediate from a mammalian virus, and provides unique insights into the virus assembly process.
Results
Cellular distribution of VEEV nucleocapsids
Electron microscopy of thin sections of BHK cells infected with VEEV TC-83 confirmed that cells had little to no virus budding 19 hr post-infection (Fig 1). However, significant quantities of VEEV pre-viral particles were observed in the cytoplasm of infected cells at 19 hr post-infection (Fig. 1B, 1D). Nucleocapsids observed in infected cells were round and occasionally appeared in small clumps around intracellular vesicles (Fig. 1B, 1D). The sizes of the nucleocapsid particles in the thin-sections ranged from 39 to 50 nm in diameter, with the average size being about 40 nm (data not shown). Immuno-gold labeling provided confirmation that the observed particles were VEEV nucleocapsids (Fig. 1D). Given the preponderance of intracellular particles relative to budding particles, the purified nucleocapsids used for cryo-EM reconstructions clearly reflect the pre-viral nucleocapsid structure prior to interactions with viral glycoproteins and encapsulation in the plasma membrane.
Figure 1.
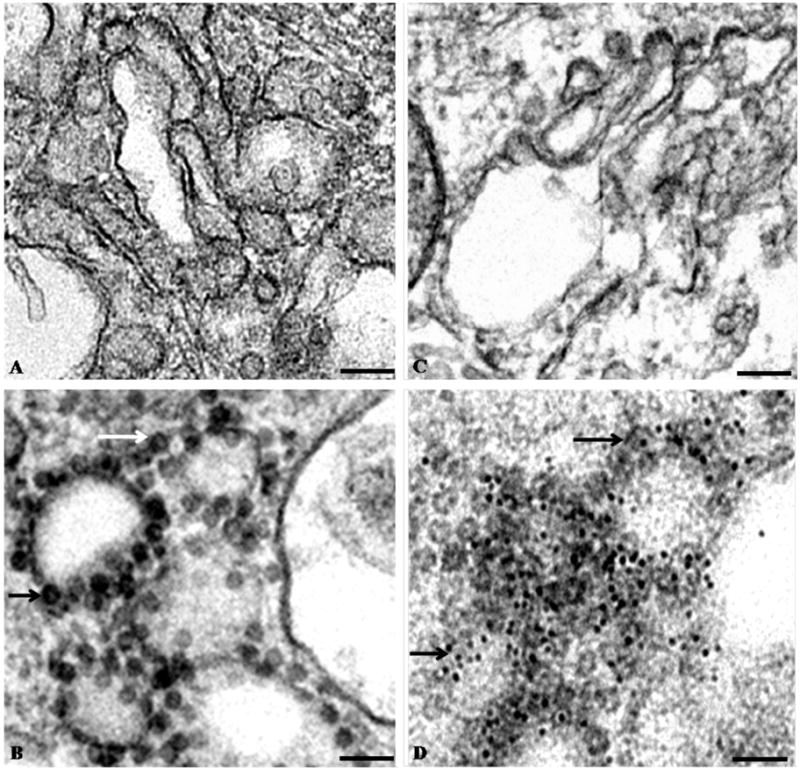
Electron micrographs of negatively stained thin-sections of BHK cells. (A) Uninfected BHK cells. (B) Sections of BHK cells 19 hr post-infection with VEEV TC-83. (C) Immuno-gold labeled thin-section of uninfected BHK cells. (D) Immuno-gold labeled thin-section of BHK cells infected with VEEV TC-83. The sections of BHK cells were labeled with primary antibody against the C-terminus of VEEV capsid protein and 5 nm gold-conjugated secondary antibody. In panels (B) and (D), the arrows point to pre-viral VEEV nucleocapsids. Scale bars = 100 nm.
Isolation of pre-viral nucleocapsids
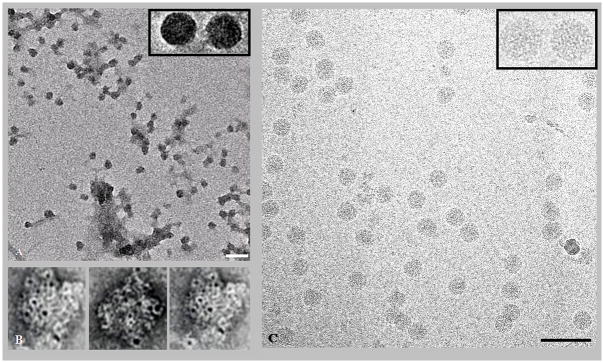
Pre-viral nucleocapsid particles were concentrated and purified using sucrose cushioning of cell supernatants and subsequent sucrose or iodixanol gradients (Fig. 2). Particles were initially observed with electron microscopy of cushion samples or gradient fractions stained with 2% uranyl-acetate. Micrographs showed large numbers of relatively homogeneous circular particles with only a few damaged/misshapen particles and little background contamination (Fig. 2A). Particles were readily labeled using immuno-gold against the VEEV capsid protein (Fig. 2B), thus confirming the identity of purified particles as VEEV pre-viral nucleocapsids. Purified samples containing VEEV pre-viral nucleocapsids were dialyzed to remove sucrose or iodixanol and observed using cryo-EM. These particles were largely circular and undamaged (Fig. 2C), although fractions recovered after gradient purification contained significantly reduced numbers of particles relative to samples recovered from the initial sucrose/iodixanol cushion (data not shown).
Figure 2.
Electron micrographs of purified pre-viral nucleocapsid particles. (A) Electron micrograph of negatively stained pre-viral nucleocapsid particles recovered from the cushion stage of particle purification. Scale bar = 100 nm. (B) Electron micrograph of selected immuno-gold labeled (anti-VEEV capsid antibody) pre-viral nucleocapsid particles recovered from the cushion stage of particle purification. Scale bar = 50 nm. (C) Electron micrograph of vitrified pre-viral nucleocapsid particles recovered from the cushion stage of particle purification. Inserts in panels A and C show representative particles enlarged for clarity. Scale bar = 100 nm.
The proteins that formed the VEEV pre-viral nucleocapsids were separated with denaturing polyacrylamide gel electrophoresis (SDS-PAGE) and visualized with Coomassie blue stain or Western blotting. Purified pre-viral nucleocapsids contained only capsid proteins; no additional contaminating viral or cellular proteins were observed in stained polyacrylamide gels (Fig. 3). The capsid protein from pre-viral nucleocapsids migrated as a dimer during separation by SDS-PAGE (Fig. 3) implying interactions between capsid proteins were stable under typically denaturing concentrations of 0.1% (w/v) SDS. This observation was consistent with previous studies that reported capsid dimerization was necessary for alphavirus nucleocapsid formation (Tellinghuisen et al., 2001). Similar capsid banding patterns were observed in particles isolated using either 30–45% iodixanol or sucrose gradients, suggesting that the stability of the capsid dimer was independent of purification conditions (Fig. 3). Interestingly, the capsid protein migrated as a monomer when isolated from mature virus (Paredes et al., 2001), which suggests that intermolecular contacts may be significantly different in the pre-viral and mature nucleocapsids.
Figure 3.
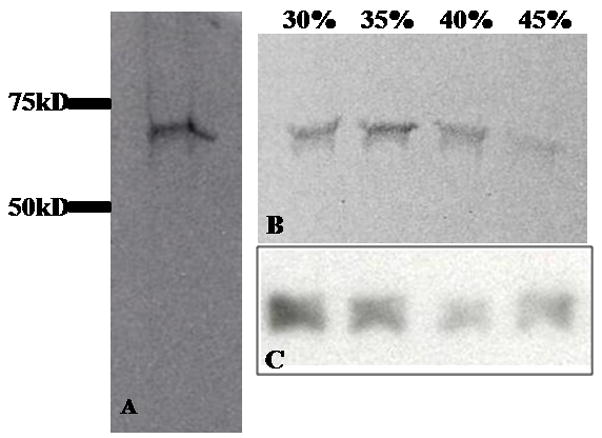
Protein composition of purified pre-viral nucleocapsids. (A) Coomassie blue staining of proteins from pre-viral nucleocapsid particles recovered from the cushion stage of particle purification and separated with SDS-PAGE. Molecular weight markers are shown along the left-hand side. (B) Coomassie blue staining of proteins from pre-viral nucleocapsid particles recovered from iodixanol gradient fractions and separated with SDS-PAGE. The iodixanol concentration of each fraction is listed above each lane. (C) Western blot (anti-VEEV capsid antibody) of proteins from pre-viral nucleocapsid particles recovered from iodixanol gradient fractions and separated with SDS-PAGE.
Pre-viral nucleocapsids showed size and density variations
The diameters of circular VEEV pre-viral nucleocapsids, isolated from cells by sucrose or iodixanol cushioning and imaged with cryo-EM, were tabulated. Particle diameters ranged between 31 and 54 nm (± 2 nm) and a frequency distribution plot of diameters closely approximated a Gaussian distribution with a mean of 43 nm (SD = 3.5 nm) (Fig. 4A). A small subpopulation of particles was observed with diameters of ~35 nm (Fig. 4A). Since the range of particle diameters isolated after sucrose cushion purification may have reflected several different pre-viral nucleocapsid assembly states, density gradients were used to separate particles based on density variations. Pre-viral particles could be separated into several fractions using sucrose or iodixanol gradients, suggesting that pre-viral nucleocapsids with different densities were present within infected cells. The average diameters for particles from the 35–45% iodixanol gradients fractions were similar, whereas particles isolated from the 30% iodixanol gradient had a lower average diameter relative to particles contained in the less dense fractions (Fig. 4B). Particles in the 30% iodixanol gradient fraction may correspond to the subpopulation of small diameter nucleocapsid particles observed during sucrose cushion isolation (Figure 4A). Moreover, the range of observed particle diameters was narrower for particles recovered from the iodixanol gradient fractions relative to pelleting onto iodixanol cushions (data not shown). Thus, several pre-viral nucleocapsid assembly states with varied diameters and densities were present within infected cells, and these physical characteristics likely reflect differences in capsid protein packing.
Figure 4.
Pre-viral nucleocapsid diameters. (A) Frequency distribution plot of diameters of pre-viral nucleocapsids (n=200) recovered from the cushion stage of purification. The dashed line shows a Gaussian curve fit to the data points. Particle diameters were measured from micrographs of vitrified particles. Measurement errors were ±1.8 nm, based on multiple diameter measurements for each particle. (B) Average diameter of pre-viral nucleocapsids (n=100) recovered from distinct iodixanol gradient fractions. Particle diameters were measured from electron micrographs of negatively stained particles. Error bars were ±2 nm, based on multiple diameter measurements for each particle.
Three-dimensional structure and characterization of a pre-viral nucleocapsid
Having established that pre-viral nucleocapsids could be isolated and purified from infected cells, we next determined the structure of a stable nucleocapsid assembly state. To maximize the number of particles available for single particle reconstruction from cryo-electron micrographs, particles were purified using centrifugation and concentrated by pelleting onto a sucrose cushion. Particles were applied to electron microscopy grids, vitrified, and imaged with a cryo-electron microscope under low dose conditions. Images were manually selected to remove particles with ice contamination, poor shape, and extreme diameters. Since the resolution of the resulting reconstruction was greater than 30 Å and defocus values ranged from ~1.5 to 3.0 microns, no CTF corrections were required to process the data since first zero of the CTF was beyond the obtained resolution and no contrast inversion was observed at that resolution.
Extreme care was taken to ensure that the final structure of the pre-viral nucleocapsid assembly state did not reflect bias from the starting model used in the reconstruction. Four distinct starting reference maps were tested, including an unbiased reference map generated with Auto3dem (Yan et al., 2007), and the T=3 red clover necrotic mosaic virus (RCNMV) (Sherman et al., 2006), the T=4 VEEV mature nucleocapsid (Paredes et al., 2001), and the T=4 Western equine encephalitis virus (WEEV) mature nucleocapsid (M. Sherman, personal communication) maps. Of special note was the Auto3dem unbiased reconstruction, where the initial reference map for subsequent alignment and refinement was generated from the dataset without using any external references (Yan et al., 2007). Parallel independent reconstructions were performed using each of the four starting models (as detailed in the Methods section). Orientation and origin search iterations, using PFTSEARCH (Baker et al., 1999), were done until no further improvement was observed, followed by iterative refinement of the parameters using OOR. At each iteration step, only images with the best correlation to the current reference map were retained. The final reconstructions were calculated using ~90 particle images. The remaining particles did not converge into any additional structures (data not shown), although significant efforts with multiple programs were expended in these attempts.
The reconstructions produced maps with clearly defined capsomers about the 3-fold and 5-fold symmetry axes (Fig. 5). The orientations of the particles used in the reconstruction evenly sampled Euler angle space in the icosahedral asymmetric unit (Fig. 5B). All four reconstructions yielded 3D maps (calculated to be ~40 Å resolution) with very similar isosurface features (Suppl. Data Fig. 1) and radial density plots through the capsid shell (Suppl. Data Fig. 2 [A–D]), independent of the starting reference map. The radial density plots of the pre-viral nucleocapsids (Supp. Data Fig. 2A–D) had more peaks than corresponding plots of the VEEV mature nucleocapsid (Supp. Data Fig. 2E). Pre-viral nucleocapsid reconstruction attempts with various subsets of particle images that did not include the above consistent set of particle images did not converge. These attempts produced practically featureless maps, clearly demonstrating that the reconstruction protocol did not generate spurious icosahedral maps from unrelated particle images.
Figure 5.
Three-dimensional reconstruction of an icosahedral VEEV pre-viral nucleocapsid assembly state. (A) Map of the pre-viral nucleocapsid viewed along an icosahedral 3-fold axis. Starting model for this reconstruction was an Auto3dem-generated map. (B) Orientations of particles used for the pre-viral nucleocapsid reconstruction plotted relative to the asymmetric unit.
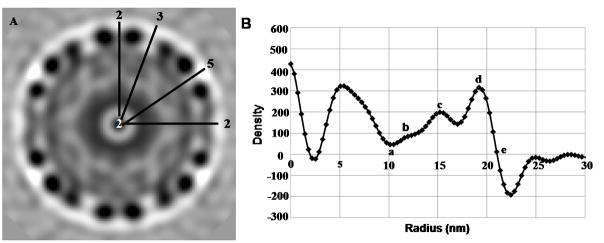
The following description of the structure of a pre-viral nucleocapsid assembly state was based on the reconstruction that used an Auto3dem-generated initial model (Fig. 5). However, the description would be equally valid for reconstructions with the other starting models (Suppl. Data Fig. 1). This assembly state of the pre-viral nucleocapsid clearly formed a 43.4 nm diameter icosahedral particle, with distinct capsomer features about the 3-fold and 5-fold axes of symmetry (Fig. 5). The 43.4 nm diameter of the reconstructed VEEV pre-viral nucleocapsid was consistent with the mean particle diameter measured for nucleocapsid particles purified by centrifugation and pelleted onto a sucrose cushion (Fig. 4A). The diameter of the pre-viral nucleocapsid was ~11% larger than the mature VEEV nucleocapsid (Paredes et al., 2001) and similar to the diameter of isolated VEEV nucleocapsids from whole virions (Paredes et al., 2003), suggesting that the nucleocapsid was compressed by the virus membrane and/or envelope proteins. A central section through the pre-viral nucleocapsid reconstruction (Fig. 6) showed the capsid shell was located at radii between 15.5 and 21.7 nm, making the capsid shell ~5% thicker in the pre-viral nucleocapsid compared to the mature VEEV nucleocapsid (Paredes et al., 2001). Assuming similar capsid masses, this suggests that the mean packing density was ~33% lower in the pre-viral nucleocapsid compared to the mature VEEV nucleocapsid. The relatively low packing density in the capsid shell of the pre-viral nucleocapsid could point to weaker intermolecular contacts, which would in turn contribute to plasticity of the pre-viral nucleocapsid. The capsid shell was penetrated by large holes, with the pentameric capsomers at the 5-fold axes containing a central hole of ~2.7 nm and the quasi-hexameric capsomers at the 3-fold axes containing a central hole of ~4.1 nm diameter. Two additional holes of ~3.8 nm diameter flanked the 2-fold axes of symmetry.
Figure 6.
Radial analysis of icosahedral pre-viral nucleocapsid particles. (A) Central section through the pre-viral nucleocapsid particle, viewed along an icosahedral 2-fold axis. (B) Radial density plot of the pre-viral nucleocapsid map.
Radial density distributions of pre-viral nucleocapsid reconstructions
3D maps (Suppl. Data Fig. 1) were produced from reconstructions that used distinctly different initial reference models (i.e., an Auto3dem-generated initial model, RCNMV, and mature VEEV and WEEV nucleocapsids). Radial density plots provided detailed descriptions of radial density distributions within the 3D reconstructions. These plots were similar for the four independent reconstructions of the pre-viral nucleocapsid (Fig. 6B, Suppl. Data Fig. 2), thereby confirming that the independent reconstructions produced consistent structures. A local density minimum at ~10 nm radius (labeled “a” in Fig. 6B) matched the boundary between disordered viral RNA and structured nucleocapsid. Three local density peaks were observed between 10 and 21.7 nm on each spherical average plot, and were located at similar radial positions. The Auto3dem reconstruction had a smaller density peak at ~11.6 nm (labeled “b” in Fig. 6B) relative to the other three reconstructions. This radial distance marked the maximum depth of the surface holes that penetrated through the capsid shell. Density peaks located at a radial distance of ~15.5 nm (labeled “c” in Fig. 6B) corresponded to the inner boundary of the capsid shell, whereas the density peak at ~19 nm (labeled “d” in Fig. 6B) was located midway through the capsid shell of the pre-viral nucleocapsid. The common zero density point at a radial distance of ~21.7 nm (labeled “e” in Fig. 6B) corresponded to the outer edge of each capsid shell. Only the inner core (radius 0–10 nm) of the pre-viral nucleocapsid, believed to contain unstructured virus RNA, showed variations in the density profiles between different reconstructions (Suppl. Data Fig. 2).
Capsid stoichiometry of a pre-viral nucleocapsid
The limited resolution of the pre-viral nucleocapsid assembly intermediate made it difficult to definitively assign capsid protein positions and stoichiometry. However, determining the stoichiometry of pre-viral nucleocapsids was important for unraveling VEEV assembly pathways and mechanisms. Two independent and complementary approaches were used to estimate the number of capsid proteins that formed the pre-viral nucleocapsid. Both approaches showed that this pre-viral nucleocapsid assembly intermediate likely contained 240 capsid proteins, identical to the number of capsid protein that form the mature VEEV nucleocapsid.
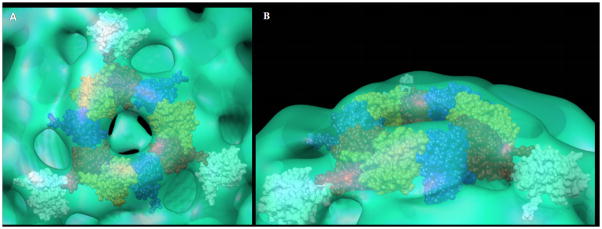
In the first approach, monomers of the C-terminal domain of VEEV capsid protein were positioned into the electron density that defined an icosahedral face of the capsid shell of the pre-viral nucleocapsid. Twelve capsid C-terminal domains, with the required 3-fold symmetry, could be contained in this density without steric clashes (i.e., intersection of protein surfaces) between subunits (Fig. 7). As a control experiment, capsid C-terminal domains were positioned into corresponding densities from the mature VEEV (Watowich and Paredes, personal communication) using equivalent software tools, electron density thresholds, and map resolution. In the control docking experiment, twelve capsid proteins could be correctly placed in the T=4 lattice described for the mature VEEV nucleocapsid (Paredes et al., 2001). Thus, these docking experiments suggested that the same number of capsid proteins formed the nucleocapsids of pre-viral assembly intermediates and mature virus.
Figure 7.
Capsid proteins modeled within the pre-viral nucleocapsid. (A) Twelve C-terminal domains of the VEEV capsid protein positioned within an icosahedral face of the pre-viral nucleocapsid particle and viewed along an icosahedral 3-fold axis. (B) Side view of the pre-viral nucleocapsid highlighting the arrangement of twelve C-terminal domains of the VEEV capsid protein within an icosahedral face of the particle.
In the second approach, the electron density volume corresponding to the capsid shell was calculated for pre-viral and mature VEEV nucleocapsids. These calculations used comparable density threshold values to delineate the protein envelope and accounted for the different density distributions in the reconstructions. A density cutoff of 2.3 σ was sufficient to enclose a volume in the mature nucleocapsid equal to the volume of 240 C-terminal domains of the VEEV capsid protein (5.08 x 106 A3). At this electron density threshold, the electron density volume of the capsid shell was larger for the pre-viral nucleocapsid compared to the mature nucleocapsid. Thus, the electron density volume of the capsid shell of the pre-viral nucleocapsid was sufficient to enclose at least 240 C-terminal domains of the VEEV capsid protein.
The limited resolution of the pre-viral nucleocapsid structure prevented definitive localization of the capsid proteins within the surface capsomers. Although the pre-viral nucleocapsid could accommodate 240 capsid proteins, these proteins did not adopt the classic T=4 capsomer lattice observed in VEEV mature nucleocapsids (Paredes et al., 2001) and VEEV nucleocapsids isolated from whole virions (Paredes et al., 2003), with quasi-hexons centered on the 2-fold axis of symmetry. Higher resolution will be necessary to determine how the 240 VEEV capsid proteins are arranged within each capsomer of the pre-viral nucleocapsid.
Discussion
VEEV pre-viral nucleocapsids were isolated from infected cells prior to virus maturation. Cryo-EM images showed that subsets of the isolated nucleocapsids were undamaged, well-formed, and homogeneous in size. A self-consistent model-independent icosahedral 3-dimensional reconstruction was generated from these image subsets. The challenging task of determining structures from subsets of available cryo-EM images was not unique to the VEEV pre-viral nucleocapsid, and had occurred for structures such as the nucleocapsids isolated from mature VEEV (Paredes et al., 2003) and Triatoma virus capsid (Estrozi et al., 2008). Moreover, the limited number of images used to determine the VEEV pre-viral nucleocapsid structure parallelled studies of other mammalian virus assembly intermediates, where virus assembly intermediates of human parvovirus B19 VP2 capsid (Chipman et al., 1996), VP60 (Barcena et al., 2004), and NT29 (Barcena et al., 2004) were determined from 37, 100, and 79 particles, respectively. These challenges have hampered high-resolution structural studies and detailed understanding of virus assembly intermediates.
The VEEV pre-viral nucleocapsid formed a unique icosahedral particle, indicating the VEEV capsid protein and vRNA were sufficient to self-assemble into an ordered structure prior to interacting with the plasma membrane and envelope proteins. It is not known if nucleocapsid self-assembly is aided by cellular factors. Unlike some viruses (e.g., turnip yellow mosaic virus; Larson et al., 2005), VEEV nucleocapsid assembly does not appear to be directed by an icosahedral viral RNA scaffold, since viral RNA purified from mature VEEV had an extended structure (as observed by negative-stained electron microscope; data not shown).
The structure of VEEV nucleocapsids purified from infected cells (i.e., pre-viral nucleocapsids) differed significantly from nucleocapsids extracted from mature VEEV (Paredes et al., 2003), and artificial nucleocapsid-like particles (CLPs) assembled from purified nucleic acid and recombinant alphavirus capsid proteins (Mukhopadhyay et al., 2002; Tellinghuisen et al., 2001). The pre-viral nucleocapsid had an icosahedral structure, but its 240 capsid proteins were not arranged in the classical T=4 lattice observed in mature (Paredes et al., 2001) and isolated (Paredes et al., 2003) nucleocapsids. The recombinant CLPs were either unstructured (Tellinghuisen et al., 2001) or formed T=4 icosahedral particles (Mukhopadhyay et al., 2002). These differences in nucleocapsid structure most likely reflect differences in particle origin and assembly environment. It is possible the pre-viral, isolated, and CLP particles might have a “pseudo-icosahedral” structure as opposed to a true icoshedral structure, and future higher resolution studies will be necessary to more fully address this prospect.
The diameter of the pre-viral nucleocapsid corresponded to the mode diameter in the nucleocapsid diameter distribution plot. Thus, the pre-viral nucleocapsid structure likely reflects a major intermediate species of the virus maturation pathway. In addition, capsid protein and viral RNA formed spherical nucleocapsids with different diameters and densities. Since these particles were recalcitrant to reconstruction, they might represent a continuum of sparsely populated pre-viral nucleocapsid structures (perhaps similar to protein-folding pathway intermediates) instead of a single unique intermediate structure (or several well-defined stable intermediate structures). Analogous assembly intermediates have been observed for other viruses, where multiple “capsid-like” particle intermediates were seen during Kaposi’s sarcoma-associated herpesvirus assembly (Deng et al., 2008) and Norwalk virus disassembly (Shoemaker et al., 2010). The detailed role of the icosahedral and other pre-viral nucleocapsid assembly intermediates in alphavirus assembly remains unknown. Since the number of capsid proteins in the pre-viral nucleocapsid structure was consistent with that of the mature nucleocapsid, the reconstructed pre-viral nucleocapsid assembly state was likely a direct precursor to the mature VEEV. The transition from the icosahedral pre-viral nucleocapsid to the mature nucleocapsid would not require rupturing the nucleocapsid but would involve local rearrangements of the capsid proteins that form the capsomers. These rearrangements would also be necessary to compress the 43.4 nm diameter pre-viral nucleocapsid into the 39 nm diameter mature virus nucleocapsid, analogous to the “expandohedra” capsomer transitions observed between different assembly states of the cowpea chlorotic mottle virus (CCMV) (Kovacs et al., 2004). Interactions between the cytoplasmic pre-viral nucleocapsid and the cell membrane and/or VEEV glycoproteins embedded in the cell membrane might provide the necessary driving force for capsid protein rearrangement and nucleocapsid compression.
The observation that pre-viral nucleocapsids isolated from infected cells had a range of diameters was consistent with previous EM studies of cells infected with Semliki Forest (Acheson and Tamm, 1967) and Western equine encephalitis viruses (Morgan et al., 1961). Those studies reported differently sized “nucleoids” (what we now term nucleocapsids) in the cytoplasm of infected cells, and concluded that the smaller “nucleoids” likely did not assemble into mature viruses (Acheson and Tamm, 1967). The ensemble of nucleocapsid particles isolated in this study may represent intermediates along a single assembly pathway that culminate in the mature nucleocapsid. Alternatively, these particles may populate independent assembly pathways that each transition to the mature nucleocapsid. If the smaller intermediates lack 240 capsid proteins, it is likely they must disassemble and reform prior to forming the mature nucleocapsids or represent “dead-end” assembly states incapable of rearranging into the mature nucleocapsid. The ability of pre-viral VEEV nucleocapsids to adopt diverse structures and rearrange its capsid proteins as it transitions to the mature virus implies self-assembly is an inefficient process (if efficiency is defined as forming a single highly stable structure that mimics the mature nucleocapsid). On the other hand, the plasticity of the nucleocapsid particle may be a fault-tolerant assembly strategy to accommodate capsid mutations before ultimately assembling into a mature infectious virus. As shown by this study and our previous work with nucleocapids extracted from mature virus (Paredes et al., 2003), the final VEEV assembly step involves significant capsid reorganization that is likely regulated by interactions between the nucleocapid particle, viral envelope proteins and/or the lipid bilayer.
Materials and Methods
Virus strain
The attenuated TC-83 vaccine strain of Trinidad donkey VEEV was used for all studies to allow experiments to be conducted in a biosafety level 2 environment. The amino acid sequences of wild-type (Trinidad donkey strain) and attenuated TC-83 VEEV are identical throughout the capsid protein, and differ by one amino acid in the E1 and NSP4 proteins and five amino acids in E2 protein (Kinney et al., 1993). The amino acid sequence identity of the capsid protein in wild-type and attenuated VEEV allowed TC-83 to serve as a VEEV surrogate in cell-based assembly experiments.
Optimal time for harvesting pre-viral nucleocapsids
To determine harvest times that provided optimal amounts of particles without observable cellular cytopathic effects, pre-viral nucleocapsids were isolated from baby hamster kidney (BHK) cells infected for 12 to 24 hours. The amount of pre-viral nucleocapsids isolated at each time point was determined by comparing purified pre-viral nucleocapsid samples against defined quantities of VEEV capsid protein using Coomassie blue staining and Western blotting of sodium dodecyl sulfate (SDS) 16%-polyacrylimide gels (data not shown). Pre-viral nucleocapsids harvested at different time points were isolated using identical protocols (described below). Pre-viral nucleocapsids harvested from cells 18–20 hours post-infection with TC-83 VEEV yielded the greatest number of particles; this time point was used in subsequent experiments.
Pre-viral nucleocapsid production and purification
BHK cells infected with VEEV TC-83 were lysed with 0.1% deoxycholic acid (Sigma-Aldrich) in isolation buffer (25 mM HEPES [Fisher] pH 7.4, 100 mM potassium acetate [Sigma-Aldrich], 1.7 mM magnesium acetate [Fluka BioChemika], Roche protease inhibitor cocktail tablet [Roche Diagnostics], and 5 mM ribonucleoside vanadyl complexes [Sigma-Aldrich] or RNAase inhibitor from human placenta [USB]). Cell lysate and debris were separated by centrifuging at 2000 rpm (Jouan CR312) at 4°C for 20 minutes. Sample supernatants were concentrated onto 25% sucrose (Fisher) or iodixanol (Sigma-Aldrich) cushions with centrifugation (Beckman L7 or Beckman L8-80M with an SW41 rotor) at 115,604 xg for 3 hours at 4°C. Pre-viral nucleocapsids were purified with density separation using a 10%–60% continuous sucrose or iodixanol gradient centrifuged at 115,604 xg for 3 hours at 4°C. Fractions containing pre-viral nucleocapsids were identified by Coomassie blue staining and Western blotting of SDS 16%-polyacrylamide gels.
Thin-sectioning of infected BHK cells
Thin sections of BHK cells were examined for virus budding around the VEEV post-infection times used to isolate nucleocapsids. Little or no budding observed at these times would suggest that particles isolated from cells were predominantly pre-viral nucleocapsids (as opposed to particles from mature virus). The thin-sectioning protocol with poly/bed 812 solutions for infiltration was adapted from previous reports (Anderson et al., 1995; Finck, 1960; Luft, 1961; Murphy and Fields, 1967). Briefly, 19 hrs post-infection, cells were fixed with freshly prepared PFGPA solution (1.25% paraformaldehyde [Sigma-Aldrich], 2.5% glutaraldehyde [Electron Microscopy Sciences], 0.03% CaCl2 [Sigma-Aldrich], 50 mM cacodylate buffer [Electron Microscopy Sciences] pH 7.3) for 1.5 hours at 4ºC, and then stored at 4ºC in 100 mM cacodylate buffer. Cells were scraped from flasks and pelleted in a benchtop centrifuge at 2000 rpm for 10 minutes at room temperature. Cell pellets were fixed with osmium tetroxide (Electron Microscopy Sciences) (Karnovsky, 1961; Murphy and Fields, 1967), followed by ethanol dehydration. Cells were gradually infiltrated with poly/bed 812 solution (Polysciences Inc.), beginning with a mixture of propylene oxide (Polysciences Inc., EM grade) and poly/bed 812 solutions (Polysciences Inc.), and followed by 100% poly/bed 812. The resin was polymerized at 37°C. Eighty to ninety nm-thick sections were cut with an ultra-microtome (Sorvall MT6000) and placed on nickel grids (formvar/carbon) (Electron Microscopy Sciences) and negatively stained with Reynolds lead citrate (from Mallinckrodt’s sodium citrate and EMS’s lead nitrate) and 2% uranyl acetate.
The protocol for immuno-labeling was adapted from Anderson et al. (Anderson et al., 1995). Briefly, cell pellets were embedded in LR white (London Resin Company, Ltd.) without post-fixation with osmium tetroxide, and thin sections were immuno-labeled with a primary polyclonal antibody against the C-terminal domain of the VEEV capsid protein (Watowich, personal communication) and secondary anti-rabbit antibody IgG (H & L) linked to 5 nm gold particles (Amersham Biosciences Auro Probe). Negative controls were run of uninfected BHK cells for both immuno and non-immuno-labeled thin sections. Thin sections were imaged using a Philips EM 201 microscope.
Confirmation of nucleocapsid identity by negative stain and immuno-gold labeling
Following pelleting, pre-viral nucleocapsid samples were dialyzed against isolation buffer to remove excess sucrose and then negatively stained with 2% uranyl acetate on copper (carbon/formvar) grids (EM Sciences). Fractions containing pre-viral nucleocapsid particles were selected for further analysis. The identity of VEEV nucleocapsids was tested using immuno-gold labeling with primary polyclonal antibody raised against the C-terminal domain of the VEEV capsid protein and secondary antibody linked to 5 nm gold particles. Images of negatively stained capsids and immuno-labeled sections were taken on a JEM 2100 electron microscope (JEOL) at 200 keV.
Vitrification of pre-viral nucleocapsids and low dose imaging
Vitrified pre-viral nucleocapsids were used for imaging and 3D reconstruction. Samples were applied to holey carbon film on copper grids (Quantifoil R2/2, Micro Tools GmbH, Jena, Germany), excess liquid removed by blotting with filter paper, and grids were plunged into liquid ethane (Lepault et al., 1983). Grids were transferred to liquid nitrogen and kept at cryogenic temperatures until examined on a cryo-electron microscope (cryo-EM). For microscopy, grids were placed in a pre-cooled cryo-specimen holder (Gatan 626) and transferred to the cryo-electron microscope. The microscope was calibrated using MAG*I*CAL (available through Electron Microscopy Sciences, www.emsdiasum.com/microscopy/products/calibration/magical.aspx) and the measurement of the helical pitch of tobacco mosaic virus as standards. Grids were maintained at −180°C during the microscopy session. Micrographs were recorded with a slow-scan CCD camera (Ultrascan 895, Gatan) using low-dose conditions (Amos et al., 1982) on a JEM 2010 (JEOL) electron microscope operating at 200 keV. Images were recorded with an electron dose of 15–20 electrons/Å2 and defocus values ranged from −1.5 to −3.0 microns. The first zero of the contrast transfer function (CTF) (Erickson and Klug, 1971) of the microscope, even at highest defocus value used, occurred at ~27 Å.
Image processing and 3D reconstruction of pre-viral nucleocapsids
Well-preserved particles were “boxed out” from micrographs using the Boxer program in the EMAN package (Ludtke et al., 1999). Based on visual inspection, particles of similar size were manually selected for 3D reconstructions. Particles were discarded if they were different in size, distorted, damaged, or had other defects. Further processing was performed with a modified version of the PFT protocol (Zhang et al., 2003). Images were normalized to have zero average value and the same standard deviation. Particle orientations and their origins were determined based on a reference 3D map generated from image data using the Auto3dem package (Yan et al., 2007). The orientations/origins were iteratively refined using either Auto3dem package or using individual refinement programs from that package or OOR program (Yan et al., 2007). After each iteration step, poorly correlating particles were discarded and the best particles retained for continued refinement. The final reconstructions used an image set containing ~90 pre-viral nucleocapsid particles. These particles correlated well with each other and were also chosen by the AUTOSELECT program. The AUTOSELECT program was unable to find well-correlating particles within data subsets that were missing the images that produced the above-converged reconstruction, and additional reconstructions were not generated from these data subsets.
To avoid model bias in the 3D map of the VEEV pre-viral nucleocapsid, additional independent reconstructions were performed using various starting maps scaled to the diameter of the VEEV pre-viral nucleocapsid. Three different capsid maps were used, corresponding to VEEV mature nucleocapsid (Paredes et al., 2001), Western equine encephalitis virus (WEEV) mature nucleocapsid (M. Sherman, personal communication), and red clover necrotic mosaic virus (RCNMV) (Sherman et al., 2006). The VEEV and WEEV mature nucleocapsid maps had T=4 symmetry, while the RCNMV map was T=3. Starting maps with different T numbers help verify that the image dataset yielded a stable consistent reconstruction regardless of the capsomer lattice used for initial particle alignment and orientation search. Resolution of the reconstructions was estimated using Fourier shell correlation with 0.5 threshold (Rosenthal and Henderson, 2003).
Volume estimation in cryo-EM maps
Pre-viral nucleocapsid volumes were calculated using the program Chimera (Pettersen et al., 2004). The outer radius was designated as the exterior edge of the nucleocapsid. The inner radius was specified to include only the part of the density map that corresponded to the C-terminal domain of the capsid protein. We compared calculated volumes from pre-viral and mature nucleocapsid maps using comparable radial cutoffs given the common features in the maps. Density thresholds were also equivalent, and comparisons between the two maps were made using a series of threshold values. Protein volume was calculated using the partial specific volume for a globular protein and the mass of the C-terminal domain of the VEEV capsid protein (PDB entry 1EP5; Watowich, personal communication).
Pseudo-atomic docking
An asymmetric unit was extracted from the pre-viral nucleocapsid electron density map using the program cut_cyl_map (M. Sherman, personal communication). Minimum and maximum radii were chosen that bounded the capsid protein shell. Chimera (Pettersen et al., 2004) was used to position monomers of the C-terminal domain of the VEEV capsid protein (PDB entry 1EP5; S. Watowich, personal communication) into density corresponding to an icosahedral face of the pre-viral nucleocapsid map. The “space-filling” viewing option of Chimera was used to visualize docked proteins. The 3-fold symmetry of the particle was used to position symmetry-related capsid proteins within the icosahedral face.
Supplementary Material
Acknowledgments
We thank Drs. V. Popov and Z. H. Zhou for providing generous access to their microscopy facilities, Dr. W. Chiu for use of electron microscopes within the National Center for Macromolecular Imaging, Dr. S. Weaver for assistance with particle purification, Dr. A. Paredes for helpful discussions, and Ms. J. Wen for assistance with imaging cellular thin-sections. Funding was provided in part by a pre-doctoral fellowship (KL) from the Keck Center for Viral Imaging, UTMB’s Sealy Center for Structural Biology, and grants from NIH/NIAID AI066160 (SJW) and the Welch Foundation (SJW).
Appendix A. Supplementary data
Supplementary data associated with this article can be found online.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acheson NH, Tamm I. Replication of Semliki Forest virus: an electron microscopic study. Virology. 1967;32:128–143. doi: 10.1016/0042-6822(67)90261-9. [DOI] [PubMed] [Google Scholar]
- Amos LA, Henderson R, Unwin PN. Three-dimensional structure determination by electron microscopy of two-dimensional crystals. Prog Biophys Mol Biol. 1982;39:183–231. doi: 10.1016/0079-6107(83)90017-2. [DOI] [PubMed] [Google Scholar]
- Anderson LE, Wang X, Gibbons JT. Three enzymes of carbon metabolism or their antigenic analogs in pea leaf nuclei. Plant Physiol. 1995;108:659–667. doi: 10.1104/pp.108.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol Mol Biol Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcena J, Verdaguer N, Roca R, Morales M, Angulo I, Risco C, Carrascosa JL, Torres JM, Caston JR. The coat protein of rabbit hemorrhagic disease virus contains a molecular switch at the N-terminal region facing the inner surface of the capsid. Virology. 2004;322:118–134. doi: 10.1016/j.virol.2004.01.021. [DOI] [PubMed] [Google Scholar]
- Bykovsky AF, Yershov FI, Zhdanov VM. Morphogenesis of Venezuelan equine encephalomyelitis virus. J Virol. 1969;4:496–504. doi: 10.1128/jvi.4.4.496-504.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng RH, Reddy VS, Olson NH, Fisher AJ, Baker TS, Johnson JE. Functional implications of quasi-equivalence in a T = 3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography. Structure. 1994;2:271–282. doi: 10.1016/s0969-2126(00)00029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman PR, Agbandje-McKenna M, Kajigaya S, Brown KE, Young NS, Baker TS, Rossmann MG. Cryo-electron microscopy studies of empty capsids of human parvovirus B19 complexed with its cellular receptor. Proc Natl Acad Sci USA. 1996;93:7502–7506. doi: 10.1073/pnas.93.15.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David AE. Lipid composition of Sindbis virus. Virology. 1971;46:711–720. doi: 10.1016/0042-6822(71)90073-0. [DOI] [PubMed] [Google Scholar]
- Deng B, O’Connor CM, Kedes DH, Zhou ZH. Cryo-electron tomography of Kaposi’s sarcoma-associated herpesvirus capsids reveals dynamic scaffolding structures essential to capsid assembly and maturation. J Struct Biol. 2008;161:419–427. doi: 10.1016/j.jsb.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrozi LF, Neumann E, Squires G, Rozas-Dennis G, Costabel M, Rey FA, Guerin DM, Navaza J. Phasing of the Triatoma virus diffraction data using a cryo-electron microscopy reconstruction. Virology. 2008;375:85–93. doi: 10.1016/j.virol.2007.12.025. [DOI] [PubMed] [Google Scholar]
- Finck H. Epoxy resins in electron microscopy. J Biophys Biochem Cytol. 1960;7:27–30. doi: 10.1083/jcb.7.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller SD, Butcher SJ, Cheng RH, Baker TS. Three-dimensional reconstruction of icosahedral particles - the uncommon line. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- Garoff H, Sjoberg M, Cheng RH. Budding of alphaviruses. Virus Res. 2004;106:103–116. doi: 10.1016/j.virusres.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Haag L, Garoff H, Xing L, Hammar L, Kan ST, Cheng RH. Acid-induced movements in the glycoprotein shell of an alphavirus turn the spikes into membrane fusion mode. EMBO J. 2002;21:4402–10. doi: 10.1093/emboj/cdf442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg CB, Robbins PW. The glycolipids and phospholipids of Sindbis virus and their relation to the lipids of the host cell plasma membrane. Virology. 1974;61:602–608. doi: 10.1016/0042-6822(74)90295-5. [DOI] [PubMed] [Google Scholar]
- Karnovsky MJ. Simple methods for “staining with lead” at high pH in electron microscopy. J Biophys Biochem Cytol. 1961;11:729–32. doi: 10.1083/jcb.11.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney RM, Chang GJ, Tsuchiya KR, Sneider JM, Roehrig JT, Woodward TM, Trent DW. Attenuation of Venezuelan equine encephalitis virus strain TC-83 is encoded by the 5′-noncoding region and the E2 envelope glycoprotein. J Virol. 1993;67:1269–1277. doi: 10.1128/jvi.67.3.1269-1277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson SB, Lucas RW, Greenwood A, McPherson A. The RNA of turnip yellow mosaic virus exhibits icosahedral order. Virology. 2005;334:245–254. doi: 10.1016/j.virol.2005.01.036. [DOI] [PubMed] [Google Scholar]
- Lepault J, Booy FP, Dubochet J. Electron microscopy of frozen biological suspensions. J Microsc. 1983;129(Pt 1):89–102. doi: 10.1111/j.1365-2818.1983.tb04163.x. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Luft JH. Improvements in epoxy resin embedding methods. J Biophys Biochem Cytol. 1961;9:409–414. doi: 10.1083/jcb.9.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini EJ, Clarke M, Gowen BE, Rutten T, Fuller SD. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol Cell. 2000;5:255–266. doi: 10.1016/s1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- Morgan C, Howe C, Rose HM. Structure and development of viruses as observed in the electron microscope. V Western equine encephalomyelitis virus. J Exp Med. 1961;113:219–234. doi: 10.1084/jem.113.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Chipman PR, Hong EM, Kuhn RJ, Rossmann MG. In vitro-assembled alphavirus core-like particles maintain a structure similar to that of nucleocapsid cores in mature virus. J Virol. 2002;76:11128–11132. doi: 10.1128/JVI.76.21.11128-11132.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy FA, Fields BN. Kern Canyon virus: electron microscopic and immunological studies. Virology. 1967;33:625–637. doi: 10.1016/0042-6822(67)90063-3. [DOI] [PubMed] [Google Scholar]
- Paredes A, Alwell-Warda K, Weaver SC, Chiu W, Watowich SJ. Venezuelan equine encephalomyelitis virus structure and its divergence from old world alphaviruses. J Virol. 2001;75:9532–9537. doi: 10.1128/JVI.75.19.9532-9537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes A, Alwell-Warda K, Weaver SC, Chiu W, Watowich SJ. Structure of isolated nucleocapsids from Venezuelan equine encephalitis virus and implications for assembly and disassembly of enveloped virus. J Virol. 2003;77:659–664. doi: 10.1128/JVI.77.1.659-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R, Weaver SC, de Siger J, Medina G, Salas RA. Emergence of a new epidemic/epizootic Venezuelan equine encephalitis virus in South America. Proc Natl Acad Sci USA. 1995;92:5278–5281. doi: 10.1073/pnas.92.12.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Schlesinger S, Schlesinger MJ. Togviridae: the viruses and their replication. In: Knipe DM, Howley PM, Griffin DE, editors. Fundamental virology. 4. Lippincott Williams & Wilkins; Philadelphia: 2001. pp. 567–588. [Google Scholar]
- Sherman MB, Guenther RH, Tama F, Sit TL, Brooks CL, Mikhailov AM, Orlova EV, Baker TS, Lommel SA. Removal of divalent cations induces structural transitions in red clover necrotic mosaic virus, revealing a potential mechanism for RNA release. J Virol. 2006;80:10395–10406. doi: 10.1128/JVI.01137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker GK, van Duijn E, Crawford SE, Uetrecht C, Baclayon M, Roos WH, Wuite GJ, Estes MK, Prasad BV, Heck AJ. Norwalk virus assembly and stability monitored by mass spectrometry. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M900620-MCP200. Epub., M900620–MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellinghuisen TL, Perera R, Kuhn RJ. In vitro assembly of Sindbis virus core-like particles from cross-linked dimers of truncated and mutant capsid proteins. J Virol. 2001;75:2810–2817. doi: 10.1128/JVI.75.6.2810-2817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel RH, Provencher SW, von Bonsdorff CH, Adrian M, Dubochet J. Envelope structure of Semliki Forest virus reconstructed from cryo-electron micrographs. Nature. 1986;320:533–535. doi: 10.1038/320533a0. [DOI] [PubMed] [Google Scholar]
- Warrier R, Linger BR, Golden BL, Kuhn RJ. Role of Sindbis virus capsid protein region II in nucleocapsid core assembly and encapsidation of genomic RNA. J Virol. 2008;82:4461–4470. doi: 10.1128/JVI.01936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Ferro C, Barrera R, Boshell J, Navarro JC. Venezuelan equine encephalitis. Annu Rev Entomol. 2004;49:141–174. doi: 10.1146/annurev.ento.49.061802.123422. [DOI] [PubMed] [Google Scholar]
- Weaver SC, Salas R, Rico-Hesse R, Ludwig GV, Oberste MS, Boshell J, Tesh RB. Re-emergence of epidemic Venezuelan equine encephalomyelitis in South America. VEE Study Group Lancet. 1996;348:436–440. doi: 10.1016/s0140-6736(96)02275-1. [DOI] [PubMed] [Google Scholar]
- Wengler G, Koschinski A, Dreyer F. Entry of alphaviruses at the plasma membrane converts the viral surface proteins into an ion-permeable pore that can be detected by electrophysiological analyses of whole-cell membrane currents. J Gen Virol. 2003;84(Pt 1):173–181. doi: 10.1099/vir.0.18696-0. [DOI] [PubMed] [Google Scholar]
- Yan X, Sinkovits RS, Baker TS. AUTO3DEM - an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol. 2007;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 A. Nat Struct Biol. 2003;10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.