Abstract
Regulators of G protein signaling (RGS) proteins are potent negative modulators of G protein signaling and have been proposed as potential targets for small-molecule inhibitor development. We report a high-throughput time-resolved fluorescence resonance energy transfer screen to identify inhibitors of RGS4 and describe the first reversible small-molecule inhibitors of an RGS protein. Two closely related compounds, typified by CCG-63802 [((2E)-2-(1,3-benzothiazol-2-yl)-3-[9-methyl-2-(3-methylphenoxy)-4-oxo-4H-pyrido[1,2-a]pyrimidin-3-yl]prop-2-enenitrile)], inhibit the interaction between RGS4 and Gαo with an IC50 value in the low micromolar range. They show selectivity among RGS proteins with a potency order of RGS 4 > 19 = 16 > 8 ≫ 7. The compounds inhibit the GTPase accelerating protein activity of RGS4, and thermal stability studies demonstrate binding to the RGS but not to Gαo. On RGS4, they depend on an interaction with one or more cysteines in a pocket that has previously been identified as an allosteric site for RGS regulation by acidic phospholipids. Unlike previous small-molecule RGS inhibitors identified to date, these compounds retain substantial activity under reducing conditions and are fully reversible on the 10-min time scale. CCG-63802 and related analogs represent a useful step toward the development of chemical tools for the study of RGS physiology.
Introduction
Networks of protein-protein interactions are crucial for efficient cellular function. There has been significant interest in developing small-molecule protein-protein interaction inhibitors (SMPPIIs) for use as research probes and potential therapeutic agents (Berg, 2003, 2008; Gadek and Nicholas, 2003; Arkin and Wells, 2004; Blazer and Neubig, 2009). The development of SMPPIIs has been difficult. One challenge has been the lack of clearly identifiable small-molecule binding sites on the relatively featureless protein-protein interaction interface. A promising approach is the use of allosteric pockets on the protein target to bypass this problem and, increasingly, there has been solid progress in SMPPII development (Berg, 2003, 2008; Arkin and Wells, 2004; Blazer and Neubig, 2009; Arkin and Whitty, 2009; Busschots et al., 2009; Niu and Chen, 2009).
RGS proteins are GTPase-accelerating proteins (GAPs) for heterotrimeric G protein α subunits (Berman et al., 1996). They increase the intrinsic rate of GTP hydrolysis by the Gα, thus reconciling the paradox of the subsecond regulation of G protein signaling in vivo versus the relatively long half-life of GTP bound to purified Gα in vitro. In mammals, there are more than 20 known RGS proteins that interact with limited selectivity to most Gα subtypes (Hollinger and Hepler, 2002; Neubig and Siderovski, 2002).
There is substantial interest in the therapeutic potential of small-molecule modulators of RGS proteins (Zhong and Neubig, 2001; Neubig and Siderovski, 2002; Riddle et al., 2005; Blazer and Neubig, 2009; Traynor et al., 2009). In brief, RGS inhibitors may potentiate signaling through GPCRs in a tissue-specific manner because of the localized expression patterns of many RGS proteins. This effect could be used to reduce side effects of clinically used GPCR agonists that stem from nontarget tissue receptor activation [e.g., μ-opioid receptor-dependent constipation during postoperative analgesia (Bueno and Fioramonti, 1988)].
To understand the physiological ramifications of inhibiting RGS protein GAP activity, we have developed two lines of mice that express mutant Gαo or Gαi2 and are insensitive to RGS effects (G184S). These mice show dramatic phenotypes, including resistance to diet-induced obesity and antidepressant-like behavioral effects (Huang et al., 2006, 2008; Talbot et al., 2010). RGS4 is up-regulated in the dorsal horn of spinal cord during the development of neuropathic pain (Garnier et al., 2003), and RGS4 can inhibit several pain-modulating receptors (e.g., μ-opioid receptor) (Garnier et al., 2003; Traynor and Neubig, 2005). Consequently, small-molecule modulators of RGS function should have utility as research tools and potentially as therapeutics. Because of the wealth of information on the structure and function of RGS4, we chose this protein as our primary target for validating the “drugability” of RGS proteins.
There have been several reported peptide inhibitors of RGS4 and related family members (Roof et al., 2006, 2008; Wang et al., 2008) and one disclosed small-molecule inhibitor (Roman et al., 2007). Because of the physical properties of the peptides, none of them function in a cellular environment unless they are introduced intracellularly [e.g., by dialysis via a patch pipette (Roof et al., 2006)]. The small-molecule compound CCG-4986 [methyl-N-[(4-chlorophenyl)sulfonyl]-4-nitrobenzenesulfinimidoate] irreversibly inhibits RGS4 by reacting with one or more cysteine residues (Kimple et al., 2007; Roman et al., 2010), and its activity is lost in the presence of free thiols. This mechanism of action makes CCG-4986 less desirable as a potential lead compound for small-molecule probe development. Consequently, we undertook this study to identify novel RGS inhibitors that retain activity under reducing conditions and ones that have a reversible mechanism of action.
This article describes the identification and characterization of the first class of reversible small-molecule inhibitors of an RGS protein. They were found in a biochemical high-throughput screen carried out in the presence of dithiothreitol (DTT). They inhibit the binding and GAP activity of RGS4 with Gαo in a reversible manner through an interaction at an allosteric regulatory site on the RGS. These compounds represent an important step toward the development of tools for the study of RGS functions in physiological and pathophysiological situations.
Materials and Methods
Reagents.
Chemicals were purchased from Sigma-Aldrich (St. Louis, MO) or Thermo Fisher Scientific (Waltham, MA) and were reagent grade or better. Alexa Fluor 488 succinimidyl ester and LanthaScreen Thiol-reactive Tb chelate were obtained from Invitrogen (Carlsbad, CA). γ[32P]GTP (10 mCi/ml) and [35S]GTPγS (12.5 mCi/ml) were obtained from PerkinElmer Life and Analytical Sciences (Waltham, MA) and isotopically diluted with unlabeled nucleotide before use. Amylose resin was purchased from New England Biolabs (Ipswich, MA). Ni-NTA resin was purchased from QIAGEN (Valencia, CA). Avidin-coated microspheres were purchased from Luminex (Austin, TX). The screening library was comprised of a commercially available subset of compounds from ChemDiv (San Diego, CA) provided through a collaboration between the University of Michigan Center for Chemical Genomics and the Novartis Institute for Biomedical Research (East Hanover, NJ). CCG-63802 [((2E)-2-(1, 3-benzothiazol-2-yl)-3-[9-methyl-2-(3-methylphenoxy)-4-oxo-4H-pyrido[1,2-a]pyrimidin-3-yl]prop-2-enenitrile)] and CCG-63808 [((2E)-2-(1,3-benzothiazol-2-yl)-3-[9-methyl-2-(4-fluorolphenoxy)-4-oxo-4H-pyrido[1,2-a]pyrimidin-3-yl]prop-2-enenitrile)] (see structures in Fig. 1) were purchased from ChemDiv, and compound identity was verified by NMR via ChemDiv and independent complete synthesis in the laboratory of Dr. Stephen M. Husbands (University of Bath).
Fig. 1.
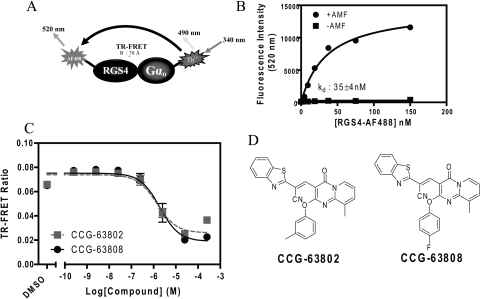
Characterization of the RGS4 TR-FRET high-throughput assay. A, schematic of RGS4-Gαo TR-FRET assay. Gαo is labeled with the LanthaScreen Tb-chelate donor fluorophore, and RGS4 is labeled with an Alexa Fluor 488 acceptor fluorophore. Excitation and emission maxima are listed for each fluorophore. B, representative data showing the AlF4−/GDP dependence of the interaction between RGS4-AF488 and 10 nM Tb-Gαo. This saturable interaction has a Kd of 35 ± 4 nM. C, two compounds identified in the high-throughput screen, CCG-63802 and CCG-63808, dose-dependently inhibit the TR-FRET signal between RGS4-AF488 and Tb-Gαo with IC50 values of 1.4 (0.76; 2.6 μM) and 1.9 μM (1.02; 3.5 μM), respectively. Data (n = 3 for all data) are presented as mean ± S.E.M. or mean (95% confidence interval) in B and C, respectively. D, the chemical structures of CCG-63802 and CCG-63808.
Compound Synthesis.
In brief, 2-hydroxy-9-methyl-4H-pyrido[1,2-α] pyrimidin-4-one was prepared by the reaction of 2-amino-3-methylpyridine with diethyl malonate according to literature methods (Ingalls and Popp, 1967). This material was first converted to 2-chloro-9-methyl-4-oxo-4H-pyrido[1,2-α]pyrimidine-3-carbaldehyde via Vilsmeier formylation, and this product was then heated with 4-fluorophenol to afford 2-(4-fluorophenoxy)-9-methyl-4-oxo-4H- pyrido[1,2-α]pyrimidine-3-carbaldehyde. Condensation of this compound with 2-benzothiazole acetonitrile using catalytic triethylamine in dichloromethane provided CCG-63808 as an orange crystalline solid (Supplemental Fig. 1). CCG-63802 was prepared in a similar manner, except 4-fluorophenol was replaced with 3-methylphenol. Synthesized compounds were verified by 1H and 13C NMR using a JEOL (Tokyo, Japan) δ-270-MHz instrument: 1H at 270 MHz, and Varian Inc. (Palo Alto, CA) Mercury-400-MHz instrument: 1H at 400 MHz, 13C at 100 MHz; d in parts per million, J in Hertz with tetramethylsilane as an internal standard, by electrospray mass spectrometry using a micrOTOF (Bruker Daltonics, Billerica, MA) and microanalysis using a PerkinElmer Life and Analytical Sciences 240C analyzer.
Protein Expression and Purification.
Human RGS4 was expressed either from the pQE80RGS4 vector, which encodes 6× histidine-tagged and N-terminally truncated form of RGS4 that lacks the first 18 residues (ΔN19RGS4), or the pKMRGS4 vector, which encodes a maltose-binding protein (MBP)-ΔN19RGS4 fusion protein. The ΔN form of RGS4 was selected because it provides better protein yield in prokaryotic expression systems. MBP-His6-RGS19ΔC11 (human), MBP-His6-RGS7 (human), MBP-His6-RGS8 (human), and MBP-His6-RGS16 (human) were expressed from constructs made with the pMALC2H10 vector as described previously (Roman et al., 2009). For the mutagenesis studies, ΔN51RGS4 (rat) wild type and cysteine → alanine mutants were expressed from the pMALC2H10 vector. Mutagenesis was performed as described elsewhere (Roman et al., 2010) using the QuikChange multi site-directed mutagenesis kit (Stratagene, La Jolla, CA) where one or more of the cysteine residues in the RGS domain of RGS4 were mutated to alanine.
All proteins were expressed in and harvested from BL21-DE3 Escherichia coli via standard transformation, growth, and lysis protocols (Lee et al., 1994; Lan et al., 1998, 2000; Roman et al., 2007; Roof et al., 2008). Histidine-tagged RGS4 was purified over a Ni-NTA affinity column (QIAGEN) followed by cation exchange chromatography and size exclusion chromatography. MBP-tagged RGS proteins were purified with an amylose affinity column followed by size exclusion chromatography. Hexahistidine-tagged rat Gαo was expressed and purified as described previously (Lee et al., 1994). G protein activity was determined by [35S]GTPγS binding (Sternweis and Robishaw, 1984). In all cases, proteins were purified to >90% homogeneity before use.
Chemical Labeling of Purified Gαo and RGS.
For Alexa Fluor 488 labeling of RGS4, ΔN19RGS4 was labeled with Alexa Fluor 488 succinimidyl ester (Invitrogen) at a 5:1 (label/protein) stoichiometry in a total volume of 2.0 ml of 50 mM HEPES, pH 8.2 at 4°C, 100 mM NaCl, and 1 mM DTT. The reaction was performed while rotating samples in the dark for 1.5 h at 4°C. The reaction was quenched by the addition of 1 mM glycine for 10 min at 4°C. Labeled RGS4 was resolved from the reaction mixture by size exclusion chromatography using a 20-ml Sephadex G-25 desalting column (GE Healthcare, Little Chalfont, Buckinghamshire, UK). Degree of labeling was determined spectroscopically to be approximately 1:1.
Tb chelate labeling of Gαo, Gαo was labeled with the LanthaScreen Tb thiol-reactive reagent (Invitrogen) at a 5:1 (label/protein) stoichiometry in a total volume of 1.0 ml of 50 mM HEPES, pH 7.25 at 4°C, 100 mM NaCl, supplemented with 10 μM GDP and 0.8 mM Tris(2-carboxyethyl)phosphine. The reaction was allowed to proceed at 4°C for 1.5 h during rotation in the dark. The reaction was quenched by the addition of 1 mM DTT for 20 min at 4°C. Labeled protein was purified from the reaction mixture by size exclusion chromatography using a Sephadex G-25 desalting column (GE Healthcare). Degree of labeling was determined spectroscopically to be approximately 1:1. The activity and effective concentration of the labeled G protein was determined by [35S]GTPγS binding as described previously (Sternweis and Robishaw, 1984).
For biotinylation of RGS proteins, RGS protein was mixed at a 3:1 (label/protein) molar ratio with biotinamidohexanoic acid N-hydroxysuccinimide ester (Sigma-Aldrich) in a buffer of 50 mM HEPES, pH 8.5 at 4°C, 100 mM NaCl, and 1 mM DTT. The reaction was allowed to proceed at 4°C while rotating for 2 h and then was quenched by the addition of a large molar excess of glycine for 10 min. Labeled protein was purified from the reaction mixture by size exclusion chromatography using a Sephadex G-25 desalting column (GE Healthcare Biosciences).
Alexa Fluor 532 labeling was performed as described previously (Roman et al., 2007). Labeled protein was purified from the reaction mixture by size exclusion chromatography using a Sephadex G-25 desalting column (GE Healthcare Biosciences).
Time-Resolved FRET.
TR-FRET experiments were performed on a PHERAstar multipurpose microplate reader (BMG Labtech GmbH, Offenberg, Germany) using the LanthaScreen filter set. These experiments were based on the method of Leifert et al. (2006). For the saturation experiments, Tb-Gαo was diluted to 20 nM in 50 mM HEPES, pH 8.0, 100 mM NaCl, 0.1% Lubrol, 30 μM GDP, 5 mM NaF, 5 mM MgCl2, and 5 μM AlCl3 and allowed to activate for 10 min on ice before use. RGS4-AF488 was serially diluted in 50 mM HEPES, pH 8.0 at room temperature, 100 mM NaCl, and 0.1% Lubrol (TR-FRET buffer). Ten microliters of the RGS4 dilution was added to a black nonstick, low-volume, 384-well plate (Corning Life Sciences, Lowell, MA) with a minimum of duplicate measurements. Ten microliters of Tb-Gαo was added (10 nM final), and the mixture was allowed to incubate at room temperature for 15 min in the dark. The nonspecific TR-FRET signal was determined by excluding AlCl3, MgCl2, and NaF from a set of samples. The fluorescence emission at both 490 and 520 nm was measured from 50 flashes of 340-nm excitation light per well. The data were collected in 10-μs bins, and the delayed emission signal was integrated from 100 to 500 μs after each flash. TR-FRET data were analyzed as the ratio of emission at 520 nm/490 nm.
High-Throughput Screening.
High-throughput screening was performed at the University of Michigan Center for Chemical Genomics. The approximately 40,000-compound screening collection was provided by the Novartis Institute for Biomedical Research and was comprised of compounds selected from the ChemDiv screening library. Five microliters of 50 mM HEPES, pH 8.0 at room temperature, 100 mM NaCl, 0.1% Lubrol, and 1 mM DTT (TR-FRET buffer) was dispensed with a Multidrop (Thermo Fisher Scientific) into every well of a black nonstick, low-volume, 384-well plate. Two hundred nanoliters of each compound (2 mM stock, 20 μM final assay concentration) or DMSO control was added to the plate with a pin tool by using a Beckman BioMek FX liquid handler (Beckman Coulter, Fullerton, CA). To this compound dilution, 5 μl of 200 nM Alexa Fluor 488-labeled RGS4 was added and incubated for 15 min at room temperature in the dark. Then, 10 μl of 20 nM Tb-labeled Gαo was added to the mixture. For this assay, the positive inhibition control (i.e., no RGS4/Gαo binding) was Tb-labeled Gαo in the inactive GDP-bound state, and the negative control (i.e., full RGS4/Gαo binding) used Gαo in the GDP/AlF4-bound state. This mixture was incubated at room temperature in the dark for 15 min before analysis with the PHERAstar plate reader. Data were compiled and analyzed by using the M-Screen database, a chemoinformatics suite developed by the Center for Chemical Genomics at the University of Michigan. Compounds that inhibited the TR-FRET signal >2 SD from the negative control were considered “actives” and were chosen for dose-response follow-up experiments.
TR-FRET Dose-Response Experiments.
Actives from the primary screen were evaluated for concentration-dependent activity in the TR-FRET assay. Compound dilutions were performed in DMSO, and 200 nl of diluted compound was spotted into the wells of a black nonstick, low-volume, 384-well plate that contained 5 μl of TR-FRET buffer. To the well, 5 μl of 200 nM Alexa Fluor 488-labeled RGS4 was added and incubated at room temperature in the dark for 15 min. Then, 10 μl of 20 nM Tb-labeled Gαo GDP/AlF4 was added to the mixture and incubated at room temperature in the dark for 30 min before analysis on the PHERAstar plate reader. Compound dilutions covered a final concentration range from 200 to 1.6 μM. Positive and negative controls were performed as in the primary screening assay. Compounds whose dose-response curves (DRCs) were not fully defined by these concentrations were repeated by using a more appropriate dilution scheme. Nonlinear least-squares regression fitting of the data were performed by using the data analysis component of the MScreen database.
Flow Cytometry Protein Interaction Assay Concentration Dependence Experiments.
Compounds that were confirmed in the follow-up TR-FRET dose-response assay were tested as described previously (Roman et al., 2007) in the flow cytometry protein interaction assay (FCPIA). This was done in part to provide a complementary set of biochemical data to filter out any compounds that might produce spectroscopic artifacts in the TR-FRET assay. In brief, biotinylated RGS proteins (5 nM, final assay concentration) were immobilized on Luminex LumAvidin beads and incubated with diluted compound in 50 mM HEPES, pH 8.0 at room temperature, 100 mM NaCl, 0.1% Lubrol, and 1 mM DTT, supplemented with 1% BSA. To each well of a 96-well PCR plate (Axygen, Union City, CA) Alexa Fluor 532-labeled Gαo was added to a final concentration of 30 nM. This mixture was incubated for 30 min at room temperature in the dark, and then it was analyzed on a Luminex 200 flow cytometer for the bead-associated fluorescence (median value). Nonlinear regression analysis of inhibition curves was performed with Prism 5.0 (GraphPad Software Inc., San Diego CA).
FCPIA Reversibility Experiments.
RGS-coated beads were prepared as above and treated with 50 μM compound or vehicle (DMSO) for 15 min at room temperature. The RGS-containing beads were then washed by resuspension in 1 ml of phosphate-buffered saline, pH 7.4 supplemented with 1% BSA, vortexing briefly, then pelleting the beads by centrifugation. This procedure was repeated a total of three times before 1000 beads were added to each quadruplicate well of a 96-well PCR plate that contained Alexa Fluor 532-labeled Gαo at a final concentration of 20 nM in the presence or absence of 50 μM test compound. The mixture was incubated for 30 min at room temperature and then analyzed on a Luminex 200 flow cytometer for bead-associated fluorescence. Data analysis was performed with Prism 5.0.
Single-Turnover GTPase Measurements.
Compounds were tested for the ability to inhibit the RGS4-stimulated increase in GTP hydrolysis by Gαo as described previously (Roof et al., 2006; Roman et al., 2007).
Thermal Stability Measurements.
Untagged ΔN19RGS4 or His6-Gαo was added to the well of a 96-well ABI Prism optical reaction plate (Applied Biosystems, Foster City CA) to a final concentration of 5 or 2.5 μM, respectively in 50 to 60 μl of 50 mM HEPES, pH 8.0 with 150 mM NaCl. Test compounds were added to the protein at the desired concentration and allowed to interact for 15 min at room temperature. To each well, Sypro Orange dye (Invitrogen) was added to a 5× final concentration (as described by the supplier), and the plate was sealed with an optically clear adhesive film. Sypro Orange fluorescence was measured continuously in an ABI HT7900 real-time PCR system during a stepwise gradient from ambient temperature to 90°C in 1°C steps lasting 30 s each. Data were analyzed by fitting the obtained curves to a Boltzmann model (eq. 1).
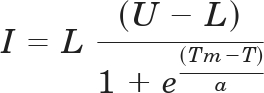 |
where I is fluorescence intensity (arbitrary units), L is the lower limit of the curve (°C), U is the upper limit of the curve (°C), T is temperature (°C), and a is a slope factor. Values obtained after the fluorescence maximum occurred were excluded from the analysis.
Results
Development of a High-Throughput TR-FRET RGS4-Gαo Interaction Screen.
We developed a biochemical TR-FRET assay by using purified human RGS4 labeled with the Alexa Fluor 488 acceptor fluorophore and purified Gαo labeled with the LanthaScreen Tb probe donor fluorophore (Fig. 1A). Using this system, we observed a saturable, aluminum fluoride-dependent interaction between RGS4 and Gα that has an affinity consistent with other reports of this PPI in the literature (Fig. 1B) (Roman et al., 2007). In collaboration with the Center for Chemical Genomics at the University of Michigan, this assay was scaled to 384-well format and used to screen ∼44,000 small molecules for inhibition of RGS4/Gαo binding in the presence of a thiol-reducing agent (Table 1). Compounds from this screen were retested in the primary screening assay to confirm the initial result and assess the concentration dependence of the inhibition using the original TR-FRET assay. Of the 162 compounds that met the 2-SD selection criteria for inhibition, 48 were either unavailable or predicted to be chemically reactive and were not followed up. The 114 selected compounds were retested in TR-FRET DRC, and 11 were confirmed as inhibitors with IC50 values <400 μM and Hill slopes <2.
TABLE 1.
RGS4/Gαo TR-FRET high-throughput screening results
Actives were determined as follows: primary screen, >2 SD from the negative control; TR-FRET DRC, IC50 value <400 μM; FCPIA DRC: IC50 value <500 μM.
| Assay | Compounds Tested | Active | Hit Rate |
|---|---|---|---|
| % | |||
| ChemDiv Library Subset | 43,878 | 162 | 0.37 |
| TR-FRET DRC | 114 | 11 | 0.025 |
| FCPIA DRC | 11 | 2 | 0.0046 |
The confirmed active compounds were obtained from the supplier as fresh powders and tested by using the FCPIA, a method that measures the binding of fluorescently tagged Gαo to an RGS protein on beads (Roman et al., 2007). Of the 11 compounds tested, 2 showed similar activity on RGS4 in both the TR-FRET dose response and FCPIA experiments (Fig. 1C). The nine compounds that did not show activity in this secondary assay are presumed to have been spectral artifacts or small-molecule aggregators that are likely to lose function in the relatively stringent conditions of the FCPIA assay buffer (50 mM HEPES, 100 mM NaCl, 1% BSA, and 0.1% Lubrol, pH 8.0).
The two active compounds that were identified from this primary screen were the closely related CCG-63808 and CCG-63802 (Fig. 1D). These compounds differ solely by the substituents on the phenyl moiety and have similar IC50 values in TR-FRET and FCPIA. The compounds also contain a vinyl cyanide moiety that may function as a reversible Michael acceptor.
CCG-63802 and CCG-63808 Selectively Inhibit Gαo-RGS Interactions.
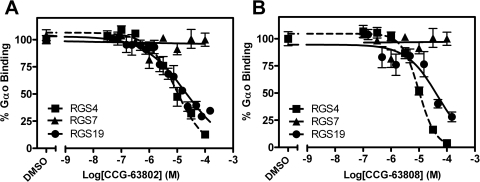
Using TR-FRET to assess the RGS4-Gαo interaction, CCG-63802 and CCG-63808 had IC50 values of 1.9 and 1.4 μM, respectively (Fig. 1C). To determine the selectivity of these compounds for different RGS proteins, they were tested in an FCPIA competition experiment against a panel of five different RGS proteins (Fig. 2; Table 2). The compounds are 6- to 7-fold less potent in blocking Gαo/RGS4 interactions when tested with FCPIA (IC50 ∼10 μM) than with the TR-FRET method. This is probably because of the high level of BSA (1%) in the FCPIA buffer sequestering compound and decreasing its apparent concentration in the assay. These compounds did not inhibit Gα binding to RGS7, which is distantly related to RGS4, and they are 2- to 10-fold more potent at RGS4 than on the other closely related R4 family members, RGS8 and RGS16 (Table 2). They are also fairly active (IC50 20–50 μM) on the one RZ family member tested, RGS19.
Fig. 2.
RGS specificity of CCG-63802 (A) and CCG-63808 (B) determined by multiplex FCPIA analysis (n ≥ 3). RGS-coated beads were treated with the indicated concentration of compound for 15 min at room temperature, after which GDP/AlF4-bound Gαo-AF532 was added and allowed to incubate with the RGS/compound mixture for 30 min before analysis. All data were calculated by using nonlinear least-squares regression with the bottom of the curves constrained to 0% binding. Data are presented as mean ± S.E.M. from at least three separate experiments.
TABLE 2.
RGS specificity of CCG-63802 and CCG-63808 determined by multiplex FCPIA analysis (n ≥ 3)
All data were calculated from at least three independent experiments using nonlinear least-squares regression with the bottom of the curves constrained to 0% binding.
| RGS Protein | CCG-63802 |
CCG-63808 |
||
|---|---|---|---|---|
| IC50 | Hill Slope | IC50 | Hill Slope | |
| μM | μM | |||
| RGS4 | 9 | −0.9 | 10 | −1.4 |
| RGS4c | >400 | −0.4 | >400 | −0.8 |
| RGS8 | 112 | −0.6 | 74 | −1.1 |
| RGS16 | 42 | −1.4 | 21 | −2.1 |
| RGS19 | 20 | −0.6 | 46 | −0.8 |
| RGS7 | N.I. | N.I. | N.I. | N.I. |
N.I., no inhibition observed at highest concentration tested (100 μM).
CCG-63802 and CCG-63808 Inhibit RGS4 GAP Activity.
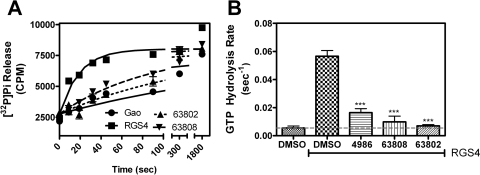
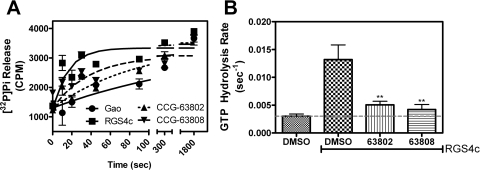
For RGS inhibitors to be functionally relevant, they need to inhibit the catalytic activity of the RGS in addition to blocking Gα/RGS binding. The two compounds inhibit the GAP activity of RGS4 as shown by measurements using the [32P]GTP single-turnover GAP assay (Fig. 3). Under these conditions, GTP hydrolysis by Gαo is accelerated ∼10-fold by the addition of wild-type RGS4, and this effect can be inhibited by the previously described RGS4 inhibitor CCG-4986 (Roman et al., 2007). At a concentration of 100 μM, CCG-63802 and CCG-63808 fully inhibit the RGS activity without affecting basal Gαo GTPase activity.
Fig. 3.
Single-turnover GAP analysis of small-molecule RGS inhibitors with RGS4. A, RGS4 treated with 100 μM CCG-4986, CCG-63808, or CCG-63802 lacks the ability to increase the intrinsic hydrolysis rate of Gαo. Representative GAP data are shown. All experiments were performed a minimum of three times. B, rate constants of GTP hydrolysis. Rate constants are presented as mean ± S.E.M. from at least three independent experiments. ***, p < 0.001 versus the DMSO-treated RGS control.
CCG-63802 and CCG-63808 Bind to RGS4 but Not to Gαo.
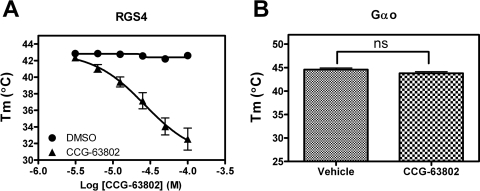
Because the studies presented so far assessed the binary interaction between two purified proteins, it was necessary to determine to which protein the compounds bound. The specificity for RGS4 over RGS7, RGS8, and RGS19 suggested, but did not prove, that the compounds bound to the RGS rather than the Gαo. To directly identify the site of action of these compounds, we developed a thermal denaturation assay to assess compound binding. This methodology is based on the principal that the stability of a protein is often altered upon ligand binding (Lo et al., 2004; Senisterra et al., 2008). For proteins that have endogenous small-molecule or peptide ligands (e.g., enzymes or receptors), binding of the ligand often increases the thermal stability. Upon binding GDP, Gαo experiences a >5°C increase in melting temperature (Tm) compared with nucleotide-free protein (Supplemental Fig. 2). This increases to a >20°C increase in Tm for Gαo binding the exceptionally high-affinity nucleotide GTPγS. Using this assay, we observed a concentration-dependent 10°C reduction (see Discussion) in the Tm of RGS4 in the presence of CCG-63802 (Fig. 4A). The concentration dependence of this effect corresponds with the IC50 values obtained in the FCPIA assay. Even at a maximal concentration of CCG-63802 (100 μM), there was no change in the Tm of Gαo (Fig. 4B).
Fig. 4.
CCG-63802 specifically binds to RGS4 and not to Gαo. A, purified RGS4 shows a dose-dependent change in melting temperature in the presence of CCG-63802 (EC50 ∼26 μM). B, a saturating concentration of CCG-63802 (100 μM) does not affect the melting temperature of Gαo. Data are presented as mean ± S.E.M. of three separate experiments.
CCG-63802 and CCG-63808 Are Reversible Inhibitors of the Gαo-RGS Interaction.
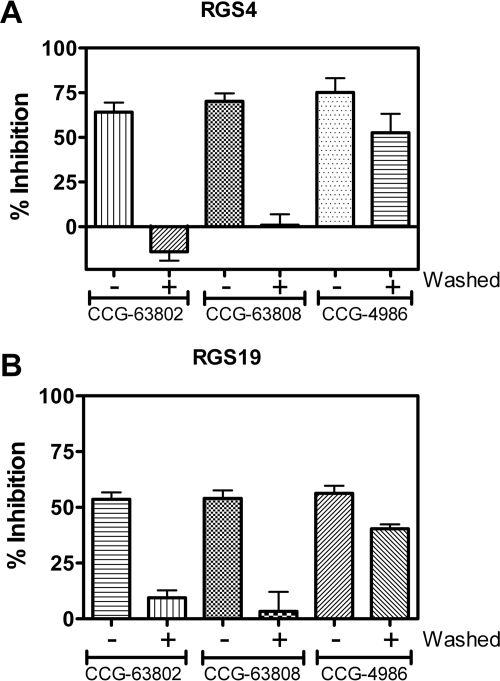
The effects of CCG-4986, our previously described RGS4 inhibitor (Roman et al., 2007), could not be reversed by dilution and washing away the compound, showing that it acts irreversibly to inhibit the function of RGS4 (Kimple et al., 2007; Roman et al., 2010) (Fig. 5). In addition, its activity was blocked in the presence of reducing agents. These effects are likely caused by the formation of a covalent adduct of the compound with a cysteine residue in the RGS (Kimple et al., 2007; Roman et al., 2010). Because our new compounds were identified through screens in the presence of DTT, we tested the reversibility of their inhibition. RGS-coated microspheres were treated with 50 μM compound or vehicle (DMSO), extensively washed (see Materials and Methods for details), and then assayed for Gαo binding (Fig. 5). In contrast to the effects of CCG-4986, full binding was restored to compound-treated RGS beads after washing (Fig. 5), showing that CCG-63802 and CCG-63808 are reversible on the 10-min time scale required for the washing procedure. Consequently, these new compounds represent the first examples of reversible small-molecule inhibitors of an RGS protein.
Fig. 5.
CCG-63802 and CCG-63808 are reversible inhibitors of RGS4 (A) and RGS19 (B). CCG-4986 is an irreversible inhibitor of RGS4 and RGS19. In all cases, RGS-coated FCPIA beads were treated with 50 μM compound (or vehicle, DMSO) and then extensively washed. The beads were then split into two groups and tested for the ability to interact with Gαo-AF532 in the presence or absence of 50 μM compound. Data shown are the mean ± S.E.M. of three separate experiments.
Cysteine Dependence of CCG-63802 and CCG-63808.
To further explore the mechanism of these compounds and the role of cysteines in their action, they were tested on a mutant of RGS4 where all cysteines in the RGS domain were mutated to alanine (RGS4c). In FCPIA measures of Gα binding to RGS4c, CCG-63802 and CCG-63808 show only modest activity, indicating a role for RGS cysteines in the actions of these compounds (Supplemental Fig. 3; Table 3). Consequently, we tested CCG-63808 and CCG-63802 with a panel of RGS4 RGS domain cysteine mutants by FCPIA (Table 3). The G protein binding affinity of these RGS mutants has been described previously (Roof et al., 2009), and the Kd values ranged from 3 to 12 nM, not drastically different from that of wild-type RGS4. No single cysteine could fully account for the effects of these compounds, but it seems that three cysteines (Cys148, Cys132, and Cys95) are important for full sensitivity to CCG-63808 and CCG-63802. Cys95 and Cys148 are located rather close to each other on RGS4; however, they are at a site distinct from the Gα interaction interface. It seems that Cys95 plays a more significant role than Cys148, possibly suggesting that the compound docks onto the RGS at a site that either is closer to this cysteine or requires this residue for proper formation of the compound binding pocket.
TABLE 3.
RGS4 cysteine mutant sensitivity to CCG-63802
Data are presented as mean ± S.E.M.
| RGS4 Mutant | IC50 | pIC50 Log(M) | Hill Slope | Inhibition at 100 μM | n |
|---|---|---|---|---|---|
| μM | % | ||||
| Wild type | 9 | 5.02 ± 0.07 | −0.86 ± 0.11 | 87 | 9 |
| C148A | 43 | 4.37 ± 0.07 | −0.95 ± 0.16 | 63 | 3 |
| C132A | 41 | 4.39 ± 0.07 | −0.97 ± 0.18 | 66 | 3 |
| C95A/C132A | 32 | 4.50 ± 0.13 | −0.78 ± 0.20 | 70 | 3 |
| C148A/C132A | 92 | 4.04 ± 0.07 | −0.75 ± 0.11 | 57 | 3 |
| C148A/C132A/C95A | ∼3000 | 2.55 ± 0.64 | −0.33 ± 0.12 | 16 | 3 |
| RGS4c | ∼8000 | 2.10 ± 1.50 | −0.36 ± 0.30 | 13 | 6 |
| A148C | ∼390 | 3.41 ± 0.17 | −0.62 ± 0.14 | 30 | 3 |
| A132C | 174 | 3.76 ± 0.19 | −0.80 ± 0.29 | 31 | 3 |
| A95C | 170 | 3.77 ± 0.23 | −1.20 ± 0.82 | 30 | 3 |
| A148C/A132C | 33 | 4.47 ± 0.05 | −1.48 ± 0.23 | 92 | 3 |
| A148C/A95C | 17 | 4.77 ± 0.12 | −1.06 ± 0.28 | 100 | 3 |
| A95C/A148C/A132C | 16 | 4.79 ± 0.12 | −0.63 ± 0.12 | 64 | 3 |
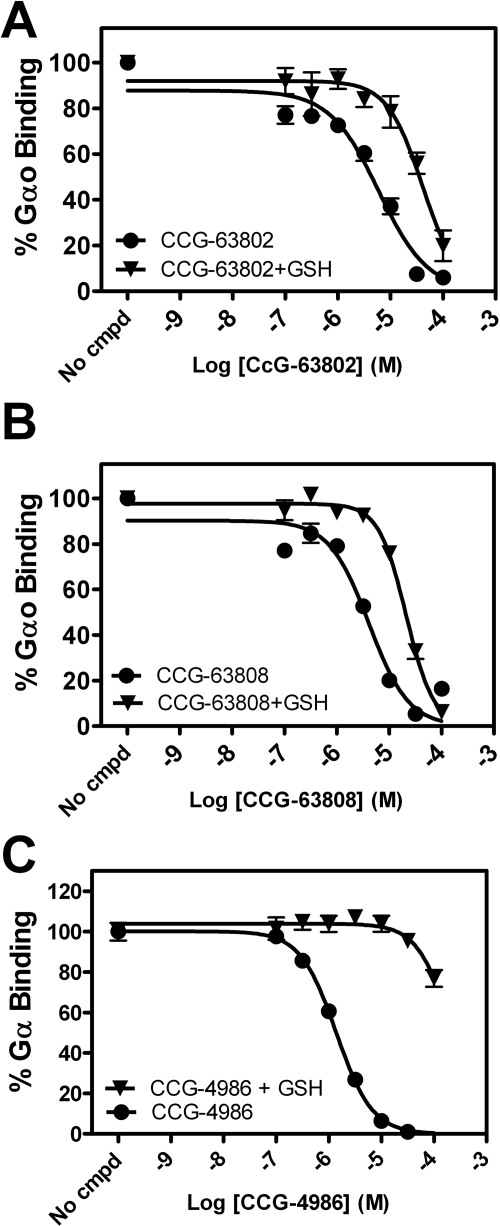
Because thiol-reactive compounds may have difficulty functioning in the reducing environment of a cell, it is important to assess the activity of any such leads under conditions mimicking the intracellular environment. Therefore, CCG-63802, CCG-63808, and CCG-4986 were tested for activity by FCPIA in the presence of 2 mM reduced glutathione (Fig. 6). This concentration of glutathione was selected because it is similar to intracellular concentrations. CCG-63802 and CCG-63808 lose approximately 0.5 to 1 Log of potency (IC50 6 → 40 μM for CCG-63802; 4 → 21 μM for CCG-63808) in the presence of 2 mM glutathione, but still retain the ability to fully inhibit the interaction between RGS4 and Gαo. In contrast, CCG-4986 loses more than 2 Logs in potency (IC50 from 1.4 → 215 μM) in the presence of 2 mM glutathione, and it is not capable of fully inhibiting the RGS-Gαo interaction up to concentrations nearing its aqueous solubility (Fig. 6).
Fig. 6.
CCG-63802 is less sensitive to glutathione than other RGS4 inhibitors. A and B, CCG-63802 (A) and CCG-63808 (B) retain full inhibitory activity in the presence of 2 mM glutathione. The potency is right-shifted by approximately 0.5–1 Log (CCG-63802: Log IC50 −5.25 ± 0.07 to −4.39 ± 0.07; CCG-63808: Log IC50 −5.39 ± 0.06 to −4.68 ± 0.03). n = 2. C, in contrast, CCG-4986 loses more than two logs of potency (Log IC50 −5.87 ± 0.03 to −3.66 ± 0.15) in the presence of glutathione. n = 3. Data presented as mean ± S.E.M.
It is noteworthy that CCG-63802 and CCG-63808 inhibit the GAP activity of the RGS4c mutant (Fig. 7) despite their much lower potency to inhibit Gαo/RGS4c binding in FCPIA (Supplemental Fig. 3). Thus, these compounds can inhibit the functional activity of the cysteine-null RGS4 mutant while having much less effect on the high-affinity binding to GDP-AMF bound Gαo (see Discussion). This inhibitory effect does not seem to be caused by compound aggregation, because it is not reversed in the presence of 0.01% Triton (data not shown), which generally blocks the activity of promiscuous small-molecule aggregators (Feng et al., 2007).
Fig. 7.
CCG-63802 and CCG-63808 inhibit the GAP activity of a cysteine-null RGS4 mutant. A, CCG-63802 or CCG-63808 (100 μM) inhibits the ability of RGS4c to accelerate the rate of GTP hydrolysis by Gαo. Representative data are shown. All experiments were performed a minimum of three times. B, rate constants of GTP hydrolysis. Rate constants are presented as mean ± S.E.M. from at least three independent experiments. **, p < 0.01 versus the DMSO-treated RGS control.
Discussion
RGS proteins play a strong modulatory role in GPCR signaling, leading to substantial interest in small-molecule inhibitors targeting this class of proteins (Zhong and Neubig, 2001; Neubig and Siderovski, 2002; Riddle et al., 2005; Blazer and Neubig, 2009; Traynor et al., 2009). The localized expression of RGS proteins (Kurrasch et al., 2004) suggested that RGS inhibitors could provide enhanced tissue specificity for GPCR agonist actions (Zhong and Neubig, 2001; Neubig and Siderovski, 2002; Blazer and Neubig, 2009). Furthermore, up-regulation of RGS proteins in various disease states, for example, RGS4 in neuropathic pain models (Garnier et al., 2003), also provides an important rationale for targeting RGS proteins. In this study, we report the second family of RGS SMPPIIs. Unlike our previously reported RGS inhibitor, CCG-4986 (Roman et al., 2007), which is irreversible and loses function in the presence of reducing agents (Kimple et al., 2007; D. L. Roman, L. L. Blazer, and R. R. Neubig, Roman et al., 2010), the new compounds identified here act reversibly and function in the presence of glutathione, a predominant intracellular reductant. These compounds, with their reversibility and activity in glutathione, therefore represent a significant step forward in the development of RGS SMPPIIs.
Similar to our original compound, CCG-63802 and CCG-63808 are relatively selective for RGS4 over other R4 family members, including the closely related RGS8 and RGS16. They have no detectable activity for the more distantly related RGS7. They also have some dependence on cysteine residues because they very weakly inhibit the cysteine-null (C → A) mutant of RGS4 (RGS4c) in the FCPIA assay. However, both compounds at 100 μM fully inhibit the GAP activity of RGS4c. There are a few potential explanations for this discrepancy. First, the compounds, which are of modest affinity (∼10 μM) in the FCPIA studies, may have a very short RGS-bound lifetime and therefore have difficulty competing with the constitutive binding of AlF4−/GDP-bound Gαo to the RGS. In the GTPase assay they may be more efficient at inhibiting the transient interaction between GTP-bound Gαo and RGS4 during the catalytic cycle. In addition, because the compounds seem to act via an allosteric site (see below), the induced conformational change in RGS4 may have a more dramatic impact on binding to or GAP activity at the Gα-GTP than for the GDP-AlF4− conformation of the Gα subunits. Indeed, it is the effects of compounds on RGS GAP activity and not on Gα binding that are most relevant in cellular or animal models.
The partial cysteine dependence of the actions of these compounds suggests a tethering model in which a reactive group binds to RGS cysteine residues. This is supported by the cysteine mutagenesis studies and also by the presence of the potential Michael acceptor functionality (vinyl cyanide) in both of the compounds. Tethered ligands can provide enhanced potency for small molecules acting on difficult targets (Erlanson et al., 2000; Arkin et al., 2003). Our ability to detect these compounds may have derived from potency enhancement from a slow off-rate caused by tethering. The reaction, however, is clearly reversible on the 10-min time scale, and attempts to demonstrate covalent binding by mass spectroscopy have been unsuccessful. Although uncommon, there are other well described examples of reversible Michael acceptor reactions with thiols (Jin et al., 2007; Ettari et al., 2008). Although most drug molecules are designed to avoid such reactive groups, there are a number of examples of clinically used drugs (e.g., omeprazole) or drug candidates [CI-1033; N-[-4-[(3-chloro-4-fluorophenyl)amino]-7-[3-(4-morpholinyl)propoxy]-6-quinazolinyl]-2-propenamide] that are thiol-reactive (Sachs et al., 1994; Ocaña and Amir, 2009). Furthermore, tethered ligands have been used to develop a structure-activity relationship in the context of the higher-affinity starting structure that is then transferred to analogs without the reactive group (Erlanson et al., 2003). The activity of CCG-63802 and CCG-63808 to inhibit the GAP activity of RGS4c suggests that this may be a reasonable approach.
The compounds described here require three cysteines for full potency of RGS4 inhibition: Cys95, Cys148, and Cys132. Cys95 and Cys148 are positioned in the “B site” of RGS proteins (Zhong and Neubig, 2001), which is proposed to participate in the allosteric modulation of RGS4 by acidic phospholipids and calmodulin (Popov et al., 2000; Ishii et al., 2005a). Cys132 is located on the outer edge of the Gα interaction interface and, at high concentrations, may react with CCG-63802 in a reversible Michael reaction to provide modest steric occlusion of the protein-protein interaction. Consequently, these compounds seem to have both allosteric and steric elements in their mechanism.
The binding of CCG-63802 induces a destabilizing effect on RGS4 in the thermal stability studies. This contrasts with the stabilizing effect observed (Grasberger et al., 2005; Wan et al., 2009) for small-molecule ligand binding to many proteins (e.g., Gαo; Supplemental Fig. 2). We observed that this family of compounds causes a left shift in the melting curve to lower temperatures. This reduced stability of the RGS4 may be related to conformational perturbation induced upon compound binding to the cysteines in the allosteric site.
In most instances, proteins with endogenous small-molecule ligands (e.g., Gα proteins) are stabilized by the presence of their ligand (Grasberger et al., 2005; Wan et al., 2009). This notion was recently borne out by the recent crystallization of several GPCRs (Cherezov et al., 2007; Rasmussen et al., 2007; Jaakola et al., 2008; Scheerer et al., 2008; Warne et al., 2008). In all cases (the notable exception being opsin), crystals were obtained only in the presence of, among other reagents, a small-molecule ligand. This strongly suggests that these ligands are important for the structural stability of this class receptor in solution. Furthermore, our data (Supplemental Fig. 2) and others (Matulis et al., 2005; Abad et al., 2008) also confirm that binding of natural or artificial ligands to sites that have evolved the capacity for small-molecule binding causes a stabilization of the protein. This stabilizing effect may be caused by the decrease in free energy derived from the binding event and also the conformational restriction required for high-affinity ligand-protein interaction. This increased protein rigidity is likely to provide a level of protection against the increasingly intense thermally induced conformational fluctuations as the temperature of the sample is raised.
On the surface, it would seem that this paradigm is contradicted by the compounds CCG-63802 and CCG-63808, which potently destabilize RGS4 even though they seem to bind close to the site on RGS4 that binds native acidic phospholipids. It is possible that these compounds bind to a site near, yet independent of, the acidic lipid site on the RGS, and binding to this non-natural site might not be expected to produce the same stabilization effect as binding of small molecules to sites that have evolved the capacity for such small-molecule-protein interactions. In addition, insertion of the compounds into the four-helix bundle, stabilized by the reversible Michael addition to a cysteine thiol, could unfold the RGS4 structure, leading to destabilization.
In this study we have identified the first examples of reversible SMPPIIs that disrupt RGS protein function. CCG-63808 and CCG-63802 are selective inhibitors of the RGS-Gα interaction and R4 family GAP activity. Their mechanism seems to, at least in part, involve an allosteric action at the B site on the RGS (Zhong and Neubig, 2001), which has been implicated in the physiological allosteric modulation of RGS proteins by acidic phospholipids and calmodulin (Ishii et al., 2005a,b). Further studies of the mechanism and structure-activity relationships for this compound class and translation to cellular and animal models of RGS function are currently underway.
Supplementary Material
Acknowledgments
We thank Roger Sunahara (University of Michigan Pharmacology) and John Tesmer (University of Michigan Pharmacology and Life Sciences Institute) for helpful discussions with this project; the staff of the University of Michigan Center for Chemical Genomics for high-throughput screening and chemoinformatics assistance; the Novartis Institute for Biomedical Research for the gift of reagents, including the screening library; and the University of Michigan Comprehensive Cancer Center for subsidizing the cost of DNA sequencing. This work used the Cell and Molecular Biology Core of the Michigan Diabetes Research and Training Center, which is funded by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases (Grant DK020572).
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the Michigan Chemistry-Biology Interface Training Program, which is funded through the National Institutes of Health National Institute of General Medical Sciences [Grant T32-GM00008597]; the National Institutes of Health National Institute on Drug Abuse [Grant R01-DA023252]; and the National Institutes of Health National Institute of General Medical Sciences [Grant F32-GM076821].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.110.065128.
- RGS
- regulator of G protein signaling
- TR-FRET
- time-resolved fluorescence resonance energy transfer
- FCPIA
- flow cytometry protein interaction assay
- SMPPII
- small-molecule protein-protein interaction inhibitor
- PPI
- protein-protein interaction
- GAP
- GTPase-accelerating protein
- GPCR
- G protein-coupled receptor
- DTT
- dithiothreitol
- MBP
- maltose-binding protein
- DMSO
- dimethyl sulfoxide
- PCR
- polymerase chain reaction
- BSA
- bovine serum albumin
- T m
- melting temperature
- DRC
- dose-response curve
- CCG-63802
- ((2E)-2-(1,3-benzothiazol-2-yl)-3-[9-methyl-2-(3-methylphenoxy)-4-oxo-4H-pyrido[1,2-a]pyrimidin-3-yl]prop-2-enenitrile)
- CCG-63808
- ((2E)-2-(1,3-benzothiazol-2-yl)-3-[9-methyl-2-(4-fluorolphenoxy)-4-oxo-4H-pyrido[1,2-a]pyrimidin-3-yl]prop-2-enenitrile)
- CCG-4986
- methyl-N-[(4-chlorophenyl)sulfonyl]-4-nitrobenzenesulfinimidoate
- CI-1033
- N-[-4-[(3-chloro-4-fluorophenyl)amino]-7-[3-(4-morpholinyl)propoxy]-6-quinazolinyl]-2-propenamide.
References
- Abad MC, Askari H, O'Neill J, Klinger AL, Milligan C, Lewandowski F, Springer B, Spurlino J, Rentzeperis D. (2008) Structural determination of estrogen-related receptor γ in the presence of phenol derivative compounds. J Steroid Biochem Mol Biol 108:44–54 [DOI] [PubMed] [Google Scholar]
- Arkin MR, Wells JA. (2004) Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov 3:301–317 [DOI] [PubMed] [Google Scholar]
- Arkin MR, Whitty A. (2009) The road less traveled: modulating signal transduction enzymes by inhibiting their protein-protein interactions. Curr Opin Chem Biol 13:284–290 [DOI] [PubMed] [Google Scholar]
- Arkin MR, Randal M, DeLano WL, Hyde J, Luong TN, Oslob JD, Raphael DR, Taylor L, Wang J, McDowell RS, et al. (2003) Binding of small molecules to an adaptive protein-protein interface. Proc Natl Acad Sci USA 100:1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T. (2003) Modulation of protein-protein interactions with small organic molecules. Angew Chem Int Ed Engl 42:2462–2481 [DOI] [PubMed] [Google Scholar]
- Berg T. (2008) Small-molecule inhibitors of protein-protein interactions. Curr Opin Drug Discov Devel 11:666–674 [PubMed] [Google Scholar]
- Berman DM, Kozasa T, Gilman AG. (1996) The GTPase-activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis. J Biol Chem 271:27209–27212 [DOI] [PubMed] [Google Scholar]
- Blazer LL, Neubig RR. (2009) Small-molecule protein-protein interaction inhibitors as CNS therapeutic agents: current progress and future hurdles. Neuropsychopharmacology 34:126–141 [DOI] [PubMed] [Google Scholar]
- Bueno L, Fioramonti J. (1988) Action of opiates on gastrointestinal function. Baillieres Clin Gastroenterol 2:123–139 [DOI] [PubMed] [Google Scholar]
- Busschots K, De Rijck J, Christ F, Debyser Z. (2009) In search of small molecules blocking interactions between HIV proteins and intracellular cofactors. Mol Biosyst 5:21–31 [DOI] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, et al. (2007) High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318:1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlanson DA, Braisted AC, Raphael DR, Randal M, Stroud RM, Gordon EM, Wells JA. (2000) Site-directed ligand discovery. Proc Natl Acad Sci USA 97:9367–9372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlanson DA, Lam JW, Wiesmann C, Luong TN, Simmons RL, DeLano WL, Choong IC, Burdett MT, Flanagan WM, Lee D, et al. (2003) In situ assembly of enzyme inhibitors using extended tethering. Nat Biotechnol 21:308–314 [DOI] [PubMed] [Google Scholar]
- Ettari R, Nizi E, Di Francesco ME, Dude MA, Pradel G, Vicík R, Schirmeister T, Micale N, Grasso S, Zappalà M. (2008) Development of peptidomimetics with a vinyl sulfone warhead as irreversible falcipain-2 inhibitors. J Med Chem 51:988–996 [DOI] [PubMed] [Google Scholar]
- Feng BY, Simeonov A, Jadhav A, Babaoglu K, Inglese J, Shoichet BK, Austin CP. (2007) A high-throughput screen for aggregation-based inhibition in a large compound library. J Med Chem 50:2385–2390 [DOI] [PubMed] [Google Scholar]
- Gadek TR, Nicholas JB. (2003) Small molecule antagonists of proteins. Biochem Pharmacol 65:1–8 [DOI] [PubMed] [Google Scholar]
- Garnier M, Zaratin PF, Ficalora G, Valente M, Fontanella L, Rhee MH, Blumer KJ, Scheideler MA. (2003) Up-regulation of regulator of G protein signaling 4 expression in a model of neuropathic pain and insensitivity to morphine. J Pharmacol Exp Ther 304:1299–1306 [DOI] [PubMed] [Google Scholar]
- Grasberger BL, Lu T, Schubert C, Parks DJ, Carver TE, Koblish HK, Cummings MD, LaFrance LV, Milkiewicz KL, Calvo RR, et al. (2005) Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells. J Med Chem 48:909–912 [DOI] [PubMed] [Google Scholar]
- Hollinger S, Hepler JR. (2002) Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol Rev 54:527–559 [DOI] [PubMed] [Google Scholar]
- Huang X, Charbeneau RA, Fu Y, Kaur K, Gerin I, MacDougald OA, Neubig RR. (2008) Resistance to diet-induced obesity and improved insulin sensitivity in mice with a regulator of G protein signaling-insensitive G184S Gnai2 allele. Diabetes 57:77–85 [DOI] [PubMed] [Google Scholar]
- Huang X, Fu Y, Charbeneau RA, Saunders TL, Taylor DK, Hankenson KD, Russell MW, D'Alecy LG, Neubig RR. (2006) Pleiotropic phenotype of a genomic knock-in of an RGS-insensitive G184S Gnai2 allele. Mol Cell Biol 26:6870–6879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingalls EA, Popp FD. (1967) The preparation, structure, and reactions of some “malonyl-α-aminopyridines.” J Heterocyclic Chem 4:523–526 [Google Scholar]
- Ishii M, Fujita S, Yamada M, Hosaka Y, Kurachi Y. (2005a) Phosphatidylinositol 3,4,5-trisphosphate and Ca2+/calmodulin competitively bind to the regulators of G-protein-signalling (RGS) domain of RGS4 and reciprocally regulate its action. Biochem J 385:65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M, Ikushima M, Kurachi Y. (2005b) In vivo interaction between RGS4 and calmodulin visualized with FRET techniques: possible involvement of lipid raft. Biochem Biophys Res Commun 338:839–846 [DOI] [PubMed] [Google Scholar]
- Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. (2008) The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 322:1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Jin XY, Jin YL, Sohn DW, Kim SA, Sohn DH, Kim YC, Kim HS. (2007) Structural requirements of 2′,4′,6′-tris(methoxymethoxy) chalcone derivatives for anti-inflammatory activity: the importance of a 2′-hydroxy moiety. Arch Pharm Res 30:1359–1367 [DOI] [PubMed] [Google Scholar]
- Kimple AJ, Willard FS, Giguère PM, Johnston CA, Mocanu V, Siderovski DP. (2007) The RGS protein inhibitor CCG-4986 is a covalent modifier of the RGS4 Gα-interaction face. Biochim Biophys Acta 1774:1213–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurrasch DM, Huang J, Wilkie TM, Repa JJ. (2004) Quantitative real-time polymerase chain reaction measurement of regulators of G-protein signaling mRNA levels in mouse tissues. Methods Enzymol 389:3–15 [DOI] [PubMed] [Google Scholar]
- Lan KL, Sarvazyan NA, Taussig R, Mackenzie RG, DiBello PR, Dohlman HG, Neubig RR. (1998) A point mutation in Gαo and Gα1 blocks interaction with regulator of G protein signaling proteins. J Biol Chem 273:12794–12797 [DOI] [PubMed] [Google Scholar]
- Lan KL, Zhong H, Nanamori M, Neubig RR. (2000) Rapid kinetics of regulator of G-protein signaling (RGS)-mediated Gαi and Gαo deactivation. Gα specificity of RGS4 and RGS7. J Biol Chem 275:33497–33503 [DOI] [PubMed] [Google Scholar]
- Lee E, Linder ME, Gilman AG. (1994) Expression of G-protein α subunits in Escherichia coli. Methods Enzymol 237:146–164 [DOI] [PubMed] [Google Scholar]
- Leifert WR, Bailey K, Cooper TH, Aloia AL, Glatz RV, McMurchie EJ. (2006) Measurement of heterotrimeric G-protein and regulators of G-protein signaling interactions by time-resolved fluorescence resonance energy transfer. Anal Biochem 355:201–212 [DOI] [PubMed] [Google Scholar]
- Lo MC, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, Ellestad G. (2004) Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem 332:153–159 [DOI] [PubMed] [Google Scholar]
- Matulis D, Kranz JK, Salemme FR, Todd MJ. (2005) Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry 44:5258–5266 [DOI] [PubMed] [Google Scholar]
- Neubig RR, Siderovski DP. (2002) Regulators of G-protein signalling as new central nervous system drug targets. Nat Rev Drug Discov 1:187–197 [DOI] [PubMed] [Google Scholar]
- Niu G, Chen X. (2009) From protein-protein interaction to therapy response: molecular imaging of heat shock proteins. Eur J Radiol 70:294–304 [DOI] [PubMed] [Google Scholar]
- Ocaña A, Amir E. (2009) Irreversible pan-ErbB tyrosine kinase inhibitors and breast cancer: current status and future directions. Cancer Treat Rev 35:685–691 [DOI] [PubMed] [Google Scholar]
- Popov SG, Krishna UM, Falck JR, Wilkie TM. (2000) Ca2+/calmodulin reverses phosphatidylinositol 3,4,5-trisphosphate-dependent inhibition of regulators of G protein-signaling GTPase-activating protein activity. J Biol Chem 275:18962–18968 [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, et al. (2007) Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature 450:383–387 [DOI] [PubMed] [Google Scholar]
- Riddle EL, Schwartzman RA, Bond M, Insel PA. (2005) Multi-tasking RGS proteins in the heart: the next therapeutic target? Circ Res 96:401–411 [DOI] [PubMed] [Google Scholar]
- Roman D, Blazer LL, Monroy CA, Neubig RR. (2010) Allosteric inhibition of the regulator of G protein signaling-G protein-protein interaction by CCG-4986. Mol Pharmacol 78:360–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman DL, Ota S, Neubig RR. (2009) Polyplexed flow cytometry protein interaction assay: a novel high-throughput screening paradigm for RGS protein inhibitors. J Biomol Screen 14:610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, Neubig RR. (2007) Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol Pharmacol 71:169–175 [DOI] [PubMed] [Google Scholar]
- Roof RA, Jin Y, Roman DL, Sunahara RK, Ishii M, Mosberg HI, Neubig RR. (2006) Mechanism of action and structural requirements of constrained peptide inhibitors of RGS proteins. Chem Biol Drug Des 67:266–274 [DOI] [PubMed] [Google Scholar]
- Roof RA, Roman DL, Clements ST, Sobczyk-Kojiro K, Blazer LL, Ota S, Mosberg HI, Neubig RR. (2009) A covalent peptide inhibitor of RGS4 identified in a focused one-bead, one compound library screen. BMC Pharmacol 9:9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof RA, Sobczyk-Kojiro K, Turbiak AJ, Roman DL, Pogozheva ID, Blazer LL, Neubig RR, Mosberg HI. (2008) Novel peptide ligands of RGS4 from a focused one-bead, one-compound library. Chem Biol Drug Des 72:111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs G, Prinz C, Loo D, Bamberg K, Besancon M, Shin JM. (1994) Gastric acid secretion: activation and inhibition. Yale J Biol Med 67:81–95 [PMC free article] [PubMed] [Google Scholar]
- Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. (2008) Crystal structure of opsin in its G-protein-interacting conformation. Nature 455:497–502 [DOI] [PubMed] [Google Scholar]
- Senisterra GA, Soo Hong B, Park HW, Vedadi M. (2008) Application of high-throughput isothermal denaturation to assess protein stability and screen for ligands. J Biomol Screen 13:337–342 [DOI] [PubMed] [Google Scholar]
- Sternweis PC, Robishaw JD. (1984) Isolation of two proteins with high affinity for guanine nucleotides from membranes of bovine brain. J Biol Chem 259:13806–13813 [PubMed] [Google Scholar]
- Talbot JN, Jutkiewicz EM, Graves SM, Clemans CF, Nicol MR, Mortensen RM, Huang X, Neubig RR, Traynor JR. (2010) RGS inhibition at Gαi2 selectively potentiates 5-HT1A-mediated antidepressant effects. Proc Natl Acad Sci USA 107:11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynor JR, Neubig RR. (2005) Regulators of G protein signaling & drugs of abuse. Mol Interv 5:30–41 [DOI] [PubMed] [Google Scholar]
- Traynor JR, Terzi D, Caldarone BJ, Zachariou V. (2009) RGS9–2: probing an intracellular modulator of behavior as a drug target. Trends Pharmacol Sci 30:105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan KF, Wang S, Brown CJ, Yu VC, Entzeroth M, Lane DP, Lee MA. (2009) Differential scanning fluorimetry as secondary screening platform for small molecule inhibitors of Bcl-XL. Cell Cycle 8:3943–3952 [DOI] [PubMed] [Google Scholar]
- Wang Y, Lee Y, Zhang J, Young KH. (2008) Identification of peptides that inhibit regulator of G protein signaling 4 function. Pharmacology 82:97–104 [DOI] [PubMed] [Google Scholar]
- Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. (2008) Structure of a β1-adrenergic G-protein-coupled receptor. Nature 454:486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Neubig RR. (2001) Regulator of G protein signaling proteins: novel multifunctional drug targets. J Pharmacol Exp Ther 297:837–845 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.