Abstract
The exposure of electrospray droplets to acid vapors can significantly affect protein charge state distributions (CSDs) derived from unbuffered solutions. Such experiments have been conducted by leaking acidic vapors into the counter-current nitrogen drying gas of an electrospray interface. Based on changes in protein CSDs, protein folding and unfolding phenomena are implicated in these studies. Additionally, non-covalently-bound complexes are preserved and transient intermediates observed, such as high charge state ions of holomyoglobin. CSDs of proteins containing disulfide bonds shift slightly, if at all, with acid vapor leak-in, but when these disulfide bonds are reduced in solution, charge states higher than the number of basic sites (Lys, Arg, His and N-terminus) are observed. Since there is no observed change in the CSD of buffered proteins exposed to acidic vapors, this novel multiple charging phenomenon is attributed to a pH effect. Thus, this acid vapor leak-in approach can be used to reverse ‘wrong-way-round’ nanoelectrospray conditions by altering solution pH in the charged droplets relative to the pH in bulk solution. In general, the exposure of electrospray droplets to acidic vapors provides means for altering protein CSDs independent of bulk unbuffered solution pH.
Keywords: Multiple Charging, Electrospray Ionization and pH, Charged Droplets
INTRODUCTION
Proteins are among the many types of biomolecules that form multiply charged gaseous ions upon electrospray ionization (ESI). The conformation of a protein in solution is one of the factors that affects the charge state distribution (CSD) generated in ESI-MS. ESI mass spectra obtained from solutions in which molecules are denatured generally exhibit higher charge states (lower m/z) than spectra obtained from solutions in which the conformations are more native1,2. Tightly folded protein molecules are expected to have a significantly smaller projected area compared to less-structured protein molecules, and thus can accommodate fewer charges on the protein surface during the ion desolvation stage3. Thus, compact folded states will generally produce ions with significantly different total charge than those that are unfolded.
A variety of methods have been described to alter protein CSDs in one direction or another. Often, this is a result of the manipulation of solution conditions, such as the use of supercharging reagents4,5, denaturing agents, different solvents6,7, or through chemical derivatization8. However, solution conditions that alter CSDs can also lead to deleterious effects such as poor ESI response, dissociation of non-covalent complexes, etc. Thus, it is desirable to be able to alter CSDs independent of initial solution conditions. For example, the reduction of absolute charge by gas-phase proton transfer has been demonstrated via ion/ion9,10 and ion/molecule11,12 reactions, while increases in absolute charge have been demonstrated via electron bombardment methods involving the ejection13 of electrons and via sequential ion/ion charge inversion reactions14,15. The latter methods operate on gas-phase ions and can be effected within the context of an MS/MS experiment. Methods that involve phenomena associated with charge droplets such as extractive electrospray ionization (EESI)16 and fused droplet-electrospray ionization (FD-ESI) 17,18 have recently been developed. In the techniques just mentioned, there is a liquid-phase interaction between the neutral analyte droplets and the charged electrospray droplets.
In this report, we relate results for proteins from the exposure of charged droplets to acidic vapors prior to ion sampling into the atmosphere/vacuum interface. Several studies have been performed using basic reagents, some in the vapor phase, to reduce CSDs to lower average charge using extractive electrospray ionization (EESI)19 and electrosonic spray ionization (ESSI)20. In contrast, in this work volatile acids were used alter CSDs after the droplets are already formed and are undergoing desolvation in the counter-current drying gas in interface region. Various protein solutions and various volatile acids have been studied to examine effects on protein CSDs over a range of conditions.
EXPERIMENTAL
Materials
Methanol, acetonitrile, ammonium hydroxide, acetic acid, formic acid, and hydrochloric acid were purchased from Mallinckrodt (Phillipsburg, NJ). Trifluoroacetic acid was purchased from Pierce Chemical (Rockford, IL). Initial concentrations of the acids present in the reagent test tubes were 11.7 M for hydrochloric acid, 13.0 M for trifluoroacetic acid, 23.6 M for formic acid, and 17.4 M for acetic acid. All proteins (bovine cytochrome c, ubiquitin from bovine red blood cells, horse heart myoglobin, hen egg white lysozyme, bovine serum albumin, and α-lactalbumin) and dithiothreitol were purchased from Sigma-Aldrich (St. Louis, MO). All samples were used without further purification. Protein solutions for both positive and negative nano-electrospray were prepared in 100% water, unless otherwise noted. When dithiothreitol (DTT) was used for protein denaturation, proteins were placed in 100 mM DTT at 40°C for several hours, and analyzed the same day. Final protein concentrations were approximately 20–50 μM.
Apparatus and Procedures
All experiments were performed using a prototype version of a QqTOF tandem mass spectrometer (Q-Star Pulsar XL, Sciex, Toronto, ON) modified to allow for ion trap CID and ion/ion reactions,21 although these types of experiments were not included in this publication. Ionization was accomplished via a nano-ESI emitter, forming either [M−nH]n− anions or [M+nH]n+ cations of the proteins. A new apparatus was designed to allow for the introduction of acidic vapors into the interface along with the curtain gas as previously described elsewhere22. Briefly, a Swagelok Tee connects a test tube containing approximately 20 μL of the acid reagent to a nitrogen line. Approximately 0.3L/min of N2 gas is directed across the test tube where head space vapors of the acid can mix with the N2 flow. These N2/acidic vapors are mixed with the curtain gas flow of the instrument (also N2) and the combined flow enters the region between the curtain plate orifice and the nozzle of the ESI interface. This near atmospheric pressure region just outside and between the curtain and orifice plates is where the molecule/droplet interaction takes place. Interactions between the acid vapors and protein ions take place in the expansion into the interface. Typically, a sample is ionized via nano-electrospray and then the metering valves are opened to introduce the acidic vapors as desired.
RESULTS AND DISCUSSION
Many factors contribute to the observed charge state distributions of proteins ionized using ESI, such as the number of ionizable groups on the molecule, solution conditions, and instrument conditions (e.g. the source and interface operating conditions). To minimize solution effects, all proteins were sprayed from 100% water when different acids were leaked in. The strengths of the different acids were then correlated with the shifts in the CSDs and, generally, the stronger the acid, the higher the CSD observed. This trend holds true except in cases in which the protein is known to fold into a molten globule state or other conformation that causes fewer basic residues to be exposed at low pH. Where relevant, the spectra obtained using this method were correlated to previous solution phase studies and the solution pH in those studies. Note that in the present arrangement, it is difficult to ensure that equal acid vapor concentrations were present when comparing the effects of the different acids. The vapor pressures of the acids (which is highest for HCl, followed by TFA, formic acid and acetic acid) is another important parameter that along with the concentrations of the acids in solution need to be considered before trying to determine the acid vapor concentration. Hence, while effects generally correlate with acid pKa, the detailed role of acid concentration cannot be determined from this work.
Cytochrome c
The ESI charge state distribution of cytochrome c has been studied extensively23,24,25. Bovine cytochrome c is a mainly helical globular protein containing 104 amino acids (23 of which are basic amino acids) and one covalently attached heme group26. The isoelectric point (pI) of cytochrome c is 10.4 and it undergoes a highly cooperative, acid-induced unfolding transition between pH= 3 and pH=227.
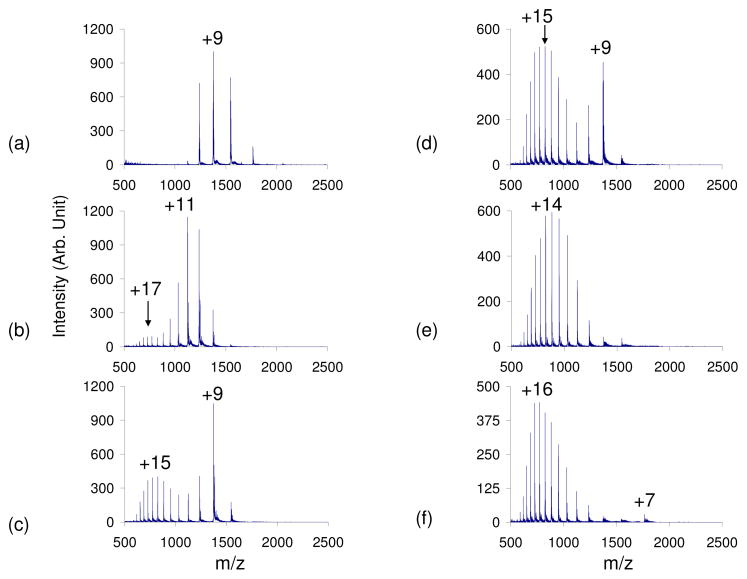
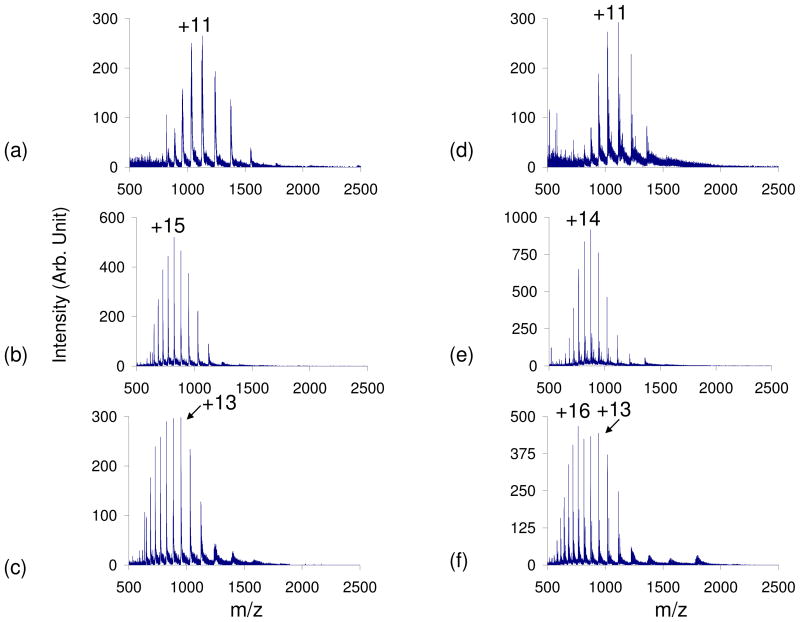
A native bovine cytochrome c sample in 100% water was subjected to positive nano-ESI. The observed CSD was from +5 to +12 with an average charge state (qave) of +8.9 (Figure 1). The average abundance weighted charge (qave) was calculated via Equation (1) where N is the number of observed analyte charge states in a given mass spectrum, qi is the net charge of the ith charge state, and Wi is the signal intensity of the ith charge state4.
Figure 1.
Positive nESI spectrum of cytochrome c in 100% H2O with a) no acid vapor, b) blank test tube in set up, c) acetic acid vapor, d) formic acid vapor, e) trifluoroacetic acid vapor, f) hydrochloric acid vapor leak-in. The strength of the acids used increases c–f.
| (1) |
Placing a blank test tube in the apparatus, which caused an increase in the total curtain gas flow, shifted the qave +2.5 charge states (Figure 1b). The shift of the cytochrome c CSD with increasing curtain gas flow has been observed previously3,28. However, this increase in CSD due to increased N2 flow was not observed for any of the other proteins in this study and therefore the effect appears to be somewhat specific for this protein. The leak-in of acetic acid (pKa=4.76) and formic acid (pKa=3.75) resulted in bimodal CSDs, while the leak-in of trifluoroacetic acid (pKa=0.52) and hydrochloric acid (pKa= −8.0) shifted the CSD to higher average charge (Figure 1c–f) 29. The leak-in of weaker acids resulted in bimodal distributions with the lower CSD centered around +9 and the higher CSD around the +15 charge state. The HCl vapor leak-in resulted in a CSD of +7 to +22 (qave=+15.4) (Figure 1f). Thus, relative to the native cytochrome c distribution, the leak-in of HCl results in the maximum charge state shifting +10 charge states and a shift of qave from 8.9 to 15.4.
The appearance of the +7 charge state, which was observed only with the leak-in of HCl, may be due to the formation of the molten globule state. Solution-phase studies have found that a refolding transition of cytochrome c has a midpoint at pH 1.0. It has been proposed that the initial unfolding is a transition from the native state to the acid-unfolded state, while the subsequent refolding is a transition from the unfolded state to the molten globule state30,31,32,33,34,35,36. Goto stated that the unfolding can be accounted for in terms of charge repulsion and anion binding. That is, when the protein is maximally charged, the addition of stronger acids adds both protons and anions to the solution. Because the protein is already maximally protonated, the addition of more protons has no effect on the ionization state. However, at high anion concentrations the internal repulsive forces are diminished, which allows for the refolding of the protein.
Ubiquitin
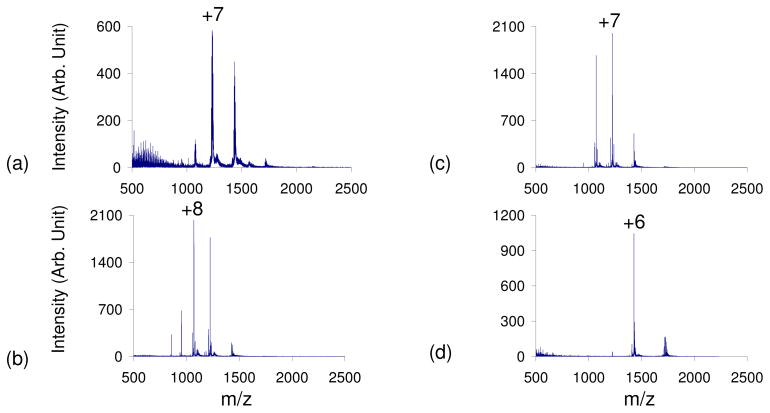
Ubiquitin, a 76-residue protein with 12 basic and 11 acidic residues with an isoelectric point of 5.2 37, was subjected to leak-in experiments following ESI from 100% water. The unfolding of ubiquitin in the solution-phase as well as its folding into the A State has been extensively studied38,39,40. The leak-in of weak acidic vapors caused a shift in the CSD to higher charge states (qave=+7 shifted to +8). This was followed by a shift to lower charge states with the leak-in of stronger acids, resulting in a qave=+6, which suggests that ubiquitin has folded into its A-State or another folded conformation. The positive nESI spectrum of ubiquitin prior to acid leak-in had a CSD from +4–+10 (qave=6.8) (Figure 2a). All of the peaks were highly adducted with Na+ and other metal ions. The leak-in of various acidic vapors cleaned up the spectrum and removed these counter-ions, with the dominant peak at each charge state now becoming the [M+nH]n+ peak. This is similar to results recently reported with this apparatus in the study of oligonucleotides18. The leak-in of weak acids aided in the unfolding of the protein, resulting in a CSD of +5–+12 and a qave =+7.8 with the leak-in of acetic acid (Figure 2b) and a qave=+7.3 with the leak-in of formic acid (Figure 2c). This slight decrease in the qave between acetic and formic acid vapor leak-in suggests that a refolding of ubiquitin is already taking place. The leak-in of TFA and HCl resulted in a distribution corresponding to the folded ubiquitin A-State or even a folding back to a more native state (N-State)36 with a CSD of +4–+8 and a qave=+5.9 in both experiments (Figure 2d). This folding is similar to that observed in solution-phase studies of ubiquitin at low pH36.
Figure 2.
Positive nESI spectrum of ubiquitin in 100% H2O with a) no acid vapor, b) acetic acid vapor, c) formic acid vapor, d) hydrochloric acid vapor leak-in. The strength of the acids used increases b–d.
Myoglobin
Myoglobin (pI = 7.2), another protein commonly featured in conformation studies, was also subjected to acid leak-in studies. The native form of myoglobin is characterized by a tightly folded conformation and a heme group that is non-covalently bound in a hydrophobic pocket41. Electrospray ionization enables the heme non-covalent interaction to be preserved42,43,44. It is widely known that acid-induced denaturation in solution results in substantial unfolding of the polypeptide chain and disruption of the non-covalent heme-protein interaction. This denaturation proceeds through a short-lived intermediate that is substantially unfolded but still retains the heme group45. The CSDs of myoglobin due to denaturation as well as the reconstitution of the acid-denatured protein in the solution-phase has been studied 46,47,48,49,50 and folding mechanisms have been proposed51,52.
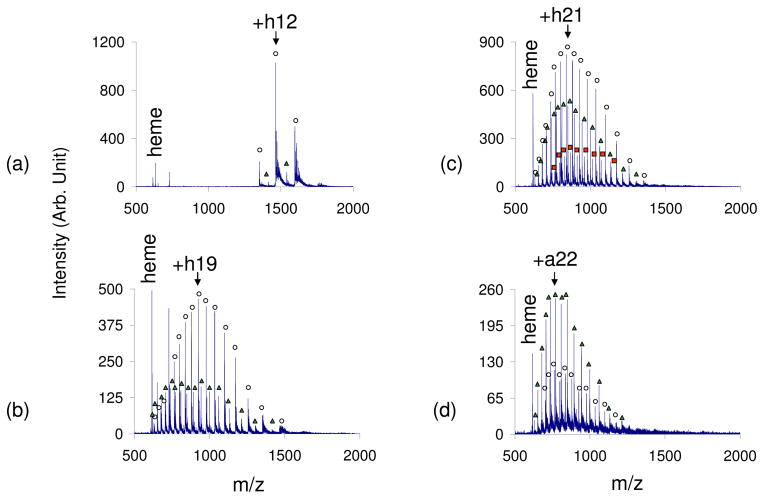
The exposure of charged droplets to acidic vapors in the interface region of the instrument results in a significant increase in the charges of holomyoglobin ions while preserving the non-covalent interaction. Because the charged droplets are treated with acidic vapors in the instrument interface, transient intermediates can be observed, presumably due to the short interaction time (μs timescale). A 100% aqueous solution of myoglobin subjected to positive nESI resulted in a CSD of holomyoglobin from +10 to +12 (qave= +11.1) (Figure 3a). Approximately 90% of the total ion abundance was due to holomyoglobin peaks, with the other 10% of the abundance corresponding to apomoyglobin peaks (also with a CSD of +10–+12). The leak-in of weak acids, such as acetic and formic acids, increased the CSD 5 charge states resulting in a qave=+16.3 for the leak-in of formic acid (see Figure 3b). The leak-in of TFA shifted the CSD of apomyoglobin to +12–+29 (qave of +20.4) and the CSD of holomyoglobin to +12–+27 (qave=+20.2) (Figure 3c). Roughly 33% of the total protein ion signal was due to apomyoglobin ions with the remainder accounted for by holomyoglobin ions. The leak-in of HCl vapors shifted the CSD even further to +12–+29 for both the apo and holo myoglobin peaks with the qave being +21.0 for apomyoglobin and +20.2 for holomyoglobin (Figure 3d). With HCl vapor leak-in, apomyoglobin ions gave rise to the most dominant peaks in the spectrum. While it is apparent that the acid vapors give rise to denaturation and a degree of heme loss, it is noteworthy that it is far from complete and that much higher charge states for holomyoglobin can be accessed in this way than by reducing the pH of the bulk solution.
Figure 3.
Positive nESI spectrum of myoglobin in 100% H2O with a) no acid vapor, b) formic acid vapor, c) trifluoroacetic acid vapor, d) hydrochloric acid vapor. The strength of the acids used increases c–f. (Peaks labeled with ○ correspond to holomyoglobin peaks, △ correspond to apomyoglobin peaks, and □ correspond to myoglobin with 2 heme groups.)
An interesting observation was also noted with the leak-in of formic acid and trifluoroacetic acid. A third charge state distribution was observed, along with the CSDs of holomyoglobin and apomyoglobin, which corresponded to a myoglobin moiety containing a second heme group (represented by □ in Figure 3c). The CSD of the two-heme myoglobin was the same as that of the apomyoglobin and holomyoglobin peaks, with this new two-heme distribution of peaks corresponding to about 10% of the total ion abundance. Heme aggregation in the presence of acid is common53 and solution-phase studies have found that the reconstitution of denatured holomyoglobin enabled the observation of myoglobin with more than one heme group54,55. Although this two-heme protein complex has previously been observed, high charge states of this complex, such as the CSD +12–+27 observed with TFA leak-in, has not been reported.
Highly charged apomyoglobin ions can be observed when denatured to pH=2.2 in solution56. Prior to leak-in, myoglobin was denatured with 1% acetic acid to a solution pH=3 and subjected to ESI resulting in a spectrum with a CSD of +10–+29 (qave= +19.9) (see Figure S1a). All of the peaks in the spectrum corresponded to charge states of apomyoglobin (no holomyoglobin ions). When HCl was leaked-in, the CSD of apomyoglobin did not change, but the qave increased to +21.9 (see Figure S1b). This result suggests that the pH of the charged droplets can be reduced from 3 to lower values by exposure to HCl vapors, which tends to weight the CSD to higher charge.
Disulfide-linked and Reduced Proteins
It is of interest to evaluate the effectiveness of the acid leak-in procedure on the CSDs of proteins that contain intramolecular disulfide bonds. It is well-known that disulfide bonds can inhibit protein unfolding, thus making it more difficult to obtain high charge states for these proteins. Hen egg white lysozyme has 129 residues and contains four disulfide bonds between cysteine residues at positions 6–127, 30–115, 64–80, and 76–9457. High charge states of this protein are observed only when the disulfide bonds are reduced 58,59,60,61 or chemically modified6 in solution. When lysozyme with intact disulfide bonds was subjected to acidic vapor leak-in, the CSD shifted from +8–+15 (qave=+11.1) with no vapor leak-in (Figure S-2a), to a CSD of +8–+11 (qave = +9.8) with formic acid leak (Figure S-2b), to one where the CSD was +6–+8 (qave=+7.5) with HCl leak-in (Figure S-2c). The leak-in of strong acidic vapors caused a shift to lower charge states and a drastic decrease in ion abundance. These results are most likely due to the folding of lysozyme. A shift of lysozyme CSD due to changing solution-phase conditions with the addition of acid has been shown previously6.
To determine the affect of the acidic vapor leak-in on denatured lysozyme, dithiothreitol was used to cleave the disulfide linkages. Prior to acid leak-in, the denatured lysozyme showed a bimodal charge state distribution with the lower CSD centered around +12 and the higher CSD around +15 (Figure S-2d). The leak-in of stronger acids caused the higher CSD to become more abundant (Figure S-2e). With TFA, the lower distribution around +11 and +12 was shifted resulting in a single CSD from +8 to +26 (qave=+17.0). The leak-in of HCl resulted in a different bimodal distribution with CSDs centered around +20 and +17 and having an overall qave=+18.2 (Figure S-2f). Lysozyme only has 18 basic amino acids (Lys, Arg, His), but the CSD of denatured lysozyme with HCl leak-in shows a maximum charge state of +26. The higher charge states can be rationalized if other less basic residues like Pro, Trp, and Gln are considered, which can be protonated in high charge state ions62. These results are consistent with the acid leak-in leading to reduced pH values in the evaporating droplets. In the protein constrained by disulfide linkages, the reduced pH apparently leads to further folding, perhaps for the same reason that the HCl leak-in resulted in a lower charge state for ubiquitin. However, when the protein can unfold at low pH values, the acid leak-in procedure can lead to significant increases in charge states.
A similar charging-up phenomenon was observed with reduced α-lactalbumin, another 14kDa protein with four disulfide bonds, and reduced bovine serum albumin (BSA), a 66 kDa protein with 17 disulfide bonds. The CSD of denatured α-lactalbumin increased from +9–+16 (qave=+12.5) with no acidic vapor leak-in (Figure S-3a) to a CSD of +10–+25 (qave=+18.0) (Figure S-3b) with HCl vapor leak-in. This protein contains 16 basic amino acid residues, with an additional 12 less basic Pro, Trp, Gln residues that can accommodate the +25 charge states observed with HCl leak-in. In the case of denatured BSA, the qave was increased from +52 prior to acidic vapor leak-in (Figure S-3c) to a qave of +77 with HCl vapor leak-in (Figure S-3d). A maximum charge state of about +100 was observed for BSA, which contains 99 basic amino acid residues (Lys, Arg, His). In general, the cleavage of disulfide bonds in solution allows for a more complete unfolding of the protein, which can accommodate the many charges introduced by the leak-in of acidic vapors.
pH Studies and the Effect on ‘Wrong Way Round’ ESI
Further support for attributing the observed effects to pH changes in the droplet was forthcoming by performing control studies with ubiquitin and cytochrome c present in ammonium acetate solutions buffered to pH=5. When the electrosprays of these solutions were exposed to acidic vapors the CSD of cytochrome c did not change (see Figure S-4).
The acid leak-in procedure was applied to unbuffered solutions generated at pH values such that the proteins of interest would be present in bulk solution as ions of polarity opposite to the ESI voltage. It is of interest to determine if the leak-in process can reverse this so-called ‘wrong-way-round’ electrospray condition61,63,64,65. Wrong-way-round conditions typically result in decreased signal abundance, with more metal counter-ion adducts observed for each charge state66. For example, proteins with of net negative charge is solution subjected to positive ESI ionized in the positive mode tend not to compete well for surface sites at the positively charged droplet surface, at least for the early generation droplets. The species in solution that most readily occupy the surface sites are concentrated in small progeny droplets and are thus depleted in the larger droplets in the Rayleigh fission process. The proteins that compete poorly for surface sites in the young droplets can better compete for surface sites in the later fission events of the larger droplet. The older droplets are also enriched in non-volatile salts and, hence, metal ion adduction is more likely. The same holds for positively charged proteins competing for surface sites on a negatively charged droplet. At pH values lower that the isoelectric point (pI) of a protein, the protein is positively charged in solution whereas the opposite is the case when the pH is greater than the protein pI.
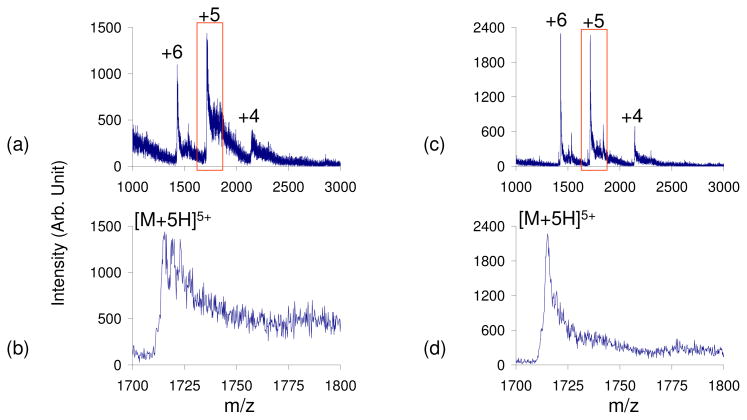
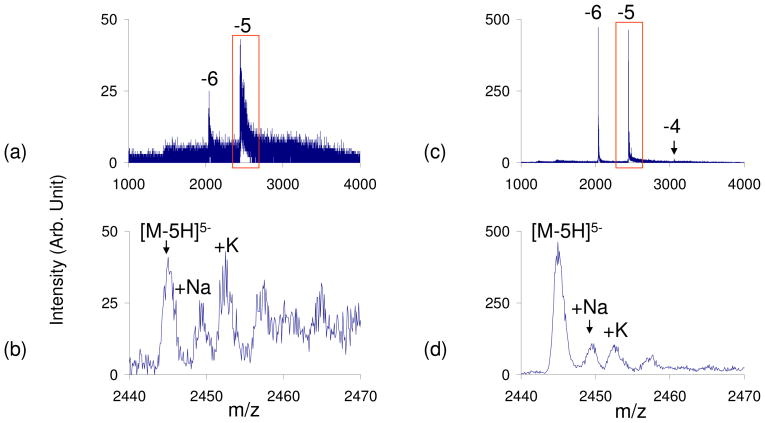
At pH=9, ubiquitin (pI=5.2) is in the wrong-way-round condition when subjected to positive ESI. In this case, the positive ESI mass spectrum shows extensive counter-ion adduction and poor signal-to-noise ratios (Figures 4a and 4b). However, the leak-in of HCl appears to reverse the wrong-way-round condition, resulting in a less adducted spectrum and improved signal-to-noise ratios (Figures 4c and 4d).
Figure 4.
Positive nESI spectrum of ubiquitin (pH=9) in the wrong-way-round condition with no vapor leak (a–b) and HCl vapor leak-in to reverse the wrong-way-round condition (c–d). b) and d) show the zoomed-in region of ubiquitin [M+5H]5+ observed in a) and c) respectively.
Similarly, a cytochrome c (pI = 10.4) solution of pH=5 subjected to negative ESI yields a mass spectrum with highly adducted peaks and poor signal-to-noise ratio (Figure 5a–b). This wrong-way-round condition can be reversed with the leak-in of ammonia generated via a ammonium hydroxide solution. Now, the main peak at each charge state is that of the [M−nH]n− ion, as observed when cytochrome c is sprayed ‘right-way-round’ (Figure 5c–d) and there is a ten-fold improvement in the signal abundance. While this paper emphasizes the exposure of charged droplets to acid vapors, the experiment of Figure 5 suggests that exposure of charged droplets to vapors of basic molecules can also be of utility. Further studies of this type will be reported elsewhere. The point here is that exposure of charged droplets to either acidic or basic vapors can alter pH conditions sufficiently to largely reverse the deleterious effects of subjecting proteins to ESI under wrong-way-round conditions, at least for unbuffered solutions.
Figure 5.
Negative nESI spectrum of cytochrome c (pH=5) in the wrong-way-round condition with no vapor leak (a–b) and NH4OH vapor leak-in to reverse the wrong-way-round condition (c–d). b) and d) show the zoomed-in region of cytochrome c [M−5H]5− observed in a) and c) respectively.
Solvent Studies
Many studies of protein conformations have examined the effect of solvent composition on protein CSDs6,7. Various hypotheses have been offered regarding the role of solvent surface tension on protein CSDs, some claiming that the maximum and average charge state observed in ESI-MS depends on the surface tension of the least volatile solvent67, while other studies claim there to be no such correlation68,69. The leak-in experiment does not address the role of solvent surface tension per se. However, it is of interest to determine the effects of exposure of droplets derived from ESI of mixed solvents on protein CSDs. The effects of methanol, acetonitrile, 1-propanol and 2-propanol as co-solvents with water in the leak-in experiments have been studied with various proteins. When either 1-propanol or 2-propanol was present as the co-solvent, protein CSDs were shifted to relatively high charge states suggesting that the proteins were mostly unfolded. Acid leak-in did not significantly change the CSDs or qave values (data not shown).
Methanol denaturation of proteins in the solution-phase has been examined70,71,72,73. In our studies, it was found that the presence of 60% methanol caused the initial CSD of the proteins to shift to higher charge states when compared to those ionized from a 100% H2O solution. When acids were leaked-in to droplets ionized from a 60% methanol protein solution, a shift in the protein CSD to higher charge states was observed, but the shift required lesser flows of acidic vapors to be observed. In other words, lower concentrations of acidic vapors or weaker acidic vapors were able to completely unfold the protein, compared to acid leak-in with 100% aqueous solutions. In the case of cytochrome c, the presence of organic solvents resulted in a higher initial CSD of the protein due to the slight unfolding as a result of the added solvents. The leaking-in of either weak or strong acids caused the CSD of cytochrome c to shift to a higher CSD when ionized from 60% acetonitrile or 60% methanol solutions (Figure 6). For both solvents, the appearance of low abundance ions at low charge states was observed with HCl leak-in, suggesting that cytochrome c was slightly refolding (Figure 6c and f).
Figure 6.
Positive nESI spectrum of cytochrome c in 60% methanol (a–c) and 60% acetonitrile (d–f) with no vapor (a & d), formic acid vapor (b & e) and HCl vapor (c & f) leak-in.
Ubiquitin unfolding with acid leak-in was also studied in the presence of different co-solvents and the results are summarized in Figure S-5. In the case of ubiquitin, which was previously observed to fold into a more condensed state with the leak-in of HCl vapors (Figure 2), the presence of 60% methanol prevented the complete refolding of the protein (Figure S-5). The CSD prior to leak-in of acids was bimodal and ranged from +5–+15, with a lower CSD around +7 and a higher CSD around +10. The leak-in of formic acid shifted the distribution of the higher charge states slightly higher, to around +11, while simultaneously shifting the distribution of lower charge states slightly lower, to around +7. The leak-in of HCl also resulted in a bimodal distribution with one distribution around +6, which is similar to the previously observed distribution in Figure 2d, and another with a CSD around +10. Analogous results were obtained with acetonitrile as the co-solvent. Thus, it is apparent that a mixture of conformations results from the leak-in experiments applied to these mixed solvent systems.
CONCLUSIONS
The exposure of electrospray droplets to acid vapors can have a significant effect on protein CSDs, particularly for unbuffered aqueous solutions. Leaking the vapors into the counter-current nitrogen drying gas provides a means for altering the pH of the droplets independent of the pH of the bulk solution. The pH change can be large enough, for example, to reverse the wrong-way-round condition. The original unaltered spectrum can be obtained simply by removing the acidic vapors. The two main experimental variables are the concentration of the acid vapor, as determined by the concentration of the acid in the solution used to generate the vapors and the flow of the head space vapors introduced into the drying gas, and the pKa of the acid. An additional characteristic of this approach is the timescale for the pH changes that take place. The evolution of the droplets and passage through the counter-current drying gas takes place on the time-scale of tens of microseconds. As a result, transient intermediates, which can be very difficult to observe, such as the highly charged holomyoglobin intermediate, can be sampled because the interaction is taking place in the interface where the ions do not have time to either fold back into a different conformation or to lose non-covalently bound groups (e.g. the heme group in the case of myoglobin).
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation under CHE-0808380 and the National Institutes of Health under Grant GM 45372.
References
- 1.Chowdhury SK, Katta V, Chait BT. J Am Chem Soc. 1990;112:9012–9013. [Google Scholar]
- 2.Kaltashov IA, Eyles SJ. Mass Spectrom Rev. 2002;21:37–71. doi: 10.1002/mas.10017. [DOI] [PubMed] [Google Scholar]
- 3.Fenn JB. J Am Soc Mass Spectrom. 1993;4:524–535. doi: 10.1016/1044-0305(93)85014-O. [DOI] [PubMed] [Google Scholar]
- 4.Iavarone AT, Jurchen JC, Williams ER. Anal Chem. 2001;73:1455–1460. doi: 10.1021/ac001251t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lomeli SH, Peng IX, Yin S, Ogorzalek Loo RR, Loo JA. J Am Soc Mass Spectrom. 2010;21:127–131. doi: 10.1016/j.jasms.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loo JA, Loo RR, Udseth HR, Edmonds CG, Smith RD. Rapid Commun Mass Spectrom. 1991;5:101–105. doi: 10.1002/rcm.1290050303. [DOI] [PubMed] [Google Scholar]
- 7.Iavarone AT, Jurchen JC, Williams ER. J Am Soc Mass Spectrom. 2000;11:976–985. doi: 10.1016/S1044-0305(00)00169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krusemark CJ, Frey BL, Belshaw PJ, Smith LM. J Am Soc Mass Spectrom. 2009;20:1617–1625. doi: 10.1016/j.jasms.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephenson JL, Jr, McLuckey SA. J Am Chem Soc. 1996;118:7390–7397. [Google Scholar]
- 10.Scalf M, Westphall MS, Krause J, Kaufman SL, Smith LM. Science. 1999;283:194–197. doi: 10.1126/science.283.5399.194. [DOI] [PubMed] [Google Scholar]
- 11.McLuckey SA, Van Berkel GJ, Glish GL. J Am Chem Soc. 1990;112:5668–5670. [Google Scholar]
- 12.Ogorzalek Loo RR, Loo JA, Udseth HR, Fulton JL, Smith RD. Rapid Commun Mass Spectrom. 1992;6:159–165. doi: 10.1002/rcm.1290060302. [DOI] [PubMed] [Google Scholar]
- 13.Zubarev RA, Nielsen ML, Budnik BA. Eur J Mass Spectrom. 2000;6:235–240. [Google Scholar]
- 14.He M, McLuckey SA. J Am Chem Soc. 2003;125:7756–7757. doi: 10.1021/ja0354521. [DOI] [PubMed] [Google Scholar]
- 15.He M, McLuckey SA. Anal Chem. 2004;76:4189–4192. doi: 10.1021/ac496087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Venter A, Cooks RG. Chem Commun. 2006;19:2042–2044. doi: 10.1039/b602614a. [DOI] [PubMed] [Google Scholar]
- 17.Chang DY, Lee CC, Shiea J. Anal Chem. 2002;74:2465–2469. doi: 10.1021/ac010788j. [DOI] [PubMed] [Google Scholar]
- 18.Shieh IF, Lee CY, Shiea J. J Proteome Res. 2005;4:606–612. doi: 10.1021/pr049765m. [DOI] [PubMed] [Google Scholar]
- 19.Chen H, Touboul D, Jecklin MC, Zheng J, Luo M, Zenobi R. Eur J Mass Spectrom. 2007;13:273–279. doi: 10.1255/ejms.879. [DOI] [PubMed] [Google Scholar]
- 20.Touboul D, Jecklin MC, Zenobi R. J Am Soc Mass Spectrom. 2008;19:455–466. doi: 10.1016/j.jasms.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 21.Xia Y, Chrisman PA, Erickson DE, Liu J, Liang X, Londry FA, Yang MJ, McLuckey SA. Anal Chem. 2006;78:4146–4154. doi: 10.1021/ac0606296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kharlamova A, Prentice BM, Huang T, McLuckey SA. Int J Mass Spectrom. 2010 in press. [Google Scholar]
- 23.Breuker K. In: Principles of Mass Spectrometry Applied to Biomolecules. Laskin J, Lifshitz C, editors. John Wiley & Sons; New York: 2006. pp. 177–212. [Google Scholar]
- 24.Grandori R, Matecko I, Muller N. J Mass Spectrom. 2002;37:191–196. doi: 10.1002/jms.272. [DOI] [PubMed] [Google Scholar]
- 25.Konermann L, Douglas DJ. Rapid Commun Mass Spectrom. 1998;12:435–442. doi: 10.1002/(SICI)1097-0231(19980430)12:8<435::AID-RCM181>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 26.Bushnell GW, Louie GV, Brayer GD. J Mol Biol. 1990;214:585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 27.Goto Y, Hagihara Y, Hamada D, Hoshino M, Nishii I. Biochemistry. 1993;32:11878–11885. doi: 10.1021/bi00095a017. [DOI] [PubMed] [Google Scholar]
- 28.Samalikova M, Matecko I, Norbert M, Grandori R. Anal Bioanal Chem. 2004;378:1112–1123. doi: 10.1007/s00216-003-2339-6. [DOI] [PubMed] [Google Scholar]
- 29.Lide DE, editor. CRC Handbook of Chemistry and Physics. CRC Press; Boca Raton, Fl: 2001–2002. [Google Scholar]
- 30.Goto Y, Calciano LJ, Fink AL. Proc Natl Acad Sci USA. 1990;87:573–577. doi: 10.1073/pnas.87.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bychkova VE, Dujsekina AE, Klenin SI, Tiktopulo EI, Uversky VN, Ptitsyn OB. Biochemistry. 1996;35:6058–6063. doi: 10.1021/bi9522460. [DOI] [PubMed] [Google Scholar]
- 32.Grandori R. Protein Science. 2002;11:453–458. doi: 10.1110/ps.45102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konermann L, Collings BA, Douglas DJ. Biochemistry. 1997;36:5554–5559. doi: 10.1021/bi970046d. [DOI] [PubMed] [Google Scholar]
- 34.Maier CS, Kim OH, Deinzer ML. Analytical Biochemistry. 1997;252:127–135. doi: 10.1006/abio.1997.2290. [DOI] [PubMed] [Google Scholar]
- 35.Mirza UA, Chait BT. Anal Chem. 1994;66:2898–2904. doi: 10.1021/ac00090a017. [DOI] [PubMed] [Google Scholar]
- 36.Ptitsyn OB. Adv Protein Chem. 1995;47:83–229. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- 37.Vijay-Kumar S, Bugg CE, Cook WJ. J Mol Biol. 1987;195:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 38.Brutscher B, Bruschweiler R, Ernst RR. Biochemistry. 1997;36:13043–13053. doi: 10.1021/bi971538t. [DOI] [PubMed] [Google Scholar]
- 39.Hoerner JK, Xiao H, Kaltashov I. Biochemistry. 2005;44:11286–11294. doi: 10.1021/bi0509548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohimen A, Dobo A, Hoerner JK, Kaltashov IA. Anal Chem. 2003;75:4139–4147. doi: 10.1021/ac034095+. [DOI] [PubMed] [Google Scholar]
- 41.Evans SV, Brayer GD. J Molec Biol. 1990;123:885–897. doi: 10.1016/S0022-2836(05)80270-0. [DOI] [PubMed] [Google Scholar]
- 42.Katta V, Chait BT. J Am Chem Soc. 1991;113:8534–8535. [Google Scholar]
- 43.Li YT, Hsieh YL, Henion JD. J Am Soc Mass Spectrom. 1993;4:631–637. doi: 10.1016/1044-0305(93)85027-U. [DOI] [PubMed] [Google Scholar]
- 44.Loo JA. Mass Spectrom Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 45.Konermann L, Rosell FI, Mauk AG, Douglas DJ. Biochemistry. 1997;36:6448–6454. doi: 10.1021/bi970353j. [DOI] [PubMed] [Google Scholar]
- 46.Dobo A, Kaltashov IA. Anal Chem. 2001;73:4763–4773. doi: 10.1021/ac010713f. [DOI] [PubMed] [Google Scholar]
- 47.Feng R, Konishi Y. J Am Soc Mass Spectrom. 1993;4:638–645. doi: 10.1016/1044-0305(93)85028-V. [DOI] [PubMed] [Google Scholar]
- 48.Konermann L, Douglas DJ. Rapid Commun Mass Spectrom. 1998;12:435–442. doi: 10.1002/(SICI)1097-0231(19980430)12:8<435::AID-RCM181>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 49.Sage JT, Morikis D, Champion PM. Biochemistry. 1991;30:1227–1237. doi: 10.1021/bi00219a010. [DOI] [PubMed] [Google Scholar]
- 50.Sogbein OO, Simmons DA, Konermann L. J Am Soc Mass Spectrom. 2000;11:312–319. doi: 10.1016/s1044-0305(99)00149-x. [DOI] [PubMed] [Google Scholar]
- 51.Gross DS, Zhao Y, Williams ER. J Am Soc Mass Spectrom. 1997;8:519–524. doi: 10.1016/S1044-0305(97)00010-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hargrove MS, Krzywda S, Wikinson AJ, Dou Y, Ikeda-Saito M, Olson JS. Biochemistry. 1994;33:11767–11775. doi: 10.1021/bi00205a012. [DOI] [PubMed] [Google Scholar]
- 53.Adams PA. Biochem J. 1976;159:371–376. doi: 10.1042/bj1590371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee VWS, Chen YL, Konermann L. Anal Chem. 1999;71:4154–4159. doi: 10.1021/ac9904664. [DOI] [PubMed] [Google Scholar]
- 55.Simmons DA, Konermann L. Biochemistry. 2002;41:1906–1914. doi: 10.1021/bi011697j. [DOI] [PubMed] [Google Scholar]
- 56.Meunier C, Jamin M, De Pauw E. Rapid Commun Mass Spectrom. 1998;12:239–245. doi: 10.1002/(SICI)1097-0231(19980314)12:5<239::AID-RCM143>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 57.Canfield RE, Liu AK. J Biol Chem. 1965;240:1997–2002. [PubMed] [Google Scholar]
- 58.Gross DS, Schnier PD, Rodriguez-Cruz SE, Fagerquist CK, Williams ER. Proc Natl Acad Sci USA. 1996;93:3143–3148. doi: 10.1073/pnas.93.7.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Konermann L, Douglas DJ. J Am Soc Mass Spectrom. 1998;9:1248–1254. doi: 10.1016/S1044-0305(98)00103-2. [DOI] [PubMed] [Google Scholar]
- 60.Loo JA, Edmonds CG, Udseth HR, Smith RD. Anal Chem. 1990;62:693–698. doi: 10.1021/ac00206a009. [DOI] [PubMed] [Google Scholar]
- 61.Reimann CT, Sullivan PA, Axelsson J, Quist AP, Altmann S, Roepstorff P, Velazquez I, Tapia O. J Am Chem Soc. 1998;120:7608–7616. [Google Scholar]
- 62.Schnier PD, Gross DS, Williams ER. J Am Soc Mass Spectrom. 1995;6:1086–1097. doi: 10.1016/1044-0305(95)00532-3. [DOI] [PubMed] [Google Scholar]
- 63.Kelly MA, Vestling MM, Fenselau CC, Smith PB. Org Mass Spectrom. 1992;27:1143–1147. [Google Scholar]
- 64.Mansoori BA, Volmer DA, Boyd RK. Rapid Commun Mass Spectrom. 1997;11:1120–1130. [Google Scholar]
- 65.Le Blanc JCY, Wang J, Guevremont R, Siu KWM. Org Mass Spectrom. 1994;29:587–593. [Google Scholar]
- 66.Pan P, Gunawardena HP, Xia Y, McLuckey SA. Anal Chem. 2004;76:1165–1174. doi: 10.1021/ac035209k. [DOI] [PubMed] [Google Scholar]
- 67.Iavarone AT, Williams ER. J Am Chem Soc. 2003;125:2319–2327. doi: 10.1021/ja021202t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Samalikova M, Grandori R. J Am Chem Soc. 2003;125:13352–13353. doi: 10.1021/ja037000u. [DOI] [PubMed] [Google Scholar]
- 69.Samalikova M, Grandori R. J Mass Spectrom. 2005;40:503–510. doi: 10.1002/jms.821. [DOI] [PubMed] [Google Scholar]
- 70.Babu KR, Douglas DJ. Biochemistry. 2000;39:14702–14710. doi: 10.1021/bi001265t. [DOI] [PubMed] [Google Scholar]
- 71.Konermann L, Douglas DJ. Biochemistry. 1997;36:12296–12302. doi: 10.1021/bi971266u. [DOI] [PubMed] [Google Scholar]
- 72.Babu KR, Moradian A, Douglas DJ. J Am Soc Mass Spectrom. 2001;12:317–328. doi: 10.1016/s1044-0305(00)00226-9. [DOI] [PubMed] [Google Scholar]
- 73.Mao D, Babu KR, Chen YL, Douglas DJ. Anal Chem. 2003;75:1325–1330. doi: 10.1021/ac020647x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.