Abstract
Only a subset of patients with newly diagnosed glioblastoma (GBM) exhibit a response to standard therapy. To date, a biomarker panel with predictive power to distinguish treatment sensitive from treatment refractory GBM tumors does not exist. An analysis was performed using GBM microarray data from 4 independent data sets. An examination of the genes consistently associated with patient outcome, revealed a consensus 38-gene survival set. Worse outcome was associated with increased expression of genes associated with mesenchymal differentiation and angiogenesis. Application to formalin fixed-paraffin embedded (FFPE) samples using real-time reverse-transcriptase polymerase chain reaction assays resulted in a 9-gene subset which appeared robust in these samples. This 9-gene set was then validated in an additional independent sample set. Multivariate analysis confirmed that the 9-gene set was an independent predictor of outcome after adjusting for clinical factors and methylation of the methyl-guanine methyltransferase promoter. The 9-gene profile was also positively associated with markers of glioma stem-like cells, including CD133 and nestin. In sum, a multigene predictor of outcome in glioblastoma was identified which appears applicable to routinely processed FFPE samples. The profile has potential clinical application both for optimization of therapy in GBM and for the identification of novel therapies targeting tumors refractory to standard therapy.
Keywords: biomarkers, MGMT promoter, glioblastoma, prognostic genes, temozolomide
Introduction
Glioblastoma (GBM) is the most common primary brain tumor in adults and is highly lethal.1 Despite increasing evidence that clinically relevant distinct molecular subtypes of GBM exist,2–7 molecular predictors currently play no role in treatment decisions.1 A recent phase III clinical trial for newly diagnosed GBM showed that a regimen consisting of radiation with concomitant/adjuvant temozolomide (TMZ) improved survival compared with radiation alone.8 In this trial, methylation of the MGMT promoter was a favorable prognostic factor, especially in the TMZ-treated arm.5 While promising, it is likely that additional biomarkers could complement MGMT status as an outcome predictor. In addition, identification of prognostic genes could point to new therapeutic approaches for tumors resistant to standard therapy.
The large number of genes investigated in expression microarray data gives rise to a multiple comparisons problem, with a resulting high false-discovery rate in individual data sets.9,10 Generalizations regarding the predictive power of genes identified from single microarray data sets must therefore be made with caution. Several studies have been published,3,7,11,12 but no consensus gene expression profile in malignant glioma reproducibly associated with patient outcome across independent data sets has been identified to date.
Methods
Gene Expression Array Data Sets and Survival Analyses
The analysis was performed using 4 previously published GBM microarray data sets.3,7,11,12 All cases were newly diagnosed GBMs as defined by the World Health Organization criteria. Clinical annotation consisted of patient age, survival time, and confirmation of no prior therapy. The median age was 54 years and the median survival time was 62 weeks. The platform for all 4 data sets was Affymetrix-based and used 2 different chip types: U95Av2 and U133A. Data between these 2 chips were merged by mapping available probe sequence data with 2 databases.13,14 Additional details are shown in Supplementary Material, Table S1. For the initial array analysis, cases were dichotomized into typical (<2 years) vs long-term (≥2 years) survival groups (TS vs LTS, respectively). Several statistical approaches15,16 were investigated for survival analyses (see Supplementary Methods). Genes were ranked according to degree of difference between TS and LTS groups. Patients were followed until death or were censored at last known follow-up.
Quantitative Reverse Transcriptase-Polymerase Chain Reaction and Immunohistochemistry
Quantitative measurement of expression of candidate survival genes from formalin-fixed, paraffin-embedded (FFPE) GBM samples was performed using TaqMan quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) assays (Supplementary Material, Table S2). None of the samples used in the validation sets were the same as those used in the microarray analysis. Details of the qRT-PCR and quality assessment are presented in Supplementary Methods. MGMT methylation was also tested by a qRT-PCR-based assay on bisulfite-treated DNA as described previously.5,17 Details of this method as well as immunohistochemistry for CD133 and nestin are presented in Supplementary Methods.
Results
Statistical Method and Concordance of Survival Association across Data Sets
Supplementary Material Fig. S1 shows the overall approach utilized for the identification of robust survival-associated genes in GBM. It is not well established which test statistic is optimal for identifying consensus genes that are significantly associated with patient outcome across independent microarray data sets.18 We reasoned that the most robust survival genes should overlap in all 4 data sets. The largest overlap was observed when the fold-change was used to rank order the genes. Other methods, such as significance analysis of microarrays (SAM),15 RankProduct,16 and Cox model, produced less overlap. As shown in Supplementary Material Fig. S2, plots of average gene rank vs standard deviation of the ranks demonstrate that ranking of genes by the ratio of the mean gene expression between TS and LTS (fold-change method) was much more stable across independent data sets than if genes were ranked by a 2-class SAM analysis. Furthermore, the standard deviation of rankings by fold-change rises rapidly beyond the top 200 genes, approaching that expected from random ranks. These findings are consistent with results from the Microarray Quality Control (MAQC) Project, which also found that gene rankings based on P-values or SAM did not generate more reproducible results compared to fold-change.18 Importantly, an examination of each consensus gene identified by the fold-change method using outcome as either a categorical variable or continuous variable showed statistical significance in all cases (not shown). The fold-change method was therefore utilized for subsequent analyses. Cross-validation approaches demonstrated that the top 200 survival genes selected using fold-change in a subset of the microarray data could predict outcome in the remaining data (Supplementary Material, Methods and Fig. S3).
Identification of a Consensus Multigene Predictor Across Independent Data Sets
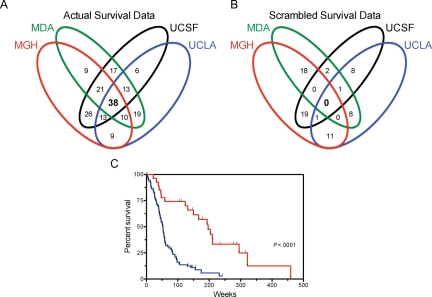
Figure 1 shows a Venn diagram of consensus genes among the top 200 genes across all institutions. There were 38 genes (Fig. 1A and Table 1) that were ranked in the top 200 in all 4 institutions, and an additional 57 genes (Fig. 1A and Supplementary Material, Table S3) that were ranked in the top 200 in 3 out of 4 institutions. Of this initial set of 38 genes, the expression of 31 were found to be associated with poor survival (higher in the TS group compared with the LTS group), while the remaining 7 were associated with favorable outcome (higher expression in LTS group compared with the TS group). The false discovery rate using the intersection of all 4 data sets was extremely low and estimated as a 0.3% chance to find 1 common gene among the 4 lists by chance, and a 99.7% chance that 0 genes would be common to the 4 lists by chance (Fig. 1B).
Fig. 1.
Identification of robust outcome-associated genes from microarray data. (A) Overlap of survival genes among 4 microarray data sets. The top 200 genes were identified for each data set individually and the overlap of the 4 lists is shown in a Venn diagram. (B) Estimation of false discovery rate. The survival data were scrambled among the samples and a list of 200 genes was generated from each data set using the scrambled survival data. (C) Survival according to metagene score. The 38 survival-associated genes common to all 4 data sets were used to calculate a metagene score for each sample. The metagene score was calculated by subtracting the sum of the values of the good-prognosis genes from the sum of the values of the poor-prognosis genes. The samples were ranked by metagene score and divided into 2 groups based on results from recursive partitioning analysis. Survival according to metagene score is shown for the group with the lower ranking metagene scores (red) vs samples with higher metagene scores (blue).
Table 1.
Survival-associated genes (n = 38) common to all 4 microarray data sets. The direction of the association to survival is shown. The 9 genes denoted by an asterisk denote those which remained robust in subsequent testing in archival FFPE samples.
| Gene Symbol | Gene Name | Expression Level in Typical vs Long-term Survivors |
|---|---|---|
| ACTN1 | Actinin, alpha 1 | Higher |
| AQP1* | Aquaporin 1 | Higher |
| CHI3L1* | YKL-40 | Higher |
| CLIC1 | Chloride intracellular channel 1 | Higher |
| COL1A2 | Collagen, type 1, alpha 2 | Higher |
| EMP3* | Epithelial membrane protein 3 | Higher |
| FABP5 | Fatty acid binding protein 5 | Higher |
| FN1 | Fibronectin 1 | Higher |
| GABBR1 | Gamma-aminobutyric acid receptor 1 | Lower |
| GPNMB* | Glycoprotein | Higher |
| GRIA2 | Glutamate receptor, ionotropic, AMPA 2 | Lower |
| IGFBP2* | Insulin-like growth factor binding protein 3 | Higher |
| IGFBP3 | Insulin-like growth factor binding protein 3 | Higher |
| KIAA0510 | Hypothetical protein | Lower |
| LDHA | Lactate dehydrogenase A | Higher |
| LGALS1 | Galectin 1 | Higher |
| LGALS3* | Galectin 3 | Higher |
| MAOB | Monoamine oxidase B | Higher |
| NNMT | Nicotinamide N-methyltransferase | Higher |
| OLIG2* | Oligodendrocyte lineage transcription factor 2 | Lower |
| OMG | Oligodendrocyte myelin glycoprotein | Lower |
| PDPN* | Podoplanin | Higher |
| PLP2 | Proteolipid protein 2 | Higher |
| RIS1 | Ras-induced senescence 1 | Higher |
| RTN1* | Reticulon 1 | Lower |
| S100A10 | S100 calcium binding protein A10 | Higher |
| SERPIN3 | Alpha-1 antiproteinase | Higher |
| SERPINE1 | Plasminogen activator inhibitor type 1 | Higher |
| SERPING1 | C1 inhibitor | Higher |
| TAGLN | Transgelin | Higher |
| TAGLN2 | Transgelin 2 | Higher |
| TCF12 | Transcription factor 12 | Lower |
| TCTE1L | T-complex-associated-testis-expressed 1-like | Higher |
| TGFBI | Transforming growth factor, beta-induced, 68 kDa | Higher |
| TIMP1 | Tissue inhibitor of metalloproteinase 1 | Higher |
| TMSB10 | Thymosin, beta 10 | Higher |
| TNC | Tenascin C | Higher |
| VEGF | Vascular endothelial growth factor | Higher |
To determine the association of this multigene set with patient outcome, the gene expression values for the 38-gene profile for each sample was condensed into a single value for each tumor, termed a metagene score, as previously described for clinical application in breast cancer.19 Each tumor was then ranked according to this metagene score. Recursive partitioning analysis using survival as a continuous variable was then used to determine the best cutoff for metagene score from these data. Use of all of the data for the recursive portioning led to 2 groups with a cutoff of approximately 75% high/unfavorable metagene scores vs the remaining 25% with low/favorable metagene scores. Patients dichotomized using this cutoff method for Kaplan–Meier analysis, not unexpectedly, showed a significant association with survival (Fig. 1C). For consistency and comparison, the identical metagene score cutoff was used in all subsequent survival analyses.
Since our prior studies indicated that favorable-prognosis GBMs have an expression profile similar to lower grade gliomas,12 we reasoned that a robust set of survival-associated genes in GBM should overlap with genes found to be differentially expressed between GBM and lower grade gliomas. We tested this hypothesis in an independent published data set of 153 glioma tumor samples of different grades20 using the data analysis tool from Oncomine. Comparing the top 2% of genes overexpressed in GBM vs lower grade gliomas in that data set with our 38-gene set, we found that 26 of our 31 poor-prognosis genes were concordant. Additionally, the results from The Cancer Genome Atlas21 (TCGA) were made available after our study was completed. To assess reproducibility of the 38 gene set, we tested whether the expression of this signature was associated with survival in the newly diagnosed GBMs from the TCGA data. Using 136 cases known to be non-recurrent, we found a significant association of the 38 gene set with survival (P < .05) in that data set. These results provided independent confirmation that our consensus gene list is likely to be a robust predictor of outcome in GBM.
Initial Validation and Further Optimization of Multigene Predictor
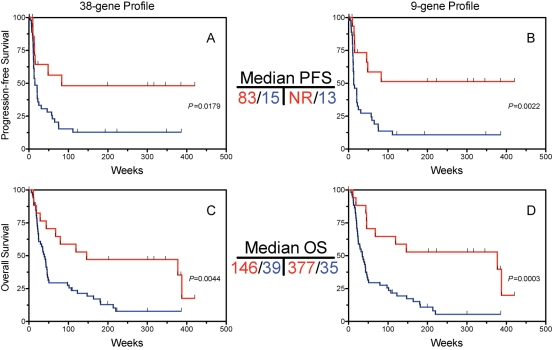
To perform initial validation of the 38-gene predictor, we utilized an independent retrospective set of FFPE tumor samples of 68 newly diagnosed GBMs. The expression of each of the 38 genes was quantified using qRT-PCR (see Supplementary Methods) compared to the average expression of 2 control genes. The resulting fold-change difference (calculated from Δ−Ct of each gene compared with the average Ct of the 2 control genes) for each gene between survival groups is summarized in Supplementary Material, Table S4. Survival analysis demonstrated that the metagene score from the entire 38-gene set, using the same cutoff as in Fig. 1, was a significant predictor of progression-free (PFS, Fig. 2A) and overall survival (OS, Fig. 2C), validating the overall 38-gene set as a survival predictor, as well as a potential predictor of PFS.
Fig. 2.
Validation of multigene predictor for overall survival in an independent sample set. A set of 68 formalin-fixed, paraffin-embedded glioblastoma samples was subject to qRT-PCR for the 38 gene set identified in Fig. 1. A metagene score was calculated as in Fig. 1 and the samples were ranked by metagene score. Patients were dichotomized into 2 groups based on metagene score using proportions identical to those in Fig. 1. Survival is shown for the lower metagene scores (red) vs the higher metagene scores (blue). Analyses were performed for the entire 38-gene set as well as a smaller 9-gene profile composed of those genes that had the highest individual survival association in the tumors and showed high technical feasibility in paraffin tissues. (A) and (B) Progression-free survival (PFS) according to the entire 38-gene set (A) as well as the 9 gene-profile (B). (C) and (D) Overall survival (OS) according to the 38-gene set (C) and 9-gene set (D). NR, median not reached.
Loss of fidelity for specific RNA species has been reported when comparing gene expression from frozen vs clinical FFPE samples, due to multiple factors including the deleterious effects of formalin fixation on mRNA quality, and variation in times of formalin exposure in clinical samples.22 To optimize the multigene predictor for application in FFPE clinical samples, the top 9 genes (Supplementary Material, Table S4) were selected on the basis of strength/significance of survival association (fold change level) as well as technical considerations (amplification consistency in FFPE samples, using a mean Ct level among the samples of ≤ 32). Hazard ratios and P-values for the 9-gene set selected using these criteria are shown in Supplementary Material, Table S5. Survival prediction based on metagene score calculated using the 9-gene profile was similar or slightly better for PFS (Fig. 2B) and OS (Fig. 2D) compared with the entire 38-gene set (Fig. 2A and C, respectively).
Validation of 9-Gene Predictor and Comparison with Known Prognostic Variables
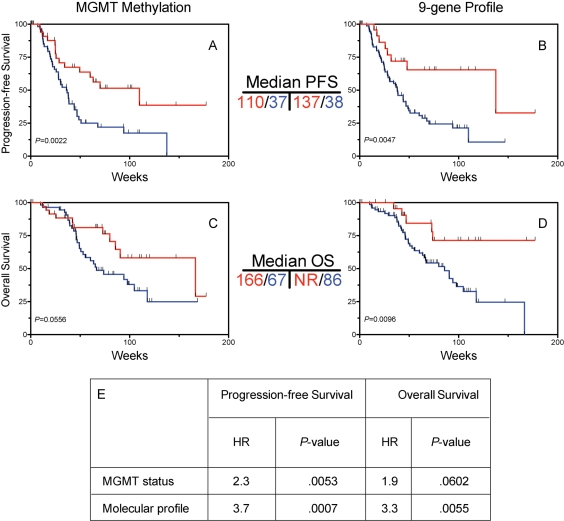
To further validate the multigene profile, an additional set of 101 FFPE tumor samples was identified from patients whose initial therapy included radiation plus concurrent and adjuvant TMZ, which is the current standard of care for GBM.8 The 9-gene profile was tested for prediction of outcome in this group. Univariate analyses showed that each of the 9 genes showed a significant or near-significant association toward overall survival when analyzed individually (Supplementary Material, Table S6). Determination of MGMT promoter methylation status, previously shown a predictor of outcome in patients treated with this regimen,5 was performed and showed an association with PFS (Fig. 3A) and OS (Fig. 3C). The 9-gene profile compared favorably to MGMT status as an outcome predictor (Fig. 3B and D). Multivariate analysis showed that the 9-gene profile was an independent predictor after adjustment for MGMT status and showed larger hazard ratios and smaller P-values for both PFS and OS compared with MGMT (Fig. 3E).
Fig. 3.
Validation of 9-gene profile in temozolomide (TMZ)-treated GBM and comparison with MGMT status. Glioblastoma samples from temozolomide-treated patients (n = 101) were tested for MGMT methylation as well as the 9-gene predictor. For all Kaplan–Meier curves, red indicates low score and blue indicates high score. Tumors were ranked by metagene score and divided into distinct metagene groups using the same cutoffs as in Fig. 1. Progression-free survival (PFS) according to the entire MGMT status (A) as well as the 9-gene profile. (C) and (D) Overall survival (OS) according to MGMT status (C) and 9-gene set (D). (E) Cox proportional hazards multivariate analysis showing that the 9-gene profile is an independent predictor of outcome in TMZ-treated patients after adjusting for MGMT status. NR, median not reached.
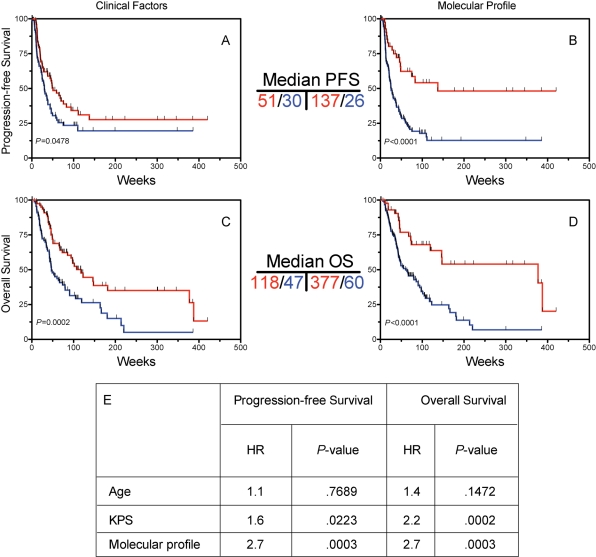
To test whether the 9-gene profile was an independent predictor after adjusting for clinical factors, multivariate analysis was performed, which included data from all 169 samples on which the qRT-PCR analysis was performed. Models that included patient age, performance status as well as molecular profile show that the molecular profile was a significant predictor of both PFS and OS. Such a model implies equal weighting of each of the genes in the multigene model, which may underestimate its predictive ability. The genes in the molecular profile were weighted according to the strength of association of survival (see Supplementary Methods). To evaluate clinical predictors, a good outcome group was defined based on patients who were both younger than the median age (57) and had a Karnofsky Performance Status (KPS) of 70 or higher. As expected, this group had favorable PFS (Fig. 4A) and OS (Fig. 4C) compared with patients older than the median age, or with KPS < 70. Kaplan–Meier curves for the optimized molecular predictor exhibited a significant association with PFS (Fig. 4B) and OS (Fig. 4D), and are shown for comparison. As was shown for the multigene set using equal weighting, the weighted multigene profile was an independent predictor of outcome in a multivariate analysis after adjusting for age and KPS (Fig. 4E). To further determine the additive predictive power of the 9-gene profile after accounting for all variables in TMZ-XRT–treated patients, Cox proportional hazards analysis was performed using MGMT methylation status, age, and performance status in the 101 patient cohort, which demonstrated that the 9-gene panel remained a significant predictor (HR = 2.39, P = .042) after accounting for relevant molecular and clinical variables in this subset of patients.
Fig. 4.
Comparison of 9-gene molecular profile with clinical variables. Cases were stratified into favorable vs unfavorable clinical groups as descrbed in the text. (A) and (B) Progression-free survival (PFS) according to clinical factors (A), as well as the 9 gene-profile. Median PFS is as shown. (C) and (D) Overall survival (OS) according to MGMT status (C) and 9-gene set (D). Median OS is as shown. (E) Cox proportional hazards multivariate analyses on all (n = 169) validation cases. To generate comparable hazard ratios, patient age was coded as above/below the median and KPS was coded as 70 or above vs <70.
Association of Molecular Profile with Mesenchymal-Angiogenic Phenotype and Glioma Stem Cell Markers
In epithelial tumors, mesenchymal transition is associated with clinical aggressiveness.23,24 In addition, mesenchymal transition has been shown to generate cells with stem-like properties.25 Stem-like cells with tumor-initiating capacity have been identified in GBM and have been associated with treatment resistance,26,27 and stem cell–related gene expression signatures have been found to correlate with treatment resistance in GBM.28 We therefore investigated whether there was an association between metagene score and stem cell markers. The expression of the stem cell genes CD133 and nestin protein was assessed semi-quantitatively in 54 and 52 tumor samples, respectively, and compared with the metagene score using the 9-gene profile (Supplementary Material, Table S7 and Fig. S4). Overexpression of CD133 and nestin were found to be highly associated with an unfavorable metagene score (both P < .01, Spearman rank), suggesting a link between markers of stem-like cells and a predictor of therapeutic resistance in GBM that identifies tumors with a mesenchymal-angiogenic phenotype.
Discussion
Identification of markers predictive of survival in GBM could optimize and individualize therapy by prospectively identifying those patients who will benefit most from standard therapy, and identifying novel therapeutic targets based on the molecular profiles of those patients refractory to standard therapy. To discover such markers, 4 independent microarray data sets were queried to identify a multigene set predictive of outcome. This multigene panel was subsequently optimized and validated in additional independent cohorts to identify the subset of genes with the most robust survival association using FFPE tissues.
While the quality of nucleic acid derived from FFPE tissue is suboptimal, FFPE samples were utilized in the validation sets to increase the feasibility of the development of a clinical test applicable to routinely available specimens. We noted that a number of the genes identified in the initial microarray analysis did not have strong survival correlations when tested in FFPE samples. This finding was not surprising based on technical considerations, given the relatively low quality of mRNA derived from FFPE tissues.22 The 9-gene profile was therefore selected based on both survival correlation and technical feasibility in FFPE tissue. This profile was then validated in a second set of patients, where all 9 remained individually predictive.
In total, the multigene panel was selected based on reproducibility in 6 independent data sets comprising a total of 279 samples. To have significant clinical utility, a biomarker or panel of biomarkers should add predictive and/or prognostic value to known clinical or molecular variables. In patients with GBM, patient age and performance status are established powerful prognostic factors. In addition, MGMT methylation status, especially in the setting of TMZ therapy, has been shown to be a predictor of outcome.5 The 9-gene profile was thus compared with both MGMT methylation status in TMZ-treated patients (Fig. 3) and clinical factors in all patients (Fig. 4) and was shown to be a significant independent predictor of outcome in multivariate analyses. These data indicate that the 9-gene profile could serve as the basis for a robust clinical test (amenable to paraffin tissue) which, along with the existing clinical and other molecular markers, could be used to optimize therapeutic choices for individual patients, analogous to the predictive test developed for optimization of patient therapy in breast cancer.29
The primary role of the 9-gene panel for optimization of therapy would be to prospectively identify GBM patients most likely to have a durable survival to standard therapy. However, the identification of specific genes with robust association with outcome can also provide insights into tumor biology that could help to identify therapies for patients resistant to standard therapy.
Examination of the known functions of the 38-gene and 9-gene sets demonstrated that better prognosis is associated with higher expression of genes associated with normal neural development (the proneural group), while poor survival is associated with increased expression of genes associated with mesenchymal tissues, angiogenesis, and extracellular matrix (the mesenchymal-angiogenic group). Immunohistochemical analyses have demonstrated that a number of these mesenchymal and angiogenic genes including YKL-40,7,30 galectin-3,31 tenascin,32 and VEGF33 are indeed expressed by GBM tumor cells. Prior unsupervised analyses by our group and others3,12 have emphasized the potential importance of this mesenchymal-angiogenic molecular phenotype in GBM. In the current study, using different methodology optimized for the identification of robust outcome-based biomarkers across multiple datasets, we find that genes characteristic of the previously defined mesenchymal and proneural represent robust biomarkers of outcome. Taken together, these data suggest that a clinically relevant mesenchymal transition occurs in GBM that is associated with increased angiogenesis and poor outcome in GBM, and is analogous to the epithelial-to-mesenchymal transition that has been described in carcinomas.34 These findings suggest that drugs targeting angiogenesis or mesenchymal transition may be promising therapies for patients with the poor prognosis molecular profile.
Recent observations demonstrate that the mesenchymal transition associated with worse outcome in breast cancer also results in tumor cells with stem-like properties.25 In GBM, stem-like cells with tumor-initiating capacity have been identified and have been implicated as a source of treatment resistance.26,27 Furthermore, a stem cell-related gene expression signature has been found to correlate with worse outcome in GBM patients receiving standard therapy.28 The association that we observed between the multigene predictor and expression of GBM stem cell markers, including CD133,27 supports the concept that the mesenchymal-angiogenic phenotype in GBM is also associated with a stem-like phenotype, and highlights the potential importance of developing therapeutic strategies that target these cells.35,36 Recent studies have demonstrated clinical activity of antiangiogenic drugs in recurrent GBM.37,38 Our data suggest that patients with the poor prognosis molecular profile may be a subset that would derive particular benefit from these therapies. Future studies will test this hypothesis and examine whether the molecular profile of GBM tumors can be used both to prospectively identify patients who will or will not respond to conventional therapy and to prospectively select a subgroup that will specifically benefit from additional agents, such as those targeting angiogenesis or tumor stem cells.
Supplementary Material
Supplementary material is available at Neuro-Oncology online (http://www.neuonc.oxfordjournals.org).
Conflict of interest statement. None declared.
Funding
Supported by grants from the V Foundation, The Dr Marnie Rose Foundation, and a SPORE grant in Brain Cancer (P50CA127001) from the National Institute of Health/ National Cancer Institute.
Supplementary Material
References
- 1.Kleihues P, Cavenee W WHO. Classification of Tumours: Pathology and Genetics of Tumours of the Nervous System. Lyon: IARC Press; 2000. [Google Scholar]
- 2.Burton EC, Lamborn KR, Feuerstein BG, et al. Genetic aberrations defined by comparative genomic hybridization distinguish long-term from typical survivors of glioblastoma. Cancer Res. 2002;62:6205–6210. [PubMed] [Google Scholar]
- 3.Freije WA, Castro-Vargas FE, Fang Z, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 4.Haas-Kogan DA, Prados MD, Lamborn KR, et al. Biomarkers to predict response to epidermal growth factor receptor inhibitors. Cell Cycle. 2005;4:1369–1372. doi: 10.4161/cc.4.10.2105. [DOI] [PubMed] [Google Scholar]
- 5.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 6.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 7.Nigro JM, Misra A, Zhang L, et al. Integrated array-comparative genomic hybridization and expression array profiles identify clinically relevant molecular subtypes of glioblastoma. Cancer Res. 2005;65:1678–1686. doi: 10.1158/0008-5472.CAN-04-2921. [DOI] [PubMed] [Google Scholar]
- 8.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 9.Ransohoff DF. Rules of evidence for cancer molecular-marker discovery and validation. Nat Rev Cancer. 2004;4:309–314. doi: 10.1038/nrc1322. [DOI] [PubMed] [Google Scholar]
- 10.Simon R. Roadmap for developing and validating therapeutically relevant genomic classifiers. J Clin Oncol. 2005;23:7332–7341. doi: 10.1200/JCO.2005.02.8712. [DOI] [PubMed] [Google Scholar]
- 11.Nutt CL, Mani DR, Betensky RA, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–1607. [PubMed] [Google Scholar]
- 12.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence project: update and current status. Nucleic Acids Res. 2003;31:34–37. doi: 10.1093/nar/gkg111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imanishi T, Itoh T, Suzuki Y, et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong F, Breitling R, McEntee CW, et al. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22:2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 17.Eads CA, Lord RV, Wickramasinghe K, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410–3418. [PubMed] [Google Scholar]
- 18.Shi L, Reid LH, Jones WD, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 20.Sun L, Hui AM, Su Q, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 21.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castiglione F, Degl'Innocenti DR, Taddei A, et al. Real-time PCR analysis of RNA extracted from formalin-fixed and paraffin-embeded tissues: effects of the fixation on outcome reliability. Appl Immunohistochem Mol Morphol. 2007;15:338–342. doi: 10.1097/01.pai.0000213119.81343.7b. [DOI] [PubMed] [Google Scholar]
- 23.Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 24.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 27.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 28.Murat A, Migliavacca E, Gorlia T, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 29.Sparano JA, Paik S. Development of the 21-gene assay and its application in clinical practice and clinical trials. J Clin Oncol. 2008;26:721–728. doi: 10.1200/JCO.2007.15.1068. [DOI] [PubMed] [Google Scholar]
- 30.Pelloski CE, Mahajan A, Maor M, et al. YKL-40 expression is associated with poorer response to radiation and shorter overall survival in glioblastoma. Clin Cancer Res. 2005;11:3326–3334. doi: 10.1158/1078-0432.CCR-04-1765. [DOI] [PubMed] [Google Scholar]
- 31.Camby I, Belot N, Rorive S, et al. Galectins are differentially expressed in supratentorial pilocytic astrocytomas, astrocytomas, anaplastic astrocytomas and glioblastomas, and significantly modulate tumor astrocyte migration. Brain Pathol. 2001;11:12–26. doi: 10.1111/j.1750-3639.2001.tb00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leins A, Riva P, Lindstedt R, et al. Expression of tenascin-C in various human brain tumors and its relevance for survival in patients with astrocytoma. Cancer. 2003;98:2430–2439. doi: 10.1002/cncr.11796. [DOI] [PubMed] [Google Scholar]
- 33.Zhou YH, Tan F, Hess KR, et al. The expression of PAX6, PTEN, vascular endothelial growth factor, and epidermal growth factor receptor in gliomas: relationship to tumor grade and survival. Clin Cancer Res. 2003;9:3369–3375. [PubMed] [Google Scholar]
- 34.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 35.Bao S, Wu Q, Sathornsumetee S, et al. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006;66:7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- 36.Calabrese C, Poppleton H, Kocak M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 37.Batchelor TT, Sorensen AG, di Tomaso E, et al. AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell. 2007;11:83–95. doi: 10.1016/j.ccr.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vredenburgh JJ, Desjardins A, Herndon JE, II, et al. Bevacizumab plus irinotecan in recurrent glioblastoma multiforme. J Clin Oncol. 2007;25:4722–4729. doi: 10.1200/JCO.2007.12.2440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.