Abstract
The flow properties of DNA are important for understanding cell division and, indirectly, cancer therapy. DNA topology controlling enzymes such as topoisomerase II are thought to play an essential role. We report experiments showing how double-strand passage facilitated by topoisomerase II controls DNA rheology. For this purpose, we have measured the elastic storage and viscous loss moduli of a model system comprising bacteriophage λ-DNA and human topoisomerase IIα using video tracking of the Brownian motion of colloidal probe particles. We found that the rheology is critically dependent on the formation of temporal entanglements among the DNA molecules with a relaxation time of ∼1 s. We observed that topoisomerase II effectively removes these entanglements and transforms the solution from an elastic physical gel to a viscous fluid depending on the consumption of ATP. A second aspect of this study is the effect of the generic topoisomerase II inhibitor adenylyl-imidodiphosphate (AMP-PNP). In mixtures of AMP-PNP and ATP, the double-strand passage reaction gets blocked and progressively fewer entanglements are relaxed. A total replacement of ATP by AMP-PNP results in a temporal increase in elasticity at higher frequencies, but no transition to an elastic gel with fixed cross-links.
Introduction
The genome is highly compacted and concentrated, yet the timescales for replication and transcription are unexpectedly fast. For instance, the segregation of the intertwined sister chromatids in the anaphase of dividing cells occurs within a minute. Topology controlling enzymes (topoisomerases) are thought to play an essential role in chromatid motion and the kinetics of chromosome condensation, but to date there are no quantitative measurements of the effect of disentanglement on the properties of the flow (viscoelasticity) (1–3). Topoisomerase II binds a DNA segment, introduces a transient break in the duplex, captures another segment from the same or another molecule, transports the captured segment through the gap of the break, reseals the break, and releases the captured segment. This enzymatic reaction requires the hydrolysis of two molecules of ATP (4,5). We report microrheology experiments showing how double-strand passage facilitated by human topoisomerase IIα affects the viscoelasticity of a model system comprising bacteriophage λ-DNA (48.5 kbp, contour length of 16.3 μm) dispersed in a physiologically relevant buffer. In particular, we explore how the disentanglement of linear DNA molecules by topoisomerase II is coupled to the dissipation of energy through the hydrolysis of ATP. A second aspect of this article is the effect of the generic topoisomerase II inhibitor adenylyl-imidodiphosphate (AMP-PNP). Topoisomerase II inhibitors constitute an important class of anticancer drugs, because they impede the division of cells. This can be due to the blocking of the double-strand passage reaction and/or the formation of cross-linking protein clamps between different DNA segments. The relative importance of these two mechanisms in controlling the viscoelasticity will be gauged with our assays.
At high concentration, linear DNA molecules become entangled and they form a dynamic network. A two-dimensional representation of a polymer network with entangled and nonentangled interaction points is shown in Fig. 1. Entanglements are topological constraints resulting from the fact that the DNA molecules cannot cross through each other. It should be noted, however, that the entanglements are separated by thousands of basepairs along the contour and distributed over relatively large distances in space as compared to the mesh size of the network. The majority of the interaction points are hence nonentangled. As a result of entanglements, concentrated solutions of DNA are viscoelastic. Viscoelasticity implies an elastic response at higher frequencies, whereas for lower frequencies and corresponding longer times, the solutions are viscous-fluid-like (6). The motion of the DNA molecules is strongly hindered by the presence of the neighboring DNA molecules and the relaxation times may become very long. DNA dynamics in the condition of entanglements can be described by the reptation model, in which the molecule is thought to move in a snakelike fashion along the axial curve (primitive path) of a tube formed by the entanglements (7–9). Dynamics of entangled DNA molecules in the presence of topoisomerases has been discussed in terms of reptation and constraint release (10). The reptation model gives specific scaling laws for the longest, global relaxation time pertaining to the motion of the DNA molecule, self-diffusion coefficient, high-frequency limiting value of the elastic storage modulus, and the zero shear limit of the viscosity. For nonentangled DNA, the concentration scaling of these transport properties is different, so that a change in scaling exponents can be used to monitor the relaxation of entanglements by topoisomerase II.
Figure 1.
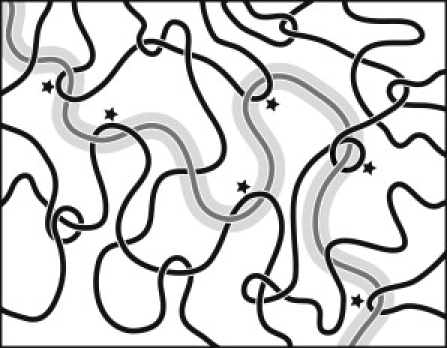
Drawing of a polymer network (not to scale). The shaded test chain can only move along the tube, because lateral displacements are prohibited by the topological constraints (indicated by the asterisks) imposed by some solid chains.
The viscoelastic properties of entangled DNA solutions have been reported in the literature before (11–15). An early attempt to quantify the effect of topoisomerase II on DNA rheology was not conclusive (16). For solutions of T2-DNA (164 kbp, contour length of 56 μm), it was shown that the zero shear viscosity obeys the same scaling law as the one for synthetic, linear polymers and that the entanglement concentration is 0.25 g of DNA/L. The relaxation times become very long on the order of 1000 s, if the concentration is increased to 1 g of DNA/L (11). For λ-DNA similar scaling behavior of the zero shear viscosity was observed, but the relaxation times are two-orders-of-magnitude shorter because of the lower molecular mass (14). The elastic storage and viscous loss moduli, G′ and G″, respectively, of λ-DNA were reported by us before (15). For concentrations exceeding the entanglement concentration, the loss modulus is crossed by the storage modulus at a crossover frequency ωc and the storage modulus levels off at a limiting high-frequency plateau value. The viscoelastic moduli as well as the self-diffusion coefficient of λ-DNA in 10 mM Tris/EDTA buffer at ambient temperature follow the reptation prediction for concentrations exceeding 0.3 g/L (15,17). For λ-DNA, the range from the nonentangled, semidilute to the moderately entangled regime with ∼12 entanglements per molecule can be covered if the concentration is increased from ∼0.1 to 1.4 g of DNA/L.
We have employed video tracking of the Brownian motion of colloidal probe particles, which requires minute samples of no more than 15 μL each (15,18). First, the effect of DNA concentration on the viscoelastic response has been investigated in the relevant conditions in terms of buffer composition and temperature. These experiments serve as a reference and were performed without topoisomerase II. We derive the number of entanglements per DNA molecule from the limiting high-frequency plateau value of the elastic storage modulus G. The longest, global relaxation time τ pertaining to the motion of a single DNA molecule is obtained from G and the low shear viscosity η as well as from the lowest crossover frequency ωc. Second, we have explored the effect of topoisomerase II on the viscoelasticity. The efficiency of topoisomerase II in the relaxation of entanglements is investigated by monitoring G after the addition of topoisomerase II to initially entangled solutions of DNA. Optimal conditions in terms of reaction time, ATP, and topoisomerase II concentrations are identified. The results for η, G, and τ are compared with the relevant scaling laws for the transport properties of (non)entangled polyelectrolyte in the condition of screened electrostatics (7–9). Third, we have investigated the effect of the inhibitor by performing a series of experiments with increasing molar fraction of AMP-PNP. The control of the viscoelasticity is discussed in terms of the double-strand transport mechanism facilitated by topoisomerase II in conjunction with the consumption of ATP or inhibition through binding of AMP-PNP.
Materials and Methods
Sample preparation
Human topoisomerase IIα was purchased from Affymetrix (Santa Clara, CA). As supplied by the manufacturer, the topoisomerase storage buffer contains 15 mM Na2HPO4, pH 7.1, 700 mM NaCl, 0.1 mM EDTA, 0.5 mM dithiothreitol, and 50% glycerol. The reaction buffer is composed of 10 mM Tris-HCl, pH 7.9, 50 mM NaCl, 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, and 15 mg/L bovine serum albumin. ATP and AMP-PNP were purchased from Sigma-Aldrich (St. Louis, MO). Water was deionized and purified by a Millipore system (Millipore, Billerica, MA) and has a conductivity <1 × 10−6 Ω−1 cm−1. Bacteriophage λ-DNA was purchased from New England Biolabs (Ipswich, MA) and used without further purification. A 12-base-long oligonucleotide (single-stranded DNA) with the complementary sequence of the right cohesive end of λ-DNA, 5′-AGGTCGCCGCCC-3′, was purchased from Sigma-Aldrich. The stock solution was concentrated to a concentration of 1.8 g of DNA/L by freeze drying and subsequently dialyzed in microdialyzers against reaction buffer. The DNA concentration was determined by ultraviolet spectrometry. The stock solution was heated to 333 K, cooled to 295 K by immersion in a water bath, and the complementary oligonucleotide was hybridized to one of the overhangs with a 20% excess molar ratio (a solution with a 100-fold excess of oligomer was also prepared). The stock solution was subsequently equilibrated for at least 48 h.
Samples were prepared by dilution of the stock solution with reaction buffer. After dilution, but before the addition of the enzyme, all samples were equilibrated for at least another 24 h. Five series of experiments were done. For the first series, ATP was added to solutions of 0.2, 0.4, 0.5, 0.7, 0.9, 1.0, and 1.2 g of DNA/L with a final concentration of 1 mM. These samples do not contain topoisomerase II. For the second series, topoisomerase II was added to solutions of 1.0 g of DNA/L in a ratio of four units per μg of DNA. The ATP concentrations are 0, 1.0, 2.5, or 4.0 mM. For the third series, topoisomerase II was added to solutions of 1.0 g of DNA/L in ratios of 1, 2, and 4 units per μg of DNA with an ATP concentration of 2.5 mM. For the fourth series, topoisomerase II was added to solutions of 0.5, 0.7, 0.9, 1.0, and 1.2 g of DNA/L in a ratio of 4 units per μg of DNA with ATP concentrations of 1.0, 1.0, 1.0, 2.5, and 4.0 mM, respectively. For the fifth series, topoisomerase II (4 units/μg) was added to solutions of 1.0 g of DNA/L including 0/2.5, 1.25/1.25, 0.2/2.3, and 2.5/0 mM AMP-PNP/ATP. We have also investigated a solution of 0.4 g of DNA/L with an AMP-PNP concentration of 2.5 mM. One unit of topoisomerase (20 ng) is the amount of enzyme required to fully relax 0.3 μg of negatively supercoiled pBR322 plasmid DNA in 15 min at 303 K under the standard assay conditions. With a molecular mass of 340 kDa, one unit per μg of λ-DNA corresponds with 1.9 dimers per DNA molecule. Just before the microrheology experiment, we added ∼2 μL of the enzyme in storage buffer to 10 μL of DNA in reaction buffer, followed by mixing the solution through gentle stirring and pipetting up and down. Shear was minimized by using pipette tips that have wide openings. All samples were spiked with polystyrene microspheres (Polysciences, Warrington, PA) of 1.83 ± 0.05 μm diameter with a final concentration of <0.1 wt %. A droplet of solution was deposited on a microscopy slide and sealed with a coverslip separated by a 0.12-mm spacer. All samples were assayed at least in duplicate. The blunting of the sticky ends was confirmed by the perfect agreement between the viscoelastic moduli obtained with 20% and 100-fold molar excess of the oligomer. Furthermore, neither degradation nor multimerization of hybridized λ-DNA was observed with field-inversion gel electrophoresis.
Microrheology
Particle tracking experiments were carried out at 310 K with a DM EP (Leica, Wetzlar, Germany) and an Eclipse 80i microscope (Nikon, Melville, NY) equipped with 50- and 100-times long-working-distance objectives. Each tracking experiment was started within 1 min after addition of the enzyme. To minimize hydrodynamic interactions, care was taken that the imaged beads are separated by at least 10 bead diameters (20 μm). Furthermore, the height level of the focal plane was adjusted so that it is situated right between slide and coverslip with maximum separation. Video was collected with a charge-coupled device camera (TK-C921EG; JVC Americas, Wayne, NJ) connected via an analog-to-digital video converter (ADVC55; Canopus, Kobe, Japan) to a computer. For each sample, at least one movie with a total duration exceeding 100 min was recorded with a rate of 25 frames/s and stored on a hard disk. Entangled solutions with topoisomerase II and AMP-PNP were investigated using a complementary metal oxide semiconductor camera (A504k; Basler, Ahrensburg, Germany) with a rate of 500 frames/s. The video was analyzed with MATLAB (The MathWorks, Natick, MA) using an avi file reader and the particle trajectories were obtained with public domain tracking software (http://physics.georgetown.edu/matlab/). To monitor the time evolution of the viscoelasticity after the addition of topoisomerase II, the trajectories of the probe beads were binned over intervals of various durations. All further data analysis was done with home-developed software scripts written in MATLAB code. The pixel resolutions in the x- and y-directions were calibrated with the help of a metric ruler. We have checked our setup by measuring the diffusion of colloidal beads dispersed in a concentrated solution of glycerol as well as by monitoring immobilized beads adsorbed at a glass slide.
Results and Discussion
Entanglements and reptation dynamics
Topoisomerase II has specific requirements for its functioning in terms of buffer composition and temperature. The reaction buffer contains a mixture of different salts as well as protein and the optimal temperature for the double-strand passage reaction is 310 K. Accordingly, we first explored the viscoelasticity of our model system and the critical concentration of DNA for the formation of entanglements in the relevant experimental conditions without the presence of the enzyme. For this purpose, we have measured the mean-square displacement 〈Δx2(t)〉 of the colloidal probe beads for a series of samples with increasing concentration of DNA in reaction buffer including 1.0 mM ATP. Some results are displayed in Fig. 2 A. The results for 0.5 and 1.2 g of DNA/L are not shown, but they fall perfectly between the marks set by the other solutions. Diffusive behavior with 〈Δx2〉 ∝ t is only observed for very long times and/or low DNA concentrations. With increasing concentration of DNA, 〈Δx2〉 decreases and the range of times with a subdiffusive scaling exponent <1 becomes wider. This behavior is typical for a viscoelastic fluid and is more conveniently discussed in terms of the elastic storage and viscous loss moduli, G′ and G″, respectively.
Figure 2.
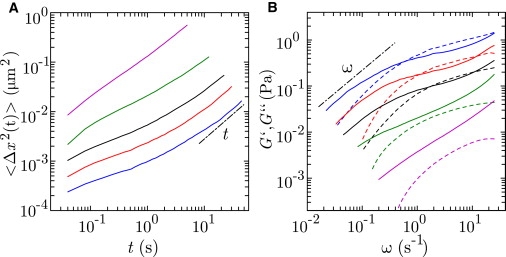
(A) Mean-square displacement 〈Δx2〉 versus time t in reaction buffer including 1.0 mM ATP at 310 K. The concentrations are 1.4 (blue), 1.0 (red), 0.7 (black), 0.4 (green), and 0.2 (magenta) g of DNA/L. (B) Elastic storage G′ (dashed) and viscous loss G″ (solid) moduli versus frequency ω. (Dashed-dotted lines in A and B) Scaling laws for the diffusive 〈Δx2〉 ∝ t and low shear G″ ∝ ω limits for long times and low frequencies, respectively.
Following the procedure as described in Zhu et al. (15), we have obtained G′(ω) and G″(ω) from the one-sided, complex Fourier transform of 〈Δx2(t)〉 and the generalized Stokes-Einstein equation. The formalism requires that the fluid can be treated as an incompressible continuum, with no-slip-boundary conditions, and that the Stokes drag can be extended over all frequencies (18). The typical mesh sizes or correlation lengths of our semidilute DNA solutions are less than 100 nm and are approximately of the DNA persistence length. The structures of the fluid, which give rise to the properties of the flow, are hence much smaller than the 1.8-μm-sized beads (19). Accordingly, the effects of a submicron scale variation in DNA density next to the colloidal beads on the viscoelastic response are vanishingly small and can be safely neglected (15,20). The viscoelastic moduli are displayed in Fig. 2 B. With increasing frequency, G″ first increases, then levels off to a certain extent depending on the concentration of DNA, and eventually increases again. Concurrently, G′ monotonously increases and eventually levels off at a constant high-frequency plateau value G. For a concentration exceeding 0.5 g of DNA/L, G′ becomes larger than G″ in an intermediate frequency range. The crossing of G′ and G″ and the development of an elastic plateau modulus are caused by the formation of entanglements (12,15). It should be noted that these entanglements are temporary and continuously disappear and reappear by the reptation dynamics of the DNA molecules (7–9). Furthermore, the critical concentration for the formation of entanglements, i.e., the entanglement concentration, is ∼10-times the overlap concentration from the dilute to the semidilute regime.
The formation of entanglements can also be inferred from the concentration scaling of the transport properties Δη, G, and τ. We obtained the low shear viscosity η from the behavior of G″ = ηω in the diffusive limit at low frequencies. The viscosity increments Δη = η – ηs, with respect to the viscosity of the buffer ηs = 0.0007 Pa s, are shown in Fig. 3 A. Note that the viscosity increment increases by two orders of magnitude if the DNA concentration is increased from 0.2 to 1.4 g/L. For concentrations exceeding 0.5 g/L, it follows a power law Δη ∝ c3.93 for entangled polyelectrolyte with screened electrostatics and Flory exponent ν = 0.588 (relevant scaling exponents are collected in Table 1). The high-frequency plateau value of the elasticity modulus is given by the density of dynamic units times the thermal energy (7–9). In the absence of entanglements, the dynamic units are the individual DNA molecules. The elasticity modulus G is then given by the Rouse modulus
with DNA density ρ (i.e., G/GR = 1). In the entangled network, the dynamic units are the DNA segments between entanglements (entanglement strands). Let Ne be the average number of entanglement points per molecule, so that the density of entanglement strands is given by [Ne + 1]ρ. For entangled DNA, the elasticity modulus hence reads
The results for G/GR are shown in Fig. 3 B. The experimental values can be considered under limits, because G′ might increase a bit for even higher frequencies outside our window of observation. At the lowest concentration of 0.2 g of DNA/L, G/GR is around unity in agreement with nonentangled dynamics. At higher concentrations, it follows the scaling law for an entangled, salted polyelectrolyte of G/GR ∝ c1.31. The longest, global relaxation time τ pertaining to the motion of the DNA molecule can be derived from the low shear viscosity and the high-frequency elasticity modulus according to Δη = π2/12 Gτ (8). For entangled solutions, τ can also be obtained from the lowest crossover frequency ωc = τ−1. As shown in Fig. 3 C, the values of τ obtained with both procedures agree within experimental accuracy. The relaxation time increases from 0.6 s for nonentangled DNA to ∼2 s for the sample with the highest concentration of DNA. With increasing concentration, τ first follows the scaling law for Rouse dynamics of a nonentangled and salted polyelectrolyte τ ∝ c0.31. We also observe the transition to reptation dynamics with τ ∝ c1.62 in the entangled regime for higher concentrations of DNA.
Figure 3.
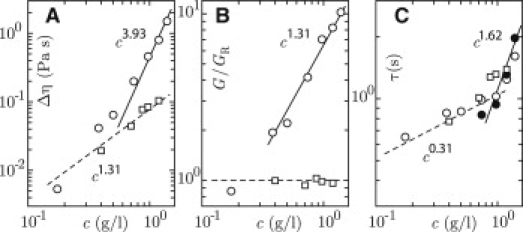
(A) Low shear viscosity increment Δη versus DNA concentration c. (B) High-frequency elasticity modulus divided by the Rouse modulus G/GR versus c. (C) Relaxation time τ versus c. (Open circles) Derived from Δη and G; (solid circles) ωc−1. (For panels A, B, and C: circles refer to solutions without enzyme, squares denote results obtained in the condition of full relaxation of entanglements, and dashed and solid lines represent scaling laws for nonentangled and entangled dynamics, respectively.)
Table 1.
Concentration scaling exponents for the viscosity increment Δη, high-frequency elasticity modulus G, and relaxation time τ (7, 9)
| Nonentangled | Entangled | |
|---|---|---|
| Δη | 1/(3ν – 1)∗ | 3/(3ν – 1) |
| G | 1 | 3ν/(3ν – 1) |
| τ | (2 – 3ν)/(3ν – 1) | (3 – 3ν)/(3ν – 1) |
ν denotes the Flory exponent (ν = 0.588).
In the relevant experimental conditions, our model system exhibits the transition from the nonentangled to the moderately entangled regime with ∼12 entanglements per DNA molecule at the highest concentration of 1.4 g of DNA/L. The entanglement concentration is in the range of previously reported values for λ-DNA in 10 mM tris-EDTA at 296 K (15,17). The increased ionic strength of the reaction buffer and the elevated temperature of 310 K result in a shift in entanglement concentration from 0.3 to 0.5 g of DNA/L, but no qualitative change in viscoelastic behavior. In the entangled regime, the concentration scaling of the transport properties also agrees with previously reported results. In the next section, we will demonstrate that the entanglements can be relaxed by topoisomerase II in conjunction with the consumption of ATP. In particular, it will be shown that Rouse dynamics with G/GR = 1 can be realized for relatively dense solutions of DNA.
Relaxation of entanglements
We have investigated the effect of topoisomerase II on the viscoelasticity of our model system in the presence of ATP. For this purpose, we have added topoisomerase II to initially entangled solutions and monitored the trajectories of the probe beads as a function of the time evolved after the addition of the enzyme. For most samples, we added four units (80 ng) of topoisomerase II per μg of DNA, which corresponds with approximately eight topoisomerase dimers per DNA molecule. The number of dimers is therefore of the same order of magnitude as the number of entanglements per molecule. To verify the effect of the concentration of topoisomerase II, we have also performed experiments with one and two units of topoisomerase II per μg of DNA. The employed ATP concentrations are 1.0, 2.5, and 4.0 mM. For full and relatively fast relaxation of entanglements in relatively concentrated solutions of 1.0 and 1.2 g of DNA/L, it was necessary to increase the ATP concentration to 2.5 and 4.0 mM, respectively. Furthermore, it was necessary to maintain the temperature of the assays at 310 K, because at ambient temperature no effect of the enzyme on the viscoelastic behavior was observed. The trajectories of the beads were binned into time intervals of various durations. We observed that the timescale of the change in viscoelasticity (minutes) is much longer than the one pertaining to the loss of correlation in the velocity of the beads (seconds). Within each interval, the system is in quasiequilibrium with Gaussian distributions in the displacements of the beads. Accordingly, for each interval 〈Δx2〉 and, subsequently, G′ and G″ were obtained as described in Zhu et al. (15). An example of the time evolution of G′ and G″ pertaining to a solution of 1.0 g of DNA/L is shown in Fig. 4. The solution is initially moderately entangled with approximately six entanglements per molecule. With increasing time, both G′ and G″ decrease until they no longer cross each other. This shows that the entanglements are relaxed and that the solution is transformed from an elastic physical gel to a viscous fluid. The time required to reach the fully relaxed state depends on the relative concentrations of ATP and topoisomerase II. For very long times, G′ and G″ increase again and eventually the original, entangled state is recovered.
Figure 4.
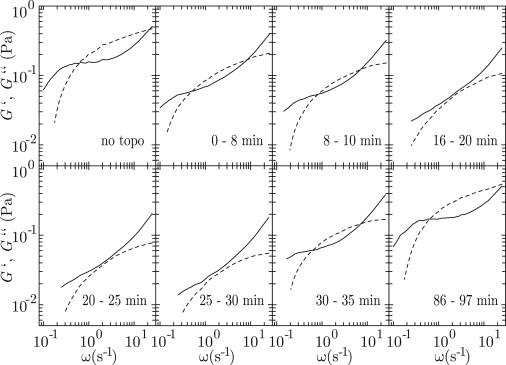
Time evolution of the elastic storage G′ (dashed curves) and viscous loss G″ (solid curves) moduli after the addition of topoisomerase II (4 units/μg) to an entangled solution of 1.0 g of DNA/L with an ATP concentration of 2.5 mM. The monitored time intervals are indicated in minutes.
Relaxation of entanglements is most conveniently followed by monitoring the high-frequency plateau value of the elastic storage modulus G. For a solution of 1.0 g of DNA/L, the time dependencies of G/GR after the addition of four units of topoisomerase II per μg of DNA (eight dimers per molecule) in the absence of ATP as well as in the presence of various concentrations of ATP are displayed in Fig. 5 A. The initial number of entanglements per molecule is ∼6. In the absence of ATP, there is clearly no activity of the enzyme and G/GR remains constant. With an initial ATP concentration of 1.0 mM, G/GR decreases, but only three out of six entanglements per molecule are relaxed. If the initial ATP concentration is increased to 2.5 mM, G/GR reduces to unity with full relaxation of entanglements within 20 min. However, for longer times G/GR increases again and eventually approaches the original value as observed before the addition of topoisomerase II. With an ATP concentration of 4.0 mM, almost all entanglements are relaxed within a similar time span followed by a faster recovery of the original, entangled state.
Figure 5.
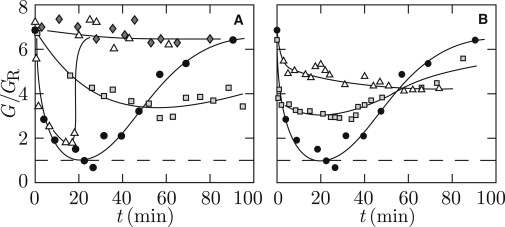
(A) High-frequency elasticity modulus divided by the Rouse modulus G/GR versus time t after the addition of topoisomerase II (4 units/μg) to a solution of 1.0 g of DNA/L without ATP (♢) or with ATP concentrations of 1.0 (□), 2.5 (●), or 4.0 (▵) mM. (B) As in panel A, but after the addition of 1 (▵), 2 (□), or 4 (●) units of topoisomerase II per μg of DNA to a solution of 1.0 g of DNA/L and 2.5 mM ATP. (For panels A and B: solid curves are guides to the eyes and dashed lines demarcate G/GR = 1, i.e., the nonentangled state.)
We have also performed experiments with topoisomerase II concentrations of one and two units per μg of DNA (two and four dimers per molecule, respectively) and an initial ATP concentration of 2.5 mM. From the behavior of G/GR shown in Fig. 5 B, it follows that approximately two and four entanglements are relaxed with one and two units of topoisomerase II per μg of DNA, respectively.
After the addition of the enzyme, there are two dynamical processes which occur simultaneously. The first process is the reptation dynamics of the DNA molecules. The characteristic timescale is the molecular relaxation time τ. A total number of entanglements per molecule Ne is renewed during the time τ (τ is also called the entanglement renewal time). The second process is the removal of entanglements by topoisomerase II, characterized by a disentanglement time τd. The disentanglement time τd includes the time for a protein dimer to find an entanglement, to bind ATP, to perform the actual double-strand passage reaction, and to disengage from the DNA molecule. Let there be Np dimers per molecule. Within time τ, the total number of disentanglements Nd executed by the enzyme is then given by Np τ/τd. Note that Nd changes over the duration of our experiment, because, among other issues, ATP is consumed. We can discern three cases:
Nd/Ne ≥ 1. All (Ne) entanglements are relaxed. The enzyme removes an entanglement at the moment it appears by reptation.
0.1 < Nd/Ne < 1. The number of entanglements which are removed by the enzyme is on the order of the number of entanglements which appear by reptation. A partial relaxation of entanglements is observed.
Nd/Ne < 0.1. Few or no entanglements are removed by the enzyme.
For full conversion of the solution from an elastic physical gel to a viscous fluid, all entanglements need to be relaxed within time τ (case i).
For a solution of 1 g of DNA/L, τ ≈ 1 s and Ne ≈ 6 (see Fig. 3). We made two key observations. First, with an initial concentration of 1 mM ATP and eight protein dimers per molecule (Np ≈ 8), we observed that three out of six entanglements are relaxed (Nd ≈ 3), so that τd ≈ 3 s. The disentanglement by the enzyme is too slow to relax all entanglements within time τ. Second, with an initial concentration of 2.5 mM ATP, all entanglements are relaxed with eight dimers per molecule. We observed, however, partial relaxation of entanglements in the cases of two and four dimers per molecule. For these subcritical topoisomerase concentrations, the number of disentanglements is approximately the same as the number of dimers per molecule, i.e., Nd ≈ Np. These results indicate that τd ≈ τ, so that τd ≈ 1 s. In this situation, a single dimer relaxes around one entanglement within the molecular relaxation time τ of 1 s. The disentanglement time is clearly dependent on the concentration of ATP, which shows that binding of ATP is the rate-limiting step in the present experimental conditions. The value of τd ≈ 1 s is in the range of previously reported values for the double-strand passage time 0.1–1 s in the context of supercoiling (21,22).
For longer times, the initial entangled state is recovered. It is our contention that this recovery is due to the consumption and eventually runs-out of ATP. Note that without topoisomerase II and/or ATP, the entangled state is established within a few seconds, because the entanglement renewal time is ∼1 s. An interesting question is whether there is enough ATP to sustain the reaction for the duration of the experiment (100 min). As an example, we consider the situation with an initial concentration of 1 mM ATP and eight dimers per DNA molecule. The number of disentanglements is Nd ≈ 3 and each disentanglement requires the hydrolysis of two ATP molecules. Accordingly, per DNA molecule, six ATP molecules are consumed during the molecular relaxation time τ of 1 s. For a concentration of 1 g of DNA/L, the reaction can proceed for 90 min. This duration is in reasonable agreement with the observation. It should be noted, however, that Nd is not constant; its value is less for shorter reaction times. Furthermore, the efficiency of the enzyme is not 100%; there can be closures of the protein dimer without transport and/or disentanglement event. Our estimation of the maximal duration is therefore an approximation. For higher concentrations of ATP, the reaction can run for similar durations. We observed, however, a faster recovery of the initial entangled state with increasing concentration of ATP. This observation can be rationalized in terms of the efficiency of ATP consumption. For the removal of supercoils by yeast topoisomerase II, it has been reported that the passage reaction becomes more efficient at low concentration of ATP (21). At high concentration of ATP, passage is fast due to rapid binding of ATP. However, more ATP is wasted by closures without capture and transport of a second DNA segment. The higher passage rate, in conjunction with a less ATP-efficient transport mechanism, results in rapid relaxation of entanglements, but also quicker run-out of ATP.
In the range of DNA concentrations used in this article, all entanglements can be relaxed with four units (80 ng) of topoisomerase II per μg of DNA (eight dimers per molecule) and a sufficiently high concentration of ATP. We have derived values of the transport properties Δη, G, and τ pertaining to the fully relaxed state as a function of the concentration of DNA. The results are also displayed in Fig. 3. The low shear viscosity increments Δη are significantly reduced with respect to the values in the absence of topoisomerase II. Furthermore, they follow the scaling law Δη ∝ c1.31 for a salted polyelectrolyte in the nonentangled regime (7–9). The ratio G/GR remains close to unity, because the entanglements are fully relaxed and the molecules move as single units. We do not observe significant changes in the molecular relaxation time τ, but they follow the relatively weak concentration dependence for Rouse dynamics τ ∝ c0.31. In the range of DNA concentrations presented in this article, the main effect of topoisomerase II on the viscoelasticity is therefore through a reduction in G rather than τ. To observe significant effects on τ, we surmise that our experiments need to be extended to higher concentrations of DNA and/or DNAs of larger molecular mass. The concentration scaling of the transport properties is in agreement with Rouse dynamics in the condition of full relaxation of entanglements by topoisomerase II.
Topoisomerase II inhibitor
To provide further proof that the control of the viscoelasticity of our model system requires the hydrolysis of ATP, we have investigated the effect of a generic topoisomerase II inhibitor. AMP-PNP is a β, γ -imido analog of ATP. It binds to topoisomerase II in the same fashion as ATP, but it cannot be hydrolyzed. Triggered by the binding of AMP-PNP, the protein dimer closes, cannot be reopened, and is converted to an annular form. The cross-sectional diameter of the hole of the closed dimer is sufficiently large for a DNA molecule to thread through (5,23,24). Because the double-strand passage activity is suppressed, we expect that entanglements are no longer relaxed. Furthermore, a cross-linking protein clamp between two DNA segments is formed, if a second segment has been captured before closure. Both the inhibition of the double-strand passage reaction and the formation of cross-linking clamps are expected to affect the viscoelasticity.
In the inhibition experiments, four units of topoisomerase II per μg of DNA (∼8 dimers per DNA molecule) were added to solutions of various DNA, ATP, and AMP-PNP concentrations. All assays were kept at 310 K. We first investigated nonentangled and entangled solutions of 0.4 and 1.0 g of DNA/L, respectively, with an initial AMP-PNP concentration of 2.5 mM. These samples do not contain ATP. Note that the experimental conditions are the same as those in the previous section, but with the 100% replacement of ATP by AMP-PNP. The time evolutions of G′ and G″ after the addition of the enzyme are displayed in Fig. 6. There is no qualitative change in the behavior of G′ and G″. In particular, we do not observe a transition to an elastic gel with fixed cross-links as characterized by parallel frequency scaling of G′ and G″ and G′ > G″ for all frequencies (6). The initially nonentangled solution of 0.4 g of DNA/L remains a viscous fluid with G′ < G″. In the case of the entangled solution of 1.0 g of DNA/L, fluidlike behavior is observed at low frequencies and corresponding long times.
Figure 6.
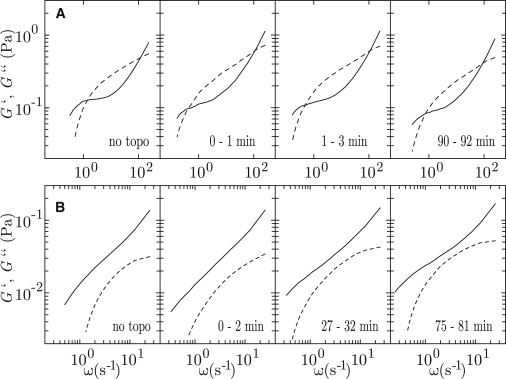
(A) Time evolution of the elastic storage G′ (dashed curves) and viscous loss G″ (solid curves) moduli after the addition of topoisomerase II (4 units/μg) to a solution of 1.0 g of DNA/L with an AMP-PNP concentration of 2.5 mM. (B) As in panel A, but for a nonentangled solution of 0.4 g of DNA/L.
The effects of the inhibitor on the elasticity modulus are most conveniently probed by monitoring G. For a series of entangled solutions of 1.0 g of DNA/L, the initial concentration of AMP-PNP was increased from 0 to 2.5 mM with a fixed 2.5 mM total concentration of ATP and AMP-PNP. The results are displayed in Fig. 7. In 2.5 mM AMP-PNP, G/GR is seen to increase immediately after the addition of the enzyme. This situation will be discussed below. In the presence of a mixture of ATP and AMP-PNP, G/GR initially decreases followed by a slow recovery to the original, entangled state. With a small amount of inhibitor, some, but not all entanglements are relaxed. For instance, in the case of initial concentrations of 0.2 mM AMP-PNP and 2.3 mM ATP, G/GR drops from ∼7 to 4, which corresponds with the relaxation of three out of six entanglements. With equal concentrations of ATP and AMP-PNP (1.25 mM each), only two out of six entanglements are relaxed. It should be noted that in the presence of ATP, each dimer can catalyze a number of transport events before it is transformed to an annular form by binding of AMP-PNP. Hence, the double-strand passage reaction gets progressively blocked, so that increasingly fewer entanglements are relaxed.
Figure 7.
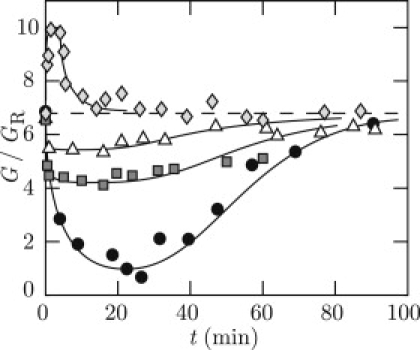
High-frequency elasticity modulus divided by the Rouse modulus G/GR versus time t after the addition of topoisomerase II (4 units/μg) to a solution of 1.0 g of DNA/L with ATP/AMP-PNP concentrations of 0/2.5 (♢), 1.25/1.25 (▵), 2.3/0.2 (□), and 2.5/0 (●) mM. (Dashed line) Initial value of G/GR.
With a total replacement of ATP by AMP-PNP, the protein can close and form a clamp only once. In the case of 2.5 mM AMP-PNP, G/GR is seen to increase followed by a decrease toward the original, entangled state. The initial increase implies that the number of constraints per molecule are enhanced rather than reduced, albeit for a short time on the order of a few minutes after the addition of the enzyme. The temporary increase in G/GR can be rationalized in terms of cross-linking protein clamps. The clamps act as dynamic constraints, similar to entanglements. However, the captured DNA segment can thread through the hole of the clamp and will eventually be released, and the probability for recapturing of another DNA molecule by a closed clamp is small (but not impossible, see (23)). The constraints formed by the protein clamps are thus short-lived with a lifetime on the order of the DNA relaxation time and they are not (or only inefficiently) renewed by reptation. As a result of clamp release by the threading motion of the linear DNA molecules, there is no transition to an elastic gel with fixed cross-links and the original, entangled state is recovered for longer times.
Conclusions
In consequence of the double-strand passage facilitated by topoisomerase II, semidilute entangled solutions of DNA are transformed from elastic physical gels to viscous fluids. This transformation has been inferred from a change in the relative frequency dependencies of the viscoelastic moduli as well as the concentration scaling of the derived transport properties. The main effect of topoisomerase II on the viscoelasticity is through the reduction in high-frequency plateau value of the elastic storage modulus rather than a change in global, longest relaxation time of the DNA molecules. In the case of semidilute nonentangled solutions, there is no significant effect on the viscoelastic response. An important feature is that the change in flow properties requires the dissipation of energy through the hydrolysis of ATP. For full relaxation of the entanglements, the initial concentration of ATP has to be sufficiently high. This shows that binding of ATP to the enzyme is the rate-limiting step. At a too-high concentration of ATP, the transport mechanism becomes less efficient in terms of consumption of ATP due to closures of the protein dimer without capture and transport of a second segment. Furthermore, for full conversion to a viscous fluid, we observed that the number of dimers has to be the same or exceed the number of entanglements per molecule. This implies that each entanglement is relaxed by a single dimer with a disentanglement time of ∼1 s and within the 1-s timescale for renewal of the entanglements by reptation. The temperature of the assays needs to be maintained at 310 K; at ambient temperature, no disentanglement by the enzyme has been observed.
A second aspect of this study is the effect of the generic topoisomerase II inhibitor AMP-PNP. In mixtures of AMP-PNP and ATP, the double-strand passage reaction gets blocked and progressively fewer entanglements are relaxed. A significant fraction of cross-linking protein clamps is formed in the condition of a total replacement of ATP by AMP-PNP. The resulting constraints are, however, dynamic and short-lived, due to the threading motion of the linear DNA molecule through the hole of the clamp and the fact that they are not (or inefficiently) renewed by reptation. Accordingly, we do not observe a transition to an elastic gel with fixed cross-links. The nonentangled solutions remain viscous fluids, and for the entangled solutions, fluidlike behavior is observed at low frequencies and corresponding longer times. For linear DNA, the main effect of the inhibitor is the blocking of the double-strand passage reaction, so that entanglements are no longer relaxed. The formation of cross-linking clamps between DNA segments is of secondary importance. This situation might be different for closed circular DNA, because a captured circle cannot be released from the clamp by threading motion (25).
Topoisomerases are mainly known for their ability to change the topological state of closed circular or looped DNA (4). We have shown that topoisomerase II can also disentangle linear, nonsupercoiled molecules with a concurrent change in the properties of the flow. We have presented conclusive evidence that the disentanglement is powered by the hydrolysis of ATP and, conversely, that it is inhibited by binding of AMP-PNP. This was made possible through microrheology. As in the case of simplification of the topology of closed circular DNA, topoisomerase II disentangles and does not tangle linear DNA. To explain disentangling, it has been proposed that the enzyme acts at a hooked juxtaposition of two DNA strands (26). Human topoisomerase IIα relaxes positively supercoiled plasmids faster than negatively supercoiled molecules (27). This selectivity is likely due to differences in processivity associated with protein binding, as observed for bacterial topoisomerase IV (28). Chiral discrimination may also result in faster removal of entanglements, if there is a preferred crossing angle of <90°. This could be investigated with topoisomerases that have no preference for positive supercoiling (e.g., human topoisomerase IIβ). The enzyme may find the randomly distributed entanglements by diffusion, possibly directed along the contour of the DNA molecule. Alternatively, an entanglement may slide along the contour by reptation until it is captured by a sharp bend induced by a DNA-bound topoisomerase (29). Disentanglement is subsequently accomplished by the catalyzed double-strand passage reaction. An important feature of the latter model is that unidirectional double-strand passage follows from the specific orientation of the enzyme with respect to the bend. Furthermore, the sliding mechanism requires the preexistence of entanglements, which naturally explains the disentangling activity of the enzyme.
Acknowledgments
We thank Jean-Louis Sikorav, Bertrand Duplantier, and Rudi Podgornik for discussions and critical reading of the manuscript. Jean-Louis Sikorav and Arach Goldar are acknowledged for gel electrophoresis.
This work was supported by grant No. R-144-000-256-112 from the National University of Singapore.
References
- 1.Sikorav J.-L., Jannink G. Kinetics of chromosome condensation in the presence of topoisomerases: a phantom chain model. Biophys. J. 1994;66:827–837. doi: 10.1016/s0006-3495(94)80859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duplantier B., Jannink G., Sikorav J.-L. Anaphase chromatid motion: involvement of type II DNA topoisomerases. Biophys. J. 1995;69:1596–1605. doi: 10.1016/S0006-3495(95)80032-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nitiss J.L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J.C. Cold Spring Harbor Laboratory Press; Woodbury, NY: 2009. Untangling the Double Helix: DNA Entanglement and the Action of the DNA Topoisomerases. [Google Scholar]
- 5.Schoeffler A.J., Berger J.M. Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochem. Soc. Trans. 2005;33:1465–1470. doi: 10.1042/BST0331465. [DOI] [PubMed] [Google Scholar]
- 6.Larson R.G. Oxford University Press; Oxford, UK: 1998. The Structure and Rheology of Complex Fluids. [Google Scholar]
- 7.de Gennes P.-G. Cornell University Press; Ithaca, NY: 1979. Scaling Concepts in Polymer Physics. [Google Scholar]
- 8.Doi M., Edwards S.F. Oxford University Press; Oxford, UK: 1986. The Theory of Polymer Dynamics. [Google Scholar]
- 9.van der Maarel J.R.C. World Scientific; Singapore: 2008. Introduction to Biopolymer Physics. [Google Scholar]
- 10.Sikorav J.-L., Jannink G. Dynamics of entangled DNA molecules in the presence of topoisomerases. C. R. Acad. Sci. IIb Mech. Phys. Chim. Astron. 1993;316:751–757. [Google Scholar]
- 11.Musti R., Sikorav J.-L., Adam M. Viscoelastic properties of entangled DNA solutions. C. R. Acad. Sci. IIb Mech. Phys. Chim. Astron. 1995;320:599–605. [Google Scholar]
- 12.Mason T.G., Dhople A., Wirtz D. Linear viscoelastic moduli of concentrated DNA solutions. Macromolecules. 1998;31:3600–3603. [Google Scholar]
- 13.Jary D., Sikorav J.-L., Lairiez D. Nonlinear viscoelasticity of entangled DNA molecules. Europhys. Lett. 1999;46:251–255. [Google Scholar]
- 14.Heo Y., Larson R.G. The scaling of zero-shear viscosities of semidilute polymer solutions with concentration. J. Rheol. (NY) 2005;49:1117–1128. [Google Scholar]
- 15.Zhu X., Kundukad B., van der Maarel J.R.C. Viscoelasticity of entangled λ-phage DNA solutions. J. Chem. Phys. 2008;129:185103. doi: 10.1063/1.3009249. [DOI] [PubMed] [Google Scholar]
- 16.Mason T.G., Dhople A., Wirtz D. Concentrated DNA rheology and microrheology. Mat. Res. Soc. Symp. Proc. 1997;463:153–158. [Google Scholar]
- 17.Smith D.E., Perkins T.T., Chu S. Self-diffusion of an entangled DNA molecule by reptation. Phys. Rev. Lett. 1995;75:4146–4149. doi: 10.1103/PhysRevLett.75.4146. [DOI] [PubMed] [Google Scholar]
- 18.Mason T.G. Estimating the viscoelastic moduli of complex fluids using the generalized Stokes-Einstein equation. Rheol. Acta. 2000;39:371–378. [Google Scholar]
- 19.Verma R., Crocker J.C., Yodh A.G. Entropic colloidal interactions in concentrated DNA solutions. Phys. Rev. Lett. 1998;81:4004–4007. [Google Scholar]
- 20.Chen D.T., Weeks E.R., Yodh A.G. Rheological microscopy: local mechanical properties from microrheology. Phys. Rev. Lett. 2003;90:108301. doi: 10.1103/PhysRevLett.90.108301. [DOI] [PubMed] [Google Scholar]
- 21.Lindsley J.E., Wang J.C. On the coupling between ATP usage and DNA transport by yeast DNA topoisomerase II. J. Biol. Chem. 1993;268:8096–8104. [PubMed] [Google Scholar]
- 22.Baird C.L., Harkins T.T., Lindsley J.E. Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc. Natl. Acad. Sci. USA. 1999;96:13685–13690. doi: 10.1073/pnas.96.24.13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roca J., Wang J.C. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell. 1992;71:833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 24.Wang J.C. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 25.Roca J., Wang J.C. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell. 1994;77:609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 26.Buck G.R., Zechiedrich E.L. DNA disentangling by type-2 topoisomerases. J. Mol. Biol. 2004;340:933–939. doi: 10.1016/j.jmb.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 27.McClendon A.K., Rodriguez A.C., Osheroff N. Human topoisomerase IIα rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J. Biol. Chem. 2005;280:39337–39345. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 28.Neuman K.C., Charvin G., Croquette V. Mechanisms of chiral discrimination by topoisomerase IV. Proc. Natl. Acad. Sci. USA. 2009;106:6986–6991. doi: 10.1073/pnas.0900574106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vologodskii A.V., Zhang W., Cozzarelli N.R. Mechanism of topology simplification by type II DNA topoisomerases. Proc. Natl. Acad. Sci. USA. 2001;98:3045–3049. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]


