Abstract
The prasinophyte order Mamiellales contains several widespread marine picophytoplankton (≤2 μm diameter) taxa, including Micromonas and Ostreococcus. Complete genome sequences are available for two Micromonas isolates, CCMP1545 and RCC299. We performed in silico analyses of nitrogen transporters and related assimilation genes in CCMP1545 and RCC299 and compared these with other green lineage organisms as well as Chromalveolata, fungi, bacteria, and archaea. Phylogenetic reconstructions of ammonium transporter (AMT) genes revealed divergent types contained within each Mamiellales genome. Some were affiliated with plant and green algal AMT1 genes and others with bacterial AMT2 genes. Land plant AMT2 genes were phylogenetically closer to archaeal transporters than to Mamiellales AMT2 genes. The Mamiellales represent the first green algal genomes to harbor AMT2 genes, which are not found in Chlorella and Chlamydomonas or the chromalveolate algae analyzed but are present in oomycetes. Fewer nitrate transporter (NRT) than AMT genes were identified in the Mamiellales. NRT1 was found in all but CCMP1545 and showed highest similarity to Mamiellales and proteobacterial NRTs. NRT2 genes formed a bootstrap-supported clade basal to other green lineage organisms. Several nitrogen-related genes were colocated, forming a nitrogen gene cluster. Overall, RCC299 showed the most divergent suite of nitrogen transporters within the various Mamiellales genomes, and we developed TaqMan quantitative polymerase chain reaction primer–probes targeting a subset of these, as well as housekeeping genes, in RCC299. All those investigated showed expression either under standard growth conditions or under nitrogen depletion. Like other recent publications, our findings show a higher degree of “mixed lineage gene affiliations” among eukaryotes than anticipated, and even the most phylogenetically anomalous versions appear to be functional. Nitrogen is often considered a regulating factor for phytoplankton populations. This study provides a springboard for exploring the use and functional diversification of inorganic nitrogen transporters and related genes in eukaryotic phytoplankton.
Keywords: nitrogen, Micromonas, Ostreococcus, green algae, green lineage, quantitative PCR, genomics, phylogeny
Introduction
Marine eukaryotic phytoplankton are phylogenetically diverse, resulting from multiple different evolutionary histories (Bhattacharya and Medlin 1998; Lane and Archibald 2008; Worden and Not 2008). Prasinophytes are thought to retain genetic information about the ancestral alga that gave rise to the extant green lineage, including land plants and green algae (Worden et al. 2009). Along with red algae, these organisms are derived from primary endosymbiosis and belong to the eukaryotic supergroup Archaeplastida (Lane and Archibald 2008). The Mamiellales is a geographically widespread order within the prasinophytes that thrives from tropical to polar waters. Three of the five genera composing the Mamiellales are very small unicellular algae, or “picophytoplankton” (≤2–3 μm diameter), specifically Bathycoccus, Micromonas, and Ostreococcus (Worden 2006; Viprey et al. 2008). In contrast, algae belonging to the Chromalveolata supergroup tend to be larger and result from secondary (or sometimes tertiary) endosymbioses (Lane and Archibald 2008). The chromalveolates include well-known episodic bloomers, such as diatoms, dinoflagellates, and coccolithophores. Gene families shared among these algae provide insights into shared ancestral characteristics, transfer events, growth regulation, and potentially niche differentiation (see, e.g., Delwiche 1999; Keeling and Palmer 2008; Worden et al. 2009).
Nitrogen is often considered a limiting resource for the phytoplankton responsible for photosynthetic marine primary production, particularly in the open ocean (Falkowski 1997). Concentrations of different nitrogen species are highest coastally but vary temporally in both coastal and open waters (Plant et al. 2009). In surface waters, variation occurs in both horizontal (coast to open ocean) and vertical dimensions (depth in the euphotic zone). Organism distributions can track these vertical gradients. For example, in the cyanobacterial photoautotroph Prochlorococcus, the genetic capacity to utilize different nitrogen sources along this gradient has been linked to speciation (Kettler et al. 2007). Fewer comprehensive studies are available on genes involved in uptake and assimilation of ammonium (and nitrate) in marine eukaryotic phytoplankton. Gene expression analyses have been performed on chromalveolates such as the pelagophyte Aureococcus anophagefferens (Berg et al. 2008) and the diatom Cylindrotheca fusiformis (Hildebrand 2005; Allen et al. 2006). With respect to the Mamiellales, little is known apart from Derelle et al. (2006) noting that Ostreococcus tauri had four ammonium transporters (AMTs), of which two seemed prokaryote-like and two green lineage related.
For photosynthetic eukaryotes in general, intracellular-reduced nitrogen in the form of ammonium is required for amino acid synthesis via the glutamine synthatase–glutamate synthase cycle (GS-GOGAT) (Mariscal et al. 2004). Nitrogen must pass two barriers before protein synthesis can occur: the plasma membrane and the chloroplast membrane. In model system land plants, such as Arabidopsis thaliana, and the model green alga Chlamydomonas reinhardtii, ammonium is taken up directly by AMTs; alternatively, nitrate transporters (NRT, also NAR) in the plasma membrane can deliver nitrate to the cytosol where it is catalytically reduced to nitrite by nitrate reductase (NIA) (Glass et al. 2002; Okamoto et al. 2006; Fernandez and Galvan 2007; Ludewig et al. 2007). Plastid-targeted nitrite transporters (NAR1) then transport nitrite into the chloroplast where it is reduced to ammonium by nitrite reductase (NII) and then synthesized into amino acids (Rexach et al. 2000). Genes-encoding aspects of these pathways are often present in multiple copies in plants and Chlamydomonas, particularly those encoding AMT and NRT transporters. In addition, low- and high-affinity transport systems have been identified for both ammonium and nitrate (Lea et al. 1992; Crawford and Glass 1998). The low-affinity transporter system (LATS) is responsible for uptake when substrate is plentiful externally, whereas the high-affinity transporter system (HATS) scavenges when substrate concentrations are low. In some cases, these transporters form phylogenetically distinct groups within a single organism (Couturier et al. 2007). Although functional assignment as a LATS or HATS requires experimental work, phylogenetically distinct copies have been shown to play either different functional roles in terms of time or location of expression or represent transporters with different substrate affinities.
Two phylogenetically unrelated NRT types are found across many lineages, including plants, both belonging to the largest family of secondary transport carriers, the major facilitator superfamily (MFS; Saier et al. 1999). Nitrate permease (typically referred to as NRT1) belongs to the MFS proton-dependent oligopeptide Transporter (POT) family (Liu and Tsay 2003). Most NRT1 genes appear to be LATSs, although some transport basic amino acids as well (Zhou et al. 1998; Rexach et al. 2000; Liu and Tsay 2003). In Arabidopsis, phosphorylation shifts the NRT gene CHL1 (also known as AtNRT1) from performing as a LATS to a HATS capable of taking up nitrate at low concentrations (Liu and Tsay 2003). NRT2 and its homologs comprise the nitrate–nitrite porter family found in eukaryotes as well as cyanobacteria and heterotrophic bacteria. In plants, NRT2 genes have been shown to encode HATSs and can be tissue specific (Okamoto et al. 2006).
The presence of two AMT types, AMT1 and AMT2, is well established in plants (Simon-Rosin et al. 2003; Loque and von Wiren 2004; Ludewig et al. 2007). The former represents the plant AMT family, whereas the latter are paralogs of bacterial and archaeal AMT genes as well as methylammonium/ammonium permeases (MEPs) in yeast. Several AMTs also transport methylammonium in plants (Shelden et al. 2001) and bacteria (Soupene et al. 1998). Plant AMTs can be expressed at different levels depending on the tissue type and can have different substrate affinities. For example in Poplar, AMT2.1 is highly expressed in leaves, whereas AMT2.2 is primarily expressed in petioles (Couturier et al. 2007). Furthermore, in Oryza sativa and Lotus japonicas, AMT2.1 genes are expressed constitutively, irrespective of the concentration of inorganic nitrogen in the case of the former (Simon-Rosin et al. 2003; Suenaga et al. 2003). Unlike plants, green algae such as Chlamydomonas have only been shown to contain AMT1 genes (Gonzalez-Ballester et al. 2004).
Here, genomes from two Micromonas species, CCMP1545 and RCC299, were analyzed for nitrogen transport and assimilation genes. The complete genomes of Micromonas (Worden et al. 2009) are less reduced in genome size and gene content than those of Ostreococcus (Derelle et al. 2006; Palenik et al. 2007). Micromonas has a broader geographical range than Ostreococcus, extending into subpolar and polar regions. We performed phylogenetic reconstructions based on NRT and AMT genes identified in silico to explore diversification and lineage affiliations of these genes. Our analyses include phylogenetic assessment of AMTs from a number of recently sequenced genomes, including the “lower” land plant Physcomitrella patens (moss) and the oomycete Phytophthora (water molds) as well as bacteria and archaea. For a subset of genes, genomes from two Ostreococcus and other green algae as well as a number of marine chromalveolate algae, including Emiliania huxleyi, A. anophagefferens and two diatoms were investigated. Finally, to facilitate future work on these physiologically important genes, quantitative polymerase chain reaction (qPCR) primers and probes were developed and validated for Micromonas sp. RCC299 in a nitrogen-depletion time course.
Methods
Gene Finding
In order to search for genes involved in nitrogen acquisition and assimilation, BlastP (Altschul et al. 1997) or TBlastN (Gertz et al. 2006) were performed against predicted protein sequences of the masked Mamiellales genomes respectively, on the US Department of Energy’s Joint Genome Institute (JGI) browser system. Genome versions were as follows: Micromonas pusilla CCMP1545 (v2.0), Micromonas sp. RCC299 (v3.0), O. tauri OTH95 (v2.0), and O. lucimarinus CCE9901 (v2.0). Known plant and bacterial protein sequences were used as queries. Searches for relevant Pfams (Finn et al. 2008) were also performed, specifically PF00909.12 AMT family, PF04898.5 glutamate synthase central domain, PF01645.8 conserved region in glutamate synthase, and PF00733.12 asparagine synthase, using Pfam annotations provided within the JGI genome browsers for each Micromonas. Micromonas gene models (Worden et al. 2009) were manually modified when computational predictions (e.g., of intron structure) appeared flawed and expressed sequence tags (ESTs) provided evidence to support the modification. Nevertheless, not all genes bore EST support, and manual modifications of 5′ and 3′ ends were then made for coding regions as possible based on homology. Predictions for subcellular targeting of these protein sequences were made by TargetP and ChloroP (Emanuelsson et al. 2000) available at http://www.cbs.dtu.dk/services as well as by Predotar and WoLF PSORT (Small et al. 2004; Horton et al. 2007).
Phylogenetic Analyses
Genes were selected from the GenBank nonredundant (nr) database for most organisms included in the NRT2 phylogeny. For the diatoms Thalassiosira pseudonana CCMP1335 (v3.0) and Phaeodactylum tricornutum CCAP1055/1 (v2.0), gene sequences were identified in the sequenced genomes (Bowler et al. 2008) using methods as above (for the Mamiellales). The NRT2 alignment was constructed using ClustalW (Thompson et al. 1994). Phylogenetic analyses used only the central region of the amino acid sequences, excluding the poorly aligned C and N termini from the alignment of 66 gene sequences, and was conducted using MrBayes 3.1 (Ronquist and Huelsenbeck 2003). For Bayesian inference trees, 5,000,000 generations and a model of evolution (RtREV + I + G + F, where G = 1.366 and I = 0.026 and F indicating the amino acid frequencies observed in the alignment should be used) selected by ProtTest (Abascal et al. 2005) were used. Neighbor Joining (NJ) and maximum parsimony (MP) trees were constructed using PAUP* (phylogenetic analysis using parsimony and other methods; Swofford 2002). The MP tree was constructed using Tree Bisection-Reconnection branch swapping after an initial tree was constructed by stepwise addition. All characters were unordered and given equal weight. A strict consensus tree was then computed from the best trees. NJ trees were constructed using distance settings calculated by Modeltest (Posada and Crandall 1998) where ties (if encountered) were broken systematically, and the distance measure was the mean character difference. Bootstrap resampling was performed for both the MP and the NJ trees using 1,000 replicates.
To ensure balanced analyses of the seemingly diverse Micromonas AMT genes, almost all eukaryotic sequences used in the general (AMT1 and AMT2 together) phylogeny were taken from sequenced genomes. This ensured that the complete suite present in other eukaryotes could be analyzed, providing more appropriate context for evaluating those in the Micromonas genomes. It also allowed selection of optimal gene models or correction of gene models as needed, generally involving extension at 5′ and 3′ regions of gene models based on available EST data. Thus, some gene sequences came directly from GenBank, whereas others were retrieved using BlastP or TBlastN against publically available genome browsers. All AMT genes were retrieved from the genomes of Candidatus Pelagibacter ubique HTCC1062, Roseobacter sp. SK209-2-6, Cyanidioschyzon merolae, as well as the following genomes sequenced at JGI: the four Mamiellales and two diatoms as above, the green alga C. reinhardtii (v4.0), the haptophyte E. huxleyi (v1.0), the pelagophyte A. anophagefferens (v1.0), the oomycetes Phytophthora ramorum (v1.1) and P. sojae (v1.1), the fungus Aspergillus niger (v3), the Poplar tree Populus trichocarpa (v1.1), and the moss Physcomitrella patens (v1.1). The general AMT alignment was performed with MUSCLE (Edgar 2004). Overall, 148 sequences were used, including fungal, bacterial, and archaeal AMTs as well as those of Arabidopsis and Populus, for the latter named as in a previous publication (Couturier et al. 2007), but with selection of alternative gene models that included 5′ and 3′ ends (to include start and stop codons). AMT alignments were manually adjusted in BioEdit and masked to exclude positions that were either ambiguously aligned or nonhomologous. An AMT2 (only) tree was built to further explore observed relationships using a greater number of bacterial taxa retrieved by blasting (BlastP) plant AMT2 genes to GenBank nr (July 2009). All plant sequences were retrieved directly from GenBank as opposed to the remodeling efforts used for some models in the general AMT tree. Initial alignments for the AMT2 tree in MUSCLE and ClustalW had different weaknesses and strengths, and the ClustalW alignment was selected. Sequences appearing too short (all from plants) due to gene modeling issues were excluded and the alignment rerun. The alignment was then manually adjusted in BioEdit, and only unambiguously aligned positions were used in phylogenetic analyses. NJ distance trees were constructed using Phylip (Felsenstien 2005), whereas maximum likelihood (ML) was performed using PhyML (Guindon and Gascuel 2003) with 100 replicates. Results from ML trees are presented. Motifs were generated for a conserved core region of the AMT protein-encoding sequences using an online server (http://weblogo.berkeley.edu/).
Gene Naming Conventions
Names assigned herein consider the relative phylogenetic position within an organism, or sometimes sistering relationships, although we did not arrive upon a truly logical system for comparison with other organisms. For both AMT and NRT2 gene sets, we used a decimal for multiple copies, although in the AMT literature in particular, use of a semicolon is a relatively common convention for multiple copies. Fungal sequences were not named as future studies will allow a more coherent integration with naming already accepted for other fungal species. Oomycete MEP sequences are named MEP purely based on their relatedness to fungal sequences, not experimental work, and lettering does not relate to that for fungi.
Cell Culture and Nitrogen-Depletion Experiments
Micromonas sp. RCC299 was grown axenically in triplicate 50 ml volumes for 10 generations in mid-exponential growth phase at 200 μEs−1m−2 and 21 °C on a diel cycle (14L:10D). Culture volumes were gradually increased to 1 l prior to initiation of a nitrogen depletion time course. Artificial seawater-based Keller (K) media was used (see http://www.mbari.org/phyto-genome/Resources.html), a common growth media that has among other constituents, 882 μM NaNO3, and 50 μM NH4Cl (final concentrations). Cultures were monitored daily starting between 9 and 10 AM (8 AM being the onset of light), before and after transfer, using flow cytometry (Coulter Epics XL). Fluoresbrite plain YG 0.75 μm diameter beads (Polysciences Inc.) were added prior to analysis for normalization purposes. The depletion time course was initiated by transferring cells into artificial seawater with all K constituents except NH4Cl and NaNO3 (herein termed “N-deplete K media”). At T0, prior to transfer, three samples for RNA extraction (50 ml each) were harvested from each replicate at 6000 × g for 20 min at 21 °C. The supernatant was removed rapidly and cell pellets were immediately frozen and stored at –80 °C until RNA extraction. In addition, three samples (5 ml each) per treatment replicate were taken for nitrate analysis using a diode array spectrophotometer (Hewlett Packard), akin to methods in a previous study (Johnson and Coletti 2002). The spectrophotometer was blanked using 18.2 MΩ H2O and calibrated using NaNO3 standards. In addition, three 5 ml samples from each replicate were analyzed for NH4+ using a conductometric technique (Plant et al. 2009). Reported nitrogen measurements represent the average and standard deviation of the biological triplicates, after evaluation of technical replicates from each flask. One milliliter samples were taken for flow cytometric analyses. The remaining culture volume (between 300 and 350 ml depending on the replicate) was then transferred into between 650 and 750 ml N-deplete K media, to bring each replicate to approximately 2,000,000 cells ml−1, and incubated in the same light and temperature conditions as above in triplicate 2-l polycarbonate bottles. At subsequent time points, cells were diluted (every 24 h) with a volume of N-deplete K media to attain 2,000,000 cells ml−1 (as had been done leading up to the time course in standard K media). Prior to transfer each day, samples were taken as outlined above for RNA extraction and nitrogen measurements. At T96, the experiment triplicates were amended with 882 and 50 μM (final concentrations) NaNO3 and NH4Cl, respectively. At T144, cells were transferred into standard K media.
RNA Extraction, cDNA Synthesis, and Gene Expression Analysis
RNA extraction was performed using the MagMAX-96 total RNA isolation kit (Ambion) following the manufacturer’s protocol. A post-extraction DNase digest was performed at 37 °C for 1 h using the TURBO DNA-free Kit (Ambion). Single-stranded cDNA was made using 10 μl of RNA (3 ng μl−1) and the Applied Biosystems (AB) High Capacity cDNA Reverse Transcription Kit, which uses random primers, following the manufacturer’s protocol. Reverse transcription controls were performed (in which template is included but no reverse transcriptase) and were negative, indicating that there was no genomic contamination. To enable future experimentation on differential expression of NRT and AMT transporters identified in Micromonas sp. RCC299, TaqMan primer and probe sets for six target genes and four commonly used endogenous control genes were designed. qPCR was performed using a 7500 Real Time PCR System (AB) in MicroAmp Optical 96-well plates with 20 μl reaction volumes consisting of 1 × TaqMan Gene Expression Master Mix (AB), 900 nM of each primer, 250 nM probe (final concentrations), and 6 ng of cDNA giving a final concentration of 0.3 ng cDNA μl−1. Cycling parameters were one cycle of 50 °C for 2 min, one cycle of 95 °C for 10 min, and 40 cycles of 95 °C for 15 s followed by 60 °C for 1 min. Primer–probe sets were verified first by running the resulting qPCR fragments on a 2% agarose gel with a 25-bp ladder (Invitrogen) to check fragment lengths Met size predictions. In addition, qPCR products were purified using a Nucleospin Extract II kit (E & K Scientific), sequenced on a 3100 Genetic Analyzer sequencer (AB), and confirmed to be from the appropriate target. The AMT2.3 fragment was cloned using the TOPO-TA Kit (Invitrogen), as direct sequencing was unsuccessful. Twenty clones were picked and sequenced.
For testing the four candidate endogenous control genes (Actin, GAPDH, 18S rDNA, and Ubiquitin), material was grown for 10 generations in mid-exponential growth phase at 200 μE s−1m−2 and 21 °C in three different media preparations all based on K media (standard K media, K media lacking NO3–, and K media lacking NH4+). Material was also used from cells kept in N-deplete K media for 5 days. The difference in threshold cycle (CT) between the four conditions was assessed using qPCR. GAPDH was used for standardizing experimental data from target genes because it showed the least change in CT between the different cell culturing conditions. Because the 18S ribosomal RNA gene sequence is not available on the JGI browser system, it was deposited under the accession HM191693.
The linear dynamic range for each primer–probe set was calculated using cDNA generated from serially diluted RNA from cells grown in K media as above and maintained in mid-exponential growth phase. For depletion experiment gene expression analyses, a RNA dilution for cDNA generation was used at which the CT for each primer–probe set was within the linear region of the curve representing a 1:1 conversion between RNA and cDNA. The efficiency of each primer–probe set was also tested using a cDNA dilution series to determine whether the sets could be assessed using the 2–ΔΔCT method (Livak and Schmittgen 2001). Depletion experiment CT data were analyzed and 2–ΔΔCT calculations performed using the 7500 System SDS Software v1.4 (AB) with GAPDH as the endogenous control and T0 as the calibrator.
Results and Discussion
Nitrate and Nitrite Transporters
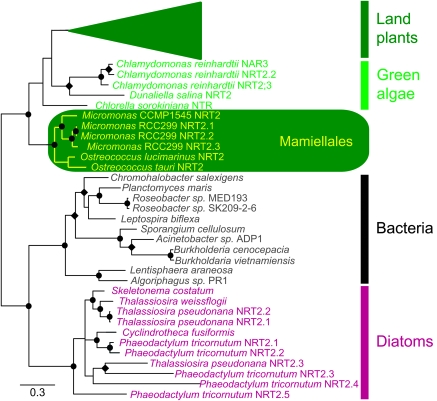
The Mamiellales contained both MFS NRT types (Table 1, supplementary table S1, Supplementary Material online). Micromonas RCC299 had six genes known to be involved in nitrate transport, including three NRT2 genes, whereas the other Mamiellales had fewer (Table 1, supplementary table S1, Supplementary Material online). In plants, NRT2 genes have been shown to serve as high-affinity NRTs and are more highly expressed under low nitrate concentrations (Galvan and Fernandez 2001; Fernandez and Galvan 2007). For the putative high-affinity NRT2 genes in RCC299, a potential duplication event appeared likely on chromosome 1, involving both the transporter and its accessory protein (NAR2). RCC299 NRT2.1 and NRT2.2 were identical at the nucleotide level as were the accessory genes (NAR2.1 and NAR2.2). NRT2.3, located on chromosome 9 differed from these at the amino acid level. Point mutations and two insertion regions were found in NRT2.3, which also lacked an associated NAR2 gene. Nevertheless, the three RCC299 NRT2 genes formed a single supported clade in phylogenetic analyses (fig. 1, supplementary table S2, Supplementary Material online). CCMP1545 (Table 1, supplementary table S1, Supplementary Material online), O. tauri, and O. lucmarinus (as well as Ostreococcus RCC809, a so-called low light–adapted strain [Rodriguez et al. 2005] with a sequenced genome [http://genome.jgi-psf.org/OstRCC809_2]), had single copies of NRT2 (supplementary table S2, Supplementary Material online) and its accessory protein NAR2. Together with those of RCC299, these formed a Mamiellales NRT2 group (fig. 1). A comprehensive phylogenetic reconstruction of 200 protein sequences, including bacteria, oomycetes, diatoms, fungi, lower and higher land plants, and a number of other eukaryotic algae recently explored NRT2 diversification (Slot et al. 2007). That analysis showed that within the Archaeplastida, NRT2 phylogeny typically tracked organismal phylogeny (Slot et al. 2007). We did not seek to replicate this work but rather to verify the placement of the Mamiellales in the context of this observation. Our analysis supported Slot et al. findings for other Archaeplastida, to the genera, and species level in the case of the Mamiellales, which fell at the base of the green lineage, forming an out-group to other green algae.
Table 1.
Ammonium, Nitrate, and Nitrite Transport Genes Identified in the Mamiellales Genomes.
| Gene/Gene Symbol | RCC299 | CCMP1545 | Otau | Oluc |
| AMT/AMT1 | 3 | 3 | 2 | 2 |
| AMT/AMT2 | 3 | 2 | 2 | 2 |
| Nitrite transporter, chloroplast targeted/NAR1 | 1 | 1 | 1 | 1 |
| Putative low-affinity NRT/NRT1 | 1 | NF | 1 | 1 |
| Putative high-affinity NRT/NRT2 | 3 | 1 | 1 | 1 |
| NRT accessory/NAR2 | 2 | 1 | 1 | 1 |
NOTE.—NF indicates not found by BlastP or TBlastN; Otau, O. tauri; Oluc, O. lucimarinus.
FIG. 1.
Unrooted Bayesian inference tree of nitrate transporter genes (NRT2) for various taxonomic groups. Symbols indicate posterior probabilities of 1 (circles) or between 0.95 and 0.99 (diamonds). The collapsed land plant group was composed of 33 NRT2 sequences from the following organisms: Arabidopsis thaliania, Brassica napus, Citris sinensis × Poncirus trifoliata, Cucumis sativus, Daucus carota, Glycine max, Hordem vulgare, Lotus japonicus, Lycopersicon esulentum, Nicotiana tabacum, Oryza sativa, Physcomitrella patens, Populus tremula × Populus tremuloides, Spinacia oleracea, Triticum aestivum, Vitis vinifera, and Zea mays. Accession numbers are provided in supplementary table S2, Supplementary Material online.
Similar to Micromonas, the diatoms also had significant differences in NRT2 numbers (fig. 1). T. pseudonana appeared to have three NRT2 genes encoded in its genome; however, P. tricornutum had six. One of these six genes (Protein ID 1040691) was not placed phylogenetically due to irresolvable model issues. This gene may be a relic or artifact or subject to genome assembly issues such as an unrecognized intron, frame shift, or other problem, which made a significant portion of the 5′ region unclear. NRT2 gene numbers in A. anophagefferens and E. huxleyi (not included in the phylogenetic analysis) are difficult to ascertain because these genomes are not completely assembled. With that caveat in mind, A. anophagefferens contained three potential NRTs, all of which gave a highest scoring pair (HSP) to NRTs from the diatoms Skeletonema costatum or P. tricornutum, as might be expected given that few algal stramenopiles have been sequenced. E. huxleyi contained nine putative NRTs, all of which returned HSPs to NRTs from either the Mamiellales or to P. patens.
Targeting predictions (supplementary tables S1, S3, and S4, Supplementary Material online) were generally consistent with localization data for other green lineage organisms (Lea et al. 1992; Mariscal et al. 2004). Chloroplast transit peptides were predicted by two methods for NII in RCC299 (supplementary table S2, Supplementary Material online) and by all four methods for CCMP1545 (supplementary table S3, Supplementary Material online) as expected given that the conversion of nitrate to nitrite performed by nitrite reductase (NII) is known to take place in the plastid (Lea et al. 1992). Likewise NAR1, the nitrite transporter showed chloroplast targeting in both CCMP1545 and RCC299.
A POT family member was also identified in three of the four Mamiellales genomes (not found in CCMP1545), likely representing a nitrate permease (NRT1). These Micromonas putative NRT1 genes (Table 1, supplementary table S1, Supplementary Material online) showed HSPs with the other three Mamiellales and then to bacterial sequences. RCC299 NRT1 showed 38% similarity to both the Ostreococcus NRT1 sequences and 36% similarity to proteobacterial sequences (e.g., Bdellovibrio bacteriovorus and Photobacterium sp. SKA34). The Ostreococcus strains had higher similarity to each other (65%), again followed by bacterial HSPs (∼34 to 35%) to the proteobacterium Sorangium cellulosum (cyanobacterial hits in the same similarity range were also seen). Using the RCC299 NRT1 gene as a query against other algal genomes (BlastP), a single gene model contaxining a putative transforming growth factor-beta receptor domain likely involved in oligopeptide transport (Paulsen and Skurray 1994; Steiner et al. 1995), was found in P. tricornutum (31% similarity, Protein ID 47218), A. anophagefferens (31% similarity, Protein ID 2185), and C. reinhardtii (38% similarity, Protein ID 136180). Putative NRT1 genes were not found via this search in T. pseudonana or E. huxleyi. Based on studies on Arabidopsis and Chlamydomonas, these genes putatively serve as LATS (Guo et al. 2001; Liu and Tsay 2003; Fernandez and Galvan 2007).
Ammonium Transporters
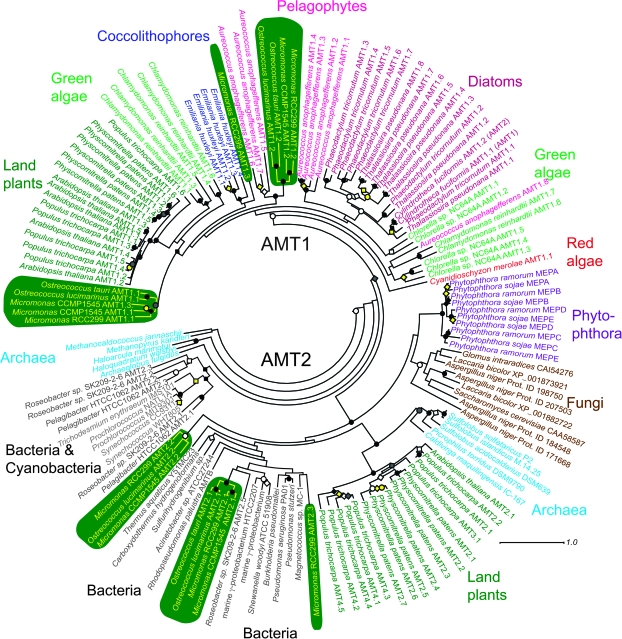
Like land plants, the four Mamiellales had AMT1 and AMT2 family genes (fig. 2). This was not the case for other algae, even other Archaeplastida algae. The green algal (Chlamydomonas and Chlorella) and red algal (Cyanidioschyzon) genomes contained only AMT1. The lack of AMT2 in Chlamydomonas has been known for some time (Suenaga et al. 2003; Gonzalez-Ballester et al. 2004), and thus, the presence/acquisition of AMT2 genes was thought unique to land plants. The chromalveolate algae, represented by genomes from two diatoms (T. pseudonana and P. tricornutum), the pelagophyte A. anophagefferens, and the haptophyte Emiliania huxyeli also lack AMT2 genes. Apart from evolutionary implications, the presence of AMT2 genes in Mamiellales green algae may also relate to substrate affinities and organism ecology.
FIG. 2.
Unrooted maximum likelihood phylogenetic tree using protein sequences from AMT1 and AMT2 gene families from a selection of eukaryotes with sequenced genomes and from bacteria and archaea (some with sequenced genomes). Note that Chlamydomonas AMT1.2 is not shown but falls within Chlamydomonas subclass II. Transcripts of this gene have been cloned and sequenced, and the gene is present in the JGI v3 Chlamydomonas genome assembly, however, is missing from v4. Also AMT2B from O. tauri was not included because the 3' end of the gene sequence was truncated by a gap (in the genome sequence). Although S. cerevisiae has three MEPs, only one ScMEP is shown, although all three Aspergillus MEPs are included. Sequences at the most basal node (not shown) were AMT2.4 Roseobacter sp. SK209-2-6 (ZP_01755968), AMT2.4 Candidatus Pelagibacter ubique HTCC1062 (AAZ22114), and Silicibacter pomeroyi DSS-3 (YP_166819). Black symbols indicate bootstrap support between 95 and 100 (circles) or between 70 and 95 (diamonds) by both ML and NJ methods. White symbols represent the same for ML support only. Gray symbols represent the same for NJ methods only. Yellow symbols indicate bootstrap support between 95 and 100 (circles) for ML and 70 and 95 by NJ (circles) or 95 and 100 NJ and 70 and 95 ML support (diamonds). Accession numbers and gene model numbers are provided in supplementary table S5, Supplementary Material online.
Different AMT genes within single genomes can have different functions, localization, and efficiencies for transport (Fernandez and Galvan 2007; Yuan et al. 2007). For example, Arabidopsis AtAMT1.1 and AtAMT1.2 are HATS genes, but AtAMT1.2 demonstrates biphasic kinetics for methylammonium, effectively operating as both a high- and a low-affinity transporter depending on substrate concentrations (Shelden et al. 2001; Sohlenkamp et al. 2002). In Chlamydomonas, CrAMT1.1 and CrAMT1.2 are strongly induced by N deficiency (Gonzalez-Ballester et al. 2005), whereas CrAMT1.4 is expressed by growth in media containing “poor N sources,” such as arginine (Kim et al. 2005). What is generally less clear is the extent to which the phylogenetically distinct subclasses relate to differing substrate affinities (as seen for AtAMT1.2).
In our analysis, land plant AMT1 genes clustered together in a single bootstrap-supported clade (fig. 2). This was similar to findings for poplar and other plants (Couturier et al. 2007) as well as tree-based inferences from analyses without statistical evaluation through bootstrapping (Simon-Rosin et al. 2003; Suenaga et al. 2003; Ludewig et al. 2007). We found that AMT1 genes of the “lower” land plant Physcomitrella formed a single supported clade within the plant AMT1 genes with less divergence than those of other plant species investigated (fig. 2). In contrast, Chlamydomonas AMT1 formed three phylogenetically distant groups (fig. 2) as reported previously (Gonzalez-Ballester et al. 2004), with subclasses I (CrAMT1.1, CrAMT1.3, and CrAMT1.5) and II (CrAMT1.4, CrAMT1.6, and CrAMT1.2) being similar to plant AMT1 genes. Chlamydomonas AMT1.2 is not shown in the figure but falls within Chlamydomonas subclass II (Gonzalez-Ballester et al. 2004). Transcripts of this gene have been cloned and sequenced, and the gene is present in the JGI v3 Chlamydomonas genome assembly; however, the gene is missing from the v4 assembly, the version analyzed herein. Subclass III (CrAMT1.7 and CrAMT1.8) was more basely positioned within AMT1, falling with one of two Chlorella AMT1 gene clades as well as the divergent Aureococcus AMT1.8. However, this node did not retain support, and long-branch attraction could have influenced positioning. The NJ tree (not shown) showed similar clade structure to the ML tree (fig. 2).
Diatom AMT1 genes formed a single bootstrap-supported group, which was surprising given how divergent these phytoplankton are from one another (Bowler et al. 2008). Still, within the diatom AMT1 group, distinct clades were identified (fig. 2). For the diatom C. fusiformis, a previous study showed differential expression for two AMT genes under the absence of nitrogen in media or single source amendments, with AMT1 being considered more efficient than AMT2 based on higher overall expression levels (Allen 2005; Hildebrand 2005). Our phylogentic analysis demonstrated that both these CfAMTs (CfAMT1 and CfAMT2) belong to the AMT1 gene family and should likely be renamed CfAMT1.1 and CfAMT1.2 to avoid confusion with membership in the AMT2 family (fig. 2). Aureococcus also contained two supported AMT1 clades. Aureococcus subclass I (AMT1.1 to 1.4) bore higher similarity to diatom AMT1s (as expected), but subclass II (AMT1.5 to 1.7) bore higher similarity to Mamiellales AMT1s, in addition to the more basal AaAMT1.8. Another Aureococcus AMT1 sequence (Protein ID 17176) was not included due to irresolvable model issues. However, the oomycete genomes (P. sojae and P. ramorum), organisms phylogenetically closest to diatoms and Aureococcus (all being stramenopiles), did not contain AMT1 genes but rather had several AMT2-like genes that formed a bootstrap-supported clade with fungal MEPs (see below). E. huxleyi contained only AMT1 genes that formed a single supported clade, similar to findings for the diatoms. Three of the seven E. huxleyi AMT sequences (Protein IDs 444314, 201096, 558249) were excluded from phylogenetic analyses as they possibly resulted from genome assembly issues or were incomplete.
The Mamiellales showed a more divergent suite of AMTs than seen in the other algae or plants analyzed to date. Micromonas RCC299 and CCMP1545 had six and five AMTs, respectively (Table 1, supplementary table S1, Supplementary Material online), most placed within four phylogenetically distinct clades (fig. 2). AMT diversification was quite unlike that for NRT2 genes (fig. 1). At the broadest level, classical AMT1 and AMT2 families were represented in all Mamiellales but had more mixed lineage affiliations than anticipated. Within AMT1, two clades were formed; subclass I (containing AMT1.1 genes, Protein IDs: RCC299 61768, CCMP1545 29536, O. tauri 29863, O. lucimarinus 28535, and AMT1.3 from CCMP1545, Protein ID 45964) was closer to land plants, forming a supported “out-group” to Chlamydomonas subclasses I and II as well as land plants. Mamiellales subclass II (containing AMT1.2 genes, Protein IDs: RCC299 92834, CCMP1545 48406, O. tauri 35731, O. lucimarinus 32264) formed a supported node sistering Aureococcus AMT1 subclass II. A cursory analysis showed that Ostreococcus RCC809 had the same subclasses as O. tauri and O. lucimarinus. AMT sequences with highest relatedness showed the same cellular localization, for example, RCC299 AMT2.1 (Protein ID 63515) and CCMP1545 AMT2.1 (Protein ID 59331) had predicted chloroplast transit peptides (supplementary table S1, Supplementary Material online). Looking at the majority predictions from the four prediction tools employed, RCC299 AMT1.2, AMT1.3, as well as AMT2.2 and AMT2.3 appear to possibly be plasma membrane associated (supplementary table S1, Supplementary Material online). Transporters associated with the plasma membrane are typically involved in uptake from the environment, whereas those that are chloroplast targeted are associated with intracellular transport across the chloroplast membrane. It should be noted that prediction tools provide only an indication of targeting. This indication is based on the validity of the gene models, which can be particularly faulty in the 5′ start region, the most essential region for successful predictions. Therefore, methods such as localization by fluorescence in situ hybridization or mass spectrometry analyses would be required to confirm the localizations. Sequence-based differences between the AMT1 genes in the Mamiellales, C. reinhardtii, Chlorella, and the chromalveolates were also visible by the motif analysis (supplementary fig. S1, Supplementary Material online).
Micromonas RCC299 deviated from the other Mamiellales in having two AMT genes that did not associate closely with other Mamiellales AMTs, one belonging to the AMT1 family and the other to AMT2. RCC299 AMT1.3 (Protein ID 92834), did not fall within either Mamiellales AMT1 subclass I or II in our analysis and had only 46% similarity to CCMP1545 AMT1.3. RCC299 AMT1.3 is located on chromosome 1 in a peculiar region of the genome designated the “low-GC region” due to its lower than average GC content (Worden et al. 2009).
Thus, far land plants were one of the few lineages known to consistently have both AMT1 and AMT2 homologs. The latter are related to AMTs in archaea and bacteria (AMT2 family), including cyanobacteria, which is evolutionarily related to the fungal MEP family (Marini et al. 1997; Soupene et al. 2002; Paz-Yepes et al. 2008); AMT1 homologs have not been observed in these lineages. Prokaryotic AMT2 and fungal MEPs have been shown to transport methylammonium, a structural analog of ammonium, in addition to transporting ammonium (e.g., Siewe et al. 1996; Soupene et al. 1998; Paz-Yepes et al. 2008). Furthermore, marine phytoplankton assemblages have been shown to take up methylammonium, although uptake is strongly inhibited in the presence of ammonium (Wheeler and McCarthy 1982). However, at least in Arabidopsis, AMT2 family members do not appear to transport methylammonium (Sohlenkamp et al. 2000). We identified two AMT2 gene clades in the Mamiellales (fig. 2, Table 1, supplementary tables S1 and S5, Supplementary Material online) falling with bacterial AMT2 genes but with low bootstrap support, leaving their placement unresolved. The Mamiellales AMT2 genes fell with heterotrophic bacteria, especially proteobacteria, not with cyanobacteria. Still, placement of AMT2 subclass II (AMT2.2 genes) did not retain support (fig. 2, supplementary fig. S3, Supplementary Material online). Mamiellales AMT2 subclass I, containing all AMT2.1 genes (Protein IDs RCC299 63515, CCMP1545 59331, O. tauri 18135, O. lucimarinus 32601), seemed possibly to be the result of a horizontal gene transfer (HGT) event. Although it is clear that the Mamiellales AMT2 genes are distinct from plant versions, the backbone nodes for the more basal bacterial clades, relative to the Mamiellales subclass I, were not supported. γ-Proteobacteria and Roseobacter AMT2.5 were placed in a supported node as a sister group to AMT2 subclass I (supplementary fig. S3, Supplementary Material online). The extent to which Mamiellales might utilize methylammonium in times of low ammonium availability is not known; however, at least one of the AMT2 genes (AMT2.2) appears to be localized to the plasma membrane (supplementary table S1, Supplementary Material online), which could facilitate uptake from the natural environment. RCC299 again had a divergent outlier (AMT2.3, Protein ID 64034, chromosome 14) not found in the other Mamiellales, which returned HSP Pseudomonas stutzeri A1501 (e-value, 1 × 10−3). This gene was also structurally quite unique (see below).
Finally, several marine bacterial AMTs showed additional domains within the same open reading frame (ORF), indicating that these genes may have specialized functions. Roseobacter sp. SK209-2-6 and C. Pelagibacter ubique (SAR11 clade) had five and four AMT genes, respectively (supplementary table S5, Supplementary Material online). The ORF encoding Roseobacter sp. SK209-2-6 AMT2.3 also contained GGDEF (linked to a wide range of nonhomologous domains in cell signaling proteins) and EAL domains. These domains are present in many bacteria and thought to be involved in regulating cell surface adhesiveness (Simm et al. 2004). Pelagibacter AMT2.2 and AMT2.3 contained the Pfam for stage II sporulation protein E (SpoIIE) required for formation of a normal polar septum during bacterial sporulation.
Land plant AMT2s formed a bootstrap-supported clade with archaeal AMT2s rather than with those of the Mamiellales (fig. 2). Thus, the Mamiellales and land plant AMT2 genes appeared to have different evolutionary histories. Differences were also visible by motif analysis (supplementary fig. S2, Supplementary Material online). To further verify this finding, we performed a second analysis (supplementary fig. S3 and table S5, Supplementary Material online) that included additional AMT2 sequences retrieved from BlastP results (GenBank nr) using plant AMT2 genes as queries. Two Trichocomaceae (fungi) and a number of plant sequences were also retrieved during this search (see Methods). These two additional fungal AMTs (Talaromyces stipiatus XP_002477878 and Penicillium marneffei XP_002145674) may have resulted from HGT based on their supported placement in a region of the tree distinct from other fungal MEPS (supplementary fig. S3, Supplementary Material online); they do not appear to be present in other sequenced fungal genomes (at least based on our cursory analysis). It should be noted that both these fungi also have more classical MEP sequences (not included in the phylogeny), which fall within the fungal/oomycete region of the tree. In this AMT2 phylogenetic analysis, three bacterial sequences fell between plant and archaeal AMT2s, two of which were from Leptospirillium (also known as Acidithiobacillus, according to the ATCC). This genus has been shown to live in extreme environments with only a few other organisms, mostly archaea, present (Tyson et al. 2004). The third sequence came from Acidimicrobium ferrooxidans DSM 10331, another extremophile (isolated from hot spring runoff). Although many bacterial HGTs seem to be intra-Kingdom, inter-Kingdom has certainly been reported (DeLong and Karl 2005). Thus, it seems possible that these three AMT2 genes are indeed more archaeal in evolutionary history than bacterial, especially given that many bacterial genomes have been sequenced, but this particular gene is generally not present. The archaeal gene cluster sistering these three bacterial AMT2s, as well as plant AMT2 sequences, was composed of Crenarchaeota genes (and one Euryarchaeota sequence), lending support to the closer evolutionary history of the crenarchaeotes with eukaryotes than other archaeal groups (Cox et al. 2008). Most sequences belonging to Euryarchaeota fell within a second more basal archaeal gene cluster (fig. 2, supplementary fig. S3, Supplementary Material online). More comprehensive phylogenies that include all available bacterial and archaeal sequences are needed to confirm these inferences. Archaeal origins of plant AMT2 genes would provide an example of an “operational” gene HGT as opposed to “information-processing system components” (e.g., core machineries of translation, transcription, and replication), the latter being the primary role ascribed to archaeal-derived genes in eukaryotes (Yutin et al. 2008). Still, given the fact that Crenarchaeota and Eukarya are considered to be evolutionarily related, the patterns seen here could simply be derived by divergence from common ancestral genes or differential loss and gain.
Our analyses of AMTs in other green algal (Chlamydomonas and Chlorella) and chromalveolate algal genomes showed only AMT1 to be present. Although Phytophthora belongs to the Chromalveolata, and specifically the heterokonts, its AMT genes were sistered by fungal MEPS, and AMT1 genes were not found. Our findings were similar to those of Slot et al. (2007), who showed that oomycete NRTs sistered fungal NRT2 homologs. The results of Slot et al. that retained bootstrap support for this relationship strongly indicated that heterokonts are paraphyletic. In our reconstructions, statistical support was not retained at this node; however, the Phytophthora genes were clearly separated from other heterokonts AMT genes, with bootstrap support for this separation. Similarities to fungi have been the basis for suggesting that HGT between these lineages may have been rampant (Andersson 2009). In a previous study, 4 of 12 genes shared between fungi and oomycetes had phylogenetic support indicating that HGT from fungi to oomycetes had occurred (Andersson 2009). Typically, the functional role of such genes has been implicated in utilization of rare metabolites. Although here lack of statistical support does not allow conclusions regarding the relationship, analyses with greater taxon representation may further resolve the relationship between oomycete and fungal MEPS.
Assimilation Genes
In addition to transporters, searches for genes involved in the assimilation of NO3– and NH4+ were performed. Different numbers of each type were found in the four Mamiellales (Table 2). When more than one version was present per genome, typically one appeared to be chloroplast targeted, for example, as seen for asparagine synthetase (ASN) and aspartate amino transferase (ASP), although not all prediction tools rendered consistent results (supplementary tables S3 and S4, Supplementary Material online). A number of these showed mixed lineage affiliations containing one or more eukaryotic-like and one or more prokaryotic-like version. For instance, ASN showed mixed affiliations similar to those of AMT.
Table 2.
Nitrogen Assimilation–Related Genes Identified in the Mamiellales Genomes.
| Gene, Gene Symbol | RCC299 | CCMP1545 | Otau | Oluc |
| Nitrate reductase, NIA | 1 | 1 | 1 | 1 |
| Nitrite reductase, NII | 1 | 1 | 1 | 1 |
| Glutamine synthetase catalytic domain, GLN | 2 | 2 | 1 | 1 |
| Glutamate synthase (Ferredonin-dependent), GLU | 1 | 1 | 1 | 1 |
| Glutamate synthase (NADH dependent), GLT | 1 | 1 | NF | NF |
| Glutamate dehydrogenase, GDH | NF | NF | NF | NF |
| Asparagine synthetase, ASN | 3 | 3 | 2 | 2 |
| Aspartate aminotransferase, ASP | 3 | 3 | 1 | 1 |
| Molybdate transporter, MoT1 | 1 | NF | 1 | 1 |
| Molybdenum biosynthesis CF, CNX1E | 1 | 1 | 1 | 1 |
| Molybdenum biosynthesis CF, CNX1G | 1 | 1 | NF | 1 |
| Molybdenum biosynthesis CF, CNX2 | 1 | 1 | 1 | 1 |
| Molybdenum biosynthesis CF, CNX3 | 1 | 1 | 1 | 1 |
| Molybdenum biosynthesis CF, CNX5 | 1 | 1 | 1 | 1 |
| Molybdenum biosynthesis CF, CNX6 | 1 | 2 | 1 | 1 |
| Molybdenum biosynthesis CF, CNX7 | 1 | 1 | 1 | 1 |
| Nitrogen-sensing protein PII homologue | 1 | 1 | NF | NF |
NOTE.—CF, cofactor. NF indicates not found by BlastP or TBlastN; Otau, O. tauri; Oluc, O. lucimarinus.
Other differences were observed between the Mamiellales genomes (Table 2). For example, although both Micromonas had NADH-dependent glutamate synthase (GLT), the two Ostreococcus genomes lacked this gene. The lack of GLT indicates that Ostreococcus relies on ferrodoxin-dependent glutamate synthase (GLU), which has been shown to be expressed at higher levels as a function of increased irradiance in Spirodela polyrhiza (giant duckweed; Teller et al. 1996). The presence of both NADH-dependent and ferrodoxin-dependent GOGAT systems in the Micromonas genomes could provide the flexibility to grow at either low irradiance (e.g., deeper in the euphotic zone in stratified waters) or high irradiance (surface waters)—conditions encountered at different times of the year in marine environments.
Differences were also observed in genes involved in transport of molybdenum, an important cofactor for NIA. We identified a transporter related to the Chlamydomonas molybdenum transporter MoT1, a gene distantly related to plant sulfate transporters (Tejada-Jimenez et al. 2007). Sulfate permease (SulP) family proteins have been shown to transport molybdenum in plants (Fitzpatrick et al. 2008). MoT1/SulP homologs were found in RCC299 and O. lucimarinus, as expected, given previous identification in O. tauri (Tejada-Jimenez et al. 2007). MoT1 was not found in the CCMP1545 genome. In almost all organisms, molybdenum cofactor biosynthesis (Moco) is a conserved pathway involving four major steps. The pathway depends on the availability of intracellular molybdenum, which is transported into the cytosol by MoT1 (Tejada-Jimenez et al. 2007). Given the importance of molybdenum and presence of molybdenum biosynthesis cofactor genes in CCMP1545 (Table 2), it seems unlikely that CCMP1545 lacks a molybdenum transport system. An alternative molybdenum transport system may be present or possibly genome assembly excluded this gene erroneously. Other mechanisms are known to be responsible for molybdenum transport. In bacteria, molybdenum is transported by ABC family transporters, an extremely broad family, members of which are responsible for transport of many substrates. Still, in eukaryotes, transport systems tend to be more specialized. In some eukaryotes, the presence of molybdate and selenate inhibits the activity of sulfate transporters (Tejada-Jimenez et al. 2007), suggesting possible substrate flexibility.
PII homologs were also identified in Micromonas (Table 2, supplementary tables S3 and S4, Supplementary Material online), akin to the homologue previously reported in Arabidopsis (Hsieh et al. 1998). This nitrogen sensing protein–encoding gene has been shown to indirectly regulate glutamine synthetase at the transcriptional and posttranslational levels in response to nitrogen limitation in Escherichia coli (Hsieh et al. 1998; Arcondeguy et al. 2001). The Arabidopsis homologue is involved in a complex signal transduction mechanism for detecting the availability of organic nitrogen by detecting internal nitrogen levels and regulating the expression of glutamine synthatase (Hsieh et al. 1998). Although present in both RCC299 and CCMP1545, PII was not identified in either Ostreococcus genome, suggesting differences in regulation of nitrogen transport and assimilation between the two genera.
Gene Structure, Arrangements, and Genome Placement
Land plant AMT1 genes have been reported to be intron free, with the exception of Lotus japonicus LjAMT1.1 (Salvemini et al. 2001; Couturier et al. 2007). In contrast, RCC299 and CCMP1545 AMT1.1 had well-supported 5′ introns, longer (323 and 406 nucleotides, respectively) than average for these genomes (195 ± 139, Simmons MP and Worden AZ, unpublished data); Chlamydomonas and Chlorella AMT1 genes also had introns. Even Physcomitrella showed introns in AMT1 genes. Thus, the lack in higher plants sequenced to date seems a more unusual feature than initially thought. In Poplar, no introns were detected in AMT1 genes, but they were present in AMT2 genes (Couturier et al. 2007). Likewise, Physcomitrella AMT2 had more introns than AMT1 genes. Micromonas AMT2 genes typically had one intron (with the exception of CCMP1545 AMT2.2, which had none), although Ostreococcus AMT2s had none. RCC299 AMT2.3 (not present in the other Mamiellales genomes) contained three introns. This gene sequence was anomalous in terms of its phylogenetic distance, and in addition to having more introns than other Mamiellales AMT2 genes, it had more than the average for all genes in the genome (0.57 introns/gene; Worden et al. 2009).
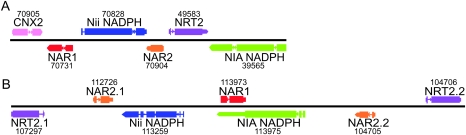
Colocalization of nitrogen transport and assimilation genes, akin to that reported for O. tauri (Derelle et al. 2006), was observed in both Micromonas genomes, although composed of different genes and gene numbers. In O. tauri, eight genes involved in nitrate transport and assimilation reportedly clustered together on chromosome 10 (Derelle et al. 2006). In CCMP1545, a cluster on scaffold 4 included six genes (fig. 3A) and just downstream, with three genes intervening (Protein IDs 57686, 57687, and 67708), was NIA1. In RCC299, seven genes were colocalized on chromosome 1 (fig. 3A). AMT1.2 (Protein ID 92834) was located downstream, separated from this cluster by three intervening genes (Protein IDs 113265, 54997, and 112698), similar to NIA1 in CCMP1545. Nitrate transport and assimilation related genes are colocated in C. reinhardtii as well (Fernandez and Galvan 2007). The presence of gene clusters has been proposed to represent selective pressure toward optimization (Trowsdale 2002). It is unclear how variations in gene arrangements seen between the four Mamiellales genomes (as well as C. reinhardtii) might relate to specific aspects of environmental optimization.
FIG. 3.
Gene arrangements showing the relative positioning of colocated nitrogen genes along the chromosome (represented by black line) within CCMP1545 on scaffold 4, nucleotide positions 1572082-1585993 (A), and within RCC299 chromosome 1, positions 483935-509879 (B), and drawn approximately to scale. Homologous genes are color coded; pink indicates a nitrogen-associated gene that does not have a colocated homolog in the other Micromonas species.
Differences in gene complements and arrangements between the four Mamiellales provide a platform for exploring the functional significance of specific gene sets. For example, the nitrogen-gene cluster and likely duplication of NRT2 on chromosome 1 in RCC299 presents two points for further consideration. First, this duplication could represent an example of dose repetition where duplication correlates with an increased production of the product (Graur and Li 2000). In addition, the chromosome 1 region in which the nitrogen-gene cluster is located is a special region, present in all the Mamiellales, with lower percentage of GC than the rest of the genome (Worden et al. 2009). Furthermore, this region shows higher gene expression levels than “normal” GC regions of the genome (Simmons and Worden, unpublished data) and has been hypothesized to be a sex chromosome (Derelle et al. 2006). Although inclusion of such essential genes in a sex chromosome seems at some level dangerous, higher expression levels might be advantageous. Micromonas RCC299 was isolated from the South Pacific Ocean, known for being a low-nutrient environment, whereas CCMP1545 was isolated from the English Channel, and both Ostreococcus strains came from coastal settings that typically have relatively high nutrient concentrations (Worden et al. 2009). If higher expression levels enable more rapid uptake, it would presumably reflect a competitive advantage in situations where nitrogen is limiting.
Verification of TaqMan Transporter Probes and Functionality
We sought to verify that the AMT and NRT genes identified in Micromonas were expressed. EST data for Micromonas RCC299 showed at least some level of expression in mid- to late-exponential phase growth for AMT and NRT transporters except for AMT2.1, AMT2.3, and NRT1 (supplementary table S1, Supplementary Material online). AMT2.2 ESTs provided evidence that at least some of the bacterial-like AMT2s were functionally active. EST support was also found for a subset of nitrogen assimilation–related genes in RCC299 (supplementary table S3, Supplementary Material online) and CCMP1545 (supplementary table S4, Supplementary Material online).
To enable future studies on how the different Mamiellales AMT and NRT genes relate to physiology, 10 primer–probe sets were developed for real-time qPCR (Table 3). RCC299 transporter genes that did not have EST support were specifically targeted (AMT2.1, AMT2.3, NRT1). Primer–probe sets were designed and tested on RCC299. These primer–probes sets are not likely to amplify corresponding genes in other Mamiellales due to mismatches (supplementary table S6, Supplementary Material online). For initial verification, products were directly sequenced, and all but one primer–probe set provided the correct sequence. The AMT2.3 product required cloning prior to successful sequencing but then rendered the expected product in two clones and ambiguous sequences (unresolved nucleotides) from 18 clones. Analyses of dynamic ranges showed that all primer–probe sets were within the linear part of the curve between 3 and 30 ng μl−1 RNA (corresponding to between ca. 3.9 × 105 and 3.9 × 106 cells ml−1), and the former concentration was used for subsequent analyses. GAPDH was selected as an endogenous control because it varied the least over the four conditions tested (supplementary table S7, Supplementary Material online). The variation seen in GAPDH gave a baseline of 1 CT, below which a difference between the gene under investigation and the endogenous control could not be detected. Target gene expression data were then normalized to GAPDH at each time point. The efficiency of all primer–probe sets was sufficiently close to 100% that the 2–ΔΔCT calculation was used for analysis (supplementary table S8, Supplementary Material online).
Table 3.
TaqMan Primer Sets and Probes Designed to Target Nitrate and AMTs in Micromonas sp. RCC299.
| Primer Set | Gene/Protein ID | Sequence (5′–3′) | Length (bp) |
| AMT2.3F | AMT2.3/64034 | GATGCTCGTCACGCAAATCG | 76 |
| AMT2.3R | GCGGCCGTAGCATGATAACT | ||
| AMT2.3probe | CCACCCGACGACTCC | ||
| AMT2.1F | AMT2.1/63515 | CGTCAACCAGGCAGACACT | 96 |
| AMT2.1R | ACGATGCCCGCGTAGAAG | ||
| AMT2.1probe | CATCTCGACCGCTCTCG | ||
| AMT1.3F | AMT1.3/92834 | ACATCTCCCAACTCGACACA | 124 |
| AMT1.3R | CCGAACAACAACCACAAAAC | ||
| AMT1.3 probe | ATGACGCCATCAACGCCGTC | ||
| NRT2.1,2F | NRT2.1/NRT2.2113915/104706 | CTCGCGGAATCGAAATTTAGCAT | 84 |
| NRT2.1,2R | GTGAGGGAAGTTGAAGGAGAAGAT | ||
| NRT2.1,2 pr. | CTGTGGATAGCGAAAACA | ||
| NRT2.3F | NRT2.3/105997 | CCGCCCACGTACTCGAT | 74 |
| NRT2.3R | GGGTGGCTGAATGAGAATATGTTGA | ||
| NRT2.3 probe | CCGTGGATAGCGAGAACA | ||
| NRT1F | NRT1/62932 | GACGTATGCGTTTTTTGGGTATTTG | 130 |
| NRT1R | CAACGCGTCGAACTCGTG | ||
| NRT1 probe | CCGCCGCTTTTCCT | ||
| GAPDHF | GAPDH/104954 | GCCGACTACATGGCCTACAT | 79 |
| GAPDHR | CCTGGGCGCCGTACTT | ||
| GAPDH probe | ACACGGTCCACGGCCAG | ||
| ACTINF | Actin/90942 | GCCCTCGTGTGCGATAAC | 89 |
| ACTINR | CCGACGATGGAGGGAAAGAC | ||
| ACTIN probe | CCGGCCTTGACCATGC | ||
| 18SF | 18S ribosomal RNA/a | GCGTTTAGCCAATGGAAGTT | 145 |
| 18SR | CATCACGACGAAATTTGGAG | ||
| 18S probe | CACGCGCGCTACACTGACGA | ||
| UBQF | Ubiquitin/60632 | AACGTCAAGGCCAAGATCCA | 73 |
| UBQR | GCTTGCCAGCGAAGATCAG | ||
| UBQ probe | CAAGGAGGGCATCCC |
NOTE.—Length refers to the overall product length.
A GenBank accession number is provided in Methods as JGI model storage only supports protein-encoding genes.
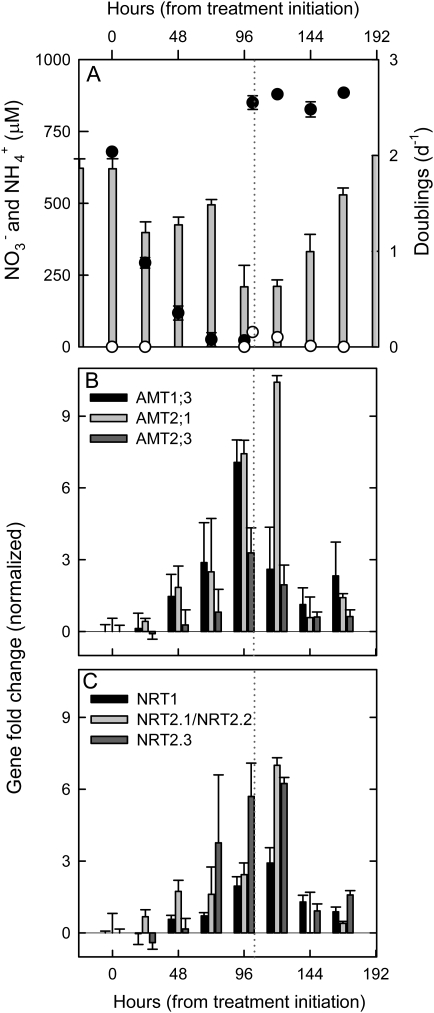
The performance of the new primer–probe sets was evaluated in a nitrogen-depletion time course. Low ammonium concentrations are not uncommon in marine environments, often less than 10 nM in oligotrophic waters, whereas coastal levels of 500 nM in the subsurface maximum up to 2000 nM are seen frequently (Plant et al. 2009). Nutrient analysis indicated that at T0, NH4+ was already below the 10 nM detection limit (fig. 4A), several orders of magnitude lower than that of the media prior to innoculation (50 μM). The depletion at T0 was due to a standard day’s growth in the period prior to treatment initiation rather than being related to the experimental manipulation (transfers in N-deplete media). NO3– was still replete (679 ± 8 μM) at the onset of the experiment. After successive transfers in N-deplete K media, NO3– gradually decreased to ∼20 μM at T96 (fig. 4A). At this point, amendment with both N sources (the “N-spike”) restored NO3– to close to the standard K media concentration, whereas NH4+ was again seen to rapidly decrease during the 24 h post each successive transfer in N-replete media.
FIG. 4.
Nitrogen-depletion experiment showing the mean nitrate (black) and ammonium (white) measurements throughout the time course (A) as well as growth rates for the triplicate RCC299 cultures shown starting 24 h prior to experiment initiation (gray bars). Normalized fold change in target gene expression, relative to expression levels at T0 of three AMT genes examined (B), and the three NRT transporter genes examined (C), with normalization as in Methods. Zero represents the time point at which cells were transferred into N-deplete K media and gray dotted line indicates amendment with NO3– and NH4+. Note: At T0, NH4+ was already below the 10 nM detection limit. In (A), (B), and (C), error bars represent the standard deviation within the triplicated biological treatments.
Analyses of the target transporter genes used T0 values as the calibrator, representing the amount of transcript expressed at T0. Thus, the data presented is fold change in gene expression normalized to GAPDH (the endogenous control) and relative to target gene expression levels at T0. The three AMT genes tested were sensitive to an apparent upregulation during the N-depletion time course (fig. 4B). AMT1.3 and AMT2.3 showed maxima at T96, just before the treatments were reamended with ammonium and nitrate. AMT1.3 is predicted to be plasma membrane associated and likely responsible for uptake from the external environment. Thus, these results make sense that gene expression increased as depletion increased and was suppressed once nitrogen was again plentiful. AMT2.3 has less clear localization data, with disagreement among prediction tools, but at least some (ChloroP and TargetP) indicating that it is not chloroplast targeted and could also be involved in environmental uptake. AMT2.1 expression was at a maximum at T120. This AMT has a predicted chloroplast target peptide, its peak expression after reamendment with nitrogen is likely to be related to cellular processes rather than uptake from the environment.
Expression quantified by the NRT2.1/NRT2;2-, NRT2.3-, and NRT1- (the putative low-/dual-affinity NRT) targeted primer–probes was also sensitive to changes during the preliminary N-depletion time course. All three targets were upregulated during the N-depletion period with a peak in fold change at T120 (fig. 4C). However, the overall extent of fold change in NRT1 was less than for NRT2.1/NRT2.2 (which amplifies two genes) and NRT2.3.
Previous studies have shown that genes unrelated to nitrogen transport can also be upregulated during nitrogen stress and recovery. For example, genes encoding proteins involved in glycolysis, trehalose-6-P metabolism, iron transport/metabolism, and sulfate uptake/reduction are all induced by addition of nitrate after a period of nitrate stress in A. thaliana (Wang et al. 2003). In addition, even putative high-affinity NRTs have been shown to have more complex roles than predicted structurally. For example, of the seven Arabidopsis NRT2 genes, AtNRT2.1 is a repressor of lateral root initiation, and this role is independent of nitrate uptake. AtNRT2.1 has now been proposed to act either as a nitrate sensor or as a signal transducer to coordinate the development of the root system with nutritional cues (Little et al. 2005). Further experimentation should clarify the extent to which Micromonas (Mamiellales) AMT gene clades and NRT genes represent transporters of differing substrate affinity levels, potentially then serving as indicators for nutritional status.
We did not explore several genes that have recently been shown to be important to nitrogen utilization in Aureococcus (Berg et al. 2008). In Aureococcus, two genes in particular showed high transcript accumulation across all nitrogen sources tested in the case of a putative purine transporter AaURA and when cells were grown on urea in the case of a putative urea transporter AaDUR3 (Berg et al. 2008). RCC299 has homologs of both these genes, Protein ID 60599 and 94458, respectively, which were expressed under standard growth conditions. However, CCMP1545 has neither and Ostreococcus only has the putative urea transporter. Other nitrogen-related transporters showed a similarly mosaic pattern among the Mamiellales, and several RCC299-specific transporters (not evaluated herein) may be of particular utility during periods of low-nitrogen availability (Worden et al. 2009).
Future Directions and Conclusions
The phylogenetic distribution of AMTs in moss, oomycetes, and marine algae add new insights to AMT evolution. It appears that the ancestor of land plants accomplished a broad expansion of these gene families. Although this expansion is not seen (or has been lost) in Chlamydomonas and Chlorella, the Mamiellales ancestor also seems to have had multiple AMT types but from a different evolutionary history than those of plants. The closer relationship observed between plant and archaeal AMT2 genes, rather than to Mamiellales AMT2s, now need exploration via more comprehensive phylogenies. In addition, similarities observed between the Aureococcus and Mamiellales AMT1 subclass II genes have implications for phytoplankton adaption and ecology depending on the events that led to their sister relationship. Given the high degree of taxon under sampling for protists, it is clear that inferences of HGT (whether in protists or plants) should be further investigated, with sampling of a greater number of taxa (see Delwiche 1999). The patterns observed here could arise from differential gain and loss of as of yet unrecognized ancestral features.
With these complexities in mind, we are now poised to investigate the physiological consequences of divergent nitrogen transport and assimilation gene clades in the Mamiellales as well as other organisms. The role of nitrogen in structuring the composition of eukaryotic phytoplankton communities in the natural environment is still not clearly understood. Seasonal successions observed in eukaryotic phytoplankton communities at sites such as the Bermuda Atlantic Time Series are presumably linked to changes in nitrogen availability (Steinberg et al. 2001). Diatoms and Aureococcus are known to assimilate diverse organic compounds (see, e.g., Gobler et al. 2002; Fan et al. 2003), whereas less is known about the extent to which the Mamiellales or other eukaryotic phytoplankton utilize organic sources. The presence of differing numbers of inorganic transport genes often leads to assumptions about the relative ecological success of phytoplankton species. A much greater level of complexity in the physiological importance and use of the phylogenetically distinct AMT and NRTs in the Mamiellales appears more likely than previously appreciated. Along with the probes developed, these findings will facilitate a more refined understanding of physiological responses to nitrogen in the environment for these primary producers.
Supplementary Material
Supplementary figures S1–S3 and tables S1–S8 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank A. Monier, E. Demir, S. Johnson, and H. Wilcox for assistance as well as AB technical staff. A. Gough kindly reshaped Figure 3, K. Johnson allowed use of his laboratory for nitrogen measurements, and M.P. Simmons provided comments on the manuscript. Aureococcus, Emiliania, and Chlorella genome data come from public JGI genome projects led by C. Gobler, B. Read, and J. Van Etten, respectively, and for the former two projects, S. Dyhrman and L. Wurch performed an initial analysis of nitrogen transporters; the data were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/ in collaboration with the user community, and the overview publications are still in preparation for these three genomes. This work was funded by a Young Investigator Award in Marine Microbiology by the Gordon and Betty Moore Foundation and National Science Foundation grant OCE-0623928/OCE-0836721 to A.Z.W., the Lucille and David Packard Foundation, and was facilitated by Department of Energy funding for sequencing of the Micromonas genomes. Author contributions: S.M.M. and A.Z.W. conceived of the study and performed preliminary gene analysis. S.M.M. performed preliminary gene trees. For final AMT analyses, A.Z.W. performed gene searches and remodeling as well as phylogenetics, interpretation, and discussion. Final NRT2 trees and other gene findings were performed by S.M.M. A.Z.W. performed motif analyses. S.M.M. developed qPCR probes as well as designed and conducted the N-depletion experiment. J.N.P. provided methods for nutrient analyses and performed the ammonium analyses. All authors contributed to writing and editing the manuscript.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Allen AE. Beyond sequence homology: redundant ammonium transporters in a marine diatom are not functionally equivalent. J Phycol. 2005;41:4–6. [Google Scholar]
- Allen AE, Vardi A, Bowler C. An ecological and evolutionary context for integrated nitrogen metabolism and related signaling pathways in marine diatoms. Curr Opin Plant Biol. 2006;9:264–273. doi: 10.1016/j.pbi.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J. Horizontal gene transfer between microbial eukaryotes. In: Gogarten MB, Gogarten JP, Olendzenski L, editors. Horizontal gene transfer: genomes in flux. New York: Humana Press; 2009. pp. 473–487. [Google Scholar]
- Arcondeguy T, Jack R, Merrick M. P(II) signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol Mol Biol Rev. 2001;65:80–105. doi: 10.1128/MMBR.65.1.80-105.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg GM, Shrager J, Glockner G, Arrigo KR, Grossman AR. Understanding nitrogen limitation in Aureococcus anophagefferens (Pelagophyceae) through cDNA and qRT-PCR analysis. J Phycol. 2008;44:1235–1249. doi: 10.1111/j.1529-8817.2008.00571.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Medlin L. Algal phylogeny and the origin of land plants. Plant Physiol. 1998;116:9–15. [Google Scholar]
- Bowler C, Allen AE, Badger JH, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Couturier J, Montanini B, Martin F, Brun A, Blaudez D, Chalot M. The expanded family of ammonium transporters in the perennial poplar plant. New Phytol. 2007;174:137–150. doi: 10.1111/j.1469-8137.2007.01992.x. [DOI] [PubMed] [Google Scholar]
- Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci U S A. 2008;105:20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM, Glass ADM. Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 1998;3:389–395. [Google Scholar]
- DeLong EF, Karl DM. Genomic perspectives in microbial oceanography. Nature. 2005;437:336–342. doi: 10.1038/nature04157. [DOI] [PubMed] [Google Scholar]
- Delwiche CF. Tracing the thread of plastid diversity through the tapestry of life. Am Nat. 1999;154:S164–S177. doi: 10.1086/303291. [DOI] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, et al. From the cover: genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci U S A. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Falkowski PG. Evolution of the nitrogen cycle and its influences on biological sequestration of CO2 in the ocean. Nature. 1997;387:272–275. [Google Scholar]
- Fan C, Glibert PM, Alexander J, Lomas MW. Characterization of urease activity in three marine phytoplankton species, Aureococcus anophagefferens, Prorocentrum minimum, and Thalassiosira weissflogii. Mar Biol. 2003;142:949–958. [Google Scholar]
- Felsenstien J. PHYLIP (phylogeny inference package) version 3.6. Distributed by the author. Seattle (WA): Department of Genome Sciences, University of Washington; 2005. [Google Scholar]
- Fernandez E, Galvan A. Inorganic nitrogen assimilation in Chlamydomonas. J Exp Bot. 2007;58:2279–2287. doi: 10.1093/jxb/erm106. [DOI] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick KL, Tyerman SD, Kaiser BN. Molybdate transport through the plant sulfate transporter SHST1. FEBS Lett. 2008;582:1508–1513. doi: 10.1016/j.febslet.2008.03.045. [DOI] [PubMed] [Google Scholar]
- Galvan A, Fernandez E. Eukaryotic nitrate and nitrite transporters. Cell Mol Life Sci. 2001;58:225–233. doi: 10.1007/PL00000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz EM, Yu Y-K, Agarwala R, Schäffer AA, Altschul SF. Composition-based statistics and translated nucleotide searches: Improving the TBLASTN module of BLAST. BMC Biology. 2006;4:41. doi: 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass ADM, Britto DT, Kaiser BN, et al. The regulation of nitrate and ammonium transport systems in plants. J Exp Bot. 2002;53:855–864. doi: 10.1093/jexbot/53.370.855. [DOI] [PubMed] [Google Scholar]
- Gobler CJ, Renaghan MJ, Buck NJ. Impacts of nutrients and grazing mortality on the abundance of Aureococcus anophagefferens during a New York brown tide bloom. Limnol Oceanogr. 2002;47:129–141. [Google Scholar]
- Gonzalez-Ballester D, Camargo A, Fernandez E. Ammonium transporter genes in Chlamydomonas: the nitrate-specific regulatory gene Nit2 is involved in Amt1; 1 expression. Plant Mol Biol. 2004;56:863–878. doi: 10.1007/s11103-004-5292-7. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ballester D, de Montaigu A, Higuera JJ, Galvan A, Fernandez E. Functional genomics of the regulation of the nitrate assimilation pathway in Chlamydomonas. Plant Physiol. 2005;137:522–533. doi: 10.1104/pp.104.050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur J, Li W-H. Fundamentals of molecular evolution. Sunderland (MA): Sinauer Associates; 2000. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guo FQ, Wang R, Chen M, Crawford NM. The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell. 2001;13:1761–1777. doi: 10.11054/TPC.010126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand M. Cloning and functional characterization of ammonium transporters from the marine diatom Cylindrotheca fusiformis (Bacillariophyceae) J Phycol. 2005;41:105–113. [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MH, Lam HM, van de Loo FJ, Coruzzi G. A PII-like protein in Arabidopsis: putative role in nitrogen sensing. Proc Natl Acad Sci U S A. 1998;95:13965–13970. doi: 10.1073/pnas.95.23.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KS, Coletti LJ. In situ ultraviolet spectrophotometry for high resolution and long term monitoring of nitrate, bromide and bisulfide in the ocean. Deep Sea Res I. 2002;49:1291–1305. [Google Scholar]
- Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- Kettler GC, Martiny AC, Huang K, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Field E, King N, Yaoi T, Kustu S, Inwood W. Spontaneous mutations in the ammonium transport gene AMT4 of Chlamydomonas reinhardtii. Genetics. 2005;170:631–644. doi: 10.1534/genetics.105.041574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane CE, Archibald JM. The eukaryotic tree of life: endosymbiosis takes its TOL. Trends Ecol Evol. 2008;23:268–275. doi: 10.1016/j.tree.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Lea PJ, Blackwell RD, Joy KW. Ammonia assimilation in higher plants. In: Mengel K, Pilbeam DJ, editors. Nitrogen metabolism of plants. Oxford: Clarendon Press; 1992. pp. 153–186. [Google Scholar]
- Little DY, Rao H, Oliva S, Daniel-Vedele F, Krapp A, Malamy JE. The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci U S A. 2005;102:13693–13698. doi: 10.1073/pnas.0504219102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Tsay YF. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. Embo J. 2003;22:1005–1013. doi: 10.1093/emboj/cdg118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loque D, von Wiren N. Regulatory levels for transport of ammonium in plant roots. J Exp Bot. 2004;55:1293–1305. doi: 10.1093/jxb/erh147. [DOI] [PubMed] [Google Scholar]
- Ludewig U, Neuhauser B, Dynowski M. Molecular mechanisms of ammonium transport and accumulation in plants. FEBS Lett. 2007;581:2301–2308. doi: 10.1016/j.febslet.2007.03.034. [DOI] [PubMed] [Google Scholar]
- Marini AM, Soussi-Boudekou S, Vissers S, Andre B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4282–4293. doi: 10.1128/mcb.17.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariscal V, Rexach J, Fernández E, Galvan A. The plastidic nitrite transporter NAR1;1 improves nitrate use efficiency for growth in Chlamydomonas. Plant Cell Environ. 2004;27:1321–1325. [Google Scholar]
- Okamoto M, Kumar A, Li WB, Wang Y, Siddiqi MY, Crawford NM, Glass ADM. High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 2006;140:1036–1046. doi: 10.1104/pp.105.074385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik B, Grimwood J, Aerts A, et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci U S A. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen IT, Skurray RA. The POT family of transport proteins. Trends Biochem Sci. 1994;19:404. doi: 10.1016/0968-0004(94)90087-6. [DOI] [PubMed] [Google Scholar]
- Paz-Yepes J, Merino-Puerto V, Herrero A, Flores E. The amt gene cluster of the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 2008;190:6534–6539. doi: 10.1128/JB.00613-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant JN, Johnson KS, Needoba JA, Coletti LJ. NH4-Digiscan: an in situ and laboratory ammonium analyzer for estuarine, coastal and shelf waters. Limnol Oceanogr Methods. 2009;7:144–156. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rexach J, Fernandez E, Galvan A. The Chlamydomonas reinhardtii Nar1 gene encodes a chloroplast membrane protein involved in nitrite transport. Plant Cell. 2000;12:1441–1453. doi: 10.1105/tpc.12.8.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez F, Derelle E, Guillou L, Le Gall F, Vaulot D, Moreau H. Ecotype diversity in the marine picoeukaryote Ostreococcus (Chlorophyta, Prasinophyceae) Environ Microbiol. 2005;7:853–859. doi: 10.1111/j.1462-2920.2005.00758.x. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Saier MH, Beatty JT, Goffeau A, et al. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- Salvemini F, Marini A, Riccio A, Patriarca EJ, Chiurazzi M. Functional characterization of an ammonium transporter gene from Lotus japonicus. Gene. 2001;270:237–243. doi: 10.1016/s0378-1119(01)00470-x. [DOI] [PubMed] [Google Scholar]
- Shelden MC, Dong B, de Bruxelles GL, Trevaskis B, Whelan J, Ryan PR, Howitt SM, Udvardi MK. Arabidopsis ammonium transporters, AtAMT1;1 and AtAMT1;2, have different biochemical properties and functional roles. Plant Soil. 2001;231:151–160. [Google Scholar]
- Siewe RM, Weil B, Burkovski A, Eikmanns BJ, Eikmanns M, Kramer R. Functional and genetic characterization of the (methyl)ammonium uptake carrier of Corynebacterium glutamicum. J Biol Chem. 1996;271:5398–5403. doi: 10.1074/jbc.271.10.5398. [DOI] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Simon-Rosin U, Wood C, Udvardi MK. Molecular and cellular characterisation of LjAMT2; 1, an ammonium transporter from the model legume Lotus japonicus. Plant Mol Biol. 2003;51:99–108. doi: 10.1023/a:1020710222298. [DOI] [PubMed] [Google Scholar]
- Slot JC, Hallstrom KN, Matheny PB, Hibbett DS. Diversification of NRT2 and the origin of its fungal homolog. Mol Biol Evol. 2007;24:1731–1743. doi: 10.1093/molbev/msm098. [DOI] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- Sohlenkamp C, Shelden M, Howitt S, Udvardi M. Characterization of Arabidopsis AtAMT2, a novel ammonium transporter in plants. FEBS Lett. 2000;467:273–278. doi: 10.1016/s0014-5793(00)01153-4. [DOI] [PubMed] [Google Scholar]
- Sohlenkamp C, Wood CC, Roeb GW, Udvardi MK. Characterization of Arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane. Plant Physiol. 2002;130:1788–1796. doi: 10.1104/pp.008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene E, He L, Yan D, Kustu S. Ammonia acquisition in enteric bacteria: physiological role of the ammonium/methylammonium transport B (AmtB) protein. Proc Natl Acad Sci U S A. 1998;95:7030–7034. doi: 10.1073/pnas.95.12.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene E, Lee H, Kustu S. Ammonium/methylammonium transport (Amt) proteins facilitate diffusion of NH3 bidirectionally. Proc Natl Acad Sci U S A. 2002;99:3926–3931. doi: 10.1073/pnas.062043799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg DK, Carlson CA, Bates NR, Johnson RJ, Michaels AF, Knap AH. Overview of the US JGOFS Bermuda Atlantic Time-series Study (BATS): a decade-scale look at ocean biology and biogeochemistry. Deep Sea Res Part II Top Stud Oceanogr. 2001;48:1405–1447. [Google Scholar]
- Steiner HY, Naider F, Becker JM. The PTR family: a new group of peptide transporters. Mol Microbiol. 1995;16:825–834. doi: 10.1111/j.1365-2958.1995.tb02310.x. [DOI] [PubMed] [Google Scholar]
- Suenaga A, Moriya K, Sonoda Y, Ikeda A, Von Wiren N, Hayakawa T, Yamaguchi J, Yamaya T. Constitutive expression of a novel-type ammonium transporter OsAMT2 in rice plants. Plant Cell Physiol. 2003;44:206–211. doi: 10.1093/pcp/pcg017. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Sunderland (MA): Sinauer Associates; 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. [Google Scholar]
- Tejada-Jimenez M, Llamas A, Sanz-Luque E, Galvan A, Fernandez E. A high-affinity molybdate transporter in eukaryotes. Proc Natl Acad Sci U S A. 2007;104:20126–20130. doi: 10.1073/pnas.0704646104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teller S, Schmidt K-H, Appenroth K-J. Ferredoxin-dependent but not NADH-dependent glutamate synthase is regulated by phytochrome and a blue/UV-A light receptor in turions of Spirodela polyrhiza. Plant Physiol Biochem. 1996;34:713–719. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J. The gentle art of gene arrangement: the meaning of gene clusters. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-comment2002. Comment 2002.1–2002.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Viprey M, Guillou L, Ferreol M, Vaulot D. Wide genetic diversity of picoplanktonic green algae (Chloroplastida) in the Mediterranean Sea uncovered by a phylum-biased PCR approach. Environ Microbiol. 2008;10:1804–1822. doi: 10.1111/j.1462-2920.2008.01602.x. [DOI] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 2003;132:556–567. doi: 10.1104/pp.103.021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler PA, McCarthy JJ. Methylammonium uptake by Chesapeake Bay phytoplankton: evaluation of the use of the ammonium analogue for field uptake measurements. Limnol Oceanogr. 1982;27:1129–1140. [Google Scholar]
- Worden AZ. Picoeukaryote diversity in coastal waters of the Pacific Ocean. Aquat Microb Ecol. 2006;43:165–175. [Google Scholar]
- Worden AZ, Lee JH, Mock T, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- Worden AZ, Not F. Ecology and diversity of picoeukaryotes. In: Kirchman DL, editor. Microbial ecology of the oceans. New York: Wiley; 2008. p. 594. [Google Scholar]
- Yuan L, Loque D, Kojima S, Rauch S, Ishiyama K, Inoue E, Takahashi H, von Wiren N. The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell. 2007;19:2636–2652. doi: 10.1105/tpc.107.052134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV. The deep archaeal roots of eukaryotes. Mol Biol Evol. 2008;25:1619–1630. doi: 10.1093/molbev/msn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JJ, Theodoulou FL, Muldin I, Ingemarsson B, Miller AJ. Cloning and functional characterization of a Brassica napus transporter that is able to transport nitrate and histidine. J Biol Chem. 1998;273:12017–12023. doi: 10.1074/jbc.273.20.12017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.