SUMMARY
To identify Pof6’s interactors, we have performed a non-stringent Pof6-TAP purification in two steps from a large scale of cells (40 L) and coupled it to MudPIT analysis (Multidimentional Protein Identification Technology).
We, consequently, decided to name the protein according to that specific feature: Sip1 for pofSix Interactor Protein 1. The domain analysis reveals the presence of a long amino-terminal stretch of HEAT repeats which makes it a candidate as a scaffolding subunit. Consequently, we propose that the three essential proteins Skp1, Pof6 and Sip1 form a ternary complex consisting of a novel type of the non-SCF F-box complex.
Important as the amount of work put on dissecting the actions required for a correct transfer of genetic material to the daughter cells, very little is known about the late phase of septation leading to the physical cells separation.
In this study we isolated by TAP purification and MudPIT analysis a novel interactor of Pof6. This uncharacterised and essential ORF was named Sip1 for PofSix interactor protein 1. Coimmunoprecipitation experiments between Pof6, Sip1 and Skp1 and native purification of Sip1-TAP confirmed the interactions between the three proteins. The isolation of a sip1 loss-of-function mutant reveals lethality in cytokinesis while GFP-Pof6 transiently accumulated at the equatorial zone in late anaphase. We suggest that Sip1, Pof6 and Skp1 play together an essential role in assembly/constriction of the CAR and subsequent cell separation.
INTRODUCTION
Cytokinesis is a critical step in the cell life, as it couples the end of mitosis to the division of the mother cell into two daughter cells. The main goal of cytokinesis is common to all organisms, and even if divergences exist, the fundamental sequences of events are universal [1]. By the end of anaphase, the fission yeasts, like animals, position an actomyosin ring (CAR, Contractile Actomyosin Ring) in the medial cortex region and constrict it to bring about partitioning of the cytoplasm [2–5]. Concomitantly, the septum is deposited on the other side of the ring to separate the incipient daughter cells. Later a secondary septum is formed and the primary membrane is digested to allow the physical separation into two daughter cells.
Not only actin and myosin play essential roles during cytokinesis, but a much larger set of well-conserved and essential proteins account for the proper assembly and activity of the CAR [2, 6, 7]. For example, the anillin-like protein Mid1, in S. pombe and other organisms, defines the position of the CAR [8, 9], and the protein phosphatase Clp1/Flp1, related to human Cdc14, coordinates cytokinesis with cell cycle progression [10]. Clp1 phosphatase fulfils its role via the Septation Initiation Network (SIN) that regulates the sequence of events in septation [11]. Being simple entities, fission yeasts possess the significant advantage of combining genetic and biochemistry techniques; such features make them a powerful model by which to study cytokinesis. Consistently, a precise timing of the process has been reported [12–14] and a plethora of mutants with defects in cytokinesis and septation have been isolated [3, 15, 16].
A role for the ubiquitin proteasome pathways during cytokinesis has been recently reported in budding yeast and may account in other eukaryotes [17]. These findings, seemingly contradictory to the idea that the APC/C (Anaphase Promoting Complex/Cyclosome) ubiquitin ligase triggers all ubiquitylation events in late/post mitosis, display facts that the SCFGrr1 (Skp1-Cullin/Cdc53-F-box protein Grr1) complex recognises substrates at the bud neck for proteasomal degradation. Originally, the F-box proteins were discovered for their essential role in cell cycle progression [18]. The F-box proteins are defined by the presence of a degenerated stretch of 40 amino acids, the F-box domain, normally on their amino-terminal portion, and contain a substrates-binding domain on their carboxy-terminal region [19]. They play the crucial role in recognising in a timely and specific manner a variety of substrates. As an adaptor subunit of the SCF complex, the F-box proteins share common core components [20]; the F-box binding protein Skp1 [21, 22], the scaffold protein Cul1/Cdc53 [22] and the RING-H2 finger protein Roc1/Rbx1 [23, 24].
Beside this well-recognised function, an increasing number of F-box proteins have been described in partnership with Skp1 as non-SCF complexes, particularly in budding yeast [25]. The proteins Skp1-Ctf13 are part of the CBF3 complex, a core DNA-binding structure of the centromere in S. cerevisiae [21, 26]. The proteins Skp1-Rcy1 are important for membrane recycling and form a portion of the SNARE complex (Soluble NSF Attachment protein Receptor) [27, 28]. In addition Skp1-Rav1 and Rav2 F-box proteins are related to the RAVE (Regulator of the H+ −ATPase of the Vacuolar and Endosomal membranes) complex involved in vacuolar ATPase assembly [29], which has recently been shown to be the case for fission yeast [30].
The S. pombe genome appears to contain 17 genes encoding F-box proteins based on computational analysis and two-hybrid assays using Skp1 as bait [25, 31]. This restricted number, in comparison to higher eukaryotes like the plant A. thaliana (approximately 700 F-box genes) or human (38 estimated genes) [32], is a genuine asset considering the diversity of the F-box proteins functionality. Among the fission yeast F-box proteins only two are essential, Pof1 and Pof6 [31, 33, 34]. Pof1, a counterpart of budding yeast Met30, has all the classical features of an F-box protein (SCFPof1): a WD40 repeat substrate binding domain in the carboxy-terminal region and poly-ubiquitylating function of a phosphorylated substrate, the transcription factor Zip1 [34]. While Pof1 follows the traditional pattern, Pof6 seems to be less canonical. Pof6 was first identified as a Skp1 interactor and no binding to a cullin was reported, bringing up the idea of a non-SCF complex [33]. In support to this, its closest ortholog in budding yeast, Rcy1, interacts with Skp1 but not cullins [28]. Further genetic characterisation of Pof6 and Skp1 revealed that both proteins are essential for cell separation in fission yeast [33].
In this study we isolated by TAP purification and MudPIT analysis a novel interactor of Pof6. This uncharacterised and essential ORF was named Sip1 for PofSix interactor protein 1. Coimmunoprecipitation experiments between Pof6, Sip1 and Skp1 and native purification of Sip1-TAP confirmed the interactions between the three proteins. The isolation of a sip1 loss-of-function mutant reveals lethality in cytokinesis while GFP-Pof6 transiently accumulated at the equatorial zone in late anaphase. We suggest that Sip1, Pof6 and Skp1 play together an essential role in assembly/constriction of the CAR and subsequent cell separation.
Experimental Procedures
Yeast strains
Yeast strains used in this study are described in Table I. Cells were grown in standard culture media, and standard yeast genetic methods were used. The carboxy- and amino-terminal epitope-tagged proteins were generated via chromosomal integration of PCR-amplified fragments [35–37]. The carboxy-terminal tagging of Sip1 by GFP compromised its function (data not shown), and as a result an amino-terminal GFP tagged strain was used for visualisation of Sip1 localisation. As for Pof6, either carboxy- or amino-terminal GFP tagging is functional and displays similar patterns of localisation (see below and data not shown).
Table I.
Strain list
| Strain name | Genotypes | Derivations |
|---|---|---|
| 513 | h− leu1 ura4 | Our stock |
| 972 | h− | Our stock |
| NSY001 | h− pof6+-2TAP-kanr | This study |
| NSY065 | h− leu1 ura4 sip1+-3HA-kanr | This study |
| NSY069 | h− or + pof6+-2TAP-hphr sip1+-3HA-kanr | This study |
| NSY110 | h−/h+ leu1/leu1 ura4/ura4 his7/his7 ade6–210/ade6–216 sip1::kanr/+ | This study |
| NSY127 | h− leu1 ura4 sip1+-2TAP-kanr | This study |
| NSY165 | h− leu1 ura4 sip1–62-HA-kanr | This study |
| NSY178 | h− leu1 ura4 myo2+-GFP-kanr | Our stock |
| NSY191 | h− or + sip1–62-HA-kanr myo2+-GFP-kanr | This study |
| NSY204 | h− or + sad1+-dsRED-leu2 kanr-nmt1-GFP-pof6+ | This study |
| NSY206 | h− or + sad1+-dsRED-leu2 kanr-nmt1-GFP-sip1+ | This study |
Construction of sip1 temperature sensitive alleles
Template DNA was prepared from a strain containing sip1+-HA linked to the G418 resistance marker gene at 3’ end. This cassette was amplified with error-prone PCR and transformed into a wild type strain. A total of 4000 to 5000 transformants was obtained on G418 plates at 27°C and temperature sensitive (ts) alleles were counter-selected at 36°C on phloxine B-containing plates. We isolated 6 ts alleles whose ts phenotypes co-segregated with G418 resistance, indicating that mutations occur in the sip1 locus. One allele, sip1–62 ts, was selected for further characterisation.
TAP purifications
The TAP purifications were mainly prepared as described previously [38]. For Pof6-TAP purification, 40 L of cells were grown in YE5S at 30°C and disrupted in liquid nitrogen. Proteins were solubilised in the lysis buffer (50 mM Tris-HCl pH8.0, 150 mM NaCl, 5 mM EDTA, 10 % glycerol, 0.2 % NP-40, 1 mM NaF, 10 mM PMSF, adjusted to pH 8.0) and loaded for a two-step TAP purification. The eluted proteins were precipitated with TCA and subjected to MudPIT analysis as described elsewhere [39]. For Sip1-TAP purification, a total of 10 L of cells were cultured in YE5S at 30°C and disrupted in liquid nitrogen. The proteins were eluted in a single-step TAP purification and precipitated in TCA for further MudPIT analysis.
Antibody preparation
DNA sequence corresponding to amino acid residues from 1 to 781 of Pof6 was PCR amplified and cloned into expression vector pET-14b (Novagen, Madison, WI). 6-His tagged Pof6 fusion protein was purified on Ni2+-NTA beads (Qiagen, Valencia, CA) as recommended by the manufacturer and rabbit polyclonal anti-Pof6 sera were prepared. Crude anti-Pof6 serum was further affinity-purified using fusion proteins. Preparation of rabbit polyclonal anti-Skp1 antibody was described previously [31].
Coimmunoprecipitation experiments
Protein were extracted in lysis buffer by breaking the cells at 4°C with glass beads (4 times for 40 s) in a FastPrep FP120 apparatus (Savant. Co, MN, USA). The protein extracts were collected after 15 min of centrifugation at 10,000 × g at 4°C. The coimmunoprecipitations were performed as described previously [40].
Synchronisation by elutriation and FACS analysis
3 L of YE5S media were inoculated with a preculture overnight. Elutriation was performed with Avanti J-20 XP (Beckman Coulter, MN, USA) by the standard procedures. Cell concentration and size were examined with a Cell Counter (Sysmex KX-21N, Kobe, Japan). The septation index was calculated by microscope observation of calcofluor-stained cells and counting over a total of 100 cells; the cell survivals were estimated by spreading a calculated number of cells on YE5S plates and incubating them at 30°C for 3–4 days. Standard methods for FACS analysis were followed using a FACScan machine (Becton Dickinson, CA, USA).
Visualisation of protein localisation
GFP-Pof6 and GFP-Sip1 were observed on lived cells at 30°C with the DeltaVision interface (Applied Precision, Washington, USA). A picture was taken every 30 s during the time-lapse experiment. For the observation of Myo2-GFP localisation, the cells were incubated at 25°C to mid-log phase. Half of the cultures were shifted to 36°C for 1 h and the other half left at 25°C. The observations were made on live cells with Volocity program (Applied Precision, Washington, USA).
Bioinformatic studies
The F-box domains of the F-box proteins Pop1, Skp2, Pof1, Pop2 and Pof6 were aligned by ClustalW alignment, default settings, in MacVector 9.0. The Skp1 interface residues and the Cul1 interface residues were marked accordingly to [41]. The protein domains of Pof6 were found described on NCBI protein website, accession number CAA21418. To identify domains in Sip1 protein, Sip1 and its closest homologue, Sls2 from Yarrowia lipolytica, were aligned and submitted to HHpred (Homology detection & structure prediction by HMM-HMM comparison) on Marx-Planck Institute for Developmental Biology Bioinformatics Toolkit website (http://toolkit.tuebingen.mpg.de/hhpred). The orthologues of Sip1 were identified by YOGI search (eukarYotic OrtholoGY) tool and KOGs classification (euKaryotic clusters of Orthologous Groups of proteins) in S. pombe Gene DB database. The resulting proteins were organised in a guide tree by using the default settings of ClustalW alignment in MacVector 9.0.
RESULTS
Identification of a novel interactor of the Pof6 F-box protein
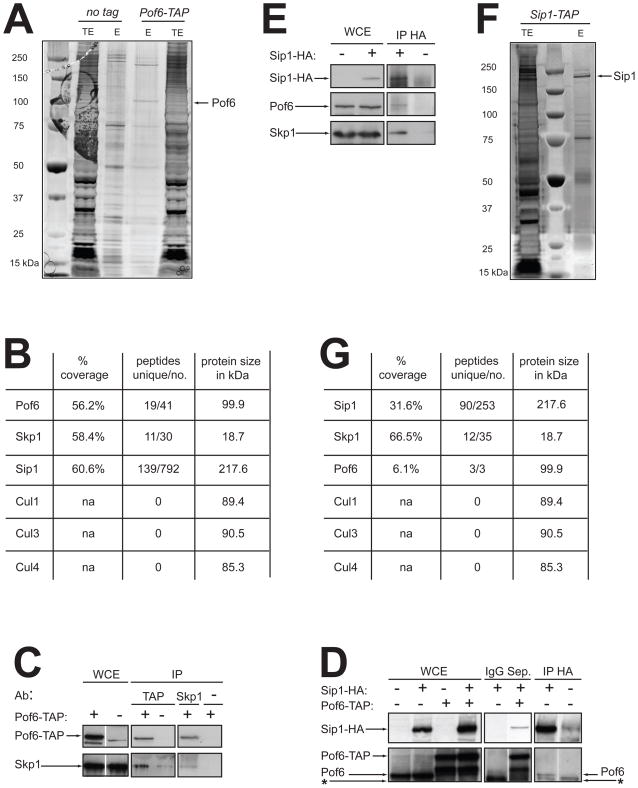
To identify Pof6 interactors, we have performed a non-stringent Pof6-TAP purification in two steps from a large scale of cells (40 L) and coupled it to MudPIT analysis (Multidimentional Protein Identification Technology). A protein migrating at the size of Pof6 is specifically detected in the silver stain of Pof6-TAP elution (Fig. 1A); the MudPIT data reveals a high coverage of Pof6 (19 unique peptides covered 56.2% the sequence) considering the size of the protein, 99.9 kDa (Fig. 1B). The protein Skp1 appears in the elution with 58.4% coverage and a total of 30 peptides (Fig. 1B). The specificity of the interaction between Pof6 and Skp1 was confirmed by coimmunoprecipitations with both proteins (Fig. 1C) as shown previously [33].
Fig. 1. Identification of a novel interactor of Pof6.
A. TAP purifications of a non-tagged control strain (972, Table 1) and Pof6-TAP. Fractions corresponding to 1/100 of each elution (E) were loaded in parallel to 100 μg of total extract (TE) and silver-stained. A band migrating around 100 kDa may correspond to Pof6 and is indicated by an arrow. B. Data of the most relevant interactors identified in Pof6-TAP’s MudPIT analysis. The percentage of protein coverage, and the number of unique peptides versus the number of total peptides with the size in kDa are shown. C. Coimmunoprecipitation of Pof6-TAP and Skp1. Proteins were Immunoprecipitated (IP) with anti-TAP, anti-Skp1 or no antibodies (α). 100 μg of whole cell extract (WCE) were loaded and 2 mg of proteins were used for each IP. Pof6-TAP was immunodetected with anti-Pof6 antibodies and Skp1 with anti-Skp1 antibodies. D. Coimmunoprecipitation of Pof6/Pof6-TAP and Sip1-HA. Proteins were immunoprecipitated with IgG Sepharose beads or anti-HA antibodies. Sip1-HA was immunodetected with anti-HA antibodies, and Pof6 with anti-Pof6 antibodies. The asterisk indicates the non-specific band of the IgG. E. Coimmunoprecipitation of Sip1-HA, Pof6 and Skp1. Sip1-HA was immunoprecipitated with anti-HA antibodies and immunodetected with the same antibodies. Pof6 was immunodetected with anti-Pof6 antibodies and Skp1 with anti-Skp1 antibodies. F. Single step TAP purification of Sip1-TAP. 1/100 of the final elution (E) was loaded on the gel in parallel to 100 μg of total extract (TE) and silver-stained. A band migrating around 220 kDa possibly represents Sip1. G. Data obtained from the Sip1-TAP’s MudPIT analysis.
An uncharacterised ORF (SPBC27B12.08) was identified as one of the most abundant protein pulled down in Pof6-TAP purification. Although the corresponding protein has the large size of 217 kDa, the peptide coverage reaches up to 60.6% of the sequence and a total of 792 peptides were isolated (Fig. 1B). The specificity of the interaction between Pof6 and this novel protein was confirmed by coimmunoprecipitation (Fig. 1D). We, consequently, decided to name the protein according to that specific feature: Sip1 (pofSix interactor protein 1).
In a reciprocal experiment, we performed a single step Sip1-TAP purification from 10 L of cells to identify its partners (Fig. 1F). One of the most abundant proteins identified by MudPIT analysis is Skp1 (Fig. 1G). The peptides cover 66.5% of Skp1 sequence and a total of 35 peptides are present. The F-box protein Pof6 is also detected as an interactor of Sip1 albeit with a much restricted coverage (6.1%). The difference between Pof6 coverage in Sip1-TAP elution and Sip1 coverage in Pof6-TAP elution may be explained by the amount of cells used for the purifications, the differences in the TAP purification protocols and the abundance/instability of each protein. To confirm the interactions observed by Sip1-TAP MudPIT analysis, Sip1-HA was immunoprecipitated (Fig. 1E). We found Pof6 and Skp1 to be specifically pulled down with Sip1-HA. It therefore appears that Sip1, Pof6 and Skp1 form a ternary complex in the cell.
None of the two MudPIT datasets contains peptides for cullins (Fig. 1B, G) confirming the absence of interaction between Pof6 and the cullins previously reported [33]. We made a careful visual inspection of the sequence corresponding to the Pof6 F-box domain and aligned it with different F-box domains present in SCF complexes [41, 42] (see Supplementary Data, Fig. S1A). The alignment shows that several residues important for the Skp1/F-box domain interface are present in Pof6 but the two residues involved in the Cul1/F-box domain core interface are not conserved; highly conserved proline is replaced by threonine in Pof6 and an acidic residue is exchanged with asparagine. Importantly, these residues have been shown to be specifically responsible for the interaction between Skp2 F-box protein and Cul1 [42].
The analysis of the Pof6 protein also reveals the presence of unusual domains in its carboxy-terminal region: two Sec10 domains and a CAAX sequence are present in the substrate-binding portion (see Supplementary Data, Fig. S1B). Such features suggest that there may be potential functional similarities between Pof6 and Rcy1 from budding yeast. Like Pof6, Rcy1 interacts with Skp1 and contains two Sec10 domains and a CAAX sequence in its carboxy-terminal region. In addition it has been suggested that Rcy1 is part of a non-SCF complex [28]. However, unlike Pof6 in fission yeast, Rcy1 is non-essential in budding yeast [33]. These results collectively suggest that Pof6 does not belong to a conventional SCF complex and instead forms a novel complex with the uncharacterised protein Sip1 and Skp1.
The sip1+ gene is essential in S. pombe and conserved amongst eukaryotes
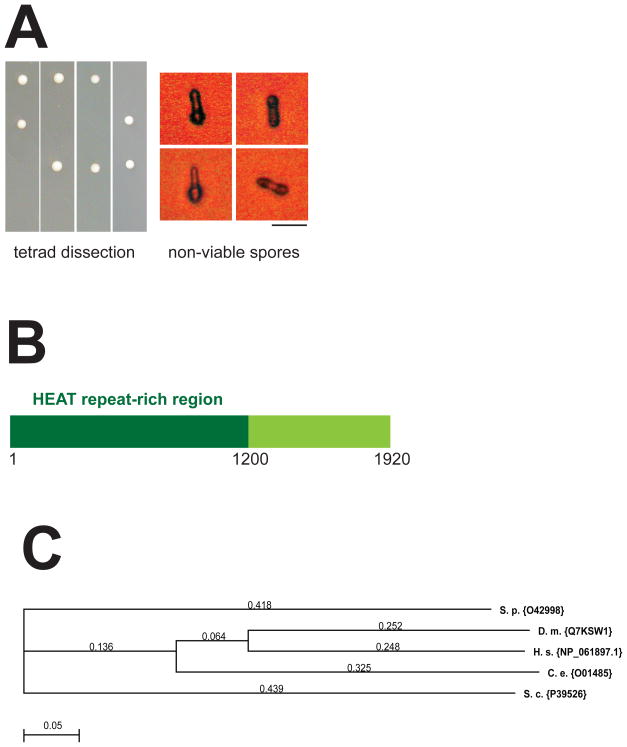
To investigate Sip1 function, we asked whether the sip1+ gene is essential in S. pombe. We deleted by PCR one copy of the gene with a G418-resistant cassette in a diploid strain. This sip1::kanr/sip1+ strain was left to sporulate and the resulting tetrads were dissected to check the viability of each haploid progeny. Only two out of four haploids grew into colonies after germination, and these viable colonies were sensitive to G418 (Fig. 2A left panel). We concluded that sip1+, like pof6+ and skp1+, is essential in S. pombe. A close observation of the non-viable spores showed germinations occur but not cell division (Fig. 2A right panel).
Fig. 2. sip1+ is an essential gene present amongst eukaryotes.
A. Tetrad dissection of asci produced by the sip1::kanr/sip1+ diploid strain. Spores were grown for 3 days at 30°C on rich media YPD (left). Phase contrast micrographs of spores showing the terminal morphology of a deduced sip1-disrupted spore (right). Bar indicates 5 μm. B. Schematic of Sip1 protein domains. Sip1 protein is 1920 amino acids long and contains an amino-terminal region rich in HEAT repeats (1–1200 aa). C. Dendogram of Sip1 orthologues identified by YOGY search. The proteins are labelled with the UniProtKB nomenclature. The ClustalW alignment default parameters of MacVector 9.0 were used to create the tree. The evolutionary distance is indicated on each branch. S.p: Schizosaccharomyces pombe; D.m: Drosophila melanogaster; H.s: Homo sapiens; C.e: Caenorhabditis elegans; S.c: Saccharomyces cerevisiae.
Next, we used the bioinformatic tool kit HH pred from the Max Planck Institute (http://toolkit.tuebingen.mpg.de/hhpred) to detect Sip1 homologues and structures by HMM-HMM prediction. The search reveals that Sip1 is structurally related to CAND1 protein (Cullin-associated and neddylation-dissociated protein 1) on its amino-terminal region [43]. The homology highlights a HEAT repeat-rich region from residue 1 to 1200, which are typically implicated in protein-protein interactions and often found in subunits of multiple-proteins complexes (Fig. 2B). Finally, Sip1 protein appears to be conserved throughout the evolution (Fig. 2C).
The sip1 ts allele is lethal during cytokinesis
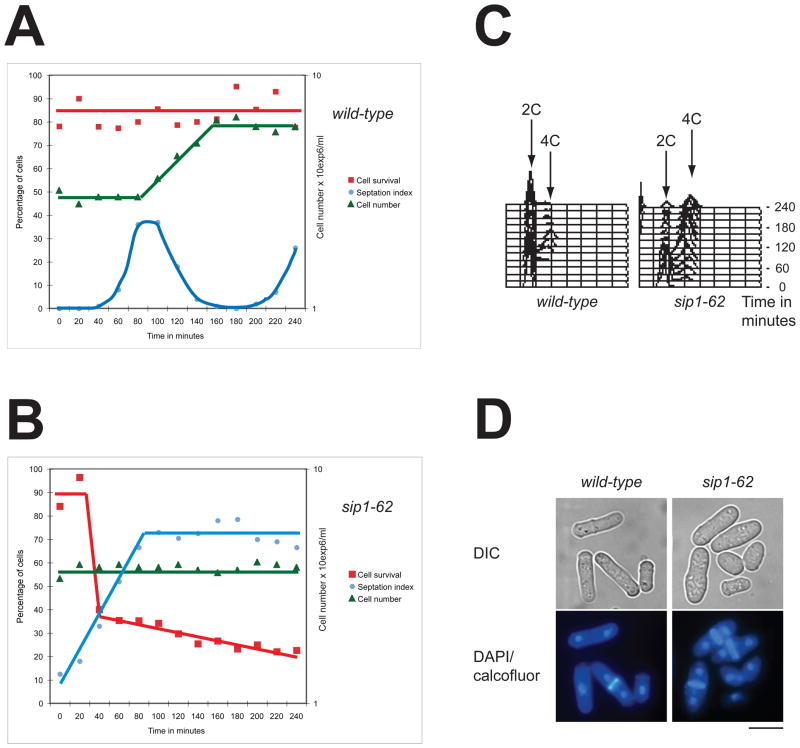
To examine the function of Sip1, we produced temperature sensitive alleles of sip1+ by mutagenic PCR (see Experimental procedures). Elutriation centrifugation was used to synchronise cell cultures in early G2 in both wild type and sip1–62 ts. The sip1–62 ts cells were grown at 25°C then shifted to 36°C where they arrested (Fig. 3B), while the wild type cells kept on dividing (Fig. 3A). Surprisingly sip1–62 ts cells lost viability dramatically concomitant with the increase of the septation index (40–60 min, Fig. 3B). The percentage of septated cells reached a plateau of 70% in 100 minutes and did not decline afterwards (Fig. 3B). The population of the binucleate cells closely followed the profile of the septated cells (data not shown). These observations suggest that sip1 ts cells proceed through the G2 phase and enter mitosis to lose viability during cytokinesis. The DNA content of the synchronised cells showed that sip1–62 ts cells arrest with a 4C DNA content, indicating a completion of DNA replication in each separated nucleus (Fig. 3C). The vast majority of the sip1–62 ts arrested cells contain two separated diploid nuclei with an abnormally thick septum in their division plane (Fig. 3D). Altogether, we concluded that Sip1 is required for the completion of cytokinesis and plays an essential role in this process. Importantly, we observed extremely few cells that complete their constriction of the equatorial plane.
Fig. 3. Characterisation of sip1–62 ts mutants.
Wild type (A) or sip1–62 cells (B) were synchronised by elutriation at 27°C, shifted to 36°C (T0) and grown for 4 h. Samples were taken every 20 min to measure cell viability, septation index and cell numbers. C. DNA content analysis of wild type and sip1–62 ts samples treated as described in A. Every 20 min a time-point was taken. D. Terminal phenotypes of sip1–62 ts cells (4 h 36°C) and wild type cells. The cells are visualised by phase contrast (upper) or by fluorescence after DAPI and calcofluor double staining (lower). Bar indicates 10 μm.
GFP-Pof6 concentrates at the equatorial region in telophase
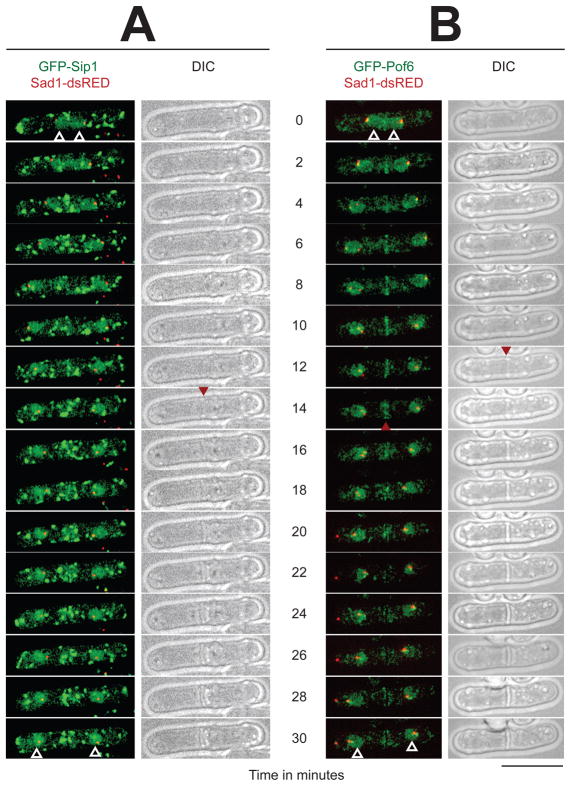
To investigate Sip1 and Pof6 localisation, we tagged them with the GFP protein on their amino-terminal end and expressed them under the thiamine-repressible nmt1 promoter (in the presence of 5 μM thiamine, see Experimental procedures). The two strains also contain a dsRED (a derivative of RFP) marker fused to Sad1 [44] to detect the spindle pole bodies simultaneously with the GFP fused proteins. We performed time-lapse imaging on live cells incubated at 30°C to visualise the protein localisation during the cell cycle. GFP-Sip1 is detected in the nuclei (Fig. 4A, white arrowheads) and as numerous large bright dots moving in the cytoplasm. These patterns do not dramatically change during the entire cell cycle. At the moment the molecular entities of these cytoplasmic structures are not known, but they may correspond to trafficking vesicles, which are recently visualised in fission yeast [45].
Fig. 4. Localisation of GFP-Sip1 and GFP-Pof6 from late mitosis.
Live-cells observations by fluorescence and phase contrast imaging. The proteins GFP-Sip1 (A) and GFP-Pof6 (B) were both expressed under the thiamine-repressible nmt1 promoter. The cells grew in minimal media containing 5 μM thiamine at 30°C. The time zero is defined as the two spindle pole bodies are already separated (Sad1-dsRED) and the nuclei start migrating from the medial region. The shots were taken every 2 min over a period of time of 30 min. The white arrows indicate the nuclei at the start and at the end of the experiment. The red arrows point the appearance of the septum (DIC columns) and the disappearance of GFP-Pof6 from the equatorial zone (B). Bar indicates 10 μm.
GFP-Pof6, like GFP-Sip1, is present in the nuclei (Fig. 4B, white arrowheads in the right panels) and as cytoplasmic dots. However compared to those of GFP-Sip1, GFP-Pof6 dots are much less intense and smaller in size. Interestingly, GFP-Pof6 also appears in the medial region during anaphase B (Fig. 4B, arrowhead in the right panels). This localisation of GFP-Pof6 occurs at the site of the incipient septum (T2–T12). GFP-Pof6 then slowly disappears from the equatorial region as the new cell wall become visible (Fig. 4A and B, arrowheads in the right DIC panels). GFP-tagging of Pof6 in the C-terminal region under the endogenous promoter also gives similar, albeit less intense, patterns. In conclusion, a live cell analysis of GFP-Sip1 and GFP-Pof6 localisation demonstrates that both proteins are present in the nuclei throughout the cell cycle. Sip1 localisation remains unchanged throughout the cell cycle, whereas Pof6 is found as a strip at the equatorial region in late anaphase/early telophase. It is of note that several proteins are transported to the equatorial region just before the appearance of the new cell wall [12]. This phenomenon coincides with Pof6 concentration to the same zone. Additionally, as soon as the septum appears by optical microscopy, GFP-Pof6 signal disappears from that region.
Sip1 is required for type II myosin to be at the contractile ring
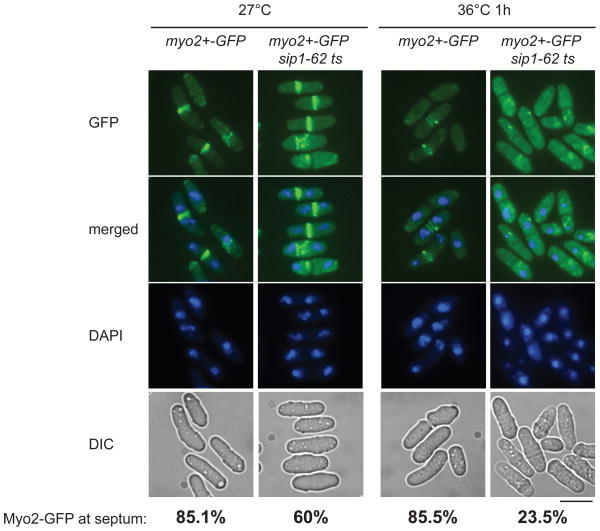
In S. pombe, like in other organisms, the CAR is assembled as cells enter mitosis and contains numerous proteins including actin, type II myosin, and various actin- and myosin-binding proteins [13, 14, 46]. The assembly of the CAR is essential for the positioning and functional integrity of the cytokinetic septum and following cell division. The type II myosin heavy chain protein, Myo2, is recruited to the division site during mitosis soon after a broad band of nodes is formed, which condenses into a sharp CAR containing actin, myosin, α-actinin and other proteins. To address the effect of Sip1 on a CAR component’s localisation, we examined myo2+-GFP in a sip1–62 ts strain. Because a myo2+-GFP sip1–62 ts strain presents marginal growth defects even at 27°C, we maintained these cells at 25°C.
We first compared the detection of Myo2-GFP in the wild type and the sip1–62 ts strains on live cells grown at 25°C over night and shifted at 27°C for 2 h (Fig. 5). Next, we shifted cultures to 36°C for 1 h to inactivate sip1–62 ts function and observed the localisation of Myo2-GFP on live cells. In that case, Myo2-GFP behaviour differs significantly between the two strains. In wild type cells at 36°C, Myo2-GFP remains at the medial zone, though the intensity of GFP signals at the medial ring somewhat plunges for unknown reasons (Fig. 5, myo2+-GFP). In contrast, under the same condition, sip1–62 ts cells showed almost complete delocalisation of Myo2-GFP, in which GFP signals appear to form undefined cytoplasmic aggregates or diffuse in the whole cytoplasm (Fig. 5, myo2+-GFP sip1–62 ts). Therefore Sip1, a newly identified conserved protein, plays an essential role in cytokinesis, in which it is required for a persistent equatorial localisation of an essential component of the CAR upon cell division.
Fig. 5. Delocalisation of the type II myosin heavy chain Myo2 in sip1–62 ts allele.
Live cells imaging of Myo2-GFP in wild type and sip1–62 ts strains. The cells were visualised for GFP fluorescence (GFP), DAPI fluorescence (DAPI), and phase contrast (DIC). The pictures of GFP and DAPI fluorescence were merged to identify the localisation of Myo2. Representative images are shown after 2 h at 27°C (A) and 1 h at 36°C (B). The percentages of binucleate cells displaying Myo2-GFP at the septum were calculated from a population of 100 cells for myo2+-GFP at 27°C and 36°C and 200 cells for myo2+-GFP sip1–62 ts at 27°C and 36°C. Bar indicates 10 μm.
DISCUSSION
In this report, we identify an uncharacterised protein, Sip1, as a novel component of the Pof6/Skp1 complex in S. pombe. The protein was detected as an interactor of Pof6 by TAP purification coupled to MudPIT analysis, and the bindings were confirmed by coimmunoprecipitation experiments. Sip1 is essential in S. pombe and conserved amongst eukaryotes. The domain analysis reveals the presence of a long amino-terminal stretch of HEAT repeats which makes it a candidate as a scaffolding subunit. Consequently, we propose that the three essential proteins Skp1, Pof6 and Sip1 form a ternary complex comprising a novel type of the non-SCF F-box complex.
Careful analysis of a sip1–62 ts strain has unravelled an essential role for Sip1 in cytokinesis and cell separation. It should be noted that a ts allele of skp1 in S. pombe, skp1–3, or pof6-deleted haploid cells germinating from heterozygous diploid strains exhibited cytokinetic defects with abnormally thick septa [33, 47], reminiscent of the terminal phenotypes of sip1–62 cells reported in this study. The GFP-Pof6 protein localises to the nuclei as reported earlier [33], and transiently accumulates at the site of the CAR in late anaphase as the nuclei migrate to opposite poles. We do not currently know the mechanism of how Pof6 alters its localisation during the cell cycle. The protein amount of Pof6 might be regulated post-translationally including proteolysis, as our preliminary data indicates that Pof6 is polyubiquitylated (N. S. and T. T., unpublished results). It is tempting to speculate the Pof6 protein is spatially and temporally regulated at the CAR in late mitosis, in which the nuclear population of Pof6 might acts as a reservoir. More analysis is required to address this point. It is of note that the cell cycle-dependent localisation of Pof6 looks similar, if not identical, to that of an anillin homologue Mid1 [8, 9], though the physiological significance of the nuclear pool of Mid1 remains elusive at the moment.
Although Sip1 coprecipitates with Pof6 (and Skp1), its cellular localisation does not parallel exactly that of Pof6. We detect GFP-Sip1 as discrete cytoplasmic dots in addition to uniform nuclear signals. The presence of Sip1 and Pof6 in the nucleus correlates with the biochemical interaction data and supports the idea that the complex exists at least in this compartment. As the size and number of Sip1 dots are large, it is not conclusive as to whether Sip1 localises to the medial region during cytokinesis. Nonetheless it is noticeable that GFP-Sip1 vesicles appear to constantly gather around the CAR in cytokinesis. To further clarify the role of Sip1 in cytokinesis, we monitored the localisation of myosin II heavy chain in a sip1–62 ts background and observed that the inactivation of Sip1 triggers Myo2-GFP delocalisation. This phenomenon can be ascribable to a defect in either Myo2 transport or its retention at the medial region. Sip1-containing vesicles might bring Pof6 to the medial ring in anaphase B and delocalise it during late telophase.
Orthologues of Pof6 and Sip1 can be found in budding yeast. Like Pof6, Rcy1 also forms a non-SCF complex and participates in protein recycling from the endosomes to the Golgi and the plasma membrane [28, 48]. However, unlike Pof6, Rcy1 is non-essential and these two proteins do not appear to perform identical molecular functions [49–51]. So far we are not able to detect Pof6 orthologs by homology search in higher eukaryotes. The identification of functional orthologues of Pof6 in higher eukaryotes would be crucial to delineate their conserved roles. In the case of Sip1, its budding yeast ortholog is called Laa1. Like Rcy1 and unlike Sip1, Laa1 is non-essential and involved in protein transport between the trans-Golgi network and endosomes [52]. Currently no functional or physical interaction between Rcy1 and Laa1 is known, though Rcy1 forms a complex with Skp1. No studies on Sip1 orthologues in other eukaryotes have been performed.
In summary, we describe the first non-SCF complex in S. pombe. We show this complex plays an essential role in cell division. Further analysis points towards the role for the Skp1-Pof6-Sip1 complex in cytokinesis and possibly protein trafficking. Having seen the highly degenerate nature of F-box sequences and the existence of Sip1 orthologues, we envision that non-SCF F-box complexes are function, the elucidation of the precise roles for the Skp1-Pof6-Sip1 complex in protein trafficking and cytokinesis and the identification of interacting proteins of Sip1 orthologues in higher eukaryotes would be the next phase of our investigation.
Supplementary Material
Acknowledgments
We appreciate the gift of a Myo2-GFP strain from Dan Mulvihill and TAP-tagging plasmids from Kathy Gould. We are grateful to Damien Hermand for exchanging unpublished information. We thank the Fermentation Unit of Cancer Research UK for preparations of large-scale fission yeast cultures. The work was supported by Cancer Research UK (T. T.) and National Institutes of Health grant P41RR011823. (J.R.Y.)
References
- 1.Barr FA, Gruneberg U. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Rajagopalan S, Wachtler V, Balasubramanian M. Trends Genet. 2003;19:403–408. doi: 10.1016/S0168-9525(03)00149-5. [DOI] [PubMed] [Google Scholar]
- 3.Guertin DA, Trautmann S, McCollum D. Microbiol Mol Biol Rev. 2002;66:155–178. doi: 10.1128/MMBR.66.2.155-178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Goff X, Utzig S, Simanis V. Curr Genet. 1999;35:571–584. doi: 10.1007/s002940050455. [DOI] [PubMed] [Google Scholar]
- 5.Feierbach B, Chang F. Curr Opin Microbiol. 2001;4:713–719. doi: 10.1016/s1369-5274(01)00273-9. [DOI] [PubMed] [Google Scholar]
- 6.Marks J, Hagan IM, Hyams JS. J Cell Sci Suppl. 1986;5:229–241. doi: 10.1242/jcs.1986.supplement_5.15. [DOI] [PubMed] [Google Scholar]
- 7.Pelham RJ, Chang F. Nature. 2002;419:82–86. doi: 10.1038/nature00999. [DOI] [PubMed] [Google Scholar]
- 8.Sohrmann M, Fankhauser C, Brodbeck C, Simanis V. Genes Dev. 1996;10:2707–2719. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- 9.Paoletti A, Chang F. Mol Biol Cell. 2000;11:2757–2773. doi: 10.1091/mbc.11.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clifford DM, Wolfe BA, Roberts-Galbraith RH, McDonald WH, Yates JR, 3rd, Gould KL. J Cell Biol. 2008;181:79–88. doi: 10.1083/jcb.200709060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Wang H, Balasubramanian MK. J Cell Sci. 2000;113 ( Pt 7):1223–1230. doi: 10.1242/jcs.113.7.1223. [DOI] [PubMed] [Google Scholar]
- 12.Wu JQ, Kuhn JR, Kovar DR, Pollard TD. Dev Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- 13.Wu JQ, Pollard TD. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 14.Vavylonis D, Wu JQ, Hao S, O'Shaughnessy B, Pollard TD. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 15.Simanis V. Semin Cell Biol. 1995;6:79–87. doi: 10.1016/1043-4682(95)90004-7. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Tang X, Liu J, Trautmann S, Balasundaram D, McCollum D, Balasubramanian MK. Mol Biol Cell. 2002;13:515–529. doi: 10.1091/mbc.01-11-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blondel M, Bach S, Bamps S, Dobbelaere J, Wiget P, Longaretti C, Barral Y, Meijer L, Peter M. EMBO J. 2005;24:1440–1452. doi: 10.1038/sj.emboj.7600627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 19.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 20.Galan JM, Peter M. Proc Natl Acad Sci U S A. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P. Mol Cell. 1999;4:21–33. doi: 10.1016/s1097-2765(00)80184-7. [DOI] [PubMed] [Google Scholar]
- 22.Patton EE, Willems AR, Sa D, Kuras L, Thomas D, Craig KL, Tyers M. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohta T, Michel JJ, Schottelius AJ, Xiong Y. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 24.Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies RJ. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hermand D. Cell Div. 2006;1:30. doi: 10.1186/1747-1028-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiederkehr A, Avaro S, Prescianotto-Baschong C, Haguenauer-Tsapis R, Riezman H. J Cell Biol. 2000;149:397–410. doi: 10.1083/jcb.149.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galan JM, Wiederkehr A, Seol JH, Haguenauer-Tsapis R, Deshaies RJ, Riezman H, Peter M. Mol Cell Biol. 2001;21:3105–3117. doi: 10.1128/MCB.21.9.3105-3117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seol JH, Shevchenko A, Deshaies RJ. Nat Cell Biol. 2001;3:384–391. doi: 10.1038/35070067. [DOI] [PubMed] [Google Scholar]
- 30.Dawson K, Toone WM, Jones N, Wilkinson CR. Eukaryot Cell. 2008;7:926–937. doi: 10.1128/EC.00037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehmann A, Katayama S, Harrison C, Dhut S, Kitamura K, McDonald N, Toda T. Genes Cells. 2004;9:367–382. doi: 10.1111/j.1356-9597.2004.00730.x. [DOI] [PubMed] [Google Scholar]
- 32.Reed SI. Results Probl Cell Differ. 2006;42:147–181. doi: 10.1007/b136681. [DOI] [PubMed] [Google Scholar]
- 33.Hermand D, Bamps S, Tafforeau L, Vandenhaute J, Makela TP. J Biol Chem. 2003;278:9671–9677. doi: 10.1074/jbc.M211358200. [DOI] [PubMed] [Google Scholar]
- 34.Harrison C, Katayama S, Dhut S, Chen D, Jones N, Bahler J, Toda T. Embo J. 2005;24:599–610. doi: 10.1038/sj.emboj.7600536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 36.Sato M, Dhut S, Toda T. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- 37.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 38.de Bruin RA, McDonald WH, Kalashnikova TI, Yates J, 3rd, Wittenberg C. Cell. 2004;117:887–898. doi: 10.1016/j.cell.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 39.McDonald WH, Yates JR., 3rd Dis Markers. 2002;18:99–105. doi: 10.1155/2002/505397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spielewoy N, Flick K, Kalashnikova TI, Walker JR, Wittenberg C. Mol Cell Biol. 2004;24:8994–9005. doi: 10.1128/MCB.24.20.8994-9005.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 42.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 43.Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 44.Hagan I, Yanagida M. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vjestica A, Tang XZ, Oliferenko S. Mol Biol Cell. 2008;19:1125–1138. doi: 10.1091/mbc.E07-07-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piekny AJ, Glotzer M. Curr Biol. 2008;18:30–36. doi: 10.1016/j.cub.2007.11.068. [DOI] [PubMed] [Google Scholar]
- 47.Kim N, Yoon H, Lee E, Song K. J Microbiol. 2006;44:641–648. [PubMed] [Google Scholar]
- 48.Lafourcade C, Galan JM, Gloor Y, Haguenauer-Tsapis R, Peter M. Mol Cell Biol. 2004;24:3815–3826. doi: 10.1128/MCB.24.9.3815-3826.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen SH, Chen S, Tokarev AA, Liu F, Jedd G, Segev N. Mol Biol Cell. 2005;16:178–192. doi: 10.1091/mbc.E04-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furuta N, Fujimura-Kamada K, Saito K, Yamamoto T, Tanaka K. Mol Biol Cell. 2007;18:295–312. doi: 10.1091/mbc.E06-05-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kus BM, Caldon CE, Andorn-Broza R, Edwards AM. Proteins. 2004;54:455–467. doi: 10.1002/prot.10620. [DOI] [PubMed] [Google Scholar]
- 52.Fernandez GE, Payne GS. Mol Biol Cell. 2006;17:3304–3317. doi: 10.1091/mbc.E06-02-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.