Summary
Human cancer genomes are characterized by widespread aberrations in DNA methylation patterns including DNA hypomethylation of mostly repetitive sequences and hypermethylation of numerous CpG islands. The analysis of DNA methylation patterns in cancer has progressed from single gene studies examining potentially important candidate genes to a more global analysis where all or almost all promoter and CpG island sequences can be analyzed. We provide a brief overview of these genome-scale methylation-profiling techniques, summarize some of the information that has been obtained with these approaches and discuss what we have learned about the specificity of methylation aberrations in cancer at a genome-wide level. The challenge is now to identify those methylation changes that are thought to be crucial for the processes of tumor initiation, tumor progression or metastasis and distinguish these from methylation changes that are merely passenger events that accompany the transformation process but have no effect per se on the process of carcinogenesis.
Introduction
DNA methylation is one of the important epigenetic mechanisms that control gene expression, chromatin structure, genome stability and X chromosome inactivation (Geiman and Robertson, 2002; Jones and Baylin, 2002). Abnormality in DNA methylation can lead to serious imbalance in normal function of cells and can promote pathological conditions. In particular, the genome of cancer cells is known to undergo substantial changes in DNA methylation (Jones and Baylin, 2002). Most notable are genome-wide hypomethylation events that preferentially target repetitive DNA elements, and gene-specific hypermethylation of CpG islands. CpG islands are sequences with greater than normal G+C DNA content (Bird, 1986). Although their exact definition varies, they are usually between 0.2 kb and 2 kb long and contain a relatively high frequency of CpG dinucleotides. CpG sequences normally are underrepresented in mammalian genomes, owing to mutational pressure and/or lack of efficient DNA repair at methylated CpGs (Pfeifer, 2006). However, in normal tissues and in the germ line, the majority of gene promoter-associated CpG islands remain unmethylated. Accordingly, they are not subject to erosion by mutational events and retain a close to expected frequency of CpG dinucleotides. Methylation of CpG islands becomes aberrant in cancer when many hundreds of CpG islands in individual tumors acquire DNA methylation.
Global DNA hypomethylation and gene-specific hypermethylation are among the prominent hallmarks of cancer genomes (Ehrlich, 2002; Ushijima, 2005). Studies of aberrant methylation emphasize the pervasiveness of these changes in tumorigenesis and tumor progression. The role of DNA hypomethylation is often considered less important due to its global nature, along with limited knowledge of specific genes and genomic regions associated with hypomethylation. However, a cancer-causing role of DNA hypomethylation is clearly suggested by studies in mice carrying hypomorphic alleles of DNA methyltransferase genes, i.e. Dnmt1 (Gaudet et al., 2003). These mice develop malignancies, in particular lymphomas and hepatocellular carcinoma but the effect of Dnmt1 loss can be complicated and may either support or inhibit tumor development (Laird et al., 1995; Gaudet et al., 2003; Yamada et al., 2005). The mechanisms how DNA hypomethylation is tumor-predisposing are unknown but it is conceivable that reactivation of methylation-silenced repetitive DNA elements and increased genomic instability are involved (Ehrlich, 2002). Most of the literature available on epigenetic factors in initiation and progression of tumorigenesis is dealing with hypermethylation of CpG islands or gene promoters and so is this review.
Aberrant DNA methylation in cancer – starting from single gene studies
DNA methylation of promoter CpG islands is strongly associated with gene silencing and is known as a frequent cause of loss of expression of, for example, tumor suppressor genes as well as other genes involved in tumor formation. Much of what is known today about the importance of DNA methylation in cancer was gained earlier through small-and moderate-scale analysis of gene promoters in different tumor types. The very first methodologies employed for the analysis of DNA methylation depended on initial digestion of DNA with methylation-sensitive restriction endonucleases followed by Southern blotting (Bird and Southern, 1978). Later on, sodium bisulfite sequencing and other methods based on that same concept became the methods of choice for single gene analysis (Frommer et al., 1992).
Initial focus on DNA methylation in tumors was centered on the question of methylation-induced silencing of known tumor suppressor genes. During tumorigenesis, both alleles of a tumor suppressor gene need to be inactivated. This can occur by chromosomal deletions or loss-of-function mutations affecting the gene's coding sequence. Alternatively, hypermethylation of CpG islands spanning the promoter regions of tumor suppressor genes (for example, RB, CDKN2A, VHL, APC, MLH1, RASSF1A and BRCA1) can lead to gene silencing and thus can be an integral mechanism in tumorigenesis equivalent to gene loss or mutation (Issa, 1999; Costello et al., 2000; Dammann et al., 2000; Jones and Baylin, 2002; Herman and Baylin, 2003; Nephew and Huang, 2003). Since hypermethylation generally leads to permanent inactivation of gene expression, and is thought to be less reversible than altered histone modifications, this epigenetic alteration is considered a key pathway for long-term silencing of genes. To give some examples on one particular type of tumor, we focus on lung cancer. Several specific CpG-island-associated gene methylation events were frequently observed including, for example, CDKN2A, RASSF1A, RARβ, MGMT, GSTP1, CDH13, APC, DAPK, TIMP3, along with many other genes (Zochbauer-Muller et al., 2001; Toyooka et al., 2003; Yanagawa et al., 2003; Franklin, 2004; Topaloglu et al., 2004; Dammann et al., 2005b; Kim et al., 2005). Genes altered by DNA methylation include those involved in important cellular pathways such as cell cycle regulation (e.g. CDKN2A, CHFR), proliferation (e.g. CDKN2A, CXCL12), DNA repair (e.g. MGMT), apoptosis (e.g. DAPK, caspase 8, FAS, TRAILR1), RAS signaling (RASSF1A, NORE1A), invasion (e.g. cadherins, ADAMTS1, TIMP3, PTGER2, laminin family) and Wnt signaling (APC, DKK1, SFRP genes). Some of these pathways affected by epigenetic change are those described as the hallmarks of cancer (Hanahan and Weinberg, 2000). Other studies of non-small cell lung cancer (NSCLC) identified many additional hypermethylated genes (e.g., ARPC1B, DNAH9, FLRT2, G0S2, IRS2, RUNX3, PKP1, SPOCK1, UCHL1, OTX1, BARHL2, MEIS1, and OC2) (Bowman et al., 2006; Rauch et al., 2007; Rauch et al., 2008; Jin et al., 2009). In the literature, the methylation frequency (i.e., the percentage of tumors analyzed that carry substantially methylated alleles) generally ranges from only a few percent for some genes to more than 80% for other genes. The reported methylation frequencies, even for the same genes, often differ substantially depending on the study population, tumor histology, and/or methodology used to assess CpG island methylation.
The choice of methylation targets analyzed in the numerous single gene studies has often been based on existing knowledge of the presumed function of a particular gene or gene family member, or it was the result of a more or less serendipitous discovery of a particular methylation event. In most cases, CpG islands overlapping the 5' gene ends or promoters of genes have been analyzed. More recent unbiased genome-wide studies, however, have revealed common tumor-associated methylation of CpG islands outside of promoter regions, and it is still unclear whether or not such methylation changes have biological consequences and what exactly these consequences are for tumor formation. Interestingly, there are often cancer-specifically methylated CpG islands not associated with any known genes at all. These CpG islands may represent remote regulatory elements or may represent functionally relevant sequences associated with non-coding RNAs.
Aberrant DNA methylation in cancer – genome-wide studies
A much better understanding of the role of DNA methylation in cancer, either as a marker of disease or as an active driver of tumorigenesis, will likely be gained from genome-wide studies of this modification in normal and malignant cells. This goal has become more reachable with the recent introduction of large-scale genome analysis methodologies. These techniques have been adopted in various ways to allow for investigation of DNA methylation of many gene loci simultaneously (Table 1). In this section, we review several technological advances in genome-wide methylation profiling.
Table 1.
Some characteristics of genome-wide DNA methylation detection techniques
| Techniquea | Sensitivity | nature of mCpGs detected | Reference |
|---|---|---|---|
| RLGS | 2–5 μg | in Not1 sites | Costello et al., 2000 |
| MS-RDA | 10 μg | in restriction sites (e.g., HpaII) | Ushijima et al., 1997 |
| DMH | 2 μg | in restriction sites (e.g. HpaII, SmaI) | Huang et al., 1999 |
| MCA | 5 μg | SmaI restriction sites | Estecio et al., 2007 |
| McrBC | 10 μg | two CpGs separated by 55 bp to 3 kb | Nouzova et al., 2004 |
| MeDIP | 2–4 μg | all, CpG density-dependent | Weber et al., 2005 |
| MIRA | 0.1–0.2 μg | all, CpG-density-dependent | Rauch et al., 2006 |
| BS | 5 μg | all | Lister et al., 2009 |
The techniques described are: RLGS, restriction landmark genomic scanning MS-RDA, methylation-sensitive representational difference analysis DMH, differential methylation hybridization McrBC, McrBC nuclease cleavage of methylated DNA MeDIP, Methylated DNA immunoprecipitation MIRA, Methylated-CpG island recovery assay BS, bisulfite sequencing
One of the earliest large-scale methylation profiling techniques developed was methylation-sensitive representational difference analysis (MS-RDA) (Ushijima et al., 1997; Smith and Kelsey, 2001; Ushijima and Yamashita, 2009). Genomic DNA is predigested using the methylation-sensitive restriction enzyme HpaII, and a mixture ratio of tester and driver DNAs is optimized to detect differences in methylation status of single copy genes between two tissue samples. Restriction endonuclease digestion-based DNA methylation analysis was modified by Huang and colleagues and developed as differential methylation hybridization (DMH) on array platforms by combining restriction endonucleases and microarrays for high-throughput analysis of the methylation status of CpG islands in human genomes (Huang et al., 1999; Yan et al., 2001; Wei et al., 2002). This method utilizes a restriction enzyme MseI, which recognizes TTAA, a sequence that is rarely present within GC-rich regions, and leaves most CpG islands intact. MseI-generated fragments are ligated to defined synthetic linkers and are further digested, for example, with BstUI, a methylation-sensitive restriction endonuclease. BstUI recognizes and digests the sequence 5'-CGCG within CpG islands when they are unmethylated. CpG islands, which are methylated, resist BstUI restriction digestion, and these methylated fragments can be subsequently amplified by linker-dependent PCR. The resulting PCR products are labeled with fluorescent dyes. To compare genome-level CpG island methylation, equal quantities of BstUI-digested amplicons from two samples (e.g., normal and cancer) are mixed and hybridized onto a microarray. The resulting ratio between the two dyes represents the methylation difference between the two samples.
There are several other methods that are based on restriction endonuclease digestion, such as classical restriction landmark genomic scanning (RLGS) (Hatada et al., 1991; Costello et al., 2000), or HpaII tiny fragment enrichment by ligation-mediated PCR (HELP) (Khulan et al., 2006; Figueroa et al., 2009). Nouzova et al (Nouzova et al., 2004) and Lippman et al (Lippman et al., 2005) developed a DNA methylation profiling technique by replacing BstUI or HpaII with McrBC, an unusual restriction enzyme that recognizes and cleaves CpG-methylated DNA. Sites on the DNA recognized by McrBC consist of two half-sites of the form (G/A)mC. These half-sites can be separated by up to 3 kb, but the optimal separation is ~50–100 base pairs. This method was used to identify a number of hypermethylated regions in an acute promyelocytic leukemia cell line compared to normal peripheral blood mononuclear cells (Nouzova et al., 2004). Irizarry et al modified the McrBC assay and developed a comprehensive high-throughput array analysis for relative methylation (CHARM) (Irizarry et al., 2008). The unmethylated or methylated fractions can be analyzed on microarray or high-throughput sequencing platforms for genome-wide identification of aberrant methylation. Methylated CpG island amplification (MCA) coupled to microarrays also is based on methylation-sensitive restriction enzymes. Target sequences are amplified by PCR using flanking primers followed by sequence analysis or microarray probing. MCA is a powerful approach for simultaneous identification of differentially methylated genomic regions (Toyota et al., 1999; Estecio et al., 2007).
Genome-scale DNA methylation analysis by bisulfite conversion of DNA has now become possible. In bisulfite conversion of DNA, treatment of DNA with sodium bisulfite converts unmethylated cytosines to uracils, whereas methylated cytosines are not affected. The bisulfite-treated samples are then PCR-amplified and unmethylated and deaminated cytosines are replaced by thymines during PCR. Then, these samples can be hybridized to microarrays for large-scale analysis of DNA methylation status (Gitan et al., 2002; Hou et al., 2004) or by Illumina sequencing (Lister et al., 2009; Gu et al., 2010; Laird, 2010). This approach still is expensive when applied to whole mammalian genomes and requires substantial computational resources. Variations of bisulfite-based approaches include analysis of sub-areas of the genome (Meissner et al., 2008; Ball et al., 2009), or a highly multiplexed PCR-based approach using the Illumina bead platform (Bibikova et al., 2006).
A third general type of high-throughput approach in methylation analysis is based on affinity-purification of methylated DNA. Methylated DNA immunoprecipitation (MeDIP) utilizes nonspecific fragmentation of the genomic DNA followed by anti-5mC antibody precipitation to enrich for methylated DNA fragments (Weber et al., 2005). The immunoprecipitated DNA, enriched in hypermethylated sequences, and total genomic DNA (as input) are labeled with fluorescent dyes Cy5 and Cy3, respectively, and cohybridized onto microarray chips or analyzed by high-throughput sequencing. MeDIP is thus a valuable general fractionation approach, compatible with different analysis platforms to query the level of methylation in genomic sequences at a level of resolution of about 100 bp. One of the crucial factors in this assay is the quality of the anti-5-methylcytosine antibody. Moreover, the MeDIP method is most sensitive for densely methylated sequences, as DNA fragments with many contiguous methylated CpGs are more efficiently precipitated. MeDIP requires effective DNA denaturation before antibody binding.
Affinity purification of methylated DNA by a protein or peptide that can specifically bind to methylated CpGs was initially reported by Cross et al (Cross et al., 1994). Among the methods most suitable for genome-wide mapping of DNA methylation, the methylated CpG island recovery assay (MIRA) represents an approach that is based on a methyl-CpG binding protein complex. MIRA depends on the fact that the methyl-CpG-binding protein MBD2B specifically recognizes methylated CpG dinucleotides (Hendrich and Bird, 1998) and that this interaction is strongly enhanced by the MBD3L1 protein (Rauch and Pfeifer, 2005; Rauch et al., 2006; Rauch et al., 2007), a heterodimerization partner of MBD2 (Jiang et al., 2004). Among all methyl-CpG-binding proteins known, MBD2B has the highest affinity for methylated DNA and displays the greatest ability to distinguish between methylated and unmethylated DNA. It recognizes a wide range of methylated CpG sequences with little sequence specificity (Fraga et al., 2003). In our lab, lack of a defined sequence specificity of the MBD2B/MBD3L1 complex was confirmed by cloning and random sequencing of MIRA-enriched DNA fragments. Pulldown of methylated fragments is most efficient when a minimum of two methylated CpG sites are present (Rauch et al., 2006). In the MIRA procedure, sonicated genomic DNA is incubated with the MBD2B/MBD3L1 protein complex. Unlike the MeDIP technique, which requires single-stranded DNA for antibody recognition, MIRA works on normal double-stranded DNA; in fact the complex does not bind to single-stranded DNA. The CpG-methylated DNA is collected from the binding reaction via the GST-tagged MBD2B and glutathione beads, linker ligated and then PCR amplified. These PCR amplified MBD-enriched DNA fractions and total genomic DNA (input) are labeled with fluorescent dyes Cy5 and Cy3, respectively, and cohybridized onto microarrays. The ratio of fluorescent intensity (Cy5 to Cy3) indicates the methylation status at each particular sequence analyzed. The MIRA-enrichment method has been proven to be compatible with several types of microarray platforms and high-throughput DNA sequencing platforms and is highly sensitive requiring only 100–200 ng of genomic DNA.
Results from DNA methylation profiling
The importance and widespread occurrence of CpG island hypermethylation in cancer is becoming increasingly recognized. In initial studies examining a limited number of loci, it has been estimated that between 0.5% and 3% of all genes carrying CpG-rich promoter sequences may be silenced by DNA methylation in several types of cancer (Costello et al., 2000; Shiraishi et al., 2002). Examining all or most CpG islands in the genome, recent reports indicate that generally several hundred to even more than a thousand CpG islands can be methylated in individual tumors (Rauch et al., 2006; Rauch et al., 2007; Dudley et al., 2008; Kuang et al., 2008; Omura et al., 2008; Rauch et al., 2008; Koga et al., 2009; Tommasi et al., 2009). Table 2 summarizes some of the recent studies describing methylation profiling of cancer genomes.
Table 2.
Genome-wide DNA methylation studies in various cancer types
| Cancer type | Technique used | Notable findings | Reference |
|---|---|---|---|
| Acute myeloid leukemia | Bisulfite Illumina bead array | Genome-wide promoter associated hypermethylation associated with improved patient survival | (Deneberg et al., 2010) |
| Brain cancer | Bisulfite lllumina bead array | Genes hypermethylated in glioblastoma were highly enriched for targets of PRC2 in embryonic stem cells | (Martinez et al., 2009) |
| Brain cancer | MIRA | Hypermethylation of neuronal differentiation genes in astrocytomas | (Wu et al., 2010) |
| Breast cancer | MIRA | Methylation of homeobox and other developmental genes regulated by the Polycomb complex | (Tommasi et al., 2009) |
| Breast cancer | MeDIP | Agglomerative epigenetic aberrations are frequent events in human breast cancer | (Novak et al., 2008) |
| Breast cancer | MeDIP | Interdependence between DNA methylome alterations and morphological changes | (Ruike et al., 2010) |
| Chronic lymphocytic leukemia | MCA | Methylation of LINE and APP was associated with shorter overall survival | (Kuang et al., 2008) |
| Chronic lymphocytic leukemia | Bisulfite Illumina bead array | Possible mechanism for pathogenesis | (Kanduri et al., 2010) |
| Colorectal cancer | MeDIP | Three different methylation epigenotypes exist in colorectal cancer | (Yagi et al., 2010) |
| Esophageal squamous cell carcinoma | HELP | Influence of genetic background on DNA methylation | (Yang et al., 2010) |
| Follicular lymphoma | MCA | Extensive hypermethylation in promoters of Polycomb target genes | (Bennett et al., 2008) |
| Haematological malignancies | MIRA | Methylation status of TFAP2A and EBF2 genes associated with advanced disease in CML | (Dunwell et al., 2010) |
| Head and neck cancer | RLGS | Methylation of genes involved in the transforming growth factor beta signaling pathway | (Bennett et al., 2008) |
| Hematologic neoplasms | Bisulfite Illumina bead array | Methylation of DBC1, DIO3, FZD9, HS3ST2, MOS, and MYOD1 and their role in development of different hematologic neoplasms | (Martin-Subero et al., 2009) |
| Hepatocellular carcinomas | MCA | DNA methylation status was correlated with the cancer-free and overall survival rates of patients | (Arai et al., 2009) |
| Lung cancer | MIRA | Biomarkers for early detection of lung cancer and extensive hypomethylation of repetitive sequences in tumors | (Rauch et al., 2008) |
| Lung cancer | MCA | Differential methylation between mesothelioma and adenocarcinoma | (Goto et al., 2009) |
| Mantle cell lymphoma | HELP | Methylation-based drug targeting | (Leshchenko et al., 2010) |
| Ovarian cancer | Bisulfite Illumina bead array | Diagnostic or risk-prediction of ovarian cancer by blood methylation profiling | (Teschendorff et al., 2009) |
| Pancreatic cancer | MCA | Identification of aberrantly methylated genes in pancreatic cancers | (Omura et al., 2008) |
| Prostate cancer | DMH | Methylation of homeobox or T-box genes | (Kron et al., 2009) |
| Skin cancer | MeDIP | Moderate increases of methylation in early and significantly in advanced-stage melanomas | (Koga et al., 2009) |
| Testicular germ cell tumors | MeDIP | Function of intergenic and intronic DMRs in the regulation of ncRNAs | (Cheung et al., 2010) |
| Urothelial cancer | MCA | DNA methylation as indicator for carcinogenetic risk estimation | (Nishiyama et al., 2010) |
Genome-wide analysis of DNA methylation of lung squamous cell carcinoma (SCC) and matching normal tissue DNA revealed a large number of lung SCC-specific hypermethylated genes. Chromosome tiling array analysis has indicated that all of them were CpG islands or CpG-rich regions, often overlapping or located in close proximity to promoter regions (Rauch et al., 2008). Islands with different CpG densities can become hypermethylated in tumors. It is clear that not all of these hundreds of methylated genes can be tumor suppressor genes. For example, substantial subsets of the methylated genes were represented by a variety of homeobox genes (Rauch et al., 2007). Homeobox gene-associated CpG islands were among the most common stage I disease DNA methylation events identified so far, i.e. this methylation event appears in almost every early stage tumor (Rauch et al., 2007; Rauch et al., 2008). Genome-wide DNA methylation analysis identified CpG island methylation, for example in proximity of the OTX1, NR2E1, PAX6, IRX2, OC2, TFAP2A, and EVX2 genes. These genes are tumor-specifically methylated with very little methylation found in normal lung tissue or in blood cell DNA (Rauch et al., 2008).
The frequent methylation of homeobox genes and other developmental genes regulated by the Polycomb complex is a phenomenon observed in many different histological types of human cancer (Rauch et al., 2006; Ohm et al., 2007; Rauch et al., 2007; Schlesinger et al., 2007; Widschwendter et al., 2007), as exemplified by several studies, which we will discuss briefly. Genome wide methylation profiling of ductal carcinoma in situ, a premalignant breast lesion with a high potential to progress towards invasive carcinoma identified 108 significant CpG islands that undergo aberrant DNA methylation in ductal carcinoma in situ and stage I breast tumors, with methylation frequencies greater than or comparable with those of more advanced invasive carcinoma (50% to 93%) (Tommasi et al., 2009). A substantial fraction of these hypermethylated CpG islands (32% of the annotated CpG islands) was associated with several homeobox genes, such as the TLX1, HOXB13, and HNF1B genes. Fifty-three percent of the genes hypermethylated in early-stage breast cancer overlapped with known Polycomb targets and included homeobox genes and other developmental transcription factors (Tommasi et al., 2009). Interestingly, one-third of the CpG islands identified by microarray analysis (26 out of the 81 annotated hits) were associated with members of various homeobox superfamilies (HOX, LHX, NKX, PAX, and so forth) and were preferential targets of de novo methylation in early-stage breast cancer (Tommasi et al., 2009). These master regulators control vital functional networks during tissue development and differentiation, and are misregulated in a variety of malignancies, including breast cancer (Abate-Shen, 2002; Coletta et al., 2004).
Large scale DNA methylation analysis of glioblastoma multiforme (GBM) identified 25 hypermethylated genes in more than 20% of the cases studied (Martinez et al., 2009). The most frequently hypermethylated genes were HOXA11, CD81, PRKCDBP, TES, MEST, TNFRSF10A and FZD9 and these were methylated in more than 50% of the samples. HOXA9, HOXA5, TFAP2C, IGFBP1 and some of the other gene were methylated to a lesser extent (between 25% and 40%) in GBM compared to controls, but were found to be methylated in various other cancers (Martinez et al., 2009). Analyzing biological features of these hypermethylated genes revealed that the group of genes hypermethylated in GBM was highly enriched (41%) for targets of the PRC2 (Polycomb repressive complex 2) in embryonic stem cells. Furthermore, this study identified promoter hypermethylation of the transcription factor gene GATA6 (occurring in 30% of GBM) that was correlated with poor patient survival (Martinez et al., 2009).
We recently completed a study on astrocytoma/glioma patients and analyzed over 28,000 CpG islands in 30 patients (Wu et al., 2010). Several hundred CpG islands undergo specific hypermethylation relative to normal brain with 428 methylation peaks common to more than 25% of the astrocytomas. Genes involved in brain development and neuronal differentiation, such as BMP4, POU4F3, GDNF, OTX2, NEFM, CNTN4, OTP, SIM1, FYN, EN1, CHAT, GSX2, NKX6-1, PAX6, RAX, and DLX2, were strongly enriched among genes frequently methylated in tumors. There was an overrepresentation of homeobox genes and 31% of the most commonly methylated genes represented targets of the Polycomb complex. We identified several chromosomal loci in which many (sometimes more than 20) consecutive CpG islands were hypermethylated in tumors. Seven of such loci were near homeobox genes, including the HOXC and HOXD clusters, and the BARHL2, DLX1, and PITX2 genes (Wu et al., 2010).
Genome-wide promoter methylation and gene expression analysis of early-passage human melanoma cell lines or tumor specimens compared with melanocytes identified a number of new hypermethylated genes on top of already known promoter-methylated genes in melanoma (e.g. RARB, RASSF1A, and PYCARD) (Spugnardi et al., 2003; Hoon et al., 2004; Furuta et al., 2006). Another study by Koga et al identified the promoter hypermethylated genes COL1A2, NPM2, HSPB6, DDIT4L, MT1G, and SOX3 and also the homeobox genes HOXB13 and HOXA7 in melanoma cells (Koga et al., 2009). This study also points out that the frequency of promoter methylation of validated hypermethylated gene promoters (COL1A2, NPM2, HSPB6, DDIT4L and MT1G) increases moderately in early and significantly in advanced-stage melanomas, using early-passage cell strains and snap-frozen tissues compared with normal melanocytes and nevi (Koga et al., 2009).
Global evaluation of DNA methylation in prostate cancer revealed a large number of hypermethylated genes that were significantly hypermethylated compared to reference samples (Kron et al., 2009). This study found that about 30% of significantly hypermethylated genes of the top 100 methylated genes in prostate cancer were homeobox or T-box genes (e.g., FOXC1, VAX1, SIX6, HOXD3, HHEX, TBX15, HOXD9, GSC, HOXC13, PROX1, TBX4, TBX3, HOXD8, PAX2, IRX6, ALX4, BARX2, BARX, PHOX2A, LBX1, DLX5, DLX6, LHX9 and HOXD8), similar to many other such methylation studies in various cancers (Rauch et al., 2006; Rauch et al., 2007; Tommasi et al., 2009).
A genome-wide screen for DNA methylation changes in head and neck squamous cell carcinoma tumors identified five candidate genes, SLC5A8, SEPT9, FUSSEL18, EBF3, and IRX1, as methylated in 27% to 67% of the HNSCC patient samples tested (Bennett et al., 2008). Genome-wide analysis of promoter-associated CpG island methylation in acute lymphoblastic leukemia identified 404 potential targets of methylation (Kuang et al., 2008). Aberrantly methylated genes identified in this study had methylation frequencies ranging from 23 to 100%. Among the genes validated in primary ALL samples were GIPC2, RSPO1, MAGI1, CAST1, ADCY5, HSPA4L, OCLN, EFNA5, MSX2, GFPT2, GNA14, SALL1, MYO5B, ZNF382 and MN1 (Kuang et al., 2008). A study of DNA hypermethylation in follicular lymphoma also discovered widespread hypermethylation of homeobox genes and previously identified targets of Polycomb repressive complex 2 (PRC2) in cell lines and primary tumors, but not in benign follicular hyperplasia (Bennett et al., 2009).
Targeted methylation of genes versus methylation of target genes or “passenger methylation” versus “driver methylation”
Some of the hypermethylated genes in cancer may be bona fide tumor suppressor genes, but it is unlikely that all of these numerous methylation changes play a causative role in tumorigenesis. Rather, many promoter CpG islands are probably methylated as a consequence of or in association with carcinogenesis (passenger methylation). It is a challenge to pinpoint those crucial genes that are susceptible to methylation-associated gene silencing and are functionally important in preventing tumorigenesis (driver methylation). The situation is perhaps analogous to the one found for mutational changes in cancer. Genome-wide DNA sequencing of either a large number of coding sequences or entire cancer genomes have revealed a staggering number of mutational changes (Pfeifer and Besaratinia, 2009). Most often, mutations in specific genes occur only a single time among a larger number of tumors sequenced and it is then difficult to determine if that particular mutation is indeed a driver mutation or just a passenger event (Carter et al., 2009). These large-scale sequencing studies have confirmed frequent mutations in known tumor suppressor genes or oncogenes, e.g. the p53 or RAS genes, but have occasionally uncovered the existence of novel and likely important driver mutations, e.g. in the BRAF gene (Davies et al., 2002) and IDH1 gene (Parsons et al., 2008).
When methylation occurs at the promoter of a known and well-established tumor suppressor gene, e.g. CDKN2A, the gene encoding the cyclin-dependent kinase inhibitor protein p16, then it is of course easy to predict that the methylation event at least has the potential to be tumor driving. However, much of the altered methylation landscape in cancers may be a phenomenon linked to “targeted methylation” whereby a particular gene or chromatin environment predisposes that gene to methylation in cancer and reflects a passenger event.
The mechanisms for aberrant CpG island methylation in cancer are mostly unknown. Generally, CpG island hypermethylation is closely linked to modification of local chromatin architecture serving as one already existing mechanism for silencing transcription. It has been proposed that gene inactivity imposed by changes in chromatin structure or histone modification predisposes to DNA methylation (Song et al., 2002; Bachman et al., 2003). Specific DNA sequences within CpG islands may be associated with the methylation process (Feltus et al., 2003; Keshet et al., 2006). Whether or not these sequences are associated with DNA binding proteins in vivo that somehow attract methylation is not clear. Feltus et al developed a method known as Pattern-based Methylation Analysis (PatMAn) based on seven short DNA sequence patterns (TCCCCCNC, TTTCCTNC, TCCNCCNCCC, GGAGNAAG, GAGANAAG, GCCACCCC, GAGGAGGNNG) that discriminated methylation-prone (MP) and methylation-resistant (MR) CpG islands. This classifier predicts CpG islands that are at higher risk for hypermethylation in cancer (Feltus et al., 2003). PatMAn predicted methylation-prone CpG islands associated with embryonic targets of Polycomb-repressive complex 2 (PRC2). McCabe et al further improved PatMAn and developed a second classifier (SUPER-PatMAn) that combines PatMAn DNA patterns with SUZ12-enriched regions as a marker of PRC2 occupancy (McCabe et al., 2009). These studies indicated that both local sequence context and a specific chromatin environment are involved in a large subset of genes undergoing hypermethylation in cancers (McCabe et al., 2009).
Among the genes targeted by Polycomb complexes are many developmental transcription factor genes including homeobox genes but also some known tumor suppressor genes, such as CDKN2A. Aberrant expression of Polycomb group (PcG) and Trithorax group (TrxG) proteins is a common event in many cancers (Pasini et al., 2004; Valk-Lingbeek et al., 2004; Raaphorst, 2005b; Esteller, 2007). Components of Polycomb repressive complex 1 (PRC1) (such as BMI1 (Valk-Lingbeek et al., 2004) and components of PRC2 (such as EZH2 (Bracken et al., 2003) are amplified and/or overexpressed in a broad spectrum of cancers. Aberrant expression of PRC components affects PcG protein complexes (Kuzmichev et al., 2005) thus potentially influencing target gene affinities (Squazzo et al., 2006). Pharmacological disruption or forced expression of PRC2 genes induces apoptosis in cancer cells and provides a proliferative advantage to primary cells, respectively (Sellers and Loda, 2002; Tan et al., 2007). Any change in function of PcG and TrxG proteins, which occur during aging (Pardal et al., 2005; Sharpless and DePinho, 2005) or inflammation (Coussens and Werb, 2002; Lu et al., 2006), may contribute to the development of cancer. However, the mechanism how Polycomb target genes undergo hypermethylation in cancer is still unknown.
It is difficult to deduce why a large number of homeobox genes become preferential targets of aberrant CpG methylation during tumorigenesis and whether this extensive methylation event can shift their finely tuned homeostasis, thus triggering tumorigenesis, or whether this process is merely associated with the neoplastic event. The widespread and recurrent nature of this phenomenon, however, seems to suggest that a common mechanistic pathway may exist in cancer cells, which promotes de novo methylation of these targets at the onset of tumor development. Paradoxically, however, several homeobox genes are upregulated rather than downregulated in breast cancer and other tumor types, suggesting that several tiers of regulation, in addition to promoter DNA methylation, may concur in determining homeobox misregulation.
Recent data have unraveled the role of Polycomb repressor complexes in targeting and modulating homeobox genes. At least six independent genome-wide studies have identified several common Polycomb targets in vertebrates and flies, most of which are represented by homeobox genes and other developmental transcription factors (Ringrose, 2007). Commonly, most of the homeobox gene-associated methylated CpG islands are embedded in regions other than promoters, consistent with the finding that the PRC2 subunit SUZ12 is distributed across large domains of developmental genes spanning from the promoter up to 2 to 35 kb into the gene (Lee et al., 2006). SUZ12 is required for the histone H3K27 methyltransferase activity and silencing function of the EED-EZH2 complex and is upregulated in different tumors (Kirmizis et al., 2003). EZH2, the PRC2 catalytic subunit exhibiting histone H3 K27 methyltransferase activity, undergoes gene amplification in several tumor types (Bracken et al., 2003) and is overexpressed in prostate cancer and breast cancer (Varambally et al., 2002; Raaphorst, 2005a; Ding and Kleer, 2006). EZH2 physically interacts with all three DNA methyltransferases in mammalian cells, and has been suggested to play a crucial role in regulating de novo DNA methylation and its maintenance at target sequences (Vire et al., 2006) although the maintenance methylation aspect is in question (McGarvey et al., 2007).
Further support for a mechanistic connection between Polycomb silencing and tumor-associated DNA methylation comes from recent studies linking Polycomb occupancy of genes in noncancerous cells and tissues (including embryonic stem cells) with cancer-associated hypermethylation events (Rauch et al., 2006; Vire et al., 2006; Eden et al., 2007; Ohm et al., 2007; Rauch et al., 2007; Schlesinger et al., 2007; Widschwendter et al., 2007; Hahn et al., 2008). Both inflammation and aging are associated with methylation of Polycomb target genes indicating that these cancer predisposing scenarios might have a specific epigenetic basis (Hahn et al., 2008; Maegawa et al., 2010).
PcG target genes may be composed of `bivalent' chromatin containing both the active histone mark H3K4me3 and the silencing mark H3K27me3 (Bernstein et al., 2006). Bracken and colleagues have suggested that, in undifferentiated cells, PcG complexes have the potential to target genes poised for silencing as well as target genes predisposed to activation (Bracken et al., 2006). The transition between alternative modes of PcG regulation may require additional signals upon differentiation (and likewise during tumorigenesis), which may include recruitment of additional transcriptional activators and/or competition with PcG antagonists, the TrxG proteins. These signals may have a counteracting effect on PcG-mediated gene repression (Bracken et al., 2006). In addition, recent studies indicate that gene silencing in cancer can occur by histone H3 lysine 27 trimethylation independent of promoter DNA methylation (Kondo et al., 2008).
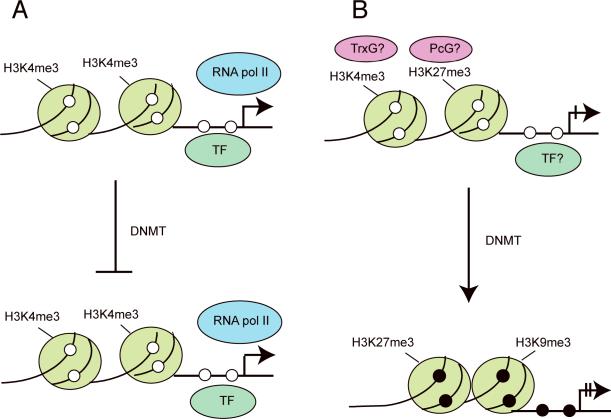
Specific histone configurations or modifications may either protect from methylation or may promote DNA methylation at CpG islands (Figure 1). One possibility is that CpG islands that do not undergo methylation in cancer carry protective factors and that methylation-prone islands lack these factors (Gebhard et al., 2010). Furthermore, it has been shown that genes with high levels of binding of RNA polymerase II, regardless of transcription levels, are resistant to induction of aberrant methylation (Takeshima et al., 2009). Trimethylation of histone H3 lysine 4 (H3K4me3) is associated with active or potentially active genes and unmethylated CpG islands (Barrera et al., 2008). This modification interferes with binding of the de novo DNA methyltransferase DNMT3L/DNMT3A complex (Jia et al., 2007; Ooi et al., 2007) and is expected to prevent methylation.
Figure 1. Targeted methylation or “passenger methylation” events.
A. Many genes are resistant to promoter methylation in cancer. Among the causes for this resistance can be association of a gene promoter with specific transcription factors or transcription factor complexes or presence of the activating histone mark H3K4me3, which interferes with DNA methylation.
B. Genes are targeted for methylation by a specific chromatin environment, for example lack of transcription and presence of the repressive Polycomb complex and the histone mark H3K27me3. These genes may initially be associated with `bivalent' chromatin characterized by both activating marks (H3K4me3 catalyzed by the Trithorax complex, TrxG) and repressive marks (H3K27me3 catalyzed by the Polycomb complex, PcG). In cancer tissue, these genes undergo DNA methylation and may be associated with other repressive marks (H3K9me3 and/or H3K27me3). The ball-shaped symbols represent nucleosome core particles over which the DNA is wrapped. Histone tail modifications are indicated and the open and closed circles represent unmethylated and methylated CpG sites, respectively.
Inactivation of important biological pathways by cancer-associated methylation
One key question is if the widespread methylation of Polycomb target genes seen in many types of caner is functionally important for tumor development. Expression of these genes is often required for certain stages of embryonic development and differentiation. However, we know very little about the importance of these developmental genes in adult somatic stem cells, the cell types which tumors are most likely derived from. One possibility is that the PcG target genes in these stem cells, e.g. homeobox genes, are already transcriptionally silent and the methylation event would have no functional consequence (passenger methylation). If the gene of interest is expressed and does have a functional role in stem cells, for example it might be important in differentiation processes, then aberrant methylation of this gene may favor the transformation process by interfering with tissue-specific differentiation pathways. In a recent study on astrocytomas, we observed that many of the tumor-specifically methylated genes had roles in neuronal differentiation supporting a model in which methylation of these genes in neural stem cells favors proliferation versus differentiation and may contribute to initiation of the malignancy (Wu et al., 2010). One key step in deciphering the role of methylation of Polycomb target genes in cancer will be the characterization of the expression and chromatin structure of these genes in adult somatic tissue stem cells.
There is evidence that methylation of genes within well-defined cellular pathways can contribute to tumorigenesis. For example, a large body of literature has established aberrant activation of Wingless-type (Wnt) signaling in various cancers such as colorectal cancer (Suzuki et al., 2004; Yue et al., 2008), head and neck carcinoma (Rhee et al., 2002), melanoma (Weeraratna et al., 2002), gastric cancer (Nojima et al., 2007), hepatocellular carcinoma (Shih et al., 2007; Takagi et al., 2008), bladder cancer (Urakami et al., 2006) and leukemia (Lu et al., 2004). The secreted frizzled-related proteins function as negative regulators of Wnt signaling and have important roles in tumorigenesis. Notably, methylation of Wnt pathway inhibitory genes, such as secreted frizzled related protein 1 and 2 (SFRP1 and SFRP2), whose inactivation enhances Wnt signaling, was observed in very early lesions of colon carcinogenesis, aberrant crypt foci (Suzuki et al., 2004). Aberrant methylation of SFRP promoters and activation of the WNT signaling pathway with excessive accumulation of beta-catenin in the nucleus was prominent in colorectal cancer (Suzuki et al., 2004) and gastric cancers (Nojima et al., 2007). Hypermethylation of SFRP genes, except for SFRP4, is frequent in hepatocellular carcinoma (HCCs). Re-activation of SFRP1 function by overexpressing SFRP1 in HCC cell lines blocked Wnt signaling and decreased abnormal accumulation of beta-catenin in the nucleus leading to arrest of cell growth. Overexpressed SFRPs downregulated T-cell factor/lymphocyte enhancer factor (TCF/LEF) transcriptional activity in HCCs (Takagi et al., 2008). siRNA mediated down regulation of SFRP1 in beta-catenin-deficient cell lines promotes cell growth by activating Wnt signaling (Shih et al., 2007). SFRP domain similarity with WNT-receptor frizzled proteins allows SFRPs to inhibit WNT receptor binding to consequently influence downstream pathway signaling during cell proliferation. Methylation silencing of other Wnt antagonists including Dickkopf 1 (DKK1) and Wnt inhibitory factor-1 (WIF-1) are also observed in different malignancies (Taniguchi et al., 2005; Aguilera et al., 2006).
SHP1 negatively regulates the Janus kinase/signal transducer and activator of transcription (Jak/STAT) signaling pathway (Chim et al., 2004a; Chim et al., 2004b). SHP1 in myeloma showed hypermethylation with constitutive STAT3 phosphorylation. Demethylated myeloma samples restored SHP1 expression with parallel down-regulation of phosphorylated STAT3 (Chim et al., 2004a). SHP1 methylation leading to epigenetic activation of the Jak/STAT pathway might have a tentative role in the pathogenesis of myeloma. Similarly frequent methylation of SHP1 was observed in mantle cell and follicular lymphoma (Chim et al., 2004c) and also in acute myeloid leukaemia (Chim et al., 2004b). Hypermethylation of SHP1 mediated activation of the Jak/STAT signaling pathway along with upregulation of cyclin D1 and BCL2 could be the basis for tumorigenicity in follicular lymphoma (Chim et al., 2004c).
One emerging cellular growth control pathway is the Hippo pathway, a proapoptotic and anti-proliferation signaling pathway initially identified in Drosophila. The pathway consists of a kinase cascade including the Drosophila Hippo kinase orthologues MST1 and MST2 and the LATS/WARTS serine/threonine kinases as well as several adapter proteins including RASSF proteins, SAV1, and MOB1 (Guo et al., 2007; Harvey and Tapon, 2007). The kinase cascade functions to inactivate the gene product of the YAP oncogene, a transcriptional co-activator of anti-apoptotic and pro-proliferative genes, by phosphorylation and cytoplasmic retention. Although YAP is overexpressed in some tumors, mutations in other components of the pathway are rare. However, several of the Hippo pathway genes including RASSF1A, MST1, MST2, and LATS1 are frequently methylated in human cancers leading to inactivation or partial dysfunction of the pathway and tumorigenesis (Dammann et al., 2005a; Takahashi et al., 2005; Seidel et al., 2007). This scenario is supported by several mouse models, in which gene targeting of Rassf1a, Mst1 and Mst2, or Lats1, leads to tumorigenesis (St John et al., 1999; Tommasi et al., 2005; Zhou et al., 2009).
The preceding paragraphs illustrate some of the potential tumor driving mechanisms that are connected to aberrant methylation of genes within specific growth control pathways. On the other hand, recent evidence also implicates growth-signaling pathways in aberrant DNA methylation patterns. One example is the significant correlation that has been reported between mutated BRAF kinase and the CpG island methylator phenotype (CIMP) in colorectal cancer (Kambara et al., 2004; Weisenberger et al., 2006). The exact mechanistic basis of this link is unclear but DNA hypermethylation of unknown targets might create a favorable context for the acquisition of mutated BRAF(V600E) in CIMP-positive colorectal cancer (Hinoue et al., 2009). These findings raise the important and unanswered question of whether genetic lesions are driving DNA methylation, or whether DNA methylation promotes or favors the selection of genetic lesions.
Methylation of non-coding RNA promoters in cancer
The emerging field of noncoding RNA adds to the complexity of cellular mechanisms that maintain normal cellular integrity. MicroRNAs (miRNAs) represent a class of small non-coding RNAs that play important roles in carcinogenesis. Aberrant miRNA expression by promoter methylation has been illustrated for several human malignancies and tumor suppressor functions have been recognized for this new class of small regulatory RNAs. Recently, miRNAs have been shown to play a role as targets in gene hypermethylation and silencing in malignant cells. Similar to protein-coding genes, an aberrant pattern of methylation of CpG islands near or within miRNAs genes could result in alterations in the expression of miRNAs that could lead to tumorigenesis (Davalos and Esteller, 2010).
Studies with myeloid leukemia showed that overexpression of miR-29b in acute myeloid leukemia cells caused significant down-regulation of DNA methyltransferases DNMT1, DNMT3A, and DNMT3B at both RNA and protein levels (Garzon et al., 2009). This decrease in expression of DNA methyltransferases resulted in a decrease of global DNA methylation and reactivation of several genes via promoter DNA hypomethylation. Yet, this miR-29b down-regulation of DNA methyltransferase was indirect by targeting Sp1, a transactivator of the DNMT1 gene (Garzon et al., 2009). A study in bacteria-induced gastric cancers highlighted the involvement of three miRNAs (miR-124a-1, miR-124a-2 and miR-124a-3). Silencing of these miRNAs due to aberrant methylation at their promoters, in addition to that of protein-coding genes, favors gastric tumorigenesis (Ando et al., 2009).
A recent study analyzing miRNAs that are aberrantly expressed in ovarian cancer identified a number of hypomethylated miRNAs genes (including miR-21, miR-203 and miR-205) with the encoded miRNAs displaying up-modulated expression (Iorio et al., 2007). Colon cancer cell line studies revealed hypermethylation of the CpG island of miR-124a in the cancer cell line but not in the normal tissue (Lujambio et al., 2007). Subsequent studies proved that miR-124a is also frequently methylated in other colon, breast and lung carcinoma cell lines, as well as in leukemias and lymphomas. Further studies showed that silencing of miR-124a by hypermethylation in cancer cells could result in increased expression of its target CDK6, an important regulator of the Rb protein. Another recent study of methylation of miRNA genes has shown hypermethylation of mir-9-1, mir-124a3, mir-148a, mir-152 and mir-663 in most of the human breast cancers tested (Lehmann et al., 2008).
Hypermethylation of CpG islands near the miR-34b, miR-137, miR-193a and miR-203 miRNA genes in oral squamous cell carcinomas resulted in silencing of these miRNAs in the cancer setting (Kozaki et al., 2008). miR-137 and miR-193a were found consistently hypermethylated in tumors, and normal expression of these two miRNAs was responsible for reduction in cell growth and proliferation factors, suggesting tumor suppressor characteristics for these miRNAs that are silenced during oral cancer progression.
Hypermethylation of the miR-34b/c CpG islands was commonly observed in colorectal cancer cell lines and in primary colorectal tumors (Toyota et al., 2008). Both miR-34b and miR-34c are part of the p53 pathway. Re-introducing miR-34b or miR-34c into colorectal cancer cells induced changes in gene expression that overlapped (Toyota et al., 2008). Hypermethylation of miR-34b/c along with miR-148a and miR-9 was observed in lymph node metastatic cancer (Lujambio et al., 2008). The reintroduction of miR-148a and miR-34b/c into cancer cells resulted in inhibition of their motility, reduced tumor growth, and inhibited metastasis formation in xenograft models, with an associated down-regulation of the miRNA target genes, such as C-MYC, E2F3, CDK6, and TGIF2 (Lujambio et al., 2008).
Experiments that could distinguish between “driver methylation” and “passenger methylation” in cancer
In order to identify driver methylation events and to set them apart from passenger methylation events, it is important to know how to define driver methylation. In simple terms, a methylation event, which promotes tumorigenesis, can be considered as driver methylation. If a methylation change is a tumor-driving or initiating event, it is more likely to occur during early stages of tumorigenesis. Using mouse models or early stage human tumor specimens and premalignant lesions, one can observe the timing of methylation changes from early, pre-neoplastic tissues to late malignant disease in established cancer models. This approach has been used successfully by Chen et al (Chen et al., 2009). Early hypermethylation of a transcription factor gene, Foxd3, influenced methylation of genes later in disease progression. Early changes are more likely to be driving the cancer phenotype, whereas later changes may simply reflect the transformed phenotype.
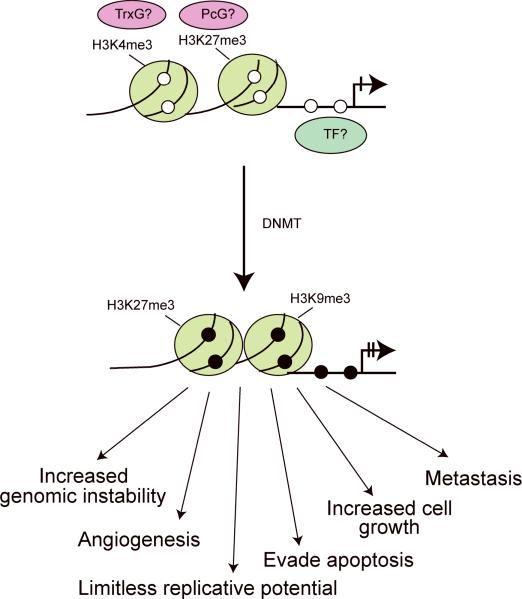
Driver methylation is not only represented by inactivation of tumor suppressor genes, but may also be equivalent to activation of oncogenes either directly or indirectly. Moreover, methylation-silenced genes may be placed into any pathway that is represented by the hallmarks of cancer (Figure 2) (Hanahan and Weinberg, 2000). Thus, methylation silencing may enable the cell to acquire properties of increased cell growth, higher potential to invade tissue, to metastasize or to initiate angiogenesis, or to evade apoptosis. In addition, there are methylation-associated events that do not immediately lead to altered cellular phenotypes; for example silencing of a DNA repair or cell cycle checkpoint gene will lead to enhanced genomic instability and that event might have secondary consequences such as promoting mutations in genes that counteract any of the hallmarks of cancer. Strong support for a cancer-driving role of a promoter methylation event will come from cancer genome sequencing data that show that the gene of interest is also mutated in a significant fraction of the tumors.
Figure 2. “Driver methylation” events.
In order to promote tumorigenesis, a driver methylation event should occur in pathways important to prevent the emergence of any of the hallmarks of cancer, for example increased cell growth, angiogenesis, metastasis and tissue invasion, evasion of apoptosis, unlimited replicative potential, or increased genomic instability.
When genome-wide screens or other discovery platforms have provided information about which genes are methylated in specific cancers, the first question to ask is whether the methylation event indeed is associated with gene silencing. Such a test is of importance inasmuch as many of the methylation events in cancer occur at genes that are already silenced in the corresponding normal tissue in which the tumor originates (Song et al., 2002; Hahn et al., 2008; Takeshima et al., 2009). Methylation-associated gene inactivation will generally be more likely for genes that are methylated in the 5' promoter region. Expression studies in tumor and normal tissue and 5-azacytidine reactivation experiments with cancer cell lines are standard approaches to address this point. Next, gene ontology analysis can be useful to determine if the gene of interest functions in a particular relevant pathway. Pathways of interest will be those potentially associated with the hallmarks of cancer or with other important cellular processes, for example defense against genome instability. If all these criteria are met, then experiments to test gene function are called for.
Usually, forced over-expression of methylation-suppressed genes in cancer cells or siRNA-mediated downregulation of the same genes in normal cells might give the first hints to differentiate between driver and passenger methylation. Forced expression of some of the frequently promoter-methylated genes in various cancer cell lines often inhibits cancer cell growth, colony formation and suppresses in vivo tumor growth in mice emphasizing the potential importance of these genes in tumorigenesis. Based on these observations it is logical to propose that genes that are frequently methylated in various types of cancer could be driver methylation events in tumorigenesis. However, to provide more definitive evidence for the causative role of a methylation-silenced gene in cancer, in vivo mouse models employing gene targeting are necessary. This proof of principal has been accomplished for a few genes that are frequently methylated in human tumors, for example HIC1, RASSF1A, SOCS3, and WIF-1 (Chen et al., 2003; Tommasi et al., 2005; Ogata et al., 2006; Kansara et al., 2009). More studies of this type should be conducted to assess the overall importance of hypermethylation of gene promoters in cancer.
In summary, genome-wide methylation profiling studies have begun to portray a much more complete picture of all the methylation changes that occur in different types of malignancies. The published data have expanded our view of the extent of these changes and it has now become a challenge to identify the critical, i.e. driver methylation events that contribute to the transformed phenotype. In analogy to the cancer genome sequencing projects, the identification of driver versus passenger events will be of importance for our understanding of cancer etiology, development of tumor-relevant biomarkers and eventually therapeutic approaches to target the driver methylation event.
Acknowledgement
Work of the authors was supported by NIH grant CA084469.
REFERENCES
- Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- Aguilera O, Fraga MF, Ballestar E, Paz MF, Herranz M, Espada J, Garcia JM, Munoz A, Esteller M, Gonzalez-Sancho JM. Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene. 2006;25:4116–4121. doi: 10.1038/sj.onc.1209439. [DOI] [PubMed] [Google Scholar]
- Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, Sugiyama T, Ushijima T. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 2009;124:2367–2374. doi: 10.1002/ijc.24219. [DOI] [PubMed] [Google Scholar]
- Arai E, Ushijima S, Gotoh M, Ojima H, Kosuge T, Hosoda F, Shibata T, Kondo T, Yokoi S, Imoto I, et al. Genome-wide DNA methylation profiles in liver tissue at the precancerous stage and in hepatocellular carcinoma. Int J Cancer. 2009;125:2854–2862. doi: 10.1002/ijc.24708. [DOI] [PubMed] [Google Scholar]
- Bachman KE, Park BH, Rhee I, Rajagopalan H, Herman JG, Baylin SB, Kinzler KW, Vogelstein B. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell. 2003;3:89–95. doi: 10.1016/s1535-6108(02)00234-9. [DOI] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera LO, Li Z, Smith AD, Arden KC, Cavenee WK, Zhang MQ, Green RD, Ren B. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res. 2008;18:46–59. doi: 10.1101/gr.6654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett KL, Karpenko M, Lin MT, Claus R, Arab K, Dyckhoff G, Plinkert P, Herpel E, Smiraglia D, Plass C. Frequently methylated tumor suppressor genes in head and neck squamous cell carcinoma. Cancer Res. 2008;68:4494–4499. doi: 10.1158/0008-5472.CAN-07-6509. [DOI] [PubMed] [Google Scholar]
- Bennett LB, Schnabel JL, Kelchen JM, Taylor KH, Guo J, Arthur GL, Papageorgio CN, Shi H, Caldwell CW. DNA hypermethylation accompanied by transcriptional repression in follicular lymphoma. Genes Chromosomes Cancer. 2009;48:828–841. doi: 10.1002/gcc.20687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird AP, Southern EM. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978;118:27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Bowman RV, Yang IA, Semmler AB, Fong KM. Epigenetics of lung cancer. Respirology. 2006;11:355–365. doi: 10.1111/j.1440-1843.2006.00859.x. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. Embo J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, Vogelstein B, Karchin R. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SS, Raval A, Johnson AJ, Hertlein E, Liu TH, Jin VX, Sherman MH, Liu SJ, Dawson DW, Williams KE, et al. Epigenetic changes during disease progression in a murine model of human chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2009;106:13433–13438. doi: 10.1073/pnas.0906455106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WY, Zeng X, Carter MG, Morrell CN, Chiu Yen RW, Esteller M, Watkins DN, Herman JG, Mankowski JL, Baylin SB. Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nature genetics. 2003;33:197–202. doi: 10.1038/ng1077. [DOI] [PubMed] [Google Scholar]
- Cheung HH, Lee TL, Davis AJ, Taft DH, Rennert OM, Chan WY. Genome-wide DNA methylation profiling reveals novel epigenetically regulated genes and non-coding RNAs in human testicular cancer. Br J Cancer. 2010;102:419–427. doi: 10.1038/sj.bjc.6605505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim CS, Fung TK, Cheung WC, Liang R, Kwong YL. SOCS1 and SHP1 hypermethylation in multiple myeloma: implications for epigenetic activation of the Jak/STAT pathway. Blood. 2004a;103:4630–4635. doi: 10.1182/blood-2003-06-2007. [DOI] [PubMed] [Google Scholar]
- Chim CS, Wong AS, Kwong YL. Epigenetic dysregulation of the Jak/STAT pathway by frequent aberrant methylation of SHP1 but not SOCS1 in acute leukaemias. Ann Hematol. 2004b;83:527–532. doi: 10.1007/s00277-004-0843-1. [DOI] [PubMed] [Google Scholar]
- Chim CS, Wong KY, Loong F, Srivastava G. SOCS1 and SHP1 hypermethylation in mantle cell lymphoma and follicular lymphoma: implications for epigenetic activation of the Jak/STAT pathway. Leukemia. 2004c;18:356–358. doi: 10.1038/sj.leu.2403216. [DOI] [PubMed] [Google Scholar]
- Coletta RD, Jedlicka P, Gutierrez-Hartmann A, Ford HL. Transcriptional control of the cell cycle in mammary gland development and tumorigenesis. J Mammary Gland Biol Neoplasia. 2004;9:39–53. doi: 10.1023/B:JOMG.0000023587.40966.f6. [DOI] [PubMed] [Google Scholar]
- Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nature genetics. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nature genetics. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nature genetics. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- Dammann R, Schagdarsurengin U, Seidel C, Strunnikova M, Rastetter M, Baier K, Pfeifer GP. The tumor suppressor RASSF1A in human carcinogenesis: an update. Histol Histopathol. 2005a;20:645–663. doi: 10.14670/HH-20.645. [DOI] [PubMed] [Google Scholar]
- Dammann R, Strunnikova M, Schagdarsurengin U, Rastetter M, Papritz M, Hattenhorst UE, Hofmann HS, Silber RE, Burdach S, Hansen G. CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur J Cancer. 2005b;41:1223–1236. doi: 10.1016/j.ejca.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Davalos V, Esteller M. MicroRNAs and cancer epigenetics: a macrorevolution. Curr Opin Oncol. 2010;22:35–45. doi: 10.1097/CCO.0b013e328333dcbb. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Deneberg S, Grovdal M, Karimi M, Jansson M, Nahi H, Corbacioglu A, Gaidzik V, Dohner K, Paul C, Ekstrom TJ, et al. Gene-specific and global methylation patterns predict outcome in patients with acute myeloid leukemia. Leukemia. 2010;24:932–941. doi: 10.1038/leu.2010.41. [DOI] [PubMed] [Google Scholar]
- Ding L, Kleer CG. Enhancer of Zeste 2 as a marker of preneoplastic progression in the breast. Cancer Res. 2006;66:9352–9355. doi: 10.1158/0008-5472.CAN-06-2384. [DOI] [PubMed] [Google Scholar]
- Dudley KJ, Revill K, Whitby P, Clayton RN, Farrell WE. Genome-wide analysis in a murine Dnmt1 knockdown model identifies epigenetically silenced genes in primary human pituitary tumors. Mol Cancer Res. 2008;6:1567–1574. doi: 10.1158/1541-7786.MCR-08-0234. [DOI] [PubMed] [Google Scholar]
- Dunwell T, Hesson L, Rauch TA, Wang L, Clark RE, Dallol A, Gentle D, Catchpoole D, Maher ER, Pfeifer GP, Latif F. A genome-wide screen identifies frequently methylated genes in haematological and epithelial cancers. Molecular cancer. 9:44. doi: 10.1186/1476-4598-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell T, Hesson L, Rauch TA, Wang L, Clark RE, Dallol A, Gentle D, Catchpoole D, Maher ER, Pfeifer GP, Latif F. A genome-wide screen identifies frequently methylated genes in haematological and epithelial cancers. Mol Cancer. 2010;9:44. doi: 10.1186/1476-4598-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- Estecio MR, Yan PS, Ibrahim AE, Tellez CS, Shen L, Huang TH, Issa JP. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res. 2007;17:1529–1536. doi: 10.1101/gr.6417007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM. Predicting aberrant CpG island methylation. Proc Natl Acad Sci U S A. 2003;100:12253–12258. doi: 10.1073/pnas.2037852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Melnick A, Greally JM. Genome-wide determination of DNA methylation by Hpa II tiny fragment enrichment by ligation-mediated PCR (HELP) for the study of acute leukemias. Methods Mol Biol. 2009;538:395–407. doi: 10.1007/978-1-59745-418-6_20. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Montoya G, Taysavang P, Wade PA, Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003;31:1765–1774. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin WA. Premalignant evolution of lung cancer: Gilles F Filley lecture. Chest. 2004;125:90S–94S. doi: 10.1378/chest.125.5_suppl.90s. [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta J, Nobeyama Y, Umebayashi Y, Otsuka F, Kikuchi K, Ushijima T. Silencing of Peroxiredoxin 2 and aberrant methylation of 33 CpG islands in putative promoter regions in human malignant melanomas. Cancer Res. 2006;66:6080–6086. doi: 10.1158/0008-5472.CAN-06-0157. [DOI] [PubMed] [Google Scholar]
- Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- Gebhard C, Benner C, Ehrich M, Schwarzfischer L, Schilling E, Klug M, Dietmaier W, Thiede C, Holler E, Andreesen R, Rehli M. General transcription factor binding at CpG islands in normal cells correlates with resistance to de novo DNA methylation in cancer cells. Cancer Res. 2010;70:1398–1407. doi: 10.1158/0008-5472.CAN-09-3406. [DOI] [PubMed] [Google Scholar]
- Geiman TM, Robertson KD. Chromatin remodeling, histone modifications, and DNA methylation-how does it all fit together? J Cell Biochem. 2002;87:117–125. doi: 10.1002/jcb.10286. [DOI] [PubMed] [Google Scholar]
- Gitan RS, Shi H, Chen CM, Yan PS, Huang TH. Methylation-specific oligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome research. 2002;12:158–164. doi: 10.1101/gr.202801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Shinjo K, Kondo Y, Shen L, Toyota M, Suzuki H, Gao W, An B, Fujii M, Murakami H, et al. Epigenetic profiles distinguish malignant pleural mesothelioma from lung adenocarcinoma. Cancer Res. 2009;69:9073–9082. doi: 10.1158/0008-5472.CAN-09-1595. [DOI] [PubMed] [Google Scholar]
- Gu H, Bock C, Mikkelsen TS, Jager N, Smith ZD, Tomazou E, Gnirke A, Lander ES, Meissner A. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods. 2010;7:133–136. doi: 10.1038/nmeth.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Tommasi S, Liu L, Yee JK, Dammann R, Pfeifer GP. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr Biol. 2007;17:700–705. doi: 10.1016/j.cub.2007.02.055. [DOI] [PubMed] [Google Scholar]
- Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, Riggs AD, Chu FF, Pfeifer GP. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68:10280–10289. doi: 10.1158/0008-5472.CAN-08-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- Hatada I, Hayashizaki Y, Hirotsune S, Komatsubara H, Mukai T. A genomic scanning method for higher organisms using restriction sites as landmarks. Proc Natl Acad Sci U S A. 1991;88:9523–9527. doi: 10.1073/pnas.88.21.9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Hinoue T, Weisenberger DJ, Pan F, Campan M, Kim M, Young J, Whitehall VL, Leggett BA, Laird PW. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS One. 2009;4:e8357. doi: 10.1371/journal.pone.0008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon DS, Spugnardi M, Kuo C, Huang SK, Morton DL, Taback B. Profiling epigenetic inactivation of tumor suppressor genes in tumors and plasma from cutaneous melanoma patients. Oncogene. 2004;23:4014–4022. doi: 10.1038/sj.onc.1207505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P, Shen JY, Ji MJ, He NY, Lu ZH. Microarray-based method for detecting methylation changes of p16(Ink4a) gene 5'-CpG islands in gastric carcinomas. World J Gastroenterol. 2004;10:3553–3558. doi: 10.3748/wjg.v10.i24.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet. 1999;8:459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, Wen B, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome research. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP. Aging, DNA methylation and cancer. Crit Rev Oncol Hematol. 1999;32:31–43. doi: 10.1016/s1040-8428(99)00019-0. [DOI] [PubMed] [Google Scholar]
- Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang CL, Jin SG, Pfeifer GP. MBD3L1 is a transcriptional repressor that interacts with methyl-CpG-binding protein 2 (MBD2) and components of the NuRD complex. J Biol Chem. 2004;279:52456–52464. doi: 10.1074/jbc.M409149200. [DOI] [PubMed] [Google Scholar]
- Jin M, Kawakami K, Fukui Y, Tsukioka S, Oda M, Watanabe G, Takechi T, Oka T, Minamoto T. Different histological types of non-small cell lung cancer have distinct folate and DNA methylation levels. Cancer Sci. 2009;100:2325–2330. doi: 10.1111/j.1349-7006.2009.01321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N, et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri M, Cahill N, Goransson H, Enstrom C, Ryan F, Isaksson A, Rosenquist R. Differential genome-wide array-based methylation profiles in prognostic subsets of chronic lymphocytic leukemia. Blood. 2010;115:296–305. doi: 10.1182/blood-2009-07-232868. [DOI] [PubMed] [Google Scholar]
- Kansara M, Tsang M, Kodjabachian L, Sims NA, Trivett MK, Ehrich M, Dobrovic A, Slavin J, Choong PF, Simmons PJ, et al. Wnt inhibitory factor 1 is epigenetically silenced in human osteosarcoma, and targeted disruption accelerates osteosarcomagenesis in mice. J Clin Invest. 2009;119:837–851. doi: 10.1172/JCI37175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, Simon I. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nature genetics. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YT, Park SJ, Lee SH, Kang HJ, Hahn S, Kang CH, Sung SW, Kim JH. Prognostic implication of aberrant promoter hypermethylation of CpG islands in adenocarcinoma of the lung. J Thorac Cardiovasc Surg. 2005;130:1378. doi: 10.1016/j.jtcvs.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Kirmizis A, Bartley SM, Farnham PJ. Identification of the polycomb group protein SU(Z)12 as a potential molecular target for human cancer therapy. Mol Cancer Ther. 2003;2:113–121. [PubMed] [Google Scholar]
- Koga Y, Pelizzola M, Cheng E, Krauthammer M, Sznol M, Ariyan S, Narayan D, Molinaro AM, Halaban R, Weissman SM. Genome-wide screen of promoter methylation identifies novel markers in melanoma. Genome Res. 2009;19:1462–1470. doi: 10.1101/gr.091447.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nature genetics. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- Kron K, Pethe V, Briollais L, Sadikovic B, Ozcelik H, Sunderji A, Venkateswaran V, Pinthus J, Fleshner N, van der Kwast T, Bapat B. Discovery of novel hypermethylated genes in prostate cancer using genomic CpG island microarrays. PLoS One. 2009;4:e4830. doi: 10.1371/journal.pone.0004830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang SQ, Tong WG, Yang H, Lin W, Lee MK, Fang ZH, Wei Y, Jelinek J, Issa JP, Garcia-Manero G. Genome-wide identification of aberrantly methylated promoter associated CpG islands in acute lymphocytic leukemia. Leukemia. 2008;22:1529–1538. doi: 10.1038/leu.2008.130. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, Reinberg D. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci U S A. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- Laird PW, Jackson-Grusby L, Fazeli A, Dickinson SL, Jung WE, Li E, Weinberg RA, Jaenisch R. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann U, Hasemeier B, Christgen M, Muller M, Romermann D, Langer F, Kreipe H. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- Leshchenko VV, Kuo PY, Shaknovich R, Yang DT, Gellen T, Petrich A, Yu Y, Remache Y, Weniger MA, Rafiq S, et al. Genome wide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2010 doi: 10.1182/blood-2009-12-257485. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Colot V, Martienssen R. Profiling DNA methylation patterns using genomic tiling microarrays. Nat Methods. 2005;2:219–224. doi: 10.1038/nmeth0305-219. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Zhao Y, Tawatao R, Cottam HB, Sen M, Leoni LM, Kipps TJ, Corr M, Carson DA. Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2004;101:3118–3123. doi: 10.1073/pnas.0308648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Ouyang W, Huang C. Inflammation, a key event in cancer development. Mol Cancer Res. 2006;4:221–233. doi: 10.1158/1541-7786.MCR-05-0261. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- Maegawa S, Hinkal G, Kim HS, Shen L, Zhang L, Zhang J, Zhang N, Liang S, Donehower LA, Issa JP. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Subero JI, Ammerpohl O, Bibikova M, Wickham-Garcia E, Agirre X, Alvarez S, Bruggemann M, Bug S, Calasanz MJ, Deckert M, et al. A comprehensive microarray-based DNA methylation study of 367 hematological neoplasms. PLoS One. 2009;4:e6986. doi: 10.1371/journal.pone.0006986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez R, Martin-Subero JI, Rohde V, Kirsch M, Alaminos M, Fernandez AF, Ropero S, Schackert G, Esteller M. A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics. 2009;4:255–264. doi: 10.4161/epi.9130. [DOI] [PubMed] [Google Scholar]
- McCabe MT, Lee EK, Vertino PM. A multifactorial signature of DNA sequence and polycomb binding predicts aberrant CpG island methylation. Cancer Res. 2009;69:282–291. doi: 10.1158/0008-5472.CAN-08-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey KM, Greene E, Fahrner JA, Jenuwein T, Baylin SB. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res. 2007;67:5097–5102. doi: 10.1158/0008-5472.CAN-06-2029. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nephew KP, Huang TH. Epigenetic gene silencing in cancer initiation and progression. Cancer Lett. 2003;190:125–133. doi: 10.1016/s0304-3835(02)00511-6. [DOI] [PubMed] [Google Scholar]
- Nishiyama N, Arai E, Chihara Y, Fujimoto H, Hosoda F, Shibata T, Kondo T, Tsukamoto T, Yokoi S, Imoto I, et al. Genome-wide DNA methylation profiles in urothelial carcinomas and urothelia at the precancerous stage. Cancer Sci. 2010;101:231–240. doi: 10.1111/j.1349-7006.2009.01330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima M, Suzuki H, Toyota M, Watanabe Y, Maruyama R, Sasaki S, Sasaki Y, Mita H, Nishikawa N, Yamaguchi K, et al. Frequent epigenetic inactivation of SFRP genes and constitutive activation of Wnt signaling in gastric cancer. Oncogene. 2007;26:4699–4713. doi: 10.1038/sj.onc.1210259. [DOI] [PubMed] [Google Scholar]
- Nouzova M, Holtan N, Oshiro MM, Isett RB, Munoz-Rodriguez JL, List AF, Narro ML, Miller SJ, Merchant NC, Futscher BW. Epigenomic changes during leukemia cell differentiation: analysis of histone acetylation and cytosine methylation using CpG island microarrays. J Pharmacol Exp Ther. 2004;311:968–981. doi: 10.1124/jpet.104.072488. [DOI] [PubMed] [Google Scholar]
- Novak P, Jensen T, Oshiro MM, Watts GS, Kim CJ, Futscher BW. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res. 2008;68:8616–8625. doi: 10.1158/0008-5472.CAN-08-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Kobayashi T, Chinen T, Takaki H, Sanada T, Minoda Y, Koga K, Takaesu G, Maehara Y, Iida M, Yoshimura A. Deletion of the SOCS3 gene in liver parenchymal cells promotes hepatitis-induced hepatocarcinogenesis. Gastroenterology. 2006;131:179–193. doi: 10.1053/j.gastro.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nature genetics. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura N, Li CP, Li A, Hong SM, Walter K, Jimeno A, Hidalgo M, Goggins M. Genome-wide profiling of methylated promoters in pancreatic adenocarcinoma. Cancer Biol Ther. 2008;7:1146–1156. doi: 10.4161/cbt.7.7.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardal R, Molofsky AV, He S, Morrison SJ. Stem cell self-renewal and cancer cell proliferation are regulated by common networks that balance the activation of proto-oncogenes and tumor suppressors. Cold Spring Harb Symp Quant Biol. 2005;70:177–185. doi: 10.1101/sqb.2005.70.057. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Helin K. Polycomb group proteins in cell cycle progression and cancer. Cell Cycle. 2004;3:396–400. [PubMed] [Google Scholar]
- Pfeifer GP. Mutagenesis at methylated CpG sequences. Curr Top Microbiol Immunol. 2006;301:259–281. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Besaratinia A. Mutational spectra of human cancer. Hum Genet. 2009;125:493–506. doi: 10.1007/s00439-009-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaphorst FM. Deregulated expression of Polycomb-group oncogenes in human malignant lymphomas and epithelial tumors. Hum Mol Genet. 2005a;14(Spec No 1):R93–R100. doi: 10.1093/hmg/ddi111. [DOI] [PubMed] [Google Scholar]
- Raaphorst FM. Of mice, flies, and man: the emerging role of polycomb-group genes in human malignant lymphomas. Int J Hematol. 2005b;81:281–287. doi: 10.1532/IJH97.05023. [DOI] [PubMed] [Google Scholar]
- Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Lab Invest. 2005;85:1172–1180. doi: 10.1038/labinvest.3700311. [DOI] [PubMed] [Google Scholar]
- Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, Kernstine KH, Riggs AD, Pfeifer GP. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, Riggs AD, Pfeifer GP. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee CS, Sen M, Lu D, Wu C, Leoni L, Rubin J, Corr M, Carson DA. Wnt and frizzled receptors as potential targets for immunotherapy in head and neck squamous cell carcinomas. Oncogene. 2002;21:6598–6605. doi: 10.1038/sj.onc.1205920. [DOI] [PubMed] [Google Scholar]
- Ringrose L. Polycomb comes of age: genome-wide profiling of target sites. Curr Opin Cell Biol. 2007;19:290–297. doi: 10.1016/j.ceb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Ruike Y, Imanaka Y, Sato F, Shimizu K, Tsujimoto G. Genome-wide analysis of aberrant methylation in human breast cancer cells using methyl-DNA immunoprecipitation combined with high-throughput sequencing. BMC Genomics. 2010;11:137. doi: 10.1186/1471-2164-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nature genetics. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Seidel C, Schagdarsurengin U, Blumke K, Wurl P, Pfeifer GP, Hauptmann S, Taubert H, Dammann R. Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma. Mol Carcinog. 2007;46:865–871. doi: 10.1002/mc.20317. [DOI] [PubMed] [Google Scholar]
- Sellers WR, Loda M. The EZH2 polycomb transcriptional repressor--a marker or mover of metastatic prostate cancer? Cancer Cell. 2002;2:349–350. doi: 10.1016/s1535-6108(02)00187-3. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. Cancer: crime and punishment. Nature. 2005;436:636–637. doi: 10.1038/436636a. [DOI] [PubMed] [Google Scholar]
- Shih YL, Hsieh CB, Lai HC, Yan MD, Hsieh TY, Chao YC, Lin YW. SFRP1 suppressed hepatoma cells growth through Wnt canonical signaling pathway. Int J Cancer. 2007;121:1028–1035. doi: 10.1002/ijc.22750. [DOI] [PubMed] [Google Scholar]
- Shiraishi M, Sekiguchi A, Terry MJ, Oates AJ, Miyamoto Y, Chuu YH, Munakata M, Sekiya T. A comprehensive catalog of CpG islands methylated in human lung adenocarcinomas for the identification of tumor suppressor genes. Oncogene. 2002;21:3804–3813. doi: 10.1038/sj.onc.1205454. [DOI] [PubMed] [Google Scholar]
- Smith RJ, Kelsey G. Identification of imprinted loci by methylation: use of methylation-sensitive representational difference analysis (Me-RDA) Methods Mol Biol. 2001;181:113–132. doi: 10.1385/1-59259-211-2:113. [DOI] [PubMed] [Google Scholar]
- Song JZ, Stirzaker C, Harrison J, Melki JR, Clark SJ. Hypermethylation trigger of the glutathione-S-transferase gene (GSTP1) in prostate cancer cells. Oncogene. 2002;21:1048–1061. doi: 10.1038/sj.onc.1205153. [DOI] [PubMed] [Google Scholar]
- Spugnardi M, Tommasi S, Dammann R, Pfeifer GP, Hoon DS. Epigenetic inactivation of RAS association domain family protein 1 (RASSF1A) in malignant cutaneous melanoma. Cancer Res. 2003;63:1639–1643. [PubMed] [Google Scholar]
- Squazzo SL, O'Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John MA, Tao W, Fei X, Fukumoto R, Carcangiu ML, Brownstein DG, Parlow AF, McGrath J, Xu T. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nature genetics. 1999;21:182–186. doi: 10.1038/5965. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nature genetics. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- Takagi H, Sasaki S, Suzuki H, Toyota M, Maruyama R, Nojima M, Yamamoto H, Omata M, Tokino T, Imai K, Shinomura Y. Frequent epigenetic inactivation of SFRP genes in hepatocellular carcinoma. J Gastroenterol. 2008;43:378–389. doi: 10.1007/s00535-008-2170-0. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Miyoshi Y, Takahata C, Irahara N, Taguchi T, Tamaki Y, Noguchi S. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin Cancer Res. 2005;11:1380–1385. doi: 10.1158/1078-0432.CCR-04-1773. [DOI] [PubMed] [Google Scholar]
- Takeshima H, Yamashita S, Shimazu T, Niwa T, Ushijima T. The presence of RNA polymerase II, active or stalled, predicts epigenetic fate of promoter CpG islands. Genome Res. 2009;19:1974–1982. doi: 10.1101/gr.093310.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, Yamamoto H, Hirata T, Miyamoto N, Oki M, Nosho K, Adachi Y, Endo T, Imai K, Shinomura Y. Frequent epigenetic inactivation of Wnt inhibitory factor-1 in human gastrointestinal cancers. Oncogene. 2005;24:7946–7952. doi: 10.1038/sj.onc.1208910. [DOI] [PubMed] [Google Scholar]
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Gayther SA, Apostolidou S, Jones A, Lechner M, Beck S, Jacobs IJ, Widschwendter M. An epigenetic signature in peripheral blood predicts active ovarian cancer. PLoS One. 2009;4:e8274. doi: 10.1371/journal.pone.0008274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasi S, Dammann R, Zhang Z, Wang Y, Liu L, Tsark WM, Wilczynski SP, Li J, You M, Pfeifer GP. Tumor susceptibility of Rassf1a knockout mice. Cancer Res. 2005;65:92–98. [PubMed] [Google Scholar]
- Tommasi S, Karm DL, Wu X, Yen Y, Pfeifer GP. Methylation of homeobox genes is a frequent and early epigenetic event in breast cancer. Breast Cancer Res. 2009;11:R14. doi: 10.1186/bcr2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topaloglu O, Hoque MO, Tokumaru Y, Lee J, Ratovitski E, Sidransky D, Moon CS. Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res. 2004;10:2284–2288. doi: 10.1158/1078-0432.ccr-1111-3. [DOI] [PubMed] [Google Scholar]
- Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, Fukuyama Y, Virmani AK, Zochbauer-Muller S, Tsukuda K, Sugio K, et al. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer. 2003;103:153–160. doi: 10.1002/ijc.10787. [DOI] [PubMed] [Google Scholar]
- Toyota M, Ho C, Ahuja N, Jair KW, Li Q, Ohe-Toyota M, Baylin SB, Issa JP. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307–2312. [PubMed] [Google Scholar]
- Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- Urakami S, Shiina H, Enokida H, Kawakami T, Tokizane T, Ogishima T, Tanaka Y, Li LC, Ribeiro-Filho LA, Terashima M, et al. Epigenetic inactivation of Wnt inhibitory factor-1 plays an important role in bladder cancer through aberrant canonical Wnt/beta-catenin signaling pathway. Clin Cancer Res. 2006;12:383–391. doi: 10.1158/1078-0432.CCR-05-1344. [DOI] [PubMed] [Google Scholar]
- Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5:223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- Ushijima T, Morimura K, Hosoya Y, Okonogi H, Tatematsu M, Sugimura T, Nagao M. Establishment of methylation-sensitive-representational difference analysis and isolation of hypo- and hypermethylated genomic fragments in mouse liver tumors. Proc Natl Acad Sci U S A. 1997;94:2284–2289. doi: 10.1073/pnas.94.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima T, Yamashita S. Methylation-sensitive representational difference analysis (MS-RDA) Methods Mol Biol. 2009;507:117–130. doi: 10.1007/978-1-59745-522-0_10. [DOI] [PubMed] [Google Scholar]
- Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature genetics. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Weeraratna AT, Jiang Y, Hostetter G, Rosenblatt K, Duray P, Bittner M, Trent JM. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1:279–288. doi: 10.1016/s1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- Wei SH, Chen CM, Strathdee G, Harnsomburana J, Shyu CR, Rahmatpanah F, Shi H, Ng SW, Yan PS, Nephew KP, et al. Methylation microarray analysis of late-stage ovarian carcinomas distinguishes progression-free survival in patients and identifies candidate epigenetic markers. Clin Cancer Res. 2002;8:2246–2252. [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nature genetics. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nature genetics. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Wu X, Rauch TA, Zhong X, Latif F, Krex D, Pfeifer GP. CpG island hypermethylation in human astrocytomas. Cancer Res. 2010;70:2718–2727. doi: 10.1158/0008-5472.CAN-09-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T, Midorikawa Y, Nishimura Y, Sakamoto H, Seto Y, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 16:21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T, Midorikawa Y, Nishimura Y, Sakamoto H, Seto Y, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010;16:21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Jackson-Grusby L, Linhart H, Meissner A, Eden A, Lin H, Jaenisch R. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci U S A. 2005;102:13580–13585. doi: 10.1073/pnas.0506612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan PS, Chen CM, Shi H, Rahmatpanah F, Wei SH, Caldwell CW, Huang TH. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001;61:8375–8380. [PubMed] [Google Scholar]
- Yanagawa N, Tamura G, Oizumi H, Takahashi N, Shimazaki Y, Motoyama T. Promoter hypermethylation of tumor suppressor and tumor-related genes in non-small cell lung cancers. Cancer Sci. 2003;94:589–592. doi: 10.1111/j.1349-7006.2003.tb01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HH, Hu N, Wang C, Ding T, Dunn BK, Goldstein AM, Taylor PR, Lee MP. Influence of genetic background and tissue types on global DNA methylation patterns. PLoS One. 2010;5:e9355. doi: 10.1371/journal.pone.0009355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue W, Sun Q, Dacic S, Landreneau RJ, Siegfried JM, Yu J, Zhang L. Downregulation of Dkk3 activates beta-catenin/TCF-4 signaling in lung cancer. Carcinogenesis. 2008;29:84–92. doi: 10.1093/carcin/bgm267. [DOI] [PubMed] [Google Scholar]
- Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, Bardeesy N. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61:249–255. [PubMed] [Google Scholar]