Abstract
The HIV gp41 N-trimer pocket region is an ideal viral target because it is extracellular, highly conserved, and essential for viral entry. Here, we report on the design of a pocket-specific d-peptide, PIE12-trimer, that is extraordinarily elusive to resistance and characterize its inhibitory and structural properties. d-Peptides (peptides composed of d-amino acids) are promising therapeutic agents due to their insensitivity to protease degradation. PIE12-trimer was designed using structure-guided mirror-image phage display and linker optimization and is the first d-peptide HIV entry inhibitor with the breadth and potency required for clinical use. PIE12-trimer has an ultrahigh affinity for the gp41 pocket, providing it with a reserve of binding energy (resistance capacitor) that yields a dramatically improved resistance profile compared to those of other fusion inhibitors. These results demonstrate that the gp41 pocket is an ideal drug target and establish PIE12-trimer as a leading anti-HIV antiviral candidate.
The HIV envelope protein (Env) mediates viral entry into cells (11). Env is cleaved into surface (gp120) and transmembrane (gp41) subunits that remain noncovalently associated to form trimeric spikes on the virion surface (16). gp120 recognizes target cells by interacting with cellular receptors, while gp41 mediates membrane fusion. Peptides derived from heptad repeats near the N and C termini of the gp41 ectodomain (N and C peptides) interact in solution to form a six-helix bundle, representing the postfusion structure (3, 55, 56). In this structure, N peptides form a central trimeric coiled coil (N trimer), creating grooves into which C peptides bind. This structure, in conjunction with the dominant-negative inhibitory properties of exogenous N and C peptides, suggests a mechanism for Env-mediated entry (10, 22, 58-60).
During entry, gp41 forms an extended prehairpin intermediate that leaves the exposed N-trimer region vulnerable to inhibition for several minutes (18, 35). This intermediate ultimately collapses as the C-peptide regions bind to the N-trimer grooves to form a trimer of hairpins (six-helix bundle), juxtaposing viral and cellular membranes and inducing fusion. Enfuvirtide (Fuzeon), the only clinically approved HIV fusion inhibitor, is a C peptide that binds to part of the N-trimer groove and prevents six-helix bundle formation in a dominant-negative manner (61). Enfuvirtide is active in patients with multidrug resistance to other classes of inhibitors and is a life-prolonging option for these patients (30, 31). However, enfuvirtide use is restricted to salvage therapy due to several limitations, including (i) high dosing requirements (90 mg, twice-daily injections), (ii) high cost (∼$30,000/year/patient in the United States), and (iii) the rapid emergence of resistant strains (21, 47).
A deep hydrophobic pocket at the base of the N-trimer groove is an especially attractive inhibitory target because of its high degree of conservation (3, 12, 48), poor tolerance to substitution (4, 34), and critical role in membrane fusion (2). Indeed, this region is conserved at both the amino acid level (for gp41 function in membrane fusion) and the nucleotide level (for the structured RNA region of the Rev-responsive element). Enfuvirtide binds to the N-trimer groove just N terminal to the pocket and is significantly more susceptible to resistance mutations than 2nd-generation C-peptide inhibitors, such as T-1249, that also bind to the pocket (8, 13, 29, 44, 46, 47, 58).
Peptide design, molecular modeling, and small-molecule screening have produced a diverse set of compounds that interact with the gp41 pocket and inhibit HIV-1 entry with modest potency, but often with significant cytotoxicity (7, 14, 15, 17, 23, 24, 26, 34, 51, 54). The first direct evidence that pocket-specific binders are sufficient to inhibit HIV entry came with the discovery of protease-resistant d-peptides identified using mirror-image phage display (12). In this technique, a phage library is screened against a mirror-image version of the target protein (synthesized using d-amino acids) (50). By symmetry, mirror images (d-peptides) of the discovered sequences will bind to the natural l-peptide target. As the mirror images of naturally occurring l-peptides, d-peptides cannot be digested by natural proteases. Protease resistance provides d-peptides theoretical treatment advantages of extended survival in the body and possible oral bioavailability (41, 42, 49).
These 1st-generation d-peptide entry inhibitors possess potency against a laboratory-adapted isolate (HXB2) at low to mid-μM concentrations (12). We previously reported an affinity-matured 2nd-generation d-peptide called PIE7, pocket-specific inhibitor of entry 7 (57). A trimeric version of PIE7 is the first high-affinity pocket-specific HIV-1 inhibitor and has potency against X4-tropic (HXB2) and R5-tropic (BaL) strains at sub-nM concentrations. However, significant further optimization is required to create a robust clinical candidate for two reasons. First, this d-peptide is much less potent (requiring high nM concentrations) against JRFL, a primary R5-tropic strain. Therefore, improved PIE potency is necessary to combat diverse primary strains. Second, by improving the affinity of our inhibitors for the pocket target, we hope to provide a reserve of binding energy that will delay the emergence of drug resistance, as described below.
We and others have reported a potency plateau for some gp41-based fusion inhibitors that is likely imposed by the transient exposure of the prehairpin intermediate (9, 27, 53, 57). For very high-affinity inhibitors, association kinetics (rather than affinity) limits potency so that two inhibitors with significantly different affinities for the prehairpin intermediate can have similar antiviral potencies. We proposed that overengineering our d-peptides with substantial affinity beyond this potency plateau would provide a reserve of binding energy that would combat affinity-disrupting resistance mutations (57). Such a resistance capacitor should also prevent the stepwise accumulation of subtle resistance mutations in Env by eliminating the selective advantage that such mutants would otherwise confer.
Here, we report on the design and characterization of a 3rd-generation pocket-specific d-peptide, PIE12-trimer, with ∼100,000-fold improved target binding compared to that of the best previous d-peptide, significantly broadened inhibitory potency, and an enhanced resistance capacitor that provides a strong barrier to viral resistance. We achieved this increased potency via structure-guided phage display and crosslinker optimization. PIE12-trimer has a dramatically improved resistance profile compared to the profiles of earlier d-peptides, as well as those of enfuvirtide and T-1249. These results validate the resistance capacitor hypothesis and establish PIE12-trimer as a leading anti-HIV therapeutic candidate.
MATERIALS AND METHODS
Peptide synthesis.
All peptides were synthesized as described previously (57). All dimers and trimers except PIE12-trimer were made essentially as described using bis-dPEG5 NHS ester (where PEG is polyethylene glycol and NHS is N-hydroxysuccinimide; catalog no. 10224; Quanta BioDesign); PIE12-trimer was synthesized using the following higher-yield protocol. PIE12-GK (2 mM) was reacted with bis-dPEG5 NHS ester crosslinker (1 M stock in dimethylacetamide) at a 1:20 (peptide/PEG) molar ratio in 100 mM HEPES (pH 7.8 to 8) for 90 s at room temperature (RT). The reaction was stopped by addition of acetic acid to 5% and 3 M guanidine HCl (GuHCl) and purified by reverse-phase high-pressure liquid chromatography (RP-HPLC; C18 column; Vydac). This product (∼3 to 5 mM) was reacted at a 2:1 molar excess with PIE12-GKK in dimethylacetamide buffered by triethylamine (pH 7.5) for 75 min and purified by RP-HPLC (C18 column; Vydac).
Phage display vector design.
Use of a commercially available phage library cloning system (NEB) allowed us to relocate cloning sites away from the flanking regions (38). We redesigned the regions immediately outside the flanking residues in our cloning vector in order to structurally isolate them and minimize any bias caused by flanking sequence randomization. Our library peptides are displayed as fusions to the phage p3 protein, which contains an N-terminal leader sequence that is cleaved by Escherichia coli secretion signal peptidases. In the original vector, the N-terminal flanking residues of the library peptides are immediately adjacent to the secretion signal. Due to proximity to the secretion signal cleavage site, it is likely that randomization of these residues would differentially affect library-p3 protein secretion and peptide presentation on the phage surface. This bias would confound the selection of N-terminal flanking sequences solely on the basis of their affinity for the N trimer. To avoid this bias, we introduced a five-amino-acid spacer to structurally isolate the cleavage site from the randomized N-terminal flanking residues. We choose the N-terminal residues (KIEEG) from maltose binding protein (MBP) as the spacer sequence, since MBP is very efficiently cleaved during secretion from E. coli.
We have observed that mutations in the C-terminal sequence that links the peptide to the phage p3 protein can also create undesirable selection bias (presumably by allowing the C terminus of the d-peptides to form a continuous helix with the N terminus of p3, thus enhancing peptide presentation to the target) (57). Therefore, a flexible GGGS spacer was inserted after the C-terminal flanking residues to structurally isolate them from the N terminus of p3.
To validate this new phage display vector, we used it to clone an earlier PIE (PIE2) along with a mutant (PIE2-AAA) which had previously been observed to enhance phage affinity for the pocket target via mutation of the linker between the library peptide and p3, although this mutation did not enhance inhibitor potency when incorporated into a d-peptide (57). We assayed the target binding affinity of the resultant phage (Φ) and compared it to that of phage produced with the previous phage vector. In the previous phage vector, PIE2-AAA-Φ “cheated” in order to bind to the target with an ∼70-fold more affinity than PIE2-Φ, but this difference was abolished in the modified vector (data not shown). Furthermore, sequencing revealed that N-terminal flanking residues from the amplified phage library prior to selection were random, indicating a lack of bias due to signal peptidase cleavage efficiency.
Phage display.
An 8-mer flanking library phage display was performed essentially as described previously (57). Four rounds of mirror-image solution-phase phage display were performed by incubating (for 2 h at RT) 1010 phage (amplified from the previous round) with 10 nM biotinylated d-IZN17 (a mimic of the d-peptide gp41 pocket target) in the presence of escalating soluble competitor (L-2K-PIE2) (10, 30, 90, and 360 μM for rounds 1 to 4, respectively) (57). Phage-bound d-IZN17 was rapidly captured from solution using Dynal T1 streptavidin-coated magnetic beads (Invitrogen) and briefly washed 3 times with 500 μl of 0.1% Tween 20 in Tris-buffered saline (wash buffer contained 100 μM d-biotin for the 1st wash). Phage was eluted in 50 μl of glycine (pH 2.2) elution buffer (10 min at RT) and neutralized with 7.5 μl of 1 M Tris, pH 9.1. The amplified phage library was sequenced prior to the first round of selection to confirm randomization, and preamplified eluted phage was sequenced following each round. All phage binding experiments were performed using the same protocol described above using 270 μM L-PIE2 soluble competitor. A 7-mer phage display was performed using a similar protocol.
Crystal growth and data collection.
The original form of PIE12 (see Table 1) contains a C-terminal GK extension and did not yield highly diffracting crystals in complex with IQN17, a gp41 pocket mimic. Variants of PIE12 instead containing an N-terminal K or KG extension (K-PIE12, KHPCDYPEWQWLCEL; KG-PIE12, KGHPCDYPEWQWLCEL) crystallized in complex with IQN17 under a variety of conditions. In each case, the reservoir (850 μl) comprised a solution from a commercially available crystallization screen, and the crystallization drop was prepared by mixing 0.3 or 0.5 μl of the IQN17-PIE12 or IQN17-PIE71 protein solution (1:1.1 molar ratio, 10 mg/ml total in water) with 0.3 μl of the reservoir solution. Crystals typically grew in 1 to 10 days. All crystals were grown by sitting-drop vapor diffusion. IQN17-PIE12 form I crystals (KG-PIE12) were grown at 21°C in Hampton Scientific condition Screen II 48 (10% PEG 20,000, 0.1 M bicine, pH 9.0, 2% dioxane). IQN17-PIE12 form II crystals (KG-PIE12) were grown at 21°C in Emerald Biosystems condition Cryo-II 37 (50% ethylene glycol, 0.1 M imidazole, pH 8.0). IQN17-PIE12 form III crystals (K-PIE12) were grown at 4°C in Emerald Biosystems condition Cryo-II 25 (40% 2-methyl-2,4-pentanediol (MPD), 0.1 M N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) [pH 10.5]). IQN17-PIE71 crystals were grown at 21°C in Qiagen PACT crystallization condition G4 (20% PEG 3350, 0.2 M potassium thiocyanate, 0.1 M bis-Tris propane, pH 7.5).
TABLE 1.
d-peptide inhibition datae
| Sample | Sequence | IC50 (nM)a |
|
|---|---|---|---|
| HXB2 | JRFL | ||
| PIE7 | KGA[PIE7]AA | 620b | 24,000b |
| PIE7-GK | GA[PIE7]AAGK | 390 | 16,000 |
| PIE7-GKK | GA[PIE7]AAGKK | 380 | 19,000 |
| PIE12 | HP[PIE7]ELGK | 37 | 580 |
| PIE13c | HP[PIE7]KL | 41 | 1,500 |
| PIE14 | HP[PIE7]RLGK | 33 | 1,100 |
| PIE15 | HA[PIE7]ELGK | 67 | 1,400 |
| N9N(PIE7)2 | (KGA[PIE7]AA)2 | 1.9b | 2,300b |
| N5C(PIE7)2 | GA[PIE7]AAGKKGA[PIE7]AA | 0.6 | 300 |
| C5C(PIE7)2 | (GA[PIE7]AAGK)2 | 0.5 | 200 |
| C5C(PIE12)2 | (HP[PIE7]ELGK)2 | 0.4 | 14 |
| N9N(PIE7)3 | (KGA[PIE7]AA)3d | 0.3b | 220b |
| C5C(PIE7)3 | (GA[PIE7]AAGK)3d | 0.1 | 6.7 |
| C5C(PIE12)3 | (HP[PIE7]ELGK)3d | 0.5 | 2.8 |
| C37 | 1.4b | 13b | |
| Enfuvirtide | 3.7b | 5.0b | |
The IC50 standard error of the mean is <25% for duplicate assays for all values.
Values are from reference 57.
PIE13 does not include a C-terminal GK extension because its C-terminal flanking sequence contains a Lys residue.
The central peptide of each trimer has two tandem Lys residues (not shown).
PIE7, CDYPEWQWLC, or PIE7 core motif.
Crystals were mounted in a nylon loop and either directly cryocooled by plunging them into liquid nitrogen or cryocooled following brief (20 s) immersion in 20 μl crystallization buffer with 30% (IQN17-PIE12) or 15% (IQN17-PIE71) added glycerol. Crystals were maintained at 100 K during data collection. Data were collected either in the laboratory using a rotating copper anode X-ray generator or at a synchrotron beam line. Data were processed using the DENZO and SCALEPACK programs (40). All structures were determined by molecular replacement using the PHASER program (33) with IQN17-PIE7 as the search model. The models were rebuilt using the O program (25) and refined against a maximum-likelihood target function using the REFMAC program (36). Structures were checked using the MolProbity program (6) (see Table 2 for data and refinement statistics).
TABLE 2.
PIE12 and PIE71 crystallographic data and refinement statistics
| Data | Result for PIE12 crystal: |
Result for PIE71 crystal | ||
|---|---|---|---|---|
| Form I | Form II | Form III | ||
| Space group | P21 | R3 | P321 | P21 |
| Resolution (Å) | 30.0-1.55 (1.61-1.55)a | 30.0-1.45 (1.50-1.45) | 30.0-1.45 (1.50-1.45) | 30.0-1.40 (1.45-1.40) |
| No. of reflections measured | 113,335 | 98,687 | 186,351 | 468,599 |
| No. of unique reflections | 25,088 | 10,475 | 14,802 | 82,774 |
| Redundancy | 4.5 | 9.4 | 12.6 | 5.7 |
| Completeness (%) | 86.5 (66.8) | 97.1 (80.1) | 99.7 (96.6) | 98.2 (97.6) |
| <I/σI>b | 18 (2.4) | 19 (3.1) | 17 (2.7) | 15 (2.0) |
| Mosaicity (degree) | 0.44 | 0.37 | 0.45 | 0.29 |
| Rsymc | 0.051 (0.250) | 0.058 (0.102) | 0.107 (0.235) | 0.052 (0.316) |
| Refinement | ||||
| Resolution (Å) | 30.0-1.55 (1.59-1.55) | 30.0-1.45 (1.49-1.45) | 30.0-1.45 (1.49-1.45) | 30.0-1.40 (1.44-1.40) |
| No. of reflections used for refinement | 23,765 | 9,448 | 13,629 | 80,532 |
| No. of reflections in Rfreed set | 1,273 | 1,026 | 1,136 | 1,654 |
| Rcryste | 0.232 (0.465) | 0.234 (0.301) | 0.243 (0.299) | 0.261 (0.306) |
| Rfree | 0.288 (0.624) | 0.264 (0.392) | 0.278 (0.350) | 0.288 (0.335) |
| RMSD bonds (Å)/angles (degrees) | 0.012/1.440 | 0.013/1.693 | 0.010/1.530 | 0.009/1.094 |
| <B>g | ||||
| All atoms (Å2)/no. of atoms | 23.7/1,172 | 31.9/384 | 29.2/384 | Molf 1, 24.3/1,555; mol 2, 36.0/1,491 |
| PIE12 molecules only (Å2)/no. of atoms | 21.3/420 | 30.8/144 | 25.9/144 | Mol 1, 18.3/368; mol 2, 39.9/322 |
| Water molecules (Å2)/no. of water atoms | 32.0/197 | 38.0/36 | 40.6/49 | 39.9/389 |
| φ/Ψh most favored (%) | 100 | 98.1 | 100 | 99.0 |
Values in parentheses refer to data in the high-resolution shell.
<I/σI>, average intensity of a group of reflections divided by the average standard deviation (sigma) of the same group of reflections.
Rsym = Σ|I − <I>|/ΣI, where I is the intensity of an individual measurement and <I> is the corresponding mean value.
Rfree is the same as Rcryst calculated with a randomly selected test set of reflections that were never used in refinement calculations.
Rcryst = Σ||Fo| − |Fc||/Σ|Fo|, where |Fo| is the observed and |Fc| is the calculated structure factor amplitude.
Mol, molecule.
<B>, temperature factor.
φ/ψ, dihedral angles.
Explanation of Lys placement.
We were concerned that direct C-terminal addition of Lys would not be well tolerated because the d-peptide C-terminal region forms an α helix critically involved in the pocket-binding interface, with the C terminus itself being amidated for helix stability. Therefore, we inserted a Gly between the original C terminus of PIE7 and the C-terminal Lys, both to cap the helix and to separate the Lys from the binding interface. Unexpectedly, PIE7-GK-monomer is slightly more potent than PIE7 (see Table 1). A version of PIE7 containing an N- and C-terminal Lys (K-PIE7-GK) has the same potency as PIE7-GK (data not shown), indicating a beneficial effect imposed by the C-terminal Gly-Lys, as opposed to a deleterious effect created by a single Lys at the N terminus. This benefit is likely the reason that the linkage consisting of an ∼22-Å cross-linker at the C terminus whose spacer arm consists of 5 PEGs (C5C) results in a potency slightly superior to that of the N5C linkage (see Table 1).
Viral infectivity assays.
Pseudovirion infectivity assays were performed as described previously (57). Purified lyophilized inhibitors were dissolved in water (monomers) or 50 mM HEPES, pH 7.5 (dimers and trimers), to make high-concentration stocks. For HEPES-containing samples, all media were adjusted so that the HEPES content matched that in the sample with the highest HEPES concentration (typically, ∼1 mM). HEPES at higher concentrations (e.g., 3 mM) enhanced infectivity up to ∼15% but had minimal effect at ≤0.5 mM. The Monogram Biosciences PhenoSense Entry and peripheral blood mononuclear cell (PBMC) assays were performed as described previously (43, 52).
CD studies.
Samples were prepared with 2 μM IZN17, a 1.1× molar ratio of inhibitor to target binding sites, phosphate-buffered saline (PBS; 50 mM sodium phosphate, 150 mM NaCl, pH 7.4), and 2 M GuHCl in a total volume of 2.5 ml. Thermal melts were performed by melting the sample in a square 1-cm cuvette from 25°C to 90°C (or 93°C for PIE12-trimer) in 2°C increments with 2 min of equilibration. To show reversibility, reverse melts were performed on each sample from 90°C to 30°C in 10°C increments with 5 min of equilibration. Data were averaged from a 30-s collection on an Aviv model 410 circular dichroism (CD) spectrapolarimeter.
For each sample, the CD data followed a smooth sigmoid transition as the sample was heated or cooled. The data were smoothed in the Kaleidagraph program (Synergy Software) using 2 points from both sides. The derivative value of the smoothed data was used to determine the point with the steepest rate of change on the melt curve, which is the melting temperature (Tm).
Passaging studies.
Laboratory-adapted HIV-1 strain NL4-3 was generated by transient transfection of proviral DNA (pNL4-3) into 293T cells using Lipofectamine (Invitrogen). Cell-free supernatants containing virus were collected 48 h posttransfection and used to infect 5 × 105 CEM-1 cells in RPMI 1640 medium (0.5 ml). Virus was serially propagated once a week by 1:5 dilution of cell-free viral supernatants into fresh CEM-1 cells (5 × 105 cells, 0.5 ml) in the absence or presence of inhibitor (PIE7-dimer, PIE12-dimer, or PIE12-trimer). Viral titers were monitored biweekly by p24 antigen enzyme-linked immunosorbent assay (PerkinElmer). The inhibitor concentration started at approximately the 50% inhibitory concentrations (IC50s; 20 nM for PIE7-dimer; 1 nM for PIE12-dimer, and PIE12-trimer) and was raised 1.5- to 2-fold when p24 antigen levels in inhibitor-containing cultures approached that in inhibitor-free cultures (usually 2 to 3 weeks for PIE7-dimer). PIE12-dimer and PIE12-trimer required a slower escalation strategy with prolonged incubation at a fixed inhibitor concentration for 5 to 15 weeks before escalation.
To identify PIE7-dimer escape mutations, viral RNA was isolated from cell-free supernatants of at least two cultures independently propagated in either the presence (resistant virus) or absence (control virus) of inhibitor (Qiagen RNA purification kit). Env cDNA was generated by reverse transcription (Eppendorf cMaster RTplus system and cMaster reverse transcription kit), amplified by PCR, and sequenced in five stretches (Thomas Jefferson University Nucleic Acid Facility). To confirm selected mutations in the gp41 N-peptide region, the cDNA segment encoding the gp41 ectodomain was reamplified by PCR and subcloned into the pAED4 vector, and the plasmid DNA from three or more individual clones was sequenced. The substitutions E560K and V570I were observed in all sequences from PIE7-dimer-resistant virus but were not observed in any sequence from control virus. An expression plasmid for HXB2 Env (pEBB_HXB2 Env) incorporating these substitutions was generated using site-directed mutagenesis (QuikChange; Stratagene) and was utilized in the pseudoviral infectivity assay described above.
Protein Data Bank accession numbers.
The Protein Data Bank (PDB) accession numbers for the PIE12-IQN17 complex are 3L35, 3L36, and 3L37 for crystal forms I, II, and III, respectively, and 3MGN for the PIE71-IQN17 complex.
RESULTS
Structure-guided phage display to optimize flanking residues.
PIE inhibitors consist of a short core sequence surrounded by a disulfide bond that imparts structural rigidity required for binding (Table 1) (12). The large jump in affinity between our 1st-generation (12) and 2nd-generation (57) inhibitors was accomplished by optimizing this core sequence. There were also four fixed flanking residues outside the disulfide that arose from phage library cloning restrictions, Gly-Ala on the N terminus and Ala-Ala on the C terminus. Interestingly, our cocrystal structures of d-peptides in complex with a mimic of its gp41 pocket target (IQN17) reveal significant contacts between these presumed inert flanking residues and the pocket (12, 57). Thus, we reasoned that their optimization would likely lead to improved d-peptide affinity for the pocket.
To optimize these flanking residues, we used a commercially available phage library cloning system (NEB) that allowed us to relocate cloning sites away from the flanking regions (38). We redesigned the regions immediately outside the flanking residues in our cloning vector in order to structurally isolate them and minimize any bias caused by flanking sequence randomization. Using this vector, we constructed a phage library that varied only these four residues in the context of our previously optimized PIE7 core sequence (XXCDYPEWQWLCXX). After four rounds of panning, our phage library showed ∼100-fold improved binding to a gp41 pocket mimic (d-IZN17) compared to that of clonal PIE7 phage with the original GA/AA flanking sequence. We extensively sequenced this phage pool to identify a consensus sequence, H(A/P)-[PIE7 core]-(R/K/E)L, as well as five dominant individual sequences. Using a phage clone binding assay, we found that these sequences bound the gp41 pocket 70- to 900-fold more tightly than PIE7, with PIE12 (HP-[PIE7 core]-EL) having the highest affinity (data not shown).
Enhanced potency of 3rd-generation d-peptides.
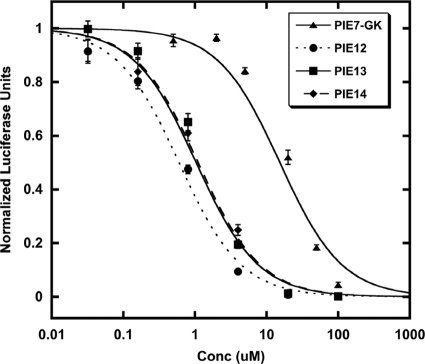
We synthesized d-peptides corresponding to the top three phage sequences in the binding assay (PIE12, PIE13, and PIE14) and tested their antiviral potencies in a pseudovirion entry assay (Table 1 and Fig. 1). Pairwise comparisons of both phage binding and inhibitor potency indicate that Pro is preferred over Ala at position 2 and Glu is preferred over Arg or Lys at position 13. As predicted from the phage binding assay, PIE12 has the best potency and is ∼40-fold more potent than PIE7 (our best previously reported monomer) against strain JRFL.
FIG. 1.
Optimization of flanking residues enhances PIE potency. Each point represents the average of quadruplicate measurements from a representative pseudovirion entry inhibition assay (JRFL strain) normalized to the measurement for an uninhibited control. Error bars represent the standard errors of the means. PIE12 is ∼2-fold more potent than PIE13 or PIE14 and is ∼25-fold more potent than PIE7-GK.
Crystal structure of PIE12.
To better understand the sources of PIE12's improved binding and potency, we crystallized PIE12 in complex with the N-trimer pocket mimic IQN17. Data were collected from three crystal forms (Table 2) at between 1.45- and 1.55-Å resolution. Each IQN17 trimer from the three crystal forms reported here and from the PIE7 structure (PDB accession number 2R5D) agreed well with one another (root mean square deviation [RMSD], 0.6 to 1.2 Å) on the basis of the least-squares overlap on all Cα atoms (residues 1 to 45 of all three chains). The structures suggest two sources of the improved affinity of PIE12 for IQN17 compared to that of PIE7. First, the new N-terminal flank residues (His1 and Pro2) form favorable ring stacking interactions with the pocket (IQN17-Trp571) (Fig. 2). Second, the substitution of Leu for Ala in the C-terminal flank sequence buries an additional ∼50-Å2 hydrophobic surface area in the pocket. Neither of these new interactions with the flanking sequence perturbs the original pocket-binding structure of the core PIE7 residues. Importantly, the structures reveal that PIE12's improved affinity does not result from new interactions with less conserved regions outside the pocket that might render PIE12 more vulnerable to resistance mutations.
FIG. 2.
Crystal structure of PIE12 binding to IQN17. Trp571 of the gp41 pocket (gray) and the N-terminal flank residues (dH1 and dP2) of PIE12 (green) appear to stabilize binding via ring-stacking interactions. The disulfide bond (yellow) is shown in the background.
Discovery and structure of a 7-mer d-peptide.
The core sequence of PIE7 and PIE12 comprises 8 residues flanked by cysteines (8-mer). Modeling based on our 8-mer d-peptide/IQN17 crystal structures suggests that a 7-mer core is compatible with pocket binding of the WXWL consensus and formation of a disulfide bond (57). Previously, we saw that decreasing the size of the PIE core (from 10 to 8 residues) led to dramatically increased pocket binding (57), so we reasoned that further decreasing the size of the core might lead to additional potency gains. To explore this alternative geometry, we used a mirror-image discovery process similar to that employed with 8-mers to identify a 7-mer, PIE71 (FVCPPEWRWLCDL). PIE71 contains the same WXWL motif found in 8-mer and 10-mer pocket binders and inhibits strain HXB2 entry with an IC50 of 410 nM (data not shown), which is ∼1.5 fold better than that of PIE7 but an order of magnitude worse than that of PIE12.
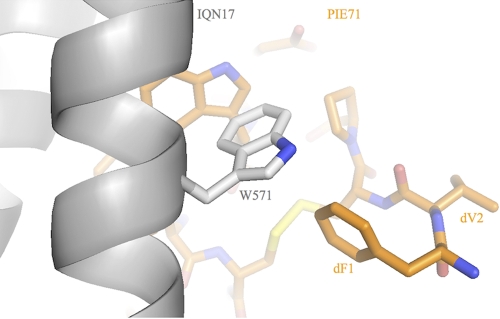
To gain a better understanding of the 7-mer binding solution, we determined a cocrystal structure of PIE71 in complex with IQN17 (Table 2). The key residues involved in the binding interface (WXWL) adopt nearly superposable conformations to those observed in PIE7 and PIE12, as do the C-terminal flank residues. However, the two structures deviate significantly at the N terminus (Fig. 2 and 3). Specifically, the 7-mer's disulfide bond is shifted much closer to the pocket, which directs the N-terminal flank residues away from the pocket region. As a result, the N-terminal flanking residues (Phe-Val) only graze the pocket, whereas PIE12's N-terminal flanking residues have an intimate interaction. So although the 7-mer is compatible with pocket binding, the smaller core is too constrained to allow optimal binding of the flank residues to the pocket. Due to this decreased binding interface and therefore decreased potency, we decided not to pursue the 7-mer geometry further.
FIG. 3.
Crystal structure of PIE71 binding to IQN17. The N-terminal flank residues (dF1 and dV2) of PIE71 (orange) are directed away from the pocket compared to the structure in PIE12 (Fig. 2). The disulfide bond (yellow) is shown in the background.
Optimization of crosslinker length and geometry.
We previously took advantage of the trimeric nature of the gp41 pocket target to geometrically increase the PIE7 binding affinity by cross-linking it into dimers and trimers (57). PIE7 has an N-terminal lysine, which furnishes a unique primary amino group (the N terminus is acetylated) and which was added for solubility. This lysine was used to produce dimers via reaction with a bis-PEG NHS ester crosslinker (NHS esters selectively react with primary amino groups). Trimers were produced by cross-linking two PIE7s to a central peptide with two lysines at the N terminus (2K-PIE7).
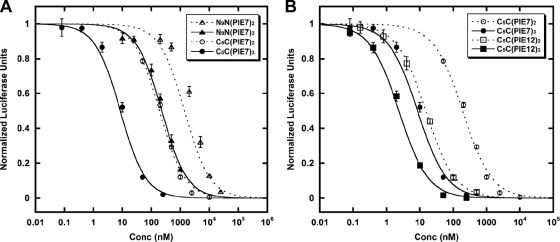
We hypothesized that the strength of the avidity effect is related to the length of the crosslinker and that shorter crosslinkers that still allow simultaneous binding to multiple pockets could strengthen potency. For the original N- to N-terminal linkage, we used a crosslinker with an ∼35-Å spacer arm consisting of 9 PEG units (N9N linkage). However, our crystal structures of d-peptides in complex with IQN17 reveal that C- to C-terminal or N- to C-terminal linkages could be significantly shorter and could be spanned by an ∼22-Å crosslinker whose spacer arm consists of 5 PEGs (C5C and N5C linkages). Therefore, we relocated Lys to the C terminus of PIE7 (PIE7-GK) in order to make the N5C heterodimers and C5C homodimers (see Materials and Methods for additional details).
The resulting N5C- and C5C-PIE7-dimers have similar potencies that are significantly enhanced compared to the potency of our previous N9N-PIE7-dimer (Table 1 and Fig. 4 A). On the basis of these data, we chose C5C connections as our standard linker, since they are simpler to produce than the hetero-N5C linkage. Here, all dimers and trimers use the C5C linkage unless otherwise specified. Combining our new optimized flanking residues and linkages, we produced PIE12-dimer and PIE12-trimer. Both are extremely potent against the difficult-to-inhibit primary strain JRFL (low-nanomolar IC50s; Fig. 4B; Table 1), being up to 2 orders of magnitude more potent than our best previously described d-peptide (N9N PIE7-trimer) (57).
FIG. 4.
Optimization of linkage geometry. Each point represents the average of quadruplicate measurements from a representative pseudovirion entry inhibition assay (JRFL strain) normalized to the measurement for the uninhibited control. Error bars represent the standard errors of the means. (A) Comparison of N9N to C5C linkages; (B) PIE7 versus PIE12-dimers and trimers (all C5C linkages).
Breadth against a diverse multiclade panel.
HIV-1 has jumped from chimpanzees to humans at least three separate times, giving rise to groups M, N, and O (19). The main group (group M) accounts for >99% of all HIV-1 infections worldwide (32). HIV's high mutation rate has led to the emergence of diverse subtypes within group M that are categorized as clades A to D, F to H, J, and K and various circulating recombinant forms (CRFs; e.g., AE and BF). In 2000, clades A to D were estimated to represent >90% of HIV infections (39); however, in recent years CRFs have become more prevalent (1). Different subtypes contain up to 35% sequence diversity in Env, often causing antibodies raised against a particular strain to be ineffective against others (20).
To ensure that our pocket-specific d-peptides are potent and broadly neutralizing against the most common subtypes of HIV, we measured the potency of PIE7-trimer, PIE12-trimer, and PIE12 (with enfuvirtide as a control inhibitor) using the PhenoSense Entry pseudovirion assay (Monogram Biosciences) (Table 3) (43). The inhibitors were tested against a panel of 23 viruses pseudotyped with clonal and polyclonal envelopes representing clades A to D, several CRFs, and enfuvirtide-resistant strains. Both PIE7 and PIE12-trimers potently inhibited all strains tested, though PIE12-trimer was generally a superior inhibitor (and in all cases more potent than enfuvirtide). While PIE12-monomer is much less potent than PIE12-trimer, it is also broadly active. Interestingly, PIE12-trimer is ∼10-fold more potent than PIE7-trimer against polyclonal virus from clades B and C (samples amplified from patient plasma), which is consistent with a resistance capacitor mechanism for maintaining potency in the presence of various Env sequences. All of the d-peptide inhibitors are unaffected by enfuvirtide resistance mutations. Additionally, lack of inhibition against a murine leukemia virus (MLV) control indicates that these inhibitors are specific and nontoxic in this assay.
TABLE 3.
PhenoSense Entry assay data
| HIV-1 isolate | Subtype | IC50 (nM) |
|||
|---|---|---|---|---|---|
| PIE7-trimer | PIE12-trimer | PIE12-monomer | Enfuvirtide | ||
| Aa | A | 5.5 | 4.1 | 2,300 | 18 |
| 92RW008 | A | 2.0 | 1.0 | 1,400 | 7.2 |
| 92UG031 | A | 18 | 4.2 | 2,600 | 20 |
| 94KE105 | AC | 16 | 0.7 | 1,900 | 13 |
| CMU02 | AE | 32 | 12 | 1,500 | 16 |
| Ba | B | 140 | 13 | 3,300 | 30 |
| 1168 | B | 54 | 31 | 4,700 | 140.0 |
| BaL | B | 2.0 | 2.5 | 1,700 | 10 |
| ENFr1a | B | 2.0 | 0.8 | 790 | 760 |
| ENFr2a | B | 0.7 | 1.0 | 300 | 5,400 |
| HXB2 | B | 0.1 | 0.3 | 50 | 2.6 |
| JRCSF | B | 13 | 3.4 | 1,100 | 14 |
| JRFL | B | 21 | 5.7 | 1,900 | 7.9 |
| NL4.3 | B | 0.3 | 0.4 | 150 | 62 |
| SF162 | B | 3.4 | 4.5 | 940 | 34 |
| 98CN009 | BC | 0.4 | 0.4 | 320 | 7.9 |
| 93BR029 | BF | 1.5 | 0.9 | 750 | 12 |
| Ca | C | 220.0 | 26 | 5,100 | 71 |
| 97ZA012 | C | 2.0 | 0.7 | 1,500 | 10 |
| 98IN022 | C | 1.1 | 1.1 | 820 | 6.9 |
| 21068 | C | 6.6 | 5.0 | 1,800 | 47 |
| Da | D | 3.1 | 3.2 | 820 | 17 |
| 92UG005 | D | 3.9 | 2.5 | 2,000 | 10 |
| aMLV | >10,000 | >10,000 | >500,000 | >15,000 | |
Polyclonal viral envelopes amplified from patient plasma.
Breadth against replication-competent primary viral isolates on PBMCs.
To more closely mimic in vivo infection and further establish inhibitory breadth, we also tested the ability of PIE7-trimer, PIE12-trimer, and PIE12 to inhibit PBMC infection by replicating primary strains, again with enfuvirtide as a control (Table 4). These data confirm the potent and broad inhibitory activities of PIE7 and PIE12-trimer against all group M strains tested, including several CRFs. Toxicity was not observed on these cells at inhibitor concentrations up to 1 μM (the highest concentration tested), demonstrating a high therapeutic index for the trimers. Interestingly, the inhibitors are more potent in this assay than in the PhenoSense Entry assay, which may be due to differential receptor expression levels between the two cell types (45).
TABLE 4.
PBMC assay data
| HIV-1 isolate | Subtype | IC50 (nM) |
|||
|---|---|---|---|---|---|
| PIE7-trimer | PIE12-trimer | PIE12-monomer | Enfuvirtide | ||
| 92UG029 | A | 1.6 | 0.7 | 290 | 190 |
| 92UG037 | A | 0.1 | 0.2 | 36 | 41 |
| 93TH073 | AE | 0.6 | 0.8 | 270 | 200 |
| CMU02 | AE | 0.2 | 0.4 | 300 | 44 |
| CMU06 | AE | 0.3 | 0.4 | 210 | 5.7 |
| IIIB | B | 0.3 | 0.8 | 140 | 28 |
| BaL | B | 0.2 | 0.3 | 72 | 20 |
| JRCSF | B | 0.1 | 0.1 | 120 | 7.0 |
| JRFL | B | 0.5 | 0.3 | 110 | 1.7 |
| 93BR019 | BF | 1.7 | 4.7 | 170 | >1,000 |
| 92BR025 | C | 15 | 5.2 | >1,000 | 310 |
| 93IN101 | C | 0.4 | 0.4 | 160 | 22 |
| 92UG001 | D | 0.8 | 4.5 | 230 | 180 |
| 92UG046 | D | 0.1 | 1.2 | 170 | 130 |
| 93BR020 | F | 0.2 | 0.4 | 190 | 59 |
| 93BR029 | F | 0.2 | 0.8 | 86 | 19 |
| G3 | G | 0.3 | 1.2 | 310 | 23 |
| RU570 | G | 0.3 | 0.4 | 480 | 37 |
| BCF01 | Group O | >1,000 | >1,000 | >1,000 | 330 |
| BCF02 | Group O | >1,000 | 440 | >1,000 | 0.4 |
Notably, two group O strains were also tested in this assay and are much less sensitive to inhibition than group M strains. Group O contains several mutations (compared to the sequence of group M) in the pocket, including Q567R, T569S, K574R, Q577R, and V580L. The crystal structures of PIE7 and PIE12 in complex with IQN17 reveal that, of these residues, the d-peptide directly interacts only with K574 (via a hydrophobic interaction) and Q577 (via hydrogen bonds). Group O gp41 has several other mutations in the groove just outside the pocket (i.e., H564E) that could also affect PIE potency (e.g., by slowing the association rate). It will be interesting to analyze the effects of these mutations in a group M (e.g., strain HXB2 or JRFL) background to see if they are responsible for the loss of potency.
Evidence for a charged resistance capacitor.
With the design of PIE12-trimer, we now observe strong evidence for a highly charged resistance capacitor in which the PIE12-trimer pocket-binding affinity vastly exceeds the inhibitory potency. Comparing PIE7 and PIE12-trimers, we observe similar potencies against pseudovirion entry (Fig. 4B; Table 1), although we expect their target affinities to be extremely different.
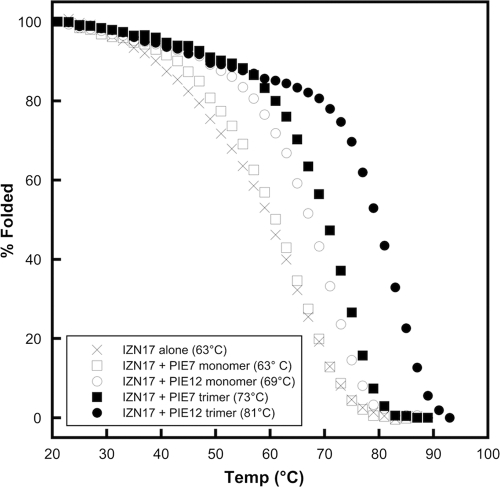
Due to extraordinarily slow off rates, direct measurements of the pocket affinities for PIE7 and PIE12-trimers via surface plasmon resonance, used for earlier d-peptides (57), were not possible. Since the binding affinity of inhibitors correlates with the stability of inhibitor-target complexes, we used thermal denaturation monitored by CD to measure the relative stabilities of each IZN17-inhibitor complex and infer the relative affinities of our ultra-high-affinity binders. The melts were performed in 2 M GuHCl to destabilize the complexes and shift their melting points into an observable range (below 100°C).
The normalized thermal melts for each IZN17-inhibitor complex are plotted in Fig. 5, with Tm values being shown in the key. As expected, PIE12-trimer forms the most stable complex and has a Tm 8°C higher than that of the next most stable inhibitor complex (PIE7-trimer). PIE12 also forms a more stable complex than PIE7, as expected. Our previous experience showed that improvements in monomer affinity translated to approximately squared and cubed improvements in the corresponding dimers and trimers (57). On the basis of PIE12-trimer's optimized C5C linkage (35-fold improved antiviral potency over that of the trimer with an N9N linkage; strain JRFL data) and the ∼25-fold difference in monomer potency between PIE7 and PIE12 (JRFL data), we estimate that PIE12-trimer binds to gp41 >105-fold (35 × 253) more tightly than N9N PIE7-trimer. This predicted binding at subfemtomolar concentrations translates to a resistance capacitor charged to ∼6 kcal/mol against strain JRFL. Interestingly, the potency plateau lies at a slightly better potency for trimers than for dimers, likely due to their faster association rates (i.e., three versus two opportunities for initial collision with the target).
FIG. 5.
Stability of d-peptide complexes. Normalized melting curves of IZN17 alone and with d-peptide inhibitors were monitored by CD in PBS-2 M GuHCl. Tm values are indicated in the key.
Selection of resistant strains.
To measure the resistance profile of our d-peptide inhibitors and test our resistance capacitor hypothesis, we conducted viral passaging studies with escalating inhibitor concentrations to select for resistant strains. These studies initially used PIE7-dimer, which was available from our previous study (57) and inhibits the parental strain, NL4-3, with an IC50 of ∼20 nM. By doubling the PIE7-dimer concentration every 2 to 3 weeks, we obtained stable viral cultures in 2,000 nM inhibitor within 20 weeks of propagation. In comparison, we were able to obtain high-level enfuvirtide resistance (>1,000-fold) in only ∼3 weeks using a similar protocol (H. K. Steger et al., submitted for publication).
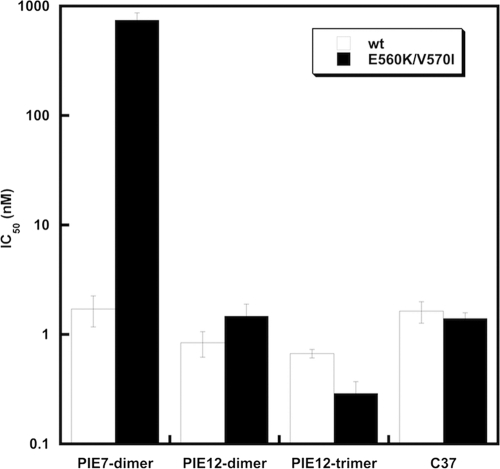
Sequencing the N-peptide region of PIE7-dimer-resistant viruses revealed two selected mutations: E560K and V570I. These substitutions in the context of HXB2 pseudovirions conferred ∼400-fold resistance to PIE7-dimer. These mutations also dramatically weaken the binding of d-peptides to the gp41 pocket but not the C-peptide inhibitor C37 (M. J. Root et al., unpublished data). It is not obvious from the PIE7 structure how these mutations weaken PIE7 binding. Despite this loss of affinity, the escape mutations had a minimal effect on the potencies of PIE12-dimer and PIE12-trimer, as well as the C37 control inhibitor (Fig. 6). This result is predicted by the resistance capacitor hypothesis: affinity-disrupting escape mutations selected in the presence of weaker-binding inhibitors should be less disruptive to the potencies of tighter-binding inhibitors.
FIG. 6.
Effect of PIE7-dimer resistance mutations on PIE7-dimer, PIE12-dimer, and PIE12-trimer potency. IC50s against wild-type (wt) and PIE7-dimer-resistant (E560K/V570I) strain HXB2 pseudovirion entry are shown. The C-peptide inhibitor C37 is included as a control. Data represent the means from at least two independent experiments. Error bars represent the standard errors of the means.
The rapid inhibitor escalation strategy utilized to generate PIE7-dimer resistance was not effective in generating HIV-1 resistant to PIE12-dimer or PIE12-trimer. Rather, the HIV-1 titer fell precipitously when inhibitor concentrations exceeded 20 nM (5 to 20 times the IC50). Instead, we switched to a much slower escalation strategy with prolonged periods at stable inhibitor concentrations (5 to 15 weeks). Resistant virus emerged after 40 weeks of propagation in PIE12-dimer and after 65 weeks of propagation in PIE12-trimer. These observations suggest that a strong resistance capacitor profoundly delays selection of resistance mutations for these optimized fusion inhibitors.
Sequencing of the pocket region of PIE12-trimer-resistant viruses reveals only one mutation, Q577R. Interestingly, this substitution is present in nearly all group O isolates (including BCF01 and BCF02; Table 4) but is rare among group M isolates. Pseudovirions bearing Q577R confirm that this mutation confers substantial resistance to PIE12-trimer (data not shown). Examination of the PIE12 crystal structure shows that Q577 makes hydrogen bonds with Glu7 and Trp10 in PIE12, which may explain the disruptive effects of this mutation. Q577R's codon is predicted to disrupt the RRE stem-loop V structure, since it base pairs with the invariant W571 codon (Trp is encoded by only one codon).
DISCUSSION
PIE12-trimer is a d-peptide entry inhibitor with ∼80-fold enhanced potency and an estimated >100,000-fold improved binding affinity compared to those of the best previously reported d-peptide. This dramatic improvement in affinity produces excellent breadth and a charged resistance capacitor to combat the emergence of resistance mutations. Indeed, PIE12-trimer was able to withstand the impact of resistance mutations to earlier d-peptides and required a much longer selection (65 weeks) to generate resistant strains. Ongoing work is exploring the mechanism of PIE7-dimer, PIE12-dimer, and PIE12-trimer resistance and its relationship to group O's insensitivity. A key question is whether HIV can develop resistance to these inhibitors independent of changes in affinity (e.g., kinetics) that are capable of maintaining viral fitness.
Viral escape affects even the newest class of FDA-approved HIV-1 drugs, integrase inhibitors. Resistance to raltegravir and corresponding treatment failure were observed in a significant subset of patients in both the phase II and III clinical studies (5), and corresponding resistance mutations can be seen within 4 weeks when resistant virus is selected in viral passaging studies (28). Our studies indicate that PIE12-trimer is a promising entry inhibitor that could overcome the limitations associated with the two currently approved entry inhibitors, enfuvirtide (high dosing, susceptibility to resistance) and maraviroc (Selzentry; effective only against R5 viruses) and may also prove to have a better resistance profile than even the newest class of HIV-1 inhibitors.
In addition to being a possible therapeutic agent, PIE12-trimer is an ideal candidate for a topical microbicide, as its protease resistance would allow it to withstand the protease-rich environment of the vaginal mucosa. In the absence of a safe and effective HIV vaccine, a topical microbicide to prevent the sexual transmission of HIV is an urgent unmet global health need. The ultimate utility of PIE12-trimer as a microbicide or therapeutic agent will be determined by advanced preclinical and clinical studies, including characterization of pharmacokinetics, in vivo toxicity, effectiveness in animal models of HIV infection (alone or in combination with other HIV inhibitors), and optimization of formulations for microbicide gels or vaginal rings.
More generally, the present work unequivocally shows that d-peptide inhibitors can be designed with high potency and specificity against natural l-protein targets. The d-peptide design methodology described here can be applied to diverse biomedical applications, particularly for the many viruses that share HIV's hairpin-closing entry mechanism (e.g., influenza virus, Ebola virus, respiratory syncytial virus, severe acute respiratory syndrome coronavirus, Dengue virus, and West Nile virus). Our resistance capacitor design strategy may also be generally applicable for treating other rapidly evolving diseases, especially when combined with recent advances in anticipating likely structural sources of drug resistance (37). Finally, the development of PIE12-trimer as a strong clinical candidate will allow d-peptide therapeutics to be evaluated in vivo to determine if their theoretical advantages warrant a prominent role as a new class of therapeutic agents.
Acknowledgments
We thank Bob Schackmann and Scott Endicott (University of Utah Peptide Synthesis Core Facility) for peptide synthesis, Yu Shi for early 7-mer phage display, and Dong Han and Pham Phung (Monogram) for technical assistance with the PhenoSense Entry assay.
PBMC assays were performed by Southern Research Institute (principal investigator, Roger Ptak), funded by contract HHSN272200700041C (from the National Institute of Allergy and Infectious Diseases, National Institutes of Health [NIH], U.S. Department of Health and Human Services). This work was supported by grants from the NIH to M.S.K. (AI076168), M.J.R. (GM066682), and C.P.H. (GM082545), as well as a University of Utah Technology Commercialization Grant to M.S.K. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL), a national user facility operated by Stanford University on behalf of the Office of Basic Energy Sciences, U.S. Department of Energy. The SSRL Structural Molecular Biology Program is supported by the Office of Biological and Environmental Research, U.S. Department of Energy, and by the National Center for Research Resources, Biomedical Technology Program, NIH, and the National Institute of General Medical Sciences.
B.D.W., D.M.E., and M.S.K. are cofounders of Kayak Biosciences. This startup company is focused on advancing d-peptide inhibitors to the clinic.
Footnotes
Published ahead of print on 18 August 2010.
REFERENCES
- 1.Buonaguro, L., M. L. Tornesello, and F. M. Buonaguro. 2007. Human immunodeficiency virus type 1 subtype distribution in the worldwide epidemic: pathogenetic and therapeutic implications. J. Virol. 81:10209-10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. U. S. A. 95:15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 4.Chinnadurai, R., D. Rajan, J. Munch, and F. Kirchhoff. 2007. Human immunodeficiency virus type 1 variants resistant to first- and second-version fusion inhibitors and cytopathic in ex vivo human lymphoid tissue. J. Virol. 81:6563-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper, D. A., R. T. Steigbigel, J. M. Gatell, J. K. Rockstroh, C. Katlama, P. Yeni, A. Lazzarin, B. Clotet, P. N. Kumar, J. E. Eron, M. Schechter, M. Markowitz, M. R. Loutfy, J. L. Lennox, J. Zhao, J. Chen, D. M. Ryan, R. R. Rhodes, J. A. Killar, L. R. Gilde, K. M. Strohmaier, A. R. Meibohm, M. D. Miller, D. J. Hazuda, M. L. Nessly, M. J. DiNubile, R. D. Isaacs, H. Teppler, and B. Y. Nguyen. 2008. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N. Engl. J. Med. 359:355-365. [DOI] [PubMed] [Google Scholar]
- 6.Davis, I. W., A. Leaver-Fay, V. B. Chen, J. N. Block, G. J. Kapral, X. Wang, L. W. Murray, W. B. Arendall III, J. Snoeyink, J. S. Richardson, and D. C. Richardson. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35:W375-W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debnath, A. K., L. Radigan, and S. Jiang. 1999. Structure-based identification of small molecule antiviral compounds targeted to the gp41 core structure of the human immunodeficiency virus type 1. J. Med. Chem. 42:3203-3209. [DOI] [PubMed] [Google Scholar]
- 8.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, Z. Zhang, W. A. O'Brien, L. Ratner, G. M. Shaw, and E. Hunter. 2001. Sensitivity of human immunodeficiency virus type 1 to fusion inhibitors targeted to the gp41 first heptad repeat involves distinct regions of gp41 and is consistently modulated by gp120 interactions with the coreceptor. J. Virol. 75:8605-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dwyer, J. J., K. L. Wilson, D. K. Davison, S. A. Freel, J. E. Seedorff, S. A. Wring, N. A. Tvermoes, T. J. Matthews, M. L. Greenberg, and M. K. Delmedico. 2007. Design of helical, oligomeric HIV-1 fusion inhibitor peptides with potent activity against enfuvirtide-resistant virus. Proc. Natl. Acad. Sci. U. S. A. 104:12772-12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckert, D. M., and P. S. Kim. 2001. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc. Natl. Acad. Sci. U. S. A. 98:11187-11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 12.Eckert, D. M., V. N. Malashkevich, L. H. Hong, P. A. Carr, and P. S. Kim. 1999. Inhibiting HIV-1 entry: discovery of d-peptide inhibitors that target the gp41 coiled-coil pocket. Cell 99:103-115. [DOI] [PubMed] [Google Scholar]
- 13.Eggink, D., C. E. Baldwin, Y. Deng, J. P. Langedijk, M. Lu, R. W. Sanders, and B. Berkhout. 2008. Selection of T1249-resistant human immunodeficiency virus type 1 variants. J. Virol. 82:6678-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst, J. T., O. Kutzki, A. K. Debnath, S. Jiang, H. Lu, and A. D. Hamilton. 2002. Design of a protein surface antagonist based on alpha-helix mimicry: inhibition of gp41 assembly and viral fusion. Angew. Chem. Int. ed. Engl. 41:278-281. [DOI] [PubMed] [Google Scholar]
- 15.Ferrer, M., T. M. Kapoor, T. Strassmaier, W. Weissenhorn, J. J. Skehel, D. Oprian, S. L. Schreiber, D. C. Wiley, and S. C. Harrison. 1999. Selection of gp41-mediated HIV-1 cell entry inhibitors from biased combinatorial libraries of non-natural binding elements. Nat. Struct. Biol. 6:953-960. [DOI] [PubMed] [Google Scholar]
- 16.Freed, E. O., and M. A. Martin. 1995. The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J. Biol. Chem. 270:23883-23886. [DOI] [PubMed] [Google Scholar]
- 17.Frey, G., S. Rits-Volloch, X. Q. Zhang, R. T. Schooley, B. Chen, and S. C. Harrison. 2006. Small molecules that bind the inner core of gp41 and inhibit HIV envelope-mediated fusion. Proc. Natl. Acad. Sci. U. S. A. 103:13938-13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276-279. [DOI] [PubMed] [Google Scholar]
- 19.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 20.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354-2360. [DOI] [PubMed] [Google Scholar]
- 21.Golding, H., M. Zaitseva, E. de Rosny, L. R. King, J. Manischewitz, I. Sidorov, M. K. Gorny, S. Zolla-Pazner, D. S. Dimitrov, and C. D. Weiss. 2002. Dissection of human immunodeficiency virus type 1 entry with neutralizing antibodies to gp41 fusion intermediates. J. Virol. 76:6780-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365:113. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, S., H. Lu, S. Liu, Q. Zhao, Y. He, and A. K. Debnath. 2004. N-substituted pyrrole derivatives as novel human immunodeficiency virus type 1 entry inhibitors that interfere with the gp41 six-helix bundle formation and block virus fusion. Antimicrob. Agents Chemother. 48:4349-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin, B. S., J. R. Ryu, K. Ahn, and Y. G. Yu. 2000. Design of a peptide inhibitor that blocks the cell fusion mediated by glycoprotein 41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 16:1797-1804. [DOI] [PubMed] [Google Scholar]
- 25.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47(Pt 2):110-119. [DOI] [PubMed] [Google Scholar]
- 26.Judice, J. K., J. Y. Tom, W. Huang, T. Wrin, J. Vennari, C. J. Petropoulos, and R. S. McDowell. 1997. Inhibition of HIV type 1 infectivity by constrained alpha-helical peptides: implications for the viral fusion mechanism. Proc. Natl. Acad. Sci. U. S. A. 94:13426-13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kahle, K. M., H. K. Steger, and M. J. Root. 2009. Asymmetric deactivation of HIV-1 gp41 following fusion inhibitor binding. PLoS Pathog. 5:e1000674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi, M., K. Nakahara, T. Seki, S. Miki, S. Kawauchi, A. Suyama, C. Wakasa-Morimoto, M. Kodama, T. Endoh, E. Oosugi, Y. Matsushita, H. Murai, T. Fujishita, T. Yoshinaga, E. Garvey, S. Foster, M. Underwood, B. Johns, A. Sato, and T. Fujiwara. 2008. Selection of diverse and clinically relevant integrase inhibitor-resistant human immunodeficiency virus type 1 mutants. Antiviral Res. 80:213-222. [DOI] [PubMed] [Google Scholar]
- 29.Lalezari, J. P., N. C. Bellos, K. Sathasivam, G. J. Richmond, C. J. Cohen, R. A. Myers, Jr., D. H. Henry, C. Raskino, T. Melby, H. Murchison, Y. Zhang, R. Spence, M. L. Greenberg, R. A. Demasi, and G. D. Miralles. 2005. T-1249 retains potent antiretroviral activity in patients who had experienced virological failure while on an enfuvirtide-containing treatment regimen. J. Infect. Dis. 191:1155-1163. [DOI] [PubMed] [Google Scholar]
- 30.Lalezari, J. P., K. Henry, M. O'Hearn, J. S. Montaner, P. J. Piliero, B. Trottier, S. Walmsley, C. Cohen, D. R. Kuritzkes, J. J. Eron, Jr., J. Chung, R. DeMasi, L. Donatacci, C. Drobnes, J. Delehanty, and M. Salgo. 2003. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N. Engl. J. Med. 348:2175-2185. [DOI] [PubMed] [Google Scholar]
- 31.Lazzarin, A., B. Clotet, D. Cooper, J. Reynes, K. Arasteh, M. Nelson, C. Katlama, H. J. Stellbrink, J. F. Delfraissy, J. Lange, L. Huson, R. DeMasi, C. Wat, J. Delehanty, C. Drobnes, and M. Salgo. 2003. Efficacy of enfuvirtide in patients infected with drug-resistant HIV-1 in Europe and Australia. N. Engl. J. Med. 348:2186-2195. [DOI] [PubMed] [Google Scholar]
- 32.Marx, P. A. 2005. Unsolved questions over the origin of HIV and AIDS. ASM News 71:15-20. [Google Scholar]
- 33.McCoy, A. J., R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni, and R. J. Read. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, M. D., R. Geleziunas, E. Bianchi, S. Lennard, R. Hrin, H. Zhang, M. Lu, Z. An, P. Ingallinella, M. Finotto, M. Mattu, A. C. Finnefrock, D. Bramhill, J. Cook, D. M. Eckert, R. Hampton, M. Patel, S. Jarantow, J. Joyce, G. Ciliberto, R. Cortese, P. Lu, W. Strohl, W. Schleif, M. McElhaugh, S. Lane, C. Lloyd, D. Lowe, J. Osbourn, T. Vaughan, E. Emini, G. Barbato, P. S. Kim, D. J. Hazuda, J. W. Shiver, and A. Pessi. 2005. A human monoclonal antibody neutralizes diverse HIV-1 isolates by binding a critical gp41 epitope. Proc. Natl. Acad. Sci. U. S. A. 102:14759-14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munoz-Barroso, I., S. Durell, K. Sakaguchi, E. Appella, and R. Blumenthal. 1998. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J. Cell Biol. 140:315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 37.Nalam, M. N., A. Ali, M. D. Altman, G. S. Reddy, S. Chellappan, V. Kairys, A. Ozen, H. Cao, M. K. Gilson, B. Tidor, T. M. Rana, and C. A. Schiffer. 2010. Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance. J. Virol. 84:5368-5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noren, K. A., and C. J. Noren. 2001. Construction of high-complexity combinatorial phage display peptide libraries. Methods 23:169-178. [DOI] [PubMed] [Google Scholar]
- 39.Osmanov, S., C. Pattou, N. Walker, B. Schwardlander, and J. Esparza. 2002. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J. Acquir. Immune Defic. Syndr. 29:184-190. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 41.Pappenheimer, J. R., C. E. Dahl, M. L. Karnovsky, and J. E. Maggio. 1994. Intestinal absorption and excretion of octapeptides composed of d amino acids. Proc. Natl. Acad. Sci. U. S. A. 91:1942-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pappenheimer, J. R., M. L. Karnovsky, and J. E. Maggio. 1997. Absorption and excretion of undegradable peptides: role of lipid solubility and net charge. J. Pharmacol. Exp. Ther. 280:292-300. [PubMed] [Google Scholar]
- 43.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray, N., J. E. Harrison, L. A. Blackburn, J. N. Martin, S. G. Deeks, and R. W. Doms. 2007. Clinical resistance to enfuvirtide does not affect susceptibility of human immunodeficiency virus type 1 to other classes of entry inhibitors. J. Virol. 81:3240-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. U. S. A. 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reeves, J. D., F. H. Lee, J. L. Miamidian, C. B. Jabara, M. M. Juntilla, and R. W. Doms. 2005. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J. Virol. 79:4991-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72:986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Root, M. J., M. S. Kay, and P. S. Kim. 2001. Protein design of an HIV-1 entry inhibitor. Science 291:884-888. [DOI] [PubMed] [Google Scholar]
- 49.Sadowski, M., J. Pankiewicz, H. Scholtzova, J. A. Ripellino, Y. Li, S. D. Schmidt, P. M. Mathews, J. D. Fryer, D. M. Holtzman, E. M. Sigurdsson, and T. Wisniewski. 2004. A synthetic peptide blocking the apolipoprotein E/beta-amyloid binding mitigates beta-amyloid toxicity and fibril formation in vitro and reduces beta-amyloid plaques in transgenic mice. Am. J. Pathol. 165:937-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schumacher, T. N., L. M. Mayr, D. L. Minor, Jr., M. A. Milhollen, M. W. Burgess, and P. S. Kim. 1996. Identification of d-peptide ligands through mirror-image phage display. Science 271:1854-1857. [DOI] [PubMed] [Google Scholar]
- 51.Sia, S. K., P. A. Carr, A. G. Cochran, V. N. Malashkevich, and P. S. Kim. 2002. Short constrained peptides that inhibit HIV-1 entry. Proc. Natl. Acad. Sci. U. S. A. 99:14664-14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Southern Research Institute. 2008, posting date. Anti-HIV evaluation assays in fresh human cells. Southern Research Institute, Birmingham, AL. http://www.southernresearch.org/contract-services/anti-hiv-evaluation-assays.html#fresh_human_cells.
- 53.Steger, H. K., and M. J. Root. 2006. Kinetic dependence to HIV-1 entry inhibition. J. Biol. Chem. 281:25813-25821. [DOI] [PubMed] [Google Scholar]
- 54.Stephens, O. M., S. Kim, B. D. Welch, M. E. Hodsdon, M. S. Kay, and A. Schepartz. 2005. Inhibiting HIV fusion with a beta-peptide foldamer. J. Am. Chem. Soc. 127:13126-13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan, K., J. Liu, J. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. U. S. A. 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 57.Welch, B. D., A. P. VanDemark, A. Heroux, C. P. Hill, and M. S. Kay. 2007. Potent d-peptide inhibitors of HIV-1 entry. Proc. Natl. Acad. Sci. U. S. A. 104:16828-16833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wild, C., J. W. Dubay, T. Greenwell, T. Baird, Jr., T. G. Oas, C. McDanal, E. Hunter, and T. Matthews. 1994. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc. Natl. Acad. Sci. U. S. A. 91:12676-12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wild, C., T. Greenwell, and T. Matthews. 1993. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res. Hum. Retroviruses 9:1051-1053. [DOI] [PubMed] [Google Scholar]
- 60.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. U. S. A. 89:10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. U. S. A. 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]