Abstract
Biomass production by plants is often negatively correlated with respiratory rate, but the value of this rate changes dramatically during diurnal cycles, and hence, biomass is the cumulative result of complex environment-dependent metabolic processes. Mitochondria in photosynthetic plant tissues undertake substantially different metabolic roles during light and dark periods that are dictated by substrate availability and the functional capacity of mitochondria defined by their protein composition. We surveyed the heterogeneity of the mitochondrial proteome and its function during a typical night and day cycle in Arabidopsis shoots. This used a staged, quantitative analysis of the proteome across 10 time points covering 24 h of the life of 3-week-old Arabidopsis shoots grown under 12-h dark and 12-h light conditions. Detailed analysis of enzyme capacities and substrate-dependent respiratory processes of isolated mitochondria were also undertaken during the same time course. Together these data reveal a range of dynamic changes in mitochondrial capacity and uncover day- and night-enhanced protein components. Clear diurnal changes were evident in mitochondrial capacities to drive the TCA cycle and to undertake functions associated with nitrogen and sulfur metabolism, redox poise, and mitochondrial antioxidant defense. These data quantify the nature and nuances of a daily rhythm in Arabidopsis mitochondrial respiratory capacity.
Biomass production by plants is by definition the remainder of the subtraction of the respiratory rate from the photosynthetic rate. The values of these rates change both in diurnal cycles and across plant development, and hence, biomass is the cumulative result of these dynamic metabolic processes. The photosynthetic rate and its underlying determinants, capacities, and limitations have been extensively investigated, quantified, and modeled in plants (1, 2). Although there have been a range of studies analyzing changes in respiratory rates in response to light, temperature, and CO2 (3–6), there has been relatively little analysis of the molecular determinants of respiratory capacity or their potential to fluctuate during the daily light and dark cycles of plant growth.
Mitochondria in photosynthetic plant tissues are known to undertake substantially different metabolic roles during light and dark periods. These changes are thought to be largely driven by fluxes in metabolism that provide different substrates to mitochondria. During the day, under photorespiratory conditions, glycine is a major substrate for mitochondrial respiration (7–9). On transfer to darkness, organic acids derived from photosynthetically derived triose phosphates are the first respiratory substrates for several minutes (10, 11); later, organic acids from the breakdown of transitory leaf starch provide the majority of respiratory substrates for hours (11). In situations of extended darkness for days, protein degradation can provide amino acids as substrates for respiration (12, 13). The need for different carbon skeleton products from mitochondria during light and dark periods is also recognized, for example as precursors for nitrogen and sulfur assimilation (14).
Measuring respiration in the light is complicated by the simultaneous operation of photosynthesis; hence, classical respiratory assays of oxygen consumption or CO2 evolution are compromised. Estimates of respiration in the light using gas exchange measurements at different light intensities or at different CO2 concentrations (15–19) suggest a lower rate of TCA cycle-linked respiration in the light than in darkness but a higher overall rate of mitochondrial activity in the light due to the glycine-dependent photorespiratory rate (20).
Recently, stable isotope labeling has predicted bidirectional, non-cyclic TCA cycle function in the light, generating 2-oxoglutarate for nitrogen assimilation and fumarate from oxaloacetate (21). Other studies have attempted to understand the effect of light on mitochondrial carbon assimilation by analysis of changes in abundance of transcripts for metabolic enzymes. For example, the abundance of mRNA encoding glycine decarboxylase (GDC)1 subunits and serine hydroxymethyltransferase (SHMT) was increased drastically in pea leaf upon exposure to light, possibly regulated by phytochrome-mediated transcriptional control of photorespiratory components (22, 23). In contrast, the expression of glutamate dehydrogenase, branched-chain α-ketoacid dehydrogenase complex (BCKDH), and electron transfer flavoprotein:ubiquinone oxidoreductase (ETFQO) was induced by sugar starvation when Arabidopsis plants were grown in extended darkness (24–26). The expression profile of the majority of genes encoding mitochondrial respiratory complexes did not show day/night differences (27), but transcripts for two alternative pathway components, nda1 and ndb2, were regulated in a diurnal manner (28). Genes in the mitochondrial genome, some of which encode respiratory components, were transcribed at a different rate during a light/dark cycle, but the overall transcript pools were maintained at a steady-state level (29). Light-induced changes of mitochondrial components may also occur at the translational and/or post-translational level that would not be detected as changes in transcript abundance. For example, mitochondrial pyruvate dehydrogenase complex can be inactivated by phosphorylation in the light (30).
The capacity of mitochondria to perform metabolism is defined by the abundance and activation state of its catalytic protein machinery and the concentration of substrates, products, and inhibitors of each reaction. Analysis of all these factors and their relative contribution to the control and regulation of the respiratory process is a daunting task. Proteome studies of plant mitochondria from various species that reveal differences in the abundance of mitochondrial proteins between different organs, for example between leaves, roots, cell culture, seeds, and storage tubers, are an important starting point (31–37). This is because protein abundance represents an upper limit constraint to function, and proteome studies can also reveal evidence for post-translation modification or degradation products from proteins. There are major differences recorded in all these plant mitochondrial studies in the abundance of GDC and SHMT. These enzymes dominate the soluble proteome in mitochondria from photosynthetic tissues but can be virtually absent from non-photosynthetic tissues. More subtle differences are noted between tissues in the abundance of mitochondrial formate dehydrogenase, proteins in amino acid metabolism, and components of the TCA cycle. From these data, it has been proposed that the mitochondrial proteome in plants is modified in different tissues to suit the prevailing metabolic requirements (31, 32, 37). Similar suggestions have been made following analysis of the heterogeneity of mitochondrial protein composition in mammalian tissues (38–40). However, although the heterogeneity in the mitochondrial proteome between plant organs is clear, it is not known to what degree short term temporal dynamics are involved in maintaining or modifying the mitochondrial proteome within photosynthetic tissues.
Here, we consider whether mitochondria from photosynthetic tissue have a static proteome during the day and night cycle or whether the proteome changes to meet the different metabolic requirements of the two periods and their transition. This analysis was undertaken by the staged quantitative analysis of the mitochondrial proteome over 10 time points covering a 24-h period in the life of 3-week-old Arabidopsis leaves. The data are coupled to detailed analysis of enzyme capacities, substrate-dependent respiratory rates, and Western blots to identify a series of specific proteins during the same time course. The combination of data reveals day- and night-enhanced protein sets and clear diurnal changes in mitochondrial capacities required to drive the TCA cycle and to undertake functions associated with nitrogen and sulfur metabolism and with cellular redox poise.
EXPERIMENTAL PROCEDURES
Isolation of Shoot Mitochondria from Arabidopsis Hydroponic Seedling Culture
Conditions for the hydroponic culture were outlined previously (32). Shoot mitochondria were isolated from 21-day-old hydroponically grown Arabidopsis using a method adapted from Day et al. (41) with slight modification. Intact plants were removed from sucrose liquid medium, and shoots were separated away from roots. All subsequent steps were carried out on ice in a cold room (4 °C) unless indicated otherwise. Approximately 300 g of shoot materials were homogenized with a Polytron blender (Kinematica, Kriens, Switzerland) in 900 ml of cold grinding medium (0.3 m sucrose, 25 mm tetrasodium pyrophosphate, 1% (w/v) PVP-40, 2 mm EDTA, 10 mm KH2PO4, 1% (w/v) BSA, 1 mm glycine, 20 mm ascorbic acid, pH 7.5) for 10 s twice with 5–10-s intervals between bursts. The homogenate was filtered through four layers of cheesecloth and centrifuged at 1,500 × g for 5 min. The supernatant was carefully decanted into a new centrifuges tube and centrifuged at 24,000 × g for 15 min. The organelle pellet was further washed by repeating the 1,500 and 24,000 × g centrifugation steps twice in sucrose wash buffer (0.3 m sucrose, 0.1% (w/v)) BSA, 1 mm glycine, 10 mm TES, pH 7.5). The pellet of crude organelles was carefully resuspended in sucrose wash buffer using a clean paintbrush and gently layered over a 35-ml continuous 28% (v/v) Percoll density gradient containing a gradient of 0–4.4% PVP-40 in sucrose wash buffer. The gradient was then centrifuged at 40,000 × g for 45 min. The mitochondrial band was seen as a band near the bottom of the tube. The upper layers of the density gradient were removed, and the mitochondrial band was collected. The mitochondrial fraction was diluted ∼5-fold with sucrose wash buffer and centrifuged at 24,000 × g for 10 min. The washed fraction was further purified with Percoll density centrifugation as described for the first gradient. The resulting mitochondrial fraction was diluted with sucrose wash buffer without BSA and centrifuged at 24,000 × g for 10 min. Isolated mitochondria were stored in sterile Eppendorf tubes at −80 °C as 500-μg protein pellets or used immediately for respiratory assays.
Gel Electrophoresis and Immunoblotting
For Western blotting with one-dimensional SDS-PAGE, Bio-Rad Criterion precast gels (10-μg samples, 10–20% (w/v) acrylamide, Tris-HCl, 1 mm, 18-well comb) were used. Gel electrophoresis was performed at 20 mA/gel for 3 h. Protein transfer onto a HybondTM-C Extra nitrocellulose membrane (GE Healthcare) was performed using a Hoefer SemiPhor (GE Healthcare) instrument according to the manufacturer's instructions. Transferred proteins were probed with primary antibodies specifically targeted to NDA1 and NDB2 (42), alternative oxidase (AOX) and porin (from Dr. Tom Elthon, Lincoln, NE), and biotin (anti-biotin-FITC antibody produced in goat, Sigma). A chemiluminescence detection linked to horseradish peroxidase was used to show the secondary antibody, and the immunoreaction was detected using the ECL Advance Western Blotting Detection kit (GE Healthcare). Images were captured using an ImageQuantTM RT ECLTM system (GE Healthcare), and the intensities of the chemiluminescence signals were quantified using ImageQuant TLTM software (version 7.0, GE Healthcare). All Western images are composites of two parts. The dark phase (0, 1, 2, 4, and 8 h) and the light phase (12, 13, 14, 16, and 20 h) were originally run on the same gel but in a different order. The compilation was done simply to order them as dark phase followed by light phase for consistency with the other data presentations. The relative signal bar graphs are based on three sets of gel images, each from a separate biological replicate gel.
Differential (DIGE) two-dimensional IEF/SDS-PAGE was performed according to Eubel et al. (43). A randomized experimental design was used to minimize the impact of preferential labeling and gel-to-gel variation on downstream statistical analysis (see supplemental Fig. 1B for experimental design). Fluorescent protein spots were visualized on a TyphoonTM laser scanner (GE Healthcare), and image comparison was done using the DeCyderTM software package (version 6.5, GE Healthcare). Gels were first analyzed using the differential in-gel analysis mode of the DeCyder (GE Healthcare) software package prior to a comprehensive biological variance analysis. Gel pictures were electronically overlaid using the ImageQuant TL software (GE Healthcare). Following scanning for fluorescent signal, gels were stained and visualized by colloidal Coomassie (G-250) for mass spectrometry analysis.
Trypsin Digestion and Mass Spectrometry Analysis
In-gel digestion of the selected gel spots was performed according to Shevchenko et al. (44). Peptides were separated by self-packed Microsorb (Varian Inc.) C18 (5 μm, 100 Å) reverse phase columns (0.5 × 50 mm) on an Agilent Technologies 1100 series capillary liquid chromatography system and analyzed using an XCT Ultra IonTrap mass spectrometer equipped with an ESI source as described previously (32). The chromatography system and mass spectrometer were controlled by Chemstation (Agilent Technologies) and MSD Trap Control version 6.0 (Build 38.15) software (Bruker Daltonik GmbH). Peptides were eluted from the column at 10 μl/min using a 9-min acetonitrile gradient (5–80% (v/v)) in 0.1% (v/v) formic acid) at a regulated temperature of 50 °C into the mass spectrometer operated in positive mode. The method used for initial ion detection utilized a mass range of 200–1400 m/z with scan mode set to standard (8100 m/z per s), ion charge control conditions set at 250,000, and three averages taken per scan. Smart mode parameter settings were used with a target of 800 m/z, a compound stability factor of 90%, a trap drive level of 80%, and “optimize” set to normal. Ions were selected for MS/MS after reaching an intensity of 80,000 counts/s, and two precursor ions were selected from the initial MS scan. MS/MS conditions used were as follows: SmartFrag for ion fragmentation, a scan range of 70–2200 m/z using an average of three scans, the exclusion of singly charged ions option, and ion charge control conditions set to 200,000 in ultrascan mode (26,000 m/z per s). Resulting MS/MS spectra were exported from the DataAnalysis for LC/MSD Trap version 3.3 (Build 149) software package (Bruker Daltonik GmbH) using default parameters for autoMS(n) and compound export. All mass spectra have been submitted to the Proteomics Identifications (PRIDE) database (http://www.ebi.ac.uk/pride/; accession numbers 10471–10525).
Results were queried against an in-house Arabidopsis database comprising ATH1.pep (release 7) from The Arabidopsis Information Resource and the Arabidopsis mitochondrial and plastid protein sets (the combined database contained a total of 30,700 protein sequences with 12,656,682 residues) using the Mascot search engine version 2.2 and utilizing error tolerances of ±1.2 Da for MS and ±0.6 Da for MS/MS, “enzyme” set to trypsin, “maximum missed cleavages” set to 1, variable modifications of oxidation (Met) and carbamidomethyl (Cys), instrument set to ESI-TRAP, and peptide charge set at 2+ and 3+. ATH1.pep is a non-redundant database with systematically named protein sequences based on Arabidopsis genome sequencing and annotation. Results were filtered using “standard scoring,” “maximum number of hits” set to “auto,” and “ion score cutoff” set at 38. In addition, only bold red peptides were considered, thus removing duplicate homologous proteins from the results. A protein match was automatically validated only when at least two unique peptides both showing an ion score higher than 38 (Mascot-defined significance/identity threshold, p ≤ 0.05) were present. For proteins identified by a single significant peptide having a score above the significance threshold, only the spectrum of the significant peptide was inspected to ensure that each peak corresponding to a fragmented ion was clearly above base-line background noise and a series of at least four continuous y or b ions were observed. To estimate the false-positive rate of our protein identification strategy, a single concatenated mgf file generated by MASCAT (Agilent Technologies), which comprised all the MS/MS output data, was then used to search against a target-decoy database using the above search strategy with decoy option selected. Using the defined search parameters, the false-positive rate in the target-decoy search was found to be ∼4.5%.
When peptides were matched to multiple members of a protein family encoded by different Arabidopsis genes, each protein match was manually inspected to identify the peptide(s) that were uniquely assigned to one gene product but not to the others. Protein isoforms that were identified by the same set of peptides were both assigned as protein matches (see supplemental Table 1). When proteins of different families were identified in a gel spot, a reference map of the Arabidopsis mitochondrial proteome (data not shown) was used to identify the match, taking into account the number of peptides with an ion score >38 and the delta mass for each peptide.
Oxygen Electrode and Spectrophotometric Measurements of Isolated Mitochondria
Oxygen consumption by plant extracts and crude and purified mitochondria was measured in a computer-controlled Clark-type O2 electrode unit (Hansatech Instruments, Pentney, UK). Calibration of the electrode was carried out by the addition of sodium dithionite to remove O2 from the electrode chamber. The air-saturated O2 concentration was assumed to be 240 μm. All reactions were carried out using 1 ml of mitochondrial reaction medium (0.3 m sucrose, 5 mm K2H2PO4, 10 mm TES, 10 mm NaCl, 4 mm MgSO4, 0.1% (w/v) BSA, pH 7.2) and 100 μg of mitochondrial sample protein. Succinate (5 mm), glutamate (10 mm), malate (10 mm), glycine (10 mm), formate (10 mm), NADH (1 mm), NAD (2 mm), CoA (12 μm), thiamine pyrophosphate (0.2 mm), and KCN (0.5 mm) were added as appropriate to modulate the O2 consumption rates of mitochondria. Purified mitochondria in sucrose wash buffer were supplemented with 1 mm glycine and 0.5 mm ATP and incubated at 4 °C for 30 min before glycine-dependent respiration measurements to ensure full activation of GDC (45). Pyruvate dehydrogenase complex (PDC), 2-oxoglutarate dehydrogenase complex (2-OGDC), and BCKDH activity was measured according to Taylor et al. (46). Formate dehydrogenase (FDH) activity was measured as described by Oliver (47). The activity of NAD-dependent malic enzyme (NAD-ME) was measured according to Jenner et al. (48). Fumarase was assayed as described by Hatch (49). The amination and deamination activity of GDH at A340 was measured according to Turano et al. (50). Citrate synthase (CS), mitochondrial malate dehydrogenase (mMDH), and aconitase (ACON) were assayed as described previously in Lee et al. (32). O-Acetylserine thiol-lyase (OAS-TL) activity was measured according to the method outlined by Winger et al. (51). NAD-dependent isocitrate dehydrogenase (IDH) was assayed by monitoring the rate of change in A340 following the reduction of NAD in the reaction mixture containing 50 mm Tris acetate (pH 7.2), 0.05% (v/v) Triton X-100, 2 mm NAD, 20 mm MnCl2, 50–100 μg of mitochondrial proteins, and 20 mm isocitrate.
Statistical Analysis
Unless stated otherwise, all data obtained from experiments are expressed as means ± S.E. to two significant figures for at least three independent biological replicates. Statistical significances were evaluated by Student's t test or one-way ANOVA and Tukey range analysis using Microsoft Excel XP (2002, SP3) or the statistical software package R (version 2.6.1).
Supplemental Data
Details of mass spectrometry-based identification of protein spots are in supplemental Table 1. Tukey range analysis of gel spot abundance changes is provided in supplemental Table 2. Tukey range analysis of enzyme assays is provided in supplemental Table 3. One-dimensional gel and two-dimensional gel DIGE images of diurnal mitochondrial protein samples are in supplemental Fig. 1. Additional immune reaction blots with selected antibodies (3-methylcrotonyl-CoA carboxylase (MCCase) α-subunit (MCCA) and porin) are included in supplemental Fig. 2. A spectrum of spot 51 (which was identified by a single peptide; supplemental Table 1) is available for reanalysis in supplemental Fig. 3 (the spectrum was extracted using MascotDatfile library (52) and manually edited to include peak masses detected).
RESULTS
Quantitative Analysis of Changes in Mitochondrial Proteome during Diurnal Cycle
Mitochondria were isolated (in triplicate) from plant shoot tissues from 21-day-old Arabidopsis shoots harvested after 0, 1, 2, 4, and 8 h of darkness and after 0, 1, 2, 4, and 8 h of illumination. To achieve this, timed mitochondrial preparations were performed on 30 different days from staggered plantings of hydroponically grown plants to generate the 30-sample time series across day 21 of Arabidopsis shoot development (supplemental Fig. 1A). We then performed a detailed differential two-dimensional (DIGE) IEF/SDS-PAGE experiment using a randomized experimental design incorporating all samples and replicates across 15 gels labeled with Cy3 or Cy5 (supplemental Fig. 1B). Cy2 was bound to a mixture of all 30 samples as an internal standard on each gel. The set of 15 gels was analyzed using the DeCyder software package, and protein spots that reproducibly changed in abundance across the time course according to a one-way ANOVA (p < 0.05) were selected for further analysis (supplemental Table 2). In total, 55 protein spots changed in abundance based on the ANOVA, and their identities were revealed using LC-MS/MS (supplemental Table 1); 45 were known mitochondrial proteins.
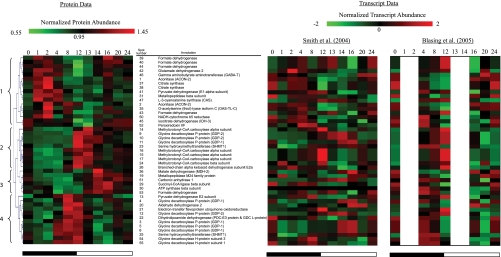
Hierarchical clustering of the abundances of the 45 mitochondrial protein spots was then performed, showing the simultaneous accumulation of protein components engaging in the same or related metabolic pathways at different times in the diurnal cycle (Fig. 1). TCA cycle enzymes peaked early in the night (cluster 1), and leucine catabolism enzymes and photorespiration enzymes peaked at or around the transition to light (clusters 2 and 4), whereas malate dehydrogenase and succinyl-CoA ligase increased during the day (cluster 3).
Fig. 1.
Changes in abundance of mitochondrial proteins and transcripts during diurnal cycle. Using The Institute for Genomic Research MultiExperiment Viewer, hierarchical clustering was performed on the normalized protein abundances for 45 mitochondrial proteins that were altered in abundance during the diurnal cycle (with one-way ANOVA p < 0.05). Hierarchical clustering was performed using average linkage and euclidean distance. On the right, the corresponding averaged and normalized (using algorithm in The Institute for Genomic Research MultiExperiment Viewer) transcript abundance data extracted from Smith et al. (53) and Bläsing et al. (27) are shown. The four major clusters observed are marked 1–4 on the far left. The heat map color gradient range is shown at the top for both transcript and protein data. The day/night cycle is indicated at the bottom of the panels by a white (day) and a black (night) bar. Mass spectrometry evidence for these identifications is shown in supplemental Table 1. A Tukey analysis of the significance of spot abundance changes over time is reported in supplemental Table 2.
The protein data were compared against the corresponding averaged transcript abundance data for each corresponding gene extracted from publicly available microarray data for diurnal changes in Arabidopsis leaves (27, 53) (Fig. 1). When comparing these profiles, although some protein abundance changes followed transcript abundance (e.g. spots 15–17, MCCase), the majority did not show clear positive correlation, suggesting that translational and post-translational responses are significantly responsible for the diurnal changes in the mitochondrial proteome. Differences in the temporal lag between transcription and translation of proteins may also be a factor in altering the correlations observed in these comparisons (e.g. GDC, spots 54 and 55). To explore the functional meaning of the differences noted from the proteome analysis, data from substrate-dependent respiratory assays, activity measurements, and Western blot analysis were integrated with the changes in protein spot abundances. Differences were explored using ANOVA and the Tukey range test (supplemental Tables 2 and 3) to justify the differences noted below. The analysis of these results is outlined below in themed sections related to mitochondrial metabolism.
Diurnal Changes in Photorespiratory Enzymes and Related Pathways
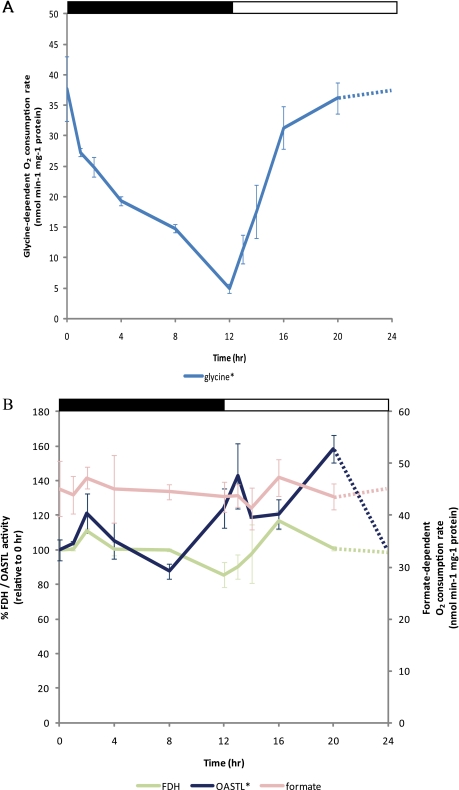
To examine the photorespiratory capacity of mitochondria during a day/night cycle, glycine-dependent oxygen consumption was measured in the freshly isolated mitochondria, which were later used for the DIGE analysis. Glycine-dependent respiratory capacity slowly declined to a minimal level at the end of the night (Fig. 2A). This was followed by an increase to the maximal rate in mitochondria isolated after the first 4 h of illumination. This correlates well with the total glycine and serine levels observed in Arabidopsis leaves (54). To our knowledge, this dramatic change in isolated mitochondrial glycine-dependent respiratory capacity across the diurnal cycle has not been reported previously.
Fig. 2.
Changes in activity of photorespiration and its related pathways in mitochondria during diurnal cycle in Arabidopsis shoot. A, oxygen consumption dependent on glycine as substrate and supplemented with appropriate cofactors was measured. B, the catalytic activity of FDH and OAS-TL and the formate-dependent oxygen consumption rate were measured. Enzyme activity profiles are plotted with respect to 0 h (100%), and glycine- and formate-dependent oxygen consumption measurements are presented as nmol min−1 mg−1 of protein. Data were compared using a one-way ANOVA cutoff of p < 0.05 with an asterisk (*) indicating significant differences between 10 data points. A Tukey analysis is shown in supplemental Table 3. The results are shown as means of biological replicates and error bars are S.E. (n = 3). The day/night cycle is indicated at the top of the plot by a white (day) and a black (night) bar.
In the DIGE analysis, we identified one isoform of SHMT (At4g37930) and several subunits of the GDC (H-protein (At1g32470 and At2g35370), L-protein (At1g48030), and P-protein (At4g26970 and At4g33010)) that were altered in abundance across the diurnal cycle. The SHMT isoform found in the major leaf form of this protein and the H, L, and P subunits are three of the four subunits of GDC. Analysis showed that these proteins clustered into two distinct groups (Fig. 1). Protein spots in cluster 4 were generally more highly abundant at the end of the night followed by a decline to the minimal levels 2–4 h after the onset of light before recovering midday (Fig. 1). The SHMT in wild-type potato leaves exhibited a similar change in abundance in the diurnal cycle (55). The pI of SHMT identified in cluster 4 (spot 25) was lower than the same protein in cluster 2 (spot 23). In comparison, cluster 2 contained protein spots that are generally more abundant in the light than in the dark with an increase of up to 3-fold detected from 4 to 12 h (Fig. 1). The GDC P-proteins identified in cluster 2 did not match to their predicted molecular weight and pI (supplemental Table 1, spots 9–12). These are both likely to be degradation products that could form in the presence of inhibitory compounds when the rate of glycine decarboxylation by the mitochondria is elevated in the light (Fig. 2A). One such compound known to damage GDC/SHMT is formaldehyde (37, 56, 57). We did not detect changes in the degradation products from other GDC subunits, but these may be too small to be identified on two-dimensional gels. At 20–24 h, the abundance of these GDC degradation products was at their lowest. Notably, this pattern inversely correlated with the level of the antioxidant protein peroxiredoxin (spot 52) toward the end of the day (Fig. 1); peroxiredoxin may help to protect proteins from damage by lowering ROS levels in mitochondria.
A number of FDH protein spots varied in abundance during the diurnal cycle, namely the accumulation of FDH proteins with different pI values (spots 39, 40, 43, and 44). It is possible that these represent isoforms of FDH with different phosphorylation status (58). However, no significant changes were observed in formate-dependent oxygen consumption or the maximal catalytic FDH activity across the time points examined (Fig. 2B). Interestingly, the changing abundance of the major FDH protein spot identified (spot 43) tightly clustered over the diurnal period with the abundance of the cysteine-synthesizing enzyme OAS-TL (Fig. 1). The abundance of this OAS-TL protein spot across the cycle was significantly different from random based on the ANOVA of the DIGE data. The OAS-TL spot abundance peaked significantly at 2 h (when compared with all other time points except 8 h) and 16 h (when compared with 13 h, the lowest protein abundance in the day) based on the Tukey range analysis (Fig. 1 and supplemental Table 2). The activity of OAS-TL measured by enzymatic assay in mitochondrial samples also changed significantly and was apparently the inverse of the spot abundance change, showing a decrease in cysteine synthesis rate during the night period followed by a ∼40% increase 8 h after light illumination (Fig. 2B and supplemental Table 3).
Changes in Entry Steps of TCA Cycle during Diurnal Cycle
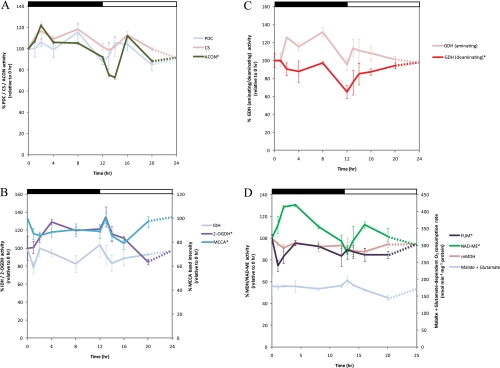
The PDC is often considered the gatekeeper controlling the entry of carbon into the TCA cycle. We identified PDC E1α (At1g59900), E2 (At1g54220), and E3 (At1g48030) protein spots to be generally higher in abundance, peaking at 2–8 h into the dark phase. No significant differences were detected in the total enzyme activity of dephosphorylated/activated PDC during the diurnal cycle (Fig. 3A). However, previous analysis of PDC activity in other plant species showed that the enzyme complex was inactivated by phosphorylation of E1α in the light and reactivated by dephosphorylation in the dark (30, 59). Therefore, the observed PDC spot abundance changes in acidic pI versions of E1α may be a consequence of such phosphorylation(s) but were not sufficient under our growth conditions to significantly change the more basic, dephosphorylated E1α spots abundances that define the activity of the enzyme.
Fig. 3.
Changes in enzyme activities focused on TCA cycle, glutamate metabolism/catabolism, and branched-chain amino acid degradation in mitochondria during diurnal cycle in Arabidopsis shoot. A, changes in the activity of entry steps of the TCA cycle in the mitochondria. Enzyme activity was measured for PDC, CS, and ACON. B, changes in the activity of decarboxylating dehydrogenases in the TCA cycle and branched-chain amino acid degradation in the mitochondria. Enzyme activity was measured for IDH and 2-oxoglutarate dehydrogenase (2-OGDH). The intensity of the band containing a biotin-containing protein, MCCA, on immunoblot over a 24-h period was also plotted (also see supplemental Fig. 2A). C, changes in the activity of aminating and deaminating activity of glutamate dehydrogenase in the mitochondria. D, analysis of the TCA cycle enzymes in the mitochondria focused on malate formation and utilization. Oxygen consumption in the presence of malate + glutamate supplemented with appropriate cofactors was measured. Enzyme activity was measured for mMDH, NAD-ME, and fumarase (FUM). All data are plotted with respect to 0 h (100% activity or band intensity) with the exception of substrate-dependent oxygen consumption measurements. The catalytic activity profiles of enzymes found to be significantly changed between data points (one-way ANOVA p < 0.05) are indicated with an asterisk (*). A Tukey analysis is shown in supplemental Table 3. The results are shown as means from three biological replicates with error bar representing relative S.E. The day/night cycle is indicated at the top of the plot by a white (day) and a black (night) bar.
The changes in abundance of the PDC subunits were clustered with two distinct groups of proteins during the diurnal cycle: the E2 subunit clustered with a group of photorespiratory proteins in cluster 4, whereas the E1α subunit was clustered with two other TCA cycle enzymes in cluster 1, CS (spots 37 and 38) and ACON (spots 1 and 2). The protein spots identified as CS were minor isoforms on preparative two-dimensional gels, and there was no significant change in the abundance of the major CS protein spot (supplemental Fig. S1B). No significant change was observed in enzymatic activity of CS in mitochondrial extracts from across the dark and light cycles (Fig. 3A). In contrast, ACON activity showed significant differences in the diurnal cycle (Fig. 3A) with the lowest activity at 1–2 h after illumination. Two protein spots containing ACON2 with different pI values were altered in abundance at distinct times of the diurnal cycle and were separated within cluster 1 (Fig. 1), indicating some differences between them. Analysis of the MS/MS spectra of the two protein spots has not yet identified a post-translational modification that could be responsible; however, peptides covering only ∼20% of the ACON2 protein sequence have so far been identified from these gels (supplemental Table 1).
Regulation of Exit and Entry of Non-decarboxylating Side of TCA Cycle via 2-Oxoglutarate
The abundance of protein spot 45, which contained IDH3 (At3g09810), declined by 2-fold at the end of the dark period (12 h) and then increased gradually to reach equilibrium during the light period (Fig. 1). A similar expression profile was observed at the transcriptional level for IDH3 (Fig. 1). Measurement of the total IDH activity showed no observable differences over the time course (Fig. 3B), possibly due to the compensation of other catalytic IDH isoforms in Arabidopsis mitochondria. Protein spot 42, containing GDH, changed in abundance in a manner resembling the change in the IDH3 protein spot (Fig. 1). Measurement of the maximal catalytic amination activity of GDH (generating glutamate) showed no significant changes during the dark/light phases (Fig. 3C). In contrast, the GDH deamination activity (generating 2-oxoglutarate) decreased by 25% during 8–12 h of the dark phase and increased rapidly in the 2 h after the dark to light transition (Fig. 3C), closely following the protein spot 42 abundance across the diurnal cycle.
2-Oxoglutarate generated from the TCA cycle or glutamate catabolism can be utilized to facilitate the degradation of branched-chain amino acids (BCAA). Seven protein spots of the BCAA degradation pathway, ETFQO, MCCA, MCCB, and BCKDH were altered in abundance during the diurnal cycle (Fig. 1). This BCAA pathway in plants contains 12 enzymes, seven of which are known to be located in mitochondria; in Arabidopsis, 14 genes encode subunits for these seven enzymes (46). The abundance of the MCCase subunits all peaked at ∼12 h in the dark or 1 h into the light (13 h), and BCKDH abundance increased gradually during the dark phase and then reached a maximum at 8–12 h of darkness, whereas ETFQO abundance peaked at around 8 h in the dark. The changing level of MCCA was independently determined by immune detection using antibodies against biotin (supplemental Fig. 2A) as it is the only known major biotin-containing plant mitochondrial protein. A 75-kDa band was detected in each mitochondrial fraction, and the intensity of all immunosignals resembled the expression profile of MCCA observed in the DIGE experiment (Figs. 1 and 3B). These results are also in concordance with elevated MCCase transcript levels detected in dark-treated plants (60) and the MCCase transcripts that peaked at the end of the night in two diurnal studies (27, 53).
If the regulation of 2-oxoglutarate generation by mitochondrial GDH and IDH affects the non-decarboxylating part of the TCA cycle, we might expect a similar or compensatory change in activity and/or abundance of 2-OGDC. The catalytic activity of 2-OGDC rose in the early night phase, peaked at 1 h after illumination, and then decreased slowly over the course of the light phase, inversely matching the deaminating reaction of GDH (Fig. 3C). The protein/activity profile of 2-OGDC shown here is similar to the metabolite profile of glutamate and GABA in young Arabidopsis leaves where both show elevated levels at night, a decline during the day, and a rise in the evening (61). Interestingly, the abundance of GABA transaminase (Fig. 1, spot 46) was higher during the day (14–24 h) than in the early morning (8–14 h), suggesting the 2-OGDC may be a preferred pathway in the morning and that GABA shunt activity is maximized later in the day. As the production of 2-oxoglutarate is also vital for metabolic processes in other cellular compartments (62), these data also suggest that 2-oxoglutarate may be used in the mitochondria for the TCA cycle or exported to the glutamine synthase/glutamine:2-oxoglutarate aminotransferase pathway at different rates during the diurnal cycle.
Diurnal Response and Fate of Malate in Mitochondria
Mitochondria freshly isolated at different time points have similar rates of malate + glutamate-dependent O2 consumption on a protein basis, and total mMDH activity in mitochondria did not change significantly during the diurnal cycle (Fig. 3D). We noted that mMDH-2 was higher in abundance during the day, reaching a maximum at 14 h (Fig. 1, spot 36, and supplemental Table 2). However, no significant diurnal change could be found in abundance of protein spots containing mMDH-1, which is the major form of mMDH in Arabidopsis shoot (supplemental Fig. 1B). The relative contributions of mMDH-2 and mMDH-1 to malate production and/or metabolism in the light remain to be resolved. The NAD-ME activity rose in the dark and peaked 2–4 h into the dark phase, fell through the night, and again increased during the transition to the day period (Fig. 3D). A role for NAD-ME in differentially contributing to nocturnal metabolism is consistent with metabolic profiling of NAD-ME knock-out plants in Arabidopsis (63). Although we did not identify fumarase as a protein spot changing in abundance in the DIGE analysis, ANOVA of fumarase activity showed significant changes (Fig. 3D and supplemental Table 3), notably a marked decrease in the 1st h of darkness, an increase over the first few hours of darkness, and a further increase at the end of the day. These data on malate synthesis and utilization enzymes indicate increased capacity for malate metabolism during the light period in contrast to the observed decreases in the capacity of the first half of the TCA cycle.
Differences in Capacity of Alternative Respiratory Chain Pathways
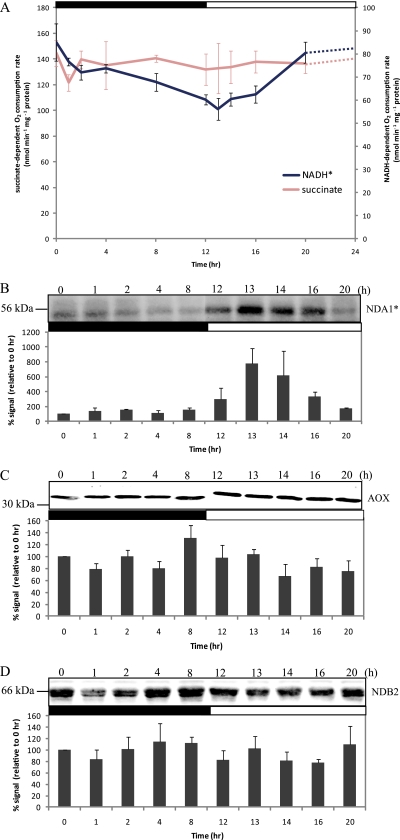
Although succinate-dependent respiration was unchanged across the time course (one-way ANOVA p > 0.05), the maximum rate of external NADH-dependent oxygen consumption of freshly isolated mitochondria showed a gradual decrease over the course of the dark period and then increased over the course of the light period (Fig. 4A and supplemental Table 3). Transcript abundance for AOX and NAD(P)H-dependent dehydrogenases in plant mitochondria has been shown to be responsive to light signals (64). To detect the abundance of these components, antibodies raised against AOX and the major internal and external NADH dehydrogenases, NDA1 and NDB2, respectively, were used (42). The protein abundance of the internal NDA1 dehydrogenase showed a marked increase at 13 h followed by progressive loss during the course of the day (Fig. 4B). This is consistent with the initial transcriptional response of NDA1 to light in Arabidopsis leaves (28, 53) and maturing potato leaves (64). In contrast, only minor changes were observed in the abundance of the AOX (Fig. 4C). Also, the intensities of the NDB2 band (Fig. 4D) were not clearly consistent with the changes in the external NADH-dependent respiration rate (Fig. 4A). Hence, the diurnal changes in NADH oxidation are either due to changes in external NADH dehydrogenase(s) other than NDB2 or are due to post-translational modification or activation of NADH dehydrogenase(s) that respond to light, photosynthetic metabolism or cytosolic redox poise.
Fig. 4.
Changes in alternative dehydrogenases and oxidases in respiratory chain during diurnal cycle. A, oxygen consumption in the presence of succinate and NADH supplemented with appropriate cofactors was measured. B–D, immunoreactivity analysis of internal NADH dehydrogenase 1 (NDA1) protein abundance (B), alternative oxidase protein abundance (C), and external NADH dehydrogenase 2 (NDB2) protein abundance (D) using specific antibodies. Equal amounts of protein were loaded in each lane (15 μg). The intensity of the protein bands was quantified by ImageQuant 7 (GE Healthcare), calculated as a percentage relative to 0 h (100%), and plotted with means and the error bars are S.E. from three biological replicates. Data were compared using a one-way ANOVA cutoff of p < 0.05 with an asterisk (*) indicating significant differences between 10 data points. The day/night cycle is indicated at the top of the plot by a white (day) and a black (night) bar.
DISCUSSION
In the present study, we identified and quantified 55 protein spots from mitochondrial extracts that were dynamically altered in abundance during a 12-h dark, 12-h light cycle (Fig. 1 and supplemental Table 1). Although the composition of the plant mitochondria proteome has been established in both dicots and monocots (65–69), in different plant tissues (31–37), and during oxidative or environmental stress (70, 71), very little is known about the temporal dynamic behavior of the mitochondrial proteome in plants. This study shows that a small but a significant set of mitochondrial proteins was altered over a daily cycle of dark and light periods, indicating that a constant rhythmic change of the organelle proteome occurs each day in plant tissues. The protein abundance changes observed in a day/night cycle were relatively subtle, likely because a high turnover rate of mitochondrial proteins would be energetically costly. The changes could reflect more minor adjustments in all mitochondria or major changes that occur in only a proportion of the cells used in the bulk tissue mitochondrial isolations. Future studies could be directed to localize these changes within the leaf architecture to address this issue. We should also note that as two-dimensional PAGE is biased toward visualization of soluble proteins and relatively higher abundance components there are likely to be other undetected changes in mitochondrial proteins during the diurnal cycle that await future identification.
It has been well established that starch synthesis and degradation follows diurnal rhythms (27, 53) and that sugar products of starch are important in controlling many diurnally regulated genes (27). ATP is required for the synthesis of sugar variants, and total ATP levels change between day and night conditions due to the operation of photosynthesis (72, 73). Given the differences in the photosynthetic rates during the diurnal cycle, adjustments of key enzymes involving in energy metabolism, amino acid metabolism, and the interaction with other organelles will be necessary to balance metabolic flow to meet the specific energetic needs of the cell. In this context, we explore below the underlying mechanisms and potential consequences of the changes in capacities of pathways observed in the mitochondria during the diurnal cycle.
Protein Inactivation and Fate in Mitochondria during Diurnal Cycle
Any rhythmic change in protein abundance or function implies mechanisms for both protein synthesis and/or activation as well as degradation and/or inactivation. The most notable response to the diurnal cycle by plant mitochondria was the change in the capacity to oxidize glycine as a respiratory substrate (Fig. 2A). At the protein level, it was not clear what was responsible for this more than 5-fold change in glycine-dependent respiratory capacity. The major protein spots of SHMT and GDC (Fig. 1, cluster 4) were slightly lower in abundance during the day, whereas the modified forms of SHMT and the GDC degradation products (Fig. 1, cluster 2) peaked during the early light phase (Fig. 1). It can be speculated that the rate of degradation and resynthesis of GDC and SHMT is accelerated in the light period to replace damaged proteins with functionally active proteins. This is consistent with the demand for photorespiration to dissipate excess redox equivalents from chloroplasts in the light (74), the light responsiveness of the GDC gene promoters (22, 23), and the light-enhanced transcription profile for GDC and SHMT during the diurnal cycle (Fig. 1). GDC is known to be susceptible to oxidative damage by ROS in mitochondria and is known to produce formaldehyde (which contains a highly reactive oxygen group that encourages peroxide generation) when methylamine is degraded in the absence of tetrahydrofolate (57, 75). It is also possible that the macromolecular organization of GDC has a diurnal rhythm and is responsible for the changes in maximal activity in isolated mitochondria, but more data would be required to evaluate such possibilities.
Aconitase activity also varied across the diurnal cycle, and ACON2 protein spots may have been modified in abundance by some sort of post-translational modification. In mammals, cytosolic SHMT has been shown to be post-translationally modified by deamidation accompanied by an overall reduced activity of the modified protein (76). A transglutaminase has also been reported in rat mitochondria to reduce ACON1 activity (77), and its activity has been shown to be affected by the ATP/GTP level and disruption of mitochondrial function in humans (78). In chloroplasts, post-translational modifications, such as phosphorylation of the damaged D1 component of the photosystem II (79, 80) and polyubiquitination of phytochrome (81), are a requirement for initiating enzymatic protein degradation. We have not uncovered the mechanism here, but we do show several targets in plant mitochondria of diurnally regulated changes in protein function.
Oxidative Damage and Antioxidant Defense Changes during Diurnal Cycle
ROS can lead to oxidative damage and subsequent degradation of mitochondrial proteins. Key TCA cycle enzymes are particularly sensitive to such damage (70, 82). ROS are produced during mitochondrial respiration throughout the diurnal cycle (83, 84); however, ROS production from plastids and peroxisomes is enhanced in the light due to photosynthesis and photorespiration (83, 85), and this is likely to have an impact on mitochondria during the day. From our data, several distinct responses in the light may reflect increased ROS load in mitochondria. In the first 1–2 h after illumination (13–14 h), the internal alternative NADH dehydrogenase (NDA1) was induced (Fig. 4B). The induction of NDA1 could help to redistribute electron transport to prevent over-reduction of complex I, one of the major ROS producers in the electron transport chain (86), especially at a time when NADH generation from photorespiratory glycine oxidation is increasing. Increases were also observed in the abundance and/or activity of FDH, peroxiredoxin, and OAS-TL. Cysteine produced from serine (from photorespiration) via OAS-TL can be exported to the cytosol and used for synthesizing glutathione (87) or used for detoxifying cyanide, an inhibitor of complex IV. Mitochondrial peroxiredoxin serves as an electron donor for the reduction of H2O2 (88). Glutathione has been shown to be the preferred electron donor for peroxiredoxin (88) and is generated during the ascorbate/glutathione cycle in mitochondria (89). The capacity of ascorbate synthesis by mitochondria has been shown to be higher in the light than in the dark (90), indicating an enhanced capacity for ascorbate/GSH-linked mitochondrial reactions in the light. FDH is also known to be induced by oxidative stress in plants (91). Formate derived from the detoxification of formaldehyde generated by GDC in the light (57) can be oxidized to generate NADH in the matrix. Each of these proteins has already been highlighted in ROS-induced stress responses observed in previous mitochondrial studies (for a review, see Refs. 92 and 93).
Impact of Sudden Light Changes on Diurnal Pattern
In this study, as is the case in most laboratory-based studies of plants, day and night were near instantaneous transitions through light cycles in growth cabinets. Some rapid changes observed may therefore be a response to the sudden change and not part of a diurnal pattern per se. Notable examples are the response of NAD-ME and aconitase activity (Fig. 3, A and C) and branched-chain amino acid metabolism components to dark-light and light-dark transitions (Fig. 1). A number of studies have demonstrated a sudden increase in CO2 release when a plant is rapidly darkened in a very short period of time in a phenomenon known as the postillumination burst (16, 94). This is then followed by a brief period of increased respiration before a steady-state level is achieved that is termed light-enhanced dark respiration (10). It has been proposed that the rate of respiration in darkness after a period of illumination reflects the amount of metabolites, especially malate and pyruvate, available to the TCA cycle in mitochondria (10, 11). So although these transitions may not reflect diurnal events per se, they do show that the abundance and activity of key mitochondrial enzymes can rapidly change in response to perturbations in metabolite concentrations and thus that the proteome is responding to the environment and is not on a prescribed 24-h rhythm.
Metabolic Phases of TCA Cycle and Related Metabolism during Night and Day
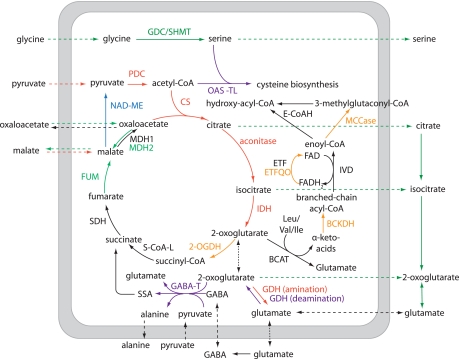
By combining the protein abundance data and activity assays presented here, we propose that there are four phases of the TCA cycle and related reactions governed by variation in the capacity of respiratory metabolism: a dark-enhanced phase (1–8 h), a midnight to early morning phase (10–14 h), a day-enhanced phase (14–20 h), and a late day phase (18–24 h) (Fig. 5). This is also supported with other literature evidence as noted below.
Fig. 5.
Metabolism scheme illustrating enzymes in mitochondrial primary metabolism with altered abundance or activity during diurnal cycle. A dark-enhanced phase (1–8 h; red), a midnight to early morning phase (10–14 h; orange), a day-enhanced phase (14–20 h; green), and a late-day phase (18–24 h; purple) are shown. One enzyme changed in dark-to-light and light-to-dark transitions (blue). BCAT, branched-chain amino acid aminotransferase; E-CoAH, enoyl-CoA hydratase; FUM, fumarase; GABA-T, GABA transaminase; S-CoA-L, succinyl-CoA ligase; IVD, isovaleryl-CoA dehydrogenase; SSA, succinic semialdehyde; ETF, electron transfer flavoprotein; MDH, malate dehydrogenase; SDH, succinate dehydrogenase; 2-OGDH, 2-oxoglutarate dehydrogenase.
In the dark (1–8 h), the enzymes of the first half of the TCA cycle (PDC, CS, ACON, and IDH) had a higher capacity. In darkened leaves, citrate is known to be catabolized through the TCA cycle, but in illuminated leaves, citrate is understood to be exported to the cytosol, bypassing the mitochondrial ACON and IDH steps, to produce 2-oxoglutarate via cytosolic reactions (95). Reduced expression of ACON has also been shown to elevate the rate of photosynthesis and increase the rate of sucrose synthesis in illuminated tomato leaves (96). Thus, lowering the ACON level in the light (Fig. 3A) could be an adaptive response to facilitate the accumulation of starch and sucrose for use in subsequent darkness.
At midnight and early morning, there was a higher capacity of the BCAA pathway and 2-OGDC, both of which use 2-oxoglutarate. The induction of Leu, Val, and Ile degradation capacity is likely linked to a decreased availability of sucrose/starch toward the end the night. A number of studies have shown that the expression of genes encoding enzymes of the BCAA catabolism are transcriptionally suppressed in the light (25, 60) and by sucrose (97). The elevated levels of Leu/Ile/Val catabolism enzymes only last a few hours after illumination (Fig. 1), presumably reflecting rapid turnover levels of these proteins when sucrose content and glycolytic intermediates begin to accumulate in the leaf.
Notably, there were differential responses of malate-forming (fumarase and mMDH) and malate-degrading (mMDH and NAD-ME) pathways across the day. Fumarase activity and mMDH-2 protein abundance is elevated during the day, whereas NAD-ME activity responds most significantly to dark-to-light transitions. These malate-linked changes could reflect alterations in the net flux of malate and oxaloacetate across the mitochondrial membrane and its use as a redox shuttle to balance the photorespiratory pathway requirements for oxidants and reductants in different cellular organelles (74).
Later in the day there was a higher abundance of GABA transaminase (Fig. 1), indicating an enhanced capacity of the GABA shunt (Fig. 5). This represents an alternative utilization of 2-oxoglutarate at the same time as 2-OGDC and BCAA metabolism pathways have reduced activity. A role for the GABA shunt in the light is consistent with the light sensitivity of Arabidopsis mutants in GABA shunt components (98). In addition, there was a change in the deaminating reactions of GDH in the latter part of the day (Fig. 3C) that could increase the 2-oxoglutarate:glutamate ratio, providing substrate for the GABA shunt and supplementing the reduced 2-oxoglutarate formation by the TCA cycle. The cysteine biosynthetic enzyme, OAS-TL, also increased later in the day, which could enhance utilization of serine from glycine oxidation and use the acetyl-CoA not incorporated into citrate due to the slower TCA cycle function during the day (Fig. 5).
Determining how all these different phases can be modeled to explain the molecular basis of modulation in respiratory CO2 release and related respiratory metabolism over the day and night will be very complex. However, these data do provide an initial systematic foundation for understanding the considerable daily rhythms in mitochondrial proteome composition and the changing capacity of plant leaf respiratory pathways that will be vital for building a complete metabolic picture of the molecular regulation of CO2 consumption in plants.
Supplementary Material
*This work was supported in part by the Australian Research Council (ARC) through the Centres of Excellence Program (Grant CE0561495).
 This article contains supplemental Figs. 1–3 and Tables 1–3.
This article contains supplemental Figs. 1–3 and Tables 1–3.
1 The abbreviations used are:
- GDC
- glycine decarboxylase
- 2-OGDC
- 2-oxoglutarate dehydrogenase complex
- ACON
- aconitase
- AOX
- alternative oxidase
- BCAA
- branched-chain amino acid(s)
- BCKDH
- branched-chain α-ketoacid dehydrogenase complex
- CS
- citrate synthase
- FDH
- formate dehydrogenase
- GDH
- glutamate dehydrogenase
- IDH
- NAD-dependent isocitrate dehydrogenase
- MCCA
- MCCase α-subunit
- MCCase
- 3-methylcrotonyl-CoA carboxylase
- MCCB
- MCCase β-subunit
- mMDH
- mitochondrial malate dehydrogenase
- NAD-ME
- NAD-dependent malic enzyme
- OAS-TL
- O-acetylserine thiol-lyase
- PDC
- pyruvate dehydrogenase complex
- ROS
- reactive oxygen species
- SHMT
- serine hydroxymethyltransferase
- TES
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid
- ANOVA
- analysis of variance
- ETFQO
- electron transfer flavoprotein:ubiquinone oxidoreductase.
REFERENCES
- 1.Farquhar G. D. (1989) Models of integrated photosynthesis of cells and leaves. Philos. Trans. R. Soc. Lond. B Biol. Sci. 323, 357–367 [Google Scholar]
- 2.Farquhar G. D., VonCaemmerer S. V., Berry J. A. (1980) A biochemical model of photosynthetic CO2 assimilation in leaves of C-3 species. Planta 149, 78–90 [DOI] [PubMed] [Google Scholar]
- 3.Atkin O. K., Westbeek M., Cambridge M. L., Lambers H., Pons T. L. (1997) Leaf respiration in light and darkness (a comparison of slow- and fast-growing Poa species). Plant Physiol. 113, 961–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Day D. A., De Vos O. C., Wilson D., Lambers H. (1985) Regulation of respiration in the leaves and roots of two Lolium perenne populations with contrasting mature leaf respiration rates and crop yields. Plant Physiol. 78, 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheurwater I., Dünnebacke M., Eising R., Lambers H. (2000) Respiratory costs and rate of protein turnover in the roots of a fast-growing (Dactylis glomerata L.) and a slow-growing (Festuca ovina L.) grass species. J. Exp. Bot. 51, 1089–1097 [PubMed] [Google Scholar]
- 6.Gonzalez-Meler M. A., Taneva L., Trueman R. J. (2004) Plant respiration and elevated atmospheric CO2 concentration: cellular responses and global significance. Ann. Bot. 94, 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arron G. P., Edwards G. E. (1980) Light-induced development of glycine oxidation by mitochondria from sunflower cotyledons. Plant Sci. Lett. 18, 229–235 [Google Scholar]
- 8.Walker J. L., Oliver D. J. (1986) Light-induced increases in the glycine decarboxylase multienzyme complex from pea leaf mitochondria. Arch. Biochem. Biophys. 248, 626–638 [DOI] [PubMed] [Google Scholar]
- 9.Lernmark U., Henricson D., Wigge B., Gardestrom P. (1990) Glycine oxidation in mitochondria isolated from light grown and etiolated plant tissue. Physiol. Plant. 82, 339–344 [Google Scholar]
- 10.Raghavendra A. S., Padmasree K., Saradadevi K. (1994) Interdependence of photosynthesis and respiration in plant cells—interactions between chloroplasts and mitochondria. Plant Sci. 97, 1–14 [Google Scholar]
- 11.Hoefnagel M. H., Atkin O. K., Wiskich J. T. (1998) Interdependence between chloroplasts and mitochondria in the light and the dark. Biochim. Biophys. Acta 1366, 235–255 [Google Scholar]
- 12.Journet E. P., Bligny R., Douce R. (1986) Biochemical changes during sucrose deprivation in higher plant cells. J. Biol. Chem. 261, 3193–3199 [PubMed] [Google Scholar]
- 13.Brouquisse R., Gaudillere J. P., Raymond P. (1998) Induction of a carbon-starvation-related proteolysis in whole maize plants submitted to light/dark cycles and to extended darkness. Plant Physiol. 117, 1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sweetlove L. J., Fait A., Nunes-Nesi A., Williams T., Fernie A. R. (2007) The mitochondrion: an integration point of cellular metabolism and signalling. Crit. Rev. Plant Sci. 26, 17–43 [Google Scholar]
- 15.Kromer S., Malmberg G., Gardestrom P. (1993) Mitochondrial contribution to photosynthetic metabolism. A study with barley (Hordeum vulgare L.) leaf protoplasts at different light intensities and CO2 concentrations. Plant Physiol. 102, 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atkin O. K., Evans J. R., Siebke K. (1998) Relationship between the inhibition of leaf respiration by light and enhancement of leaf dark respiration following light treatment. Aust. J. Plant Physiol. 25, 437–443 [Google Scholar]
- 17.Griffin K. L., Anderson O. R., Gastrich M. D., Lewis J. D., Lin G., Schuster W., Seemann J. R., Tissue D. T., Turnbull M. H., Whitehead D. (2001) Plant growth in elevated CO2 alters mitochondrial number and chloroplast fine structure. Proc. Natl. Acad. Sci. U.S.A. 98, 2473–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tcherkez G., Cornic G., Bligny R., Gout E., Ghashghaie J. (2005) In vivo respiratory metabolism of illuminated leaves. Plant Physiol. 138, 1596–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tcherkez G., Bligny R., Gout E., Mahé A., Hodges M., Cornic G. (2008) Respiratory metabolism of illuminated leaves depends on CO2 and O2 conditions. Proc. Natl. Acad. Sci. U.S.A. 105, 797–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Padmasree K., Padmavathi L., Raghavendra A. S. (2002) Essentiality of mitochondrial oxidative metabolism for photosynthesis: optimization of carbon assimilation and protection against photoinhibition. Crit. Rev. Biochem. Mol. Biol. 37, 71–119 [DOI] [PubMed] [Google Scholar]
- 21.Tcherkez G., Mahé A., Gauthier P., Mauve C., Gout E., Bligny R., Cornic G., Hodges M. (2009) In folio respiratory fluxomics revealed by 13C isotopic labeling and H/D isotope effects highlight the noncyclic nature of the tricarboxylic acid “cycle” in illuminated leaves. Plant Physiol. 151, 620–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner S. R., Hellens R., Ireland R., Ellis N., Rawsthorne S. (1993) The organization and expression of the gene encoding the mitochondrial glycine decarboxylase complex and serine hydroxymethyltransferase in pea (Pisum sativum). Mol. Gen. Genet. 236, 402–408 [DOI] [PubMed] [Google Scholar]
- 23.Tepperman J. M., Zhu T., Chang H. S., Wang X., Quail P. H. (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. U.S.A. 98, 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujiki Y., Sato T., Ito M., Watanabe A. (2000) Isolation and characterization of cDNA for the E1-beta and E2 subunits of the branched-chain alpha-ketoacid dehydrogenase complex in Arabidopsis. J. Biol. Chem. 275, 6007–6013 [DOI] [PubMed] [Google Scholar]
- 25.Ishizaki K., Larson T. R., Schauer N., Fernie A. R., Graham I. A., Leaver C. J. (2005) The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17, 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyashita Y., Good A. G. (2008) NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. J. Exp. Bot. 59, 667–680 [DOI] [PubMed] [Google Scholar]
- 27.Bläsing O. E., Gibon Y., Günther M., Höhne M., Morcuende R., Osuna D., Thimm O., Usadel B., Scheible W. R., Stitt M. (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17, 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elhafez D., Murcha M. W., Clifton R., Soole K. L., Day D. A., Whelan J. (2006) Characterization of mitochondrial alternative NAD(P)H dehydrogenases in Arabidopsis: intraorganelle location and expression. Plant Cell Physiol. 47, 43–54 [DOI] [PubMed] [Google Scholar]
- 29.Okada S., Brennicke A. (2006) Transcript levels in plant mitochondria show a tight homeostasis during day and night. Mol. Genet. Genomics 276, 71–78 [DOI] [PubMed] [Google Scholar]
- 30.Tovar-Méndez A., Miernyk J. A., Randall D. D. (2003) Regulation of pyruvate dehydrogenase complex activity in plant cells. Eur. J. Biochem. 270, 1043–1049 [DOI] [PubMed] [Google Scholar]
- 31.des Francs-Small C. C., Ambard-Bretteville F., Darpas A., Sallantin M., Huet J. C., Pernollet J. C., Rémy R. (1992) Variation of the polypeptide composition of mitochondria isolated from different potato tissues. Plant Physiol. 98, 273–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee C. P., Eubel H., O'Toole N., Millar A. H. (2008) Heterogeneity of the mitochondrial proteome for photosynthetic and non-photosynthetic Arabidopsis metabolism. Mol. Cell. Proteomics 7, 1297–1316 [DOI] [PubMed] [Google Scholar]
- 33.Lind C., Hallden C., Moller I. M. (1991) Protein synthesis in mitochondria purified from roots, leaves and flowers of sugar beet. Physiol. Plant. 83, 7–16 [Google Scholar]
- 34.Newton K. J., Walbot V. (1985) Maize mitochondria synthesize organ-specific polypeptides. Proc. Natl. Acad. Sci. U.S.A. 82, 6879–6883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Remy R., Ambard-Bretteville F., Colas des Francs C. (1987) Analysis by two-dimensional gel electrophoresis of the polypeptide composition of pea mitochondria isolated from different tissues. Electrophoresis 8, 528–532 [Google Scholar]
- 36.Sachlstrom S., Ericson I. (1984) Comparative electrophoretic studies of polypeptides in leaf, petiole and root mitochondria from spinach. Physiol. Plant. 61, 45–50 [Google Scholar]
- 37.Bardel J., Louwagie M., Jaquinod M., Jourdain A., Luche S., Rabilloud T., Macherel D., Garin J., Bourguignon J. (2002) A survey of the plant mitochondrial proteome in relation to development. Proteomics 2, 880–898 [DOI] [PubMed] [Google Scholar]
- 38.Mootha V. K., Bunkenborg J., Olsen J. V., Hjerrild M., Wisniewski J. R., Stahl E., Bolouri M. S., Ray H. N., Sihag S., Kamal M., Patterson N., Lander E. S., Mann M. (2003) Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115, 629–640 [DOI] [PubMed] [Google Scholar]
- 39.Forner F., Foster L. J., Campanaro S., Valle G., Mann M. (2006) Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol. Cell. Proteomics 5, 608–619 [DOI] [PubMed] [Google Scholar]
- 40.Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., Ong S. E., Walford G. A., Sugiana C., Boneh A., Chen W. K., Hill D. E., Vidal M., Evans J. G., Thorburn D. R., Carr S. A., Mootha V. K. (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Day D. A., Neuburger M., Douce R. (1985) Biochemical characterization of chlorophyll-free mitochondria from pea leaves. Aust. J. Plant Physiol. 12, 219–228 [Google Scholar]
- 42.Carrie C., Murcha M. W., Kuehn K., Duncan O., Barthet M., Smith P. M., Eubel H., Meyer E., Day D. A., Millar A. H., Whelan J. (2008) Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett. 582, 3073–3079 [DOI] [PubMed] [Google Scholar]
- 43.Eubel H., Lee C. P., Kuo J., Meyer E. H., Taylor N. L., Millar A. H. (2007) Free flow electrophoresis for purification of plant mitochondria by surface charge. Plant J. 52, 583–594 [DOI] [PubMed] [Google Scholar]
- 44.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins from silvered-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 45.Zhang Q., Wiskich J. T. (1995) Activation of glycine decarboxylase in pea leaf mitochondria by ATP. Arch. Biochem. Biophys. 320, 250–256 [DOI] [PubMed] [Google Scholar]
- 46.Taylor N. L., Heazlewood J. L., Day D. A., Millar A. H. (2004) Lipoic acid-dependent oxidative catabolism of alpha-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol. 134, 838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oliver D. J. (1981) Formate oxidation and oxygen reduction by leaf mitochondria. Plant Physiol. 68, 703–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jenner H. L., Winning B. M., Millar A. H., Tomlinson K. L., Leaver C. J., Hill S. A. (2001) NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol. 126, 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hatch M. D. (1978) A simple spectrophotometric assay for fumarate hydratase in crude tissue extracts. Anal. Biochem. 85, 271–275 [DOI] [PubMed] [Google Scholar]
- 50.Turano F. J., Dashner R., Upadhyaya A., Caldwell C. R. (1996) Purification of mitochondrial glutamate dehydrogenase from dark-grown soybean seedlings. Plant Physiol. 112, 1357–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winger A. M., Taylor N. L., Heazlewood J. L., Day D. A., Millar A. H. (2007) Identification of intra- and intermolecular disulphide bonding in the plant mitochondrial proteome by diagonal gel electrophoresis. Proteomics 7, 4158–4170 [DOI] [PubMed] [Google Scholar]
- 52.Helsens K., Martens L., Vandekerckhove J., Gevaert K. (2007) MascotDatfile: An open-source library to fully parse and analyse MASCOT MS/MS search results. Proteomics 7, 364–366 [DOI] [PubMed] [Google Scholar]
- 53.Smith S. M., Fulton D. C., Chia T., Thorneycroft D., Chapple A., Dunstan H., Hylton C., Zeeman S. C., Smith A. M. (2004) Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 136, 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gibon Y., Usadel B., Blaesing O. E., Kamlage B., Hoehne M., Trethewey R., Stitt M. (2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol. 7, R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schjoerring J. K., Mäck G., Nielsen K. H., Husted S., Suzuki A., Driscoll S., Boldt R., Bauwe H. (2006) Antisense reduction of serine hydroxymethyltransferase results in diurnal displacement of NH4+ assimilation in leaves of Solanum tuberosum. Plant J. 45, 71–82 [DOI] [PubMed] [Google Scholar]
- 56.Guilhaudis L., Simorre J. P., Blackledge M., Marion D., Gans P., Neuburger M., Douce R. (2000) Combined structural and biochemical analysis of the H-T complex in the glycine decarboxylase cycle: evidence for a destabilization mechanism of the H-protein. Biochemistry 39, 4259–4266 [DOI] [PubMed] [Google Scholar]
- 57.Douce R., Bourguignon J., Neuburger M., Rébeillé F. (2001) The glycine decarboxylase system: a fascinating complex. Trends Plant Sci. 6, 167–176 [DOI] [PubMed] [Google Scholar]
- 58.Bykova N. V., Stensballe A., Egsgaard H., Jensen O. N., Moller I. M. (2003) Phosphorylation of formate dehydrogenase in potato tuber mitochondria. J. Biol. Chem. 278, 26021–26030 [DOI] [PubMed] [Google Scholar]
- 59.Budde R. J., Randall D. D. (1990) Pea leaf mitochondrial pyruvate dehydrogenase complex is inactivated in vivo in a light-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 87, 673–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ishizaki K., Schauer N., Larson T. R., Graham I. A., Fernie A. R., Leaver C. J. (2006) The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J. 47, 751–760 [DOI] [PubMed] [Google Scholar]
- 61.Allan W. L., Shelp B. J. (2006) Fluctuations of gamma-aminobutyrate, gamma-hydroxybutyrate, and related amino acids in Arabidopsis leaves as a function of the light-dark cycle, leaf age, and N stress. Can. J. Bot. 84, 1339–1346 [Google Scholar]
- 62.Hodges M. (2002) Enzyme redundancy and the importance of 2-oxoglutarate in plant ammonium assimilation. J. Exp. Bot. 53, 905–916 [DOI] [PubMed] [Google Scholar]
- 63.Tronconi M. A., Fahnenstich H., Gerrard Weehler M. C., Andreo C. S., Flügge U. I., Drincovich M. F., Maurino V. G. (2008) Arabidopsis NAD-malic enzyme functions as a homodimer and heterodimer and has a major impact on nocturnal metabolism. Plant Physiol. 146, 1540–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Svensson A. S., Rasmusson A. G. (2001) Light-dependent gene expression for proteins in the respiratory chain of potato leaves. Plant J. 28, 73–82 [DOI] [PubMed] [Google Scholar]
- 65.Millar A. H., Sweetlove L. J., Giegé P., Leaver C. J. (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 127, 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- 66.Kruft V., Eubel H., Jänsch L., Werhahn W., Braun H. P. (2001) Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 127, 1694–1710 [PMC free article] [PubMed] [Google Scholar]
- 67.Heazlewood J. L., Howell K. A., Whelan J., Millar A. H. (2003) Towards an analysis of the rice mitochondrial proteome. Plant Physiol. 132, 230–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heazlewood J. L., Tonti-Filippini J. S., Gout A. M., Day D. A., Whelan J., Millar A. H. (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16, 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang S., Taylor N. L., Narsai R., Eubel H., Whelan J., Millar A. H. (2009) Experimental analysis of the rice mitochondrial proteome, its biogenesis, and heterogeneity. Plant Physiol. 149, 719–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sweetlove L. J., Heazlewood J. L., Herald V., Holtzapffel R., Day D. A., Leaver C. J., Millar A. H. (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 32, 891–904 [DOI] [PubMed] [Google Scholar]
- 71.Taylor N. L., Heazlewood J. L., Day D. A., Millar A. H. (2005) Differential impact of environmental stresses on the pea mitochondrial proteome. Mol. Cell. Proteomics 4, 1122–1133 [DOI] [PubMed] [Google Scholar]
- 72.Krömer S., Heldt H. W. (1991) On the role of mitochondrial oxidative phosphorylation in photosynthesis metabolism as studied by the effect of oligomycin on photosynthesis in protoplasts and leaves of barley (Hordeum vulgare). Plant Physiol. 95, 1270–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kromer S. (1995) Respiration during photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 45–70 [Google Scholar]
- 74.Raghavendra A. S., Padmasree K. (2003) Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 8, 546–553 [DOI] [PubMed] [Google Scholar]
- 75.Taylor N. L., Day D. A., Millar A. H. (2002) Environmental stress causes oxidative damage to plant mitochondria leading to inhibition of glycine decarboxylase. J. Biol. Chem. 277, 42663–42668 [DOI] [PubMed] [Google Scholar]
- 76.di Salvo M. L., Delle Fratte S., Maras B., Bossa F., Wright H. T., Schirch V. (1999) Deamidation of asparagine residues in a recombinant serine hydroxymethyltransferase. Arch. Biochem. Biophys. 372, 271–279 [DOI] [PubMed] [Google Scholar]
- 77.Kim S. Y., Marekov L., Bubber P., Browne S. E., Stavrovskaya I., Lee J., Steinert P. M., Blass J. P., Beal M. F., Gibson G. E., Cooper A. J. (2005) Mitochondrial aconitase is a transglutaminase 2 substrate: transglutamination is a probable mechanism contributing to high-molecular-weight aggregates of aconitase and loss of aconitase activity in Huntington disease brain. Neurochem. Res. 30, 1245–1255 [DOI] [PubMed] [Google Scholar]
- 78.Lesort M., Tucholski J., Zhang J., Johnson G. V. (2000) Impaired mitochondrial function results in increased tissue transglutaminase activity in situ. J. Neurochem. 75, 1951–1961 [DOI] [PubMed] [Google Scholar]
- 79.Koivuniemi A., Aro E. M., Andersson B. (1995) Degradation of the D1- and D2-proteins of photosystem II in higher plants is regulated by reversible phosphorylation. Biochemistry 34, 16022–16029 [DOI] [PubMed] [Google Scholar]
- 80.Tikkanen M., Nurmi M., Kangasjärvi S., Aro E. M. (2008) Core protein phosphorylation facilitates the repair of photodamaged photosystem II at high light. Biochim. Biophys. Acta 1777, 1432–1437 [DOI] [PubMed] [Google Scholar]
- 81.Jabben M., Shanklin J., Vierstra R. D. (1989) Red light-induced accumulation of ubiquitin-phytochrome conjugates in both monocots and dicots. Plant Physiol. 90, 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kristensen B. K., Askerlund P., Bykova N. V., Egsgaard H., Møller I. M. (2004) Identification of oxidised proteins in the matrix of rice leaf mitochondria by immunoprecipitation and two-dimensional liquid chromatography-tandem mass spectrometry. Phytochemistry 65, 1839–1851 [DOI] [PubMed] [Google Scholar]
- 83.Foyer C. H., Noctor G. (2003) Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol. Plant. 119, 355–364 [Google Scholar]
- 84.Dutilleul C., Garmier M., Noctor G., Mathieu C., Chétrit P., Foyer C. H., de Paepe R. (2003) Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15, 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim C., Meskauskiene R., Apel K., Laloi C. (2008) No single way to understand singlet oxygen signalling in plants. EMBO Rep. 9, 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moller I. M. (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 569–591 [DOI] [PubMed] [Google Scholar]
- 87.Heeg C., Kruse C., Jost R., Gutensohn M., Ruppert T., Wirtz M., Hell R. (2008) Analysis of the Arabidopsis O-acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. Plant Cell 20, 168–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Finkemeier I., Goodman M., Lamkemeyer P., Kandlbinder A., Sweetlove L. J., Dietz K. J. (2005) The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J. Biol. Chem. 280, 12168–12180 [DOI] [PubMed] [Google Scholar]
- 89.Chew O., Whelan J., Millar A. H. (2003) Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 278, 46869–46877 [DOI] [PubMed] [Google Scholar]
- 90.Bartoli C. G., Yu J., Gómez F., Fernández L., McIntosh L., Foyer C. H. (2006) Inter-relationships between light and respiration in the control of ascorbic acid synthesis and accumulation in Arabidopsis thaliana leaves. J. Exp. Bot. 57, 1621–1631 [DOI] [PubMed] [Google Scholar]
- 91.Hourton-Cabassa C., Ambard-Bretteville F., Moreau F., Davy de Virville J., Rémy R., Francs-Small C. C. (1998) Stress induction of mitochondrial formate dehydrogenase in potato leaves. Plant Physiol. 116, 627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rhoads D. M., Umbach A. L., Subbaiah C. C., Siedow J. N. (2006) Mitochondrial reactive oxygen species. Contribution to oxidative stress and interorganellar signaling. Plant Physiol. 141, 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moller I. M., Kristensen B. K. (2004) Protein oxidation in plant mitochondria as a stress indicator. Photochem. Photobiol. Sci. 3, 730–735 [DOI] [PubMed] [Google Scholar]
- 94.Laisk A., Kiirats O., Oja V. (1984) Assimilatory power (postillumination CO2 uptake) in leaves—measurement, environmental dependencies, and kinetic properties. Plant Physiol. 76, 723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hanning I., Heldt H. W. (1993) On the function of mitochondrial metabolism during photosynthesis in spinach (Spinacia oleracea L.) leaves. (Partitioning between respiration and export of redox equivalents and precursors for nitrate assimilation products). Plant Physiol. 103, 1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carrari F., Nunes-Nesi A., Gibon Y., Lytovchenko A., Loureiro M. E., Fernie A. R. (2003) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol. 133, 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Che P., Wurtele E. S., Nikolau B. J. (2002) Metabolic and environmental regulation of 3-methylcrotonyl-coenzyme A carboxylase expression in Arabidopsis. Plant Physiol. 129, 625–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bouché N., Fait A., Bouchez D., Møller S. G., Fromm H. (2003) Mitochondrial succinic-semialdehyde dehydrogenase of the gamma-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proc. Natl. Acad. Sci. U.S.A. 100, 6843–6848 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.