Summary
Nedd4 family interacting protein-1 (Ndfip1) is a protein whose only known function is that it binds Nedd4, a HECT-type E3 ubiquitin ligase. Here we show that mice lacking Ndfip1 developed severe inflammation of the skin and lung and died prematurely. This condition was due to a defect in Ndfip1−/− T cells. Ndfip1−/− T cells were activated, and they proliferated and adopted a T helper 2 (Th2) phenotype more readily than did their Ndfip1+/+ counterparts. This phenotype resembled that of Itchy mutant mice, suggesting that Ndfip1 might affect the function of Itch, an E3 ubiquitin ligase. We show that T cell activation promoted both Ndfip1 expression and its association with Itch. In the absence of Ndfip1, JunB half-life was prolonged after T cell activation. Thus, in the absence of Ndfip1, Itch is inactive and JunB accumulates. As a result, T cells produce Th2 cytokines and promote Th2-mediated inflammatory disease.
Introduction
Ubiquitin was first discovered nearly 30 years ago as a lymphocyte differentiation-promoting factor (Goldstein et al., 1975). Since then, accumulating evidence suggests that, among other functions, ubiquitin ligation is used to regulate both innate and adaptive immune responses (Coscoy and Ganem, 2003; Heissmeyer et al., 2004; Jeon et al., 2004; Liu et al., 2005; Uchida et al., 2004). Although hundreds of proteins have been identified that act directly as enzymes in the ubiquitination process, regulation of these proteins is not well understood.
Protein ubiquitination is a highly ordered process, the net result of which is the covalent binding of one or more ubiquitin moieties to a protein substrate (Liu, 2004). Ubiquitin conjugation can have one of several consequences for the protein, targeting it for degradation, changing its subcellular location, or altering its activation status. Among the proteins responsible for these complex series of events, the E3 ubiquitin ligases are key in determining which proteins are targeted. E3 ubiquitin ligases are classified into three families based on their structures: the homology to the E6-associated protein carboxyl terminus (HECT) domain-containing E3 ubiquitin ligases (Huibregtse et al., 1995), the really interesting new gene (RING) domain E3 ubiquitin ligases (Freemont, 2000), and the U-box E3 ubiquitin ligases (Hatakeyama et al., 2001).
HECT-type E3 ubiquitin ligases have several shared features. As their name implies, they all have HECT domains that facilitate the transfer of ubiquitin to the substrate (Huibregtse et al., 1995). Additionally, HECT-type E3 ubiquitin ligases all have multiple WW domains that mediate protein-protein interactions. WW domains have been shown to bind proline-rich domains (Pirozzi et al., 1997) and phosphoserine and phosphothreonine residues (Lu et al., 1999; Staub et al., 1996; Sudol, 1996) and thus support a stable interaction with binding partners.
The most well-characterized WW domain-containing HECT-type E3 ubiquitin ligases are Nedd4-1 and Nedd4-2. Nedd4-2 has been shown to regulate epithelial Na+ channels in the kidney and other tissues (Abriel et al., 2000; Dinudom et al., 1998; Harvey et al., 1999; Staub et al., 1996, 1997). Nedd4-1 has been implicated in lymphocyte function and tolerance (Heissmeyer et al., 2004, 2005; Magnifico et al., 2003; Scharschmidt et al., 2004). Because of their potential importance, several groups have focused on identifying targets of Nedd4 ubiquitin ligase activity. These studies used the WW domains of either Nedd4-1 or Nedd4-2 to identify associated proteins (Ingham et al., 2005; Jolliffe et al., 2000; Kurakin and Bredesen, 2002; Murillas et al., 2002).
Nedd4 family interacting protein 1 (Ndfip1, N4WBP5) was recently identified by one of these screens because of its ability to bind the WW domains of Nedd4-1 (Jolliffe et al., 2000). When overexpressed in COS cells, Ndfip1 was also shown to interact with the WW domains of other HECT-type family members, including K1AA0332, WWP2, AIP-4, and Itch (Harvey et al., 2002). Whether and under what circumstances any of these proteins interact in vivo is not known. Furthermore, the biologic outcome of such interactions is also unknown except that it leads to the ubiquitination of Ndfip1 (Harvey et al., 2002). This observation might suggest that Ndfip1 is simply a target of Nedd4’s ubiquitination activity. However, data from yeast suggest that Ndfip1 may have broader effects on E3 ubiquitin ligase activities.
In yeast, the Ndfip1 ortholog Bsd2 acts as an adaptor, linking the yeast Nedd4 ortholog Rsp5 to its substrates (Hettema et al., 2004). In Bsd2 mutant strains, several targets of Rsp5 were not properly degraded. This finding led the authors to propose that Bsd2 plays an important role in the recognition and removal of misfolded membrane proteins by Rsp5 (Hettema et al., 2004).
To better understand the interaction between Ndfip1 and the HECT-type E3 ligases, we generated mice lacking Ndfip1. Ndfip1−/− mice develop a severe inflammatory disease at 6 weeks of age, and few animals survive beyond 12 weeks of age. This phenotype is, at least in part, due to a defect in T cells that, in vivo, results in their spontaneous activation. Based on data we present here, we propose that Ndfip1 binds Itch and promotes Itch function. Association between Ndfip1 and Itch facilitates the degradation of a well-described Itch target, JunB, and prevents JunB from driving T helper 2 (Th2) cytokine production.
Results
The Generation of Mice that Are Homozygous for a Disrupted Ndfip1 Locus
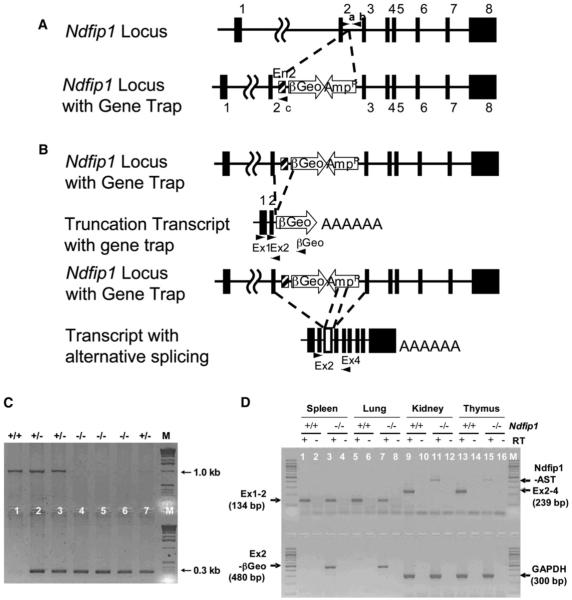
ES cells harboring a disruption of the Ndfip1 gene were obtained from BayGenomics (cell line code RRD002). The targeted ES cells contain a gene-trapping vector that was inserted within intron 2 of the gene encoding Ndfip1 (Stryke et al., 2003). The gene trap vector is composed of an artificial intron (En2), a splice acceptor site, and a βGeo cassette (Figure 1A). This disruption of the Ndfip1 gene results in a truncation of the mRNA transcript just beyond exon 2 (Figure 1B). To confirm the presence of the gene trap vector, ES cells were tested by PCR. PCR with primers “a” and “b” (Figure 1A) produces the 1.0 kb bp band, indicating the presence of the wild-type locus. In contrast, PCR with primers “a” and “c” yielded a band of 0.3 bp, indicating disruption of the Ndfip1 locus. ES cells carrying this mutation were injected into mouse blastocysts to generate chimeras as described previously (McDonald et al., 1999). Two male chimeras transmitted to the germline. The resulting agouti progeny were tested for the presence of the disrupted Ndfip1 allele by PCR (data not shown).
Figure 1.
The Generation of Mice Homozygous for a Disrupted Ndfip1 Gene
(A) The location of the gene trap vector within the Ndfip1 locus is shown. The Ndfip1 locus contains 8 exons spanning about 45 Kb of genomic DNA.
(B) The gene trap vector was inserted into the second intron and results in the production of a truncated Ndfip1 mRNA that encodes exons 1 and 2.
(C) Offspring resulting from the intercross between Ndfip1+/− mice were tested by PCR to identify Ndfip1−/− animals via the primer pairs shown in Figure 1A.
(D) To confirm the disruption of Ndfip1, RNA was isolated from Ndfip1+/+ and Ndfip1−/− mice and RT-PCR was performed. We detect the presence of two transcripts in Ndfip1−/− tissues, Ndfip1-AST and Ex2-βGEO. GADPH is shown as a reverse transcriptase and loading control.
Mice heterozygous for the disrupted locus were inter-crossed to produce homozygous Ndfip1−/− animals. The PCR protocol described above was used to genotype the resulting progeny (Figure 1C). Once identified, homozygous mice were tested by RT-PCR to see whether they expressed any full-length Ndfip1 mRNA (Figure 1D). These data show that two kinds of transcripts were produced in Ndfip1−/− tissues. One of them (EX2-βGeo) was a truncated transcript that consisted of exons 1 and 2 and βGeo. The second one (Ndfip1-AST), based on mRNA sequencing, was an alternatively spliced transcript consisting of the full-length Ndfip1 with 206 bp from the ampicillin resistance gene inserted in the reverse orientation between exons 2 and 3 (data not shown). The βGeo was not included in this transcript. This Amp fragment introduced a translation stop site in each of the three possible reading frames. Taken together, these data suggest that insertion of the gene trap vector into the Ndfip1 locus results in a disruption of the Ndfip1 gene.
Mice Lacking Ndfip1 Develop Spontaneous Inflammation of the Skin and Die Prematurely
Ndfip1−/− mice appeared normal at birth. Furthermore, the number of Ndfip1−/− mice produced from inter-crosses of Ndfip1+/− animals conformed, for the most part, to normal Mendelian expectations (see Table S1 in the Supplemental Data available online). At 6 weeks, Ndfip1−/− began to develop skin lesions on their ears (data not shown), and by 8 weeks of age, all Ndfip1−/− mice had these lesions. Gross inspection of the mice revealed a profound hepatomegally and splenomegally. Organ size was increased from a liver to body weight ratio of 48 ± 4 mg/g for Ndfip1+/+ animals to 101 ± 11 mg/g for Ndfip1−/− mice (p < 0.008) and from a spleen to body weight ratio of 3.4 ± 0.5 mg/g for Ndfip1+/+ mice to 16.9 ± 2.7 mg/g for Ndfip1−/− animals (p < 0.003). Additionally, over time, the tails of Ndfip1−/− became segmented in appearance and tended to be shorter then the tails of their Ndfip1+/+ littermates (data not shown).
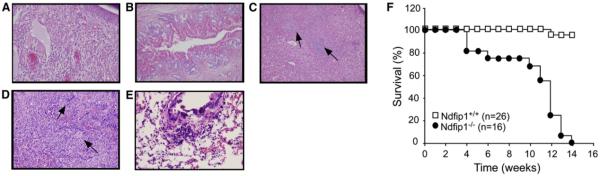
In an effort to determine the underlying cause of the increased spleen and liver size and inflammation of the ear, tissue sections were examined. Hematoxylin and eosin (H&E) staining of paraffin-embedded sections of organs from Ndfip1−/− mice revealed multiple defects. Ear sections revealed a high degree of inflammation with a predominantly eosinophilic and lymphocytic infiltrate (Figure 2A). The liver contained intrahepatic bile ducts (Figure 2B). Apparent in both liver and spleen were extensive foci of extramedullary hematopoiesis (Figures 2C and 2D). Lungs of Ndfip1−/− mice also displayed signs of inflammation with goblet cell hyperplasia and an inflammatory infiltrate in the perivascular regions (Figure 2E). Kidneys in the Ndfip1−/− mice appeared normal (data not shown).
Figure 2.
Loss of Ndfip1 Leads to Inflammatory Disease, Splenomegally, Hepatomegally, and Extramedullary Hematopoiesis of the Spleen and Liver
(A–D) H&E-stained tissue sections from 8-week-old Ndfip1−/− ear (A), liver (B and C), and spleen (D).
(E) PAS-stained sections of Ndfip1−/− lung.
(F) The number of live Ndfip1−/− and Ndfip1+/+ mice was measured over time.
Because the phenotype showed both an alteration in hematopoiesis and was inflammatory in nature, we characterized hematopoietic-derived cells of primary and secondary lymphoid organs by flow cytometry. We found that Ndfip1−/− mice had fewer B cells (B220+) and more myeloid lineage cells (GR1+) in their bone marrow as compared to age-matched Ndfip1+/+ animals, whereas pre-erythroid cells (ter119+) were equal in number (Figure S1A). The spleens of mice lacking Ndfip1 also showed elevated numbers of myeloid lineage cells and, in keeping with histological evidence of splenic hematopoiesis, pre-erythroid cells (Figure S1B). T cells in Ndfip1−/− animals, particularly those that expressed CD4, were increased in number and were activated as shown by their increased expression of CD44.
Although some Ndfip1−/− mice died soon after weaning, many of the mice survived longer (Figure 2F). The persistent inflammation of the ear resulted in destruction of much of the ear tissue, and once inflammation was established, mice began to appear cachectic. Beginning at 10 weeks, there was a dramatic decrease in the survival of Ndfip1−/− mice, and none of these mice survived beyond 14 weeks of age.
The Ndfip−/− Inflammatory Phenotype Is Due to a Defect in Cells of the Hematopoietic Lineage
Flow cytometric analysis revealed multiple changes in cells from the hematopoietic lineage; however, these changes either could have been due to a primary defect caused by the loss of Ndfip1 or could have been caused by inflammation. To find out whether Ndfip1 deficiency causes a defect in bone marrow-derived cells that initiates inflammation, we transferred Ndfip1−/− or Ndfip1+/+ bone marrow cells into lethally irradiated C57BL/6 recipients and monitored the mice for signs of inflammation.
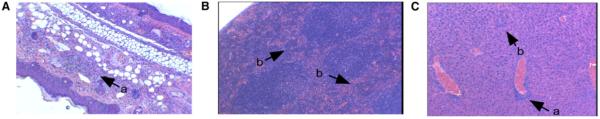
Mice receiving Ndfip1−/− cells, but not those that were reconstituted with Ndfip1+/+ cells, developed skin lesions beginning approximately 5 weeks postreconstitution and, like Ndfip1−/− mice, died within 8 weeks of the onset of inflammation. Recipients of Ndfip1−/− bone marrow cells also developed splenomegaly and hepatomegaly (data not shown). Again, the inflammatory infiltrate in the skin was predominantly lymphocytic and eosinophilic (Figure 3A), and extramedullary hematopoiesis was observed in the enlarged spleen and liver (Figures 3B and 3C). However, some of the characteristics of the Ndfip1−/− mice were less severe or not recapitulated in the bone marrow chimeras. Splenomegally and hepatomegally was less pronounced in the chimeras, and the reconstituted mice did not develop a segmented tail (data not shown) or intrahepatic bile ducts (Figure 3C). Thus, nonhematopoietic cells are required for these phenotypes in the Ndfip1−/− mice.
Figure 3.
Reconstitution of Wild-Type Mice with Ndfip1−/− Marrow Leads to Inflammatory Disease
C57BL6 mice were lethally irradiated and then reconstituted with Ndfip1−/− bone marrow cells. The resulting mice developed skin lesions between 6 and 8 weeks of age.
(A) The skin lesions showed an infiltration of inflammatory cells.
(B and C) Histological findings in spleen (B) and liver (C) were similar to those seen in Ndfip1−/− mice. a, inflammation; b, extramedullary hematopoiesis.
However, these data show that bone marrow-derived cells are responsible for the inflammatory disease and premature deaths observed in Ndfip1−/− mice.
T Cells Lacking Ndfip1 Are Increased in Number and Are Activated
To find out which bone marrow-derived cells were responsible for promoting inflammation in the Ndfip1−/− mice, we reconstituted lethally irradiated mice with a mixture of equal numbers of GFP+ (Schaefer et al., 2001b) Ndfip1+/+ and GFP− Ndfip1−/− bone marrow. This experimental design allowed study of Ndfip1+/+ and Ndfip1−/− cells exposed to the same inflammatory conditions. Additionally, this experiment allowed us to distinguish between primary events (which would occur only in the Ndfip1−/− cells) and secondary events (which would affect both Ndfip1−/− and Ndfip1+/+ cells).
Mice reconstituted with Ndfip1−/− bone marrow developed disease approximately 6 weeks after bone marrow transfer (see above), and therefore we chose to analyze the mixed chimeras between 5 and 6 weeks after reconstitution. This time frame allowed us to see changes in immune system cells that preceded any outward signs of inflammation. Because GFP expression in the Ndfip1+/+ cells limited the fluorimeter channels available to characterize the cells, we sorted live GFP+ and live GFP− cells from each tissue and stained cells with the various antibodies, including some that would register in the same channel as GFP. We then treated each sample with saponin, thereby releasing all of the GFP from the cells, which allowed us to detect antibody staining that would otherwise be obscured.
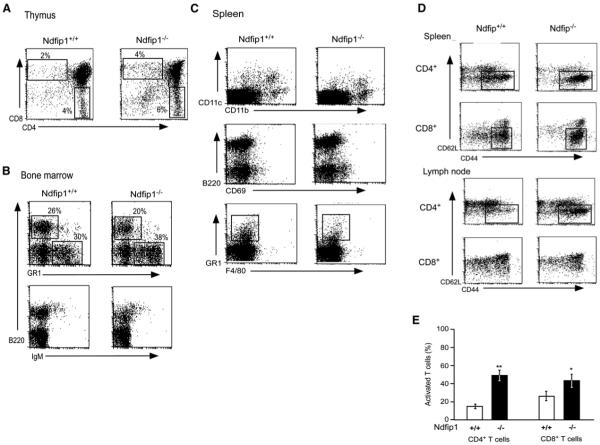
With this technique, data were collected from six mice, and in each case, Ndfip1+/+ cells were compared to Ndfip1−/− cells isolated from the same mouse. The percentages of the Ndfip1+/+ and Ndfip1−/− cells in the thymuses and bone marrow of the mixed chimeras were similar (Figures 4A and 4B). Likewise, in the spleens of the chimeras, the numbers of macrophages (CD11b+), a subset of dendritic cells (CD11b+ and CD11c+), and B cells of both origins were comparable and there was no evidence that B cells of either type were abnormally activated, as tested by expression of CD69 (Figure 4C). However, a greater percentage of the Ndfip1−/− cells in spleens (and lymph nodes, data not shown) were T cells (15.5% ± 3% of Ndfip1−/− cells versus 9.2% ± 1.8% of Ndfip1+/+ cells). In addition, many more of the Ndfip1−/− cells were activated, as defined by increased expression of CD44 and Ly6c (data not shown). This difference was true for both CD4+ and CD8+ T cells (Figures 4D and 4E). Thus, T cells lacking Ndfip1 were activated prior to any outward signs of inflammation.
Figure 4.
Mixed Bone Marrow Chimeras Reveal a Profound Defect in Ndfip1−/− T Cells
C57BL/6 mice were lethally irradiated and reconstituted with a mixture of equal numbers of Ndfip1−/− and Ndfip1+/+ (GFP-expressing) bone marrow. Primary and secondary lymphoid organs of the resulting mice were analyzed 5–6 weeks after the bone marrow transfer. Cells were sorted based on whether or not they expressed GFP, stained, and then treated with saponin to allow release of GFP from the cells.
(A) Cells were isolated from the thymus and stained for CD4 and CD8 single-positive populations.
(B) Cells from the bone marrow were stained for B cell markers (B220 and IgM) or myeloid markers (GR1).
(C) Cells were isolated from the spleen and stained for B cell (B220), dendritic cell (CD11b and CD11c), or myeloid (GR1 and F4/80) markers or for markers indicating activation (CD69).
(D) Spleen cells were stained for T cell-specific markers (CD4 and CD8) or markers indicating activation status (CD62L and CD44).
(E) The percent of activated cells among the different populations in both the spleen and lymph nodes were quantified. Open bars represent Ndfip1+/+ T cells, whereas the black bars represent Ndfip1−/− T cells. Error bars depict standard errors (n = 8). Both CD4+ and CD8+ T cells lacking Ndfip1 were significantly more likely to show an activated phenotype. *p < 0.05; **p < 0.01 (paired t test).
To ensure that the T cell defects were not due to differences in the genetic background between the Ndfip1−/− (C57BL/6 and 129.ola) and GFP Tg. mice (C57BL/6), we compared these mixed chimeras to those made with a mixture of cells from GFP Tg. mice and Ndfip1+/− mice. The Ndfip1+/− mice have a mixed genetic background that is similar to that of the Ndfip1−/− mice. Comparison of these two sets of mixed chimeras revealed that the T cell defect was a direct consequence of loss of Ndfip1 (Figure S2).
Based on these data, we could not rule out the possibility that cells other then T cells are affected by the loss of Ndfip1. However, our data clearly indicate that T cells lacking Ndfip1 are activated before any outward signs of disease and that this activation is intrinsic to the mutant T cell.
Ndfip1−/− T Cells Proliferate More and Readily Produce IL-4 In Vitro
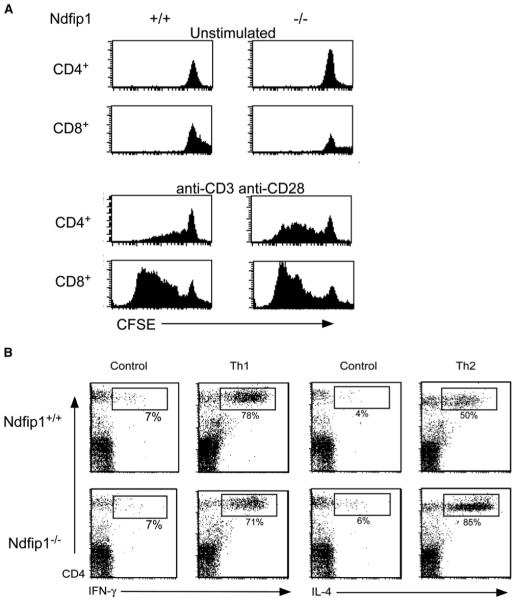
Inflammatory disorders of the skin, particularly those with eosinophilic involvement, are often potentiated by Th2 CD4+ T cells (Del Prete, 1992; Ricci et al., 1994; Romagnani et al., 1991). Accordingly, we tested whether Ndfip1−/− T cells were capable of responding effectively to TCR-mediated signals that lead to proliferation and/or the production of the Th2 cytokine, IL-4, or the Th1 cytokine, IFN-γ. We again used T cells isolated from mixed chimera mice to ensure that the T cells were exposed to the same environment prior to analysis. T cells from the mixed chimeras were sorted for GFP expression, labeled with CFSE, and cultured for 3 days in the presence or absence of the TCR-stimulating reagents, anti-CD3 and anti-CD28. We then stained cells with antibodies against CD4 and CD8 and treated the cells with saponin to remove GFP.
Unstimulated cells did not divide regardless of Ndfip1 expression, demonstrating that Ndfip1−/− cells were still dependent on TCR stimulation to divide. On the other hand, when cells were stimulated, Ndfip1−/− CD4+ T cells proliferated more readily than wild-type cells (Figure 5A). These data imply that Ndfip1 might affect how T cells respond to activation signals.
Figure 5.
T Cells Lacking Ndfip1 Are More Likely to Proliferate and Produce Th2 Cytokines
(A) Spleen cells from mixed chimeras, 5–6 weeks posttransfer, were sorted based on GFP expression, labeled with CFSE, and stimulated with anti-CD3 (plate-bound) and anti-CD28, or left unstimulated. After 3 days, cells were stained, treated with saponin, and analyzed for loss of CFSE.
(B) Spleen and lymph node cells from 5- to 6-week-old Ndfip1−/− and Ndfip1+/+ mice were isolated and depleted of CD44+ cells and CD8+ cells. Cells were then stimulated with anti-CD3 and anti-CD28 or stimulated in the presence of Th1 or Th2 differentiating conditions. The percentages represent cytokine-producing cells as a percent of the total CD4+ cells. Data compiled from three independent experiments show that the percent of IL-4 producers averaged 39.8% ± 5.5% for Ndfip1+/+ and 72.9% ± 8.1% for Ndfip1−/− T cells; p < 0.04 paired Student’s t test. IFN-γ production was not statistically different between the two genotypes.
We then wanted to see whether Ndfip1−/− T cells were capable of producing cytokines after culture in Th1 or Th2-polarizing conditions. T cells were isolated from the spleens of 5- to 6-week-old Ndfip1+/+ and Ndfip1−/− mice, and activated T cells (CD44+) were depleted from each sample. Cells were then cultured for 6 days under either Th1- or Th2-polarizing conditions or activated in the absence of cytokine polarization.
When cells were activated in the absence of polarizing conditions (control), neither type of cell produced much IL-4 or IFN-γ (Figure 5B). Furthermore, when cells were cultured under Th1-polarizing conditions, Ndfip1−/− T cells were no more likely to produce IFN-γ than control cells. In contrast, when cells were cultured in Th2-polarizing conditions, Ndfip1−/− T cells were much more likely to make IL-4. These data support the hypothesis that loss of Ndfip1 biases T cells toward a Th2 phenotype and might help to explain why mice lacking Ndfip1 are prone to develop an inflammatory condition with high numbers of infiltrating eosinophils.
Ndfip1−/− T Cells Are Much More Likely to Drive a Th2 Response In Vivo
The presence of eosinophils at the inflammatory sites suggests that Ndfip1−/− mice develop a Th2-mediated disease. Knowing that loss of Ndfip1 led to a defect in T cells suggested to us that these T cells might drive disease because of an uncontrolled bias toward production of Th2 cytokines. Thus, we wished to test whether Ndfip1−/− T cells were Th2 biased in vivo and whether this bias resulted in increased Th2-dependent immunoglobulin switching.
For this experiment, we made bone marrow chimera mice to study a large number of animals that were healthy at the time of immunization. We immunized the mice with ovalbumin (OVA) mixed with an adjuvant that induces either a Th2-polarized response (Alum) or a Th1-polarized response (complete Freund’s adjuvant, CFA). Mice reconstituted with Ndfip1−/− bone marrow typically began to show signs of inflammation 6 weeks after the transfer of bone marrow, and their condition worsened over the next 4-6 weeks. We found that when these same mice were immunized with Alum 5 weeks after reconstitution, they became very sick within 8 days such that the experiment had to be terminated prematurely. Chimeras made from Ndfip1+/+ bone marrow remained healthy even after they were immunized with OVA + Alum and they showed no signs of inflammation in either their skin or lung (Figures S2A and S2B). In contrast, chimeras made from Ndfip1−/− bone marrow that received OVA + Alum had visible lesions on their skin (data not shown) and inflammation in the skin and lungs (Figures S2C and S2D).
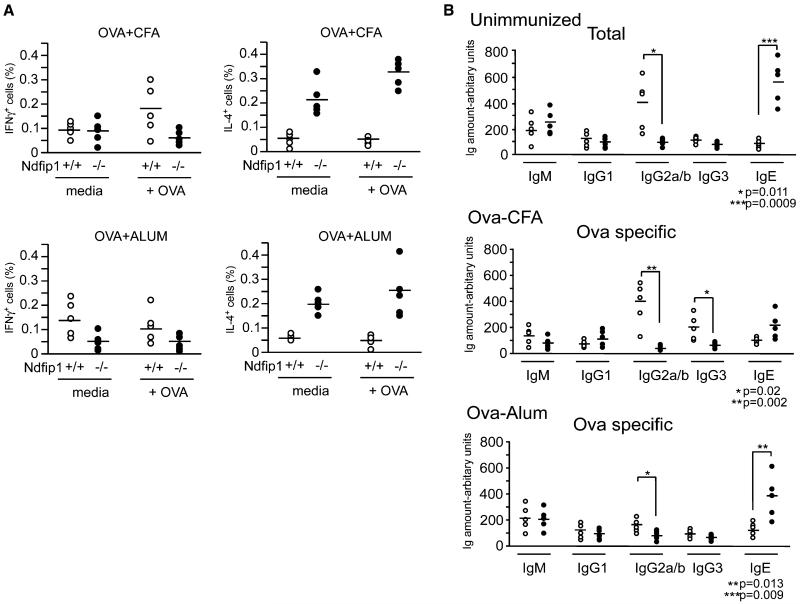
T cells from all groups were analyzed 8 days after antigen stimulation for cytokine production. Very few T cells from Ndfip1+/+ bone marrow chimeras made IL-4 in response to in vitro challenge with antigen (Figure 6A). This was true regardless of the adjuvant used and is probably due to the short duration of the experiment. In contrast, the Ndfip1+/+ cells were able to produce IFN-γ in response to ex vivo antigen exposure but only if they had come from animals immunized with OVA + CFA, in line with the known Th1-polarizing capacity of this adjuvant. In contrast to these results, T cells lacking Ndfip1 were consistently less likely to produce IFN-γ as compared to wild-type cells and instead produced IL-4. This was true regardless of the adjuvant used to prime the animal and occurred even in the absence of antigen challenge in vitro.
Figure 6.
Immunization of Ndfip1−/− Mice with OVA+CFA or OVA+ALUM Leads to IL-4 Production and Increased Antigen-Specific IgE Ndfip1+/+ and Ndfip1−/− bone marrow chimeras were immunized with OVA+CFA or OVA+ALUM 5 weeks after reconstitution.
(A) 8 days later, cells were isolated from spleens and lymph nodes and cultured in media alone or in the presence of OVA peptide (OVA323-339). Effector CD4+ T cells (CD4+, CD44hi, CD62Llo) were then analyzed for intracellular IL-4 or IFN-γ.
(B) Sera were collected from the mice 8 days after immunization and analyzed for total and OVA-specific immunoglobulin levels by ELISA. Each circle represents the percent of (A) CD44+ T cells producing cytokine or (B) the Ig amounts from an individual mouse. Open circles indicate that the chimera was Ndfip1+/+, whereas closed circles represent Ndfip1−/− chimeras.
To find out whether the Th2 bias of Ndfip1−/− T cells were reflected in the antibody isotypes generated in mice containing these cells, we measured the amounts of various immunoglobulin isotypes in unimmunized chimeras and the isotypes of ovalbumin antibodies in the immunized animals (Figure 6B). In comparison with Ndfip1+/+ chimeras, Ndfip1−/− chimeras contained high amounts of the Th2-dependent immunoglobulin class IgE and low amounts of the Th1-dependent isotypes IgG2a/b. Similar results were observed for ovalbumin antibodies after immunization with OVA + CFA or OVA + Alum, with amounts of IgE higher, and levels of IgG2a/b and IgG3 lower in the Ndfip1−/− chimeras than the Ndfip1+/+ mice.
These results support the conclusion that lack of Ndfip1 predisposes T cells toward a Th2 phenotype, regardless of the conditions under which they are activated.
Ndfip1 Binds Itch after T Cell Stimulation and Promotes Itch Function
The phenotype of the Ndfip1−/− mice is reminiscent of that described for the Itchy mice that lack functional Itch protein (Hustad et al., 1995; Perry et al., 1998). It has been shown that Itch ubiquitinates Jun proteins (Gallagher et al., 2006; Gao et al., 2004) and Jun protein amounts are increased in Itchy animals (Fang et al., 2002). Jun proteins promote IL-4 synthesis (Hartenstein et al., 2002; Li et al., 1999) and thus could cause T cells to become Th2 biased.
Given that Ndfip1−/− and Itch mutant mice have a similar phenotype and because a WW domain portion of Itch has been shown to bind Ndfip1 in vitro (Harvey et al., 2002), we postulated that Ndfip1 might regulate Itch. To test this idea, we first needed to know whether Ndfip1 protein is expressed in T cells and whether its expression affects Itch expression.
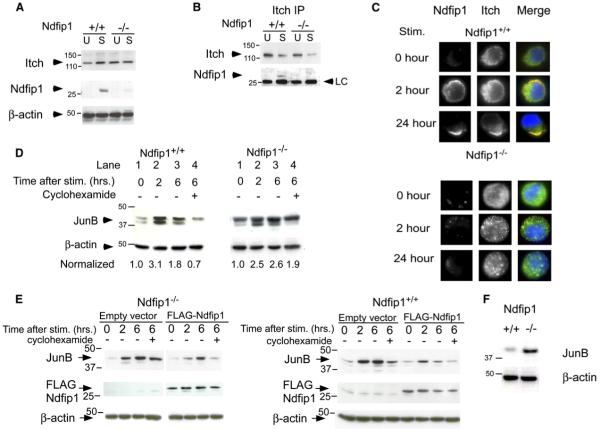
T cells were isolated from 6- to 8-week-old Ndfip+/+ and Ndfip1−/− mice, the cells were cultured in media or stimulated for 24 hr, and their whole-cell lysates were analyzed by immunoblot for expression of Itch and Ndfip1. Ndfip1+/+ and Ndfip1−/− T cells contained equivalent amounts of Itch, indicating that expression of Ndfip1 does not regulate Itch expression in T cells.
Unstimulated T cells expressed negligible amounts of Ndfip1 protein. After 2 hr of stimulation, Ndfip1 protein increased in amount (Figure 7A), suggesting that Ndfip1 function may be particularly relevant in activated T cells.
Figure 7.
Ndfip1 Interacts with Itch and Promotes Degradation of JunB
(A) T cells were isolated from the spleens and lymph nodes of Ndfip1+/+ and Ndfip1−/− mice and cultured in media alone or stimulated 24 hr with anti-CD3 and anti-CD28. Cell lysates were immunoblotted with antibodies against Itch, Ndfip1, or β-actin.
(B) Ndfip1+/+ and Ndfip1−/− T cells were lysed directly after isolation (U) or cultured in the presence of anti-CD3 and anti-CD28 (S). Lysates were immunoprecipitated with an anti-Itch antibody and blots were probed for Itch and Ndfip1.
(C) Ndfip1+/+ and Ndfip1−/− T cells were stimulated for the times points indicated and then fixed, permeabilized, and stained for intracellular Itch and Ndfip1.
(D) Ndfip1+/+ and Ndfip1−/− T cells were isolated and depleted for CD44+ cells. Cells were then cultured in media alone or stimulated for 2 or 6 hr or stimulated for 6 hr with cyclohexamide added for the last 4 hr of stimulation. Numbers represent JunB amounts normalized to unstimulated controls for each genotype. Degradation of JunB is measured by comparing lane 4 versus lane 2 for each genotype.
(E) Ndfip1−/− and Ndfip1+/+ T cell lines were transduced with the empty MIG retrovirus (MMLV-IRES-eGFP) or a Flag-tagged Ndfip1 containing MIG retrovirus. GFP+ cells were sorted and JunB degradation was analyzed as described in (D).
(F) T cells were directly isolated from an 8- to 10-week-old Ndfip1+/+ and Ndfip1−/− mice and WCL were analyzed for JunB protein amounts. β-actin is shown as a control for protein loading. LC, light chain.
To find out whether Ndfip1 could physically associate with Itch, we immunoprecipitated Itch from lysates of T cells that were unstimulated or stimulated for 24 hr. We found that isolates of Itch contained Ndfip1 in stimulated T cells (Figure 7B). This was specific for the Itch IP and did not occur in isotype controls (Figure S4); thus, Ndfip1 does bind Itch in activated T cells.
To determine whether these interactions could occur after lysis, we chose to look at whether the proteins colocalized in activated T cells. Itch and Ndfip1 localization was examined in unstimulated T cells or in cells that had been stimulated for 2 or 24 hr. In unstimulated cells, Ndfip1 was not expressed, and Itch was found in intracellular vesicles (Figure 7C). 2 hr after stimulation, Ndfip1 could be detected and was localized near the plasma membrane. Because we did not see staining with this antibody in nonpermeabilized cells (data not shown), we believe this region to represent cytoplasm near the plasma membrane. At this time point, some of the Itch colocalized near the plasma membrane with Ndfip1. Colocalization of Itch with Ndfip1 was more evident by 24 hr when nearly all the Itch and Ndfip1 polarized into a region near the inner surface of the cell. Interestingly, in cells lacking Ndfip1, Itch remained localized within the cytoplasmic vesicles for the duration of this experiment. This would suggest that Ndfip1 is required to recruit Itch to a discrete region within the cell.
That Itch and Ndfip1 are physically associated after T cell stimulation supports the hypothesis that Ndfip1 might promote Itch function. One well-described function of Itch is ubiquitination of JunB, a phenomenon that leads to degradation of the protein. JunB expression is increased 1–2 hr after T cell stimulation and then wanes (Foletta et al., 1998). This timing is consistent with expression of Ndfip1 and its colocalization with Itch. Therefore, we postulated that Ndfip1 might promote Itch-dependent degradation of JunB. This would predict that JunB could have a longer half-life in cells lacking Ndfip1.
To test this idea, JunB expression was measured in unstimulated T cells, in T cells that had been stimulated for 2 or 6 hr, and in T cells that had been stimulated for 6 hr, but incubated in cyclohexamide for the last 4 of these 6 hr, to block protein synthesis. As predicted by previous reports, JunB amounts increased after 2 hr of stimulation, and this was also true in cells lacking Ndfip1 (Figure 7D, compare lanes 1 and 2). Amounts of JunB subsequently declined in Ndfip1+/+ cells (Figure 7D), but this decline did not occur in cells lacking Ndfip1. The maintenance of JunB in Ndfip1−/− cells was mainly due to lack of JunB degradation, rather than increased synthesis of the protein because amounts of JunB remained high in these cells even if the cells were cultured in cyclohexamide. Thus, Ndfip1 controls amounts of JunB in activated T cells by inducing its degradation, probably via association of Ndfip1 with Itch.
To determine whether the reduced JunB degradation was a direct result of the loss of Ndfip1 rather than a by-product of the activation status of the cells, we retrovirally re-expressed Ndfip1 in an Ndfip1−/− T cell line. As was the case in primary T cells that lack Ndfip1, cells from an Ndfip1−/− T cell line that were transduced with an empty vector showed prolonged JunB expression after stimulation (Figure 7E, top left). In contrast, cells transduced with an Ndfip1-containing vector degraded JunB to the same extent as did Ndfip1+/+ cells.
We also wanted to know whether increasing Ndfip1 in wild-type cells would alter their JunB degradation. To do this, we overexpressed Ndfip1 in an Ndfip1+/+ T cell line, again via the retroviral system. Like primary T cells, cells from the Ndfip1+/+ cell line transduced with an empty vector show degradation of JunB 6 hr after stimulation (Figure 7E, bottom left). When Ndfip1 expression was increased in these cells, by expressing a Flag-tagged Ndfip1, JunB expression was reduced. Cells that expressed the Flag-tagged Ndfip1 contained less JunB protein 2 hr after stimulation when compared to empty vector controls. 6 hr after stimulation, JunB expression had returned to prestimulation amounts in cells overexpressing Ndfip1, while their wild-type counterparts continued to express elevated amounts of JunB.
These data predict that JunB expression might be unusually high in T cells from mice lacking Ndfip1. To test this, we isolated T cells from 8- to 10-week-old Ndfip1+/+ and Ndfip1−/− mice and tested their cell lysates for JunB by immunoblot. JunB expression was increased in T cells lacking Ndfip1 (Figure 7F). These amounts were quantified in several different experiments, normalized to β-actin, and compared to Ndfip1+/+ T cells (normalizing wild-type to 1). We found that Ndfip1−/− T cells contained approximately 5-fold more JunB than wild-type cells; it is possible, however, that some of the increased JunB in these cells results from their increased activation status. Taken together, these data support our hypothesis that the loss of Ndfip1 results in reduced degradation of JunB, likely the result of reduced Itch function.
Discussion
Ndfip1 was recently described to be a novel membrane-associated protein whose only known function was that it binds to, and is ubiquitinated by, Nedd4 (Harvey et al., 2002). The data we present here reveal that Ndfip1 plays a prominent role in T cell function and prevents spontaneous inflammation. This is illustrated by the fact that Ndfip1−/− mice have an inflammatory disease characterized by skin lesions that resemble the human condition known as atopic dermatitis. T cells from Ndfip1−/− mice are increased in number and appear activated prior to the onset of disease. This phenotype is directly attributable to the loss of Ndfip1 expression in T cells, because wild-type T cells within the same mouse are much less likely to display an activated phenotype. Therefore, in wild-type T cells, Ndfip1 acts to control T cell activity and thus prevent inflammation and Th2-mediated disease.
The phenotype we observed in Ndfip1−/− mice was nearly identical to that described for Itchy mutant mice, suggesting that Ndfip1 and Itch might interact. Two independent lines of evidence, colocalization of Ndfip1 and Itch and coimmunoprecipitation of Ndfip1 with Itch, supported this hypothesis.
Itch has been implicated in the management of T cell tolerance. Itch has been shown to regulate the response of committed Th2 cells (Venuprasad et al., 2006). Additionally, Itch expression is induced in anergic T cells (Heissmeyer et al., 2005), positioning Itch to control checkpoints in newly activated T cells. Furthermore, the E3 ligase activity of Itch increases after T cell activation, a process that requires the serine/threonine kinase JNK (Gao et al., 2004). JNK phosphorylates Jun proteins and also phosphorylates Itch. Phosphorylated Itch then causes the ubiqutination of JunB. In the absence of Itch, JunB builds up in T cells, contributing to their Th2 bias (Fang et al., 2002; Gao et al., 2004). Here we show that, in the absence of Ndfip1, JunB half life is lengthened and the amount of JunB increases, suggesting that Ndfip1 is needed for Itch to catalyze ubiqutination and turnover of the protein. Our data suggest that Ndfip1 might cause relocalization of Itch in a manner that facilitates this interaction. Thus, whether Ndfip1 promotes ubiquitination by bringing Itch together with its target proteins, or enhances Itch activity in another way, has yet to be determined. It seems likely that Ndfip1 could affect the ubiquitination of Itch targets other than JunB. Furthermore, in addition to Itch, Ndfip1 may alter the function of other HECT-type E3 ligases, particularly those of the Nedd4 family, thus contributing to the acute onset of disease in Ndfip1−/− mice.
Based on these data, we propose a novel control mechanism for Itch activation, in which T cell stimulation increases Ndfip1 expression, thereby allowing Ndfip1 to bind Itch and promote Itch function, ultimately resulting in JunB degradation. This process could act to suspend the T cell in a state of active quiescence, in which proteins required for effector function and cytokine secretion are actively produced but degraded and the cell awaits further instruction.
Experimental Procedures
Gene-Targeted ES Cells
The ES cell line with a disrupted Ndfip1 gene (RRD002) was obtained from BayGenomics. The gene-trapping vector that caused the disruption was inserted within intron 2 according the sequences obtained from 5′ RACE (Stryke et al., 2003). The ES cells were injected into mouse blastocysts to generate chimeras as described previously (McDonald et al., 1999). Five chimeras, four males and one female, were generated and germline transmission was obtained from two of the male chimeras.
Genotyping of Mice
Intron 2 of the mouse Ndfip1 gene is about 5 Kb long. A common reverse primer against the artificial intron (En2) within the gene-trapping vector, 5′ GTT GCA CCA CAG ATG AAA CG 3′, and five forward primers (equally distributed within the intron) were designed and used for amplification to determine the site of vector insertion within this intron. Upon identification of the insertion site, other primers were used for genotyping. The following two primers amplified a 466 bp fragment from the insertional allele of Ndfip1: forward, 5′ TAG GCC AAG GTG AAA ACT GG 3′; and reverse, 5′ AGT GCG GTA CCA GAC TCT CC 3′. The same forward primer paired with the following reverse primer amplified a 1010 bp fragment from the wild-type allele: 5′ AGA GGT GGG TTC AAC AGT GG 3′.
RT-PCR
Total RNA was isolated from liver, spleen, kidney, heart, thymus, and lymph nodes of both wild-type and knockout mice with Tri-Reagent (Sigma Chemical Company, St. Louis, MO) according to the manufacturer’s instructions. Three sets of primers were designed to identify the presence of different parts of the transcript, the upstream (exons 1 and 2) and downstream (exons 4 and 7) coding regions, and the coding region across the insertion site (exons 2 and 3): Ndfip1Ex1f, 5′ GCC CGA TCA GCT CTC TCG 3′, and Ndfip1Ex2r, 5′ CAG GCT CCT CTT CAT TCT GC 3′, amplify 134 bp fragment from cDNA; Ndfip1Ex2f, 5′ ATG CTC CTC CAC CAT ACA GC 3′, and Ndfip1Ex3r, 5′ GGA ACC AAA GGG ATC GTA GC 3′, amplify 166 bp fragment from cDNA; Ndfip1Ex5f, 5′ CCA GCT GAG GAT AGG AAA CG 3′, and Ndfip1Ex7r, 5′ GGC ATC TTC CGA ACT TTT GC 3′, amplify 294 bp fragment from cDNA.
Mice
Ubi-GFP mice have been previously described (Schaefer et al., 2001b). C57BL/6 and mice were purchased from The Jackson Laboratory. All mice were maintained in a specific pathogen-free (SPF) barrier facility. Care of the mice used in the experiments met the standards set forth by the National Institutes of Health in their guidelines for the care and use of experimental animals.
T Cell Isolation, Cell Culture, Stimulation, and JunB Degradation
For most experiments, T cells were isolated from lymph nodes and/or spleen by nylon wool. To make cell lines, we cultured cells, alternating every 4 days between stimulating conditions (25 μg/ml plate-bound anti-CD3 and 25 mg/ml anti-CD28) and resting conditions (20 ng/ml IL-2) for two rounds and then maintaining the cells in IL-2. To measure JunB degradation, we stimulated cells as described above for 2 or 6 hr or we stimulated cells for 2 hr, added 20 μg/ml cyclohexamide to block protein synthesis, and then continued stimulation for the remaining 4 hr.
T Cell Polarization and Cytokine Staining
For in vitro polarization, T cells were cultured in either Th1-polarizing media (10% FCS, 25 μg/ml plate-bound anti-CD3, 25 μg/ml plate-bound anti-CD28, 1:50 dilution of IL-2 conditioned media, 5 ng/ml IL-12, 3 μg/ml anti-IL4) or Th2-polarizing media (10% FCS, 25 μg/ml plate-bound anti-CD3, 25 μg/ml plate-bound anti-CD28, 1:50 dilution of IL-2 conditioned media, 5 ng/ml IL-4, 5 μg/ml anti-IFN-γ) for 7 days. The cells were then washed and incubated for 4 hr with 25 μg/ml plate-bound anti-CD3 and anti-CD28 and media containing Brefeldin A. The cells were surface stained with anti-CD4 antibody diluted in 2.4G2 conditioned supernatant to block Fc receptors. Cells were washed, fixed, and permeabilized according to manufacturer’s instructions (Cytofix/Cytoperm Plus Kit, BD Biosciences) and incubated with anti-IL-4 and anti-IFNγ antibodies for 1 hr at room temperature. Data were acquired on a FACScalibur and analyzed by CellQuestPro (Beckton Dickenson). To polarize cells in vivo, mice were immunized with 100 mg OVA + either 2 mg Alum or 7 μg CFA subcutaneously in the hind leg. 8 days after immunization, spleen were taken and single-cell suspensions prepared. Splenocytes were activated with OVA323-339 peptide at either 10 or 100 μg/ml or in the absence of peptide. 72 hr later, Brefeldin A was added to the cultures and cells were then incubated for a further 6 hr. The cells were then harvested, washed, incubated with 2.4G2 conditioned supernatant and stained with anti-CD4-APC-Cy7, CD44-Alexa 488, and CD62L-APC for 15 min. Cells were washed, fixed, and permeabilized and then stained with anti-IL-4-PE or anti-IFNγ-PE as above. Data were acquired on a CYAN (Cytomation) and analyzed by FlowJO (Treestar).
Antibodies, Antibody Production, and Flow Cytometry
Cells were isolated and then incubated with various combinations of the following antibodies diluted in 2.4G2 (anti-FcRγ antibody) containing media. Antibodies used were purchased from BD Biosciences with the following exceptions: F4/80 (Serotec), TCRβ (Ham597) (McCormack et al., 1994). Flow cytometry was performed on a FACScalibur instrument (Beckton Dickenson) or a Cyan (Cytomation), and samples were analyzed with CellQuestPro software (Beckton Dickenson) or by FlowJo (Treestar). The Ndfip1 antibody (12-22) was made by immunizing hamsters with a synthetic peptide corresponding to the N-terminal portion of Ndfip1 (VEPACGSG YQQLQNEEPGE) coupled to KLH. After boosting, sera were collected and tested in an ELISA by means of the Ndfip1-NTP coupled to ovalbumin. Antibody-producing hybridomas were made as previously described (Pullen et al., 1988), and their specificity was tested by western blot (Figure 7) and ELISA.
Itch Immunoprecipitation and Western Blotting
Cells were washed once in cold phosphate-buffered saline, lysed with 500 μl cold immunoprecipitation buffer (50 mM Tris [pH 7.5], 10% glycerol, 1% Nonidet-P40, 137 mM NaCl, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 mM PMSF, 2 mM NaF, 1 mM Na3VO4), and then centrifuged at 15,000 rpm for 10 min.
Protein was quantified with a micro BCA kit and the lysates were precleared with protein-A Sepharose beads for 30 min at 4°C. Lysates were immunoprecipitated with Itch antibody (BD Biosciences) and protein-A Sepharose beads for 2 hr at 4°C. Beads were washed and then boiled in Laemmli sample buffer containing 20 mM DTT for 5 min at 100°C. Samples were subjected to SDS-PAGE and transferred to nitrocellulose. Membranes were blocked with 5% milk in Tris-buffered saline (20 mM Tris [pH 7.5], 137 mM NaCl) with 0.5%(v/v) Tween 20 (TTBS) for 1 hr at room temperature. Membranes were then immunoblotted with anti-Itch (BD Biosciences), anti-Jun B (Santa-Cruz), anti-Ndfip (described above), or anti-Ubiquitin (Cell Signaling). Secondary antibodies were horseradish peroxidase linked, and the detecting reagent was ECL.
Mixed Bone Marrow Chimeras and Cell Sorting
Bone marrow was flushed from the femurs of the various mice, and the red blood cells (RBC) were lysed with buffered ammonium chloride. Cells were washed once and resuspended in PBS. Recipient mice were lethally irradiated with either a single dose of 1000 rads or a split dose of 800 and 400 rads, and 1–2 hr later, mice received an equal mix of Ndfip1+/+ (Ubi-GFP) and Ndfip1−/− cells or Ndfip1+/− and a total of 5 × 106 bone marrow cells by tail vein injection. To prepare cells for analysis, spleen and lymph node cells were isolated and sorted for live, GFP+ or live, GFP− cells. Cells were surface stained and then permeabilized with 0.1% saponin to release GFP prior to flow cytometry analysis.
Retroviral Expression of Vectors
Ndfip1 cDNA was amplified from a Ndfip1-containing vector (ATCC) by PCR via a forward primer 5′ GCG CAG ATC TAT GCC TTG GCG TTG GCG GCG CTG G 3′ and a reverse primer 5′ GCG CAG ATC TAA TAA ATA AAG AGA ACT CTG GTC C 3′. A Bgl II site was introduced within each primer (underlined), and the Bgl II fragment including Ndfip1 was subcloned into expression vector pCMV-Tag1 (Stratagene). Sequencing confirmed that this fused an in-frame FLAG epitope tag to the N-terminal of the Ndfip1 protein. This Flag-tagged Ndfip1 was subsequently subcloned into pMIG (MMLV-IRES-eGFP) (Schaefer et al., 2001a). Ecotropic Phoenix cells were transduced (lipofectamine, Invitrogen) with either empty pMIG or the Flag-tagged Ndfip1-containing vector and pCL-Eco. Ndfip1+/+ and Ndfip1−/− cell lines were transduced by spinfection (Schaefer et al., 2001a). GFP+ cells were sorted and tested for Flag expression (Figure 7E).
Supplementary Material
Acknowledgments
The authors wish to thank A. Schlueter (Department of Pathology, University of Iowa) for helpful discussion of this project. The skillful technical assistance of J. Loomis, E. Sweezer, T. Kinney, and J. Guo is gratefully acknowledged. Mouse husbandry was supported by the Gene Targeting Core Facility at the University of Iowa. This work was supported by UPHSC grants P50 DK52617 and AI-22295 and AI-52225.
Footnotes
Supplemental Data
Supplemental Data include four figures and one table and can be found with this article online at http://www.immunity.com/cgi/content/full/25/6/929/DC1/.
References
- Abriel H, Kamynina E, Horisberger JD, Staub O. Regulation of the cardiac voltage-gated Na+ channel (H1) by the ubiquitin-protein ligase Nedd4. FEBS Lett. 2000;466:377–380. doi: 10.1016/s0014-5793(00)01098-x. [DOI] [PubMed] [Google Scholar]
- Coscoy L, Ganem D. PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol. 2003;13:7–12. doi: 10.1016/s0962-8924(02)00005-3. [DOI] [PubMed] [Google Scholar]
- Del Prete G. Human Th1 and Th2 lymphocytes: their role in the pathophysiology of atopy. Allergy. 1992;47:450–455. doi: 10.1111/j.1398-9995.1992.tb00662.x. [DOI] [PubMed] [Google Scholar]
- Dinudom A, Harvey KF, Komwatana P, Young JA, Kumar S, Cook DI. Nedd4 mediates control of an epithelial Na+ channel in salivary duct cells by cytosolic Na+ Proc. Natl. Acad. Sci. USA. 1998;95:7169–7173. doi: 10.1073/pnas.95.12.7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Elly C, Gao B, Fang N, Altman Y, Joazeiro C, Hunter T, Copeland N, Jenkins N, Liu YC. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat. Immunol. 2002;3:281–287. doi: 10.1038/ni763. [DOI] [PubMed] [Google Scholar]
- Foletta VC, Segal DH, Cohen DR. Transcriptional regulation in the immune system: all roads lead to AP-1. J. Leukoc. Biol. 1998;63:139–152. doi: 10.1002/jlb.63.2.139. [DOI] [PubMed] [Google Scholar]
- Freemont PS. RING for destruction? Curr. Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- Gallagher E, Gao M, Liu YC, Karin M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc. Natl. Acad. Sci. USA. 2006;103:1717–1722. doi: 10.1073/pnas.0510664103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Labuda T, Xia Y, Gallagher E, Fang D, Liu YC, Karin M. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- Goldstein G, Scheid M, Hammerling U, Schlesinger DH, Niall HD, Boyse EA. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc. Natl. Acad. Sci. USA. 1975;72:11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein B, Teurich S, Hess J, Schenkel J, Schorpp-Kistner M, Angel P. Th2 cell-specific cytokine expression and allergen-induced airway inflammation depend on JunB. EMBO J. 2002;21:6321–6329. doi: 10.1093/emboj/cdf648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF, Dinudom A, Komwatana P, Jolliffe CN, Day ML, Parasivam G, Cook DI, Kumar S. All three WW domains of murine Nedd4 are involved in the regulation of epithelial sodium channels by intracellular Na+ J. Biol. Chem. 1999;274:12525–12530. doi: 10.1074/jbc.274.18.12525. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Shearwin-Whyatt LM, Fotia A, Parton RG, Kumar S. N4WBP5, a potential target for ubiquitination by the Nedd4 family of proteins, is a novel Golgi-associated protein. J. Biol. Chem. 2002;277:9307–9317. doi: 10.1074/jbc.M110443200. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- Heissmeyer V, Macian F, Im SH, Varma R, Feske S, Venuprasad K, Gu H, Liu YC, Dustin ML, Rao A. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat. Immunol. 2004;5:255–265. doi: 10.1038/ni1047. [DOI] [PubMed] [Google Scholar]
- Heissmeyer V, Macian F, Varma R, Im SH, Garcia-Cozar F, Horton HF, Byrne MC, Feske S, Venuprasad K, Gu H, et al. A molecular dissection of lymphocyte unresponsiveness induced by sustained calcium signalling. Novartis Found. Symp. 2005;267:165–174. [PubMed] [Google Scholar]
- Hettema EH, Valdez-Taubas J, Pelham HR. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 2004;23:1279–1288. doi: 10.1038/sj.emboj.7600137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustad CM, Perry WL, Siracusa LD, Rasberry C, Cobb L, Cattanach BM, Kovatch R, Copeland NG, Jenkins NA. Molecular genetic characterization of six recessive viable alleles of the mouse agouti locus. Genetics. 1995;140:255–265. doi: 10.1093/genetics/140.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CS, Yu J, Hersi K, Raaijmakers J, Gish G, Mbamalu G, et al. WW domains provide a platform for the assembly of multiprotein networks. Mol. Cell. Biol. 2005;25:7092–7106. doi: 10.1128/MCB.25.16.7092-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon MS, Atfield A, Venuprasad K, Krawczyk C, Sarao R, Elly C, Yang C, Arya S, Bachmaier K, Su L, et al. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity. 2004;21:167–177. doi: 10.1016/j.immuni.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Jolliffe CN, Harvey KF, Haines BP, Parasivam G, Kumar S. Identification of multiple proteins expressed in murine embryos as binding partners for the WW domains of the ubiquitin-protein ligase Nedd4. Biochem. J. 2000;351:557–565. [PMC free article] [PubMed] [Google Scholar]
- Kurakin A, Bredesen D. Target-assisted iterative screening reveals novel interactors for PSD95, Nedd4, Src, Abl and Crk proteins. J. Biomol. Struct. Dyn. 2002;19:1015–1029. doi: 10.1080/07391102.2002.10506805. [DOI] [PubMed] [Google Scholar]
- Li B, Tournier C, Davis RJ, Flavell RA. Regulation of IL-4 xpression by the transcription factor JunB during T helper cell differentiation. EMBO J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC. Ubiquitin ligases and the immune response. Annu. Rev. Immunol. 2004;22:81–127. doi: 10.1146/annurev.immunol.22.012703.104813. [DOI] [PubMed] [Google Scholar]
- Liu YC, Penninger J, Karin M. Immunity by ubiquitylation: a reversible process of modification. Nat. Rev. Immunol. 2005;5:941–952. doi: 10.1038/nri1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature. 1999;399:784–788. doi: 10.1038/21650. [DOI] [PubMed] [Google Scholar]
- Magnifico A, Ettenberg S, Yang C, Mariano J, Tiwari S, Fang S, Lipkowitz S, Weissman AM. WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J. Biol. Chem. 2003;278:43169–43177. doi: 10.1074/jbc.M308009200. [DOI] [PubMed] [Google Scholar]
- McCormack JE, Kappler J, Marrack P. Stimulation with specific antigen can block superantigen-mediated deletion of T cells in vivo. Proc. Natl. Acad. Sci. USA. 1994;91:2086–2090. doi: 10.1073/pnas.91.6.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald FJ, Yang B, Hrstka RF, Drummond HA, Tarr DE, McCray PB, Jr., Stokes JB, Welsh MJ, Williamson RA. Disruption of the beta subunit of the epithelial Na+ channel in mice: hyperkalemia and neonatal death associated with a pseudohypoaldosteronism phenotype. Proc. Natl. Acad. Sci. USA. 1999;96:1727–1731. doi: 10.1073/pnas.96.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillas R, Simms KS, Hatakeyama S, Weissman AM, Kuehn MR. Identification of developmentally expressed proteins that functionally interact with Nedd4 ubiquitin ligase. J. Biol. Chem. 2002;277:2897–2907. doi: 10.1074/jbc.M110047200. [DOI] [PubMed] [Google Scholar]
- Perry WL, Hustad CM, Swing DA, O’Sullivan TN, Jenkins NA, Copeland NG. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat. Genet. 1998;18:143–146. doi: 10.1038/ng0298-143. [DOI] [PubMed] [Google Scholar]
- Pirozzi G, McConnell SJ, Uveges AJ, Carter JM, Sparks AB, Kay BK, Fowlkes DM. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J. Biol. Chem. 1997;272:14611–14616. doi: 10.1074/jbc.272.23.14611. [DOI] [PubMed] [Google Scholar]
- Pullen AM, Marrack P, Kappler JW. The T-cell repertoire is heavily influenced by tolerance to polymorphic self-antigens. Nature. 1988;335:796–801. doi: 10.1038/335796a0. [DOI] [PubMed] [Google Scholar]
- Ricci M, Matucci A, Rossi O. T cells, cytokines, IgE and allergic airways inflammation. J. Investig. Allergol. Clin. Immunol. 1994;4:214–220. [PubMed] [Google Scholar]
- Romagnani S, Maggi E, Parronchi P, Macchia D, Piccinni MP, Ricci M. Increased numbers of Th2-like CD4+ T cells in target organs and in the allergen-specific repertoire of allergic patients. Possible role of IL-4 roduced by non-T cells. Int. Arch. Allergy Appl. Immunol. 1991;94:133–136. doi: 10.1159/000235344. [DOI] [PubMed] [Google Scholar]
- Schaefer BC, Mitchell TC, Kappler JW, Marrack P. A novel family of retroviral vectors for the rapid production of complex stable cell lines. Anal. Biochem. 2001a;297:86–93. doi: 10.1006/abio.2001.5327. [DOI] [PubMed] [Google Scholar]
- Schaefer BC, Schaefer ML, Kappler JW, Marrack P, Kedl RM. Observation of antigen-dependent CD8+ T-cell/ dendritic cell interactions in vivo. Cell. Immunol. 2001b;214:110–122. doi: 10.1006/cimm.2001.1895. [DOI] [PubMed] [Google Scholar]
- Scharschmidt E, Wegener E, Heissmeyer V, Rao A, Krappmann D. Degradation of Bcl10 induced by T-cell activation negatively regulates NF-kappa B signaling. Mol. Cell. Biol. 2004;24:3860–3873. doi: 10.1128/MCB.24.9.3860-3873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle’s syndrome. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- Staub O, Gautschi I, Ishikawa T, Breitschopf K, Ciechanover A, Schild L, Rotin D. Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J. 1997;16:6325–6336. doi: 10.1093/emboj/16.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryke D, Kawamoto M, Huang CC, Johns SJ, King LA, Harper CA, Meng EC, Lee RE, Yee A, L’Italien L, et al. BayGenomics: a resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 2003;31:278–281. doi: 10.1093/nar/gkg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudol M. The WW module competes with the SH3 domain? Trends Biochem. Sci. 1996;21:161–163. [PubMed] [Google Scholar]
- Uchida D, Hatakeyama S, Matsushima A, Han H, Ishido S, Hotta H, Kudoh J, Shimizu N, Doucas V, Nakayama KI, et al. AIRE functions as an E3 ubiquitin ligase. J. Exp. Med. 2004;199:167–172. doi: 10.1084/jem.20031291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venuprasad K, Elly C, Gao M, Salek-Ardakani S, Harada Y, Luo JL, Yang C, Croft M, Inoue K, Karin M, Liu YC. Convergence of Itch-induced ubiquitination with MEKK1-JNK signaling in Th2 tolerance and airway inflammation. J. Clin. Invest. 2006;116:1117–1126. doi: 10.1172/JCI26858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.