Abstract
S-Palmitoylation is rapidly emerging as an important post-translational mechanism to regulate ion channels. We have previously demonstrated that large conductance calcium- and voltage-activated potassium (BK) channels are palmitoylated within an alternatively spliced (STREX) insert. However, these studies also revealed that additional site(s) for palmitoylation must exist outside of the STREX insert, although the identity or the functional significance of these palmitoylated cysteine residues are unknown. Here, we demonstrate that BK channels are palmitoylated at a cluster of evolutionary conserved cysteine residues (Cys-53, Cys-54, and Cys-56) within the intracellular linker between the S0 and S1 transmembrane domains. Mutation of Cys-53, Cys-54, and Cys-56 completely abolished palmitoylation of BK channels lacking the STREX insert (ZERO variant). Palmitoylation allows the S0-S1 linker to associate with the plasma membrane but has no effect on single channel conductance or the calcium/voltage sensitivity. Rather, S0-S1 linker palmitoylation is a critical determinant of cell surface expression of BK channels, as steady state surface expression levels are reduced by ∼55% in the C53:54:56A mutant. STREX variant channels that could not be palmitoylated in the S0-S1 linker also displayed significantly reduced cell surface expression even though STREX insert palmitoylation was unaffected. Thus our work reveals the functional independence of two distinct palmitoylation-dependent membrane interaction domains within the same channel protein and demonstrates the critical role of S0-S1 linker palmitoylation in the control of BK channel cell surface expression.
Keywords: Ion Channels, Membrane Proteins, Membrane Trafficking, Potassium Channels, Protein Palmitoylation
Introduction
Large conductance calcium- and voltage-gated potassium (BK) channels play an important role in regulating diverse physiological processes from neuronal excitability (1, 2) to the control of blood flow (3, 4). Dysfunction of the BK channel has been implicated in a number of disorders including epilepsy (5, 6), cerebellar ataxia (2), hypertension (3, 4), incontinence (7), and tumor cell proliferation (8, 9).
The BK channel pore-forming α-subunit, encoded by a single gene KCNMA1 (10), assembles as tetramers forming a K+-selective channel. Functional diversity of BK channels is achieved by association with β-subunits (3) and other proteins (11), alternative splicing (12–14), and post-translational modifications such as phosphorylation (15).
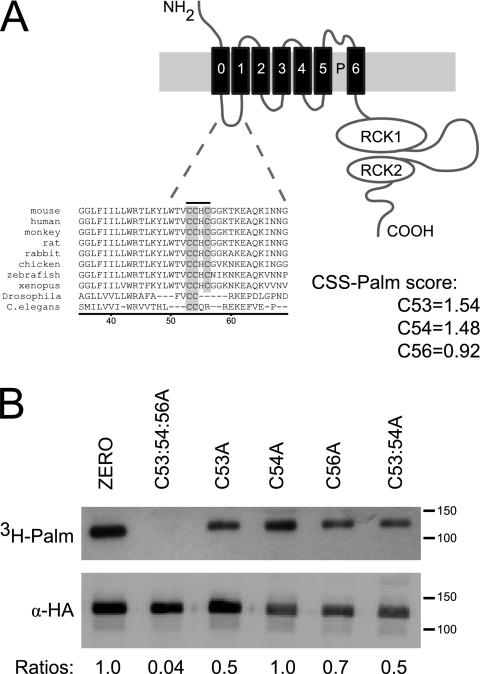
We have demonstrated previously that an alternatively spliced variant of the BK channel, the Stress-regulated exon (STREX),3 is palmitoylated within the STREX insert and regulates protein kinase A-mediated inhibition of STREX channels (16). These studies also revealed that channels lacking the STREX insert (ZERO variant) could also be palmitoylated indicating that additional cysteine residues are targets for palmitoylation (16). Furthermore, BK channels were identified in a proteomic screen for palmitoylated proteins in adult rat brain (17), a tissue with generally low level expression of STREX channels when compared with the ZERO channel variant (5, 18). Moreover, a protein palmitoylation prediction algorithm (CSS-Palm) (19, 20) indicates that the BK channel contains several evolutionary conserved cysteine residues in the intracellular linker that links the S0 and S1 transmembrane domains (S0-S1 linker) that can be palmitoylated (Fig. 1A). Whether these residues are in fact palmitoylated is not known and the functional role of palmitoylation of the BK channel outside of the STREX insert has not been addressed.
FIGURE 1.
The S0-S1 linker is palmitoylated in BK channels. A, schematic illustrating the topology of the BK channel pore-forming α-subunit. Sequence alignments of cysteine residues in the S0-S1 linker indicate evolutionary conservation (gray box) across vertebrates, Drosophila and C. elegans. Murine sequence numbered starting from the initiation methionine (MDALI), accession number: AF156674. CSS-Palm prediction scores were determined with the CSS-Palm v2.0 platform. B, representative fluorographs (upper) and Western blots (lower) of full-length ZERO-HA channels and ZERO channels with mutation of key cysteine residues in the S0-S1 linker, expressed in HEK293 cells. Constructs were labeled with [3H]palmitate for 4 h and immunoprecipitated (IP) by using α-HA magnetic microbeads and detected by fluorography. Ratios (normalized to the wild-type ZERO channel) of [3H]palmitate detection in comparison to total protein expression are included.
Increasing evidence points to an important role for palmitoylation in the dynamic control of function, assembly or trafficking of membrane proteins, including ion channels. Palmitoylation increases protein hydrophobicity by post-translational thioester linkage of a saturated 16-carbon palmitic acid to specific cysteine residues, a reversible process that is dependent on a large family of protein palmitoyltransferases (DHHCs) and thioesterases (21). Palmitoylation can play a diverse role in controlling ion channel function including: the modulation of voltage sensing in Kv1.1 channels (22); control of phosphorylation status of BK channels (16) and the GluR6 receptor (23); assembly of sodium channels (24); cell surface stability of GABAA receptors (25); and trafficking of AMPA and NMDA receptors (26, 27).
Here, we have identified evolutionary conserved cysteine residues in the intracellular N-terminal S0-S1 linker of BK channels that can be palmitoylated and that allows the domain to associate with the plasma membrane modulating cell surface expression of the channel. Importantly, the functional role of S0-S1 linker palmitoylation is distinct from that conferred by palmitoylation of the previously characterized C-terminal alternatively spliced STREX insert (16). Thus our work demonstrates functionally distinct palmitoylation-dependent membrane interaction domains within the same channel protein and that palmitoylation of the BK channel S0-S1 linker is an important determinant of cell surface expression.
EXPERIMENTAL PROCEDURES
Channel Constructs and Expression
The subcloning and site directed mutagenesis was performed on the murine BK channel α-subunit as has been described previously (13, 28). All amino acid numbering in full-length channel constructs is based on the murine (mSlo) BK channel sequence with start methionine at MDALI., accession number: AF156674. The S0-S1 linker -YFP fusion constructs used in Fig. 2 (S0-S1-YFP) incorporated the entire intracellular linker between the two predicted transmembrane domains, S0 and S1, beginning at residue Arg-44 and ending at residue Arg-114 cloned into a pEYFP-N1 vector backbone. Extracellular N-terminal Flag tag and intracellular C- terminal HA tag constructs of full-length channels were created in pcDNA3.1 as described (13, 27). Site-directed mutations were generated by standard procedures using the QuikChange system (Stratagene).
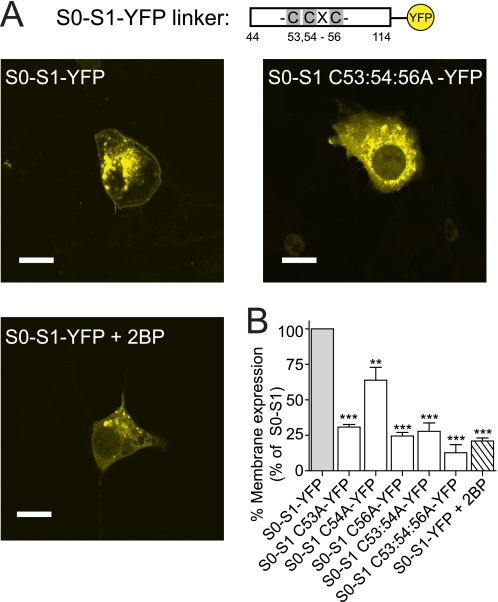
FIGURE 2.
Palmitoylation targets the S0-S1 linker to the plasma membrane. A schematic of the short S0-S1 linker YFP fusion construct (S0-S1-YFP), that encodes the 70 amino acid intracellular S0-S1 linker between amino acids Arg-44 and Arg-11 fused in-frame with YFP, and relative position of the palmitoylated cysteine residues. A, representative single cell confocal images of the S0-S1-YFP linker, S0-S1 C53:54:56A-YFP linker, and the S0-S1-YFP linker fusion proteins after incubation with the palmitoylation inhibitor 2BP (100 μm) for 24 h, expressed in HEK293 cells (scale bars: 10 μm). B, summary bar graph illustrates the effect of site-directed mutagenesis of cysteine residues in the S0-S1 linker on localization of the respective S0-S1-YFP fusion protein at the plasma membrane expressed as a percentage of the wild-type S0-S1-YFP fusion protein (where wild-type S0-S1-YFP membrane expression is normalized to 100%). (For all S0-S1 linker constructs, N>3, n>330 cells analyzed). **, p < 0.01; ***, p < 0.001 compared with wild-type S0-S1 (ANOVA with Tukey post hoc test).
HEK293 Cell Culture and Immunofluorescence
HEK293 cells were maintained and transfected as described (13, 28). Cell surface labeling of the N-terminal Flag tag epitope of BK channels in non-permeabilized HEK293 cells (28) was performed using mouse monoclonal anti-Flag M2 antibody (5 μg/μl, Sigma) and Alexa-594-conjugated anti-mouse rabbit IgG (Molecular Probes). Cells were then fixed in 4% paraformaldehyde, permeabilized with 0.3% Triton X-100 in phosphate buffered saline (PBS) for 10 min and blocked with 0.05% Triton X-100 in PBS containing 3% BSA for 30 min. The intracellular C-terminal HA epitope tag was detected using 1.25 mg/ml anti-HA polyclonal rabbit antibody (Zymed Laboratories Inc.) and Alexa-488-conjugated anti-rabbit chicken IgG (Molecular Probes) and cells mounted using Mowiol. Confocal images were acquired on a Zeiss LSM510 laser scanning microscope, using a 63× oil Plan Apochromat (NA = 1.4) objective lens, in multitracking mode (16). Quantification of Flag surface expression was done in two ways: (i) using a threshold method to detect total number of transfected cells that displayed Flag surface expression in each group; and (ii) using absolute measures based on ratios of surface Flag (extracellular) fluorescence to intracellular signal (HA) in a random subset of all cells analyzed using Image J. 1.42q (Wayne Rasband, NIH). Data were then normalized to the corresponding control group (100%) as indicated in the respective figure legend. In these experimental paradigms the data obtained for relative surface expression using the threshold method was quantitatively the same as using absolute ratio measures.
CSS-Palm Prediction
We used the published CSS-Palm palmitoylation algorithm (19, 20), to predict cysteine residues within the entire coding sequence of the murine BK channel α-subunit. Sequences were analyzed with the CSS-Palm v2.0 web interface. The palmitoylation prediction threshold was set to the highest cutoff.
[3H]Palmitic Acid Incorporation
HEK293 cells were transiently transfected in 6-well cluster dishes (≈3 × 106 cells per well) with full-length channel constructs as previously described (16). Briefly, 48 h after transfection, cells were washed and 1 ml of fresh DMEM containing 10 mg/ml fatty acid-free BSA was added for 30 min at 37 °C. Cells were incubated in DMEM/BSA containing 0.5 mCi/ml [3H]palmitic acid for 4 h at 37 °C, and then the medium containing the free label was removed. Cells were lysed in 150 mm NaCl, 50 mm Tris-Cl, 1% Triton X-100 (pH 8.0), and channel fusion proteins were captured by using magnetic microbeads coupled to HA/GFP antibody (μMACS epitope tag isolation kits, Miltenyi Biotec). Captured proteins were eluted in SDS/PAGE sample buffer (50 mm Tris-HCl, pH 6.8, 5 mm DTT, 1% SDS, 1 mm EDTA, 0.005% bromphenol blue, 10% glycerol) prewarmed to 95 °C. The recovered samples were separated by SDS/PAGE, transferred to nitrocellulose membranes, and probed with a polyclonal HA antibody (1:1,000; Zymed Laboratories Inc.). A duplicate membrane was dried, sprayed with En3hance fluorographic spray (PerkinElmer-Cetus), and exposed to light-sensitive film at −80 °C by using a Kodak Biomax transcreen LE (Amersham Biosciences).
Cell Surface Biotinylation Assay
Plasmids expressing HA-tagged BK channels were transiently transfected into HEK293 cells with Exgen 500 (Fermentas). 48 h post-transfection, cells were washed 3× with Hank's buffered salt solution (HBSS), and incubated on ice for 2 h in the presence of 5 μg/ml of Sulfo-NHS-LC-biotin (Pierce). Cells were lysed in NLB buffer with a protease inhibitor mixture after washing in ice-cold 100 mm glycine in HBSS (Roche, Germany). Biotinylated cell lysates were incubated with streptavidin-immobilized beads (Pierce) overnight at 4 °C and washed 3× with cold HBSS and once with water. The biotinylated membrane BK channel proteins were removed from the beads by incubating at 45 °C for 15 min in 2× Laemmli protein sample buffer, separated by SDS-PAGE and detected with anti-HA antibody using Western blotting. Parallel control biotinylation assays were conducted with mock transfected cells.
Electrophysiology
Single channel recordings and macropatch recordings were performed in the excised inside-out configuration of the patch-clamp technique at room temperature. The pipette solution (extracellular) contained 140 mm NaCl, 5 mm KCl, 0.1 mm CaCl2, 1 mm MgCl2, 20 mm glucose, 10 mm Hepes (pH 7.3). The bath solution (intracellular) contained 140 mm KCl, 5 mm NaCl, 1 mm MgCl2, 1 mm BAPTA, 20 mm glucose, 10 mm Hepes, (pH 7.3) with free calcium [Ca2+]i buffered from 0.33 to 10 μm, as indicated. Channel activity was determined during 100 ms depolarizations over a voltage range from −120 mV to +120 mV in 20 mV increments from a holding potential of −80 mV. Tail currents were then examined and normalized to the peak current (100%) in 1 μm Ca2+ and plotted as G/GMAX corresponding to channel activity. Voltage sensitivity was determined by a logarithmic transformation linearizing the activating component of the normalized (G/GMAX) curves in 1 μm Ca2+. Data acquisition and voltage protocols were controlled by an Axopatch 200 B amplifier and pCLAMP9 software (Axon Instruments). All recordings were sampled at 10 kHz and filtered at 2 kHz. Channel activity was allowed to stabilize for at least 10 min after patch excision before recording.
Fluorescent Membrane Potential Assay
Membrane potential assays were performed in transfected HEK293 cells incubated with FLIPR® Membrane Potential Blue Dye (Molecular Devices, Sunnyvale, CA) as described (29). Briefly, cells were plated in black walled, clear bottom 96-well plates and loaded with dye for 30 min at 37 °C to allow dye loading into the cell membrane. Assays were performed as room temperature using the Flexstation R II system (Molecular Devices) and channels activated by 1 μm ionomycin (a calcium ionophore), 16 s after measurement began. Fluorescent changes were read over a period of 3 min as relative fluorescent units (RFU). An increase in BK channel activity caused a hyperpolarization which was detected as a decrease in fluorescence. To compare between mutants the RFU were determined at t = 70 s, and the response of each channel mutant was then isolated by subtracting the control HEK depolarizing response and normalizing the hyperpolarization of the wild-type ZERO or STREX channel to 100%.
Statistics
All statistical analysis was performed using GraphPad Prism using 1-way ANOVA with Tukey post hoc test for significance between groups.
RESULTS
The S0-S1 Linker Is Palmitoylated in BK Channels
Using a palmitoylation prediction algorithm (CSS-Palm) (19, 20), the murine ZERO BK channel variant sequence revealed three cysteine residues, Cys-53, Cys-54, and Cys-56 that scored highly as predicted palmitoylation sites within the BK channel, scoring, 1.54, 1.48, and 0.92, respectively (Fig. 1A).
Sequence alignment of the three cysteine residues in the S0-S1 linker (Cys-53, Cys-54, and Cys-56) indicate strong evolutionary conservation across vertebrates with conservation of double cysteines (Cys-53 and Cys-54) extending across Drosophila and Caenorhabditis elegans, suggesting that this region may be of functional significance to the channel (Fig. 1A).
To directly assess whether the S0-S1 linker is in fact palmitoylated in the ZERO channel we assayed [3H]palmitate incorporation into full-length channel proteins in HEK293 cells (Fig. 1B). Full-length ZERO channels were robustly palmitoylated by endogenous palmitoyltransferases. Importantly, mutation of all three cysteine residues in the S0-S1 linker to alanine (ZERO C53:54:56A) completely abolished [3H]palmitate incorporation. Individual point mutation of cysteine residues demonstrated a reduced [3H]palmitate incorporation but did not abolish it, thus suggesting that the triple cysteine mutant (ZERO C53:54:56A) is the key determinant for palmitoylation of the ZERO channel. Robust expression in Western blot analysis suggested that a change in palmitoylation status of the channels was not the result of decreased synthesis or degradation of channel protein (Fig. 1B). Together these data suggest that C53:54:56 is the only site that is palmitoylated in the ZERO channel.
Palmitoylation Targets the S0-S1 linker to the Plasma Membrane
Palmitoylation increases the hydrophobicity of a protein facilitating interaction with the intracellular plasma membrane. To examine whether the S0-S1 linker alone may target to the plasma membrane, constructs that incorporated the 70 amino acid S0-S1 linker fused in-frame with YFP (S0-S1-YFP construct, Fig. 2A) were created to determine the functional contribution of each cysteine residue within the identified region. Transient expression of the S0-S1 linker constructs in HEK293 cells resulted in robust expression at the plasma membrane as well as fusion protein trapped in intracellular compartments (Fig. 2A). Site-directed mutagenesis of the cysteine residues predicted to be palmitoylated in the S0-S1 linker to alanine resulted in significantly decreased expression at the plasma membrane. Mutation of individual cysteine residues resulted in significantly decreased plasma membrane targeting of between ∼40–70% in relation to the S0-S1-YFP linker construct. Mutation of the double cysteines, conserved from human to C.elegans, also significantly decreased expression at the plasma membrane. Mutation of all three cysteine residues (S0-S1 C53:54:56A-YFP construct) resulted in near abolition (>90% decrease) of membrane targeting of the S0-S1 linker fusion protein (Fig. 2B).
We demonstrated previously that palmitoylation-dependent membrane association of C-terminal constructs containing the alternatively spliced STREX insert of the BK channel could be controlled by phosphorylation of neighboring consensus phosphorylation sites (16). Phosphoproteomic analysis of BK channels has demonstrated that serine residues Ser-70 and Ser-71, that are immediately downstream of the S0-S1 palmitoylated cysteine residues, are phosphorylated in vivo (30). However, site-directed mutagenesis of these residues to either phosphomimetic (S70:71E) or phosphornull (S70:71A) had no significant effect on membrane association of the S0-S1-YFP fusion protein at the cell surface (3 ± 6.4% and 3 ± 9.5% reduction of wild-type S0-S1 membrane expression, respectively). This suggests that the phosphorylation status of these residues does not determine S0-S1 linker membrane association.
The effect of inhibition of endogenous palmitoylation on localization of the S0-S1 linker at the plasma membrane was examined with cells incubated with a palmitoylation inhibitor, 2-bromopalmitate (2-BP, 100 μm) for 24 h. This reduced association of the S0-S1 linker at the plasma membrane by ∼80% (Fig. 2B). Together these data support the mutagenesis studies that targeting of the S0-S1 linker to the plasma membrane is controlled through palmitoylation of key cysteine residues.
S0-S1 Palmitoylation Has No Effect on Intrinsic Channel Activity
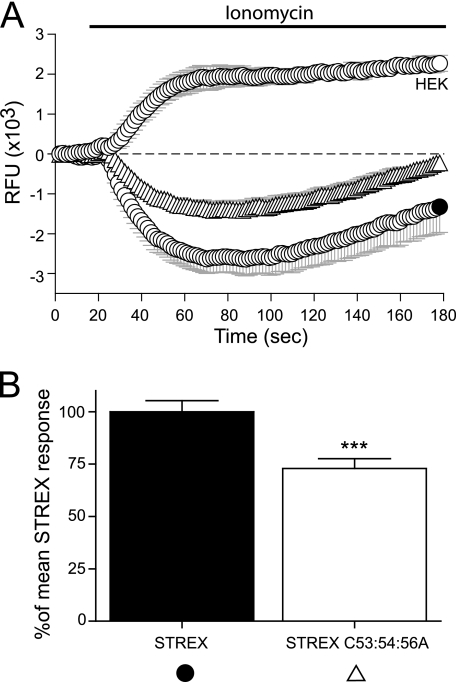
To determine whether palmitoylation of S0-S1 cysteine residues influences channel function we used a membrane-potential assay that has been previously shown to discriminate between BK channel splice variants with different calcium sensitivities (29). In this assay, BK channel activity is driven by ionomycin-induced calcium influx and is reported by movement of a fluorescent voltage-sensitive dye across the cell membrane in response to changes in cell membrane potential. Activation of BK channels results in membrane hyperpolarization revealed by a decrease in relative fluorescence. We used full-length ZERO variant channels with cysteine mutations in the S0-S1 linker. The peak wild-type ZERO channel response at the 70 s time point was taken as 100%, to which mutant channels could then be compared (Fig. 3A). Mutation of the triple (C53:54:56A) cysteine residues within the identified palmitoylation site, significantly attenuated the activation driven by ionomycin of mutant channels by ∼60% when compared with the wild-type ZERO channel (Fig. 3B). Mutation of individual and double cysteine residues (C53A, C54A, C56A, C53:54A) had no significant effect.
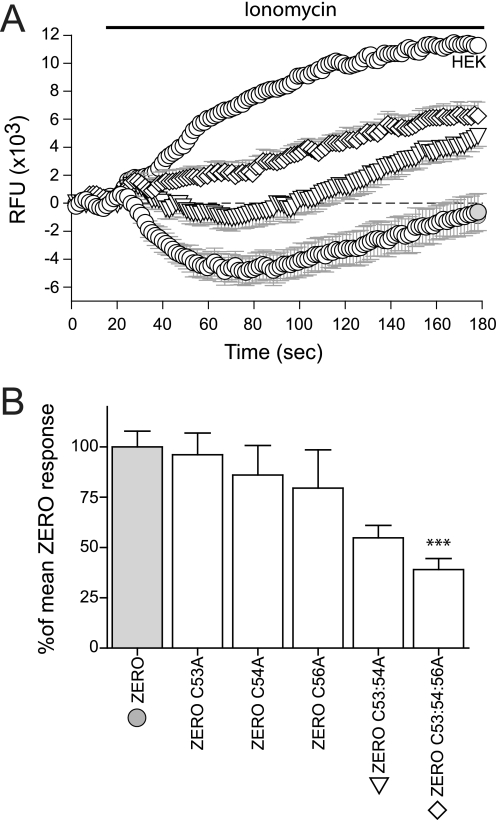
FIGURE 3.
Ionomycin-driven activation of BK channels is attenuated in S0-S1 mutant channels. A, representative time course plots of mean change in relative fluorescence units (RFU) of the FLIPR-blue membrane potential dye in HEK293 cells expressing ZERO (closed gray circles), ZERO C53:54A (inverted triangles,▽), ZERO C53:54:56A (diamonds,♢), and mock-transfected HEK293 (open circles,○), in response to calcium influx induced by 1 μm ionomycin. B, summary bar chart of the membrane potential change for each construct expressed as a percentage of the maximal hyperpolarization, following subtraction of the HEK293 response, in the ZERO (gray) variant (where the ZERO response is normalized to 100%). Data were determined at the maximum hyperpolarizing response in the wild-type ZERO channel (t = 70 s) in the time course plots in A. All data are means ± S.E. (n = 3, n>24), ***, p < 0.001, compared with ZERO (ANOVA with Tukey post hoc test).
The membrane potential assay cannot discriminate between: shifts in the calcium/voltage sensitivity of the channel; altered channel conductance; or changes in expression of functional channels at the plasma membrane. As the C53:54:56A mutant channel showed the greatest change in membrane potential, palmitoylation incorporation, and in the imaging assays, the functional impact of the ZERO C53:54:56A mutant channel was compared with that of the wild-type ZERO variant channel hereafter.
Single channel slope conductance in excised inside-out patches was unchanged between ZERO and ZERO C53:54:56A channels (ZERO, 231 ± 3.9 pS and ZERO C53:54:56A, 227 ± 5.4 pS) determined in symmetrical potassium gradients (Fig. 4, A and B). Macropatch recordings over a range of calcium (0.33–10 μm [Ca2+]i) showed no significant difference in the half maximal voltage for activation of the channel (Fig. 4, C–E) and voltage dependence of the ZERO C53:54:56A channel was not significantly changed from wild-type ZERO channels (data not shown). These data suggest that neither changes in single channel conductance nor channel voltage and calcium sensitivity underlie the significantly reduced channel activity of the C53:54:56A mutant channels observed in the membrane potential assays. However, we noted that there appeared to be fewer mutant channels (ZERO C53:54:56A channels were reduced by ∼70%, n = 26) at the plasma membrane compared with wild-type ZERO channels (n = 12) in our electrophysiology assays (data not shown). Therefore, we examined cell surface expression of the ZERO channel and the mutant (ZERO C53:54:56A) channel in quantitative immunofluorescence and cell surface biotinylation assays.
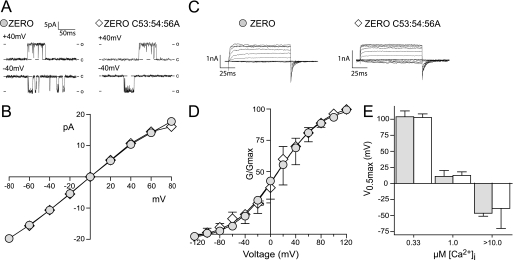
FIGURE 4.
The intrinsic channel properties are un-affected in de-palmitoylated BK channels. A, representative single channel conductance recordings of excised inside-out patches at +40 and −40 mV in 0.33 μm Ca2+. B, current (pA) voltage (V) plot for ZERO channels (closed gray circles) and ZERO C53:54:56A (diamonds,♢), showing that single channel conductance is unaffected in 0.33 μm Ca2+ (n = 3). C, representative macropatch recordings traces showing BK currents in response to a depolarizing voltage step protocol (−120 mV to +120 mV) from a holding potential of −80mV from excised inside-out patch recordings in equimolar potassium gradients and 1 μm free Ca2+ (scale bars: 1 pA/25 ms). D, G/GMAX conductance curves show no change in channel activation at 1 μm free Ca2+ between ZERO (closed gray circles) and ZERO C53:54:56A (diamonds,♢). E, summary bar graph illustrates no significant changes in V0.5max across the physiological calcium range 0.33–10 μm free calcium. All data are means ± S.E. (n >3).
Palmitoylation Is an Important Determinant of BK Channel Cell Surface Expression
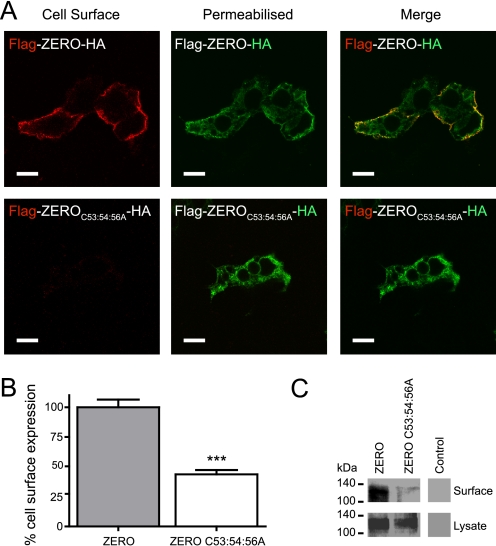
Mutation of the C53:54:56 palmitoylation site in the S0-S1 linker of the BK channel might disrupt normal expression at the plasma membrane. Channel constructs were created with extracellular N-terminal FLAG tag epitopes that enabled detection on the extracellular surface of transfected HEK293 cells and with intracellular C-terminal HA tag epitopes, to determine total protein expression. In non-permeabilized cells, ZERO channels could be detected at the plasma membrane however, in the palmitoylation-deficient channels (ZERO C53:54:56A) surface expression was significantly reduced despite total protein expression being unaffected. Quantitative immunofluorescence analysis of N-terminal Flag-tagged ZERO C53:54:56A mutant channels, showed a significant ∼55% decrease in cell surface expression in relation to the ZERO channel (Fig. 5, A and B). A similar reduction in cell surface expression of the C53:54:56A mutant channels was also observed using cell surface biotinylation assays (Fig. 5C).
FIGURE 5.
Palmitoylation of the S0-S1 linker regulates cell surface expression of BK channels. A, representative confocal images of HEK293 cells expressing Flag-ZERO-HA (top panels), and Flag-ZERO C53:54:56A-HA (bottom panels). The extracellular Flag epitope was labeled (red) under non-permeabilized conditions (cell surface) with the C-terminal HA epitope tag (green) labeled following cell permeabilization. Flag and HA labeling from the same cell are then overlaid (merge) (scale bars: 10 μm). B, quantification of cell surface expression between ZERO (gray bars) and ZERO C53:54:56A (white bars). Data are means ± S.E. (n >3). **, p < 0.01, ANOVA with post hoc Tukey test compared with Flag-ZERO-HA construct. C, representative Western blots of HA immunoreactivity from cell surface biotinylation assays (top panels) and corresponding whole cell lysates (bottom panels) in HEK293 cells expressing ZERO-HA and ZERO C53:54:56A-HA channels and control mock transfected cells.
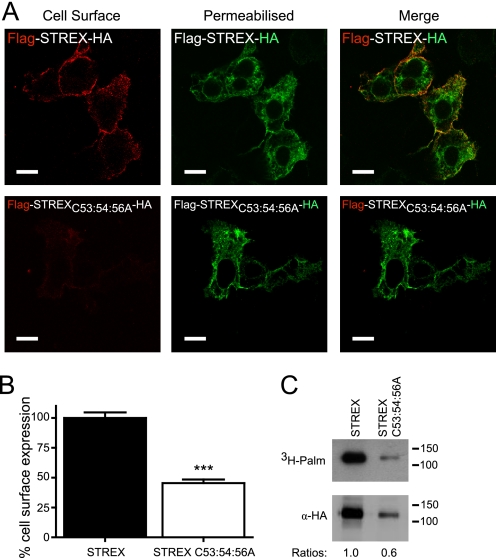
To investigate whether palmitoylation of the S0-S1 linker controls cell surface expression in channels that are also palmitoylated at the distinct C-terminal alternatively spliced STREX insert (16), STREX channels were also examined with mutations in the S0-S1 palmitoylation site. Firstly, we examined the S0-S1 linker mutation (C53:54:56A) in the STREX channel using the membrane-potential assay as before. Mutation of the S0-S1 palmitoylation site in STREX channel showed a ∼30% attenuation of the ionomycin-induced activation of the channels, similar to that observed for the ZERO channel (Fig. 6, A and B). Therefore, to determine whether the S0-S1 linker palmitoylation site also controls cell surface expression of the STREX splice variant of the BK channel quantitative immunofluorescence analysis was performed. N-terminal Flag-tagged STREX C53:54:56A mutant channels also showed a significant reduction in channel surface expression of ∼55% when compared with wild-type STREX channels (Fig. 7, A and B). STREX channels that could not be palmitoylated at the S0-S1 linker (STREX C53:54:56A) could still be palmitoylated via the STREX insert as predicted (Fig. 7C). The reduction in cell surface expression was further recapitulated in cell surface biotinylation assays (data not shown). Taken together, these data suggest that the S0-S1 linker palmitoylation site functions independently of the C-terminal palmitoylation site in the STREX insert and that the palmitoylation status of the S0-S1 linker is a critical determinant of cell surface channel expression irrespective of C-terminal splice variation.
FIGURE 6.
Disruption of the S0-S1 linker palmitoylation site in STREX splice variant channels also attenuates the ionomycin-driven channel activation. A, representative time course plots of mean change in relative fluorescence units (RFU) of the FLIPR-blue membrane potential dye in HEK293 cells expressing STREX (closed black circles, ●), STREX C53:54:56A (upright triangles, △), and mock-transfected HEK293 (open circles, ○), in response to calcium influx induced by 1 μm ionomycin. B, summary bar chart of the membrane potential change for each construct expressed as a percentage of the maximal hyperpolarization, following subtraction of the HEK293 response, in the STREX (black) variant (where the STREX response is normalized to 100%). Data were determined at the maximum hyperpolarizing response in STREX (t = 70 s) in the time course plots in A. All data are means ± S.E. (n = 3, n > 24), ***, p < 0.001, compared with STREX (ANOVA with Tukey post hoc test).
FIGURE 7.
S0-S1 palmitoylation site functions independently of additional STREX splice variant palmitoylation site to control BK channel surface expression. A, representative confocal images of HEK293 cells expressing Flag-STREX-HA (top panels), and Flag- STREX C53:54:56A -HA (bottom panels). The extracellular Flag epitope was labeled (red) under non-permeabilized conditions (cell surface) with the C-terminal HA epitope tag (green) labeled following cell permeabilization. Flag and HA labeling from the same cell are then overlaid (merge) (scale bars: 10 μm). B, quantification of cell surface expression between STREX (black bars) and STREX C53:54:56A (white bars). Data are means ± S.E. (n >3). *, p < 0.05, ANOVA with post hoc Tukey test compared with Flag-STREX-HA construct. C, representative fluorographs (upper) and Western blots (lower) of full-length STREX-HA channels and STREX C53:54:56A channels expressed in HEK293 cells. Constructs were labeled with [3H]palmitate for 4 h and immunoprecipitated (IP) by using α-HA magnetic microbeads and detected by fluorography. In this particular experiment protein expression of the two constructs was not equivalent; however, the C53:54:56A mutations do not compromise STREX expression per se. These data reveal the residual palmitoylation of the channel mediated via the STREX insert in contrast to the ZERO variant (see Fig. 1). Ratios (normalized to the wild-type STREX channel) of palmitate incorporation relative to total protein expression are included.
DISCUSSION
We have identified an evolutionary conserved palmitoylation site within the intracellular N-terminal S0-S1 linker of BK channels that plays a critical role in the control of channel cell surface expression. Palmitoylation of the S0-S1 linker allowed the association of this N-terminal intracellular domain with the plasma membrane and controlled surface expression even in channel variants that express an additional palmitoylation-dependent membrane association domain (STREX) in the intracellular C terminus of the channel (16). As STREX palmitoylation controls channel regulation by phosphorylation (16), rather than surface expression, as demonstrated here for the S0-S1 linker, our work demonstrates functionally distinct palmitoylation-dependent membrane interaction domains within the same channel protein.
The identified triple cysteine palmitoylation residues in the S0-S1 linker (C53:54:56) are conserved across the vertebrate phylum and are largely conserved in Drosophila and C. elegans, suggesting evolutionary retention of a functionally important region. This site would therefore be present in all functionally expressed BK channel proteins and is not predicted to be excluded by alternative splicing events. Indeed, proteomic screens identified BK channels as palmitoylated in the adult rat brain (17), a tissue in which the only other known site for BK channel palmitoylation is located within the STREX insert, which is expressed at relatively low levels. As protein palmitoylation is a highly dynamic and reversible process this would suggest that palmitoylation of the S0-S1 linker may be an important determinant in controlling BK channel cell surface expression under different physiological demands. For example, cell surface expression of BK channels is modified in aging coronary arteries (31), in smooth muscle cells of the uterus during pregnancy (32), colonic epithelia in response to aldosterone (33), and during malignant glioma tumor cell proliferation (9). While multiple mechanisms may control steady state surface expression the control of BK channel expression at the plasma membrane by palmitoylation may be an important determinant of a wide range of physiological functions.
Palmitoylation of the S0-S1 linker could modulate surface expression by multiple mechanisms including: (i) facilitation of channel assembly or ER export, (ii) stabilization at the plasma membrane, (iii) reduced retrieval from the plasma membrane, (iv) increased recycling, or (v) reduced channel degradation. Examination of the role of palmitoylation in each of these mechanisms warrants further investigation. Importantly, the S0-S1 linker per se appears to play an important role in controlling channel cell surface expression as revealed from analysis of alternative splice variants that may be included in this region. For example, a human splice variant of the BK channel called mk44, introduces a 44 amino acid sequence immediately downstream of the C53:54:56 palmitoylation site in the S0-S1 intracellular linker (34). This mk44 splice variant introduces a motif for endoproteolytic digestion and a site for N-myristoylation. N-Myristoylation of mk44 results in trapping of the channel in the ER. However, endoproteolytic cleavage allows the S0 transmembrane domain to traffic independently of the rest of the transmembrane and C-terminal domain of the channel to the plasma membrane (34). Another splice variant in the human BK channel, SV1, has been implicated in ER retention. The SV1 splice variant introduces 33 amino acids to the end of the S0-S1 linker introducing an ER retention-retrieval motif, CVLF (35). Although inclusion of these splice variants is not predicted, using the CSS-Palm algorithm, to significantly reduce palmitoylation of the C53:54:56 site,4 whether these inserts disrupt palmitoylation of S0-S1 linker or its functional regulation of channel cell surface expression remains to be explored. The S0 transmembrane domain of BK channels is also an important determinant of BK channel α-subunit assembly with the regulatory β-subunits (36–40). Indeed, recent structural studies examining intra-α subunit di-sulfide cross-linking has demonstrated that the S0 transmembrane domain is located outside of the voltage sensing S1-S4 domains (36) and can form a major contact with the transmembrane domain 2 (TM2) of regulatory β1 (36) and β4 (40) subunits. Co-expression studies have also suggested a role for β-subunits in controlling BK channel cell surface expression. Indeed, β1-subunits have been suggested to decrease surface expression (41), and the β2-subunit also appears to modulate BK surface expression via a similar mechanism (42). Therefore, increasing evidence supports an important role for the S0-S1 linker and surrounding transmembrane domains in controlling BK channel cell surface expression.
Palmitoylation of the S0-S1 linker had no significant effect on the intrinsic single channel conductance or calcium/voltage sensitivity of BK channels. Moreover, mutation of the S0-S1 linker cysteines C53:54:56 to alanine also reduced cell surface expression of the STREX variant of the BK channel. We have previously demonstrated that the C terminus alternatively spliced STREX insert is palmitoylated and targets the STREX insert to the plasma membrane (16). Palmitoylation status of the STREX insert has no significant effect on channel cell surface expression however, it affects the calcium/voltage sensitivity of the channel and more importantly determines whether the STREX channel is inhibited by protein kinase A (PKA)-dependent phosphorylation (16). In the latter case, PKA phosphorylation dissociates the STREX domain from the plasma membrane through phosphorylation of an upstream serine residue. In contrast, phosphomimetic or phosphornull mutations of the S0-S1 linker at a tandem serine motif, shown to be phosphorylated in vivo (30), immediately downstream of the palmitoylated cysteine residues, have no effect of S0-S1 linker interaction with the plasma membrane. Taken together, these data demonstrate that BK channels may express two functionally distinct palmitoylation-dependent membrane interaction domains: the C-terminal alternatively spliced STREX insert and the constitutively expressed S0-S1 linker. The distinct functional consequence of the palmitoylation status of these two separate palmitoylated domains within the same channel protein also suggests that palmitoylation of S0-S1 and STREX may be independently regulated.
Acknowledgments
We thank Trudi Gillespie and the IMPACT imaging facility for assistance in confocal imaging assays.
This work was supported by the Wellcome Trust.
O. Jeffries and M. J. Shipston, unpublished data.
- STREX
- stress-regulated exon
- HEK293
- human embryonic kidney 293 cell.
REFERENCES
- 1.Raffaelli G., Saviane C., Mohajerani M. H., Pedarzani P., Cherubini E. (2004) J. Physiol. 557, 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sausbier M., Hu H., Arntz C., Feil S., Kamm S., Adelsberger H., Sausbier U., Sailer C. A., Feil R., Hofmann F., Korth M., Shipston M. J., Knaus H. G., Wolfer D. P., Pedroarena C. M., Storm J. F., Ruth P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9474–9478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner R., Peréz G. J., Bonev A. D., Eckman D. M., Kosek J. C., Wiler S. W., Patterson A. J., Nelson M. T., Aldrich R. W. (2000) Nature 407, 870–876 [DOI] [PubMed] [Google Scholar]
- 4.Sausbier M., Arntz C., Bucurenciu I., Zhao H., Zhou X. B., Sausbier U., Feil S., Kamm S., Essin K., Sailer C. A., Abdullah U., Krippeit-Drews P., Feil R., Hofmann F., Knaus H. G., Kenyon C., Shipston M. J., Storm J. F., Neuhuber W., Korth M., Schubert R., Gollasch M., Ruth P. (2005) Circulation 112, 60–68 [DOI] [PubMed] [Google Scholar]
- 5.Brenner R., Chen Q. H., Vilaythong A., Toney G. M., Noebels J. L., Aldrich R. W. (2005) Nat. Neurosci. 8, 1752–1759 [DOI] [PubMed] [Google Scholar]
- 6.Du W., Bautista J. F., Yang H., Diez-Sampedro A., You S. A., Wang L., Kotagal P., Lüders H. O., Shi J., Cui J., Richerson G. B., Wang Q. K. (2005) Nat. Genet. 37, 733–738 [DOI] [PubMed] [Google Scholar]
- 7.Meredith A. L., Thorneloe K. S., Werner M. E., Nelson M. T., Aldrich R. W. (2004) J. Biol. Chem. 279, 36746–36752 [DOI] [PubMed] [Google Scholar]
- 8.Bloch M., Ousingsawat J., Simon R., Schraml P., Gasser T. C., Mihatsch M. J., Kunzelmann K., Bubendorf L. (2007) Oncogene 26, 2525–2534 [DOI] [PubMed] [Google Scholar]
- 9.Weaver A. K., Liu X., Sontheimer H. (2004) J. Neurosci. Res. 78, 224–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler A., Tsunoda S., McCobb D. P., Wei A., Salkoff L. (1993) Science 261, 221–224 [DOI] [PubMed] [Google Scholar]
- 11.Tian L., Chen L., McClafferty H., Sailer C. A., Ruth P., Knaus H. G., Shipston M. J. (2006) Faseb. J. 20, 2588–2590 [DOI] [PubMed] [Google Scholar]
- 12.Shipston M. J. (2001) Trends Cell Biol. 11, 353–358 [DOI] [PubMed] [Google Scholar]
- 13.Tian L., Duncan R. R., Hammond M. S., Coghill L. S., Wen H., Rusinova R., Clark A. G., Levitan I. B., Shipston M. J. (2001) J. Biol. Chem. 276, 7717–7720 [DOI] [PubMed] [Google Scholar]
- 14.Xie J., McCobb D. P. (1998) Science 280, 443–446 [DOI] [PubMed] [Google Scholar]
- 15.Tian L., Coghill L. S., McClafferty H., MacDonald S. H., Antoni F. A., Ruth P., Knaus H. G., Shipston M. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11897–11902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian L., Jeffries O., McClafferty H., Molyvdas A., Rowe I. C., Saleem F., Chen L., Greaves J., Chamberlain L. H., Knaus H. G., Ruth P., Shipston M. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 21006–21011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., Thompson J. X., Roth A. F., Drisdel R. C., Mastro R., Green W. N., Yates J. R., 3rd, Davis N. G., El-Husseini A. (2008) Nature 456, 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacDonald S. H., Ruth P., Knaus H. G., Shipston M. J. (2006) BMC Dev. Biol. 6, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. (2008) Protein Eng. Des. Sel. 21, 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou F., Xue Y., Yao X., Xu Y. (2006) Bioinformatics 22, 894–896 [DOI] [PubMed] [Google Scholar]
- 21.Fukata Y., Iwanaga T., Fukata M. (2006) Methods 40, 177–182 [DOI] [PubMed] [Google Scholar]
- 22.Gubitosi-Klug R. A., Mancuso D. J., Gross R. W. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5964–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pickering D. S., Taverna F. A., Salter M. W., Hampson D. R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 12090–12094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt J. W., Catterall W. A. (1987) J. Biol. Chem. 262, 13713–13723 [PubMed] [Google Scholar]
- 25.Rathenberg J., Kittler J. T., Moss S. J. (2004) Mol. Cell Neurosci. 26, 251–257 [DOI] [PubMed] [Google Scholar]
- 26.Hayashi T., Thomas G. M., Huganir R. L. (2009) Neuron 64, 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi T., Rumbaugh G., Huganir R. L. (2005) Neuron 47, 709–723 [DOI] [PubMed] [Google Scholar]
- 28.Chen L., Tian L., MacDonald S. H., McClafferty H., Hammond M. S., Huibant J. M., Ruth P., Knaus H. G., Shipston M. J. (2005) J. Biol. Chem. 280, 33599–33609 [DOI] [PubMed] [Google Scholar]
- 29.Saleem F., Rowe I. C., Shipston M. J. (2009) Br. J. Pharmacol. 156, 143–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan J., Olsen J. V., Park K. S., Li W., Bildl W., Schulte U., Aldrich R. W., Fakler B., Trimmer J. S. (2008) Mol. Cell Proteomics 7, 2188–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marijic J., Li Q., Song M., Nishimaru K., Stefani E., Toro L. (2001) Circ. Res. 88, 210–216 [DOI] [PubMed] [Google Scholar]
- 32.Song M., Zhu N., Olcese R., Barila B., Toro L., Stefani E. (1999) FEBS Lett. 460, 427–432 [DOI] [PubMed] [Google Scholar]
- 33.Sørensen M. V., Matos J. E., Sausbier M., Sausbier U., Ruth P., Praetorius H. A., Leipziger J. (2008) J. Physiol. 586, 4251–4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korovkina V. P., Fergus D. J., Holdiman A. J., England S. K. (2001) Am. J. Physiol. Cell Physiol. 281, C361–C367 [DOI] [PubMed] [Google Scholar]
- 35.Zarei M. M., Eghbali M., Alioua A., Song M., Knaus H. G., Stefani E., Toro L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10072–10077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu G., Niu X., Wu R. S., Chudasama N., Yao Y., Jin X., Weinberg R., Zakharov S. I., Motoike H., Marx S. O., Karlin A. (2010) J. Gen. Physiol. 135, 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu G., Zakharov S. I., Yang L., Deng S. X., Landry D. W., Karlin A., Marx S. O. (2008) J. Gen. Physiol. 131, 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meera P., Wallner M., Song M., Toro L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 14066–14071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wallner M., Meera P., Toro L. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14922–14927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu R. S., Chudasama N., Zakharov S. I., Doshi D., Motoike H., Liu G., Yao Y., Niu X., Deng S. X., Landry D. W., Karlin A., Marx S. O. (2009) J. Neurosci. 29, 8321–8328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toro B., Cox N., Wilson R. J., Garrido-Sanabria E., Stefani E., Toro L., Zarei M. M. (2006) Neuroscience 142, 661–669 [DOI] [PubMed] [Google Scholar]
- 42.Zarei M. M., Song M., Wilson R. J., Cox N., Colom L. V., Knaus H. G., Stefani E., Toro L. (2007) Neuroscience 147, 80–89 [DOI] [PubMed] [Google Scholar]