Abstract
Ribosomes are vital for cell growth and survival. Until recently, it was believed that mutations in ribosomes or ribosome biogenesis factors would be lethal, due to the essential nature of these complexes. However, in the last few decades, a number of diseases of ribosome biogenesis have been discovered. It remains a challenge in the field to elucidate the molecular mechanisms underlying them.
Introduction
Ribosomes are the cellular factories responsible for making proteins. Ribosome biogenesis, the process of making ribosomes, is a complex and energy intensive process that involves several hundred factors. These factors include multiple enzymes, transcription regulators, chaperones, nuclear export proteins, and more, all of which ensure that the ribosomal RNA (rRNA) is properly folded and assembled with the ribosomal proteins (r-proteins). Much of the progress in elucidating the factors involved in making ribosomes has been carried out in the single-celled model organism, Saccharomyces cerevisiae. Because of its genetic tractability, the yeast is ideal for high-throughput studies, including yeast two-hybrid, microarrays, and affinity purification followed by mass spectrometry; these techniques have enabled interaction maps of ribosome biogenesis factors to be created,1–4 furthering our understanding of this complex process. Furthermore, due to the high conservation of both the RNA and protein moieties of ribosomes and of the ribosome biogenesis machinery from yeast to humans, in general what we have learned in yeast also applies to humans (for recent reviews see ref. 5,6). Ribosome biogenesis is so important for cell growth that a growing yeast cell synthesizes approximately 2000 ribosomes every minute, requiring 60% of total cellular transcription.7 In mammalian cells, this number is even higher; for example, a HeLa cell makes 7500 ribosomal subunits per minute.8
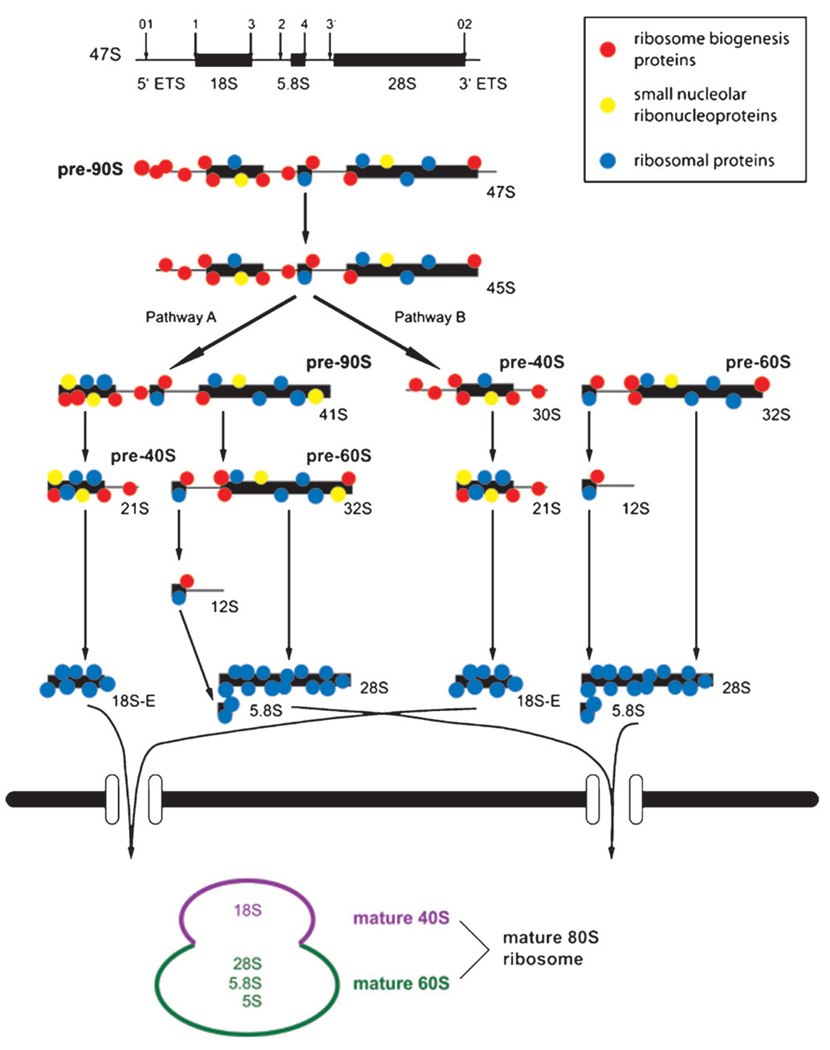
In eukaryotic cells, ribosome biogenesis begins with the transcription of the ribosomal DNA (rDNA) into a 47S (35S in yeast) polycistronic precursor by RNA polymerase I (Pol I) in the nucleolus (Fig. 1). Hundreds of r-proteins, non-ribosomal proteins involved in processing, and small nucleolar RNAs (snoRNAs) associate co-transcriptionally with the pre-rRNA forming the 90S pre-ribosome.9,10 In the same transcription unit but in the opposite direction, the 5S rRNA is transcribed by RNA polymerase III (Pol III) and may associate with the 90S pre-ribosome.6 The 47S pre-rRNA undergoes multiple cleavage11–13 and modification14–16 events, and after association with r-proteins, gives rise to the pre-40S small subunit (SSU) and the pre-60S large subunit (LSU). These subunits are then exported to the cytoplasm where final maturation takes place. The mature ribosome consists of the SSU, which contains the 18S rRNA associated with about 30 r-proteins, and the LSU, which contains the 28S (25S in yeast), 5.8S, and 5S rRNAs and about 45 r-proteins, depending on the species.
Fig. 1.
Model of pre-rRNA processing in human cells. The primary pre-rRNA is transcribed and associates co-transcriptionally with processing factors. Processing can occur by either of two pathways, beginning with cleavage at site 1 or site 2, respectively. Final maturation takes place in the cytoplasm.
Nearly all ribosome biogenesis factors, as well as the r-proteins themselves, are essential for cell survival. It is therefore somewhat surprising that some defects in ribosome biogenesis lead to detectable human disease, rather than embryonic lethality. Nonetheless, over the last few decades, a growing variety of diseases linked to ribosomes and ribosome biogenesis have been reported (Tables 1–3). Furthermore, all of these diseases show some degree of tissue-proclivity, which is unexpected given that ribosomes are present in all tissues in the body.
Table 1.
Genetic diseases of ribosome biogenesis proteins
| Gene |
Impaired molecular function |
Disease | Clinical manifestations | Inheritance and incidence (if known) |
Online Mendelian Inheritence in Man (OMIM) # |
References | |
|---|---|---|---|---|---|---|---|
| human | yeast | ||||||
| TCOF1 | — | rDNA transcription and methylation of 18S rRNA |
Treacher Collins syndrome (TCS) |
craniofacial abnormalities | autosomal dominant; incidence: 1 in 10,000 to 50,000 live births |
#154500 | 21,24,154 |
| UTP14c | UTP14 | maturation of the 18S rRNA of the 40S ribosomal subunit |
male infertility | severe oligospermia or azoospermia |
unkown | N/A | 9,26 |
| CIRH1A | UTP4 | maturation of the 18S rRNA of the 40S ribosomal subunit |
Native American Indian childhood cirrhosis (NAIC) |
neonatal jaundice progressing to biliary cirrhosis; lethal by adolescence without liver transplant |
autosomal recessive; <100 cases reported |
#604901 | 9,28,31 |
| EMG1 | EMG1 | maturation of the 40S ribosomal subunit |
Bowen-Conradi syndrome (BCS) |
growth retardation, psychomotor delay, skeletal abnormalities; lethal by early childhood |
autosomal recessive; incidence: 1 in 355 live births in the Hutterite population |
#211180 | 33 |
| RBM28 | NOP4 | maturation of the 60S ribosomal subunit |
alopecia, neurological defects, and endocrinopathy syndrome (ANE syndrome) |
growth retardation, loss of motor ability, mental retardation, skeletal and skin abnormalities, hair loss, central adrenal insufficiency |
autosomal recessive | #612079 | 37 |
| SBDS | SDO1 | maturation and export of the 60S ribosomal subunit |
Shwachman- Diamond syndrome (SDS) |
growth retardation, exocrine pancreas insufficiency, hematologic defects, skeletal abnormalities, cancer predisposition |
autosomal recessive; incidence: 1 in 76,000 live births |
#260400 | 39,44,155 |
| WDR36 | UTP21 | maturation of the 18S rRNA of the 40S ribosomal subunit |
candidate gene for primary open angle glaucoma (POAG) |
degeneration of optical nerve leading to blindness |
multifactorial | #137760 | 9,48 |
| HCA66 | UTP6 | maturation of the 18S rRNA of the 40S ribosomal subunit |
modifier of neurofibromatosis type 1 (NF1) |
mental retardation, craniofacial and connective tissue abnormalities, congenital heart defects, tumor predilection |
autosomal dominant | #162200 | 9,53 |
Table 3.
Genetic diseases of ribosomal proteins
| Gene |
Impaired function |
Disease | Clinical manifestations | Inheritance and incidence (if known) |
OMIM # | References | |
|---|---|---|---|---|---|---|---|
| human | yeast | ||||||
| RPS7 |
RPS7A RPS7B |
5′ -end processing of 18S rRNA |
Diamond Blackfan anemia (DBA) |
anemia, bone marrow failure, craniofacial abnormalities, cardiac defects, limb and urogenital abnormalities, cancer predisposition |
sporadic, autosomal dominant (40–45%); incidence: 1 in 100,000 to 200,000 live births |
#105650 | 104,107–112 |
| RPS17 |
RPS17A RPS17B |
3′ -end processing of the 18S rRNA |
as above | as above | as above | as above | as above |
| RPS19 |
RPS19A RPS19B |
3′ -end processing of the 18S rRNA |
as above | as above | as above | as above | as above |
| RPS24 |
RPS24A RPS24B |
5′ -end processing of 18S rRNA |
as above | as above | as above | as above | as above |
| RPL5 | RPL5 | processing of pre-rRNA of the LSU |
as above | as above | as above | as above | as above |
| RPL11 |
RPL11A RPL11B |
processing of pre-rRNA of the LSU |
as above | as above | as above | as above | as above |
| RPL35A |
RPL33A RPL33A |
processing of pre-rRNA of the LSU |
as above | as above | as above | as above | as above |
| RPS14 |
RPS14A RPS14B |
pre-18S rRNA processing |
5q− syndrome | severe macrocytic anemia, myelodysplastic syndrome, cancer predisposition |
unknown | #153550 | 131 |
Genetic diseases of ribosome biogenesis proteins
The small subunit (SSU) processome is a large ribonucleo-protein (RNP) that is required for the maturation of the 18S rRNA of the SSU of the ribosome. It was first discovered in yeast through a combination of directed9 and high throughput3 techniques, and later was also described in humans.17 This complex contains the U3 snoRNA and over 40 proteins, many of which first assemble into subcomplexes.3,9,17,18 In addition to the SSU processome proteins, many other proteins are required for the maturation of the large and small ribosomal subunits. The majority of these processing proteins are not present in the mature ribosome. Almost all of these proteins are essential in yeast9 and several have also been shown to be essential in mammals,19,20 suggesting conservation from yeast to humans. Although complete loss of these proteins is expected to be lethal, mutations in ribosome biogenesis factors can act either as the causative agents of disease or, in rare cases, as modifying agents, by increasing the severity of a disease caused by a mutation in a non-pre-ribosome associated protein (Table 1).
Mutations in small ribosomal subunit biogenesis proteins that cause disease
Treacle – Treacher Collins syndrome
Treacher Collins syndrome (TCS) is an autosomal dominant craniofacial disorder which varies widely in disease severity, but usually includes abnormalities of the ears, eyes, and facial bones, particularly the lower jaw and cheek.21 Five mutations in the gene encoding Treacle (TCOF1) were first reported to cause TCS over 10 years ago21 and since that time, more than 200 mutations have been reported in the TCOF1 database (www.genoma.ib.usp.br/TCOF1_database). The mutations include primarily nonsense mutations, insertions, deletions, and alternative splicing, all of which lead to truncation of the protein. There is no correlation between genotype and disease severity, which may be explained by the observation that genetic background greatly alters the phenotype in mouse models.22 Treacle is a putative nucleolar phosphoprotein that plays a role in rDNA transcription23 and methylation of the 18S rRNA.24
All TCS patients are heterozygous for a TCOF1 mutation and the disease appears to be due to haploinsufficiency rather than dominant negative effects.23 Extensive work in the mouse has shown that Tcof1 is required for neural crest cell formation and proliferation. In Tcof1+/− mice, an increase in caspase3-mediated apoptosis is observed specifically in the neuroepithilium, resulting in a smaller starting population of migrating neural crest cells. This diminished population of cells then proliferates more slowly than cells expressing wild-type Tcof1, further reducing the number of neural crest cells and leading to the hypoplasia observed in TCS. Both the apoptosis and the slow growth rate are likely due to a lack of mature ribosomes; indeed fewer ribosomes are observed specifically in the neuroepithelium of Tcof1+/− mice.25 The cause of the neuroepithelium-specific defect remains to be elucidated since TCOF1 is expressed in a wide variety of fetal and adult tissues21 and ribosomes are required in all cells in all tissue types.
UTP14 – male infertility
In yeast, Utp14 is an SSU processome protein and is essential for 18S rRNA maturation; however, its specific function is unknown.9 In humans, there are three genes encoding UTP14: UTP14a, UTP14b, and UTP14c.26 UTP14a is the X-linked ancestral copy of the gene and is ubiquitously expressed across tissue types. UTP14b is a degraded retroposon located within ACSL3 and is not expressed in human tissue. UTP14c is an active retroposon that inserted into the 3′ UTR of GT8 (a putative glycosyl transferase-containing gene) and is only expressed in the testis and ovary.
Mutations in Utp14 were first identified in a naturally occurring infertile male mouse, the jsd mouse.27 A subsequent screen for UTP14 mutations in infertile men found a heterozygous mutation that resulted in truncation of the UTP14c protein by 28 amino acids (Y738X) in three patients.26 Two of the patients presented with nonobstructive azoospermia and one presented with severe oligospermia. A testicular biopsy was available for one of the patients with azoospermia and showed that germ cells were arrested at the early stage of pachytene spermatocytes. Hormone levels were normal. These findings suggest that the failure in spermatogenesis is due to haploinsufficiency of UTP14c: not enough UTP14c protein is being produced to support the large demand for ribosomes during sperm development. Alternately, the production of a truncated UTP14c protein could have a dominant negative effect if its incorporation into the SSU processome prevents proper formation of the complex.
Cirhin – North American Indian childhood cirrhosis (NAIC)
North American Indian childhood cirrhosis is an autosomal recessive disorder found in the Ojibway-Cree population in northwestern Quebec.28 The disease presents as neonatal jaundice and then progresses to biliary cirrhosis. The only treatment is liver transplantation, which is required by early adolescence.29 Chagnon et al. reported that a missense mutation in the C-terminus of Cirhin causes NAIC, with all patients carrying two copies of the R565W mutation.28 Cirhin/UTP4 is a member of the t-Utp subcomplex of the SSU processome,3,30 a subcomplex that is required for optimal transcription of the rDNA in both yeast30 and human cells.31 Although Cirhin is known to be required for ribosome biogenesis,9,31 little is known about the molecular mechanism(s) that leads to this disease.
EMG1 – Bowen-Conradi syndrome (BCS)
Bowen-Conradi syndrome (BCS) is a lethal, autosomal recessive syndrome described primarily in the Hutterite population. Patients with BCS exhibit a variety of symptoms including pre- and post-natal growth retardation, psychomotor delay, microcephaly and multiple joint abnormalities.32 Patients rarely survive beyond the first year. Recently, a missense mutation in EMG1 was reported to be the cause of BCS.33 EMG1 is a putative methyltransferase that is required for biogenesis of the 40S subunit of the ribosome.34–36
Modeling of the EMG1 protein suggests that the D86G substitution found in BCS patients may interfere with the formation of a salt bridge, leading to aggregation and subsequent degradation of EMG1. This hypothesis is further supported by two results: (1) the D86G mutation increases dimerization of EMG1 in the yeast two-hybrid system; and (2) a dramatic reduction in EMG1 protein levels is observed in BCS fibroblasts, while EMG1 mRNA expression remains unchanged between patient and control samples.33 Thus, partial loss of EMG1 protein expression and a subsequent reduction in ribosome biogenesis provide a plausible molecular mechanism for Bowen-Conradi syndrome.
Mutations in large ribosomal subunit biogenesis proteins that cause disease
RBM28 – alopecia, neurological defects, and endocrinopathy syndrome (ANE syndrome)
Alopecia, neurological defects, and endocrinopathy syndrome (ANE syndrome) is an autosomal recessive disease that is clinically heterogeneous. ANE syndrome patients display multiple signs including a varied amount of hair loss, mental retardation, progressive loss of motor ability beginning in the second decade of life, hypogonadism, central adrenal insufficiency, short stature, microcephaly, and several other skeletal and skin abnormalities.37 A missense mutation in RBM28, a protein required for biogenesis of the 60S subunit of the ribosome,38 has been reported to cause ANE syndrome.37 Similar to EMG1, the L351P mutation in RBM28 leads to a nearly complete loss of protein expression in patient cells, although the mRNA level remains constant. This loss of RBM28 protein results in a decrease in the number of ribosomes in patient fibroblasts,37 likely due to a defect in 60S ribosomal subunit biogenesis.
SBDS – Shwachman-Bodian-Diamond syndrome (SDS)
Similar to ANE syndrome, Shwachman-Bodian-Diamond syndrome (SDS) is an autosomal recessive disorder that is pleiotropic.39 Symptoms include pancreatic insufficiency likely caused by replacement of pancreatic tissue with adipose tissue, hematologic defects including neutropenia and occasionally also anemia and/or thrombocytopenia, skeletal abnormalities, short stature, enlarged liver with elevated liver enzymes, endocrine abnormalities, defects in T- and B-cell function, and predisposition to leukemia.40 Up to 90% of cases of SDS are reported to be caused by mutations in SBDS, a protein that has an unknown specific function, but is involved in maturation and export of the ribosomal 60S subunit.41,42 SBDS may also play a role in gene expression at both the transcriptional and translational levels.43,44
The vast majority of mutations found in SDS patients are due to conversions between SBDS and a pseudogene copy of SBDS that share 97% sequence identity. The two most common mutations result in a truncated SBDS protein, due to the introduction of an inframe stop codon (K62X), or a donor splice-site mutation with a subsequent frameshift (84Cfs3), respectively. While the 84Cfs3 mutation is found in both heterozygous and homozygous states, the K62X mutation is always heterozygous suggesting that it is a null mutation and is lethal when homozygous.39 Studies on cells from SDS patients show that SBDS mutations lead to a decrease in newly synthesized rRNA with no corresponding change in polysome profile,41 shortening of telomeres,45 and an increase in Fas-mediated apoptosis.46 It is unclear whether these are primary or secondary effects of reduced SBDS levels. Nonetheless, defects in ribosome biogenesis should be considered as an etiologic agent for SDS.
Mutations in ribosome biogenesis proteins that modify disease
WDR36 – modifier/cause of primary open angle glaucoma (POAG)
Primary open angle glaucoma (POAG), a complex disorder that arises as the result of multiple genetic and environmental factors, is the leading cause of blindness worldwide. POAG usually presents in individuals over age 35 and includes degeneration of the optic nerve and often increased intraocular pressure.47 Unlike the diseases discussed above, no single gene has been identified in the etiology of POAG; however, there are three main candidates.47 One of these candidates is WDR36/UTP21, which encodes a protein of unknown specific function, but is a member of the UtpB subcomplex of the SSU processome.3 Genotyping of 130 families with POAG revealed that 9 families had WDR36 variants, all resulting in single amino acid substitutions in the protein, that segregate with the disease. These variants were divided into “disease causing” and “disease susceptibility” mutations based on their frequency of occurrence in POAG versus control populations.48 A subsequent study found that POAG patients with WDR36 variants had a more severe form of the disease than patients without WDR36 variants, suggesting that WDR36 is a modifier gene of POAG.49 Most variants are heterozygous, suggesting that haploinsufficiency of WDR36 could either cause or modify the manifestation of POAG. However, since numerous other studies find the same percentage of WDR36 variants in both POAG and control populations,50,51 the role of WDR36 in glaucoma remains unclear.
HCA66 – modifier of neurofibromatosis type 1 (NF1)
Neurofibromatosis type 1 (NF1) is an autosomal dominant tumor predilection syndrome caused by deletion of the NF1 gene. In approximately 5% of cases, a heterozygous microdeletion of genes surrounding NF1 also occurs. In such cases, the disease is more severe and includes early onset and increased number of neurofibromas, mental retardation, congenital heart defects, dysmorphic features of the head and face, and connective tissue dysplasia.52 There are four candidate genes in the microdeletion region including HCA66/UTP6,53 which encodes a protein that is also a member of the UtpB subcomplex of the SSU processome.3 While the specific function of UTP6 in ribosome biogenesis is unknown, UTP6 has been reported to play a role in promoting apoptosis via an interaction with APAF-1.54 The additional clinical features seen in the microdeletion syndrome may be due to haploin-sufficiency of UTP6, with the increase in tumors resulting from a decrease in UTP6/APAF-1 mediated apoptosis and all other signs resulting from a decrease in ribosome biogenesis.
Genetic diseases of small nucleolar ribonucleoproteins
Small nucleolar ribonucleoproteins (snoRNPs) are complexes composed of both RNA and proteins that localize to the nucleolus. Three major types that are involved in ribosome biogenesis can be distinguished by their different RNA and protein components: RNase MRP, box H/ACA snoRNPs, and box C/D snoRNPs.55 These snoRNPs perform enzymatic functions, including endonucleolytic cleavage and chemical modifications of the pre-rRNA, and also function as chaperones to facilitate folding of the pre-rRNA. Mutations in genes encoding either the non-coding RNA components or the protein components of these RNPs can result in disease. To date, diseases caused by mutations in all three classes of snoRNPs have been identified (Table 2).
Table 2.
Genetic diseases of small nucleolar ribonucleoproteins
| Gene |
Mutated protein/RNA |
Impaired molecular function |
Disease | Clinical manifestations |
Inheritance and incidence (if known) |
OMIM # | References | |
|---|---|---|---|---|---|---|---|---|
| human | yeast | |||||||
| RMRP | NME1 | noncoding RNA component of RNase MRP |
maturation of 5.8S rRNA of 60S ribosomal subunit |
Anauxetic dysplasia (AD) |
extreme short stature, hypodontia, mild mental retardation, severely reduced number of chondrocytes |
autosomal recessive; <200,000 cases in the US |
#607095 | 63,156,157 |
| RMRP | NME1 | as above | maturation of 5.8S rRNA of 60S ribosomal subunit; degradation of cell-cycle regulated RNAs; mitochondrial DNA replication |
Cartilage-hair dysplasia (also called metaphyseal chondrodysplasia McKusick type; CHH) |
disproportional short stature, metaphyseal dysplasia, hair hypoplasia, anemia, immunodeficiency, Hirschprung disease, cancer predisposition |
autosomal recessive; <200,000 cases in the US |
#250250 | 59,62 |
| RMRP | NME1 | as above | as above | Metaphyseal dysplasia without hypotrichosis (MDWH) |
short stature, metaphyseal skeletal changes |
autosomal recessive; <200,000 cases in the US |
#250460 | 59,157,158 |
| DKC1 | CBF5 | pseudour- idine synthase of box H/ACA RNPs (snoRNPs, scaRNPs, and telomerase) |
telomerase deficiency, disease aggravated by box H/ACA snoRNP pseudouridiylation defects |
Dyskeratosis congenita (also called Zinsser-Engman- Cole syndrome) |
mucocutaneous abnormalities, bone marrow failure, cancer predisposition |
X-linked recessive; prevalence: 1 in 1,000,000 |
#305000 | 70,72,78 |
| DKC1 | CBF5 | as above | as above | Hoyeraal- Hreidarsson syndrome (some cases are severe variants of Dyskeratosis congenita) |
severe growth re- tardation, bone mar- row failure, immunodeficiency, microcephaly, cerebellar hypoplasia |
X-linked recessive; prevalence: 1 in 1,000,000 |
#300240 | 70,71 |
| NOP10 | NOP10 | protein component of box H/ACA RNPs (snoRNPs, scaRNPs, and telomerase) |
telomerase deficiency, disease aggravated by box H/ACA snoRNP pseudouridiylation defects |
Dyskeratosis congenita (also called Zinsser-Engman- Cole syndrome) |
mucocutaneous abnormalities, bone marrow failure, cancer predisposition |
autosomal recessive; prevalence: 1 in 1,000,000 |
#224230 | 70,159 |
| NHP2 | NHP2 | protein component of box H/ACA RNPs (snoRNPs, scaRNPs, and telomerase) |
telomerase deficiency, disease aggravated by box H/ACA snoRNP pseudouridiylation defects |
Dyskeratosis congenita (also called Zinsser- Engman-Cole syndrome) |
mucocutaneous abnormalities, bone marrow failure, cancer predisposition |
autosomal recessive; prevalence: 1 in 1,000,000 |
#224230 | 70,160 |
| HBII-85 | — | deleted box C/D snoRNA cluster |
specific function unknown |
Prader-Willi syndrome (PWS) |
neonatal hypotonia, short stature, hyperphagia, obesity, mental retardation, hypogonadism |
autosomal, paternal deletion or maternal disomy of Chr. 15q12, imprinting defects; incidence: 1 in 10,000 to 15,000 in live births |
#176270 | 90 |
RNase MRP snoRNA – skeletal dysplasias
RNase MRP (mitochondrial RNA processing) is an RNP composed of the non-coding MRP RNA and at least 10 different proteins. Studies in yeast and mammalian cells show that RNase MRP is essential and has multiple functions within cells: (1) it is involved in ribosome biogenesis, being an endonuclease that cleaves the pre-rRNA and separates the precursors of the SSU from the LSU pre-rRNA and is specifically required for the maturation of the 5.8S rRNA;13 (2) it is involved in cell cycle regulation by cleaving the cyclin B2 mRNA;56 and (3) it is involved in mitochondrial DNA replication.57,58
A variety of mutations in the gene for the non-coding RNA component of RNase MRP have been linked to genetically inherited skeletal dysplasias. Depending on the severity of the skeletal dysplasia and the accompanying extraskeletal manifestations, they are clinically classified into: metaphyseal dysplasia without hypotrichosis (MDWH) with mild dysplasia, cartilage-hair dysplasia (CHH) with moderate dysplasia, and anauxetic dysplasia (AD) with severe dysplasia. CHH is also associated with a variety of extraskeletal abnormalities, including hypotrichosis, hypoplastic anemia, immunodeficiency, and an increased predisposition to cancer.59
Mutations associated with these diseases all affect the MRP gene and cluster in the promoter region, where they can result in a decreased expression of the encoded RNA, or in evolutionarily highly conserved regions of the MRP RNA.60–62 Recent phenotype-genotype correlation studies indicate that mutations that impair the rRNA processing function of RNase MRP more severely affect the skeletal system and are less frequently associated with extraskeletal manifestations (as in AD). Conversely mutations that affect both rRNA processing and mRNA processing are observed in patients with less severe skeletal abnormalities but with extraskeletal symptoms (as in CHH).63,64 Thus, different mutations in RNase MRP appear to separate the different functions of RNase MRP genetically, resulting in a varied spectrum of clinical manifestations. While so far disease causing mutations have been found only in the RNA component of RNase MRP, it remains to be seen whether future studies and genetic analysis of more patients with these clinical signs will also reveal mutations in genes encoding the protein components of RNase MRP that impair function.
The molecular mechanisms by which these pathogenic mutations affect the different functions of RNase MRP remain poorly understood. Several scenarios, some of which are supported by the literature, can be imagined: (1) mutations in structurally important regions of the MRP RNA can result in RNA structural changes that may interfere with RNP assembly;65 (2) reduced transcription of the MRP RNA due to promoter mutations60,61 and instability of the MRP RNA transcript66 may reduce steady state levels of both the RNA and the RNase MRP snoRNP below a threshold required for normal cellular function; and (3) altered MRP RNA and RNP may interfere with substrate recognition or catalytic activity itself. A combined effort of genetic (including mouse models), biochemical, and structural studies will be necessary to better understand the mutant RNase MRP complex and the underlying molecular mechanisms resulting in these diseases.
Box H/ACA snoRNPs – Dyskeratosis congenita (DC)
Dyskeratosis congenita (DC) is a genetic disease characterized by genetic as well as symptomatic heterogeneity. Clinically, patients can present with mucocutaneous abnormalities, a predisposition to a variety of cancers, and bone marrow failure, the latter being the leading cause of mortality.67–70 In severe cases, DC can also be associated with immunodeficiency, growth retardation and neurological symptoms (Hoyeraal-Hreidarsson syndrome).71 Genetic inheritance occurs as either X-linked recessive or autosomal dominant or recessive. Hence, DC can be caused by mutations in a variety of different genes: the X-linked form of DC is caused by mutations in the dyskerin gene, encoding an essential component of box H/ACA RNPs (including snoRNPs, scaRNPs [small Cajal body-specific RNPs], and telomerase); the autosomal recessive form can be caused either by mutations in NHP2, NOP10 (both encoded proteins are also essential components of box H/ACA RNPs), or TERT (gene encoding the reverse transcriptase of the telomerase RNP); and the autosomal dominant form is associated with mutations in the telomerase RNA or TERT.68,70
The original discovery of mutations in the dyskerin gene suggested that impaired ribosome biogenesis is the etiological molecular defect, since dyskerin is an essential protein component of box H/ACA snoRNPs, which catalyze pseudouridylation of rRNAs and are required for pre-rRNA processing.72 This conclusion is supported by results from studies of mouse models of the X-linked recessive form of DC, which target the dyskerin gene and reproduce some of the clinical phenotypes of the disease including the ribosome biogenesis defect, although to varying degrees.73–76 Furthermore, it has been shown that ribosomes in DC patient cells are functionally distinct from normal ribosomes, since IRES-dependent translation of some mRNAs is reduced.77 However, the subsequent discovery of mutations in telomerase RNA or the gene for telomerase reverse transcriptase (TERT) and the fact that telomerase is a box H/ACA RNP containing all protein components that are found in box H/ACA snoRNPs suggests instead that DC is primarily caused by impaired function of telomerase.70,78 This hypothesis is confirmed by the observation of shortened telomeres, reduced telomerase levels and activity, but normal rRNA processing in DC patient cells.79,80
So what is the underlying molecular etiology of DC? Given the genetic and clinical heterogeneity of DC, it is most likely that, at least in some cases, the disease is caused by a combined effect of impaired telomerase, box H/ACA snoRNP and box H/ACA scaRNP functions, especially in the case of the X-linked form. Interestingly, the X-linked form of DC caused by mutations in the dyskerin gene typically presents with more severe symptoms than the autosomal dominant form of DC that is caused by mutations in telomerase RNA or the TERT gene directly. The more severe disease caused by dyskerin mutations could be explained by a combined effect of dysfunction in both telomere maintenance and ribosome biogenesis. Future studies will be necessary to uncouple both functions of H/ACA RNP components better genetically. Until then, a contribution of ribosome biogenesis defects to the molecular pathogenesis of DC, especially the X-linked form, should not be neglected.
Box C/D snoRNPs – Prader-Willi syndrome (PWS)
Prader-Willi syndrome (PWS) is a complex disease characterized by neonatal hypotonia, short stature, hyperphagia and obesity, hypogonadism and characteristic facial features.81 Genetic mapping linked PWS to chromosomal region 15q11-q13. Gene expression in this region is regulated by imprinting, and PWS results from loss of expression of genes normally only expressed from the paternal chromosome.82 Most cases of PWS result from parental deletions or maternal disomy of chromosome 15q11-q13, and in a some cases imprinting defects are responsible for silencing of the genes. Interestingly, while the PWS locus contains few protein encoding genes, it encodes numerous non-coding RNAs of the box C/D type. Fine mapping studies have shown that a loss of expression of the HBII-85 box C/D snoRNA cluster is responsible for most of the symptoms observed in PWS patients.83–90 Consistent with this, mouse models that include deletion of these snoRNAs of the PWS locus recapitulate some of the pheno-types observed in PWS patients.91–93 However, a contribution of other box C/D snoRNA genes to the complex clinical manifestations observed in PWS patients cannot be completely ruled out at this point.
Box C/D snoRNAs in humans associate with the conserved proteins, 15.5 K, NOP56, NOP58, and fibrillarin to form box C/D type RNPs.55 Box C/D snoRNPs typically catalyze 2′-O-ribose methylation of ribosomal RNA, participate in pre-rRNA folding, and some are also essential for pre-rRNA processing.94 The specificity of these events is dictated by base pairing of the guide sequences of box C/D snoRNAs with the substrate RNAs. Besides their function in ribosome biogenesis, box C/D type RNPs have also been shown to perform 2′-O-ribose methylations of snRNAs (box C/D scaRNPs) and at least one mRNA; the latter has been shown to regulate mRNA splicing and mRNA editing.95–98 Very recently, box C/D type RNAs have also been suggested to be precursors for microRNA-like small RNAs.99,100 Thus, box C/D type RNAs and RNPs function in a variety of different aspects of RNA processing.
Box C/D snoRNAs encoded by the PWS locus most likely assemble into box C/D RNPs since they can be immunoprecipitated with antibodies against fibrillarin.83 However, the target RNAs of the snoRNAs encoded in the PWS locus are largely unknown, with the exception of the HBII-52 snoRNAs and a few yet unvalidated snoRNAs predicted to target ribosomal RNAs.83,96–98,101 Unfortunately, the targets of the HBII-85 snoRNA cluster remain completely obscure, and hence it is unkown how loss of expression of these snoRNAs results in PWS. Identification of their target RNAs as well as defining the function of these RNAs will be crucial for understanding the molecular mechanism underlying this disease.
Genetic diseases of ribosomal proteins
In mammals there are about 80 ribosomal proteins (r-proteins) that are structural components of the ribosome, with some of them also participating in the control of the translation process. Many r-proteins have roles in the processing of pre-rRNA during ribosome biogenesis and are important for the assembly of the ribosomal subunits. Some have additionally been shown to have extraribosomal functions, but in general, their known functions are in ribosome biogenesis or in its surveillance.102,103 Surprisingly, it was discovered that haploinsufficiency of some r-proteins leads to diseases of the bone marrow such as Diamond Blackfan anemia and the 5q− syndrome (Table 3).
Diamond Blackfan anemia (DBA)
Diamond-Blackfan anemia (DBA) is an inherited bone marrow failure syndrome of children characterized by proapoptotic hematopoiesis, bone marrow failure, birth defects and a predisposition to cancer. About 40% of the patients also present with craniofacial, cardiac, limb, and urogenital abnormalities.104 Recently, a number of reviews have discussed different aspects of the disease, its diagnosis, treatment, and molecular pathogenesis.104–106 Unexpectedly, all the genes currently shown to be involved in DBA are r-protein genes and account for about 50% of DBA cases.107 Haploinsufficiency of these r-proteins is thus likely to be the basis for DBA. Different types of mutations have been found in genes corresponding to both small and large subunit r-proteins: RPS19 (25%), RPL5 (6.6%), RPL11 (4.8%), RPL35A (3%), RPS24 (2%), RPS17 (1%), and RPS7 (<1%).108–112 Possible mutations were also reported for RPS15, RPS27A, and RPL36.112 Mutations in the most commonly affected gene, RPS19, include nonsense and missense mutations, small insertions and deletions, splice site defects and large deletions, and rearrangements.113
Correlations between some of the symptoms observed in DBA and the mutated r-protein are emerging. Physical malformations are more often associated with RPL5 and RPL11 mutations (~70%) compared to RPS19 (~46%).112 Furthermore, mutations in RPL5 seem to give rise to a more severe manifestation and mutations in only this gene are associated with cleft lip and/or cleft palate abnormalities in DBA.
RPS7, RPS17, RPS19, and RPS24 are involved in the maturation of the 18S rRNA and are required for the assembly of the 40S subunit.112,114–116 Interestingly, Rps7 was also shown to be a component of the SSU processome in yeast.117 Similarly, the large ribosomal subunit proteins RPL5, RPL11 and RPL35A participate in the processing of the rRNAs that compose the large subunit and are essential for the assembly of the mature large subunit.110,112,118 Defects in pre-rRNA processing were observed in primary fibroblasts and bone marrow progenitor cells from patients with DBA for all of the r-proteins mentioned,111,112,119–121 confirming that defects in ribosome biogenesis contribute to DBA.
The r-proteins involved in DBA also have other functions that could contribute to the development of the disease, and many of these functions could contribute to the predisposition to cancer observed in DBA patients. For example, RPS19 interacts with the PIM-1 oncoprotein (a serine-threonine kinase which phosphorylates proteins like CDC25C and c-MYC) in vivo and PIM-1 phosphorylates RPS19 in vitro.122 RPL11 can modulate the activity of both the tumor suppressor p53123 and c-MYC, an oncoprotein and transcription factor.124 Similarly, RPS7, RPL5 and RPL23 can also modify p53 activity.123,125
All the mutations found so far in DBA patients have been in genes encoding r-proteins, including components of both small and large ribosomal subunits, making it very likely that DBA is a ribosome biogenesis disease. All of the r-proteins identified are required for both pre-rRNA processing and ribosome assembly. Interestingly, not all of them affect the same step in ribosome biogenesis, and they can affect the formation of the small or large subunit. To complicate the issue even further, often times when the level of expression of one r-protein gene is reduced other r-proteins are expressed at lower levels also.116 To explain the pathogenesis of DBA, the emerging consensus is that ribosome biogenesis defects lead to a reduced number of ribosomes available to the cell and consequently to reduced translation, growth in cell size and cell division. During the proliferation and differentiation of red blood cell precursors there is a high demand for ribosomes, since red blood cells proliferate very rapidly. It has been proposed that because these processes require very high translation rates, they present an elevated sensitivity to the effects of diminished ribosome production.106 Another hypothesis involves the connection between ribosome biogenesis and the tumor suppressor p53: the “ribosomal stress hypothesis”.104,105 In particular, aberrant ribosome biogenesis as a result of a decrease in an r-protein would activate p53-mediated cell cycle arrest and apoptosis. Studies in zebrafish and mouse models support this hypothesis. In zebrafish, p53 activation is involved in the mechanism by which rps19 haploinsufficiency gives rise to a reduction in erythrocytes and physical abnormalities,126 and loss of Rpl11 affects embryonic development.127 A new mouse model with a mutation in Rps19 presented with some of the DBA signs, including decreased red blood cells and reticulocyte levels and increased apoptosis.128 Somewhat puzzling is that no defects in the bone marrow were observed in these mice. More details on the links between ribosome biogenesis and the p53 pathway are presented in the section “Cancer and ribosome biogenesis”.
5q− syndrome
The 5q− syndrome is a myelodysplastic syndrome, preponderant in adult females, characterized by a defect in erythroid differentiation and associated with progression to acute myeloid leukemia (AML). As the name implies, the 5q− syndrome is characterized by a deletion in the long arm of chromosome 5.129 The critical region for the 5q− syndrome contains 40 genes,130 including RPS14. Recently, a study that used an RNA interference-based functional screen showed that RPS14 is the haploinsufficient tumor suppressor gene associated with the syndrome.131 Rps14 is a component of both the small subunit of the ribosome and of the SSU processome in yeast,117 and has been shown to be required for processing of the 18S rRNA and ribosome assembly.131,132 To further confirm the importance of ribosome biogenesis for 5q− syndrome, about 55 ribosomal- and translation- related factors were found to be differentially expressed in cells of patients with 5q−, including some r-proteins.133
Notably, there are some striking similarities between the 5q− syndrome and DBA (Table 4). Even though the age of onset is quite different, as the 5q− syndrome appears in adulthood and DBA in early childhood, they both present with macrocytic anemia, and both result in a predilection for AML. For both, the underlying molecular defect is a defective or absent r-protein. It is thus plausible that a similar type of molecular mechanism acts in both diseases.
Table 4.
Comparison of Diamond Blackfan anemia and 5q− syndrome characteristics
| Characteristic | Diamond Blackfan anemia | 5q− syndrome |
|---|---|---|
| Age and gender predominance | Younger than a year, both sexes equally affected | Older adults, female predominance |
| Macrocytic anemia | Present | Present |
| Cancer predisposition | Increased (AML, solid tumors) | Can progress to AML |
| Congenital abnormalities | Frequently present | Not present |
| Treatment | Transfusions, corticosteroids, spontaneous remission frequent | Transfusions, lenalidomide |
Cancer and ribosome biogenesis
The links between ribosome biogenesis and cancer are complex and not yet well understood. Ribosome biogenesis and nuclear structure are altered significantly to meet the needs of cancer cells,134,135 which are usually characterized by high protein synthesis and rRNA transcription rates. The nucleolar number, size and morphology are convenient diagnostic markers for cancer.134,135 Dysregulation of ribosome biogenesis has been associated with alterations in cell cycle, cell proliferation and cell growth and often contributes to increased susceptibility to cancer.134,136 It is not surprising, therefore, that ribosome biogenesis and ribosome function have become targets for anticancer therapies.137
R-proteins can regulate the c-MYC oncogene and the p53 tumor suppressor, which in turn both function as ribosome synthesis regulators.138,139 One of the mechanisms by which the activity of the transcription factor p53 can be regulated by r-proteins is through binding to MDM2, the main E3-ligase of p53 which ubiquitinates p53 and promotes its degradation.140 Three large subunit r-proteins, RPL5, RPL11 and RPL23,123,141,142 and a small subunit r-protein, RPS7,123,125,143 bind MDM2 and activate p53 upon ribosomal stress. Recently, it was also proposed that RPL5 and RPL11 act together in this function.144 A number of other r-proteins were also proposed to regulate the activity of p53, and although their mechanism of action is not known, it was proposed that they do not necessarily act through MDM2.145 RPL26 binds to the 5′ and/or 3′UTR of human p53 mRNA and selectively promotes p53 translation,146 and haploinsufficiency of RPS6 activates the p53 tumor suppressor.147,148 Recently, it was proposed that reduction in the expression of small subunit r-proteins, in particular RPS6 and RPS7, and subsequent disruption of 40S ribosome biogenesis will activate p53 by an RPL11-translation mechanism.147 Specifically, in the aforementioned conditions, the translation of mRNAs with a polypyrimidine tract at the transcriptional start (5′-TOP mRNAs) will be upregulated, regardless of the decrease in global translation. Since all r-proteins have a 5′-TOP, RPL11 would be overexpressed, inhibit MDM2, and activate p53.147
The c-MYC oncogene is a transcription factor that activates genes involved in processes including cell division, apoptosis and angiogenesis. Furthermore, c-MYC has an important role in ribosome biogenesis, since it regulates transcription by RNA polymerases.138,149 Haploinsufficiency of some r-proteins has been shown to suppress c-MYC oncogenic activity,124,150 which is unexpected given that haploinsufficiency of r-proteins often leads to an increase in tumors. Interestingly, RPL11 is one of the r-proteins that inhibits c-MYC activity and does so through a negative feedback mechanism.124,151
Several r-proteins are overexpressed in a variety of tumors,134,135,137 but it is not yet clear if these alterations are a cause or a result of tumorigenesis. In addition, it has also been shown that reduced ribosome biogenesis or loss of function of r-protein genes can be correlated with malignant transformation in zebrafish,152 while overexpression of r-protein genes can prevent tumor growth.
Conclusions and perspective
Ribosomes are essential for life. If functional, mature ribosomes are not synthesized, a decrease in protein translation occurs, resulting in reduced cell growth and cell death. Therefore, it is expected that mutations in ribosomal proteins or ribosome biogenesis factors would result in embryonic lethality; surprisingly this is not always the case. Over the last few decades, multiple rare, genetic diseases have been attributed to defects in ribosome function or ribosome biogenesis. With the current advances in whole genome sequencing, more diseases of ribosome biogenesis are likely to be discovered.
While diseases of ribosome biogenesis span a wide range of symptoms, impaired molecular processes, and modes of inheritance, some common patterns between diseases can be observed (see Tables 1–3). First, nearly all of the described diseases are the result of haploinsufficiency (e.g. Treacher Collins syndrome and 5q− syndrome) or partial loss of protein expression (e.g. Bowen-Conradi syndrome), supporting the idea that homozygous null mutations in ribosome biogenesis factors are embryonic lethal. Second, although no one symptom is present in all diseases, many diseases share clinical features such as short stature, mental retardation, joint abnormalities, anemia and predisposition to cancer. Third, mutations in many ribosomal and ribosome biogenesis proteins seem to activate the p53 pathway, providing an underlying mechanism for cell death and perhaps partially explaining the connection between ribosome biogenesis and cancer, as discussed earlier.
Taken together, the most perplexing feature of all of the ribosome biogenesis diseases is the tissue proclivity of the manifestations. Ribosomes are present in all cells in all tissues in the body and are required for cell survival, cellular growth and division; therefore, all tissues should be equally affected by defects in ribosome biogenesis. However, this is not the case. For some diseases, such as male infertility, the tissue proclivity is easy to explain since UTP14c expression is under the control of a testis-specific promoter.26 However, in most cases, the cause of tissue proclivity remains somewhat of a mystery. It has been proposed that highly proliferating tissues, such as the bone marrow, would have a high need for ribosomes, and would be more affected than the other organ systems.106 Indeed, this is observed in DBA and the 5q− syndrome. Nonetheless, this hypothesis cannot entirely account for the tissue proclivity since in a developing embryo, all tissues are dividing rapidly and therefore they should all be affected if adequate numbers of ribosomes cannot be made. Furthermore, the bone marrow is not the only self-renewing organ system in the adult human.
So how does tissue specificity arise? Although little published evidence exists, we can speculate on several possibilities: the disease-associated proteins may have tissue-specific interaction partners, resulting in impaired function in only particular tissue types. Or, rather than tissue-specific interacting proteins, their gene expression may be regulated by tissue-specific microRNAs. Furthermore, the encoded mRNAs may have tissue-specific alternatively spliced forms, an appealing possibility given that almost 95% of human genes are alternatively spliced153 and that tissue-specific transcripts have been observed for some of the mRNAs that encode ribosome biogenesis factors (e.g. see ref. 21,39). Another possibility is that these proteins have extra-ribosomal functions, as is observed with some of the r-proteins implicated in Diamond Blackfan anemia,122,123,151 and it is these other functions that confer tissue proclivity. Elucidation of the molecular mechanisms of ribosome biogenesis diseases, and in particular the reasons for tissue proclivity, remain some of the main challenges for the future.
Acknowledgements
This work was supported by NIH grant R01GM52581 (S.J.B.), Yale Liver Center Grant NIDDK P30-34989 (S.J.B.), the Anna Fuller Fund Fellowship in Molecular Oncology (L.M.D.) and NIH 5 T32 HD07149-32. We would like to thank E. Champion and A. Szekely for their help and contributions.
Biographies

Emily F. Freed
Emily F. Freed received her BS from Stanford University. She is now a PhD candidate at Yale studying how ribosome dysfunction leads to disease.

Franziska Bleichert
Franziska Bleichert, a PhD student at Yale University, is studying small ribonucleo-proteins involved in ribosome biogenesis. Her work focuses on understanding the architecture and structure of these ribonucleoproteins and how this structure dictates the functions of these complexes.

Laura M. Dutca studied chemistry at West University of Timisoara, Romania. In 2006 she graduated with a PhD in Chemistry from Iowa State University, where she developed an interest in the study of large macromolecular assemblies. As a postdoctoral researcher at Yale University she is developing biochemical methods to investigate structural changes in pre-ribosomal RNA and formation of macro-molecular complexes during ribosome biogenesis in yeast.

Susan J. Baserga, MD, PhD, received her degrees from Yale University and is a Professor of Molecular Biophysics & Biochemistry, Genetics and Therapeutic Radiology at Yale.
References
- 1.Gavin AC, et al. Nature. 2002;415:141. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 2.Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1143. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krogan NJ, et al. Mol. Cell. 2004;13:225. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 4.Peng WT, et al. Cell. 2003;113:919. doi: 10.1016/s0092-8674(03)00466-5. [DOI] [PubMed] [Google Scholar]
- 5.Granneman S, Baserga SJ. Exp. Cell Res. 2004;296:43. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 6.Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. Cell. Mol. Life Sci. 2008;65:2334. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warner JR, Vilardell J, Sohn JH. Cold Spring Harbor Symp. Quant. Biol. 2001;66:567. doi: 10.1101/sqb.2001.66.567. [DOI] [PubMed] [Google Scholar]
- 8.Lewis JD, Tollervey D. Science. 2000;288:1385. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- 9.Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, Beyer AL, Hunt DF, Baserga SJ. Nature. 2002;417:967. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schäfer T, Kuster B, Tschochner H, Tollervey D, Gavin AC, Hurt E. Mol. Cell. 2002;10:105. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 11.Bleichert F, Granneman S, Osheim YN, Beyer AL, Baserga SJ. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9464. doi: 10.1073/pnas.0603673103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elela SA, Igel H, Ares M., Jr Cell. 1996;85:115. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 13.Schmitt ME, Clayton DA. Mol. Cell Biol. 1993;13:7935. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganot P, Bortolin ML, Kiss T. Cell. 1997;89:799. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 15.Kiss-László Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Cell. 1996;85:1077. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 16.Nicoloso M, Qu LH, Michot B, Bachellerie JP. J. Mol. Biol. 1996;260:178. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- 17.Turner AJ, Knox AA, Prieto JL, McStay B, Watkins NJ. Mol. Cell. Biol. 2009;29:3007. doi: 10.1128/MCB.00029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pérez-Fernández J, Román A, De Las Rivas J, Bustelo XR, Dosil M. Mol. Cell. Biol. 2007;27:5414. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richter A, Mitchell GA, Rasquin A. Med. Sci. (Paris) 2007;23:1002. doi: 10.1051/medsci/200723111002. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S, Shi M, Hui CC, Rommens JM. Mol. Cell. Biol. 2006;26:6656. doi: 10.1128/MCB.00091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The Treacher Collins Syndrom Collaborative Group. Nat. Genet. 1996;12:130. doi: 10.1038/ng0296-130. [DOI] [PubMed] [Google Scholar]
- 22.Dixon J, Dixon MJ. Dev. Dyn. 2004;229:907. doi: 10.1002/dvdy.20004. [DOI] [PubMed] [Google Scholar]
- 23.Valdez BC, Henning D, So RB, Dixon J, Dixon MJ. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10709. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzales B, Henning D, So RB, Dixon J, Dixon MJ, Valdez BC. Hum. Mol. Genet. 2005;14:2035. doi: 10.1093/hmg/ddi208. [DOI] [PubMed] [Google Scholar]
- 25.Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, Rey JP, Dixon MJ, Trainor PA. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13403. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohozinski J, Lamb DJ, Bishop CE. Biol. Reprod. 2006;74:644. doi: 10.1095/biolreprod.105.046698. [DOI] [PubMed] [Google Scholar]
- 27.Rohozinski J, Bishop CE. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11695. doi: 10.1073/pnas.0401130101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chagnon P, Michaud J, Mitchell G, Mercier J, Marion JF, Drouin E, Rasquin-Weber A, Hudson TJ, Richter A. Am. J. Hum. Genet. 2002;71:1443. doi: 10.1086/344580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bétard C, Rasquin-Weber A, Brewer C, Drouin E, Clark S, Verner A, Darmond-Zwaig C, Fortin J, Mercier J, Chagnon P, Fujiwara TM, Morgan K, Richter A, Hudson TJ, Mitchell GA. Am. J. Hum. Genet. 2000;67:222. doi: 10.1086/302993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. Genes Dev. 2004;18:2506. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prieto JL, McStay B. Genes Dev. 2007;21:2041. doi: 10.1101/gad.436707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowen P, Conradi GJ. Birth Defects Orig. Artic. Ser. 1976;12:101. [PubMed] [Google Scholar]
- 33.Armistead J, et al. Am. J. Hum. Genet. 2009;84:728. doi: 10.1016/j.ajhg.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eschrich D, Buchhaupt M, Kötter P, Entian KD. Curr. Genet. 2002;40:326. doi: 10.1007/s00294-001-0269-4. [DOI] [PubMed] [Google Scholar]
- 35.Leulliot N, Bohnsack MT, Graille M, Tollervey D, Van Tilbeurgh H. Nucleic Acids Res. 2008;36:629. doi: 10.1093/nar/gkm1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu PC, Thiele DJ. Mol. Biol. Cell. 2001;12:3644. doi: 10.1091/mbc.12.11.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nousbeck J, et al. Am. J. Hum. Genet. 2008;82:1114. doi: 10.1016/j.ajhg.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun C, Woolford JL., Jr EMBO J. 1994;13:3127. doi: 10.1002/j.1460-2075.1994.tb06611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM. Nat. Genet. 2003;33:97. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 40.Shimamura A. Semin. Hematol. 2006;43:178. doi: 10.1053/j.seminhematol.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 41.Ganapathi KA, Austin KM, Lee CS, Dias A, Malsch MM, Reed R, Shimamura A. Blood. 2007;110:1458. doi: 10.1182/blood-2007-02-075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menne TF, Goyenechea B, Sánchez-Puig N, Wong CC, Tonkin LM, Ancliff PJ, Brost RL, Costanzo M, Boone C, Warren AJ. Nat. Genet. 2007;39:486. doi: 10.1038/ng1994. [DOI] [PubMed] [Google Scholar]
- 43.Hesling C, Oliveira CC, Castilho BA, Zanchin NI. Exp. Cell Res. 2007;313:4180. doi: 10.1016/j.yexcr.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 44.Rujkijyanont P, Adams SL, Beyene J, Dror Y. Br. J. Haematol. 2009;145(6):806–815. doi: 10.1111/j.1365-2141.2009.07692.x. [DOI] [PubMed] [Google Scholar]
- 45.Calado RT, Graf SA, Wilkerson KL, Kajigaya S, Ancliff PJ, Dror Y, Chanock SJ, Lansdorp PM, Young NS. Blood. 2007;110:1141. doi: 10.1182/blood-2007-03-080044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rujkijyanont P, Watanabe K, Ambekar C, Wang H, Schimmer A, Beyene J, Dror Y. Haematologica. 2008;93:363. doi: 10.3324/haematol.11579. [DOI] [PubMed] [Google Scholar]
- 47.Allingham RR, Liu Y, Rhee DJ. Exp. Eye Res. 2009;88:837. doi: 10.1016/j.exer.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monemi S, Spaeth G, DaSilva A, Popinchalk S, Ilitchev E, Liebmann J, Ritch R, Héon E, Crick RP, Child A, Sarfarazi M. Hum. Mol. Genet. 2005;14:725. doi: 10.1093/hmg/ddi068. [DOI] [PubMed] [Google Scholar]
- 49.Hauser MA, Allingham RR, Linkroum K, Wang J, LaRocque-Abramson K, Figueiredo D, Santiago-Turla C, del Bono EA, Haines JL, Pericak-Vance MA, Wiggs JL. Invest. Ophthalmol. Visual Sci. 2006;47:2542. doi: 10.1167/iovs.05-1476. [DOI] [PubMed] [Google Scholar]
- 50.Fingert JH, Alward WL, Kwon YH, Shankar SP, Andorf JL, Mackey DA, Sheffield VC, Stone EM. Arch. Ophthalmol. 2007;125:434. doi: 10.1001/archopht.125.3.434-b. [DOI] [PubMed] [Google Scholar]
- 51.Hewitt AW, Dimasi DP, Mackey DA, Craig JE. Am. J. Ophthalmol. 2006;142:324. doi: 10.1016/j.ajo.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 52.Mensink KA, Ketterling RP, Flynn HC, Knudson RA, Lindor NM, Heese BA, Spinner RJ, Babovic-Vuksanovic D. J. Med. Genet. 2006;43:e08. doi: 10.1136/jmg.2005.034256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartelt-Kirbach B, Wuepping M, Dodrimont-Lattke M, Kaufmann D. Neurogenetics. 2009;10:79. doi: 10.1007/s10048-008-0154-0. [DOI] [PubMed] [Google Scholar]
- 54.Piddubnyak V, Rigou P, Michel L, Rain JC, Geneste O, Wolkenstein P, Vidaud D, Hickman JA, Mauviel A, Poyet JL. Cell Death Differ. 2007;14:1222. doi: 10.1038/sj.cdd.4402122. [DOI] [PubMed] [Google Scholar]
- 55.Matera AG, Terns RM, Terns MP. Nat. Rev. Mol. Cell Biol. 2007;8:209. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 56.Gill T, Cai T, Aulds J, Wierzbicki S, Schmitt ME. Mol. Cell. Biol. 2004;24:945. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang DD, Clayton DA. Science. 1987;235:1178. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- 58.Chang DD, Clayton DA. EMBO J. 1987;6:409. doi: 10.1002/j.1460-2075.1987.tb04770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin AN, Li Y. Cell Res. 2007;17:219. doi: 10.1038/sj.cr.7310120. [DOI] [PubMed] [Google Scholar]
- 60.Bonafé L, Dermitzakis ET, Unger S, Greenberg CR, Campos-Xavier BA, Zankl A, Ucla C, Antonarakis SE, Superti-Furga A, Reymond A. PLoS Genet. 2005;1:e47. doi: 10.1371/journal.pgen.0010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hermanns P, Bertuch AA, Bertin TK, Dawson B, Schmitt ME, Shaw C, Zabel B, Lee B. Hum. Mol. Genet. 2005;14:3723. doi: 10.1093/hmg/ddi403. [DOI] [PubMed] [Google Scholar]
- 62.Ridanpää M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, vanVenrooij W, Pruijn G, Salmela R, Rockas S, Mäkitie O, Kaitila I, de la Chapelle A. Cell. 2001;104:195. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 63.Thiel CT, Horn D, Zabel B, Ekici AB, Salinas K, Gebhart E, Rüschendorf F, Sticht H, Spranger J, Müller D, Zweier C, Schmitt ME, Reis A, Rauch A. Am. J. Hum. Genet. 2005;77:795. doi: 10.1086/497708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thiel CT, Mortier G, Kaitila I, Reis A, Rauch A. Am. J. Hum. Genet. 2007;81:519. doi: 10.1086/521034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Welting TJ, Mattijssen S, Peters FM, van Doorn NL, Dekkers L, van Venrooij WJ, Heus HA, Bonafé L, Pruijn GJ. Biochim. Biophys. Acta. Mol. Cell Res. 2008;1783:455. doi: 10.1016/j.bbamcr.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 66.Nakashima E, Tran JR, Welting TJ, Pruijn GJ, Hirose Y, Nishimura G, Ohashi H, Schurman SH, Cheng J, Candotti F, Nagaraja R, Ikegawa S, Schlessinger D. Am. J. Med. Genet., Part A. 2007;143a:2675. doi: 10.1002/ajmg.a.32053. [DOI] [PubMed] [Google Scholar]
- 67.Kirwan M, Dokal I. Clin. Genet. 2008;73:103. doi: 10.1111/j.1399-0004.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 68.Savage SA, Alter BP. Hematol./Oncol. Clin. North Am. 2009;23:215. doi: 10.1016/j.hoc.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walne AJ, Dokal I. Mech. Ageing Dev. 2008;129:48. doi: 10.1016/j.mad.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 70.Walne AJ, Dokal I. Br. J. Haematol. 2009;145:164. doi: 10.1111/j.1365-2141.2009.07598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yaghmai R, Kimyai-Asadi A, Rostamiani K, Heiss NS, Poustka A, Eyaid W, Bodurtha J, Nousari HC, Hamosh A, Metzenberg A. J. Pediatr. 2000;136:390. doi: 10.1067/mpd.2000.104295. [DOI] [PubMed] [Google Scholar]
- 72.Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. Nat. Genet. 1998;19:32. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 73.Gu BW, Bessler M, Mason PJ. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10173. doi: 10.1073/pnas.0803559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He J, Navarrete S, Jasinski M, Vulliamy T, Dokal I, Bessler M, Mason PJ. Oncogene. 2002;21:7740. doi: 10.1038/sj.onc.1205969. [DOI] [PubMed] [Google Scholar]
- 75.Mochizuki Y, He J, Kulkarni S, Bessler M, Mason PJ. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10756. doi: 10.1073/pnas.0402560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP. Science. 2003;299:259. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- 77.Yoon A, Peng G, Brandenburger Y, Brandenburg Y, Zollo O, Xu W, Rego E, Ruggero D. Science. 2006;312:902. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 78.Mitchell JR, Wood E, Collins K. Nature. 1999;402:551. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 79.Montanaro L, Chillá A, Trerè D, Pession A, Govoni M, Tazzari PL, Derenzini M. J. Invest. Dermatol. 2002;118:193. doi: 10.1046/j.0022-202x.2001.01634.x. [DOI] [PubMed] [Google Scholar]
- 80.Wong JM, Collins K. Genes Dev. 2006;20:2848. doi: 10.1101/gad.1476206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bittel DC, Butler MG. Expert Rev. Mol. Med. 2005;7:1. doi: 10.1017/S1462399405009531. DOI: 10.1017/S1462399405009531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peters J. Nat. Genet. 2008;40:688. doi: 10.1038/ng0608-688. [DOI] [PubMed] [Google Scholar]
- 83.Cavaillé J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Hüttenhofer A. Proc. Natl. Acad. Sci. U. S. A. 2000;97:14311. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de los Santos T, Schweizer J, Rees CA, Francke U. Am. J. Hum. Genet. 2000;67:1067. doi: 10.1086/303106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Smith JA, et al. Hum. Mol. Genet. 2009 [Google Scholar]
- 86.Kanber D, Giltay J, Wieczorek D, Zogel C, Hochstenbach R, Caliebe A, Kuechler A, Horsthemke B, Buiting K. Eur. J. Hum. Genet. 2009;17:582. doi: 10.1038/ejhg.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meguro M, Mitsuya K, Nomura N, Kohda M, Kashiwagi A, Nishigaki R, Yoshioka H, Nakao M, Oishi M, Oshimura M. Hum. Mol. Genet. 2001;10:383. doi: 10.1093/hmg/10.4.383. [DOI] [PubMed] [Google Scholar]
- 88.Runte M, Hüttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. Hum. Mol. Genet. 2001;10:2687. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 89.Runte M, Varon R, Horn D, Horsthemke B, Buiting K. Hum. Genet. 2005;116:228. doi: 10.1007/s00439-004-1219-2. [DOI] [PubMed] [Google Scholar]
- 90.Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. Nat. Genet. 2008;40:719. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ding F, Li HH, Zhang S, Solomon NM, Camper SA, Cohen P, Francke U. PLoS One. 2008;3:e1709. doi: 10.1371/journal.pone.0001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Skryabin BV, Gubar LV, Seeger B, Pfeiffer J, Handel S, Robeck T, Karpova E, Rozhdestvensky TS, Brosius J. PLoS Genet. 2007;3:e235. doi: 10.1371/journal.pgen.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsai TF, Jiang YH, Bressler J, Armstrong D, Beaudet AL. Hum. Mol. Genet. 1999;8:1357. doi: 10.1093/hmg/8.8.1357. [DOI] [PubMed] [Google Scholar]
- 94.Reichow SL, Hamma T, Ferré-D’Amaré AR, Varani G. Nucleic Acids Res. 2007;35:1452. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Doe CM, Relkovic D, Garfield AS, Dalley JW, Theobald DE, Humby T, Wilkinson LS, Isles AR. Hum. Mol. Genet. 2009;18:2140. doi: 10.1093/hmg/ddp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kishore S, Stamm S. Cold Spring Harbor Symp. Quant. Biol. 2006;71:329. doi: 10.1101/sqb.2006.71.024. [DOI] [PubMed] [Google Scholar]
- 97.Kishore S, Stamm S. Science. 2006;311:230. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 98.Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaillé J, Huttenhofer A. J. Cell Biol. 2005;169:745. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. Mol. Cell. 2008;32:519. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 100.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. RNA. 2009;15:1233. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sridhar P, Gan HH, Schlick T. J. Biomed. Sci. 2008;15:697. doi: 10.1007/s11373-008-9271-x. [DOI] [PubMed] [Google Scholar]
- 102.Caldarola S, De Stefano MC, Amaldi F, Loreni F. FEBS J. 2009;276:3199. doi: 10.1111/j.1742-4658.2009.07036.x. [DOI] [PubMed] [Google Scholar]
- 103.Warner JR, McIntosh KB. Mol. Cell. 2009;34:3. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lipton JM, Ellis SR. Hematol./Oncol. Clin. North Am. 2009;23:261. doi: 10.1016/j.hoc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dianzani I, Loreni F. Haematologica. 2008;93:1601. doi: 10.3324/haematol.2008.000513. [DOI] [PubMed] [Google Scholar]
- 106.Ellis SR, Lipton JM. Curr. Top. Dev. Biol. 2008;82:217. doi: 10.1016/S0070-2153(07)00008-7. [DOI] [PubMed] [Google Scholar]
- 107.Vlachos A, et al. Br. J. Haematol. 2008;142:859. doi: 10.1111/j.1365-2141.2008.07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Hum. Mutat. 2007;28:1178. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- 109.Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, Tentler D, Mohandas N, Carlsson B, Dahl N. Nat. Genet. 1999;21:169. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 110.Farrar JE, et al. Blood. 2008;112:1582. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gazda HT, et al. Am. J. Hum. Genet. 2006;79:1110. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gazda HT, et al. Am. J. Hum. Genet. 2008;83:769. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boria I, Quarello P, Avondo F, Garelli E, Aspesi A, Carando A, Campagnoli MF, Dianzani I, Ramenghi U. Hum. Mutat. 2008;29:E263. doi: 10.1002/humu.20864. [DOI] [PubMed] [Google Scholar]
- 114.Choesmel V, Fribourg S, Aguissa-Touré AH, Pinaud N, Legrand P, Gazda HT, Gleizes PE. Hum. Mol. Genet. 2008;17:1253. doi: 10.1093/hmg/ddn015. [DOI] [PubMed] [Google Scholar]
- 115.Léger-Silvestre I, Caffrey JM, Dawaliby R, Alvarez-Arias DA, Gas N, Bertolone SJ, Gleizes PE, Ellis SR. J. Biol. Chem. 2005;280:38177. doi: 10.1074/jbc.M506916200. [DOI] [PubMed] [Google Scholar]
- 116.Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M. RNA. 2008;14:1918. doi: 10.1261/rna.1132008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bernstein KA, Gallagher JE, Mitchell BM, Granneman S, Baserga SJ. Eukaryotic Cell. 2004;3:1619. doi: 10.1128/EC.3.6.1619-1626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martín-Marcos P, Hinnebusch AG, Tamame M. Mol. Cell. Biol. 2007;27:5968. doi: 10.1128/MCB.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Crétien A, Leblanc T, Tchernia G, Da Costa L, Gleizes PE. Blood. 2007;109:1275. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, Karlsson S, Ellis SR. Blood. 2007;109:980. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Idol RA, Robledo S, Du HY, Crimmins DL, Wilson DB, Ladenson JH, Bessler M, Mason PJ. Blood Cells, Mol. Dis. 2007;39:35. doi: 10.1016/j.bcmd.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 122.Chiocchetti A, Gibello L, Carando A, Aspesi A, Secco P, Garelli E, Loreni F, Angelini M, Biava A, Dahl N, Dianzani U, Ramenghi U, Santoro C, Dianzani I. Haematologica. 2005;90:1453. [PubMed] [Google Scholar]
- 123.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Mol. Cell. Biol. 2004;24:7654. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dai MS, Arnold H, Sun XX, Sears R, Lu H. EMBO J. 2007;26:3332. doi: 10.1038/sj.emboj.7601776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C. Mol. Cell. 2009;35:316. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Uechi T, Nakajima Y, Chakraborty A, Torihara H, Higa S, Kenmochi N. Hum. Mol. Genet. 2008;17:3204. doi: 10.1093/hmg/ddn216. [DOI] [PubMed] [Google Scholar]
- 127.Chakraborty A, Uechi T, Higa S, Torihara H, Kenmochi N. PLoS One. 2009;4:e4152. doi: 10.1371/journal.pone.0004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, Attardi LD, Barsh GS. Nat. Genet. 2008;40:963. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ebert BL. Leukemia. 2009;23(7):1252–1256. doi: 10.1038/leu.2009.53. [DOI] [PubMed] [Google Scholar]
- 130.Boultwood J, Fidler C, Strickson AJ, Watkins F, Gama S, Kearney L, Tosi S, Kasprzyk A, Cheng JF, Jaju RJ, Wainscoat JS. Blood. 2002;99:4638. doi: 10.1182/blood.v99.12.4638. [DOI] [PubMed] [Google Scholar]
- 131.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, Golub TR. Nature. 2008;451:335. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ferreira-Cerca S, Pöll G, Kühn H, Neueder A, Jakob S, Tschochner H, Milkereit P. Mol. Cell. 2007;28:446. doi: 10.1016/j.molcel.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 133.Pellagatti A, Hellström-Lindberg E, Giagounidis A, Perry J, Malcovati L, Della Porta MG, Jädersten M, Killick S, Fidler C, Cazzola M, Wainscoat JS, Boultwood J. Br. J. Haematol. 2008;142:57. doi: 10.1111/j.1365-2141.2008.07178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ruggero D, Pandolfi PP. Nat. Rev. Cancer. 2003;3:179. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 135.Maggi LB, Jr, Weber JD. Cancer Invest. 2005;23:599. doi: 10.1080/07357900500283085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Montanaro L, Treré D, Derenzini M. Am. J. Pathol. 2008;173:301. doi: 10.2353/ajpath.2008.070752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bilanges B, Stokoe D. Oncogene. 2007;26:5973. doi: 10.1038/sj.onc.1210431. [DOI] [PubMed] [Google Scholar]
- 138.Oskarsson T, Trumpp A. Nat. Cell Biol. 2005;7:215. doi: 10.1038/ncb0305-215. [DOI] [PubMed] [Google Scholar]
- 139.White RJ. Nat. Rev. Mol. Cell Biol. 2005;6:69. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- 140.Kruse JP, Gu W. Cell. 2009;137:609. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dai MS, Lu H. J. Biol. Chem. 2004;279:44475. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 142.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Mol. Cell. Biol. 2003;23:8902. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R. Oncogene. 2007;26:5029. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 144.Horn HF, Vousden KH. Oncogene. 2008;27:5774. doi: 10.1038/onc.2008.189. [DOI] [PubMed] [Google Scholar]
- 145.Castro ME, Leal JF, Lleonart ME, Ramon Cajal YS, Carnero A. Carcinogenesis. 2008;29:1343. doi: 10.1093/carcin/bgm302. [DOI] [PubMed] [Google Scholar]
- 146.Takagi M, Absalon MJ, McLure KG, Kastan MB. Cell. 2005;123:49. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 147.Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, Babcock GF, Bernardi R, Pandolfi PP, Thomas G. Nat. Cell Biol. 2009;11:501. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma SC, Thomas G. Science. 2000;288:2045. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 149.Adhikary S, Eilers M. Nat. Rev. Mol. Cell Biol. 2005;6:635. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 150.Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D. Nature. 2008;456:971. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dai MS, Sears R, Lu H. Cell Cycle. 2007;6:2735. doi: 10.4161/cc.6.22.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. PLoS Biol. 2004;2:e139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Nature. 2008;456:470. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Passos-Bueno MR, Ornelas CC, Fanganiello RD. Am. J. Med. Genet., Part A. 2009;149a:1853. doi: 10.1002/ajmg.a.32950. [DOI] [PubMed] [Google Scholar]
- 155.Goobie S, Popovic M, Morrison J, Ellis L, Ginzberg H, Boocock GR, Ehtesham N, Bétard C, Brewer CG, Roslin NM, Hudson TJ, Morgan K, Fujiwara TM, Durie PR, Rommens JM. Am. J. Hum. Genet. 2001;68:1048. doi: 10.1086/319505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Horn D, Rupprecht E, Kunze J, Spranger J. J. Med. Genet. 2001;38:262. doi: 10.1136/jmg.38.4.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Verloes A, Pierard GE, Le Merrer M, Maroteaux P. J. Med. Genet. 1990;27:693. doi: 10.1136/jmg.27.11.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bonafé L, Schmitt K, Eich G, Giedion A, Superti-Furga A. Clin. Genet. 2002;61:146. doi: 10.1034/j.1399-0004.2002.610210.x. [DOI] [PubMed] [Google Scholar]
- 159.Walne AJ, Vulliamy T, Marrone A, Beswick R, Kirwan M, Masunari Y, Al-Qurashi FH, Aljurf M, Dokal I. Hum. Mol. Genet. 2007;16:1619. doi: 10.1093/hmg/ddm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Vulliamy T, Beswick R, Kirwan M, Marrone A, Digweed M, Walne A, Dokal I. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8073. doi: 10.1073/pnas.0800042105. [DOI] [PMC free article] [PubMed] [Google Scholar]