Abstract
It is well established that bacteria are able to respond to temporal gradients (e.g., by chemotaxis). However, it is widely held that prokaryotes are too small to sense spatial gradients. This contradicts the common observation that the vast majority of bacteria live on the surface of a solid substrate (e.g., as a biofilm). Herein we report direct experimental evidence that the nonmotile bacterium Staphylococcus aureus possesses a tactile response, or primitive sense of touch, that allows it to respond to spatial gradients. Attached cells recognize their substrate interface and localize adhesins toward that region. Braille-like avidity maps reflect a cell's biochemical sensory response and reveal ultrastructural regions defined by the actual binding activity of specific proteins.
Introduction
Prokaryotes often encounter physical and chemical gradients in nature. These simple cells are known to respond to gradients. Chemotaxis is arguably the most frequently documented form of gradient response in bacteria. This form of taxis is the directed movement of a motile bacterium toward (or away from) a chemical signal. Although a chemical gradient is typically a spatial phenomenon, bacteria respond to temporal rather than spatial cues. A bacterium senses a change in the concentration of a chemical outside the cell over time and responds by actively swimming toward or away from the chemical. A central tenet or dogma of microbiology is that prokaryotes, unlike larger eukaryotic cells, are too small to sense a gradient along their body length (1).
This contradicts the common observation that most bacteria live on the surface of a solid substrate (e.g., as a biofilm). It seems reasonable to infer that bacteria evolved a mechanism for recognizing surfaces, considering that an overwhelming 97% of all prokaryotes (motile and nonmotile) live in close proximity to solids (2). Indeed, studies have presented theoretical (3) and indirect experimental (4,5) evidence suggesting that some motile bacteria can sense gradients across their body lengths.
Here, we report direct experimental evidence that the nonmotile microorganism Staphylococcus aureus possesses a tactile response (i.e., a primitive sense of touch) by which attached cells recognize the steep gradient near their substrate interface and localize substrate-specific biomolecules toward that region accordingly. S. aureus is a Gram-positive bacterium that is part of the normal microbial flora of humans. This microorganism can form infectious biofilms on the surface of host tissues or implanted medical devices if it enters the human bloodstream (6,7). These biofilms begin to form when a protein receptor on the outer cell wall of S. aureus forms a bond with host ligands that form the outer coating on virtually all surfaces within humans (e.g., host tissue or foreign implants). The initial bacteria-host bond is often mediated through the interaction of human fibronectin (Fn) and Fn-binding protein (FnBP) expressed on S. aureus (8–10). We used atomic force microscopy (AFM) to measure binding forces between an Fn-coated AFM tip and living S. aureus. A distinct, sawtooth-shaped force signature was observed when Fn formed a bond with putative FnBP on S. aureus. Analysis of the force traces with the worm-like chain (WLC) model was consistent with a zipper-like binding of multiple parallel domains of Fn and FnBP. We also observed a linear increase of the rupture force with the logarithm of the loading rate, suggesting a specific interaction between Fn on the tip and FnBP on S. aureus. We were able to image the spatial localization of putative FnBP on living S. aureus by mapping the position of the Fn-FnBP binding signature. These avidity maps reveal ultrastructural regions on S. aureus defined by the activity and/or bond resilience of binding receptors on living bacteria. We found that S. aureus responds very differently to three dissimilar substrates, suggesting that at least some bacteria have the ability to perceive spatial differences across their body length.
Materials and Methods
Bacteria specimens and growth conditions
For our experiments we used S. aureus I399 (11). This bacterium was isolated from a patient at Duke University Medical Center who had a confirmed S. aureus infection of an implanted cardiac device. A polymerase chain reaction was used to confirm the presence of the gene coding for FnBP A (fnbA; accession number J04151). Western blot analysis confirmed that FnBP was localized to the cell wall of S. aureus I399.
Two laboratory-derived S. aureus mutants (DU5883 and DU5883+pFNBA4) and one laboratory-derived Lactococcus lactis mutant (pOri23-fnbA) were used in control experiments. S. aureus DU5883 lacks the fnbA gene and has lost the ability to attach to surface-bound Fn (12). S. aureus DU5883+pFNBA4 contains the shuttle plasmid for the fnbA gene, which restores the adhesion-defective phenotype of DU5883 to Fn-coated substrates (12). L. lactis was selected as a control bacterium because although it is a Gram-positive bacterium like S. aureus, it does not naturally adhere to Fn (13). The recombinant L. lactis pOri23-fnbA expresses functional FnBP on its outer surface, which allows it to adhere to Fn-coated substrates (13–15).
Growth cultures for AFM analysis were started from cryogenically preserved samples. S. aureus was cultured to exponential phase (OD600 ∼ 0.50) at 37°C in tryptic soy broth containing 0.2% dextrose. S. aureus is known to express FnBP when cultured in this media and harvested at this particular growth stage (6,11). Antibiotics were added to the broth of the two mutant strains of S. aureus according to published methods (12,13). L. lactis was grown to exponential phase at 30°C in M17 medium supplemented with the appropriate antibiotics according to previous studies (14,15). Under such conditions, L. lactis pOri23-fnbA constitutively expresses FnBP (14,15).
AFM measurements
Silicon nitride AFM cantilevers were coated with Fn (Sigma-Aldrich, St. Louis, MO) according to published methods (11,16). A total of 10 different Fn-coated tips (nominal tip radius 20 nm and measured spring constant 0.02 nN nm−1) were used in these AFM experiments. In some control experiments, AFM tips were coated with bovine serum albumin (BSA) as described previously (17). We also created blunt AFM tips by repeatedly imaging a sharp, pyramidal AFM tip on an alumina substrate. Because of the hardness difference between silicon nitride and alumina, we were able to grind several AFM tips into flattened squares roughly 100–500 nm on a side. The blunt AFM tips were coated with Fn and used to pluck cells off of a coverslip so that force curves could be collected with S. aureus cells linked to the end of an AFM tip.
Force measurements were performed using a Veeco/Digital Instruments (Santa Barabara, CA) Bioscope AFM and Nanoscope IV controller. The AFM was not engaged in image mode to find bacteria cells on a glass slide before force measurements were obtained, because this could have contaminated the AFM tip. Rather, an inverted optical microscope (Axiovert 200M; Zeiss, Oberkochen, Germany) was used to position an AFM tip over a binary fission pair or a small patch of four cells on a glass coverslip. The probe was brought into contact with a bacterium and pushed against the cell wall until the cantilever flexed 100 nm. The probe was then retracted away from the bacterium until the probe was completely separated from the cell. For most force curves, the approach-retraction cycle took 2 s (i.e., 0.5 Hz scan rate). For loading rate experiments, the scan rate varied from 0.03 to 2 Hz.
AFM was also operated in force-volume mode to map receptor-ligand sites on surfaces. This form of AFM was previously developed to map eukaryotic macromolecules on cells, membranes, and inorganic substrates (18–23). More recently, this elegant technique has been applied to microorganisms (24,25). For our experiments, AFM probes were baited with Fn and then used on S. aureus I399 cells deposited onto three different solid substrates: clean glass, unclean glass, and Fn-coated glass. The clean glass substrate was prepared by soaking zinc titania glass (i.e., a typical coverslip) in piranha solution (26) and rinsing 10 times in MilliQ (Billerica, MA) water (18.2 MΩ cm). The unclean glass was simply a coverslip that was used as supplied by the manufacturer (Corning Inc., Corning, NY). This coverglass has a proprietary coating that prevents sticking of adjacent coverslips within the packaging container. The Fn-coated glass (BD Biosciences, Franklin Lakes, NJ) was chosen to represent a high-affinity surface. To best observe the native function and activity of cell wall macromolecules, the adhesion of S. aureus to all three substrates was allowed to occur naturally in a phosphate buffered saline (PBS) solution at circumneutral pH without further experimental influence. For the force-volume experiments, an Fn-coated tip was repeatedly brought into and out of contact with different regions on a bacterium to fish for a reaction with putative FnBPs expressed on a cell. The scan rate was 1.0 Hz over 1.5 μm extension, and the tip was pushed against the sample until the flexible cantilever deflected 10–20 nm.
Results and Discussion
S. aureus biofilms—bonds between a bacterial protein and human protein
S. aureus is a nonmotile, Gram-positive bacterium that is an opportunistic pathogen. It is part of the normal flora of microorganisms that live on and in the human body. This bacterium commonly grows as a biofilm on surfaces such as the skin or the mucous membrane of the anterior nares (1,27). If it enters the bloodstream, S. aureus can form infectious biofilms on the surface of implanted medical devices (6,7). In the medical community, S. aureus is a significant concern because it is the leading cause of infection of prosthetic implants, such as cardiac valves (7,28).
The initial step of biofilm formation is mediated at least in part by FnBP expressed on the external cell wall of S. aureus (29,30). FnBP is a covalently anchored transmembrane protein that is part of a family of molecules called microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (9,10). MSCRAMMs form bonds with human protein ligands, such as Fn, fibrinogen, and collagen, that are common constituents of the human bloodstream. These human proteins typically coat the surface of an implanted medical device and in turn serve as attachment sites for S. aureus (12,31). Fn is the predominant ligand-promoting attachment molecule for implants that remain in the body for extended periods of time, such as cardiac devices (31,32). Therefore, we focused on bonds that form between a substrate coated with Fn and FnBP on S. aureus.
The structure and binding activity of Fn and S. aureus FnBP are well documented in the literature. Fig. 1 highlights the binding regions along each of these two proteins. The N- and C-termini of these two proteins are shown opposite each other to better illustrate the binding sites in each molecule. The ∼29 kD N-terminal region of Fn contains a string of FI modules that bind to FnBP (9,33–35). Other regions of Fn, such as the GBF or FnIII heparin-binding module, have also been shown to bind to FnBP (34,36). The Fn-binding regions within FnBP include the D1–D4 region (9,12,35), the Du and B1–B2 domains (13), and a string of 11 subdomains between the A and D regions within FnBP (30,36,37).
Figure 1.
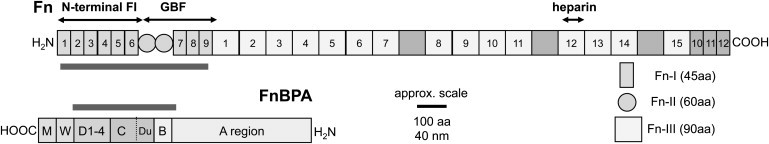
Primary structure of Fn (top) and FnBPA (bottom) drawn to approximate scale. Highlighted are regions on Fn (N-terminal Fn-I and Fn-II domains) and FnBP (A, B, Du, C, and D1–D4) that are important in binding. Fn is ∼2000 amino acids or 800 nm long, and FnBPA is ∼1000 amino acids or 400 nm long. Modified from Buck et al. (74).
AFM force measurements between Fn on a solid substrate and FnBP expressed by living bacteria
It is well documented that the continual passage of bacteria, such as type strains, through liquid growth media can transform a biofilm-forming pathogen into a planktonic lab-adapted strain. Costerton et al. (38) noted that “these lab-adapted cultures are really not good models for the study of diseases that have been shown unequivocally to be caused by bacteria growing in biofilms”. Therefore, for our experiments we used a biofilm-forming strain isolated from an actual clinical setting. S. aureus I399 was obtained from a patient at Duke University Medical Center who had an infected cardiac device. This isolate was stored cryogenically and passed through only one or two flasks of liquid media before each AFM experiment.
A total of 4170 force curves were collected on 21 different cells prepared from four different culture flasks of S. aureus I399. Fig. 2 shows randomly selected force curves collected as an Fn-coated AFM tip was pulled from contact with the surface of S. aureus I399 in PBS solution. We frequently found that the binding forces rose up to several hundreds of piconewtons, and unbinding occurred as either one district step or two to three sequential steps, each resulting in nonlinear, sawtooth-shaped force profiles. These sawtooth-shaped profiles were observed in ∼50% of the traces for an Fn-coated tip on S. aureus I399 cells, which were harvested at the exponential phase when FnBPs are expressed in S. aureus (6).
Figure 2.
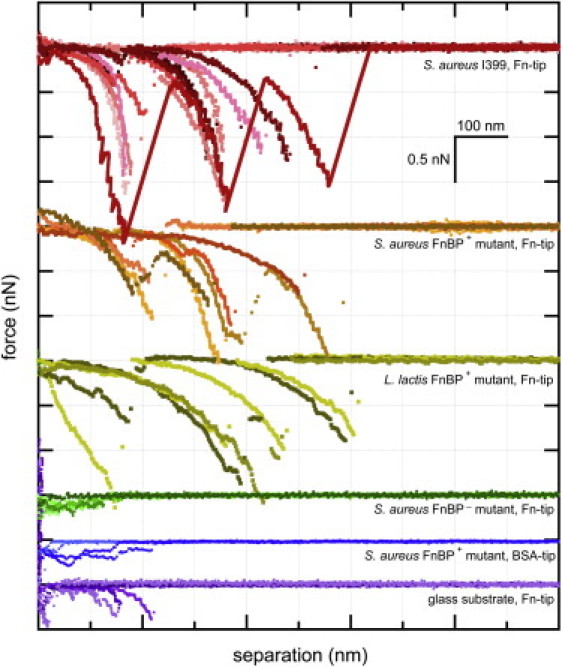
Force spectra collected by AFM in a buffer solution. Shown are randomly selected retraction traces for the following pairs (from top to bottom): Fn-coated tip on a clinical isolate of S. aureus (topmost curves), Fn-coated tip on a mutant strain of S. aureus that overexpresses FnBP on its outer surface, Fn-coated tip on a mutant strain of L. lactis that overexpresses FnBP on its outer surface, Fn-coated tip on a mutant strain of S. aureus that cannot express FnBP on its outer surface, BSA-coated tip on a mutant strain of S. aureus that overexpresses FnBP on its outer surface, and Fn-coated tip on a glass slide (bottom-most curves). See Buck et al. (74) for a complementary study. (Color figure online.)
The sawtooth-shaped force-distance trajectory is consistent with the unfolding/extension of a protein and suggests a specific binding event rather than generalized adhesion due to nonspecific contact forces (39–41) (see Fig. S1 in the Supporting Material). Ideally, we would have injected free antibody (e.g., anti-FnBP) into the experiment to quench the Fn-FnBP bond and confirm the specificity of the observed binding event. However, others have shown that anti-MSCRAMM antibodies (e.g., anti-FnBP) react with the adhesin but do not inhibit Fn binding (42). This is due at least in part to the fact that FnBP has multiple reactive sites along its length rather than a single binding pocket (e.g., see Fig. 1). Therefore, we took a slightly different approach to determine whether the AFM force signatures were due to binding events between Fn on the AFM tip and FnBP on S. aureus. We collected force measurements on some elegantly constructed mutants that either express or do not express FnBP on their outer surface. These mutant strains have already been described in the literature (12,14,15).
Fig. 2 shows retraction traces for an Fn-coated tip on the following mutant strains: S. aureus that overexpresses FnBP on its exterior cell wall (S. aureus FnBP+), S. aureus that cannot express FnBP on its cell wall (S. aureus FnBP−), and a recombinant strain of L. lactis that expresses FnBP (L. lactis FnBP+). The sawtooth-shaped binding events were largely absent from S. aureus FnBP−, whereas S. aureus FnBP+ and L. lactis FnBP+ exhibited these distinct binding events. L. lactis is particularly useful because it is a Gram-positive bacterium, like S. aureus, but natural strains of L. lactis do not have the genes to produce the MSCRAMM proteins (e.g., FnBP) that are present in S. aureus. A comparison of the traces for S. aureus I399 and the three mutant strains (see Fig. 2) suggests that the sawtooth-shaped binding events involve proteins on the outer surface of S. aureus that bind to Fn (e.g., FnBPs).
Fig. 2 also shows retraction profiles for a BSA-coated tip on the mutant strain of S. aureus that overexpresses FnBP, and an Fn-coated tip on a glass slide. In similarity to the measurements with the S. aureus that cannot express FnBP, the BSA-coated tip did not normally elicit the characteristic sawtooth-shaped profiles even when it was used on S. aureus that overexpress FnBP (Fig. 2). The Fn-coated tip showed some general adhesion force when it was pulled from the glass slide. In a few instances, the Fn-coated tip also exhibited a relatively weak, short-range, sawtooth-shaped profile when it was pulled from contact with a glass slide (Fig. 2). This is consistent with other studies that used AFM to unravel a molecule of Fn tethered between the tip and a glass slide (16,43,44).
Confirmation that the bacteria were alive during AFM experiments
The conditions of our AFM experiments were similar to those employed by Touhami et al. (45), who used AFM to image the binary fission of living S. aureus cells in real time. We performed two additional experiments to confirm that we were performing AFM on living cells. First, we exchanged the PBS solution in the fluid cell of the AFM with growth media after the AFM experiments. The optical microscope, which formed the base of our AFM, was used to observe S. aureus cells on the microscope slide growing into a layer of cells and eventually a thick biofilm. Second, we used a blunt, Fn-coated tip to pick up two cells (a binary fission pair) from the microscope slide after the AFM experiments. The AFM tip was aseptically transferred to a flask of liquid growth media (supplemented with the appropriate antibiotics for the mutant strains of S. aureus). Growth of S. aureus cells was observed in the culture flasks when the cells on the AFM tip were used as the inoculum. No growth was observed for a control flask that was inoculated with an Fn-coated tip used in the AFM to probe a plain glass slide (i.e., no S. aureus cells). These experiments confirm that our AFM measurements were likely performed on living bacteria.
WLC model of an Fn-FnBP binding event
The sawtooth-shaped profiles observed in the AFM force curves are consistent with the unfolding/extension of a protein that forms a bond between two surfaces (40,41). The forced extension of a linear polymer (e.g., a protein) can be described with the WLC model (46,47). This theory has been used to interpret AFM force measurements on isolated protein molecules purified from bacteria (48,49), as well as proteins on the outer surface of living bacteria (50,51). The Marko-Siggia WLC equation is given as (52,53):
| (1) |
where F (in Newtons) is the force associated with the mechanical unfolding or elongation of a protein to distance x (in meters), kB is the Boltzmann's constant (kB = 1.381 × 10−23 J K−1), and T is temperature (in Kelvins). The final two parameters of the WLC model are the persistence length (p) and the contour length (L). The persistence length is a measure of the bending rigidity or stiffness of a polypeptide chain. For an ideal single protein molecule, the persistence length is between 0.1 nm and 2.0 nm (48,54–57). This dimension is similar to the physical length of 0.4 nm between Cα atoms in the backbone of a protein (58). The contour length is the extended length of either an entire protein molecule or a structural domain within a protein.
Fig. 3 shows the theoretical force-extension relationship for a protein with a persistence length of 0.4 nm (i.e., the physical length of an amino acid) and contour length of 210 nm. This corresponds to the 525 amino acids on the N-terminal region of a single molecule of Fn (see Fig. 1 for reference). This represents the region on Fn that binds to FnBP from S. aureus (9,34,35). As described above, AFM was used to measure binding forces on Fn molecules tethered between the AFM tip and a glass slide. Most of the retraction traces showed only a jump-from-contact feature, which is not unexpected since the substrate was a glass slide. However, some of the traces exhibit a relatively weak, sawtooth-shaped binding event near 200 nm (see Fig. 3). This is consistent with the modeled extension of the 525 amino acids that make up the FnI and FnII domains on Fn.
Figure 3.
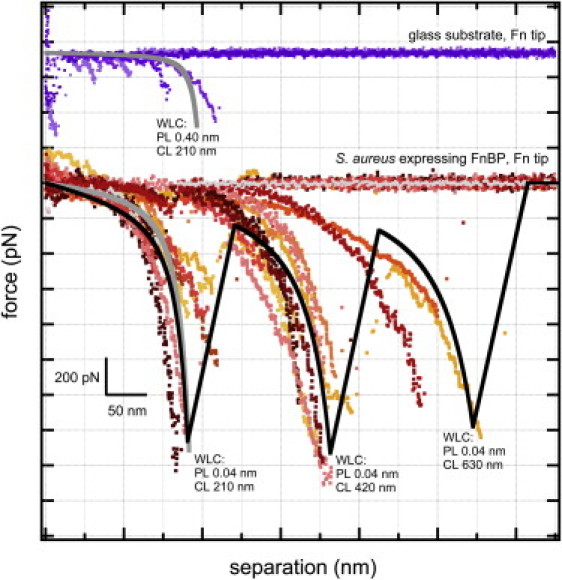
Experimentally measured force spectra for Fn on glass (top series of traces) and Fn on the clinical strain of S. aureus I399 as well as the mutant strain of S. aureus that overexpresses FnBP (bottom series of curves). The WLC model was used to determine theoretical force-distance profiles (shown as gray or black curves). The top gray-colored curve corresponds to the unfolding of the N-terminal FnI and FnII domains on Fn (p = 0.4 nm; L = 210 nm). The bottom gray-colored curve corresponds to the unfolding of one FnBP in parallel with nine Fn molecules (p = 0.04 nm; L = 210 nm). The solid black trace corresponds to the zipper-like unfolding of three discrete events, each involving a total of 10 parallel bonded molecules of Fn and FnBP (p = 0.04 nm; L = 210, 420, and 630 nm). (Color figure online.)
The WLC model can also be used on multiple protein molecules that form parallel bonds between two surfaces (54,59–62) (for example, a molecule of FnBP on S. aureus that has formed bonds along the N-terminal domain of an Fn molecule on the AFM tip). This approximation is accomplished by a simple summation of the individual WLC expressions for each protein molecule (63). For example, 10 identical protein chains in parallel would exert a force 10 times that of a single chain for a given extension length. Mathematically, the summation of 10 identical protein chains (e.g., 10 molecules each with p = 0.4 and L = 210) will yield the same force-extension relationship as one protein chain with the same contour length but a persistence length that is one-tenth as long (e.g., p = 0.04 and L = 210).
This application of the WLC model is particularly useful for S. aureus because each FnBP can bind up to nine different molecules of Fn (35,36,64). Fig. 3 shows the overall WLC force-extension relationship of 10 protein molecules each with a persistence length of 0.4 nm and a contour length of 210 nm. This corresponds to one FnBP on S. aureus that has formed bonds along the length of the 525 amino acids on the N-terminus from each of nine Fn molecules. The WLC model predicts an increasingly nonlinear response that reaches a maximum force near their extended length of ∼200 nm. There is a remarkable similarity between the theoretical force profile and the ∼200 nm sawtooth observed in the AFM traces of Fn on S. aureus I399, as well as on the mutants that overexpress FnBP (see Fig. 3). This similarity is consistent with binding of an FnBP on S. aureus to the N-terminal region of multiple molecules of Fn on the AFM tip.
In addition to the sawtooth at ∼200 nm, at least two longer-range sawteeth are consistently observed for S. aureus I399 and the mutants that overexpress FnBP (see Figs. 2 and 3). These traces can be modeled as 10 parallel-bonded protein molecules, each with p = 0.4 nm, and contour lengths that are multiples of 210 nm (i.e., L = 420 and 630 nm). Fig. 3 shows the summative WLC model for the sequential unfolding of three discrete binding events, each of which involves the unfolding of 10 protein molecules bound in parallel (i.e., the summation of 10 chains each with p = 0.4, which is mathematically equivalent to one chain with p = 0.04). As shown in this figure, there is a striking correlation between the WLC model and the observed retraction traces. One possible explanation is a zipper array of bonds where force is applied to the lead bond; once that bond fails, force propagates to the next bond, and so on. This is similar to the recently proposed structure-based model of the Fn-FnBP complex. Nuclear magnetic resonance and x-ray diffraction were used on purified segments of Fn and FnBP to reveal a tandem, β-zipper type of interaction in repeat units (30,37). Our observation of binding events at repetitive intervals is also consistent with the proposal of Casolini et al. (42) that FnBP takes on an organized conformational structure in repeat units upon binding to its ligand, Fn.
At this point, we cannot assign with absolute certainty a particular sawtooth to a specific mechanical (or structural) domain within Fn or FnBP. This would require a number of additional AFM experiments with truncated forms of both Fn and FnBP expressed on the outer surfaces of S. aureus and L. lactis. Nonetheless, we can say with certainty that the sawtooth events observed in the AFM traces represent a distinct force signature that corresponds to the binding of Fn on the AFM tip with proteins on S. aureus that have an affinity for Fn. These force signatures are most likely due to FnBP on S. aureus (e.g., compare the force traces with the mutants that express FnBP or cannot express FnBP). However, we cannot categorically rule out the possibility that other cell wall proteins participate in bonding with Fn. Indeed, others have shown some cross-reactivity between MSCRAMMs such as FnBP on S. aureus and human ligands such as Fn, fibrinogen, and collagen (65).
Loading-rate experiments for the bond between Fn and FnBP on S. aureus
A final series of AFM experiments were conducted to determine the specificity of the interactions between Fn on the AFM tip and putative FnBPs on S. aureus. The rupture force of the Fn-FnBP bond was measured at different loading rates (N s−1) according to standard protocol (17,66,67). For a specific ligand-receptor interaction, the rupture force should increase with the loading rate as shown in the following equation (68):
| (2) |
where xβ is the separation distance between the bound and transition states along the reaction coordinate (m), r is the loading rate (N s−1), koff is the dissociation rate constant for the ligand-receptor pair (s−1), and all other parameters are as described in Eq. 1. For a specific ligand-receptor pair, a linear relationship should be evident in a semilog plot of rupture force versus loading rate. The slope equals kBT / xβ, and extrapolating to F = 0 gives a value of koff = rxβ / kBT.
As discussed above, FnBP has the capacity to bind up to nine different molecules of Fn. However, Eq. 2 is ideally suited for single ligand-receptor pair (68). Therefore, for the loading-rate experiments we included only those traces that could be fit with a WLC model corresponding to one Fn-FnBP pair linked in parallel (i.e., with a persistence length of 0.2 nm). Fig. 4 shows the loading-rate measurements for an Fn-coated tip on S. aureus expressing FnBP. The rupture force increases with the loading rate, consistent with a specific interaction. Furthermore, our measurements agree with those of Bustanji et al. (69), who used AFM to measure the rupture force between Fn and Fn-binding adhesins on another species of Staphylococcus, S. epidermis. Fig. 4 plots the data from our study relative to their results (xβ = 3.3 Å and koff = 4.8 s−1 (69)).
Figure 4.
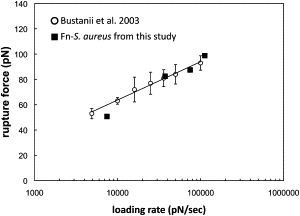
The rupture force as a function of the loading rate for an Fn-coated tip on S. aureus. Also shown are data from Bustanji et al. (69), who measured the binding force between an Fn-coated AFM tip and adhesins on the outer surface of the bacterium S. epidermis.
Avidity maps of putative FnBPs on living S. aureus deposited on different substrates
Binding events between the Fn-coated tip and an S. aureus cell expressing FnBP resulted in distinct sawtooth-shaped, force-separation traces that disappeared when either Fn or FnBP was absent in the AFM experiments (Fig. 2). The observed force-distance trajectories are consistent with the theoretical WLC unfolding/extension of Fn molecules bound in parallel to FnBP on S. aureus (Fig. 3). Finally, the bond between Fn and FnBP on S. aureus appears to be a specific interaction as the rupture force increases with the bond's loading rate (Fig. 4). Taken together, these force measurements suggest that the AFM can be used to map the location of specific Fn-binding proteins on living cells of S. aureus by tuning into this force signature. This form of AFM mapping, sometimes called force-volume imaging, has already been used with success on both eukaryotic (19,39,70) and prokaryotic (24,25,71) cells.
We attempted to use this approach on the clinical isolate of S. aureus (I399). The bacterium was cultured to the exponential stage, when it expresses FnBP on its cell wall. The cells were then deposited onto three different surfaces: an Fn-coated glass slide, an unclean glass slide, and a clean glass slide. The adhesion of S. aureus to all three substrates was allowed to occur naturally in a PBS solution at circumneutral pH without further experimental influence, aside from the subsequent prodding of the AFM tip. This provided the best chance of observing the native function and activity of cell wall macromolecules on living S. aureus. We then used AFM to translate an Fn-coated tip across each sample, collecting a two-dimensional array of force curves across cells and surrounding substrate. In essence, we used Fn as molecular bait to fish for a binding reaction with putative FnBPs expressed on a cell.
Fig. 5 A shows the force curves for S. aureus I399 on each of the three substrates. Again, the sawtooth-shaped binding traces were observed in the retraction curves. Bond rupture lengths and forces were consistent and reproducible for a specific substrate, but varied significantly among the three surfaces (see Fig. 5 and inset table). Although the extension profiles taken from the clean and unclean glass substrates generally exhibited one sawtooth signature, the cell on Fn-coated glass substrate consistently exhibited two to four sawteeth, suggesting that larger protein segments with multiple domains were unfolding.
Figure 5.
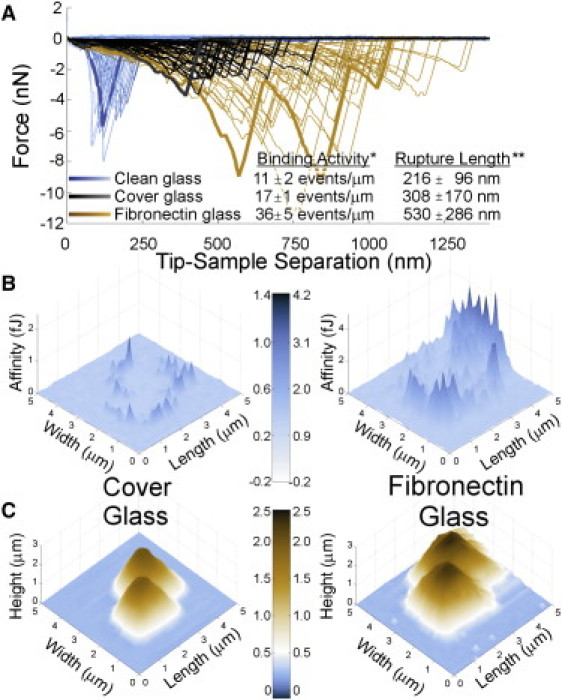
AFM force curves (A), avidity maps (B), and topographic maps (C) of living S. aureus cells on clean glass, unclean glass, or Fn-coated glass immersed in aqueous solution. Force-volume data were taken on a 32 × 32 grid over a 5 μm × 5 μm area. For the inset table in A, binding activity is normalized to cell perimeter, per 1024 fishing attempts over the scan area, and rupture length is the average length of extension at bond rupture. The avidity maps (B) show the position of putative FnBP on the pair of S. aureus cells shown in the complementary topography images (C).
These dissimilar extension characteristics demonstrate that the S. aureus binding proteins function differently in response to similar force stimuli exerted by the tip. Additionally, specific binding events occurred two to three times more frequently for cells attached to Fn-coated glass than for the other substrates (see inset table in Fig. 5), suggesting a higher activity and/or that more proteins were available for binding. All differences in cell response can be attributed to the substrate, since the measurements involved one bacterial strain that was grown, harvested, and imaged under identical conditions.
We also performed force-volume measurements with bare tips (i.e., no Fn) on S. aureus bacteria (harvested at the exponential stage) that were deposited onto the three substrates. Although some retraction traces showed short-range, jump-from-contact adhesion (see Fig. S1), the sawtooth-shaped binding signature was absent from all traces even though the S. aureus should have been expressing FnBP on their outer surface. This control experiment confirms that the force signatures observed for the Fn-coated tip on S. aureus (Fig. 5 A) are due to binding events between Fn and macromolecules on S. aureus that bind to Fn.
These types of force profiles can be consolidated to form maps of a cell surface defined not by topography but by the binding activity and/or resilience of bonds between Fn and putative FnBPs. To produce these maps, we determined the work required to separate the tip from the sample surface. The work was calculated by integrating force over separation to a distance of ∼1400 nm. Fig. 5 B shows the images for S. aureus I399 on a glass coverslip versus an Fn-coated glass coverslip. These maps reflect the cells' biochemical sensory response to the substrate and delve beyond the morphological details evident in topographic images (Fig. 5 C). As discussed above, the force curves likely involved the interactions between multiple molecules of Fn and/or FnBP rather than a single Fn-FnBP bond. Therefore, Fig. 5 B represents an avidity map rather than an affinity map per se.
Independently of substrate, the regions of greatest Fn activity were always along the cells' perimeter (see Fig. 5, B and C). Specific binding events were not observed at the tops of the cells far from the substrate interface. We believe that the activity along the perimeter reflects localization of binding proteins between the cells and the substrate, which would then be accessible to external probes such as AFM only at the cell edges. To determine whether FnBPs were present under a S. aureus cell, we used a blunt Fn-coated AFM tip (∼500 × 500 nm flattened, square tip) to pluck a pair of S. aureus cells from an Fn-coated coverslip. The cells on the AFM tip were immediately used on an adjacent region of the coverslip that was coated with Fn. The distinct sawtooth-shaped binding events were observed repeatedly when the S. aureus cells on the AFM tip were retracted from contact with the Fn-coated slide. This supports the notion that the FnBPs were localized at the interface between the S. aureus and the substrate. We also considered the possibility that increased contact area between the cell and the pyramidal AFM tips could enhance the apparent activity along the cells' perimeter. However, this effect cannot account for the abrupt decrease in affinity away from the bacterium-substrate interface (see Fig. S2).
Conclusions
The data presented here suggest that small prokaryotic cells possess the ability to sense and respond to spatial gradients. Nonmotile S. aureus were shown to recognize a substrate surface and localize substrate-dependent adhesins to the interface. These adhesins were most likely FnBPs; however, we cannot rule out the possibility that other proteins on the outer surface of S. aureus were also involved in these reactions. This would not be entirely unexpected, as there is some cross-reactivity between MSCRAMMs on S. aureus and human ligands such as Fn (65). At the same time, this could be one of the most important attributes of avidity (or affinity) maps like those shown in Fig. 5 B. That is, the avidity maps show regions defined by the native activity of binding proteins toward particular ligands regardless of whether they have been classified as FnBP. These observations are not limited only to Gram-positive bacteria, as some studies with a Gram-negative microorganism have suggested that similar behavior is exhibited by motile organisms that are attached to a substrate (71). Others have suggested that physical adsorption of bacterial cells promotes structural rearrangements (72,73), but the substrate's influence on protein quantity, function, and localization cumulatively suggests that more complex mechanisms are also involved. Presumably, complex and interconnected mechanisms exist for recognizing a surface, controlling the function and quantity of proteins used for binding, and localizing proteins to the substrate interface. For example, S. aureus appears to localize small numbers of Fn-binding proteins at its interface when it is near substrates that are not coated with Fn (see Fig. 5 B, left). The production and localization of significant numbers of Fn-binding proteins on S. aureus (see Fig. 5 B, right) could be an inducible response triggered by an external stimulus, such as a substrate that is actively expressing Fn on its surface (e.g., a human host cell). Further work will be required to elucidate the spatial-sensing mechanism(s), tentatively dubbed NERVS (for networked ensemble response to vicinal surfaces), and determine whether this is a universal bacterial response.
Acknowledgments
We thank T. Foster and Y. A. Que for providing the mutant strains of bacteria, E. D. Williams and T. L. Clanton for constructive comments, and P. McPhail and M. Newell from Veeco Instruments for technical assistance. S.K.L. acknowledges the support of J. Tak.
This work was supported by the National Institutes of Health (grant HL086593) and the National Science Foundation (grant EAR0745808).
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article is dedicated to deceased colleague Terry Beveridge.
Supporting Material
References
- 1.Madigan M.T., Martinko J.M., Parker J. Prentice Hall; Upper Saddle River, NJ: 2003. Brock Biology of Microorganisms. [Google Scholar]
- 2.Whitman W.B., Coleman D.C., Wiebe W.J. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dusenbery D.B. Spatial sensing of stimulus gradients can be superior to temporal sensing for free-swimming bacteria. Biophys. J. 1998;74:2272–2277. doi: 10.1016/S0006-3495(98)77936-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otto K., Silhavy T.J. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA. 2002;99:2287–2292. doi: 10.1073/pnas.042521699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thar R., Kuhl M. Bacteria are not too small for spatial sensing of chemical gradients: an experimental evidence. Proc. Natl. Acad. Sci. USA. 2003;100:5748–5753. doi: 10.1073/pnas.1030795100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowy F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 7.Darouiche R.O. Treatment of infections associated with surgical implants. N. Engl. J. Med. 2004;350:1422–1429. doi: 10.1056/NEJMra035415. [DOI] [PubMed] [Google Scholar]
- 8.Proctor R.A., Mosher D.F., Olbrantz P.J. Fibronectin binding to Staphylococcus aureus. J. Biol. Chem. 1982;257:14788–14794. [PubMed] [Google Scholar]
- 9.Patti J.M., Allen B.L., Höök M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 10.Foster T.J., Höök M. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 1998;6:484–488. doi: 10.1016/s0966-842x(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 11.Yongsunthon R., Fowler V.G., Jr., Lower S.K. Correlation between fundamental binding forces and clinical prognosis of Staphylococcus aureus infections of medical implants. Langmuir. 2007;23:2289–2292. doi: 10.1021/la063117v. [DOI] [PubMed] [Google Scholar]
- 12.Greene C., McDevitt D., Foster T.J. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 1995;17:1143–1152. doi: 10.1111/j.1365-2958.1995.mmi_17061143.x. [DOI] [PubMed] [Google Scholar]
- 13.Massey R.C., Kantzanou M.N., Peacock S.J. Fibronectin-binding protein A of Staphylococcus aureus has multiple, substituting, binding regions that mediate adherence to fibronectin and invasion of endothelial cells. Cell. Microbiol. 2001;3:839–851. doi: 10.1046/j.1462-5822.2001.00157.x. [DOI] [PubMed] [Google Scholar]
- 14.Que Y.A., Haefliger J.A., Moreillon P. Expression of Staphylococcus aureus clumping factor A in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infect. Immun. 2000;68:3516–3522. doi: 10.1128/iai.68.6.3516-3522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Que Y.A., François P., Moreillon P. Reassessing the role of Staphylococcus aureus clumping factor and fibronectin-binding protein by expression in Lactococcus lactis. Infect. Immun. 2001;69:6296–6302. doi: 10.1128/IAI.69.10.6296-6302.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oberdorfer Y., Fuchs H., Janshoff A. Conformational analysis of native fibronectin by means of force spectroscopy. Langmuir. 2000;16:9955–9958. [Google Scholar]
- 17.Hanley W., McCarty O., Konstantopoulos K. Single molecule characterization of P-selectin/ligand binding. J. Biol. Chem. 2003;278:10556–10561. doi: 10.1074/jbc.M213233200. [DOI] [PubMed] [Google Scholar]
- 18.Radmacher M., Fritz M., Hansma P.K. Imaging adhesion forces and elasticity of lysozyme adsorbed on mica with the atomic-force microscope. Langmuir. 1994;10:3809–3814. [Google Scholar]
- 19.Hinterdorfer P., Baumgartner W., Schindler H. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 1996;93:3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rotsch C., Braet F., Radmacher M. AFM imaging and elasticity measurements on living rat liver macrophages. Cell Biol. Int. 1997;21:685–696. doi: 10.1006/cbir.1997.0213. [DOI] [PubMed] [Google Scholar]
- 21.Rotsch C., Radmacher M. Mapping local electrostatic forces with the atomic force microscope. Langmuir. 1997;13:2825–2832. [Google Scholar]
- 22.Rotsch C., Jacobson K., Radmacher M. Dimensional and mechanical dynamics of active and stable edges in motile fibroblasts investigated by using atomic force microscopy. Proc. Natl. Acad. Sci. USA. 1999;96:921–926. doi: 10.1073/pnas.96.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stroh C.M., Ebner A., Hinterdorfer P. Simultaneous topography and recognition imaging using force microscopy. Biophys. J. 2004;87:1981–1990. doi: 10.1529/biophysj.104.043331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dague E., Alsteens D., Dufrêne Y.F. High-resolution cell surface dynamics of germinating Aspergillus fumigatus conidia. Biophys. J. 2008;94:656–660. doi: 10.1529/biophysj.107.116491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arce F.T., Carlson R., Avci R. Nanoscale structural and mechanical properties of nontypeable Haemophilus influenzae biofilms. J. Bacteriol. 2009;191:2512–2520. doi: 10.1128/JB.01596-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lo Y.S., Huefner N.D., Beebe T.P. Organic and inorganic contamination on commercial AFM cantilevers. Langmuir. 1999;15:6522–6526. [Google Scholar]
- 27.von Eiff C., Becker K., Peters G., Study Group Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 2001;344:11–16. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 28.Xiong Y.Q., Fowler V.G., Bayer A.S. Phenotypic and genotypic characteristics of persistent methicillin-resistant Staphylococcus aureus bacteremia in vitro and in an experimental endocarditis model. J. Infect. Dis. 2009;199:201–208. doi: 10.1086/595738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Neill E., Pozzi C., O'Gara J.P. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 2008;190:3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bingham R.J., Rudiño-Piñera E., Potts J.R. Crystal structures of fibronectin-binding sites from Staphylococcus aureus FnBPA in complex with fibronectin domains. Proc. Natl. Acad. Sci. USA. 2008;105:12254–12258. doi: 10.1073/pnas.0803556105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foster T.J. Staphylococcus. In: Baron S., editor. Medical Microbiology. University of Texas Medical Branch at Galveston; Galveston, Texas: 1996. [PubMed] [Google Scholar]
- 32.Arrecubieta C., Asai T., Lowy F.D. The role of Staphylococcus aureus adhesins in the pathogenesis of ventricular assist device-related infections. J. Infect. Dis. 2006;193:1109–1119. doi: 10.1086/501366. [DOI] [PubMed] [Google Scholar]
- 33.Kuusela P. Fibronectin binds Staphylococcus aureus. Nature. 1978;276:718–720. doi: 10.1038/276718a0. [DOI] [PubMed] [Google Scholar]
- 34.Bozzini S., Visai L., Speziale P. Multiple binding-sites in fibronectin and the Staphylococcal fibronectin receptor. Eur. J. Biochem. 1992;207:327–333. doi: 10.1111/j.1432-1033.1992.tb17054.x. [DOI] [PubMed] [Google Scholar]
- 35.Huff S., Matsuka Y.V., Ingham K.C. Interaction of N-terminal fragments of fibronectin with synthetic and recombinant D motifs from its binding protein on Staphylococcus aureus studied using fluorescence anisotropy. J. Biol. Chem. 1994;269:15563–15570. [PubMed] [Google Scholar]
- 36.Schwarz-Linek U., Höök M., Potts J.R. The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol. Microbiol. 2004;52:631–641. doi: 10.1111/j.1365-2958.2004.04027.x. [DOI] [PubMed] [Google Scholar]
- 37.Schwarz-Linek U., Werner J.M., Potts J.R. Pathogenic bacteria attach to human fibronectin through a tandem β-zipper. Nature. 2003;423:177–181. doi: 10.1038/nature01589. [DOI] [PubMed] [Google Scholar]
- 38.Costerton W., Veeh R., Ehrlich G. The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Invest. 2003;112:1466–1477. doi: 10.1172/JCI20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willemsen O.H., Snel M.M.E., Figdor C.G. Simultaneous height and adhesion imaging of antibody-antigen interactions by atomic force microscopy. Biophys. J. 1998;75:2220–2228. doi: 10.1016/S0006-3495(98)77666-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willemsen O.H., Snel M.M.E., De Grooth B.G. A physical approach to reduce nonspecific adhesion in molecular recognition atomic force microscopy. Biophys. J. 1999;76:716–724. doi: 10.1016/S0006-3495(99)77238-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lower B.H., Yongsunthon R., Lower S.K. Simultaneous force and fluorescence measurements of a protein that forms a bond between a living bacterium and a solid surface. J. Bacteriol. 2005;187:2127–2137. doi: 10.1128/JB.187.6.2127-2137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casolini F., Visai L., Speziale P. Antibody response to fibronectin-binding adhesin FnbpA in patients with Staphylococcus aureus infections. Infect. Immun. 1998;66:5433–5442. doi: 10.1128/iai.66.11.5433-5442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oberhauser A.F., Badilla-Fernandez C., Fernandez J.M. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J. Mol. Biol. 2002;319:433–447. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- 44.Craig D., Gao M., Vogel V. Tuning the mechanical stability of fibronectin type III modules through sequence variations. Structure. 2004;12:21–30. doi: 10.1016/j.str.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 45.Touhami A., Jericho M.H., Beveridge T.J. Atomic force microscopy of cell growth and division in Staphylococcus aureus. J. Bacteriol. 2004;186:3286–3295. doi: 10.1128/JB.186.11.3286-3295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flory P.J. Hanser Publishers; New York: 1989. Statistical Mechanics of Chain Molecules. [Google Scholar]
- 47.Marko J.F., Siggia E.D. Statistical mechanics of supercoiled DNA. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 1995;52:2912–2938. doi: 10.1103/physreve.52.2912. [DOI] [PubMed] [Google Scholar]
- 48.Müller D.J., Baumeister W., Engel A. Controlled unzipping of a bacterial surface layer with atomic force microscopy. Proc. Natl. Acad. Sci. USA. 1999;96:13170–13174. doi: 10.1073/pnas.96.23.13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lower B.H., Shi L., Lower S.K. Specific bonds between an iron oxide surface and outer membrane cytochromes MtrC and OmcA from Shewanella oneidensis MR-1. J. Bacteriol. 2007;189:4944–4952. doi: 10.1128/JB.01518-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lower S.K., Hochella M.F., Jr., Beveridge T.J. Bacterial recognition of mineral surfaces: nanoscale interactions between Shewanella and α-FeOOH. Science. 2001;292:1360–1363. doi: 10.1126/science.1059567. [DOI] [PubMed] [Google Scholar]
- 51.Lower B.H., Hochella M.F., Lower S.K. Putative mineral-specific proteins synthesized by a metal reducing bacterium. Am. J. Sci. 2005;305:687–710. [Google Scholar]
- 52.Marko J.F., Siggia E.D. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 53.Wang M.D., Yin H., Block S.M. Stretching DNA with optical tweezers. Biophys. J. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kellermayer M.S.Z., Smith S.B., Bustamante C. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 55.Carrion-Vazquez M., Oberhauser A.F., Fernandez J.M. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tskhovrebova L., Trinick J., Simmons R.M. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 1997;387:308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 57.Oberhauser A.F., Marszalek P.E., Fernandez J.M. Single protein misfolding events captured by atomic force microscopy. Nat. Struct. Biol. 1999;6:1025–1028. doi: 10.1038/14907. [DOI] [PubMed] [Google Scholar]
- 58.Mueller H., Butt H.J., Bamberg E. Force measurements on myelin basic protein adsorbed to mica and lipid bilayer surfaces done with the atomic force microscope. Biophys. J. 1999;76:1072–1079. doi: 10.1016/S0006-3495(99)77272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bemis J.E., Akhremitchev B.B., Walker G.C. Single polymer chain elongation by atomic force microscopy. Langmuir. 1999;15:2799–2805. [Google Scholar]
- 60.Dugdale T.M., Dagastine R., Wetherbee R. Single adhesive nanofibers from a live diatom have the signature fingerprint of modular proteins. Biophys. J. 2005;89:4252–4260. doi: 10.1529/biophysj.105.062489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dugdale T.M., Dagastine R., Wetherbee R. Diatom adhesive mucilage contains distinct supramolecular assemblies of a single modular protein. Biophys. J. 2006;90:2987–2993. doi: 10.1529/biophysj.105.079129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee G., Abdi K., Marszalek P.E. Nanospring behaviour of ankyrin repeats. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- 63.Zhang B., Evans J.S. Modeling AFM-induced PEVK extension and the reversible unfolding of Ig/FNIII domains in single and multiple titin molecules. Biophys. J. 2001;80:597–605. doi: 10.1016/S0006-3495(01)76040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fröman G., Switalski L.M., Höök M. Isolation and characterization of a fibronectin receptor from Staphylococcus aureus. J. Biol. Chem. 1987;262:6564–6571. [PubMed] [Google Scholar]
- 65.Wann E.R., Gurusiddappa S., Hook M. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J. Biol. Chem. 2000;275:13863–13871. doi: 10.1074/jbc.275.18.13863. [DOI] [PubMed] [Google Scholar]
- 66.Benoit M., Gabriel D., Gaub H.E. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nat. Cell Biol. 2000;2:313–317. doi: 10.1038/35014000. [DOI] [PubMed] [Google Scholar]
- 67.Schwesinger F., Ros R., Pluckthun A. Unbinding forces of single antibody-antigen complexes correlate with their thermal dissociation rates. Proc. Natl. Acad. Sci. USA. 2000;97:9972–9977. doi: 10.1073/pnas.97.18.9972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Evans E. Probing the relation between force-lifetime and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 69.Bustanji Y., Arciola C.R., Samorí B. Dynamics of the interaction between a fibronectin molecule and a living bacterium under mechanical force. Proc. Natl. Acad. Sci. USA. 2003;100:13292–13297. doi: 10.1073/pnas.1735343100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Almqvist N., Bhatia R., Lal R. Elasticity and adhesion force mapping reveals real-time clustering of growth factor receptors and associated changes in local cellular rheological properties. Biophys. J. 2004;86:1753–1762. doi: 10.1016/S0006-3495(04)74243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lower B.H., Yongsunthon R., Lower S.K. Antibody recognition force microscopy shows that outer membrane cytochromes OmcA and MtrC are expressed on the exterior surface of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 2009;75:2931–2935. doi: 10.1128/AEM.02108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Camesano T.A., Natan M.J., Logan B.E. Observation of changes in bacterial cell morphology using tapping mode atomic force microscopy. Langmuir. 2000;16:4563–4572. [Google Scholar]
- 73.Vadillo-Rodríguez V., Busscher H.J., Van Der Mei H.C. Comparison of atomic force microscopy interaction forces between bacteria and silicon nitride substrata for three commonly used immobilization methods. Appl. Environ. Microbiol. 2004;70:5441–5446. doi: 10.1128/AEM.70.9.5441-5446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buck A.W., Fowler V.G., Jr., Lower S.K. Bonds between fibronectin and fibronectin-binding proteins on Staphylococcus aureus and Lactococcus lactis. Langmuir. 2010;26:10764–10770. doi: 10.1021/la100549u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


