Abstract
Fertilization in angiosperms depends on a complex cellular “courtship” between haploid pollen and diploid pistil. These pollen-pistil interactions are regulated by a diversity of molecules, many of which remain to be identified and characterized. Thus, it is unclear to what extent these processes are conserved among angiosperms, a fact confounded by limited sampling across taxa. Here, we report the analysis of pistil-expressed genes in Senecio squalidus (Asteraceae), a species from euasterid II, a major clade for which there are currently no data on pistil-expressed genes. Species from the Asteraceae characteristically have a “semidry stigma,” intermediate between the “wet” and “dry” stigmas typical of the majority of angiosperms. Construction of pistil-enriched cDNA libraries for S. squalidus allowed us to address two hypotheses: (1) stigmas of S. squalidus will express genes common to wet and dry stigmas and genes specific to the semidry stigma characteristic of the Asteraceae; and (2) genes potentially essential for pistil function will be conserved between diverse angiosperm groups and therefore common to all currently available pistil transcriptome data sets, including S. squalidus. Our data support both these hypotheses. The S. squalidus pistil transcriptome contains novel genes and genes previously identified in pistils of species with dry stigmas and wet stigmas. Comparative analysis of the five pistil transcriptomes currently available (Oryza sativa, Crocus sativus, Arabidopsis thaliana, Nicotiana tabacum, and S. squalidus), representing four major angiosperm clades and the three stigma states, identified novel genes and conserved genes potentially regulating pollen-pistil interaction pathways common to monocots and eudicots.
Rapid and reliable communication between the male (pollen) and female (pistil) reproductive tissues is essential for successful fertilization in angiosperms. The tissue of the pistil acts as a physical and chemical interface between the male and female gametophytes, beginning at the stigma surface during pollen germination and continuing until successful fertilization at the ovule (Hiscock and Allen, 2008). Many different processes occur simultaneously in pistil tissues, including species recognition, self-incompatibility (SI), pollen hydration, pollen tube growth, and pathogen defense (for review, see Swanson et al., 2004; Malhó et al., 2006; Wilsen and Hepler, 2007; Hiscock and Allen, 2008). It is clear that a high diversity of both pollen and pistil molecules mediate the complex interactions between these two tissues; however, with the exception of SI, relatively little is known about the molecular pathways involved (Edlund et al., 2004; Sanchez et al., 2004; Swanson et al., 2004).
In addition to the large range of molecules present in the pistil tissues of a given species, it is likely that a number of specific molecules have diversified between taxa (Swanson et al., 2004). There are two reasons for this: first, genes that are involved in regulating sexual reproduction are likely to evolve at a higher rate than those controlling housekeeping processes (Swanson and Vacquier, 2002); second, genes involved in maintaining species boundaries will be, by their nature, species specific and hence highly diverse between species (Edlund et al., 2004; Swanson et al., 2004). To date, much research into pollen-pistil interactions has focused on SI, and these studies have shown that a high diversity of molecules and processes are employed for the same role of recognizing and rejecting self-pollen (for review, see Hiscock and McInnis, 2003; Takayama and Isogai, 2005; Franklin-Tong, 2008). This suggests that a similar or greater diversity of molecules is likely to regulate compatible pollen-pistil interactions. Certainly, despite the identification of an increasing number of molecules implicated in compatible pollen-pistil interactions, there are few examples of shared genes between species (Lord, 2003; Hiscock and Allen, 2008). However, these types of studies have focused on specific genes in model species, and the low consensus so far observed between species is largely a consequence of this limited sampling. It is important, therefore, to extend studies of genes and proteins regulating pollen-pistil interactions to include other species from diverse families of flowering plants. One approach to identifying more genes involved in pollen-pistil interactions in diverse angiosperm species is through the generation of pistil-enriched cDNA libraries, which allows comparison of tissue-specific transcriptomes between species.
Recently, several studies have identified genes expressed in the reproductive tissues of Arabidopsis thaliana (Arabidopsis), Oryza sativa (rice), Crocus sativus (crocus), and Nicotiana tabacum (tobacco), representing three major angiosperm clades: monocots (rice and crocus), rosids (Arabidopsis), and asterid I (tobacco; Angiosperm Phylogeny Group, 2003). Large data sets have been generated to represent the transcriptomes of pollen and/or pistil tissues at different stages of development (Becker et al., 2003; Swanson et al., 2005; Tung et al., 2005; D’Agostino et al., 2007; Li et al., 2007; Quiapim et al., 2009).
Angiosperm stigmas can be broadly classified as either “wet” or “dry,” depending on the presence or absence of secretions at the stigma surface (Heslop-Harrison and Shivanna, 1977). Taxa possessing wet stigmas typically secrete an exudate, which allows pollen hydration and germination to occur, even with pollen from other species. By contrast, in species with dry stigmas, pollen adhesion and recognition often precede hydration, and these processes are highly regulated (Dickinson, 1995). Arabidopsis, rice, and crocus possess dry stigmas, whereas tobacco has a wet stigma (Heslop-Harrison and Shivanna, 1977; Supplemental Fig. S1).
To expand phylogenetic sampling of pistil-enriched/pistil-specific transcriptome data sets to include the asterid II clade, we have used suppression subtractive hybridization (SSH) to construct pistil-enriched cDNA libraries for Senecio squalidus (Oxford ragwort [Asteraceae]). Importantly, S. squalidus possesses a “semidry” stigma that shows features intermediate between dry and wet stigmas (Hiscock et al., 2002). Like species with dry stigmas, the S. squalidus stigmatic papillae possess a surface cuticle, but unlike the typical dry stigma (e.g. Brassica sp.), this is not continuous and does not extend to the base of the papillae, and when stigmas reach maturity a small amount of extracellular secretion is present in these basal regions between papillae (Hiscock et al., 2002). The semidry stigma state, which appears to be common to all members of the Asteraceae (Hiscock et al., 2002; Allen and Hiscock, 2008), the second largest family of flowering plants, has not been well studied at the molecular level, and its evolutionary relationship with wet and dry stigmas is unknown.
The availability of five pistil transcriptome data sets from four major angiosperm clades that together contain more than 90% of flowering plants has allowed us to compare the diversity and conservation of pistil-expressed genes in species with wet, dry, and semidry stigmas across the monocots (crocus [Liliales] and rice [Poales]) and eudicots (Arabidopsis [Brassicales], tobacco [Solanales], and S. squalidus [Asterales]; Supplemental Fig. S1). Using these data, we investigated two hypotheses: (1) the semidry stigma of S. squalidus will express genes common to species with both wet and dry stigmas as well as expressing genes specific to the semidry stigma of the Asteraceae; and (2) specific classes of genes potentially essential for pistil function will be conserved between diverse angiosperm groups and therefore common to the five pistil transcriptomes sampled.
RESULTS
Identification and Functional Classification of Putative Pistil-Specific Genes
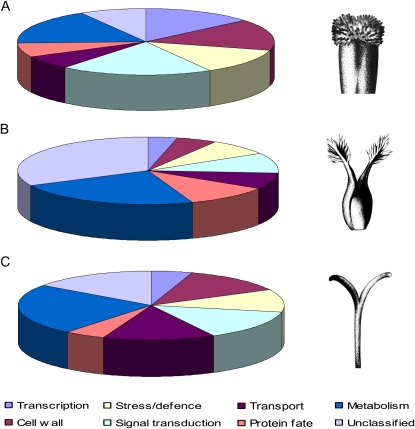
Three cDNA libraries were created from the cloned products of SSH, each corresponding to pistil-expressed sequences from three different S-genotypes (S1S2, S1S3, S1S4) of S. squalidus. Approximately 80 cDNA clones from each library were selected on the basis of the differential screening results (hybridization to subtracted pistil probe versus no hybridization to leaf probe) and sequenced. Functional annotation of the putative pistil-specific genes was performed using the BLAST-X algorithm. To generate a representative data set of the S. squalidus transcriptome, the three subtracted libraries were combined and assigned functional categories (Table I). A total of 174 cDNA clones was identified, and of these, 86% (150 cDNA clones) could be assigned a putative function according to the Gene Ontology database. The 174 cDNA clones corresponded to 115 different genes, with several genes present in multiple copies in the cDNA libraries. Over 50% of the genes identified were assigned functions in just four functional categories: metabolism (22%), transport (15%), signaling (11%), and cell wall related (10%; Fig. 1).
Table I. Candidate stigma-specific genes from S. squalidus.
A total of 174 genes were isolated by SSH and assigned a putative function using BLAST. Genes highlighted in boldface represent novel pistil-specific transcripts. ID, Identification code.
| Functional Category | Description | No. of cDNAs Isolated | Accession No. | Arabidopsis ID |
| Cell wall related | ||||
| Pi-Tubulin | 3 | GO255121 | ||
| β-Tubulin | 1 | GO255103 | ||
| Extensin (class I) | 10 | GO255084 | AT1G26250 | |
| Structural molecule | 1 | GO255092 | AT5G54110 | |
| Integral membrane family protein | 1 | GO255089 | AT4G25040 | |
| Pectinesterase family protein | 1 | GO255167 | AT5G62350 | |
| α-l-Arabinofuranosidase | 1 | GO255144 | AT5G49360 | |
| β-Galactosidase precursor | 2 | GO255246 | AT5G56870 | |
| β-Xylosidase | 3 | GO255211 | AT5G49360 | |
| Pro-rich extensin-like family | 1 | GO255237 | AT5G35190 | |
| Extensin-like protein | 1 | GO255222 | ||
| Stress/defense | ||||
| Stigma-specific peroxidase precursor | 5 | GO255081 | ||
| Putative nematode resistance protein | 1 | GO255085 | AT3G55840 | |
| Nodulin MTN3 | 1 | GO255182 | AT4G10850 | |
| Senescence-associated nodulin 1A | 3 | GO255150 | AT3G19000 | |
| Dehydration-responsive protein | 2 | GO255187 | AT3G51070 | |
| EDGP precursor | 3 | GO255139 | AT1G03230 | |
| Aluminum-induced protein | 2 | GO255210 | AT5G43830 | |
| Copper/zinc-superoxide dismutase | 2 | GO255230 | ||
| Hormone related | ||||
| Abscisic acid-inducible protein | 3 | GO255142 | AT5G38760 | |
| Ethylene-forming enzyme | 1 | GO255174 | ||
| Auxin/indole-3-acetic acid | 1 | GO255241 | AT3G23050 | |
| Metabolism | ||||
| β-Fructofuranosidase | 3 | GO255090 | AT1G12240 | |
| GDSL-motif lipase/hydrolase family | 2 | GO255184 | AT5G45670 | |
| Mandelonitrile lyase | 6 | GO255125 | AT1G73050 | |
| Galactokinase-like protein | 2 | GO255086 | AT4G16130 | |
| Fru-bisP aldolase | 3 | GO255113 | AT5G03690 | |
| Putative phosphatase | 1 | GO255111 | AT1G17710 | |
| GMC oxidoreductase | 3 | GO255209 | AT1G73050 | |
| Short-chain alcohol dehydrogenase | 1 | GO255135 | AT3G55310 | |
| Cytochrome P450 | 4 | GO255238 | AT3G19270 | |
| 3-Ketoacyl-CoA thiolase | 1 | GO255148 | AT2G33150 | |
| Vacuolar H+-ATPase subunit | 2 | GO255206 | ||
| Ser hydroxymethyltransferase | 1 | GO255188 | AT5G61820 | |
| Glycerophosphoryl diester | 1 | GO255129 | AT5G43300 | |
| Hypothetical protein OsJ_006695 | 1 | GO255175 | AT5G43300 | |
| 2OG-Fe(II) oxygenase | 1 | GO255176 | AT3G11180 | |
| NEC1 | 1 | GO255138 | AT5G23660 | |
| Prephenate dehydratase | 2 | GO255163 | AT1G08250 | |
| Xyloglucan endotransglycosylase | 1 | GO255161 | AT4G14130 | |
| UDP-Gal | 1 | GO255126 | AT1G12780 | |
| Lipase, putative | 1 | GO255164 | AT1G28600 | |
| MIOX1 (myoinositol oxygenase) | 1 | GO255130 | AT2G19800 | |
| Glycosyl hydrolase family 3 protein | 1 | GO255211 | AT5G49360 | |
| Enoyl-CoA hydratase/isomerase | 1 | GO255232 | AT3G60510 | |
| Acetoacetyl-CoA thiolase | 1 | GO255227 | AT2G33150 | |
| 3-Hydroxyisobutyryl-CoA | 1 | GO255194 | AT2G30660 | |
| Glycosyl transferase, family 48 | 1 | GO255243 | AT1G05570 | |
| Protein fate | ||||
| Putative polyubiquitin | 1 | GO255110 | AT1G65350 | |
| Seven in absentia family protein | 1 | GO255172 | AT4G27880 | |
| Endoplasmic reticulum retrieval | 1 | GO255137 | AT2G21600 | |
| Structural constituent of ribosome | 1 | GO255147 | AT3G43980 | |
| Binding protein | 1 | GO255200 | AT3G13330 | |
| Cys protease | 1 | GO255232 | AT4G31810 | |
| UDP-glucuronate decarboxylase 1 | 1 | GO255100 | AT5G59290 | |
| Ser carboxypeptidase-like 42 | 1 | GO255215 | AT5G42240 | |
| Signaling | ||||
| Kinase-interacting family protein | 1 | GO255171 | AT1G09720 | |
| GASA-like protein | 4 | GO255165 | AT3G02885 | |
| Shaggy-related protein kinase 1 | 1 | GO255160 | AT3G05840 | |
| Protein kinase, MAPK | 2 | GO255162 | AT3G18750 | |
| Calcium-binding protein; kinase | 1 | GO255154 | AT3G48260 | |
| α-Kinase | 1 | GO255168 | ||
| Leu-rich repeat protein kinase | 1 | GO255220 | AT5G40340 | |
| Somatic embryogenesis receptor kinase | 2 | GO255098 | AT1G71830 | |
| Putative AMP-binding protein | 1 | GO255112 | AT5G16340 | |
| Putative protein kinase; resistance gene | 1 | GO255082 | AT4G35600 | |
| Receptor protein kinase | 2 | GO255115 | AT1G63430 | |
| Protein kinase; adipokinetic hormone | 1 | GO255095 | AT1G06840 | |
| Transcription | ||||
| Transcription factor/regulator | 1 | GO255078 | AT2G47270 | |
| Leu-rich repeat | 1 | GO255114 | AT5G66650 | |
| Nucleic acid binding | 1 | GO255153 | AT2G02570 | |
| Zinc finger protein | 1 | GO255216 | AT5G04390 | |
| Sunflower 16 protein (SF16) | 2 | GO255239 | AT1G01110 | |
| Ser/Thr protein phosphatase | 1 | GO255235 | AT1G56440 | |
| RNA-binding region RNP-1 | 2 | GO255195 | AT2G43370 | |
| Putative reverse transcriptase | 6 | GO255236 | ||
| Transport | ||||
| Iron transporter protein IRT1 | 1 | GO255096 | AT4G19690 | |
| Putative auxin efflux carrier protein 9 | 1 | GO255104 | AT1G73590 | |
| Heavy metal transport detox protein | 2 | GO255118 | AT1G12520 | |
| FAD-binding domain-containing protein | 1 | GO255105 | AT4G20830 | |
| Stigma/style ABC transporter | 2 | GO255101 | AT5G13580 | |
| Membrane-associated protein | 1 | GO255107 | AT5G54110 | |
| Calcium-binding EF hand family protein | 2 | GO255128 | AT2G34030 | |
| E-class P450, group I | 2 | GO255238 | AT3G26330 | |
| Lipid transfer protein | 2 | GO255151 | AT5G59310 | |
| Amino acid carrier | 1 | GO255185 | AT5G09220 | |
| PIP1 aquaporin | 1 | GO255146 | AT4G00430 | |
| Endoplasmic reticulum ATPase | 1 | GO255244 | AT5G03340 | |
| Cytochrome oxidase subunit I | 1 | GO255196 | AT3G57450 | |
| Cys proteinase | 1 | GO255193 | ||
| FAD-linked oxidase, N terminal | 1 | GO255221 | AT3G63440 | |
| Ras-related GTP-binding protein | 1 | GO255224 | AT1G02130 | |
| Zinc ion binding | 1 | GO255248 | AT3G60520 | |
| Unclassified | ||||
| Sunflower 21 protein (SF21) | 2 | GQ227732 | AT2G19620 | |
| Putative Pro-rich protein | 1 | GO255157 | AT5G45670 | |
| Hypothetical protein | 1 | GO255217 | ||
| Unknown protein | 2 | GO255213 | AT3G46070 | |
| Cytokine-induced apoptosis inhibitor | 1 | GO255116 | AT5G18400 | |
| Cupin family protein | 1 | GO255124 | ||
| Hypothetical protein | 1 | GO255119 | ||
| Uncharacterized Cys-rich domain | 1 | GO255145 | AT2G37110 | |
| Unknown protein | 1 | GO255192 | AT4G35710 | |
| Expressed protein (rice) | 1 | GO255156 | ||
| Hypothetical protein | 1 | GO255178 | AT5G06380 | |
| LIM domain protein PLIM-2 | 1 | GO255173 | AT1G01780 | |
| Gag polyprotein | 1 | GO255127 | ||
| CBS domain-containing protein | 1 | GO255201 | AT3G48530 | |
| Sunflower 3 protein (SF3) | 1 | GO255242 | AT1G10200 | |
| Unknown protein | 2 | GO255233 | AT1G54320 | |
| Hypothetical protein | 1 | GO255205 | AT3G22850 | |
| GEG protein | 1 | GO255228 | AT2G18420 | |
| Unknown protein | 1 | GO255207 | AT3G57450 | |
| Hypothetical protein | 1 | GO255247 | AT3G52710 | |
| DENN domain-containing protein | 1 | GO255219 | AT5G35560 | |
| Hypothetical protein | 1 | GO255189 | AT2G19800 |
Figure 1.
Functional classification of the pistil-specific genes from the data sets of Arabidopsis (A; 501 genes; Swanson et al., 2005; Tung et al., 2005), rice (B; 115 genes; Li et al., 2007), and S. squalidus (C; 115 genes; this study). Functions were assigned according to the Gene Ontology database. Drawings at right illustrate the structure of the pistil of each species.
Confirmation of Pistil-Specific Expression by Northern Hybridization
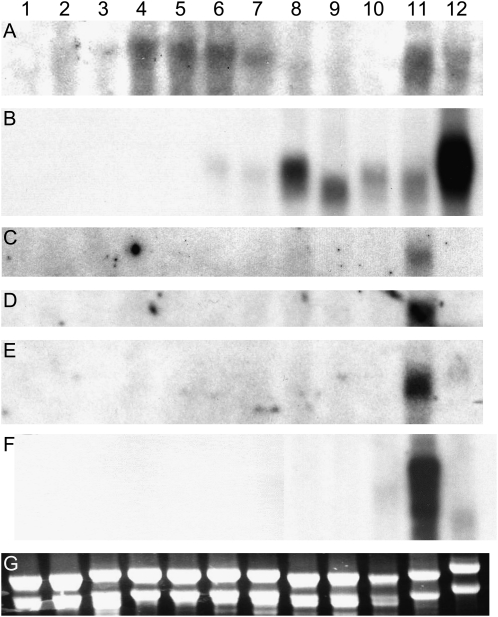
To confirm the expression of candidate genes in pistil tissues, northern hybridization was performed on a subset of genes in the data set (Fig. 2). All six cDNA clones were expressed in pistil tissue but not in leaf tissue, indicating that the suppression subtraction had worked efficiently. Four of the clones (encoding a nodulin protein, a membrane-associated protein, a myoinositol oxygenase protein, and a nematode resistance protein) were expressed exclusively in the pistil (Fig. 2, C–F), and two clones (encoding a cytochrome P450 and a calcium-binding kinase) were also expressed in pollen (Fig. 2, A and B). The nematode resistance gene was expressed as several different sized transcripts in the pistil.
Figure 2.
Developmental northern-blot analysis of candidate stigma-specific gene expression. Total RNA was extracted from S. squalidus tissues; each lane contains 10 μg of total RNA. Lane 1, root; lane 2, leaf; lane 3, stem; lane 4, small capitulum bud (2–3 mm); lane 5, medium capitulum bud (5–6 mm); lane 6, large capitulum bud (8–9 mm); lane 7, open capitulum; lane 8, floret buds; lane 9, mix of open florets and buds; lane 10, florets all open; lane 11, mature stigmas; lane 12, pollen. The RNA was probed separately with cytochrome P450 (A; GO255238), calcium-binding kinase (B; GO255154), nodulin (C; GO255182), myoinositol oxygenase (D; GO255123), membrane-associated protein (E; GO255107), and nematode resistance protein (F; GO255085). G represents a loading control.
Comparison of the S. squalidus, Arabidopsis, Rice, Tobacco, and Crocus Pistil Transcriptomes
Pistil-specific data sets have so far been generated in just two species: Arabidopsis (Swanson et al., 2005; Tung et al., 2005) and rice (Li et al., 2007). The Arabidopsis data sets were generated from microarray analysis and cDNA subtraction of stigma tissue (Swanson et al., 2005) and microarray analysis of stigma/style tissue (Tung et al., 2005). The rice data set was produced from analysis of an Affymetrix rice whole-genome array and cDNA microarray comparisons of stigma tissue (Li et al., 2007). Additional data for pistil transcriptomes have also been generated recently in crocus (D’Agostino et al., 2007) and tobacco (Quiapim et al., 2009). The tobacco pistil EST data set was generated from stigma and style cDNA (Quiapim et al., 2009), and the crocus EST data set from stigma cDNA (D’Agostino et al., 2007). Comparisons of the results generated by these separate studies with our data set from S. squalidus showed broad correlations in the functional gene classes identified (Fig. 1; Table II). General classification of the stigma-enriched data sets of S. squalidus, Arabidopsis, and rice revealed that the proportions of genes in each functional class were similar. The proportions of genes in the functional classes transcription, cell wall, stress/defense, signal transduction, and unclassified from S. squalidus were comparable to those of Arabidopsis and rice, but S. squalidus had the highest proportions of genes involved in transport and metabolism. A Wilcoxon signed-rank test on the proportion data confirmed that the results were not significantly different from each other (S. squalidus/Arabidopsis, P = 1.00; S. squalidus/rice, P = 0.484; Arabidopsis/rice, P = 0.674).
Table II. S. squalidus pistil-expressed genes that have been identified in at least two of the pistil preferential data sets from different species.
The study species used were Arabidopsis (Swanson et al., 2005; Tung et al., 2005), rice (Li et al., 2007), tobacco (Quiapim et al., 2009), and crocus (D’Agostino et al., 2007). Presence (+)/absence (−) is indicated for each species, followed by transcript numbers in parentheses.
| S. squalidus Genes | Arabidopsis | Rice | Crocus | Tobacco |
| Cytochrome P450 | + (6) | + (8) | + (6) | + (1) |
| LTP | + (4) | + (1) | + (2) | + (1) |
| 3-Ketoacyl-CoA synthase | − | + (1) | + (1) | − |
| Acyl-CoA-binding protein | + (1) | + (1) | + (1) | − |
| β-Xylosidase | − | + (3) | + (1) | − |
| β-Galactosidase precursor | − | + (3) | + (1) | − |
| UDP-glycosyltransferase | + (4) | + (1) | + (5) | − |
| Zinc finger protein | − | + (2) | + (6) | + (1) |
| Peroxidase | + (3) | + (3) | + (1) | − |
| Aquaporin | + (1) | + (1) | + (1) | − |
| FAD oxidoreductase | + (1) | − | + (1) | − |
| ABC transporter family | + (1) | + (3) | + (2) | + (3) |
| Extensin-like protein | + (1) | − | + (1) | + (7) |
| Cys protease | + (1) | + (1) | + (4) | − |
| Mandelonitrile lyase | + (2) | − | − | − |
| Pectinesterase | + (2) | + (3) | + (1) | − |
| Ser carboxypeptidase | + (2) | + (1) | − | − |
| Receptor protein kinase | + (2) | + (5) | + (4) | + (4) |
| Disease resistance | + (1) | + (5) | + (1) | + (1) |
| Lipase/hydrolase | + (1) | + (1) | − | − |
| Cys-rich protein | + (1) | + (1) | − | − |
| Nodulin/mtn3 | + (3) | + (2) | − | + (2) |
Two independent studies in Arabidopsis have indicated that the categories of metabolism, stress/defense, signaling, and cell wall related contain a large proportion of pistil-specific genes (Swanson et al., 2005; Tung et al., 2005). A similar study in rice identified the categories of cell wall related, stress/defense, and signal transduction as being the largest categories and also reported a large number of genes involved in transcription (Li et al., 2007). Correlations between the S. squalidus, Arabidopsis, and rice data sets have highlighted certain functional groups that contain high numbers of pistil-specific genes in all three species. In particular, these were cell wall related and signaling, with the categories of transport, stress/defense, and metabolism also containing a high percentage of pistil-specific genes.
Analysis of Conserved Pistil-Specific Genes
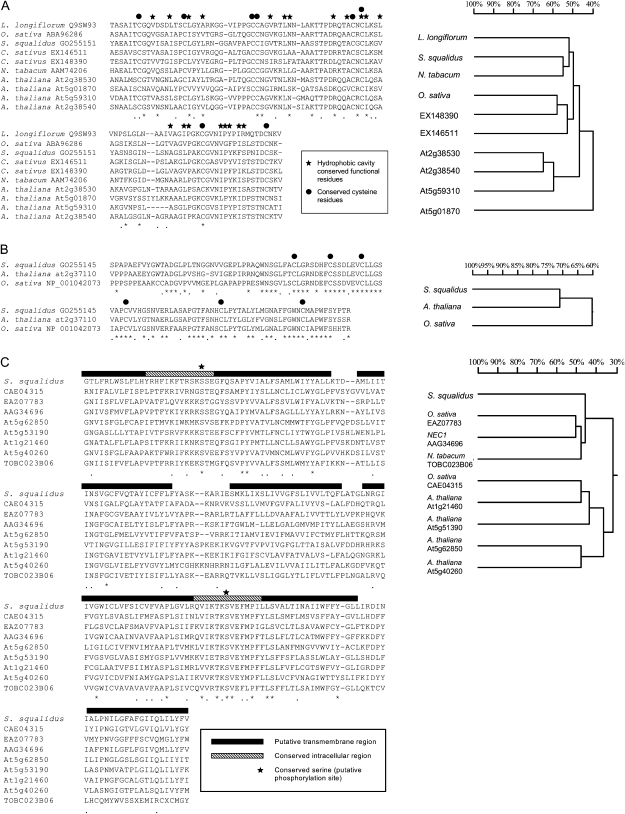
In addition to the similarities in the patterns of functional annotation of the pistil-enriched data sets, a number of pistil-enriched gene classes were common to three or more different species (Table II). Gene families that were consistently detected in the pistil tissues included cytochrome P450, ATP-binding cassette (ABC) transporters, and lipid transfer proteins (LTPs). A greater number of shared genes were identified between S. squalidus and the dry stigma species (Arabidopsis, rice, and crocus) than with the wet stigma species (tobacco). This may be a consequence of the smaller tobacco pistil preferential data set or may reflect fundamental differences between wet and dry stigmas. Sequence analysis of three conserved proteins indicated structural and functional similarity (Fig. 3). Lipid-transfer proteins identified in the pistil-enriched data sets of Arabidopsis (At2g38530, At2g38540, At5g59310, At5g01870), rice (AK105838), crocus (EX146511, EX1483990), and S. squalidus showed sequence homology to stigma/style Cys-rich adhesin (SCA; Q9SW93; approximately 50% protein identity), a LTP identified in lily (Lilium longiflorum; Park and Lord, 2003) and shown to function in pollen tube adhesion and guidance (Chae et al., 2007; TableII; Fig. 3A). All 20 feature residues of the hydrophobic cavity characteristic of this gene family were conserved in the S. squalidus sequence. Additionally, there were eight Cys residues conserved between the sequences from all species (Fig. 3A). A further Cys-rich protein of unknown function was also identified in three out of five of the study species, S. squalidus, Arabidopsis (At2g37110), and rice (NP_001042073; Table II), with a high level of sequence conservation (60% identity between rice and Arabidopsis, 71% identity between Arabidopsis and S. squalidus; Fig. 3B).
Figure 3.
Sequence alignments and homology trees of conserved pistil-specific proteins from S. squalidus, rice, Arabidopsis, tobacco, and crocus. A, LTP, orthologous to SCA (L. longiflorum). B, Cys-rich protein. C, Nodulin/mtn3 family protein, orthologous to mtn3 (Medicago trunculata) and NEC1 (P. hybrida).
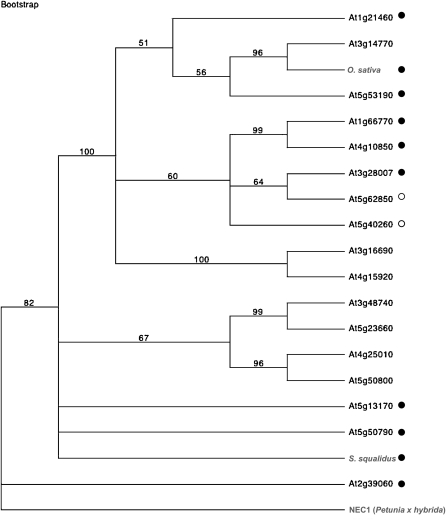
A pistil-specific gene showing sequence similarity to a member of the nodulin/mtn3 gene family was identified in separate studies of Arabidopsis (At1g21460, At5g53190), rice (CAE04315), tobacco (TOBC023B06), and S. squalidus. Seventeen copies of this gene have been identified in Arabidopsis (Guan et al., 2008) and 18 in rice genomes (Chu et al., 2006; Yang et al., 2006). Two of the Arabidopsis orthologs were shown to be pistil specific (At1g21460, At5g51390) and a further two to be pollen specific (At5g62850, At5g40260). Interestingly, the S. squalidus protein exhibits higher homology to the tobacco (C023B06, C061G07) and rice (CAE04315) proteins than any of the Arabidopsis proteins. All the nodulin/mtn3-like protein sequences share seven transmembrane regions and two conserved intracellular regions but have more variable extracellular regions (Fig. 4). Further analysis of this protein family in Arabidopsis indicated a complex pattern of evolution of pistil- and pollen-specific proteins, with the S. squalidus and rice pistil-specific proteins allying to different Arabidopsis clades (Fig. 4). The S. squalidus pistil-specific nodulin shares greater sequence identity with the tobacco sequence (55%) than those from Arabidopsis and rice (35% and 31%, respectively).
Figure 4.
Phylogenetic relationship between nodulin/mtn3 genes in Arabidopsis and pistil-specific homologs in rice (BAG89875) and S. squalidus (GO255182). The parsimonious tree was constructed in PAUP version 4.0b 10 using a heuristic search method executed on a protein sequence alignment and rooted with the NEC1 sequence from P. hybrida (AAG34696). Bootstrap support values are indicated next to nodes, based on 1,000 replicates. Pistil-expressed genes are indicated by black circles; pollen-expressed genes are indicated by white circles.
Novel Pistil-Specific Genes in S. squalidus
The pistil cDNA libraries in S. squalidus contain a number of novel genes not previously identified as pistil specific in other species and not present in our S. squalidus floral database (www.seneciodb.org). These include a WNK (for With No K/Lys) kinase with a putative calcium-binding domain, a membrane-associated protein, a nematode resistance protein, and several hypothetical proteins of unknown function (Table I). The identification of novel pistil-specific genes in S. squalidus is particularly interesting, as this species possesses a sporophytic self-incompatibility (SSI) system, which operates through a different mechanism from the well-characterized SSI system found in the Brassicaceae (Hiscock et al., 2003; Tabah et al., 2004). Therefore, the S. squalidus pistil data set is expected to contain genes potentially involved in mediating the female side of SSI, including primary S-recognition genes.
DISCUSSION
The S. squalidus Pistil-Enriched Transcriptome
SSH was used successfully to isolate pistil-enriched transcripts from cDNA libraries constructed for three different S-genotypes of S. squalidus. When combined, these yielded 115 different candidates for pistil-specific genes for S. squalidus. Differential screening of the cDNA libraries confirmed the expression of clones in pistil tissue. Northern-blot analysis, performed on a subset of cDNA clones, revealed that expression of the majority of these genes was exclusive to pistil tissue. Both these methods of screening illustrated the efficiency of SSH for isolating tissue-specific genes. Pistil-specific genes are likely to play important roles in many stigma functions, including defense, pollen adhesion and hydration, pollen tube guidance and structural support within the extracellular matrix, and SI. The pistil-enriched libraries created for S. squalidus have identified potential components of all these systems and may be used to compliment and confirm information from other species for which pistil transcriptome data are available. Therefore, we compared all five available pistil transcriptomes, rice and crocus (monocots), Arabidopsis (rosid clade), tobacco (euasterid I clade), and S. squalidus (euasterid II clade), spanning four major angiosperm clades that contain more than 90% of all flowering plants.
All five pistil transcriptomes show broad similarities in the proportion of genes in particular functional classes, the types of genes identified, and sequence similarities between protein products. Additionally, the S. squalidus SSH libraries contained a number of genes showing orthology to pistil-specific genes previously identified in other species (Park and Lord, 2003; Otsu et al., 2004; McInnis et al., 2005). Comparisons with data from other species and extensive S. squalidus EST databases (www.seneciodb.org) also identified a number of novel genes in the S. squalidus pistil cDNA libraries (Table I). These are likely to correspond to rare transcripts in the pistil transcriptome and highlight the usefulness of SSH as a technique that is able to identify genes expressed specifically in the tissue of interest.
Comparisons of the currently available pistil data sets highlighted differences between results from whole-pistil and pistil-specific studies. The mature stigma data set from crocus contained a large number of genes involved in metabolism, transport, and transcription (D’Agostino et al., 2007; 157 genes). The overrepresentation of metabolism genes in crocus compared with the other dry stigma species may be a consequence of the data set being a sample of the unsubtracted pistil transcriptome, not just pistil-specific transcripts. Alternatively, these differences may reflect the high levels of carotenoid metabolism in the highly specialized stigma of crocus (D’Agostino et al., 2007). A comparison of the functional categories of the Arabidopsis pistil data set at different resolutions of pistil specificity revealed differences in proportions of functional classes, highlighting classes that were important in general cellular function (plastid-related genes) and those required for pistil-specific processes (cell wall, endoplasmic reticulum, response to stress, plasma membrane; Hiscock and Allen, 2008).
Comparative Analyses of Wet, Dry, and Semidry Stigma/Style Transcriptomes
When the data generated by this study were compared with data from Arabidopsis, tobacco, rice, and crocus, similarities and differences were revealed (Swanson et al., 2005; Tung et al., 2005; D’Agostino et al., 2007; Li et al., 2007; Quiapim et al., 2009). The study species differ in relatedness, pistil structure and function, and specific pollen-pistil processes, allowing the opportunity to assess consensus across diverse taxa (Supplemental Fig. S1). Differences were observed between the dry stigma species (Arabidopsis, rice, crocus) and wet stigma species (tobacco) data sets, most notably in the proportions of genes in each functional class and in the types of genes expressed exclusively in the pistil (Table II). The largest functional categories in the tobacco pistil transcriptome were posttranslational modification/protein turnover, translation, and energy production and conversion, suggesting that the tobacco pistil is composed of highly active metabolic cells, perhaps reflecting the active production of wet surface secretion, a defining feature of wet stigma species (Quiapim et al., 2009).
Data from dry stigma species consistently highlighted categories of cell wall related and signaling as containing large numbers of genes and being overrepresented in the dry stigma (Swanson et al., 2005; Tung et al., 2005; Li et al., 2007; Hiscock and Allen, 2008). A detailed comparison of the Arabidopsis and tobacco data sets revealed a low percentage of homologous sequences between the two species (Quiapim et al., 2009). In contrast, comparisons of the rice and Arabidopsis data sets revealed 83 similar sequences, most of which belonged to cell wall-related and signal transduction groups, indicating conservation of these functions in the dry stigma. The S. squalidus data set shared a greater number of homologous genes with the dry stigma species (Arabidopsis, 37; rice, 46; crocus, 39) than the wet stigma species (tobacco, eight), although a more thorough comparison involving additional species would be needed to confirm these results. However, similarities were also detected between the tobacco and S. squalidus pistil data sets, most notably a high number of extensin-like gene transcripts, a class of gene that has been well studied in the tobacco pistil (de Graaf et al., 2003). It is interesting to observe similarities between S. squalidus and species with wet stigmas and dry stigmas, because the semidry stigma of S. squalidus (Hiscock et al., 2002) may represent a derived form of either.
Evidence of Conserved Pistil-Specific Genes in Diverse Angiosperm Groups
Despite the differences in gross pistil morphology between these diverse species, certain features of the pollination process appear to be shared: in particular, the presence of lipids at the pollen-stigma interface, the involvement of small Cys-rich proteins in pollen-stigma interactions, and the role of water as a directional cue for the pollen tube (Hiscock and Allen, 2008). This study was able to identify additional potential correlations between the pistil data sets and individual pistil-specific genes, with several genes appearing to be restricted to wet, dry, or semidry stigma transcriptomes (detailed below). In particular, the protein sequences of three genes (a LTP, a Cys-rich protein, and a nodulin/mtn3 protein) from different species were aligned and exhibited sequence similarity and conservation of functional residues (Fig. 3). These examples of conserved pistil-specific genes may represent ancient pistil processes that have been maintained in diverse angiosperm species across the monocot-eudicot divide. Alternatively, these genes may be evidence of the convergent evolution of classes of genes to acquire pistil-specific functions. There is a clear need for studies of the pistil transcriptomes of lower eudicots and basal angiosperms to answer these critical evolutionary questions.
Cys-Rich Proteins
Our comparative transcriptome analysis provides further support for the hypothesis that Cys-rich proteins play important and varied roles during the pollen-pistil interaction (Doughty et al., 1998; Takayama et al., 2000; Tang et al., 2004; Verhoeven et al., 2005; Chae et al., 2007). Several Cys-rich proteins were identified in the pistil-enriched data sets of S. squalidus, Arabidopsis, rice, tobacco, and crocus. Two of these, a Cys-rich protein of unknown function and a LTP, were aligned to demonstrate sequence similarity and conservation of functional residues between the different species, suggesting a potential conserved role of these proteins in pistil function (Fig. 3). The Cys-rich protein of unknown function exhibited particularly high sequence similarity between the dry/semidry stigma species S. squalidus, Arabidopsis, and rice (60%–70% identity). This protein was not detected in the tobacco pistil transcriptome, suggesting a role specific to dry/semidry stigmas only. The LTP was detected in all species studied, and typically was expressed at high levels in the pistil (Swanson et al., 2005; Tung et al., 2005; Quiapim et al., 2009). The tobacco sequence exhibits very high sequence identity (83.4%) to LTP (Q03461), a protein that has been shown to mediate cell wall-loosening activity in the stigma exudate (Nieuwland et al., 2005). These LTPs are also related to another LTP, SCA (Q9SW93), which is involved in pollen tube adhesion and guidance in lily (Park and Lord, 2003; Kim et al., 2006; Chae et al., 2007).
Cell Wall-Related Proteins
Within the cell wall-related functional category, several classes of genes were present in data sets from all five study species. These included extensin-like proteins and Hyp-rich glycoproteins, consistent with previous work that has shown these classes of protein to be ubiquitous components of the transmitting tissue of the pistil through which pollen tubes grow and navigate to the ovules (Wu et al., 2001). Both the S. squalidus and tobacco data sets contained large numbers of extensin-like genes, suggesting that this class of gene is particularly important in wet and semidry stigmas, in contrast to the Arabidopsis, rice, and crocus data sets, which each contained just one extensin-like gene. In tobacco, these proteins have been implicated to function in a range of different processes within the pistil, including pollen tube guidance and SI (Cheung et al., 1993; Wu et al., 1995; de Graaf et al., 2003; Hancock et al., 2005). Another putative pistil-specific cell wall component identified in S. squalidus was a pectinesterase, a class of enzymes also present in the data sets of rice, Arabidopsis, and crocus, highlighting their importance in dry and semidry stigma function. Pectinesterase-like proteins are hypothesized to function in enabling pollen tube growth through the papilla cell wall by mediating cell wall loosening and expansion (Bosch et al., 2005; Bosch and Hepler, 2006).
Signaling Genes
In the signaling class of pistil-specific proteins, a relatively large number of receptor-like protein kinases are present in the S. squalidus data set, consistent with results from other species (Swanson et al., 2005; Tung et al., 2005; D’Agostino et al., 2007; Li et al., 2007; Quiapim et al., 2009). It is likely that these pistil-specific kinases are specialized to coordinate pistil development, defense responses, and to facilitate communication between the pollen and pistil, with downstream implications on pollen tube growth (Johnson and Preuss, 2003). This class of proteins have been studied in both dry and wet stigmas of the Brassicaceae and Solanaceae, respectively. The most extensively studied example of a stigma-specific receptor kinase is the S-receptor kinase from the Brassicaceae, which regulates the female response in SSI (Takayama and Isogai, 2005). Pollen-specific receptor-like kinases (Muschietti et al., 1998) have been shown to interact with corresponding pistil Cys-rich protein ligands LAT52 (Tang et al., 2002) and LeSTig1 (Tang et al., 2004) and are thought to mediate pistil response to pollen tube growth in tomato (Solanum lycopersicum).
Transport Genes
The S. squalidus, Arabidopsis, and rice pistil-enriched data sets contain a significant proportion of genes that potentially play a role in transport within the pistil, suggesting that this function is particularly important in dry and semidry stigmas. All five species data sets contained at least one member of the ABC transporter family, a family of proteins that function in the transport of a wide variety of substrates across extracellular and intracellular membranes, including metabolic products, lipids, and sterols (Sidler et al., 1998). The S. squalidus and Arabidopsis (NP_181467) sequences exhibited high sequence similarity (74% and 73% identity, respectively) with a pistil-specific ABC transporter gene, NtWBC1 (for tobacco ABC transporter of the white-brown complex subfamily [AAR06252]), which has been characterized in tobacco (Otsu et al., 2004). Expression of NtWBC1 is localized to the stigmatic secretory zone, to the cells that produce the stigmatic exudate, and it is thought that this protein may be involved in the transfer of lipids to the stigmatic exudate (Otsu et al., 2004). It may be hypothesized that the ortholog of NtWBC1 acts in a similar way in the semidry stigma of S. squalidus, where secretion of the surface exudate is enhanced in stigmatic papillae after pollination (Hiscock et al., 2002).
Another common feature of pistil function is the presence of a stigmatic water gradient acting as an initial directional cue for pollen tube growth (Hiscock and Allen, 2008). In dry stigma species, the presence of water at the stigma surface is highly regulated (Dickinson, 1995). In Brassica, a stigma-specific aquaporin (MIP-MOD [AAB61378]) has been implicated in the regulation of water flow into the pollen grain during grain hydration (Dixit et al., 2001). The pistil-specific aquaporins identified in S. squalidus, Arabidopsis, and rice, therefore, may have a similar role. In support of this suggestion, the S. squalidus pistil-specific aquaporin shares 85% sequence identity with the Brassica MIP-MOD protein. This putative role of aquaporins in pollen hydration, therefore, may explain why they were not detected in the tobacco pistil preferential data set, since the presence of the secreted stigmatic exudate in tobacco precludes the need for control of water flow to the stigma surface (Wolters-Arts et al., 2002).
Stress/Defense Genes
Our comparative transcriptome analyses have identified a number of genes encoding proteins potentially involved in stress/defense responses. This is particularly interesting as there is evidence that the molecules involved in pollination and stress/defense responses may be related evolutionally and functionally (Vogt et al., 1994; Li et al., 2007). Indeed, many authors have proposed that certain SI mechanisms may have arisen through the modification of preexisting pathogen defense mechanisms (de Nettancourt, 1977; Hodgkin et al., 1988; Dickinson, 1995; Elleman and Dickinson, 1999). Several of the genes involved in stress/defense responses identified in the S. squalidus pistil data set may be involved in reactive oxygen species (ROS) signaling. Stigmatic tissues have been shown to accumulate high levels of ROS constitutively, particularly hydrogen peroxide (H2O2), and it has been hypothesized that the high levels of ROS/H2O2 may protect stigmas against pathogen attack (McInnis et al., 2006b). ROS respond to external stimuli, acting as early messengers in signaling cascades by inducing the expression of a number of genes including pathogenesis-related protein genes (Sepúlveda-Jiménez et al., 2005). For instance, in the leaves of Beta vulgaris, a UDP-glucosyltransferase is induced by high levels of ROS, which accumulate as a response to wounding and bacterial infection (Sepúlveda-Jiménez et al., 2005). Therefore, the S. squalidus pistil-specific UDP-glucosyltransferase may respond in a similar way to the high levels of ROS in the stigmatic tissue and act downstream by glucosylating hormones and secondary metabolites. UDP-glucosyltransferases were detected in the pistil data sets of all the dry stigma species, suggesting conservation of their function in pistils.
Another gene identified in the S. squalidus pistil data set showed high sequence similarity (71% protein identity) to an extracellular dermal glycoprotein (EDGP [BAA03413]) from carrot (Daucus carota; Shang et al., 2005) and the Nectarin IV protein (67% sequence identity [AAX81588]) from tobacco (Naqvi et al., 2005). Both EDGP and Nectarin IV belong to a newly identified superfamily of inhibitor proteins (Naqvi et al., 2005). Nectarin IV is expressed in the nectary of ornamental tobacco plants during anthesis until after fertilization, when expression peaks (Naqvi et al., 2005). Analogies have been made between the high levels of ROS and H2O2 detected in the stigma and the equally high levels in nectar, which are hypothesized to protect against pathogen attack (Carter and Thornburg, 2004; McInnis et al., 2006a). Recent studies in S. squalidus have shown that levels of ROS/H2O2 were reduced in stigmatic papillae to which pollen grains had adhered, suggesting that nitric oxide from pollen may be acting to reduce ROS/H2O2 abundance in stigmatic papillae, potentially to allow pollen to be distinguished from fungal pathogens. The presence of catalase transcripts in the S. squalidus pistil libraries indicates that the pistil cells are capable of actively breaking down H2O2.
In addition to the putative EDGP/Nectarin IV ortholog, a gene showing sequence similarity (59% identity) to Nectarin1 from Petunia × hybrida (NEC1 [AAG34696]) was also identified in the S. squalidus pistil-enriched data sets. NEC1 belongs to the mtn3/saliva family of proteins (Ge et al., 2000). A homolog of another member of this family, nodulin/mtn3, was also identified in the S. squalidus, Arabidopsis, tobacco, and rice pistils. Members of this protein family have been implicated in diverse cellular processes, including disease resistance and pollen development, although their specific function is unknown (Yang et al., 2006; Guan et al., 2008). In the Arabidopsis and rice genomes, nodulin/mtn3 proteins are present in high copy number (17 and 18 genes, respectively). In Arabidopsis, at least four members of this gene family are expressed exclusively in reproductive tissues (Fig. 4). Identification of pistil-specific proteins in four of the pistil preferential data sets (Table II; Fig. 3) highlighted the potential importance of these genes in reproductive tissues, where their function may be conserved across diverse taxa. Sequence analysis showed the S. squalidus protein to lie within a subclade of this family, which also contained NEC1 and Xa13/Os8N3 (ABD78942), a disease resistance gene for bacterial blight of rice (Fig. 4; Chu et al., 2006).
CONCLUSION
The aim of this study was to expand upon currently available data for genes expressed in pistils by analyzing the pistil transcriptome of a species from the Asteraceae (S. squalidus). S. squalidus possesses a semidry stigma, intermediate between the wet and dry stigmas typical of most angiosperms and all species previously analyzed at the level of pistil gene expression. By selecting a species from the Asteraceae for analysis, we were also able to sample a representative of the asterid II clade, a major angiosperm clade for which, to our knowledge, there were no data on pistil-expressed genes prior to our study. Therefore, we were able to explore two key hypotheses: (1) that the semidry stigma of S. squalidus will express genes common to species with wet and dry stigmas as well as expressing genes specific to the semidry stigma of the Asteraceae; and (2) that certain classes of genes potentially essential for pistil function will be conserved between diverse angiosperm groups and therefore common to the pistil transcriptomes of the five angiosperm species sampled to date. Overall, our findings support both these hypotheses. With respect to hypothesis 1, our data show that the pistil transcriptome of S. squalidus is generally more similar to the dry stigma transcriptome of Arabidopsis but also contains genes expressed in wet stigmas. S. squalidus pistils also express a number of unique genes that could potentially be involved in the novel mechanism of SSI found in the Asteraceae.
With respect to hypothesis 2, our comparison of the five available pistil transcriptomes, rice and crocus, representing monocots, and Arabidopsis, tobacco, and S. squalidus, representing three divergent clades of eudicots, identified genes encoding a number of potentially orthologous proteins, most notably a LTP, a Cys-rich protein, and a nodulin/mtn3 protein. It is likely that these proteins play similar roles in their respective species and may represent components of the pollen-pistil interaction machinery system common to the majority of the angiosperms. This suggests that some of the complex interactions underlying pistil function in diverse species with wet, dry, and semidry stigmas are shared and have been inherited from the common ancestor of monocots and eudicots (Hiscock and Allen, 2008).
MATERIALS AND METHODS
Plant Material
All Senecio squalidus plants were grown in glasshouse conditions, according to Hiscock (2000). S-genotyped individuals (S1S2, S1S3, and S1S4; Brennan et al., 2010) were used for RNA and DNA extraction.
SSH
Total RNA was extracted from leaf and pistil tissue from plants of three different S-genotypes (S1S2, S1S3, and S1S4) using the TRIzol reagent according to the manufacturer’s instructions (Invitrogen). cDNA was then synthesized from total pistil and leaf RNA using the SMART PCR cDNA Synthesis kit (Clontech). Subtraction was performed separately for each genotype using cDNA from pistil and leaf tissue using the PCR-Select cDNA Subtraction kit (Clontech). This technique includes a normalization step to equalize the abundance of transcripts, allowing comparisons of copy number to be made. Reverse subtractions were also performed for each genotype for the purposes of differential screening. The subtracted PCR products were cloned using the TOPO TA-Cloning kit (Invitrogen) and screened for inserts. All colonies containing inserts were picked and transferred to separate wells of a 96-well plate containing Luria-Bertani broth.
Colony arrays were created by dot blotting cultures of each clone onto a nylon membrane (Hybond-NX; GE Healthcare), placed on the surface of a plate of Luria-Bertani agar containing 100 μg mL−1 ampicillin, and incubated at 37°C overnight. The membrane was denatured and neutralized, and DNA was fixed to the membrane by baking for 1.5 h at 80°C. The resulting subtracted libraries were differentially screened using total leaf cDNA, total pistil cDNA, subtracted leaf cDNA, and subtracted pistil cDNA as probes. Probes were prepared using Ready-To-Go DNA Labeling Beads (GE Healthcare) and labeled with [α-32P]dCTP (Amersham Biosciences). Hybridization was performed in Southern hybridization solution (300 mm NaPO4 buffer, 7% SDS, 1 mm EDTA, and 10 mg mL−1 bovine serum albumin) at 65°C. Following hybridization, the membranes were washed in four changes of 0.2× SSC/0.5% SDS buffer at 65°C before being exposed to BioMax MS-1 Autorad film (Kodak). cDNA clones, which hybridized strongly to subtracted pistil cDNA but not to leaf cDNA, were identified as pistil expressed and sequenced by Geneservice (University of Oxford) using the M13 forward universal primer. Nucleotide sequences were identified using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST/) with default parameters. Functional annotation was assigned according to the Gene Ontology database. The equality of proportions of functional categories was tested by the nonparametric Kruskal-Wallis test as implemented in the Statistical Package for the Social Sciences program (SPSS).
Northern-Blot Analysis
Total RNA was extracted from vegetative tissues (root, leaf, stem, capitulum bud, flower bud, and pistil tissue) using the Plant RNeasy kit (Qiagen) and from pollen using TRI Reagent (Molecular Research Centre) according to the manufacturer’s instructions. Total RNA (10 μg) from each tissue was separated on a 1.2% agarose formaldehyde gel at 80 V for 4 h and blotted onto Hybond-NX membrane (GE Healthcare) using standard procedures (Sambrook et al., 1989). Following transfer, RNA was immobilized onto the membrane using a CL-1000 Ultraviolet Crosslinker (UVP) set at 120 mJ cm−2. Probes were prepared using Ready-To-Go DNA Labeling Beads (GE Healthcare) and labeled with [α-32P]dCTP (Amersham Biosciences). Northern hybridization was performed at 42°C according to Sambrook et al. (1989). After hybridization, the membranes were washed in four changes of 1× SSC/0.1% SDS buffer at 42°C, before being exposed to BioMax MS-1 Autorad film (Kodak).
Comparison of Pistil Data Sets from Different Species
Pistil-enriched data sets from Arabidopsis (Arabidopsis thaliana; Swanson et al., 2005; Tung et al., 2005), rice (Oryza sativa; Li et al., 2007), crocus (Crocus sativus; D’Agostino et al., 2007), and tobacco (Nicotiana tabacum; Quiapim et al., 2009) were used for comparison with the S. squalidus data set. Functional annotation was assigned for genes in each data set according to the Gene Ontology database. Common genes between data sets were identified using keyword searches. Of these, genes possessing high levels of sequence homology were identified by aligning sequences (see below) and calculating the percentage of sequence identity.
Sequence Alignment
DNA and protein sequences were aligned using DNAMAN (Lynnon Corp.). Homology trees were created from multiple alignments using default parameters. The DNAsp package was used to calculate summary statistics of polymorphism data. Phylogenetic trees were generated from alignments using PAUP version 4.0b 10 (Swofford, 2003). Parsimonius trees were generated via a heuristic search with branch swapping set at 1,000 rearrangements. Bootstrap calculations were based on 1,000 replicates.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers given in Table I.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The phylogenetic distribution of species most widely used in studies of pollen-pistil interactions.
Supplementary Material
Acknowledgments
We thank Matthew Hegarty, Tom Batstone, and Christopher Thorogood for constructive comments on earlier drafts of the manuscript, Gary Barker for bioinformatic assistance, Andrew Hughes for plant maintenance, and Christopher Thorogood for the stigma illustrations.
References
- Allen AM, Hiscock SJ. (2008) Evolution and phylogeny of self-incompatibility systems in angiosperms. VE Franklin-Tong, ed, Self-Incompatibility in Flowering Plants: Evolution, Diversity, and Mechanisms. Springer-Verlag, Berlin, pp 73–101 [Google Scholar]
- Angiosperm Phylogeny Group (2003) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. Bot J Linn Soc 141: 399–436 [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA. (2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol 133: 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Cheung AY, Hepler PK. (2005) Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol 138: 1334–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Hepler PK. (2006) Silencing of the tobacco pollen pectin methylesterase NtPPME1 results in retarded in vivo pollen tube growth. Planta 223: 736–745 [DOI] [PubMed] [Google Scholar]
- Brennan AC, Tabah DA, Harris SA, Hiscock SJ. (2010) Sporophytic self-incompatibility in Senecio squalidus (Asteraceae): S allele dominance interactions and modifiers of cross-compatibility and selfing rates. Heredity (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Thornburg RW. (2004) Is the nectar redox cycle a floral defense against microbial attack? Trends Plant Sci 9: 320–324 [DOI] [PubMed] [Google Scholar]
- Chae K, Zhang K, Zhang L, Morikis D, Kim ST, Mollet JC, de la Rosa N, Tan K, Lord EM. (2007) Two SCA (stigma/style cysteine-rich adhesin) isoforms show structural differences that correlate with their levels of in vitro pollen tube adhesion activity. J Biol Chem 282: 33845–33858 [DOI] [PubMed] [Google Scholar]
- Cheung AY, May B, Kawata EE, Gu Q, Wu HM. (1993) Characterization of cDNAs for stylar transmitting tissue-specific proline-rich proteins in tobacco. Plant J 3: 151–160 [PubMed] [Google Scholar]
- Chu Z, Fu B, Yang H, Xu C, Li Z, Sanchez A, Park YJ, Bennetzen JL, Zhang Q, Wang S. (2006) Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor Appl Genet 112: 455–461 [DOI] [PubMed] [Google Scholar]
- D’Agostino N, Pizzichini D, Chiusano ML, Giuliano G. (2007) An EST database from saffron stigmas. BMC Plant Biol 7: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf BHJ, Knuiman BA, Derksen J, Mariani C. (2003) Characterization and localization of the transmitting tissue-specific PELPIII proteins of Nicotiana tabacum. J Exp Bot 54: 55–63 [DOI] [PubMed] [Google Scholar]
- de Nettancourt D. (1977) Incompatibility in Angiosperms. Springer-Verlag, Berlin [Google Scholar]
- Dickinson HG. (1995) Dry stigmas, water and self-incompatibility in Brassica. Sex Plant Reprod 8: 1–10 [Google Scholar]
- Dixit R, Rizzo C, Nasrallah M, Nasrallah J. (2001) The Brassica MIP-MOD gene encodes a functional water channel that is expressed in the stigma epidermis. Plant Mol Biol 45: 51–62 [DOI] [PubMed] [Google Scholar]
- Doughty J, Dixon S, Hiscock SJ, Willis AC, Parkin IAP, Dickinson HG. (1998) PCP-A1, a defensin-like Brassica pollen coat protein that binds the S locus glycoprotein, is the product of gametophytic gene expression. Plant Cell 10: 1333–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund AF, Swanson R, Preuss D. (2004) Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell (Suppl) 16: S84–S97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleman CJ, Dickinson HG. (1999) Commonalities between pollen/stigma and host/pathogen interactions: calcium accumulation during stigmatic penetration by Brassica oleracea pollen tubes. Sex Plant Reprod 12: 194–202 [Google Scholar]
- Franklin-Tong VE. editor (2008) Self-Incompatibility in Flowering Plants: Evolution, Diversity, and Mechanisms. Springer-Verlag, Berlin [Google Scholar]
- Ge YX, Angenent GC, Wittich PE, Peters J, Franken J, Busscher M, Zhang LM, Dahlhaus E, Kater MM, Wullems GJ, et al. (2000) NEC1, a novel gene, highly expressed in nectary tissue of Petunia hybrida. Plant J 24: 725–734 [DOI] [PubMed] [Google Scholar]
- Guan YF, Huang XY, Zhu J, Gao JF, Zhang HX, Yang ZN. (2008) RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol 147: 852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock CN, Kent L, McClure BA. (2005) The stylar 120 kDa glycoprotein is required for S-specific pollen rejection in Nicotiana. Plant J 43: 716–723 [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison Y, Shivanna KR. (1977) The receptive surface of the angiosperm stigma. Ann Bot (Lond) 41: 1233–1258 [Google Scholar]
- Hiscock SJ. (2000) Self-incompatibility in Senecio squalidus L. (Asteraceae). Ann Bot (Lond) 85: 181–190 [DOI] [PubMed] [Google Scholar]
- Hiscock SJ, Allen AM. (2008) Diverse cell signalling pathways regulate pollen-stigma interactions: the search for consensus. New Phytol 179: 286–317 [DOI] [PubMed] [Google Scholar]
- Hiscock SJ, Hoedemaekers K, Friedman WE, Dickinson HG. (2002) The stigma surface and pollen-stigma interactions in Senecio squalidus L. (Asteraceae) following cross (compatible) and self (incompatible) pollinations. Int J Plant Sci 163: 1–16 [Google Scholar]
- Hiscock SJ, McInnis SM. (2003) The diversity of self-incompatibility systems in flowering plants. Plant Biol 5: 23–32 [Google Scholar]
- Hiscock SJ, McInnis SM, Tabah DA, Henderson CA, Brennan AC. (2003) Sporophytic self-incompatibility in Senecio squalidus L (Asteraceae): the search for S. J Exp Bot 54: 169–174 [DOI] [PubMed] [Google Scholar]
- Hodgkin T, Lyon GD, Dickinson HG. (1988) Recognition in flowering plants: a comparison of the Brassica self-incompatibility system and plant pathogen interactions. New Phytol 110: 557–569 [Google Scholar]
- Johnson MA, Preuss D. (2003) On your mark, get set, GROW! LePRK2-LAT52 interactions regulate pollen tube growth. Trends Plant Sci 8: 97–99 [DOI] [PubMed] [Google Scholar]
- Kim ST, Zhang K, Dong J, Lord EM. (2006) Exogenous free ubiquitin enhances lily pollen tube adhesion to an in vitro stylar matrix and may facilitate endocytosis of SCA. Plant Physiol 142: 1397–141116998086 [Google Scholar]
- Li M, Xu W, Yang W, Kong Z, Xue Y. (2007) Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice. Plant Physiol 144: 1797–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord EM. (2003) Adhesion and guidance in compatible pollination. J Exp Bot 54: 47–54 [DOI] [PubMed] [Google Scholar]
- Malhó R, Liu Q, Monteiro D, Rato C, Camacho L, Dinis A. (2006) Signalling pathways in pollen germination and tube growth. Protoplasma 228: 21–30 [DOI] [PubMed] [Google Scholar]
- McInnis SM, Costa LM, Gutiérrez-Marcos JF, Henderson CA, Hiscock SJ. (2005) Isolation and characterization of a polymorphic stigma-specific class III peroxidase gene from Senecio squalidus L. (Asteraceae). Plant Mol Biol 57: 659–677 [DOI] [PubMed] [Google Scholar]
- McInnis SM, Desikan R, Hancock JT, Hiscock SJ. (2006a) Production of reactive oxygen species and reactive nitrogen species by angiosperm stigmas and pollen: potential signalling crosstalk? New Phytol 172: 221–228 [DOI] [PubMed] [Google Scholar]
- McInnis SM, Emery DC, Porter R, Desikan R, Hancock JT, Hiscock SJ. (2006b) The role of stigma peroxidases in flowering plants: insights from further characterization of a stigma-specific peroxidase (SSP) from Senecio squalidus (Asteraceae). J Exp Bot 57: 1835–1846 [DOI] [PubMed] [Google Scholar]
- Muschietti J, Eyal Y, McCormick S. (1998) Pollen tube localization implies a role in pollen-pistil interactions for the tomato receptor-like protein kinases LePRK1 and LePRK2. Plant Cell 10: 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi SMS, Harper A, Carter C, Ren G, Guirgis A, York WS, Thornburg RW. (2005) Nectarin IV, a potent endoglucanase inhibitor secreted into the nectar of ornamental tobacco plants: isolation, cloning, and characterization. Plant Physiol 139: 1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwland J, Feron R, Huisman BAH, Fasolino A, Hilbers CW, Derksen J, Mariani C. (2005) Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell 17: 2009–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu CT, daSilva I, de Molfetta JB, da Silva LR, de Almeida-Engler J, Engler G, Torraca PC, Goldman GH, Goldman MHS. (2004) NtWBC1, an ABC transporter gene specifically expressed in tobacco reproductive organs. J Exp Bot 55: 1643–1654 [DOI] [PubMed] [Google Scholar]
- Park SY, Lord EM. (2003) Expression studies of SCA in lily and confirmation of its role in pollen tube adhesion. Plant Mol Biol 51: 183–189 [DOI] [PubMed] [Google Scholar]
- Quiapim AC, Brito MS, Bernardes LA, Dasilva I, Malavazi I, DePaoli HC, Molfetta-Machado JB, Giuliatti S, Goldman GH, Goldman MH. (2009) Analysis of the Nicotiana tabacum stigma/style transcriptome reveals gene expression differences between wet and dry stigma species. Plant Physiol 149: 1211–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sanchez AM, Bosch M, Bots M, Nieuwland J, Feron R, Mariani C. (2004) Pistil factors controlling pollination. Plant Cell (Suppl) 16: S98–S106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepúlveda-Jiménez G, Rueda-Benítez P, Porta H, Rocha-Sosa M. (2005) A red beet (Beta vulgaris) UDP-glucosyltransferase gene induced by wounding, bacterial infiltration and oxidative stress. J Exp Bot 56: 605–611 [DOI] [PubMed] [Google Scholar]
- Shang C, Sassa H, Hirano H. (2005) The role of glycosylation in the function of a 48-kDa glycoprotein from carrot. Biochem Biophys Res Commun 328: 144–149 [DOI] [PubMed] [Google Scholar]
- Sidler M, Hassa P, Hasan S, Ringli C, Dudler R. (1998) Involvement of an ABC transporter in a developmental pathway regulating hypocotyl cell elongation in the light. Plant Cell 10: 1623–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Clark T, Preuss D. (2005) Expression profiling of Arabidopsis stigma tissue identifies stigma-specific genes. Sex Plant Reprod 18: 163–171 [Google Scholar]
- Swanson R, Edlund AF, Preuss D. (2004) Species specificity in pollen-pistil interactions. Annu Rev Genet 38: 793–818 [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. (2002) The rapid evolution of reproductive proteins. Nat Rev Genet 3: 137–144 [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2003) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- Tabah DA, McInnis SM, Hiscock SJ. (2004) Members of the S-receptor kinase multigene family in Senecio squalidus L. (Asteraceae), a species with sporophytic self-incompatibility. Sex Plant Reprod 17: 131–140 [Google Scholar]
- Takayama S, Isogai A. (2005) Self-incompatibility in plants. Annu Rev Plant Biol 56: 467–489 [DOI] [PubMed] [Google Scholar]
- Takayama S, Shiba H, Iwano M, Asano K, Hara M, Che FS, Watanabe M, Hinata K, Isogai A. (2000) Isolation and characterization of pollen coat proteins of Brassica campestris that interact with S locus-related glycoprotein 1 involved in pollen-stigma adhesion. Proc Natl Acad Sci USA 97: 3765–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Ezcurra I, Muschietti J, McCormick S. (2002) A cysteine-rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell 14: 2277–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kelley D, Ezcurra I, Cotter R, McCormick S. (2004) LeSTIG1, an extracellular binding partner for the pollen receptor kinases LePRK1 and LePRK2, promotes pollen tube growth in vitro. Plant J 39: 343–353 [DOI] [PubMed] [Google Scholar]
- Tung CW, Dwyer KG, Nasrallah ME, Nasrallah JB. (2005) Genome-wide identification of genes expressed in Arabidopsis pistils specifically along the path of pollen tube growth. Plant Physiol 138: 977–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven T, Feron R, Wolters-Arts M, Edqvist J, Gerats T, Derksen J, Mariani C. (2005) STIG1 controls exudate secretion in the pistil of petunia and tobacco. Plant Physiol 138: 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T, Pollak P, Tarlyn N, Taylor LP. (1994) Pollination- or wound-induced kaempferol accumulation in petunia stigmas enhances seed production. Plant Cell 6: 11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsen KL, Hepler PK. (2007) Sperm delivery in flowering plants: the control of pollen tube growth. Bioscience 57: 835–844 [Google Scholar]
- Wolters-Arts M, Van der Weerd L, Van Aeist AC, Van der Weerd J, Van As H, Mariani C. (2002) Water conducting properties of lipids during pollen hydration. Plant Cell Environ 25: 513–519 [Google Scholar]
- Wu H, de Graaf B, Mariani C, Cheung AY. (2001) Hydroxyproline-rich glycoproteins in plant reproductive tissues: structure, functions and regulation. Cell Mol Life Sci 58: 1418–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HM, Wang H, Cheung AY. (1995) A pollen tube growth stimulatory glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower. Cell 82: 395–403 [DOI] [PubMed] [Google Scholar]
- Yang B, Sugio A, White FF. (2006) Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci USA 103: 10503–10508 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.