Abstract
Because DNA damage is so rare, DNA glycosylases interact for the most part with undamaged DNA. Whereas the structural basis for recognition of DNA lesions by glycosylases has been studied extensively, less is known about the nature of the interaction between these proteins and undamaged DNA. Here we report the crystal structures of the DNA glycosylase AlkA in complex with undamaged DNA. The structures revealed a recognition mode in which the DNA is nearly straight, with no amino acid side chains inserted into the duplex, and the target base pair is fully intrahelical. A comparison of the present structures with that of AlkA recognizing an extrahelical lesion revealed conformational changes in both the DNA and protein as the glycosylase transitions from the interrogation of undamaged DNA to catalysis of nucleobase excision. Modeling studies with the cytotoxic lesion 3-methyladenine and accompanying biochemical experiments suggested that AlkA actively interrogates the minor groove of the DNA while probing for the presence of lesions.
Keywords: Crystal Structure, DNA-binding Protein, DNA-Protein Interaction, DNA Repair, Enzyme Structure, Nucleic Acid Enzymology, Nucleic Acid Structure, Protein-DNA Interaction, DNA Damage, Molecular Recognition
Introduction
The integrity of covalent structure in the genome is essential for normal cellular function and for faithful transmission of the heritable information reposited therein. DNA inside cells is under constant attack by exogenous environmental toxins and endogenous reactive cellular constituents, giving rise to nucleobase modifications such as oxidation, hydrolytic deamination, and alkylation (1, 2). If left uncorrected, these lesions and the products of their mismanagement by the cell can cause mutations and also interfere with essential cellular processes including transcription, recombination, and DNA replication, events that are causally linked with cancer (3, 4).
Although several repair pathways exist for eliminating aberrant nucleobases from the genome, BER4 is the primary cellular response for the repair of single lesion bases in DNA. The proteins responsible for initiating BER are DNA glycosylases, enzymes that recognize aberrant nucleoside lesions and catalyze scission of their glycosidic linkage. Most BER glycosylases are characterized by high specificity for one particular lesion, or at most a few closely related ones, with this specificity arising from binding interactions between the lesion nucleobase and residues within the enzyme active site (5, 6). There does exist, however, a class of BER enzymes, the 3-methyladenine DNA glycosylases, many members of which exhibit the ability to repair a highly diverse array of nucleoside lesions (7) mostly resulting from DNA alkylation. Prototypical members of this class of enzymes include the Saccharomyces cerevisiae Mag1, human alkyladenine glycosylase AAG, and Escherichia coli 3-methyladenine glycosylase II AlkA.
In E. coli, the expression of AlkA is under the control of the adaptive response and the ada regulon, which induces transcription of the alkA gene some hundredfold upon exposure to certain DNA alkylating agents (8–10). Among the remarkably broad range of AlkA substrates are the alkylated purines N3- and N7-methylguanine and -adenine and O2-methylpyrimidines (11, 12). In addition, AlkA can recognize cyclic nucleobases such as 1,N6-ethenoadenine, deaminated and electron-deficient nucleobases such as hypoxanthine, and even undamaged nucleobases (preferably purines) that are in mismatched base pairs (13–15). Biochemical studies looking at the rates of excision of various lesions by AlkA have suggested that there is an energetic barrier to extrusion of the lesion nucleobase from DNA, a step that necessarily precedes glycosidic bond cleavage, and that this explains at least in part why lesions that cannot form stable Watson-Crick base pairs are processed up to 2 orders of magnitude more quickly than those that do form stable base pairs (15). It has furthermore been proposed that the preferential excision of alkylated and electron-deficient nucleobases by AlkA is not so much the result of specific recognition of these lesions by the enzyme active site but is instead due to the weakened glycosidic bond of these electron deficient substrates (15). Much of what is currently known about the mechanism of DNA lesion recognition and subsequent base excision has been inferred from the crystal structure of AlkA bound to DNA containing a transition state mimic of glycosidic bond cleavage, 1-azaribose; this structure of a lesion recognition complex is referred to hereafter as the LRC (16).
High resolution structures of many BER glycosylases, both in unliganded form and in complex with lesion-containing DNA, have provided a great deal of insight into the fundamental mechanisms of DNA lesion recognition and catalysis. However, there remains relatively scant structural information on how BER glycosylases distinguish rare lesions from the greater than millionfold excess of undamaged genomic DNA. Headway on this issue could in principle be gained by solving high resolution structures of DNA glycosylases bound to undamaged DNA, but doing so has been hampered by the difficulty of crystallizing such nonspecific complexes, a problem that arises from the inherent inhomogeneity of nonspecific protein-DNA complexes. Progress on this front has recently been made with the finding that the roaming range of a DNA glycosylase on undamaged DNA can be restricted through the introduction of an intermolecular disulfide cross-link into the protein-DNA interface and that the resulting complexes can be crystallized and characterized structurally at high resolution. Using this strategy, it has been shown that the 8-oxoguanine DNA glycosylase MutM is able to remodel drastically the conformation of undamaged DNA (17). There is no reason to expect, however, that the mode of interaction of MutM with undamaged DNA will be conserved in the cases of other DNA glycosylases, given that the structural and energetic demands of lesion extrusion from DNA and extrahelical lesion recognition clearly differ from lesion to lesion and that the structures of these proteins vary widely. To cite just one example, MutM induces severe bending in DNA while binding its extrahelical 8-oxoguanine lesion, whereas the DNA shows little bending in the alkyladenine glycosylase LRC (18, 19). Thus, each DNA glycosylase can be expected to follow a particular extrusion pathway that has been optimized to the particular features and demands of that enzyme and its DNA substrate.
To understand the nature of the interaction of a 3-methyladenine glycosylase with undamaged DNA at atomic resolution, we utilized DXL to obtain several crystal structures of AlkA in complex with non-lesion-containing oligonucleotides. Unlike the aforementioned structures of MutM interrogating undamaged DNA, the present structures reveal a less invasive search intermediate with AlkA, one in which the DNA is nearly unbent and no amino acid side chains are inserted into the DNA helix. To our knowledge, these structures are the first to have captured a DNA glycosylase bound to unbent, undamaged DNA, and therefore we refer to the structures as undamaged DNA complexes (UDCs). A comparison of the UDCs with the previously published LRC (16) reveals the nature of the conformational changes in both the DNA and protein as the glycosylase transitions to its catalytically active state. Modeling of the cytotoxic lesion m3A into the UDCs in combination with biochemical studies indicates that AlkA utilizes the interaction between its interrogating residue, Leu-125, and the methyl group of m3A to recognize and extrude from DNA the 3-methylated genotoxic variants of guanine and adenine.
EXPERIMENTAL PROCEDURES
AlkA Expression, Purification, and Crystallization
Point mutants were constructed using the QuikChange site-directed mutagenesis kit (Stratagene) and mutations confirmed by sequencing. Wild-type AlkA and point mutants were purified as described previously (20). Oligonucleotides were synthesized using an automated synthesis procedure and purified by PAGE. The 5-bromo-2′-deoxyuridine, H-phosphonate, O6-phenyl-deoxyinosine, 3-deaza-2′-deoxyadenosine, and 2-fluoro-2′-deoxyinosine phosphoramidites were purchased from Glen Research. The 3-deaza-3-methyl-2′-deoxyadenosine was purchased from Berry and Associates. The G* modification with the two-carbon thiol tether was done according to the protocol provided by Glen Research. The backbone DXL addition of the two-carbon thiol tether was performed as described previously (21). The sequence of the thiol tether-containing DNA strand is 5′-GCAG*TCATGTCA-3′, where G* is the location of the modified base for the UDC0 structure, and 5′-GGCATT*CATGTCA-3′, where T* is the location of the modified base for the UDC1 and UDC2 structures. The complementary strand is 5′-GACABrUGACBrUGCCT-3′ (where BrU is the brominated uridine), 5′-ACABrUGAABrUGCCT-3′, and 5′-GACABrUGAABrUGCCT-3′ for the UDC0, UDC1, and UDC2 structures, respectively. DXL reactions were performed in 20 mm Tris-HCl, pH 7.4, and 50 mm NaCl with 17 μm purified AlkA and 14 μm of modified DNA at 25 °C for 1–4 days. The complementary strand was added to the reaction and allowed to anneal for 24 h before being further purified on a Mono Q (GE Healthcare) ion-exchange column. The AlkA double-stranded DNA complex was placed in a buffer containing 10 mm Tris-HCl, pH 7.4, and 50 mm NaCl and subsequently concentrated to an A260 of 0.8–0.9 prior to crystallization.
Crystals of the UDCs were grown at 25 °C using the sitting drop vapor diffusion method, with the drop consisting of a 1:1 ratio of the stock protein-DNA solution and a reservoir solution of 25–29% polyethylene glycol 3350, 100 mm bis-Tris, pH 6.0–6.6, 200 mm Li2S04, and 3% 6-aminocaproic acid. Prior to data collection the crystals were flash-frozen in liquid nitrogen.
Structure Determination
Data for the UDC structures were collected on beamline 24-ID at the Northeastern Collaborative Access Team (NE-CAT) at the Advanced Photon Source (APS), Argonne National Laboratory. Data were processed with HKL2000 and merged with SCALEPACK (22). The crystals have one protein-DNA complex in the asymmetric unit and belong to the tetragonal space group I4122. Molecular replacement, using the unliganded AlkA monomer (Protein Data Bank accession code 1DIZ) (16) as a search model, was performed on each UDC using PHASER (23) as part of the CCP4 program suite (24). 2Fo − Fc and Fo − Fc maps showed clear density for the DNA, although the register for the DNA could not be assigned unambiguously. Anomalous data collected at the bromine edge wavelength on crystals containing 5-bromodeoxyuridine were used to calculate a phased anomalous map using the CCP4 program suite (24), which allowed identification of the bromine atom positions within the asymmetric unit and the proper assignment of the nucleotide positions. The DNA was fitted to the electron density using the program COOT (25), and the model was refined in CNS (crystallography and NMR system) (26) and PHENIX (27). Crystallographic statistics for the UDC crystal structures are presented in Table 1. Coordinates have been deposited under Protein Data Bank accession codes 3OGD, 3OH6, and 3OH9.
TABLE 1.
Data collection and refinement statistics
| UDC0a | UDC1a | UDC2a | |
|---|---|---|---|
| Data collection | |||
| Space group | I4122 | I4122 | I4122 |
| Cell dimensions | |||
| a, b, c (Å) | 139.7, 139.7, 95.3 | 139.9, 139.9, 91.1 | 140.3, 140.3, 94.8 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.0-2.8 (2.9-2.8) | 50.0-2.8 (2.9-2.8) | 50.0-2.9 (3.0-2.9) |
| Rsymb (%) | 5.8 (45.6) | 6.5 (48.3) | 8.5 (43.5) |
| I/σI | 22.9 (3.3) | 27.4 (2.9) | 32.6 (4.4) |
| Completeness (%) | 99.5 (100.0) | 95.8 (96.9) | 99.8 (100.0) |
| Refinement | |||
| Resolution (Å) | 50.0-2.80 | 50.0-2.8 | 50.0-2.9 |
| No. reflections | 21,391 | 19,529 | 19,097 |
| Rworkc/Rfreed (%) | 21.5/26.9 | 20.5/25.7 | 22.7/26.9 |
| No. atoms | 2622 | 2701 | 2719 |
| r.m.s.e deviations | |||
| Bond lengths (Å) | 0.006 | 0.007 | 0.006 |
| Bond angles (°) | 1.017 | 1.047 | 1.044 |
| Ramachandran statistics | |||
| Favored (%) | 93.4 | 93.7 | 92.7 |
| Allowed (%) | 5.9 | 5.6 | 6.6 |
| Outlier (%) | 0.7 | 0.7 | 0.7 |
a Data were collected from a single crystal. Values in parentheses are for the highest resolution shell.
b Rsym = Σ|I − 〈I〉|/ΣI, where I is the integrated intensity of a given reflection.
c Rwork = Σ|F(obs) − F(calc) |/ΣF(obs), where F(obs) and F(calc) are the observed and calculated structure factor amplitudes, respectively.
d Rfree = Σ|F(obs) − F(calc) |/ΣF(obs), calculated using 5% of the data omitted from the refinement.
e r.m.s., root mean square.
Cleavage Assay
The sequences of the oligos used in the assay are 5′-CGATAGCATCCTYCCTTCTCTCCAT-3′, where Y is the location of the lesion base, and 5′-ATGGAGAGAAGGZAGGATGCTATCG-3′ for the complementary strand, where Z is the base opposite the lesion. The lesion strands were end-labeled using T4 polynucleotide kinase and [γ-32P]ATP followed by annealing with the excess unlabeled complementary strand by heating to 95 °C followed by slow cooling to 4 °C. The labeled duplex DNA was incubated with excess AlkA in cleavage buffer (50 mm Tris-HCl, pH 8.5, 1 mm EDTA, 100 mm NaCl, and 0.1 mg/ml bovine serum albumin) at 37 °C for 24 h. The reaction was quenched with 0.2 m NaOH and heated to 70 °C for 30 min to cleave the abasic DNA sites produced by AlkA. Samples were mixed with formamide loading buffer and resolved by 20% PAGE. The bands were visualized with a phosphorimaging system and quantified using ImageQuant TL (v2003.02). The percent cleavage represents the intensity of the expected 12-mer cleavage product divided by the total radioactivity intensity. The results were graphed with Microsoft Excel and represent the average of three independent experiments. Error bars denote the standard deviation from the mean.
DNA Melting Temperature
The oligos used in the UV thermal denaturation experiments were the same as in the cleavage assay. UV thermal denaturation curves were acquired on a DU800 spectrophotometer (Beckman Coulter). All measurements were performed in 50 mm bis-Tris, pH 6.0, or 50 mm Tris-HCl, pH 8.5, with 1 mm EDTA and 100 mm NaCl. Absorbance versus temperature spectra were collected at 260 nm over a range of 10 to 85 °C, with the reverse experiment from 85 to 10 °C also done for consistency. The heating/cooling rate was 1 °C/min, and the melting temperature (Tm) values were determined with a first derivative analysis of the curves using the software package provided by the spectrophotometer manufacturer. All Tm values are the average of four independent experiments. The reported plots were generated using Microsoft Excel.
Illustrations and Modeling
The coordinates for m3A were generated using PRODRG (28). Modeling was done by aligning the coordinates of the m3A base with C20 using PyMOL. Figs. 2, 3, and 4 were computed using PyMOL as a renderer.
FIGURE 2.
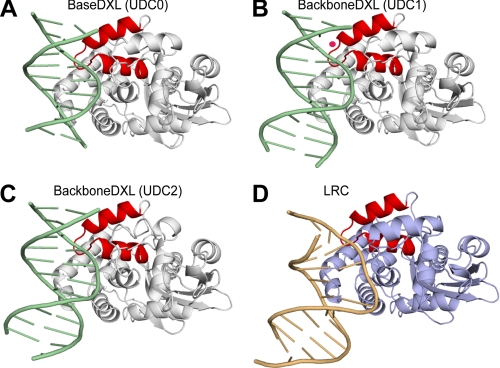
Structure of the AlkA-DNA undamaged DNA and lesion recognition complexes. A–C, ribbon representation of the UDC structures with AlkA colored white, the DNA colored green, and the HhH colored red. In B, the ordered water molecule is pink. D, ribbon representation of the LRC with the AlkA protein colored light blue, the DNA colored brown, and the HhH colored red.
FIGURE 3.
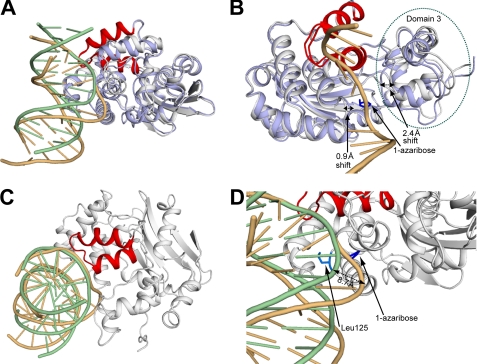
Cα superposition of the AlkA undamaged DNA complex with the lesion recognition complex. A, structural comparison between the AlkA undamaged DNA and lesion recognition complexes. The color scheme is the same as described for Fig. 2. B, differences in protein conformation between the UDC and LRC structures. The dotted black lines illustrate the important loop and domain movements. The 1-azaribose lesion (blue)-containing DNA strand is shown for reference. Domain 3, which contains the catalytic residue and many key active site residues, is circled by a green dotted line. C, view down the axis of the DNA duplex illustrating the overlap of the phosphate backbone between the DNA of the UDC and LRC structures within the region of the HhH motif. The AlkA monomer from the LRC has been omitted for clarity. D, a zoomed-in view (from A) into the active site of AlkA from the UDC and LRC structures. The dotted black line denotes the 8.7 Å shift in the path of the DNA between the two structures.
FIGURE 4.
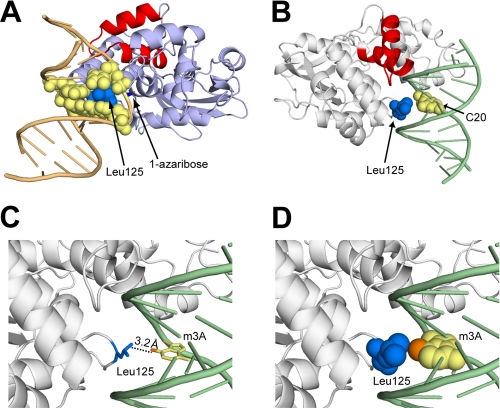
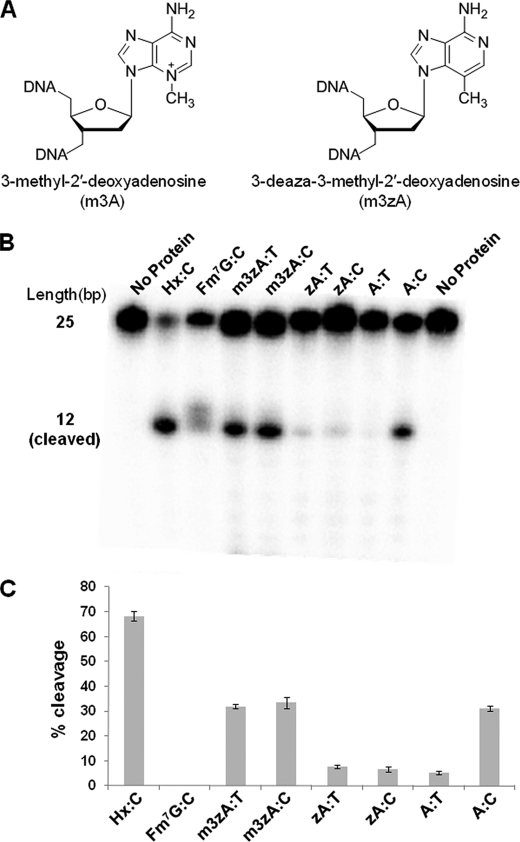
Role of Leu-125 in the undamaged DNA and lesion recognition complexes. A, the LRC of AlkA with a space-filling model of Leu-125 (blue) and the nucleobases (yellow) that are in van der Waals contact. B, view of the minor groove of the UDC with Leu-125 and the interrogated base C20 shown as a space-filling model. C, same view as in B, with the base of m3A modeled in place of C20. The N3-methyl group is shown in orange. D, same view as in C, with Leu-125 and the base of m3A shown as a space-filling model, illustrating the van der Waals contact between the Leu-125 side chain and the N3-methyl adduct (orange).
RESULTS
Crystal Structure of Complexes Having AlkA Bound to Undamaged DNA
AlkA has been shown to possess weaker affinity for lesion-containing DNA than most glycosylases and to exhibit little thermodynamic preference for lesion-containing versus undamaged DNA (15).5 Additionally, AlkA shows a propensity to bind to the ends of DNA, a property observed directly in crystallographic structures and used as the basis of a host-guest system for structural elucidation of diverse DNA lesions (20). These properties of the AlkA-DNA interactions suggested the need for a strategy by which to restrict the roaming range of AlkA to the central portion of a DNA duplex if we aimed to capture the enzyme in the mode of interrogating undamaged duplex DNA. In previous work, we had used DXL to trap otherwise unstable or fleeting states of DNA interaction by DNA-binding proteins, and we and others have shown this strategy to be effective in producing complexes of sufficient homogeneity to crystallize and yield to structural elucidation (17, 21, 29–36). We therefore turned to DXL for the present study, specifically to employ this technology to trap AlkA at the stage of interrogating an undamaged DNA duplex.
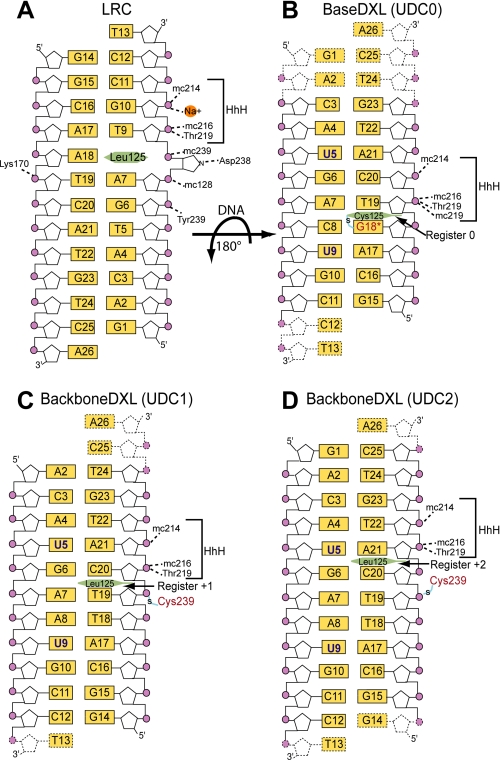
Potential cross-linking sites were chosen on the basis of the 1-azaribose LRC structure (16). We immediately focused our attention on Leu-125, because this residue penetrates deeply into the DNA helix in the LRC and thus could be expected to lie near the DNA surface at all stages of the search and extrusion process. Furthermore, in previously published studies on human 8-oxoguanine glycosylase (hOgg1), the DNA-penetrating residue of that protein proved to be an excellent choice of DXL attachment site (35). We therefore mutated Leu-125 to Cys in AlkA, and we replaced adenine 18 (A18) in the LRC with a modified guanine (G*18) containing an ethanethiol tether attached to the minor groove N2-exocyclic amine (supplemental Fig. 1A and Fig. 1, A and B). Additionally, the 1-azaribose lesion in the LRC was replaced by a cytosine residue (C8), which was expected to base pair with the thiol-tethered residue G*18. Diffraction quality co-crystals of this complex were obtained but only in conditions quite different from those used to crystallize the LRC (see “Experimental Procedures”). The structure of this DXL complex was solved by molecular replacement to 2.8 Å resolution (Table 1), but the quality of the electron density map was insufficient to assign unambiguously the register of the DNA. We therefore substituted the bases corresponding to T5 and T9 in the LRC with 5-bromo-2′-deoxyuridine and used the anomalous scattering peaks from the bromine atoms as landmarks to accurately model the positions of the nucleobases within the DNA (Fig. 1, A and B). Having thus conclusively established the orientation of the AlkA and DNA in our complex, we made the unexpected and interesting observation that the DNA in this structure (hereafter referred to as UDC0) is rotated 180° about the axis of the cross-link with respect to its orientation in the LRC structure (Fig. 1, A and B). By virtue of this rotation, the DNA strand most proximal to AlkA in the LRC (the lesion-containing or target strand) is distal to the protein in UDC0 and vice versa. This swapping of strands also transposes the cross-linking site from the non-target (LRC) to the target strand (UDC0), such that in UDC0, Cys-125 now lies adjacent to the engineered G*18 residue (Fig. 1, A and B). The base pair bearing the DXL attachment site, G*18:C8, is fully intrahelical and maintains normal Watson-Crick hydrogen bonding. To facilitate comparison with other UDC structures, we arbitrarily assigned the register of AlkA with respect to DNA in UDC0 as register 0.
FIGURE 1.
Schematic representation of the AlkA-DNA interactions. The orientation of the DNA in the UDCs are rotated 180° with respect to the LRC. In the UDC structures, residues in the HhH motif are solely responsible for hydrogen bonding with the phosphate (purple circles) backbone with main chain atoms, designated by prefix mc. A, in the LRC structure, the lesion 1-azaribose is depicted as an extrahelical sugar interacting with Asp-238. In B, the 5-bromo-2′-deoxyuridine nucleobase is designated as a blue U, and the modified guanine, designated G18*, is shown in red. In B–D, the two-carbon thiol tether is shown as an S with a blue line through it, the interrogating residue of AlkA is a green hexagon, and the disordered nucleotides are shown as dotted lines.
As the key helix-penetrating residue of AlkA, Leu-125 obviously plays an important role and would preferably be retained in our structure. We therefore used the structure of UDC0 to choose an appropriate DXL site at a remote place in the protein-DNA interface, one that would keep Leu-125 intact (supplemental Fig. 1B). This two-step strategy of performing DXL on a key residue to obtain an initial structure, which is then used as the basis for selection of a more remote cross-linking site, followed by determination of a structure that is disulfide cross-linked at the remote site, is similar to that first reported by He and colleagues (31) in studies on AlkB and its human homologue, ABH2. We chose the position occupied by Tyr-239 in wild-type AlkA as the remote DXL site, as this residue has no interactions with the DNA in the UDC0 structure (Fig. 1, A and B) and contributes only one hydrogen-bonding interaction with a DNA backbone phosphate in the LRC. Based on the structure of UDC0, an ethanethiol tether was introduced as a backbone N-alkylphosphoramidate 5′ to C20 (supplemental Fig. 1B and Fig. 1, C and D). The Y239C mutant of AlkA and backbone-tethered oligonucleotide were found to undergo efficient DXL formation, and following purification the resulting complex furnished diffraction quality crystals under the same conditions used to crystallize UDC0. Two variants of these crystals were grown, the only difference between them being the presence or absence of a G nucleotide on the 5′-end of the un-cross-linked (non-target) strand (Fig. 1, C and D); the structures of these complexes were refined to 2.9 and 2.8 Å, respectively (Table 1). As in the case of the UDC0 structure, the register of the DNA was determined by the anomalous peaks of bromine residing on 5-bromo-2′-deoxyuridine substituted at positions 5 and 9 in the DNA duplex (Fig. 1, C and D). The two backbone-cross-linked structures have the same DNA orientation as in UDC0, but interestingly in them AlkA has translocated by either 1 or 2 base pair registers relative to that in UDC0. Specifically, in the structure that has a 12-mer non-target strand, hereafter referred to as UDC1, Leu-125 is positioned in the minor groove proximal to the T19:A7 base pair, a shift in register of +1 from the UDC0 structure (Fig. 1, B and C). In the structure containing a 13-mer non-target strand (UDC2), the additional nucleotide resulted in a 4 Å expansion of the unit cell along the axis of DNA packing (Table 1) and another 1-base pair shift in the register of the protein on DNA, such that Leu-125 is now proximal to C20:G6, a shift of +2 from UDC0 and +1 from UDC1 (Fig. 1, B–D). In both the UDC1 and UDC2 structures, the interrogated base pair is intrahelical and maintains normal Watson-Crick hydrogen bonding.
Analysis of the three UDCs reveals that despite the variations in cross-linking strategies used to obtain them, and apart from the differences in register of the complexes and DNA length, the structures are nearly identical (Cα root mean square deviation = 0.3 Å; Fig. 1, B–D, and Fig. 2, A–C). As mentioned above, the interrogated base pairs in all three UDCs are intrahelical. Additionally, the DNA duplexes are nearly straight with little to no bend angle (Fig. 2, A–C), in contrast to the LRC, which has a pronounced 66° bend (Fig. 2D). The footprint of AlkA on the DNA is small (520 Å2 of buried surface area) with the primary protein-DNA contact region being localized to the HhH DNA-binding motif. Hydrogen bonding between AlkA and the phosphate backbone is mediated mainly through interactions with the protein main chain, although the side chain of Thr-219 also contributes a hydrogen bond. The residues on AlkA engaged in interactions with DNA are identical in all three UDC structures (although the main chain of Thr-219 also contributes an additional hydrogen bond in the UDC0 structure; Fig. 1B). Although the patterns of amino acid/phosphate contacts are virtually identical in all three structures, the identity of the particular phosphate moieties in DNA that make protein contacts varies among the UDCs because of their differences in binding register to AlkA (Fig. 1, B–D). The similarity of the three structures, despite their substantial differences in cross-link configuration, lends credence to the notion that these structures represent physiologically relevant states of the AlkA-DNA interaction rather than being artifacts of DXL. Of the three UDCs, the UDC0 structure contains more disordered bases and has higher temperature factors for the DNA than UDC1 and UDC2. UDC1 and UDC2 are of comparably high quality, although UDC2 has more ordered bases; consequently, the structural analysis that follows was performed using the UDC2 structure.
Comparison of the Undamaged DNA and Lesion Recognition Complexes
The conformation of AlkA is largely unchanged between UDC2 and the LRC (Fig. 3A), with noteworthy differences evident in domain 3 of AlkA, which contains most of the active site residues, including the essential catalytic acid Asp-238. In the LRC, domain 3 is shifted 2.4 Å toward the lesion strand of the DNA duplex with respect to its position in the UDC2 structure, thus bringing the key residues of AlkA involved in active site recognition and base excision in closer proximity to the extrahelical lesion (Fig. 3B). Additionally, the loop that contains the interrogating residue, Leu-125, which is located opposite domain 3, is shifted 0.9 Å closer to the lesion strand (Fig. 3B). The combined effect of these conformational changes is to “sandwich” the lesion in the LRC, thereby providing additional protein contacts to the DNA and stabilizing the extrahelical conformation of the lesion in the enzyme active site. The UDC structures, in contrast, have an active site entrance that is open and poised to accommodate a lesion base upon its extrusion from the duplex.
As mentioned above, AlkA interacts with the DNA in a nonspecific manner through interactions with the phosphate backbone. Therefore, despite the fact that the orientation of the DNA is different between the LRC and UDC, it is possible to compare the backbone phosphate contacts in these two types of complexes by frame-shifting them on the DNA sequence. The difference in bend angle between the nearly straight DNA in the UDC structures and the large 66° bend in the LRC results in significant positional differences of the atoms in the DNA upon Cα superposition of the structures (Fig. 3A). Despite these differences, the phosphate backbones nearly superimpose on each other in the region contacting the HhH (Fig. 3C). In fact, the residues of the HhH motif that form hydrogen bonds to the DNA phosphate backbone are identical between the UDC structures and the LRC (Fig. 2). The similarity in HhH binding to the DNA between the UDC and LRC structures suggests that the role of this motif is to provide nonspecific DNA contacts, and it does not appear to function in distorting the DNA. Similar nonspecific protein-DNA interactions between the HhH and the DNA were also seen in the host-guest complex structures of AlkA in which the HhH motif anchors AlkA to the ends of a DNA duplex (20). In the LRC, a sodium ion was found to provide an additional contact to the DNA (Fig. 1A) (16). In the UDC complexes, weak difference density appeared in a similar location to the sodium ion in the LRC. However, in the UDC0 and UDC2 structures, modeling of this density resulted in a worsening of the refinement statistics, and hence it was omitted from the final refinement. In the UDC1 structure, modeling of this density as a water molecule resulted in an improvement of the refinement statistics, and it was included in the final model (Fig. 2B).
The most significant divergence in the path of the DNA between the LRC and UDC structures is in the area centered around the interrogation site, occupied by the lesion in the LRC or by an intact base pair in the UDCs (Fig. 3, A and D). In the LRC, the extrusion of the lesion base into the active site results in an 8.7 Å shift of the DNA phosphate backbone with respect to the UDC structure (Fig. 3D). This shift in the lesion strand brings the phosphate backbone closer to the loop containing the interrogating residue, Leu-125, allowing its intercalation into the DNA duplex (see below). The shift also enables the formation of two additional hydrogen bonds in the LRC, those involving the phosphate backbone and the main chain of Val-128 plus the side chain of Tyr-239 (Fig. 1A). Additionally, the lesion strand is severely bent as it exits the active site (Fig. 3D). In contrast, the DNA in the UDC structure abuts the entrance of the active site of AlkA, making no additional contacts with AlkA as it diverges from the protein in a nearly straight manner (Fig. 3D).
As a consequence of the conformational changes in both the protein and DNA upon lesion binding to the active site of AlkA, Leu-125 inserts itself through the minor groove into the space vacated by the extruded lesion (Fig. 4A). The hydrophobic side chain of leucine forms nonspecific interactions with the neighboring base pairs as well as the estranged base opposite the extruded lesion; it is likely that these interactions contribute significantly to stabilizing the severely bent conformation of the DNA (Fig. 4A). In the UDC structures, Leu-125 also lies in the minor groove of the DNA; however, unlike in the LRC, there are no interactions with the DNA (Fig. 4B), or more specifically, with the interrogated base pair. Leu-125 thus gives the appearance of scanning the minor groove as it searches for a lesion, rather than actively probing each base pair by aggressive contact. This suggests that Leu-125 does not directly insert itself into the DNA duplex as it searches for lesions but instead is poised to enter the duplex upon base extrusion.
Modeling of 3-Methyladenine into the Undamaged DNA Complexes
The N3-methyl variants of adenine and guanine are considered particularly deleterious lesions due to their cytotoxic effects. 3-Methyladenine has been shown to inhibit DNA synthesis by preventing the necessary interactions between DNA polymerase and the minor groove of the duplex DNA, thereby inducing S-phase cell cycle arrest (3, 37–42). Additionally, cells exposed to chemical agents that selectively lead to m3A lesion formation exhibit enhanced apoptosis in multicellular organisms (3, 43).
The m3A lesion is so unstable chemically that it has never been incorporated site-specifically into DNA, and hence it has not been characterized structurally. We therefore modeled the lesion into the UDC structures by superimposing m3A onto the interrogated base (Fig. 4, C and D). The results of this modeling exercise with UDC2 revealed a distance of 3.2 Å between the N3-methyl group of m3A and the Leu-125 side chain (Fig. 4C). The modeling studies suggest potential van der Waals interactions between the N3-methyl group on the m3A lesion and the side chain of Leu-125, which could in turn signal to AlkA the presence of this cytotoxic lesion in the DNA (Fig. 4D). It is impossible to tell whether the interaction between the N3-methyl group and the Leu-125 side chain is attractive or sterically repulsive, although it is worth noting that an ethyl group would clearly clash sterically with Leu-125 and AlkA excises N3-ethyl adenine at a comparable or slightly faster rate than m3A (44). Nevertheless, the striking physical proximity of the N3-methyl group on m3A and the interrogating Leu-125 side chain of AlkA are certainly suggestive of a functional role for this interaction in lesion recognition and extrusion from DNA.
Biochemical Studies on 3-Methyladenine Cleavage
We sought to devise a test for the hypothesis that AlkA can recognize the m3A lesion via its N3-methyl group. This is not straightforward, however, because as mentioned earlier, m3A cannot be stably incorporated into DNA. We therefore turned to the use of an isosteric analog, m3zA (Fig. 5A), which is identical to m3A save for the replacement of the N3 nitrogen atom in m3A with a carbon atom in m3zA. This substitution decreases the electron deficiency of the purine ring, thereby stabilizing the glycosidic bond of m3zA relative to that of m3A; hence, the analog can be stably incorporated into DNA. Given that the rates of base excision by AlkA are strongly influenced by the stability of the target glycosidic bond (15), it is reasonable to suppose that m3zA is less efficiently cleaved by AlkA than is m3A, although there is presently no way to test this notion. To isolate the effect of the methyl group in m3zA alone, we decided to compare the rate of AlkA cleavage of m3zA with its des-methyl version, namely N3-deazaadenine.
FIGURE 5.
Biochemistry of AlkA lesion cleavage. A, chemical structures of 3-methyl-2′-deoxyadenosine and 3-deaza-3-methyl-2′-deoxyadenosine. B, cleavage assay for AlkA illustrating cleavage of the m3A lesion when paired with thymine but no cleavage of adenine when paired with thymine. There is a faint delocalized band in the Fm7G lane, which represents a breakdown of the lesion base that occurs under basic conditions rather than specific cleavage of the lesion. C, quantification of the cleavage product for each lesion tested in B. Hx, hypoxanthine.
The interpretation of these experiments is potentially complicated by the fact that deazapurines have altered Watson-Crick hydrogen bonding characteristics relative to purines, presumably because the pKa of the N1 atom that participates in Watson-Crick base-pairing is modulated by the nature of the substituent at the 3-position (45). Melting temperature analyses have shown that the deaza substitution in m3zA substantially weakens base-pairing with T at pH 6.0, the optimal pH determined previously for base excision by AlkA (15) (data not shown). However, we found that m3zA and zA form a stable base pair with either T or C at pH 8.5 (supplemental Fig. 2), conditions under which AlkA retains substantial catalytic activity. Consequently, our analysis of AlkA cleavage of m3zA and zA was performed at pH 8.5 (see “Experimental Procedures”).
The results of the cleavage assay are shown in Fig. 5, B and C. As a positive control for robust AlkA cleavage, we employed hypoxanthine, which is known to be a good substrate lesion for AlkA. Not surprisingly, we found that it undergoes ready cleavage by AlkA in the present assay system. AlkA is known to be capable of cleaving adenines engaged in mispairs, although at a modest rate, whereas the enzyme has little activity on correctly base-paired A (15). Indeed, we observed that AlkA cleaved A in the A:C mismatch but not the A:T base pair (Fig. 5, B and C). As a negative control, we examined the lesion analog Fm7G, which is known to act as an inhibitor of the AlkA glycosylase reaction (46); as shown in Fig. 5B, Fm7G is not cleaved to any appreciable extent by AlkA. To examine the ability of AlkA to cleave m3zA, we compared oligonucleotides that incorporated this lesion opposite a T, the expected partner of m3A in the cell, and opposite a C. We compared the rates of cleavage of m3zA in DNA to that of zA, and normal A paired opposite T or C. We found that the lesion analog m3zA was cleaved to the same extent regardless of whether it was paired with C or T, whereas the des-methyl version, zA, was not substantially cleaved opposite either of the pyrimidines (Fig. 5, B and C). To examine the possibility that these differences in cleavage rate stemmed from differences in helix stability, we performed melting curve analyses, which revealed that m3zA and zA both stably base pair with both T and C at pH 8.5 (supplemental Fig. 2), with the presence of the methyl group having a negligible effect on helix stability. Thus it appears that the ability of AlkA to cleave the “lesion” in the m3zA:T base pair but not the zA:T (or zA:C) base pair under the same conditions (Fig. 5, B and C) strongly suggests that AlkA can respond in terms of catalysis to the presence of a single methyl group at the N3 position.
DISCUSSION
Single molecule studies have shown that glycosylases diffuse along short stretches of DNA in search of their target lesion nucleobases; and they have further shown that, at least in the case of human 8-oxoguanine glycosylase, this diffusion mode exhibits reptation, indicative of tracking along the grooved surface in DNA (47, 48). Additionally, evidence has been presented that glycosylases undergo microscopic dissociation or “hopping” to avoid protein “obstacles” on the DNA (49). The fleeting nature of these translocation intermediates has rendered them elusive from the vantage point of structural characterization at high resolution. This situation has changed only very recently, with the development of the DXL strategy to enable structural characterization of such intermediates. In the first instance, DXL was used to obtain a high resolution structure of the bacterial 8-oxoguanine repair protein, MutM, interrogating normal DNA while searching for a target lesion (21). In those structures the DNA is drastically bent, with a helix-interrogating residue, Phe-114, fully inserted into the helical stack at the site of the target base pair, which is severely buckled. Computational simulations established that this highly active mode of DNA interrogation is associated with significant lowering of the activation barrier for extrusion of the target nucleoside from DNA (30). It is not known at present whether this structure represents that of MutM as it translocates rapidly and processively along DNA or whether it represents a nonprocessive state of active helix interrogation. Importantly, atomic force microscopy studies have revealed that MutM employs two modes of interaction with undamaged DNA, one having a drastically bent duplex and the other having an unbent duplex (50), only the former of which is consistent with the states of MutM-DNA interaction thus far observed crystallographically.
Here we have captured, for the first time, a DNA glycosylase tracking the groove surface of an undamaged DNA duplex. In the state observed here, the DNA is unbent, and AlkA has a limited number of direct DNA contacts, these being limited to the signature HhH DNA-binding motif common to all members of the HhH glycosylase structural superfamily to which AlkA belongs. The completely noninvasive nature of the AlkA interaction with the target base pair in the UDC structures stands in stark contrast to the highly invasive interaction observed in the structures of MutM bound to undamaged DNA. It is of course possible that a search intermediate exists of AlkA in which the DNA is highly bent, with Leu-125 being intercalated into the helical stack, perhaps buckling an intact target base pair. Inspection of the structures of AlkA and MutM bound to DNA lesions suggests it is unlikely that Leu-125 of AlkA can intercalate without displacing the target base. Whereas the intercalating helix-probe residue of MutM, Phe-114, inserts itself into the helical stack on the complementary strand across from the target base, Leu-125 of AlkA actually takes the place of the extruded target base. Furthermore, in the UDC structures, Leu-125 lies directly in the plane of the target base such that any movement that would draw the DNA closer to AlkA, as is necessary to form a lesion recognition complex, would necessarily produce a severe steric clash between Leu-125 and the target nucleobase, forcing extrusion of the nucleobase. It is also noteworthy that the protein-DNA contacts around the target nucleobase are much more extensive with MutM than AlkA, making MutM more capable of stabilizing the highly distorted DNA conformation seen in the structure of the fully intrahelical search intermediate (21).
The differences in the search mechanism between these two different classes of glycosylases may be due to the different challenges that these two enzymes face in locating their cognate forms of damage. MutM must identify a lesion that differs from an undamaged guanine only by a single oxygen atom and one lone pair of electrons, an alteration that has a negligible effect on the helical conformation of DNA and little effect on base-pairing energetics. By contrast, AlkA has to recognize numerous different forms of DNA damage, most of which do not form stable base pairs in DNA and hence have little energetic penalty to extrusion. Whether AlkA locates these lesions through capture of lesions that have spontaneously become extrahelical (51) or encounters them in the intrahelical state and promotes extrusion of the lesion is unknown at present. We hypothesize that many of the lesions recognized by AlkA are identified using either of these search mechanisms. That weakening of the target base pair accelerates the rate of glycolytic cleavage by AlkA is well established (15).
Understanding the strategy that AlkA employs to locate its two most abundant lesions, N7-methylguanine and N3-methyladenine, poses a more difficult challenge. N7-methylguanine has no discernible effect on the conformation of duplex DNA (46) and slightly stabilizes it thermodynamically (52). The structural and thermodynamic effects of m3A are not known, but there is no reason to expect them to be significant, because m3A has the same base-pairing functionality as A, and the methyl group creates no obvious clash. In the case of m3A, modeling the lesion into the target site of a UDC suggests a potential interaction between the methyl group on m3A and the side chain Leu-125, which, although not overtly repulsive, could signal to AlkA the presence of this lesion. The notion that AlkA can detect the presence of a methyl group at this position was supported by our biochemical studies on DNA containing 3-deaza analogs. The mechanism by which AlkA identifies N7-methylguanine, a lesion with a methyl group in the major groove, is unknown. Interrogation of the minor groove of the DNA by AlkA would not be expected to result in an interaction with the N7-methyl that would signal the presence of this lesion. Enzyme kinetic experiments show that AlkA exhibits similar rate constants (kcat) for cleavage of N7-methylguanine and m3A when paired with C and T, respectively (14, 15). However, the kcat/Km values, a measure of the specificity of AlkA for the methylated bases, indicate that AlkA exhibits a near hundredfold higher specificity for the m3A lesion when in a Watson-Crick base pair with T than does the N7-methylguanine paired with C (15). The greater specificity of AlkA for the m3A lesion could be due to greater accessibility to the m3A lesion through its facilitated extrusion from the DNA duplex via steric interactions between the N3-methyl and Leu-125 rather than because of lesion specific interactions within the AlkA active site.
Both N7-methylguanine and N3-methyladenine are repaired at a faster rate than uncharged lesions because of their weakened glycosidic bond. However, given the known cytotoxic effects of the N3-methylated bases, it is in the best interest of the cell to rid itself of these lesions as quickly as possible. Indeed, E. coli constitutively expresses another 3-methyladenine glycosylase, Tag1, which exhibits a narrow substrate range specific to m3A (53, 54). We propose that the primary role of AlkA is to scan the genome actively probing the minor groove of the DNA with its interrogating residue, Leu-125, thereby facilitating the extrusion of cytotoxic N3-modified purines from the duplex. During this search process, AlkA could promote the extrusion of helix-destabilizing lesions, as proposed for MutM (17, 21), or the enzyme might merely capture spontaneously extrahelical ones, as proposed for uracil-DNA glycosylase (51). In either case, once the extrahelical lesion is inserted into the enzyme active site, catalysis ensues at a rate dictated primarily by the chemical stability of the lesion glycosidic linkage. AlkA thus appears to be capable of using both passive and active lesion search processes to locate aberrant bases within the genome, affording the cell maximum protection from the varying types of lesions that could be encountered under conditions of environmental stress.
Supplementary Material
Acknowledgments
We thank Frank Murphy, Narayanasami Sukumar, and all of the staff of the Northeastern Collaborative Access Team (NE-CAT) at the Advanced Photon Source for assistance during data collection. We also thank Yevgeny Brudno for help with the melting temperature studies and Hariharan Jayaram, Danaya Pakotiprapha, Marie Spong, Rou-Jia Sung, Yan Qi, and members of the Verdine laboratory for useful discussions. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357.
This work was supported, in whole or in part, by National Institutes of Health Grant GM044853 (to G. L. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
The atomic coordinates and structure factors (codes 3OGD, 3OH6, and 3OH9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
B. R. Bowman, unpublished results.
- BER
- base excision repair
- LRC
- lesion recognition complex
- DXL
- disulfide cross-linking
- UDC
- undamaged DNA complex
- m3A
- 3-methyladenine
- m3zA
- 3-deaza-3-methyladenine
- zA
- N3-deazaadenine
- HhH
- helix-hairpin-helix
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Fm7G
- 7-methyl-2′-fluorodeoxyguanosine.
REFERENCES
- 1.Friedberg E. C., Walker G. C., Siede W. (1995) DNA Repair and Mutagenesis, American Society for Microbiology, Washington D. C [Google Scholar]
- 2.Lindahl T. (2004) Nature 427, 598. [DOI] [PubMed] [Google Scholar]
- 3.Engelward B. P., Allan J. M., Dreslin A. J., Kelly J. D., Wu M. M., Gold B., Samson L. D. (1998) J. Biol. Chem. 273, 5412–5418 [DOI] [PubMed] [Google Scholar]
- 4.Jelinsky S. A., Samson L. D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1486–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCullough A. K., Dodson M. L., Lloyd R. S. (1999) Annu. Rev. Biochem. 68, 255–285 [DOI] [PubMed] [Google Scholar]
- 6.Mol C. D., Parikh S. S., Putnam C. D., Lo T. P., Tainer J. A. (1999) Annu. Rev. Biophys. Biomol. Struct. 28, 101–128 [DOI] [PubMed] [Google Scholar]
- 7.Wyatt M. D., Allan J. M., Lau A. Y., Ellenberger T. E., Samson L. D. (1999) Bioessays 21, 668–676 [DOI] [PubMed] [Google Scholar]
- 8.Samson L., Cairns J. (1977) Nature 267, 281–283 [DOI] [PubMed] [Google Scholar]
- 9.Evensen G., Seeberg E. (1982) Nature 296, 773–775 [DOI] [PubMed] [Google Scholar]
- 10.Nakabeppu Y., Miyata T., Kondo H., Iwanaga S., Sekiguchi M. (1984) J. Biol. Chem. 259, 13730–13736 [PubMed] [Google Scholar]
- 11.Bjelland S., Bjørås M., Seeberg E. (1993) Nucleic Acids Res. 21, 2045–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjelland S., Birkeland N. K., Benneche T., Volden G., Seeberg E. (1994) J. Biol. Chem. 269, 30489–30495 [PubMed] [Google Scholar]
- 13.Saparbaev M., Kleibl K., Laval J. (1995) Nucleic Acids Res. 23, 3750–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saparbaev M., Laval J. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5873–5877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Brien P. J., Ellenberger T. (2004) J. Biol. Chem. 279, 26876–26884 [DOI] [PubMed] [Google Scholar]
- 16.Hollis T., Ichikawa Y., Ellenberger T. (2000) EMBO J. 19, 758–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi Y., Spong M. C., Nam K., Banerjee A., Jiralerspong S., Karplus M., Verdine G. L. (2009) Nature 462, 762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fromme J. C., Verdine G. L. (2003) J. Biol. Chem. 278, 51543–51548 [DOI] [PubMed] [Google Scholar]
- 19.Lau A. Y., Wyatt M. D., Glassner B. J., Samson L. D., Ellenberger T. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13573–13578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowman B. R., Lee S., Wang S., Verdine G. L. (2008) Structure 16, 1166–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerjee A., Santos W. L., Verdine G. L. (2006) Science 311, 1153–1157 [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 23.Read R. J. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 1373–1382 [DOI] [PubMed] [Google Scholar]
- 24.Collaborative Computational Project (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 25.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 26.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 27.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 28.Schüttelkopf A. W., van Aalten D. M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 29.Huang H., Chopra R., Verdine G. L., Harrison S. C. (1998) Science 282, 1669–1675 [DOI] [PubMed] [Google Scholar]
- 30.Banerjee A., Yang W., Karplus M., Verdine G. L. (2005) Nature 434, 612–618 [DOI] [PubMed] [Google Scholar]
- 31.Yang C. G., Yi C., Duguid E. M., Sullivan C. T., Jian X., Rice P. A., He C. (2008) Nature 452, 961–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corn J. E., Pelton J. G., Berger J. M. (2008) Nat. Struct. Mol. Biol. 15, 163–169 [DOI] [PubMed] [Google Scholar]
- 33.Duguid E. M., Rice P. A., He C. (2005) J. Mol. Biol. 350, 657–666 [DOI] [PubMed] [Google Scholar]
- 34.Fromme J. C., Banerjee A., Huang S. J., Verdine G. L. (2004) Nature 427, 652–656 [DOI] [PubMed] [Google Scholar]
- 35.Banerjee A., Verdine G. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15020–15025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radom C. T., Banerjee A., Verdine G. L. (2007) J. Biol. Chem. 282, 9182–9194 [DOI] [PubMed] [Google Scholar]
- 37.Larson K., Sahm J., Shenkar R., Strauss B. (1985) Mutat. Res. 150, 77–84 [DOI] [PubMed] [Google Scholar]
- 38.Doublié S., Tabor S., Long A. M., Richardson C. C., Ellenberger T. (1998) Nature 391, 251–258 [DOI] [PubMed] [Google Scholar]
- 39.Eom S. H., Wang J., Steitz T. A. (1996) Nature 382, 278–281 [DOI] [PubMed] [Google Scholar]
- 40.Pelletier H., Sawaya M. R., Kumar A., Wilson S. H., Kraut J. (1994) Science 264, 1891–1903 [PubMed] [Google Scholar]
- 41.Beard W. A., Osheroff W. P., Prasad R., Sawaya M. R., Jaju M., Wood T. G., Kraut J., Kunkel T. A., Wilson S. H. (1996) J. Biol. Chem. 271, 12141–12144 [DOI] [PubMed] [Google Scholar]
- 42.Paulovich A. G., Hartwell L. H. (1995) Cell 82, 841–847 [DOI] [PubMed] [Google Scholar]
- 43.Allan J. M., Engelward B. P., Dreslin A. J., Wyatt M. D., Tomasz M., Samson L. D. (1998) Cancer Res. 58, 3965–3973 [PubMed] [Google Scholar]
- 44.Habraken Y., Ludlum D. B. (1989) Carcinogenesis 10, 489–492 [DOI] [PubMed] [Google Scholar]
- 45.Peng X., Li H., Seela F. (2006) Nucleic Acids Res. 34, 5987–6000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee S., Bowman B. R., Ueno Y., Wang S., Verdine G. L. (2008) J. Am. Chem. Soc. 130, 11570–11571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blainey P. C., van Oijen A. M., Banerjee A., Verdine G. L., Xie X. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5752–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blainey P. C., Luo G., Kou S. C., Mangel W. F., Verdine G. L., Bagchi B., Xie X. S. (2009) Nat. Struct. Mol. Biol. 16, 1224–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hedglin M., O'Brien P. J. (2010) ACS Chem. Biol. 5, 427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen L., Haushalter K. A., Lieber C. M., Verdine G. L. (2002) Chem. Biol. 9, 345–350 [DOI] [PubMed] [Google Scholar]
- 51.Parker J. B., Bianchet M. A., Krosky D. J., Friedman J. I., Amzel L. M., Stivers J. T. (2007) Nature 449, 433–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ezaz-Nikpay K., Verdine G. L. (1994) Chem. Biol. 1, 235–240 [DOI] [PubMed] [Google Scholar]
- 53.Thomas L., Yang C. H., Goldthwait D. A. (1982) Biochemistry 21, 1162–1169 [DOI] [PubMed] [Google Scholar]
- 54.Metz A. H., Hollis T., Eichman B. F. (2007) EMBO J. 26, 2411–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.