Abstract
In budding yeast, silent chromatin is defined at the region of telomeres, rDNA loci, and silent mating loci. Although the silent chromatin at different loci shows structural similarity, the underlying mechanism to establish, maintain, and inherit these structures may be fundamentally different. In this study, we found two arginine residues within histone H2B, which are specifically required to maintain either the telomeric or the rDNA silenct chromatin. Arginine 95 (R95) plays a specific role at telomeres, whereas arginine 102 (R102) is required to maintain the silent chromatin at rDNA and to ensure the integrity of rDNA loci by suppressing recombination between rDNA repeats. R95 mutants show enhanced rDNA silencing but a paradoxically low Sir2 protein abundance. Furthermore weakened silencing at telomeres in R95 mutants can be suppressed by a specific SIR3 allele, SIR3–D205N, which increases the affinity of Sir proteins to telomeres, suggesting H2B–R95 may directly mediate telomeric Sir protein–nucleosome interactions. Double mutations of R95 and R102 lead to desilencing of both rDNA and telomeres, indicating both arginines are necessary to ensure integrity of silent chromatin at these loci. Furthermore, mutations of R102 cause accumulation of extrachromosomal rDNA circles and reduce life span, suggesting that histone H2B contributes to longevity.
IN budding yeast Saccharomyces cerevisiae, silent mating loci [hidden MAT left (HML) and hidden MAT right (HMR)] and telomere regions are akin to heterochromatin in higher eukaryotes, forming a compacted chromatin structure that represses transcription. A form of silent chromatin is also defined at regions of ribosomal DNA (rDNA) gene arrays, in which reporter genes transcribed by RNA polymerase II are silenced if inserted (Rusche et al. 2003). Despite their similarity in repressing transcription, the underlying mechanisms of HM/telomeric and rDNA silencing are apparently distinct, given their requirement for almost completely different protein complexes (Huang and Moazed 2003; Huang et al. 2006). In fact the molecular basis for rDNA silencing remains poorly understood although recent evidence suggests that the biological basis for rDNA silencing lies in rDNA copy number control (Kobayashi and Ganley 2005).
In addition to repressing transcription, the rDNA silent locus also suppresses homologous recombination (HR) within this otherwise highly recombinogenic repetitive region. Failure to suppress HR at rDNA regions leads to the formation and accumulation of extrachromosomal rDNA circles (ERCs), a phenomenon associated with aged cells postulated as a factor limiting longevity in budding yeast (Sinclair and Guarente 1997). Deletion of SIR2, the only SIR gene required for both rDNA and HM/telomere silencing, also activates rDNA recombination, resulting in accumulation of ERCs and a shorter life span, (Sinclair and Guarente 1997). Besides ERCs, caloric restriction and TOR-dependent signaling define two alternative genetic pathways that modulate replicative life span, both of which seem to be SIR2 independent (Agarwal et al. 2005; Kaeberlein et al. 2005). Additional pathways/genes that regulate replicative life span, are still emerging. Histones are known to be important for silencing at both HM/telomeres and rDNA regions. Silencing can be affected either by changing the balance of expression of the four core histones or by the modification status on these histones. In addition to the loss of rDNA silencing (LRS) nucleosome surface identified previously (Park et al. 2002), additional residues were shown to be important for silencing in recent mutagenesis studies of histones H3 and H4 (Buchberger et al. 2008; Dai et al. 2008). More interestingly, we found a large number of histone H3 tail deletions compromise silencing at rDNA but not at telomeres or HM loci, suggesting a possibly specific function of the H3 tail on rDNA silencing. Several lines of evidence suggest that histone H2A and histone H2B are also involved in regulating silencing: (1) Deletion of one of the two H2A and H2B gene pairs in yeast, HTA1–HTB1 causes activation of Ty1 transposition, which otherwise was suppressed when inserted in the rDNA (Bryk et al. 1997); (2) mutations in HIR3, which regulates H2A/H2B expression, result in increasing rDNA silencing (Smith et al. 1999); (3) UBC2/RAD6, an E2 ubiquitin-conjugating enzyme required for ubiquitylation of lysine 123 in histone H2B is also involved in telomeric silencing (Huang et al. 1997); and (4) sumoylation on histone H2B is enriched at the regions close to telomeres (Nathan et al. 2006). However, systematic studies on the functional analysis of histone H2A and histone H2B residues in transcription silencing are lacking. Moreover, there is no published evidence thus far that histones might be directly involved in mechanisms that ensure rDNA integrity and thereby affect replicative life span.
Here we systematically generated a collection of histone H2A and H2B mutants at residues suspected to be modified, to study the function of these potential modifications in budding yeast. For each mutant, we tested (1) sensitivity to genotoxic reagents such as hydroxyurea (HU), camptothecin (CPT), and methyl methanesulfonate (MMS), and (2) transcriptional repression of reporter genes inserted at the silent chromatin regions. Mutations at two histone H2B residues are analyzed in detail: arginine 95 (R95) and arginine 102 (R102) as substitutions at these sites show strong dominant loss of silencing phenotypes. We provide evidence that these residues play specific and opposite roles in controlling silencing at different loci. Furthermore, our data indicate that H2B R95 and R102 function in distinct ways to affect rDNA recombination, formation, and accumulation of ERCs and replicative life span in budding yeast.
MATERIALS AND METHODS
Yeast strains, plasmids, and media:
All strains used in this work are described in Table 1. The yeast strain used for telomeric silencing assays (JDY76) was derived from UCC3505 (Singer and Gottschling 1994). JDY78, which contains both HIS3–mURA3 and MET15 reporters in the rDNA was derived from YNB9, the parental strain of JPY12 described previously (Park et al. 2002). Both copies of the HTA–HTB cassette were knocked out using a one-step PCR method. The complete deletion of each cassette was verified by PCR. In each strain, a CEN TRP1 plasmid carrying HTA2–HTB2 (pJD82) was transformed before knocking out HTA1–HTB1 cassette to support cell viability. pJD82 was constructed by inserting the chromosome copy of HTA2–HTB2 into pRS414 using NotI and SacI restriction sites. The mutant plasmids were generated by a standard two-step PCR method (Muhlrad et al. 1992) and sequenced to verify the presence of desired mutations. Rich medium (YPD) was used in all cell cultures except where otherwise mentioned. MLA medium was made as described previously (Park et al. 2002).
TABLE 1.
Yeast strains used in this study
| Strain name | Genotype | Source |
|---|---|---|
| Figure 1Aa | ||
| JDY88 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 sir2∷KanMX6 [CEN TRP1 HTA1-HTB1] | This work |
| JDY230 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN LEU2 HTA1-HTB1] | This work |
| JDY231 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN LEU2 HTA1-HTB1-R95A] | This work |
| JDY232 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN LEU2 HTA1-HTB1-R95K] | This work |
| JDY233 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN LEU2 HTA1-HTB1-R102A] | This work |
| JDY234 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN LEU2 HTA1-HTB1-R102K] | This work |
| Figure 1Bb | ||
| JDY235 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR [CEN TRP1 HTA1-HTB1] | This work |
| JDY236 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR [CEN TRP1 HTA1-HTB1-R95A] | This work |
| JDY237 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR [CEN TRP1 HTA1-HTB1-R95K] | This work |
| JDY238 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR [CEN TRP1 HTA1-HTB1-R102A] | This work |
| JDY239 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR [CEN TRP1 HTA1-HTB1-R102K] | This work |
| JDY240 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR [CEN TRP1 HHT2-HHF2] | This work |
| JDY241 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR [CEN TRP1 HHT2-HHF2-K16A] | This work |
| JDY242 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR [CEN TRP1 HHT2-A75V-HHF2] | This work |
| JDY243 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR [CEN TRP1 HHT2-K79R-HHF2] | This work |
| JDY244 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR [CEN TRP1 HHT2-HHF2-R45C] | This work |
| Figure 1Cc | ||
| JDY89 | MATa his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 sir2∷KanMX6 [CEN TRP1 HTA2-HTB2] | This work |
| JDY245 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN LEU2 HTA1-HTB1] | This work |
| JDY246 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN LEU2 HTA1-HTB1-R95A] | This work |
| JDY247 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN LEU2 HTA1-HTB1-R95K] | This work |
| JDY248 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN LEU2 HTA1-HTB1-R102A] | This work |
| JDY249 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN LEU2 HTA1-HTB1-R102K] | This work |
| Figure 1Dd | ||
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | |
| JDY89 | See Figure 1C | |
| JDY245 | See Figure 1C | |
| JDY246 | See Figure 1C | |
| JDY247 | See Figure 1C | |
| JDY248 | See Figure 1C | |
| JDY249 | See Figure 1C | |
| Figure 2A | ||
| JDY89 | See Figure 1D | |
| JDY245 | See Figure 1C | |
| JDY246 | See Figure 1C | |
| JDY247 | See Figure 1C | |
| JDY248 | See Figure 1C | |
| JDY249 | See Figure 1C | |
| JDY250 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN LEU2 HTA1-HTB1-R95A,R102A] | |
| Figure 2B | ||
| JDY251 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1] [pRS425 2μ LEU2] | This work |
| JDY252 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [pJD84 CEN TRP1 HTA1-HTB1] [2μ LEU2 SIR2] | This work |
| JDY253 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [pJD107 CEN TRP1 HTA1-HTB1-R95A] [pRS425 2μ LEU2] | This work |
| JDY254 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [pJD107 CEN TRP1 HTA1-HTB1-R95A] [2μ LEU2 SIR2] | This work |
| JDY255 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [pJD109 CEN TRP1 HTA1-HTB1-R102A] [pRS425 2μ LEU2] | This work |
| JDY256 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [pJD109 CEN TRP1 HTA1-HTB1-R102A] [2μ LEU2 SIR2] | This work |
| Figure 2C | ||
| JD257 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1] [pRS425 2μ LEU2] | This work |
| JD258 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R95A] [pRS425 2μ LEU2] | This work |
| JD259 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R95K] [pRS425 2μ LEU2] | This work |
| JD260 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R102A] [pRS425 2μ LEU2] | This work |
| JD261 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R102K] [pRS425 2μ LEU2] | This work |
| JD262 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1] [2μ LEU2 SIR2] | This work |
| JD263 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R95A] [2μ LEU2 SIR2] | This work |
| JD264 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R95K] [2μ LEU2 SIR2] | This work |
| JD265 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R102A] [2μ LEU2 SIR2] | This work |
| JD266 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R102K] [2μ LEU2 SIR2] | This work |
| Figure 2D | ||
| JDY245 | See Figure 1C | |
| JDY246 | See Figure 1C | |
| JDY248 | See Figure 1C | |
| JDY250 | See Figure 2A | |
| Figure 3A | ||
| JD267 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1] [CEN LEU2 SIR3] | This work |
| JD268 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R95A] [CEN LEU2 SIR3] | This work |
| JD269 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R95K] [CEN LEU2 SIR3] | This work |
| JD270 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R102A] [CEN LEU2 SIR3] | This work |
| JD271 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R102K] [CEN LEU2 SIR3] | This work |
| JD272 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1] [CEN LEU2 SIR3-D205N] | This work |
| JD273 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R95A] [CEN LEU2 SIR3-D205N] | This work |
| JD274 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R95K] [CEN LEU2 SIR3-D205N] | This work |
| JD275 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R102A] [CEN LEU2 SIR3-D205N] | This work |
| JD276 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R102K] [CEN LEU2 SIR3-D205N] | This work |
| Figure 3B | ||
| JD277 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1] [CEN LEU2 SIR3] | This work |
| JD278 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R95A] [CEN LEU2 SIR3] | This work |
| JD279 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R95K] [CEN LEU2 SIR3] | This work |
| JD280 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R102A] [CEN LEU2 SIR3] | This work |
| JD281 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R102K] [CEN LEU2 SIR3] | This work |
| JD282 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1] [CEN LEU2 SIR3-D205N] | This work |
| JD283 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R95A] [CEN LEU2 SIR3-D205N] | This work |
| JD284 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R95K] [CEN LEU2 SIR3-D205N] | This work |
| JD285 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R102A] [CEN LEU2 SIR3-D205N] | This work |
| JD286 | MATα his3Δ200 leu2Δ1 trp1Δ63 lys2Δ0 ura3-167 met15Δ0 ade2∷his RDN1∷mURA3/HIS3 RDN1∷Ty1-MET15 TELV∷ADE2 hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R102K] [CEN LEU2 SIR3-D205N] | This work |
| Figure 3C | ||
| JD277 | See Figure 3B | |
| JD278 | See Figure 3B | |
| JD279 | See Figure 3B | |
| JD280 | See Figure 3B | |
| JD281 | See Figure 3B | |
| JD282 | See Figure 3B | |
| JD283 | See Figure 3B | |
| JD284 | See Figure 3B | |
| JD285 | See Figure 3B | |
| JD286 | See Figure 3B | |
| Figure 4 | ||
| JDY245 | See Figure 1C | |
| JDY246 | See Figure 1C | |
| JDY248 | See Figure 1C | |
| Figure 5A | ||
| JDY230 | See Figure 1A | |
| JDY231 | See Figure 1A | |
| JDY232 | See Figure 1A | |
| JDY233 | See Figure 1A | |
| JDY234 | See Figure 1A | |
| JDY287 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 ppr1∷HIS3 adh4∷URA3-TEL-VIIL ADE2-TEL-VR hta2-htb2∷HygMX4 hta1-htb1∷NatMX4 [CEN TRP1 HTA1-HTB1-R95A,R102A] | |
| Figure 5B | ||
| JDY245 | See Figure 1C | |
| JDY246 | See Figure 1C | |
| JDY247 | See Figure 1C | |
| JDY248 | See Figure 1C | |
| JDY249 | See Figure 1C | |
| JDY250 | See Figure 2B | |
| Figure 5C | ||
| JDY245 | See Figure 1C | |
| JDY246 | See Figure 1C | |
| JDY247 | See Figure 1C | |
| JDY248 | See Figure 1C | |
| JDY249 | See Figure 1C | |
| JDY250 | See Figure 2B | |
| Figure 6A | ||
| BY4741 sir2∷KanMX4 | ||
| BY4741 fob1∷KanMX4 | ||
| JDY289 | MATα his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0 hta2-htb2∷HygMX4 hta1-htb1∷G418 [CEN LEU2 HTA1-HTB1] | This work |
| JDY290 | MATα his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0 hta2-htb2∷HygMX4 hta1-htb1∷G418 [CEN LEU2 HTA1-HTB1-R95A] | This work |
| JDY291 | MATα his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0 hta2-htb2∷HygMX4 hta1-htb1∷G418 [CEN LEU2 HTA1-HTB1-R102A] | This work |
| JDY292 | MATα his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0 hta2-htb2∷HygMX4 hta1-htb1∷G418 [CEN LEU2 HTA1-HTB1-R95A,R102A] | This work |
| JDY296 | MATα his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0 hta2-htb2∷HygMX4 hta1-htb1∷G418 [CEN LEU2 HTA1-HTB1-R95K] | This work |
| JDY297 | MATα his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0 hta2-htb2∷HygMX4 hta1-htb1∷G418 [CEN LEU2 HTA1-HTB1-R102K] | This work |
| Figure 6Bd | ||
| BY4741 sir2∷KanMX4 | ||
| JDY289 | See Figure 6A | |
| JDY290 | See Figure 6A | |
| JDY291 | See Figure 6A | |
| JDY292 | See Figure 6A | |
All these strains were derived from UCC3505 and were generated by PCR-mediated gene replacement.
These strains are UCC3505 transformed with an extra plasmid-borne copy of the indicated histones in addition to the two endogenous gene copies.
All these strains are in the GRF167 background and were generated by PCR-mediated gene replacement.
All these strains are in the BY background.
Silencing assays:
Silencing at telomere or rDNA was assayed as described. Cells grown overnight in YPD medium at 30° were first diluted to an A600 = 1.0, then serially diluted tenfold and spotted onto either nonselective or selective media. Pictures were taken after the plates were incubated at 30° for 2–3 days. To assay colony color using the ADE2 marker, plates were incubated at 30° for 2 days and then stored at 4° for 1 week before pictures were taken.
Chromatin immunoprecipitation analysis:
Chromatin immunoprecipitation (ChIP) analysis was performed as described after cross-linking at room temperature for 15 min (Braunstein et al. 1993). Rabbit polyclonal anti-Sir2p antibody was purchased from Santa Cruz Biotechnology (sc-25753) and used at 10 μl per immunoprecipitation (IP). The amount of DNA resulting from each IP was analyzed using the Applied Biosystems SYBR green RT–PCR system. Each IP was normalized to the control PHO5 locus and input DNA and wild-type H2B, as described previously (Norris et al. 2008). Primers used for telomere and PHO5 were as in Norris et al. (2008). Primer pair 21 (NTS2) in Huang et al. (2006) was used to quantify rDNA ChIP.
Transcript analysis:
Transcript microarray analysis was performed in triplicates with the Yeast Genome 2.0 array from Affymetrix. Total RNA was extracted using hot phenol–chloroform, followed by RNeasy (Qiagen) cleanup. Statistical analysis of microarray data was performed on the basis of QT normalization.
Extrachromosomal circles and replicative life span analysis:
ERCs and replicative life span were analyzed as described (Medvedik and Sinclair 2007). To avoid bias, the person who performed replicative life-span analysis was blinded to strain identity during the experiments.
RESULTS
Previously, we described systematic mutagenesis analysis of histone H3 and histone H4 focused on the modifiable residues (Hyland et al. 2005). Recently, more comprehensive mutant libraries were built to dissect the function of every residue on all four core histones (Matsubara et al. 2007; Dai et al. 2008; Nakanishi et al. 2008). To expand knowledge about modification on histones H2A and H2B, we systematically generated a new mutant library. Similar to our first version of the histone H3 and histone H4 library, we mutated all putatively modified yeast residues on the basis of a set of conserved residues in calf histones that were reportedly modified (Zhang et al. 2003). Using the same strategy described previously (Hyland et al. 2005), amino acid substitution was used to mimic either modified or unmodified status as much as is possible with native amino acid substitutions. We tested the sensitivity of yeast strains bearing histone mutations to a variety of genome-damaging reagents including HU, MMS, benomyl, CPT, and ultraviolet (UV) irradiation. For most mutations on histone H2A and histone H2B, we found few significant changes in damage sensitivity. However, upon screening the library for silencing defects we found many mutants that compromised silencing at either telomeric or rDNA loci (all data were deposited into the histonehits database at http://www.histonehits.org (Huang et al. 2009); the subset of mutants with phenotypes is also listed in Table 2). Consistent with a previous report that deletion of RAD6, the H2B K123 E2 ligase, causes defects in telomeric silencing (Huang et al. 1997), we found all mutations of lysine 123 in histone H2B resulted in a strong loss of silencing at both telomere and rDNA (data not shown). Furthermore, mutations of H2B K123 caused pleiotropic phenotypes, presumably as a result of perturbing nucleosome ubiquitylation, which in turn causes loss of methylation on histone H3 K4 and K79. In this report, from this point on, we focus on mutations on residues R95 and R102 in histone H2B, which had strong dominant silencing phenotypes.
TABLE 2.
Summary of phenotypes observed for indicated histone alleles
| Phenotypesa | H2A alleles | H2B alleles |
|---|---|---|
| Loss of telomeric silencing (LTS) | R18A | R75A; R75K; R75L; R75M; K89A; R95A; R95K; K111Q; R119K; K123A; K123R; K123Q |
| Increase of telomeric silencing (ITS) | R102A; R102K | |
| Loss of rDNA silencing (LRS) | K21A; R75A; R102A; R102K; K111R; R119K; K123A; K123R; K123Q | |
| Increase of rDNA silencing (IRS) | R18A; R36A; K75R; R78K; K96A; K119Q | K6Q; K11A; K11Q; K16A; K16Q; K34A; K34Q; K46A; K46Q; K89A; K89R; K89Q; R95A; R95K |
| Sensitive to HU | K76A; K76Q; R78A | 111Q; K123A; K123R; K123Q |
| Sensitive to MMS | R18A; K76A; K76Q; R78A | |
| Sensitive to CPT | R78A | |
| Temperature sensitivity (39°) | R18A; K76A; K76Q; R78A; R78K | R95A; R102K; R119K; K123A; K123R; K123Q |
Residues covered in this test include H2A: K4, K7, K13, R18, R36, K75, K76, R78, K96, K119, K123, K126 and H2B: K3, K6, K11, K16, K21, K22, K34, K46, R75, K88, K89, R95, R102, K111, R119, K123. Each lysine was mutated to alanine, arginine, and glutamine and each arginine was mutated to lysine and glutamine. H2B K75 was also mutated to leucine and methionine. The mutants were analyzed and their phenotypes were compared to wild type and scored arbitrarily.
Other phenotypes tested include sensitivity to 6-azauracil (6-AU), benomyl, UV irradiation, and temperature sensitivity at 37° and 16°, which none of the mutants have.
Locus-specific regulation of silencing by histone H2B mutants:
To evaluate the function of histone H2A and H2B mutations on silencing, we used two reporter strains. One strain (JDY76) is derived from UCC3505 and carries the URA3 reporter gene at telomere TELVIIL and an ADE2 reporter gene at telomere TELVR (Singer and Gottschling 1994). Telomeric silencing was measured by cells' ability to grow on synthetic medium lacking uracil (SC −Ura) or synthetic medium containing 5-fluoroorotic acid (5-FOA) and by colony color. The other strain (JDY78) is similar to one described previously (Park et al. 2002), a derivative of YNB12 in the GRF167 background, in which two reporter genes, MET15 and mURA3–HIS3 were inserted into distinct rDNA repeats. In this strain, silencing at the rDNA region can be monitored by growth on synthetic medium containing 5-FOA (mURA3 reporter) or by colony color on a Modified Lead Agar (MLA) plate (MET15 reporter). In both strains, the two copies of HTA–HTB genes were knocked out and H2A and H2B were supplied from a single copy TRP1 plasmid (pJD78 CEN TRP1HTA2–HTB2) to support viability. A second plasmid (CEN LEU2HTA1–HTB1) carrying either wild-type sequence or a point mutation in the histones was introduced to replace pJD78 (see materials and methods for details).
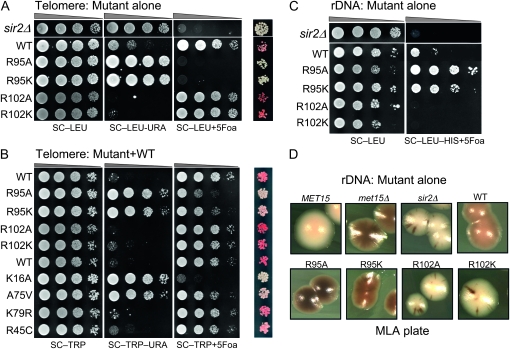
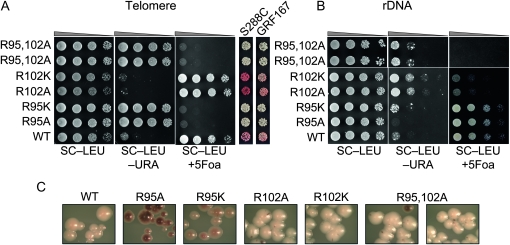
Figure 1A shows that in JDY76 bearing wild-type histones H2A and H2B, the telomeric URA3 reporter was largely silenced. This is indicated by the fact that only a few cells can grow on SC −Ura and most cells are able to grow on media containing 5-FOA. However, in R95A or R95K mutants, this silencing is completely abolished and no cells can grow on 5-FOA medium. The loss of telomeric silencing is also shown by restored expression of ADE2, resulting in white colonies. Interestingly, strains containing mutations in R102 displayed a distinct phenotype. No obvious loss of telomeric silencing was observed. Instead, the R102 mutants showed modestly but reproducibly enhanced telomeric silencing as indicated by the complete lack of growth on SC−Ura and a deeper red color than the wild type (Figure 1A). The R102A mutant seems to have even more strengthened telomeric silencing than R102K, whereas there is no obvious difference between the R95A and R95K alleles in loss of telomeric silencing. Silencing at HM loci was not tested directly. From the transcription profile of the mutants (MATα strain, see below), we found transcription of HMRa1, which is normally silenced at HMR, is upregulated, suggesting perturbed HMR silencing. We further tested silencing at HM loci by mating in the mutant strains. Unexpectedly, we made the observation that R95A compromised mating ability but only in the MATa strain. Only a slight mating defect was observed in R95K; no mating defects were observed in R102 mutants.
Figure 1.—
Locus-specific regulation of silent chromatin by R95 and R102 mutants. (A) Telomeric silencing of the URA3 reporter at telomere VIIR was measured by growth on synthetic medium lacking uracil (SC−Ura) or SC medium containing 5-FOA (SC +5-FOA). Silencing of the ADE2 reporter at telomere VL was measured by colony color. Strains tested only contain a single copy of either the wild-type or the indicated mutant histone H2A and histone H2B genes. (B) Telomeric silencing was measured as in A, but the strains contained intact histone genes with an additional copy of the mutant genes. (C) rDNA silencing of URA3 reporter was measured by growth of cells on SC +5-FOA. (D) rDNA silencing of the MET15 reporter was measured by color of colonies on MLA plates.
Another interesting phenotype of both these mutants is that the silencing defects are dominant. Wild-type strains containing an extra copy of either of the histone H2B R95 mutants showed almost complete loss of telomeric silencing (growth on SC −Ura and white colony color in Figure 1B). When we performed comparative silencing assays with a few known histone H3 mutants that affect telomeric silencing, we found that K16A and A75V also caused significant dominant loss of telomeric silencing, whereas K79R and R45C had relatively little dominant effect on silencing. Thus these mutant phenotypes are as severe as those of the strongest H3/H4 mutants known.
In contrast to telomeric and HM silencing, which require all four Sir proteins, silencing at the rDNA requires only one of the Sir proteins, Sir2p. In addition, rDNA is only silenced for Pol II transcription, whereas Pol I and Pol III transcription remain active (Rusche et al. 2003). In the H2B arginine mutations, we found R95 mutants showed increased silencing at rDNA loci as indicated by both increased growth on SC +5-FOA (Figure 1C) and the darker color of colonies on the MLA plate (Figure 1D). On the other hand the R102 mutants completely lost rDNA silencing (Figure 1, C and D). Furthermore, in the R102 mutants, the recombination frequency at rDNA loci was dramatically increased, as revealed by the dramatically sectored colonies on MLA plates. This phenotype is similar to that of a sir2Δ strain, which not only loses rDNA silencing but also shows elevated rDNA recombination frequency. We also tested whether these mutants had dominant effects on rDNA silencing; none were dominant (data not shown).
In summary, we identified mutations in two arginine residues in histone H2B with distinct functions in regulating gene silencing. Interestingly, mutation of these neighboring residues shows distinct effects on telomeric and rDNA silent chromatin. Furthermore, these silencing phenotypes observed are complementary, suggesting that these residues underlie a yin/yang relationship controlling silencing at the rDNA and telomeres.
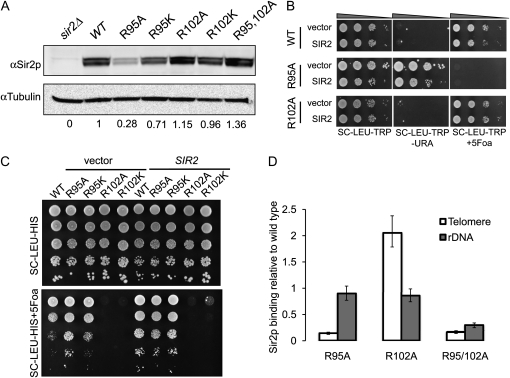
Sir2p expression is reduced in R95A but not R102A mutant:
In light of the fact that only Sir2p is required for rDNA silencing, we tested whether the H2B arginine mutants were indirectly affecting rDNA silencing by altering the expression of Sir2p, using immunoblotting. To our surprise, we observed a substantial loss of Sir2p protein in the R95A mutant, which paradoxically displays increased rDNA silencing (Figures 1A and 2A). Sir2p expression is also weakened in R95K mutants but to a lesser extent. No significant change of Sir2p expression was observed in R102A or R102K mutants. The double mutant R95A R102A restores the Sir2p level (Figure 2A). A possible explanation for R95A phenotype is that when Sir2p expression is reduced in R95 mutants, the limited amount of remaining Sir2p available preferentially binds to rDNA, enhancing silencing there, and this leads to a loss of Sir2p at telomeres, similar to what has been observed in older cells (Kennedy et al. 1997). If this were correct, overexpression of Sir2p might restore telomeric silencing in R95 mutants. To test this hypothesis, we introduced a 2-μm plasmid containing SIR2. The silencing phenotypes of these transformants were tested and compared to an empty vector control. As shown in Figure 2B, overexpression of Sir2p fails to complement this silencing defect, suggesting that loss of telomeric silencing in these mutants transcends the Sir2p expression defect. To rule out the possibility that the loss of silencing was due to the loss of another Sir protein besides Sir2p, we also overexpressed Sir3p or Sir4p in these mutants. None of these protein overexpression constructs restored telomeric silencing in the histone mutants (data not shown). Consistent with a previous study (Smith et al. 1998), overexpression of Sir2p itself leads to increased silencing at rDNA as shown in Figure 2C when compared to wild-type strains transformed with either pRS425 or pRS425–SIR2. However overexpression of Sir2p did not further increase rDNA silencing in R95 mutants and cannot restore rDNA silencing to R102 mutants (Figure 2C), although restored expression of Sir2p in the test strains was confirmed by immunoblot (data not shown). Therefore, we conclude that the silencing defects were not due to decreased Sir2p abundance; instead we suspect the altered nucleosome structure is directly responsible. The restoration of Sir2p expression in the double mutant is correlated with the loss of rDNA silencing in the double mutant; we propose a model tying together these observations in the discussion.
Figure 2.—
Sir2p expression and binding in R95 and R102 mutants. (A) Steady-state Sir2p level in strains bearing wild-type histone H2B and H2B mutant alleles as determined by immunoblotting. Tubulin was used as the loading control. Quantification was done by BioRad Quantity One with three biological replicates. Telomeric silencing (B) or rDNA silencing (C) was measured as in Figure 1 with strains overexpressing either empty vector or SIR2. (D) ChIP of Sir2p to telomere or rDNA. Values were normalized to input DNA and the PHO5 locus and wild-type histone H2B (Norris et al. 2008). Data were obtained from three replicates; the bar represents the standard error.
If the silencing defect is not caused by reduced Sir protein abundance in these histone mutants, it suggests that histone mutants might directly affect the binding of Sir (or other) proteins to silent regions. To test this hypothesis, we performed ChIP experiments using anti-Sir2p antibody in either wild-type or the mutants. At subtelomeric regions, we observed a large reduction of Sir2p binding in the R95A mutant while the association of Sir2p increased significantly in R102A mutants (Figure 2D). The occupancy by Sir2p correlates well with the silencing phenotypes observed in these mutants, suggesting silencing complex occupancy underlies the phenotypes. At the rDNA locus, the amount of Sir2p was not altered significantly in either R95A or R102A mutants, despite the reduced overall total Sir2p level in the R95A mutant. Together, these data suggest that R95 and R102 might be required for specific binding of Sir2p-containing silencing complexes to either telomeric or rDNA silent chromatin. In support of this idea, as described below, when both R95 and R102 are mutated to alanine, Sir2p binding at both telomeres and rDNA loci is lost (Figure 2D), in agreement with the loss of silencing at both loci in the double mutant.
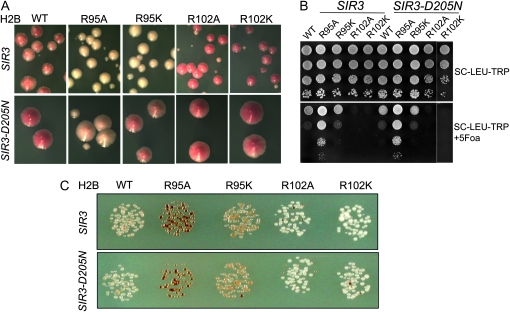
Suppression of telomeric silencing defect in R95 mutants by SIR3 suppressor alleles:
If the silencing defects are due to the failure to assemble silent chromatin in these histone mutants because of the reduced affinity of Sir proteins, certain Sir mutant alleles that have increased affinity to chromatin might suppress the silencing defect in these mutants. One such dominant allele, SIR3–D205N, suppresses a variety of silencing defects (Johnson et al. 1990; Liu and Lustig 1996; Connelly et al. 2006; Onishi et al. 2007). Recently, we showed that SIR3–D205N could also suppress the telomeric silencing defect in several LRS mutants of histones H3 and H4 (Norris et al. 2008). In addition, we also showed that binding of Sir proteins (Sir2p and Sir3p) was greatly increased at telomeres in strains containing SIR3–D205N (Norris et al. 2008). Therefore, we tested whether SIR3–D205N could also suppress the silencing defect in the histone H2B mutants.
An extra copy of either wild-type SIR3 or SIR3–D205N was introduced into the yeast strains carrying these histone H2B mutations. As a control, we showed that an additional copy of SIR3–D205N in the cells showed increased silencing at telomere in the strain containing wild-type histones, consistent with a previous observation (Norris et al. 2008). Furthermore we found that SIR3–D205N partially suppressed the telomeric silencing defect in both R95 mutants, with a more pronounced effect on R95K (Figure 3A, note darker red color). This finding is consistent with a recent study, where SIR3–D205N increased telomeric Sir3p binding, enhancing telomeric silencing (Norris et al. 2008). We did not expect to observe increased telomeric silencing in R102 mutants since telomeric silencing is already enhanced in these mutants. As expected, there is no significant effect on rDNA silencing by introducing an extra copy of SIR3 or SIR3–D205N since Sir3p is not directly involved in rDNA silencing (Figure 3, B and C). Together, these data support the hypothesis that the loss of silencing in these histone H2B mutants may result from weakened interaction between the Sir proteins and the nucleosome, and silencing can be partially restored by increasing the affinity of Sir proteins to the telomeric region.
Figure 3.—
Partial suppression of silencing defect by SIR3–D205N allele. (A) Telomeric silencing of the ADE2 reporter was measured by colony color. (B) rDNA silencing of URA3 reporter was measured by growth of cells on SC +5-FOA media. (C) rDNA silencing of the MET15 reporter was evaluated by colony color on MLA medium.
Transcriptional profile of R95A and R102A:
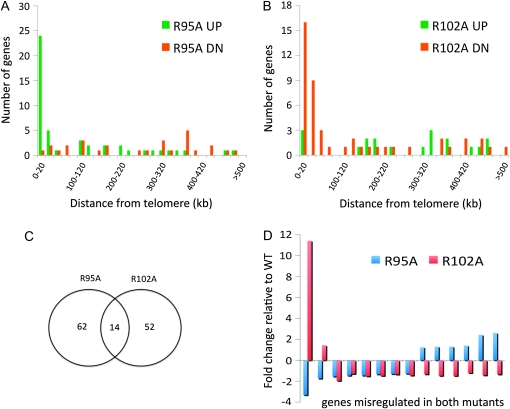
To determine whether the H2B arginine substitutions were globally affecting gene expression or specifically affecting transcription at heterochromatic silent regions, we analyzed transcriptional profiles of R95A and R102A mutants and compared them to an isogenic strain carrying wild-type histones. In the R102A mutant, only 66 genes were misregulated [false discovery rate (FDR) ≤ 0.1]. Among these genes, only 18 genes showed a change of ≥1.5-fold and 7 of these 18 genes encoded proteins/putative proteins of unknown function (supporting information, Table S1). MET15 and ADE2, the two reporters integrated in the silent chromatin, were among the 18 genes, consistent with the above results from silencing assays. In the R95A mutant, only 76 genes were identified as statistically significantly deregulated; 34 of these were affected >1.5-fold relative to the wild type. About one-quarter of these were genes of unknown function. Similar to R102, the rDNA-silencing marker MET15 was identified as one of the most downregulated genes, in agreement with the silencing phenotypes. Furthermore, we found the SIR2 transcript level was significantly downregulated in the R95 mutant, explaining the lower Sir2p level. The fact that only a very small number of genes were affected in these mutants suggests the regulation of these genes by both residues may be specific.
Furthermore, we analyzed the genomic distribution of genes that showed significant differential expression (FDR ≤ 0.1) in mutant vs. wild type. We measured the distance of each gene from its closest telomere and correlated it with transcriptional regulation. We found the majority of the genes upregulated in R95A are located within 20 kb of the telomere, whereas only one of the downregulated genes resided there and these genes were spread throughout 12 chromosomes (Figure 4A). Conversely, 24% (16) of genes downregulated in R102A were located within 20 kb of telomeres (on 11 chromosomes) and ∼42% (28) of them are within 60 kb (Figure 4B). In addition, only 14 genes are misregulated in both R95A and R102A (Figure 4C). Strikingly, almost all of the differentially regulated genes common to both R95A and R102A are localized within 20 kb of telomeres, excepting MET15 at the rDNA and PRP11, a subunit of the SF3a splicing factor complex (Hodges and Beggs 1994). On the other hand, among 6 genes repressed in both mutants, only 1 resides within 20 kb of the telomere (Figure 4D). These data were consistent with the behavior of the reporter genes inserted at silent chromatin regions, suggesting that the regulation of silent chromatin in these mutants was specific and occurred at many telomeres.
Figure 4.—
Transcription profile of R95A and R102A mutants. (A) Enrichment of genes that are differentially regulated at telomere regions in R95A mutant relative to wild type. (B) Same as A but in R102A mutant. (C) Venn diagram indicating the degree of overlap between gene expression in R95A or R102A mutant strains relative to wild type. (D) Fold change of transcription relative to wild type of genes misregulated in both R95A and R102A mutant strains. The genes were ordered by relative transcript abundance in R95A mutant.
R95, R102A double mutant loses silencing at both telomere and rDNA:
R95 mutants are defective at telomeric silencing but enhance rDNA silencing, whereas in R102 mutants, telomeric silencing is increased and rDNA silencing is disrupted. This raises an interesting question—How do the two residues differentiate rDNA and telomeric silencing and how do they interact?
To address this question, we combined the alanine substitutions for both R95 and R102 in the same H2B gene and investigated the silencing status at telomere and rDNA loci. As shown in Figure 5A, at telomeres the double mutant has similar phenotypes as R95A mutants with a strong loss of silencing. Whereas the double mutant displays the same phenotype as R95 mutants at telomeres, it does not have the same phenotype as R95 at rDNA loci. Instead, rDNA silencing is completely lost in the double mutant, which is the same as the R102 mutants as assayed using the RDN1∷URA3 and RDN1∷MET15 reporters (Figure 5B). Furthermore, it also showed the persistence of hyperrecombination phenotype at the rDNA, similar to that observed in R102 mutants (Figure 5C). Together, these data suggest that R95 and R102 function in distinct pathways to modulate silencing at telomere and rDNA. Both are required to preserve the integrity of silent chromatin.
Figure 5.—
Silencing phenotypes in R95 102A double mutant. (A) Telomeric silencing as measured in Figure 1A in strains containing either wild-type or mutant histones. (B) rDNA silencing as measured in Figure 1C in strains containing either wild-type or mutant histones. (C) rDNA silencing of MET15 reporter was measured by colony color on MLA medium.
R95 and R102 regulate formation and accumulation of ERCs and replicative life span in yeast:
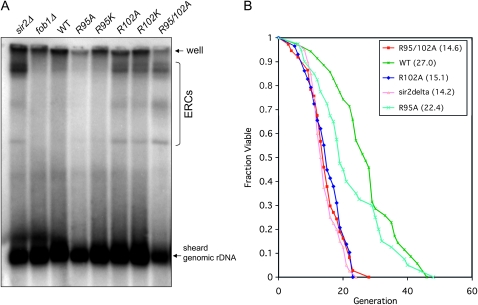
Sir2p suppresses the formation of ERCs, which has been proposed to lead to reduced replicative life span, in yeast (Cohen et al. 2004). Given the similarity between the R102 histone H2B mutants and a sir2Δ strain, we first tested whether ERCs can accumulate in these mutants, following protocol formulated by Medvedik and Sinclair (2007). As shown in Figure 6A, the amount of ERCs increased dramatically in the sir2Δ strain and decreased in the fob1Δ strain, consistent with previous observations. In R102A and R102K mutants, we also observed significant increases of ERCs. On the other hand, the amount of ERCs was slightly reduced in R95A and R95K mutants compared to wild-type strains. This result suggested that in strains with wild-type histones, rDNA recombination is largely inhibited and the amount of ERCs is low. However, when R102 is mutated, the cells lose their ability to suppress recombination and result in the formation and accumulation of ERCs.
Figure 6.—
Analysis of ERCs and replicative life span, of histone mutants. (A) Analysis of rDNA and ERCs isolated from wild-type or histone mutant strains. Probe was amplified from genomic DNA specific to 25S rDNA. ERCs are indicated by a bracket. The arrow indicates sheared genome rDNA. (B) Replicative life span analysis was performed in either wild-type or sir2Δ or histone mutant strains. The strains used for replicative life span are the same as that in A.
Next, we asked whether the histone mutants also have reduced replicative life span. We performed the analysis on these mutants and compared them to the wild type or sir2Δ. We found that replicative life span, was greatly reduced in the R95 102A double mutant and the R102A mutant (Figure 6B). Strikingly, we found that the life span, of the R102A mutant or the R95 102A double mutant was not distinguishable from sir2Δ, with a mean life span, of ∼14 generations. For the strain carrying the R95A mutant, the life span, was slightly, but not significantly decreased when compared to wild type (P = 0.056). The reduced life span, in both R102A mutant and R95 102A double mutants correlates well with the hyperrecombination phenotype and the level of ERCs in these mutants, consistent with the idea that an elevated level of ERCs formed by rDNA recombination can lead to replicative senescence (Sinclair and Guarente 1997). Overall, our data suggest that H2B R102 plays an important role in ensuring the integrity of rDNA silent chromatin by suppressing homologous recombination and thereby enhancing survival.
DISCUSSION
In this article, we systematically studied the function of the possibly modified residues on histone H2A and histone H2B in yeast related to DNA damage and heterochromatic gene silencing. Unlike histone H3 and histone H4, in which we found quite a large number of residues sensitive to certain DNA damage reagents, as reported previously (Dai et al. 2008), very few mutations in histone H2A and histone H2B showed sensitivity to these reagents, suggesting a lesser role of these two histones in DNA metabolism. This may be because (1) the coverage of the mutants tested was limited and deeper testing will reveal a distinct outcome or (2) the nucleosomal position of histone H2A and histone H2B may limit their impact on repair processes. Increasing evidence suggests that residues at the DNA entry site and dyad axis are critical for DNA-related activities since mutations in these residues cause cells to either lose viability or show sensitivities to multiple DNA damage reagents (Matsubara et al. 2007). These regions are mainly occupied by histone H3 and histone H4 (Luo et al. 2010).
Regulating heterochromatic gene silencing by core histones:
Given the fact that histones are key architectural proteins of chromatin, it is not surprising that mutations on histones may alter the structure of silent chromatin, which can be easily monitored by assaying the expression of reporter genes inserted at these regions. Most studies thus far have reported on histones H3 and H4. Analysis of a comprehensive histone H3/H4 mutant library showed >30% of the mutants displaying altered silencing in rDNA and/or telomeric loci (Dai et al. 2008), giving a more complete view of critical residues in histones H3 and H4. In contrast, little is known about how histones H2A and H2B contribute to silent chromatin. One residue, lysine 123 on histone H2B, is known to be ubiquitylated. Monoubiquitylation of K123 in histone H2B is required for di- and trimethylation on histone H3 K4 and K79, and K79 mutants show silencing defects. This seems to be the case since all K123 mutations tested caused loss of silencing at both rDNA and telomeres (data not shown). Of 89 histone H2A and histone H2B mutants, a total of 29 (32.6%) and 15 (16.8%), respectively, conferred altered rDNA and/or telomeric silencing to different extents. The ratios are lower compared to those for histone H3/H4 mutants, suggesting a somewhat smaller effect of these histones on silent chromatin structure. More residues may emerge as important for heterochromatic gene silencing when the size of our mutant library gets bigger. The basis for the dominant nature of these mutants (and certain H3 and H4 mutants) on telomeric silencing is unknown. It could reflect a stronger silencing phenotype overall for the dominant mutants, or perhaps the dominant mutants might have a specific effect on spreading of silent chromatin.
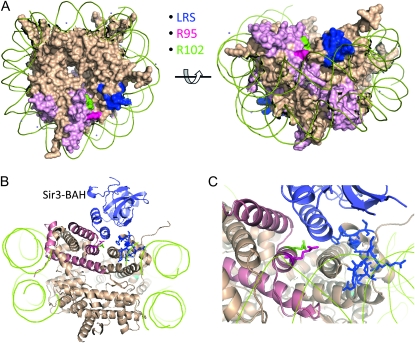
The histone H2B arginine substitutions analyzed here are interesting in their ability to distinguish rDNA and telomeric silent chromatin, consistent with the distinct mechanisms and protein complexes yeast use to confer silencing at these loci (Rusche et al. 2003). Therefore, a straightforward model is that these two residues provide critical binding sites for at least two locus-specific silencing complexes. On the nucleosome, both R95 and R102 reside on the “cliff” of a deep groove between histone H2B and histone H3 and lie proximal to the previously described LRS surface (Figure 7A) (Park et al. 2002), which can serve as the binding site for either Sir3p or Dot1p, depending on the chromosome location (Norris et al. 2008). If this model is correct, Sir3p may be the telomere-specific silencing factor that interacts with R95. In support of this, R95 and several other residues in the histone H2B C terminus, including R102, are within 5Å of the Sir3p BAH domain as predicted by the docking model of Norris et al. (2008) and Norris and Boeke (2010) (Figure 7B). Furthermore, of >500 histone H3/H4 mutants studied, we found that in histone H3, only K79R has similar silencing phenotypes as H2B R95 mutants (Park et al. 2002), whereas E73A and E73D phenocopy the R102 mutant. Consistent with this, both K79 and E73 residues critical for silencing on histone H3 are within or adjacent to the LRS surface. Whereas it is clear that Sir3p binds this surface at telomeres via its BAH domain (Buchberger et al. 2008; Norris et al. 2008), it is still not clear which protein or protein complex interacts with this surface at the rDNA locus. Possible candidates include members of the RENT complex, the fork-blocking protein Fob1p, and other BAH-containing complexes such as RSC and ORC. Interestingly, rsc2 mutants alter HM locus silencing (Jambunathan et al. 2005) and ORC binding is also important for HM silencing (Fox et al. 1995; Palacios DeBeer et al. 2003).
Figure 7.—
Histone H2B R95 and R102 are located at nucleosome disc surface. (A) Disc face representation of the nucleosome 1ID3 (White et al. 2001) rendered using PyMOL. H2B chains are highlighted in deep salmon. LRS residues are shown in blue, H2B R95 in red, and R102 in green. H2B R95 and R102 are on the nucleosome surface and adjacent to the LRS residues. (B) The same model for Sir3p BAH domain binding to the LRS surface as described previously (Norris et al. 2008). The Sir3–BAH domain is shown in light blue. Others are as in A. (C) Same as B but the interaction region between the Sir3–BAH domain and nucleosome surface is enlarged.
Both R95 and R102 are among the most highly conserved residues in histone H2B and invariant across phyla. Although R102 was shown to be methylated in calf histone H2B, and R95 was listed as a possibly methylated residue (Zhang et al. 2003), we could not find evidence these residues were modified in yeast, despite extensive efforts and our ability to detect many other known H2B modifications in yeast (J. Dai, unpublished data). However, we cannot exclude the possibility that modification on these residues can be found only at specific silent chromatin regions, making it extremely difficult to identify due to low abundance.
rDNA silencing and Sir2p expression:
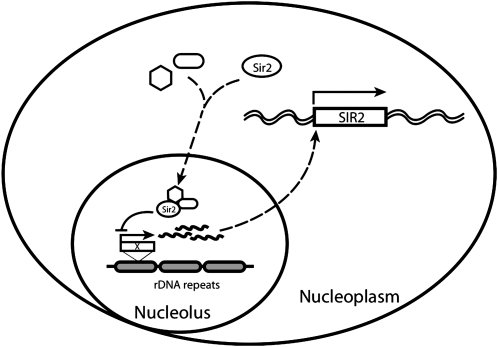
Sir2p is the only Sir protein required for silencing at both rDNA and telomeres. It is also the only Sir protein that has been shown to have enzymatic activity and its NAD+-dependent histone deacetylase activity is required for silencing at both loci. The bulk of Sir2p is localized in nucleolus and preferentially associates with rDNA (Gotta et al. 1997). Both dosage and distribution of Sir2p have been demonstrated to be critical for rDNA silencing previously and are regulated by Sir4p but not Sir3p (Bryk et al. 1997; Smith et al. 1998). In addition, Sir2p expression in the cells has to be tightly regulated since overexpression of Sir2p is toxic (Holmes et al. 1997). Furthermore, previous study showed that Sir2 is downregulated in strains containing short rDNA repeats and Sir2p can also negatively regulate its own expression through an unknown mechanism (Michel et al. 2005). However, we did not find an obvious change in the copy number of rDNA repeats in the R95A mutant strain (data not shown). These observations lead us to hypothesize the existence of an unknown cellular factor X (perhaps a noncoding RNA transcript) that might sense the strength of rDNA silencing and regulate Sir2p expression accordingly (Figure 8). When rDNA is strongly silenced such as in the case of R95A mutant, or when rDNA copy number decreases, the transcription of X is reduced, which reduces SIR2 transcription and decreases Sir2p expression. Consistent with this, overexpression of Sir2p increases rDNA silencing, which in turn, represses X transcription and leads to downregulation of Sir2p expression. A recent study showed that depletion of Nrd1p or mutation in one of the exosome components (rrp4-1) resulted in the accumulation of noncoding RNA transcribed from IGS1 and the loss of rDNA silencing. Surprisingly to the authors, but consistent with this model, these mutants cause increased Sir2 transcription (Vasiljeva et al. 2008). We are now performing experiments to test our hypothesis directly; these results will be reported elsewhere.
Figure 8.—
Model for SIR2 transcriptional regulation. The nucleolar rDNA copies are presumed to be a part of a global silencing “sensor” that signals to control SIR2 expression. This signal is capable of integrating information on the total number of rDNA copies (Michel et al. 2005) as well as the overall strength of rDNA silencing as in the R95A mutant. The wavy lines might represent the Pol II transcripts described by Kobayashi and Ganley (2005) and Ide et al. (2010) to play a role in rDNA copy number control or might be some other signaling molecule; the dashed lines imply that there may be intermediate steps in the regulatory paths.
rDNA recombination, replicative life span, and histone mutants:
In budding yeast, the highly repetitive rDNA locus must be efficiently repressed to avoid genome instability caused by homologous recombination. Failure to repress results in the formation and accumulation of ERCs, which in turn leads to a shortened replicative life span, (Sinclair and Guarente 1997). This is supported by our finding that all the hyperrecombinogenic mutants tested also have shortened life span (R95K, R102A, and R102K in Figure 6B). How recombination is repressed at rDNA is still poorly understood. One possible model, supported by multiple studies on SIR2, suggests that the establishment of silent chromatin in the rDNA may create a specialized chromatin structure that prevents recombination. However, emerging evidence indicates that rDNA recombination can be decoupled from silencing since numerous mutants have been identified that only affect rDNA silencing and not recombination. For all the histone H2A and histone H2B mutants analyzed in this report and histone H3 and histone H4 mutants studied previously, we also found a large number of them that derepress rDNA silencing but do not affect recombination. It is striking that the R102 mutants display hyperrecombinogenic phenotypes, to a level comparable to sir2Δ (Figure 1D), suggesting a specific role of histone H2B in regulating rDNA recombination. These findings highlight the importance of the less-well studied H2A and H2B histones and the possibility of generating a lot more interesting data from a global mutagenesis study.
Acknowledgments
We thank Haiping Hao at the Johns Hopkins Medical Institutions microarray facility core for microarray hybridization and Chunfa Jie for bioinformatics analyses. We thank Qing Huang for help in life-span analysis. We thank Michelle Tang and James Lin for assistance with mutagenesis.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.118489/DC1.
References
- Agarwal, S., S. Sharma, V. Agrawal and N. Roy, 2005. Caloric restriction augments ROS defense in S. cerevisiae, by a Sir2p independent mechanism. Free Radic. Res. 39 55–62. [DOI] [PubMed] [Google Scholar]
- Braunstein, M., A. B. Rose, S. G. Holmes, C. D. Allis and J. R. Broach, 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7 592–604. [DOI] [PubMed] [Google Scholar]
- Bryk, M., M. Banerjee, M. Murphy, K. E. Knudsen, D. J. Garfinkel et al., 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11 255–269. [DOI] [PubMed] [Google Scholar]
- Buchberger, J. R., M. Onishi, G. Li, J. Seebacher, A. D. Rudner et al., 2008. Sir3-nucleosome interactions in spreading of silent chromatin in Saccharomyces cerevisiae. Mol. Cell. Biol. 28 6903–6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, H. Y., C. Miller, K. J. Bitterman, N. R. Wall, B. Hekking et al., 2004. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305 390–392. [DOI] [PubMed] [Google Scholar]
- Connelly, J. J., P. Yuan, H. C. Hsu, Z. Li, R. M. Xu et al., 2006. Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain. Mol. Cell. Biol. 26 3256–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, J., E. M. Hyland, D. S. Yuan, H. Huang, J. S. Bader et al., 2008. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell 134 1066–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, C. A., S. Loo, A. Dillin and J. Rine, 1995. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev. 9 911–924. [DOI] [PubMed] [Google Scholar]
- Gotta, M., S. Strahl-Bolsinger, H. Renauld, T. Laroche, B. K. Kennedy et al., 1997. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 16 3243–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges, P. E., and J. D. Beggs, 1994. RNA splicing. U2 fulfils a commitment. Curr. Biol. 4 264–267. [DOI] [PubMed] [Google Scholar]
- Holmes, S. G., A. B. Rose, K. Steuerle, E. Saez, S. Sayegh et al., 1997. Hyperactivation of the silencing proteins, Sir2p and Sir3p, causes chromosome loss. Genetics 145 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., A. Kahana, D. E. Gottschling, L. Prakash and S. W. Liebman, 1997. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 17 6693–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., A. M. Maertens, E. M. Hyland, J. Dai, A. Norris et al., 2009. HistoneHits: a database for histone mutations and their phenotypes. Genome Res 19674–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., and D. Moazed, 2003. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 17 2162–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., I. L. Brito, J. Villen, S. P. Gygi, A. Amon et al., 2006. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 20 2887–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland, E. M., M. S. Cosgrove, H. Molina, D. Wang, A. Pandey et al., 2005. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol. Cell. Biol. 25 10060–10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide, S., T. Miyazaki, H. Maki and T. Kobayashi, 2010. Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327 693–696. [DOI] [PubMed] [Google Scholar]
- Jambunathan, N., A. W. Martinez, E. C. Robert, N. B. Agochukwu, M. E. Ibos et al., 2005. Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics 171 913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, L. M., P. S. Kayne, E. S. Kahn and M. Grunstein, 1990. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 87 6286–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein, M., R. W. Powers, 3rd, K. K. Steffen, E. A. Westman, D. Hu et al., 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310 1193–1196. [DOI] [PubMed] [Google Scholar]
- Kennedy, B. K., M. Gotta, D. A. Sinclair, K. Mills, D. S. McNabb et al., 1997. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell 89 381–391. [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., and A. R. Ganley, 2005. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309 1581–1584. [DOI] [PubMed] [Google Scholar]
- Liu, C., and A. J. Lustig, 1996. Genetic analysis of Rap1p/Sir3p interactions in telomeric and HML silencing in Saccharomyces cerevisiae. Genetics 143 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, J., X. Xu, H. Hall, E. M. Hyland, J. D. Boeke et al., 2010. Histone H3 exerts a key function in mitotic checkpoint control. Mol. Cell Biol. 30 537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara, K., N. Sano, T. Umehara and M. Horikoshi, 2007. Global analysis of functional surfaces of core histones with comprehensive point mutants. Genes Cells 12 13–33. [DOI] [PubMed] [Google Scholar]
- Medvedik, O., and D. A. Sinclair, 2007. Caloric restriction and life span determination of yeast cells. Methods Mol. Biol. 371 97–109. [DOI] [PubMed] [Google Scholar]
- Michel, A. H., B. Kornmann, K. Dubrana and D. Shore, 2005. Spontaneous rDNA copy number variation modulates Sir2 levels and epigenetic gene silencing. Genes Dev. 19 1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., R. Hunter and R. Parker, 1992. A rapid method for localized mutagenesis of yeast genes. Yeast 8 79–82. [DOI] [PubMed] [Google Scholar]
- Nakanishi, S., B. W. Sanderson, K. M. Delventhal, W. D. Bradford, K. Staehling-Hampton et al., 2008. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 15 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan, D., K. Ingvarsdottir, D. E. Sterner, G. R. Bylebyl, M. Dokmanovic et al., 2006. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 20 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris, A., and J. D. Boeke, 2010. Silent information regulator 3: the Goldilocks of the silencing complex. Genes Dev. 24 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris, A., M. A. Bianchet and J. D. Boeke, 2008. Compensatory interactions between Sir3p and the nucleosomal LRS surface imply their direct interaction. PLoS Genet. 4 e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi, M., G. G. Liou, J. R. Buchberger, T. Walz and D. Moazed, 2007. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell 28 1015–1028. [DOI] [PubMed] [Google Scholar]
- Palacios DeBeer, M. A., U. Muller and C. A. Fox, 2003. Differential DNA affinity specifies roles for the origin recognition complex in budding yeast heterochromatin. Genes Dev. 17 1817–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. H., M. S. Cosgrove, E. Youngman, C. Wolberger and J. D. Boeke, 2002. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 32 273–279. [DOI] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72 481–516. [DOI] [PubMed] [Google Scholar]
- Sinclair, D. A., and L. Guarente, 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91 1033–1042. [DOI] [PubMed] [Google Scholar]
- Singer, M. S., and D. E. Gottschling, 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266 404–409. [DOI] [PubMed] [Google Scholar]
- Smith, J. S., C. B. Brachmann, L. Pillus and J. D. Boeke, 1998. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics 149 1205–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. S., E. Caputo and J. D. Boeke, 1999. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19 3184–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva, L., M. Kim, N. Terzi, L. M. Soares and S. Buratowski, 2008. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol. Cell 29 313–323. [DOI] [PubMed] [Google Scholar]
- White, C. L., R. K. Suto and K. Luger, 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20 5207–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., E. E. Eugeni, M. R. Parthun and M. A. Freitas, 2003. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma 112 77–86. [DOI] [PubMed] [Google Scholar]