Abstract
The essential yeast protein Cdc13 facilitates chromosome end replication by recruiting telomerase to telomeres, and together with its interacting partners Stn1 and Ten1, it protects chromosome ends from nucleolytic attack, thus contributing to genome integrity. Although Cdc13 has been studied extensively, the precise role of its N-terminal domain (Cdc13N) in telomere length regulation remains unclear. Here we present a structural, biochemical, and functional characterization of Cdc13N. The structure reveals that this domain comprises an oligonucleotide/oligosaccharide binding (OB) fold and is involved in Cdc13 dimerization. Biochemical data show that Cdc13N weakly binds long, single-stranded, telomeric DNA in a fashion that is directly dependent on domain oligomerization. When introduced into full-length Cdc13 in vivo, point mutations that prevented Cdc13N dimerization or DNA binding caused telomere shortening or lengthening, respectively. The multiple DNA binding domains and dimeric nature of Cdc13 offer unique insights into how it coordinates the recruitment and regulation of telomerase access to the telomeres.
Telomeres are composed of protein-bound guanine (G)-rich DNA repeats that protect the ends of linear chromosomes and are replicated by telomerase, a ribonucleoprotein reverse transcriptase that uses an integral RNA template for DNA synthesis (5, 18, 22). Appropriate telomere length regulation and protection are necessary for chromosome maintenance and cellular viability (1, 23, 40). Single-stranded DNA (ssDNA) binding proteins, such as Cdc13 in Saccharomyces cerevisiae or the Pot1-TPP1 complex in Schizosaccharomyces pombe and in humans, play a critical role in telomere maintenance by regulating telomere length in a telomerase-dependent manner (36, 45, 57), and also by protecting telomere overhangs from being recognized improperly and repaired as DNA breaks (16, 19), a switch mediated by the Hsp90 chaperone Hsp82 (10). Cdc13-dependent capping is mediated in concert with the Cdc13-interacting complex Stn1-Ten1 (also known as the CST [Cdc13/Stn1/Ten1] complex) (20, 21, 44, 50, 58, 61). Capping requires Cdc13 localization to the ends of the chromosomes, a process mediated principally by the C-terminal oligonucleotide/oligosaccharide binding (OB) fold (also known as the DNA binding domain [DBD]) of Cdc13, which has a high affinity for short, single-stranded, telomeric DNAs (3, 42, 43). OB folds have been identified in a number of Cdc13 structural homologues, including the ciliate Oxytricha nova telomere end binding protein (OnTEBP) and Pot1 (8, 27, 33, 48). There is emerging evidence that single-stranded telomere binding proteins may act on telomeres via multiple OB folds (56). For example, Oxytricha nova TEBP and the Pot1-TPP1 complex contain several OB folds, some of which are involved in DNA binding, while others mediate protein-protein interactions (45, 57).
In addition to its direct role in telomere capping as part of the CST complex (44, 54, 58), Cdc13 is also known to recruit a number of proteins to the telomeres via its N-terminal 340 amino acids, including the telomerase component Est1, Pol1, Imp4, Sir4, and Zds2 (28, 53). Cdc13-dependent recruitment of telomerase to the telomeres is mediated by Est1, a polypeptide associated with the RNA component of the telomerase dimer (13, 37, 38, 53). The Est1 binding site of Cdc13, which overlaps with that of Stn1/Ten1, is located near the N-terminal portion of the protein (between amino acids 190 and 340) and downstream of the DNA polymerase alpha interacting site (11, 13, 14, 35, 50). Binding of Est1 over Stn1/Ten1 to Cdc13 is dependent on Cdk1 phosphorylation of conserved serine and threonine residues that comprise this site (35). There is also limited evidence that implicates the N-terminal 257 residues of Cdc13 in telomere binding (6). Mutations within the N-terminal 200 residues of Cdc13 lead to the upregulation of telomere length (53), a characteristic shared with the N-terminal OB fold of human Pot1 (39), suggesting that the N-terminal portion of this family of proteins may be involved directly in telomere length control.
Cdc13 is an essential yeast protein that maintains the length and integrity of the telomeric DNA via telomerase regulation and chromosome end capping (13, 15, 16, 35, 41). Protecting the ends of chromosomes while allowing telomerase access to the telomeric overhang likely comprises complex mechanistic events that involve the coordination of multiple functional domains. To better understand the action of Cdc13 at telomeres, we solved the structure of its N-terminal portion (Cdc13N) to 2.5-Å resolution. Structural and biochemical analyses revealed an OB fold that binds long, single-stranded, telomeric DNAs, and this is supported by genetic analysis (6). Our data also show that Cdc13N is involved in Cdc13 dimerization and that its DNA binding activity is directly dependent on Cdc13 oligomerization. Furthermore, functional assays suggest that the proper regulation of telomere length depends on the dimerization and DNA binding activities of Cdc13N. Together, these data provide novel insights into the mechanism of Cdc13 assembly and telomerase recruitment and regulation.
MATERIALS AND METHODS
Protein expression and purification.
The Saccharomyces cerevisiae Cdc13 N-terminal domain, comprising residues 13 to 227, was identified via limited proteolysis and cloned into a pET28b vector with a cleavable hexahistidine tag at the amino terminus. The full-length Cdc13 protein and the wild-type and Cdc13N mutant proteins were overexpressed in Escherichia coli BL21-CodonPlus(DE3)-RIPL cells (Stratagene) at 37°C for 4 h, using 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Gold Biotechnology). The cells, which were harvested by centrifugation, were resuspended and lysed in a buffer containing 25 mM Tris, 0.5 M KCl, 5 mM 2-mercaptoethanol, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 mM benzamidine (pH 7.5) via sonication. The proteins first were purified over a Ni-nitrilotriacetic acid (Ni-NTA) column, followed by tobacco etch virus (TEV) cleavage of the hexahistidine tag overnight at 4°C. The TEV-treated proteins were further purified on a Poros-HS column (PerSpective Biosystems) to remove trace amounts of protein contaminants carried over from the Ni-NTA purification step. For the final stage of purification, the proteins were passed over a Superdex S200 sizing column (GE Healthcare) equilibrated with 25 mM Tris, 0.5 M KCl, 1 mM tris (2-carboxyethyl)phosphine (TCEP), and 10% glycerol (pH 7.5).
Protein crystallization and data collection.
The purified protein (20 mg/ml) was dialyzed in 5 mM Tris-HCl, 100 mM KCl, and 1 mM TCEP (pH 7.5) prior to crystallization trials. Initial Cdc13N crystal screening using commercial Qiagen and Hampton Research crystallization screens produced two crystal forms, one of which belonged to the orthorhombic P21212 space group and the other of which belonged to the triclinic (P1) space group. The P1 crystal form was prepared by mixing 1 volume of the dialyzed protein with 1 volume of 10% polyethylene glycol 3350 (PEG 3350), 0.01 M magnesium chloride hexahydrate, and 50 mM HEPES (pH 7.0) at 18°C. The P21212 crystals were prepared by mixing 1 volume of protein with 1 volume of 0.2 M potassium chloride, 0.1 M magnesium acetate tetrahydrate, 0.05 M sodium cacodylate trihydrate, pH 6.5, and 10% (wt/vol) PEG 8000 at 18°C. The P1 crystals were harvested in cryoprotectant consisting of 10% PEG 3350, 0.01 M magnesium chloride hexahydrate, 50 mM HEPES (pH 7.0), and 25% PEG 400. The P21212 crystals were harvested in cryoprotectant consisting of 0.2 M potassium chloride, 0.1 M magnesium acetate tetrahydrate, 0.05 M sodium cacodylate trihydrate, pH 6.5, 10% (wt/vol) PEG 8000, and 25% PEG 400. Data were collected at the National Synchrotron Light Source (NSLS), using beam lines X6A and X25, and were processed with HKL2000 or MOSFLM (34), as implemented in ELVES (26) (see Table S1 in the supplemental material). Crystals of the P21212 space group contained one monomer in the asymmetric unit, while the P1 crystals contained two dimers.
Structure determination and refinement.
Initial phases for the P21212 crystal form were obtained by the method of single isomorphous replacement with anomalous signal (SIRAS) to 3.5-Å resolution, using a single mercury derivative (Hg). The derivative was prepared by soaking the crystals with 5 mM methylmercury chloride (MeHgCl2) for 15 min. Heavy atom sites were located using SOLVE (55), and the sites were refined and new phases calculated with MLPHARE (9), as implemented in ELVES (26) (see Table S1 in the supplemental material). The model was built in COOT (12), using the mercury-labeled cysteines as guideposts to trace the amino acid sequence. The model was refined using both CNS-SOLVE (7) and REFMAC5 (46). The last cycles of refinement were carried out with TLS restraints, as implemented in REFMAC5 (see Table S1 in the supplemental material). The refined P21212 model was subsequently used for molecular replacement (PHASER) to solve the P1 crystal form. Figures were prepared in PyMOL (www.pymol.org), and electrostatic surfaces were determined in APBS (4).
Gel shift assays.
Single-stranded telomeric DNA oligomers (Eurofins MWG Operon) were radiolabeled with γ-ATP (10 μCi/μl) by use of T4 polynucleotide kinase (New England Biolabs). The labeling reaction, which was carried out at 37°C for 1 h, was stopped with 10 mM EDTA, and the excess nucleotides were removed using a Microspin G-25 column (GE Healthcare). The radiolabeled DNA was used at a concentration of 1 nM with various protein concentrations (0.1, 0.3, 0.4, 0.5, 0.6, 0.8, 1, 1.5, 2, and 2.5 μM) in the presence of 1.7 nM single-stranded poly(dT) (50 bases long) (Eurofins MWG Operon), 40% binding reaction buffer (20 mM Tris-HCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 50 mM NaCl, 2 mM MgCl2, and 10% glycerol [pH 8]), and 5% (DNase-free, RNase-free) sterile water. Reaction mixtures were incubated on ice for 1 h to allow for protein-nucleic acid association and were run in a 6% DNA retardation gel (Invitrogen) with 0.5× Tris-borate-EDTA (TBE) buffer on ice for 30 min at 200 V. The gels were prerun with the binding buffer for 30 min prior to loading the reaction mix. Gels then were dried and exposed overnight on a storage phosphor screen (GE Healthcare), and the bands were analyzed using ImageQuant software (Molecular Dynamics).
Native gel analysis.
Stocks of wild-type and dimerization mutant Cdc13N were diluted to 0.5 mg/ml in buffer A containing 20 mM Tris-HCl, 100 mM NaCl, 1 mM DTT, and 10% glycerol (pH 8) prior to being loaded on a native gel at 4°C. Homemade gels containing 5.5% 40:1 acrylamide-bisacrylamide were prerun with buffer A for 30 min before loading of the samples. Samples were run in 1× TBE buffer at 4°C for 2 h (25 mA) and then stained with Coomassie blue for protein detection.
Sedimentation equilibrium analysis.
Sedimentation equilibrium experiments were performed with an Optima XL-I analytical ultracentrifuge (Beckman Coulter) using interference optics to measure the protein concentration gradient. The cells were loaded with 110 ml of sample or reference buffer. For each experiment, a minimum of three different initial protein concentrations (0.15, 0.3, and 0.6 mg/ml) were used. A blank scan of distilled water was taken before the run to correct for the effects of window distortion of the fringe displacement data (63). Experiments were performed at 4 and 25°C, using speeds of 13,000 and 18,400 rpm. Fringe displacement data were collected every 4 h until attainment of equilibrium was reached (as determined by comparison of successive scans, using WinMatch, version 0.99). Data were edited using WinReed, version 0.99. Analysis of the sedimentation equilibrium data was performed using the program Winnl, version 1.06 (30). WinMatch, WinReed, and Winnl are available from the Analytical Ultracentrifugation Facility at the University of Connecticut via their FTP site (http://biotech.uconn.edu/auf). The program SEDNTERP, version 1.08 (written by T. Laue, J. Hayes, and J. Philo and available at http://www.rasmb.bbri.org), was used to calculate M and ῡ from the amino acid composition of the protein and the r value of the solvent. At least three data sets for different loading concentrations were fitted simultaneously. Goodness of fit was determined by examination of the residuals and minimization of the variance.
Yeast strains and plasmids.
Experiments were performed in the BY4743 background of S. cerevisiae. The various alanine point mutations in Cdc13 were introduced into a pRS415-based plasmid expressing CDC13 under the control of its own promoter (a derivative of pVL1091, a kind gift of V. Lundblad [13], from which we deleted the EST1 open reading frame [ORF]) by use of QuikChange site-directed mutagenesis (Stratagene). For Fig. 5, CDC13/cdc13Δ:kanMX diploid cells were transformed with the wild-type or various mutant CDC13 plasmids and sporulated, and cdc13Δ cells containing plasmid were obtained by tetrad dissection. Cells were cultured in yeast extract-peptone-dextrose (YPD), yeast extract-peptone-galactose (YPGal), or synthetic complete medium lacking Leu (SC−Leu) as described in reference 2. Plasmid transformations and gene replacements were performed using standard genetic methods (2).
Telomere Southern blots.
Telomere Southern analysis was performed as described previously (31). Briefly, cultures were grown overnight and DNA was extracted using a standard phenol-chloroform-isoamyl alcohol-based method. XhoI-digested genomic DNAs were separated in a 1% agarose gel, denatured under alkaline conditions, and transferred to a Hybond-XL membrane. An S. cerevisiae TG1-3 telomere repeat probe was radiolabeled by random priming with [α-32P]dCTP and was used to detect total telomeric TG repeat DNA. Southern blots were imaged with a Molecular Dynamics phosphorimager.
Protein structure accession numbers.
Atomic coordinates and structure factors for the P1 and P21212 structures have been deposited in the Protein Data Bank (PDB) under PDB codes 3NWS and 3NWT, respectively.
RESULTS
Structure overview.
To investigate the role of the N-terminal domain of S. cerevisiae Cdc13, we determined the structure of a construct containing residues 13 to 227 (Cdc13N) (Fig. 1 A and B), identified by limited proteolysis. Phases were obtained by the method of SIRAS, using a mercury derivative (see Table S1 in the supplemental material).
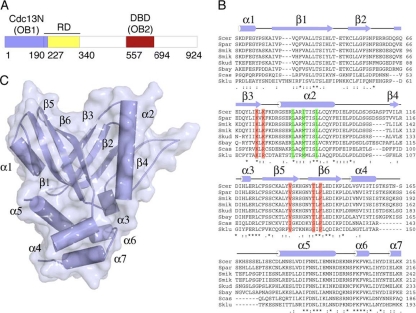
FIG. 1.
Domain organization, sequence alignment, and structure of the N-terminal domain of S. cerevisiae Cdc13. (A) Schematic of S. cerevisiae Cdc13 primary structure. Cdc13N (OB fold 1 and part of the recruitment domain [RD]) is shown in blue, RD is shown in yellow, and the DBD (OB fold 2) is shown in red. (B) Alignment of yeast Cdc13N sequences (S. cerevisiae [Scer], Saccharomyces mikatae [Smik], Saccharomyces paradoxus [Spar], Saccharomyces bayanus [Sbay], Saccharomyces castellii [Scas], Saccharomyces kluyveri [Sklu], and Saccharomyces kudriavzevii [Skud]) (sequences are available online at http://www.yeastgenome.org/cache/fungi/YDL220C.html) showing secondary structure and the mutants tested here for DNA binding (red) and Cdc13N dimerization (green). (C) Structure of Cdc13N. Secondary structure elements are labeled.
The N-terminal 150-residue peptide of the structure consists of six antiparallel β-strands organized into a β-barrel that belongs to the family of OB fold proteins (Fig. 1C). The organization of the β-strands creates a well-defined indentation on the surface of the protein that most likely comprises the nucleic acid binding site of this domain. A survey of the PDB database by use of the Dali server (DaliLite v.3) (24, 25) showed that the Cdc13N OB fold is most similar to the DNA binding domains of Cdc13 (42, 43) (see Fig. S1B in the supplemental material), human and fission yeast Pot1 (32, 33) (see Fig. S1C), and TEBPα from Oxytricha nova (8, 27) (see Fig. S1D). Within the Cdc13N OB fold are two α-helices that localize on opposite sides of the domain and sandwich this motif (α1 and α2). The C-terminal portion of the molecule consists of four α-helices (α4 to α7) organized into a helical bundle and contains part of the Cdc13 recruiting domain (Fig. 1A and C) (28, 35, 51). The organization of the N- and C-terminal subdomains of Cdc13N places the helical bundle and the surface indentation formed by the OB fold strands at opposite ends of the molecule (Fig. 1C). Both the OB fold and the helical bundle of this structure contain several long, partly untraceable loops that could become ordered upon substrate binding.
Cdc13N promotes full-length Cdc13 dimerization.
A striking feature of the Cdc13N structure is a long helix (helix α2) located on one side of the β-barrel (Fig. 1C). Helix α2 spans the entire length of the OB fold and consists of several conserved (Fig. 1B and 2 A) and solvent-exposed hydrophobic residues, suggesting that this motif may be involved in protein-protein interactions. Moreover, helix α2, together with part of the OB fold, forms a large hydrophobic groove on the surface of the protein that is solvent accessible for peptide binding (Fig. 2B). It is worth noting that the overall organization of the dimer places the N termini of the two protomers on the same side of the molecule (Fig. 2B), an arrangement that may have significant implications for nucleic acid binding. Several lines of evidence indicate that the observed crystal dimer is biologically relevant. First, contacts between the two protomers are extensive (960 Å2) and hydrophobic in nature and are mediated by conserved residues (Fig. 1A). Second, both size-exclusion chromatography (see Fig. S2B in the supplemental material) and dynamic light scattering (see Fig. S2C) support the existence of a dimer in solution. Third, a different crystal form (P21212) (see Table S1) of the same protein consists of dimers of precisely the same configuration as that for the P1 crystal form (see Fig. S2D). To further examine Cdc13N oligomerization in solution, we performed sedimentation equilibrium experiments at various concentrations, temperatures, and speeds (Fig. 2C). In all cases, the Cdc13N dimerization Kd (dissociation constant) was measured to be approximately 9 nM, which supports the existence of a tight Cdc13N dimer in solution. To determine whether the full-length Cdc13 protein is also a dimer, we purified (see Fig. S3A) the protein and showed that it binds single-stranded telomeric DNA with a high affinity (see Fig. S3B). We then examined the oligomeric state of the protein by size-exclusion chromatography (see Fig. S3C) and dynamic light scattering (see Fig. S3D). Both approaches indicated that the protein is a stable dimer in solution.
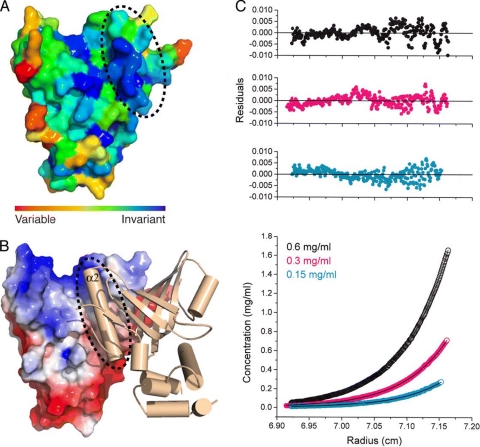
FIG. 2.
Structural and biophysical evidence for Cdc13N dimerization. (A) Surface conservation of Cdc13N showing invariant residues in dark blue and the interacting region of helix α2 with a dashed circle. (B) Cdc13N dimer. The monomer on the left (monomer A) is shown by surface charge, while the monomer on the right (monomer B) is shown in cartoon representation. A dashed circle shows the hydrophobic cleft of monomer A and where helix α2 of monomer B is docked. (C) Sedimentation equilibrium analysis of Cdc13N. The bottom panel shows the raw data (circles) and the global fit to a dimer model (line). The top three panels show the residuals of the fitted curve for a dimer model of the data points for each of the three different concentrations (0.15, 0.3, and 0.6 mg/ml) at which the experiments were carried out.
Cdc13N binds single-stranded telomeric DNA.
The Cdc13N domain belongs to the OB fold family of proteins and is structurally similar to the DBD of Cdc13, the N-terminal OB fold of Pot1, and the OB folds of OnTEBPα, all of which bind single-stranded telomeric DNA. To determine whether Cdc13N has affinity for single-stranded telomeres, we screened single-stranded DNA oligonucleotides containing anywhere from two to six S. cerevisiae telomeric repeats, using electrophoretic mobility shift assays (EMSAs). We decided to test substrates of various lengths because we were curious whether the Cdc13N dimer (contains two OB folds) binds two single strands of telomeric DNA independent of each other (in which case an 11-mer oligonucleotide could be sufficient) or one single strand of DNA, in which case a much longer piece of DNA would be required for binding. Modeling the 11-mer DNA to our structure by using the DBD DNA structure (PDB code 1S40) allowed us to determine the length of ssDNA that would span both OB folds to be approximately 35 bases long. Specifically, we tested the 11-mer DNA known to associate with the Cdc13 DBD with a high affinity (47), an 18-mer DNA, a 26-mer DNA, a 37-mer DNA, and a 43-mer DNA (see Table S2 in the supplemental material). Surprisingly, the 11-mer DNA did not bind Cdc13N, even at a concentration of 2.5 μM (Fig. 3 A), while the 18-mer and 26-mer DNAs showed poor affinity for this domain (Kd > 2.5 μM). Further binding experiments with Cdc13N and longer DNA substrates, such as a 37-mer DNA and a 43-mer DNA (Fig. 3B), showed an increase in Cdc13N affinity (Kd, ∼600 nM) with oligomer length, with specificity for single-stranded telomeric DNA (see Fig. S4B, C, and D in the supplemental material). The specificity of Cdc13N for telomeric DNA is further enhanced by localization of the full-length protein to the telomeres via the DBD, which has a high affinity and specificity for single-stranded telomeric DNA.
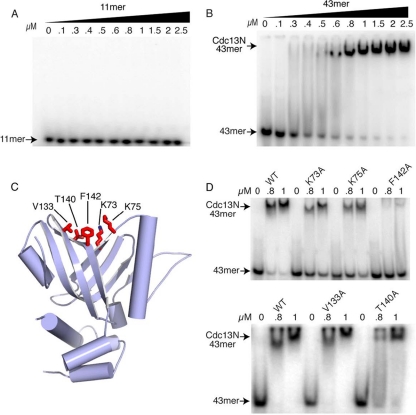
FIG. 3.
DNA binding assays of wild-type (WT) and mutant Cdc13N. (A) EMSA of wild-type Cdc13N with the 11-mer DNA that binds the DBD of Cdc13 with a 3 pM affinity. (B) EMSA of WT Cdc13N with a 43-mer single-stranded yeast telomeric DNA. (C) Structure of Cdc13N showing single alanine mutants tested for DNA binding as red stick models. (D) EMSA of WT Cdc13N and K73A, K75A, V133A, T140A, and F142A single mutants with the 43-mer DNA.
Cdc13N mutants that affect DNA binding.
To identify the DNA binding site of Cdc13N, we made single alanine mutations of conserved residues (K73A, K75A, V133A, T140A, and F142A) (Fig. 1B) that are solvent accessible and localize to the cleft formed by the OB fold (Fig. 3C), where the Cdc13N model (based on the DBD 11-mer structure [PDB code 1S40]) (see Fig. S5A and B in the supplemental material) supports DNA binding. The stable Cdc13N mutants showed a 30 to 80% decrease in DNA binding affinity for the 43-mer DNA (Fig. 3D), suggesting that these residues make direct contacts with the nucleic acid substrate. Both K73 and K75 localize at the base of the OB cleft, and their positively charged side chains are solvent exposed for coordination with the backbone of the incoming DNA substrate (Fig. 3C). V133, T140, and F142 are located in the vicinity of K73 and K75, and their side chains are accessible, possibly for stacking interactions with a free purine or pyrimidine base of the single-stranded telomeric DNA. The F142A mutant showed a severe loss of DNA binding affinity, while the effects of V133A and T140A mutations were modest (Fig. 3D) in comparison, clearly indicating that these residues are also involved in DNA binding. The weak effect observed for the V133A and T140A mutants was not surprising considering that their short, branched side chains were converted to alanine. Interestingly, both the DBD of Cdc13 and the N-terminal OB fold of human Pot1 also contain a hydrophobic side chain (I138 in Cdc13 and I96 in Pot1) in this usually positively charged cleft and where F142 is located. In the case of Pot1, the isoleucine makes stacking interactions with a base of the incoming telomeric DNA. It is worth noting that residues identified previously to interact with Pol1 (53) are located away from and do not form part of the DNA binding pocket of Cdc13N.
Cdc13N dimerization mutants show reduced telomeric ssDNA binding.
To better understand the role of the Cdc13N-dependent Cdc13 dimerization, we made single and double alanine mutations of conserved residues (L84A, I87A, L84A/I87A, and L91A) (Fig. 1B) that form part of helix α2 and are involved in dimer formation (Fig. 4 A and B). The L91A mutant resulted in significant disruption of the dimer, while the single L84A and I87A mutants did not have a direct effect, as indicated by native gels (Fig. 4C). The lack of effect of the L84A and I87A mutants in Cdc13N dimer disruption was not surprising considering the extensive contacts between the two monomers. To address this issue and to interrogate the role of these two conserved residues in Cdc13 dimerization, we made an L84A/I87A double mutant, and this mutant, like the L91A mutant, severely disrupted Cdc13N dimerization (Fig. 4C). All three residues (L84, I87, and L91) localize on the same face of helix α2 and are accessible for direct contacts with the adjacent protomer (Fig. 4B). In particular, the side chains of L84 and I87 are buried in the hydrophobic cleft of the adjacent protomer, located at the interface of helix α2 and the proximal side of the OB fold. L84 interacts with residues L74, A85, and T88, while the shorter side chain of I87 is located between L84 and L91 and makes limited contacts with the side chains of T88 and L143 of the adjacent protomer (Fig. 4B). Unlike the case for L84 and I87, the interactions between L91 and the adjacent protomer are extensive and involve residues T88, L91, L92, I146, and P148 (Fig. 4B). Extensive interactions between these residues provide the stability required for a tight Cdc13 dimer and most likely explain the increased effect of the L91A mutant in disrupting Cdc13N dimerization (Fig. 4C).
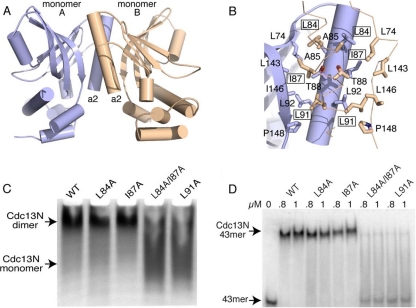
FIG. 4.
Cdc13N dimerization contacts and mutants. (A) Structure of the Cdc13N dimer, with protomer A shown in blue and protomer B shown in wheat color. (B) Zoomed-in area showing dimerization site of Cdc13N. Residues involved in dimer formation between protomers are shown as stick models. Single alanine mutants that affect dimer formation and DNA binding are boxed. (C) Native gel of WT Cdc13N and of the single and double alanine mutants tested for disruption of Cdc13N oligomerization and DNA binding. (D) EMSA of the WT and the L84A, I87A, L84A/I87A, and L91A mutants with the 43-mer DNA.
We also wanted to determine whether disruption of Cdc13N dimerization affected the ability of Cdc13N to associate with single-stranded telomeric DNA. Interestingly, EMSA of either the L91A or L84A/I87A mutant with the 43-mer DNA showed an 80 to 90% loss of DNA binding affinity, while the L84A or I87A mutant showed only a subtle effect (Fig. 4D). Because the residues are buried and therefore not accessible for direct contacts with the DNA substrate and are away from the identified DNA binding pocket, these observations clearly suggest that Cdc13N DNA binding is directly linked to dimer formation. The fact that the L84A and I87A mutants also showed a loss of DNA binding affinity (although subtle), even though these mutants did not appear to disrupt dimer formation, suggests that these mutations likely destabilize the dimer interface, which in turn leads to subtle reorganization of the two protomers, thus affecting DNA binding. Again, it is worth noting that residues identified previously to interact with Pol1 (53) are located away from and do not form part of the dimerization site of Cdc13N.
Cdc13N dimerization and DNA binding mutants affect telomere length regulation.
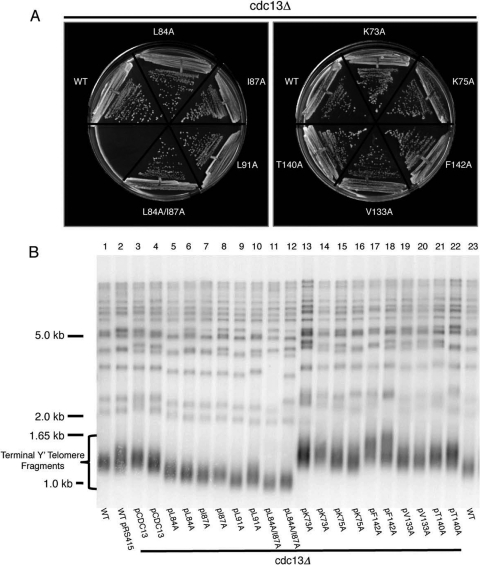
To assess whether the Cdc13N mutants would affect telomere maintenance in vivo, we engineered eight single point mutations (K73A, K75A, L84A, I87A, L91A, V133A, T140A, and F142A) and a double point mutation (L84A/I87A) into full-length Cdc13 expressed from the CDC13 promoter in ARS/CEN plasmids (see Table S3 in the supplemental material). These or vector alone was introduced into CDC13/cdc13Δ diploids, which were then sporulated to obtain haploid cdc13Δ cells containing each plasmid. Although the vector did not support growth (not shown), cells carrying each of the mutants grew as well as those carrying wild-type Cdc13 after more than 90 generations of growth from spore germination (Fig. 5 A).
FIG. 5.
Functional assays of Cdc13N dimerization and DNA binding mutants. (A) Cdc13N dimerization and DNA binding mutants expressed from the CDC13 promoter support normal growth in cdc13Δ mutants. CDC13/cdc13Δ diploids were transformed with the indicated plasmids, and haploid cdc13Δ cells were obtained via sporulation. The cells shown had grown for ∼90 generations from spore germination. WT, cdc13Δ cells carrying the wild-type CDC13 plasmid. (B) Telomere Southern blot analysis of Cdc13N point mutants that affect dimerization or DNA binding. XhoI-digested DNA was blotted and probed for telomere repeat DNA. Cells were obtained as described above, and two independent spore products were examined for each plasmid. Also shown are samples from wild-type cells (the same sample is loaded in lanes 1 and 23) and wild-type cells carrying the vector (lane 2).
To determine if the Cdc13N mutants affected recruitment or regulation of telomerase, we used Southern analysis to examine telomere length in the same cdc13Δ strains that carried plasmids expressing wild-type Cdc13 or the point mutants, using cells that had grown for ∼90 generations from spore germination. For the mutants that disrupted DNA binding (K73A, K75A, V133A, T140A, and F142A), telomeres were significantly lengthened (Fig. 5B, lanes 13 to 22 versus lanes 1 to 4 and 23), which suggests a defect in Cdc13-dependent telomerase inhibition. In contrast, the L84A and I87A dimerization mutants, and in particular the L91A and L84A/I87A mutants, caused a decrease in telomere length in cdc13Δ strains (Fig. 5B, lanes 5 to 12 versus lanes 1 to 4 and 23), which suggests a failure of mutant Cdc13 to efficiently recruit telomerase to the telomeres.
DISCUSSION
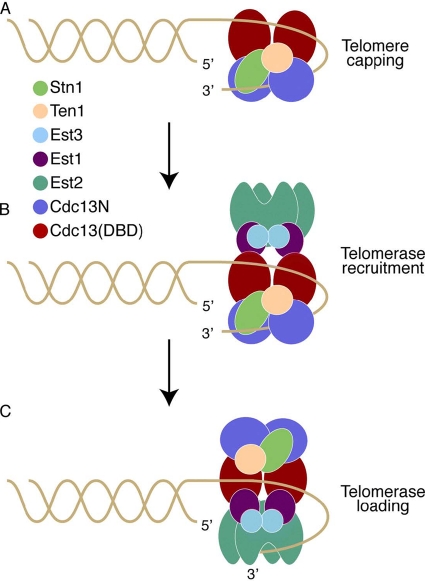
The CST complex, a replication protein A (RPA)-like complex now identified in many species (44), caps the ends of chromosomes, protecting them from exonucleolytic degradation and from being recognized and repaired as DNA breaks (Fig. 6 A) (16, 36, 49). Cdc13 also recruits telomerase and regulates its access to the telomeres via its interaction with Est1 (Fig. 6B and C), a process that requires displacement of Stn1 from the Cdc13 recruitment domain—a switch mediated by Cdk1-dependent phosphorylation of this site (13, 35, 38, 53). Protecting the ends of the chromosomes while allowing telomerase access to the telomeric overhang is most likely a complex mechanistic event that involves the coordination of multiple functional domains. For example, it would be necessary for the CST cap to be removed first from the end of the chromosome to allow for telomerase access to the telomeric overhang while simultaneously keeping the Cdc13-Est1-telomerase ternary complex attached to the chromosomes so that telomerase can then be loaded onto the telomeric overhang (Fig. 6C). Cdc13 most likely achieves this complex process via its multiple DNA binding domains, with one keeping the protein associated with the chromosomes, thus allowing for telomerase recruitment to the telomeres, and the second being involved in chromosome end capping (Fig. 6B). Once telomerase is in position to be loaded onto the telomeric overhang, the portion of the CST cap that sequesters the telomeric overhang and prevents access to telomerase is removed via the Hsp90 chaperone Hsp82, which alters the DNA binding activity of Cdc13 (10), thus allowing for efficient telomerase loading (Fig. 6C). The results presented here, together with existing biochemical data, clearly show that Cdc13, like its structural homologues OnTEBP and Pot1, contains multiple OB folds involved in telomeric DNA binding that may be involved in the above processes. The first identified OB fold of Cdc13 (DBD) is located toward the C-terminal portion of the protein and has a high affinity for telomeric DNA (3 pM) (3), comparable to that of the full-length protein (29). The fact that the DBD binds telomeric DNA with such high affinity suggests that it would require significant energy for its dissociation from the telomeres. In contrast, the N-terminal OB fold of Cdc13 (Cdc13N), presented here, has a weak DNA binding affinity (Kd, ∼600 nM) compared to that of the DBD, a characteristic that makes this domain ideal for fast and easy association with and dissociation from the telomeric overhang. Moreover, Cdc13N is located in proximity to the Est1-interacting site of Cdc13 (Fig. 1A) (47, 50), and therefore a free telomeric end would be easily accessible to telomerase. All of these characteristics suggest that the DBD of Cdc13 may act as an anchor that keeps the protein attached to the telomeres during telomerase recruitment and loading onto the telomeres, while Cdc13N may regulate telomerase access to the telomeres (Fig. 6). This hypothesis is supported by existing data showing that mutations of the Cdc13N domain lead to upregulation of telomere length but do not disrupt telomere binding (29, 53), a notion that is further supported by the in vivo data presented here, which also show that mutants of Cdc13N that disrupt DNA binding lead to telomere length upregulation (Fig. 5). Interestingly, mutants of the N-terminal OB fold of human Pot1 also lead to rapid telomere lengthening (39). Upregulation of telomere length upon disruption of the N-terminal DNA binding function of Cdc13 and Pot1 suggests increased access of active telomerase to this region for telomere elongation. Clearly, there is emerging evidence which suggests that protecting the ends of the chromosomes while regulating telomerase access to the telomeres involves the coordination of multiple OB folds provided by multisubunit protein complexes, such as the CST complex in yeast or the Pot1-TPP1 complex in humans (57), and this process appears to be conserved across species.
FIG. 6.
Model of Cdc13 action at the telomeres. (A) The Cdc13 dimer, together with Stn1 and Ten1, binds the single-stranded telomeric overhang and caps the ends of the chromosomes, most likely in late S phase, when the telomeric overhangs are long. Cdc13N (blue spheres) caps the chromosome end, while the DBD (red ovals) binds telomeric DNA upstream of this site. (B) Cdc13 recruits telomerase (Est2; green claw shapes) to the telomeres via its interaction with Est1 (magenta spheres). (C) Cdc13N dissociates from the telomeric overhang to allow telomerase access to the 3′ end of the DNA and initiation of telomere replication, while Cdc13 remains tethered to the telomeres via the DBD-DNA interaction.
Another interesting finding of this study is that Cdc13, like its structural homolog OnTEBPα, forms a stable homodimer. Interestingly, the CST structural homologue RPA does not form such an oligomeric structure. This could be explained by the fact that although the CST and RPA complexes share core structural similarities, they also contain significant sequence-related differences that most likely reflect the distinct functions of the two assemblies. For example, the RPA complex binds ssDNA with a high affinity and low specificity and is essential for various DNA transactions in eukaryotic cells (60). The CST complex localizes exclusively to the ends of the chromosomes and binds single-stranded telomeric DNA with a high affinity and specificity (3, 47). The CST complex is further distinguished from the RPA complex by the acquisition of an additional domain at the C terminus of Stn1, which confers a telomere-specific function (17). Cdc13 also contains a telomerase recruitment domain, located at the N terminus of the protein and adjacent to Cdc13N (50). The Cdc13N domain, which is absent in the RPA complex, is involved in Cdc13 dimerization.
Cdc13 dimerization appears to be related directly to N-terminal telomeric DNA binding (Fig. 4D). The Cdc13N dimer contains two OB folds, and therefore two DNA binding sites, which raises an important question: does the Cdc13N dimer bind one or two chromosome ends? The data presented here show that unlike the monomeric DBD of Cdc13, the Cdc13N dimer has a preference for long (>35 bases), single-stranded telomeric DNAs (Fig. 3). Moreover, disruption of the Cdc13N dimer via single or double alanine mutation resulted in an ∼20 to 90% loss of DNA binding affinity (Fig. 4C), clearly indicating that Cdc13N-dependent telomere binding is directly linked to oligomerization. Furthermore, the overall organization of the DNA binding clefts (both face the same side of the dimer), between the two OB folds of the Cdc13N dimer, constrains the distance between the 3′ and 5′ ends of the adjacent DNA binding sites to 35 Å (see Fig. S6 in the supplemental material). This means that a 12- to 13-base nucleotide would be required, at minimum, to span the two adjacent DNA binding clefts, making 35 nucleotides the total length of telomeric DNA required to bind between the monomers of a Cdc13N dimer. All of these characteristics suggest that a Cdc13 dimer binds a single strand of telomeric overhang (see Fig. S6), which in turn suggests that Cdc13N may act in late S phase, when the yeast telomere tails are of sufficient length for Cdc13N binding (59). It is worth noting that the yeast telomerase and its interacting partner Est3 function as a dimer (52, 62) and therefore contain two possible sites of interaction with Cdc13 via Est1 (Fig. 6B). The requirement of a Cdc13 dimer at the chromosome ends could therefore be explained by the fact that telomerase acts as a dimer and thus its efficient recruitment to the telomeres could be achieved only through dimeric Cdc13. This notion is supported by the fact that functional assays of single alanine mutants that disrupt Cdc13N dimerization led to telomere shortening, which suggests a failure of Cdc13 to efficiently recruit telomerase to the telomeres.
Supplementary Material
Acknowledgments
This project was funded by the Pennsylvania Department of Health, The Ellison Medical Foundation, The Emerald Foundation, and the NIA.
None of the authors of this work have a financial interest related to this work.
Footnotes
Published ahead of print on 13 September 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Allsopp, R.C., et al. 1992. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 89:10114-10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amberg, D. C., D. J. Burke, and J. N. Strathern. 2005. Methods in yeast genetics 2005. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 3.Anderson, E. M., W. A. Halsey, and D. S. Wuttke. 2002. Delineation of the high-affinity single-stranded telomeric DNA-binding domain of Saccharomyces cerevisiae Cdc13. Nucleic Acids Res. 30:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, N. A., D. Sept, S. Joseph, M. J. Holst, and J. A. McCammon. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackburn, E. H., and J. G. Gall. 1978. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 120:33-53. [DOI] [PubMed] [Google Scholar]
- 6.Bourns, B. D., M. K. Alexander, A. M. Smith, and V. A. Zakian. 1998. Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo. Mol. Cell. Biol. 18:5600-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunger, A. T., et al. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 8.Classen, S., J. A. Ruggles, and S. C. Schultz. 2001. Crystal structure of the N-terminal domain of Oxytricha nova telomere end-binding protein alpha subunit both uncomplexed and complexed with telomeric ssDNA. J. Mol. Biol. 314:1113-1125. [DOI] [PubMed] [Google Scholar]
- 9.Collaborative Computational Project 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 10.DeZwaan, D. C., O. A. Toogun, F. J. Echtenkamp, and B. C. Freeman. 2009. The Hsp82 molecular chaperone promotes a switch between unextendable and extendable telomere states. Nat. Struct. Mol. Biol. 16:711-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeZwaan, D. C., and B. C. Freeman. 2009. The conserved Est1 protein stimulates telomerase DNA extension activity. Proc. Natl. Acad. Sci. U. S. A. 106:17337-17342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 13.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 14.Evans, S. K., and V. Lundblad. 2002. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics 162:1101-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, S. K., and V. Lundblad. 2000. Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 113:3357-3364. [DOI] [PubMed] [Google Scholar]
- 16.Garvik, B., M. Carson, and L. Hartwell. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15:6128-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gelinas, A. D., et al. 2009. Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc. Natl. Acad. Sci. U. S. A. 106:19298-19303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillis, A. J., A. P. Schuller, and E. Skordalakes. 2008. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 455:633-637. [DOI] [PubMed] [Google Scholar]
- 19.Grandin, N., C. Damon, and M. Charbonneau. 2001. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J. 20:6127-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grandin, N., C. Damon, and M. Charbonneau. 2001. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 20:1173-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grandin, N., S. I. Reed, and M. Charbonneau. 1997. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 11:512-527. [DOI] [PubMed] [Google Scholar]
- 22.Greider, C. W., and E. H. Blackburn. 1987. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 51:887-898. [DOI] [PubMed] [Google Scholar]
- 23.Harley, C. B., A. B. Futcher, and C. W. Greider. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345:458-460. [DOI] [PubMed] [Google Scholar]
- 24.Holm, L., and J. Park. 2000. DaliLite workbench for protein structure comparison. Bioinformatics 16:566-567. [DOI] [PubMed] [Google Scholar]
- 25.Holm, L., S. Kaariainen, P. Rosenstrom, and A. Schenkel. 2008. Searching protein structure databases with DaliLite v. 3. Bioinformatics 24:2780-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holton, J., and T. Alber. 2004. Automated protein crystal structure determination using ELVES. Proc. Natl. Acad. Sci. U. S. A. 101:1537-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horvath, M. P., V. L. Schweiker, J. M. Bevilacqua, J. A. Ruggles, and S. C. Schultz. 1998. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell 95:963-974. [DOI] [PubMed] [Google Scholar]
- 28.Hsu, C. L., et al. 2004. Interaction of Saccharomyces Cdc13p with Pol1p, Imp4p, Sir4p and Zds2p is involved in telomere replication, telomere maintenance and cell growth control. Nucleic Acids Res. 32:511-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes, T. R., R. G. Weilbaecher, M. Walterscheid, and V. Lundblad. 2000. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl. Acad. Sci. U. S. A. 97:6457-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, M. L., J. J. Correia, D. A. Yphantis, and H. R. Halvorson. 1981. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys. J. 36:575-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, J. Y., J. L. Mogen, A. Chavez, and F. B. Johnson. 2008. Sgs1 RecQ helicase inhibits survival of Saccharomyces cerevisiae cells lacking telomerase and homologous recombination. J. Biol. Chem. 283:29847-29858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei, M., E. R. Podell, P. Baumann, and T. R. Cech. 2003. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature 426:198-203. [DOI] [PubMed] [Google Scholar]
- 33.Lei, M., E. R. Podell, and T. R. Cech. 2004. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 11:1223-1229. [DOI] [PubMed] [Google Scholar]
- 34.Leslie, A. G. W. 1992. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 ESF-EAMCB Newsl. Protein Crystallogr. no. 26.
- 35.Li, S., et al. 2009. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell 136:50-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, J. J., and V. A. Zakian. 1996. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl. Acad. Sci. U. S. A. 93:13760-13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, J. J., and V. A. Zakian. 1995. An in vitro assay for Saccharomyces telomerase requires EST1. Cell 81:1127-1135. [DOI] [PubMed] [Google Scholar]
- 38.Livengood, A. J., A. J. Zaug, and T. R. Cech. 2002. Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol. Cell. Biol. 22:2366-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loayza, D., and T. De Lange. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 423:1013-1018. [DOI] [PubMed] [Google Scholar]
- 40.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 41.Lustig, A. J. 2001. Cdc13 subcomplexes regulate multiple telomere functions. Nat. Struct. Biol. 8:297-299. [DOI] [PubMed] [Google Scholar]
- 42.Mitton-Fry, R. M., E. M. Anderson, T. R. Hughes, V. Lundblad, and D. S. Wuttke. 2002. Conserved structure for single-stranded telomeric DNA recognition. Science 296:145-147. [DOI] [PubMed] [Google Scholar]
- 43.Mitton-Fry, R. M., E. M. Anderson, D. L. Theobald, L. W. Glustrom, and D. S. Wuttke. 2004. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J. Mol. Biol. 338:241-255. [DOI] [PubMed] [Google Scholar]
- 44.Miyake, Y., et al. 2009. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 36:193-206. [DOI] [PubMed] [Google Scholar]
- 45.Miyoshi, T., J. Kanoh, M. Saito, and F. Ishikawa. 2008. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 320:1341-1344. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 47.Nugent, C. I., T. R. Hughes, N. F. Lue, and V. Lundblad. 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274:249-252. [DOI] [PubMed] [Google Scholar]
- 48.Paeschke, K., et al. 2008. Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat. Struct. Mol. Biol. 15:598-604. [DOI] [PubMed] [Google Scholar]
- 49.Paulovich, A. G., R. U. Margulies, B. M. Garvik, and L. H. Hartwell. 1997. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics 145:45-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 51.Petreaca, R. C., et al. 2006. Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat. Cell Biol. 8:748-755. [DOI] [PubMed] [Google Scholar]
- 52.Prescott, J., and E. H. Blackburn. 1997. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 11:2790-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qi, H., and V. A. Zakian. 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated Est1 protein. Genes Dev. 14:1777-1788. [PMC free article] [PubMed] [Google Scholar]
- 54.Surovtseva, Y. V., et al. 2009. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36:207-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terwilliger, T. C. 2003. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374:22-37. [DOI] [PubMed] [Google Scholar]
- 56.Theobald, D. L., and D. S. Wuttke. 2004. Prediction of multiple tandem OB-fold domains in telomere end-binding proteins Pot1 and Cdc13. Structure 12:1877-1879. [DOI] [PubMed] [Google Scholar]
- 57.Wang, F., et al. 2007. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445:506-510. [DOI] [PubMed] [Google Scholar]
- 58.Wellinger, R. J. 2009. The CST complex and telomere maintenance: the exception becomes the rule. Mol. Cell 36:168-169. [DOI] [PubMed] [Google Scholar]
- 59.Wellinger, R. J., A. J. Wolf, and V. A. Zakian. 1993. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72:51-60. [DOI] [PubMed] [Google Scholar]
- 60.Wold, M. S. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66:61-92. [DOI] [PubMed] [Google Scholar]
- 61.Xu, L., R. C. Petreaca, H. J. Gasparyan, S. Vu, and C. I. Nugent. 2009. TEN1 is essential for CDC13-mediated telomere capping. Genetics 183:793-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang, C. P., Y. B. Chen, F. L. Meng, and J. Q. Zhou. 2006. Saccharomyces cerevisiae Est3p dimerizes in vitro and dimerization contributes to efficient telomere replication in vivo. Nucleic Acids Res. 34:407-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yphantis, D. A. 1964. Equilibrium ultracentrifugation of dilute solutions. Biochemistry 3:297-317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.