Abstract
The miR-15/107 group of microRNA (miRNA) genes is increasingly appreciated to serve key functions in humans. These miRNAs regulate gene expression involved in cell division, metabolism, stress response, and angiogenesis in vertebrate species. The miR-15/107 group has also been implicated in human cancers, cardiovascular disease, and neurodegenerative diseases including Alzheimer’s disease. Here, we provide an overview of (1) the evolution of miR-15/107 group member genes, (2) the expression levels of the miRNAs in mammalian tissues, (3) evidence for overlapping gene regulatory functions by the different miRNAs, (4) the normal biochemical pathways regulated by miR-15/107 group miRNAs, and (5) the roles played by these miRNAs in human diseases. Membership in this group is defined on the basis of sequence similarity near the mature miRNAs’ 5′ end: all include the sequence AGCAGC. Phylogeny of this group of miRNAs is incomplete so a definitive taxonomic classification (for example, designation as a “superfamily”) is currently not possible. While all vertebrates studied to date express miR-15a, -15b, -16, -103, and -107, mammals alone are known to express miR-195, -424, -497, -503, and -646. Multiple different miRNAs in the miR-15/107 group are expressed at moderate-to-high levels in human tissues. We present data on the expression of all known miR-15/107 group members in human cerebral cortical gray and white matter using new miRNA profiling microarrays. There is extensive overlap in the mRNAs targeted by miR-15/107 group members. We show new data from cultured H4 cancer cells that demonstrate similarities in mRNAs targeted by miR-16 and miR-103, and also support the importance of the mature miRNAs’ 5′ seed region in mRNA target recognition. In conclusion, the miR-15/107 group of miRNA genes is a fascinating topic of study for evolutionary biologists, miRNA biochemists, and clinically-oriented translational researchers alike.
Introduction
Here we describe a group of microRNA (miRNA) genes with strong influences on human biology in health and disease: the miR-15/107 gene group. Basic information about miRNA genomics, biogenesis, processing, and mRNA target recognition will not be described here but many other sources are available1; 2; 3; 4; 5; 6; 7; 8. New data will be presented that focuses on specific biological aspects of miR-15/107 gene group members including the expression of individual miRNAs in human brain and their functional overlap in cultured cells. To provide introductory information about these homologous genes, we will first review prior studies related to miRNA evolutionary biology.
Heterogeneity and evolution of the miR-15/107 group of miRNA genes
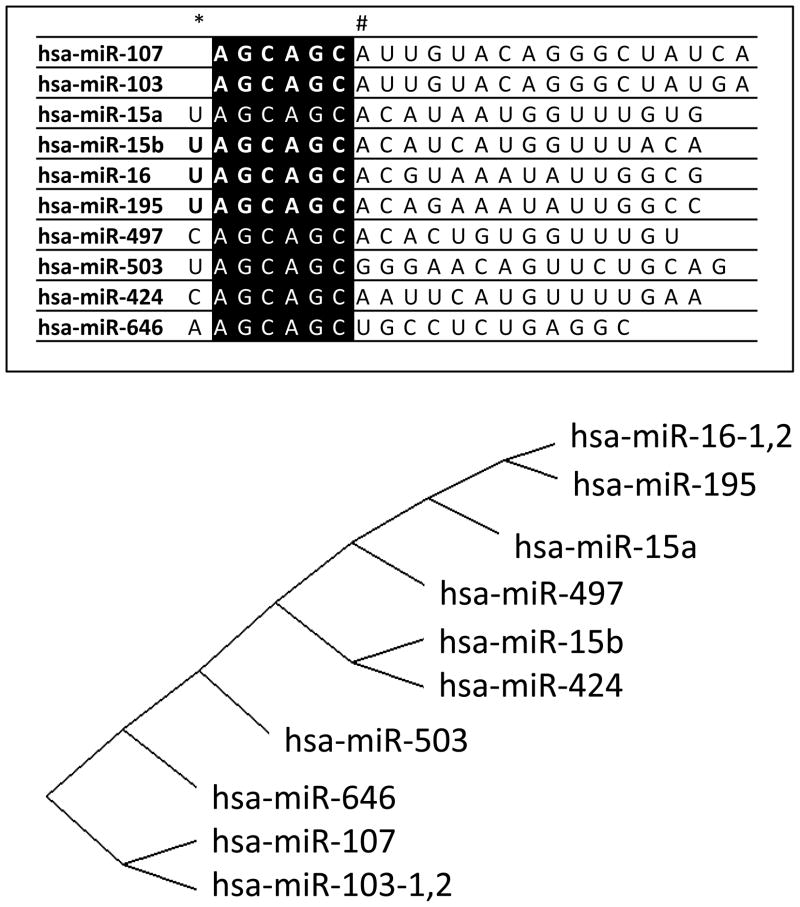
The 5′ end-portion of miRNAs is particularly important determinant of miRNA function 9; 10; 11; 12, and a group of vertebrate miRNAs shows 5′ end sequence similarity and also many common targets. There are 12 annotated human genes in the miRBase miRNA Registry13 with the sequence AGCAGC starting at either the first or second nucleotide (nt) from the 5′ end of the mature (~22nt single-stranded) miRNA, a motif which we refer to as AGCx2. The human AGCx2 miRNAs are shown in Figure 1, along with a diagram that demonstrates how the mature miRNAs cluster hierarchically according to sequence similarity. Half of the AGCx2 human genes (hsa-miR-15a14, hsa-miR-16-114, hsa-miR-16-215, hsa-miR-103-115, hsa-miR-103-215, and hsa-miR-10715) were originally identified based on studies in cultured HeLa cervical cancer cells16. The remaining six AGCx2 miRNA genes were identified in other human-derived cancer cell lines (hsa-miR-42417, and hsa-miR-64618) or from non-neoplastic tissues (hsa-miR-15b19, hsa-miR-19520, hsa-miR-49721, and hsa-miR-50321; 22).
Figure 1.
Members of the miR-15/107 miRNA gene group that are known to be expressed in humans. The list of genes (top) shows the 5′ AGCAGC sequence highlighted in black. The 5′ portion of mature miRNAs is known to confer specificity in terms of regulated mRNA targets. Note that the AGCAGC sequence starts at the first nucleotide (nt) of miR-103 and miR-107 but the second nt for the other genes in the group (*). Also note, most of the miRNAs highly-expressed in humans incorporate a 7-nt common sequence AGCAGCA (#). The sequences of miR-15/107 group members were compared using a web-based alignment tool (GeneBee; http://www.genebee.msu.su/services/phtree_reduced.html using the “Cluster” algorithm) that provide an unbiased graphic illustration of their sequence similarity (bottom).
Reconstructing the functional evolution of medically-relevant genes can help to predict the clinical consequences of the sequence variation we observe. However, we are unable to clearly delineate a taxonomy for the AGCx2 miRNA genes. A gene family is a group of genes with a common phylogenetic origin and possible functional homology23; 24. As a practical matter, the reconstruction of gene phylogenies is nearly always based on incomplete information. The miRBase miRNA Registry25 provides the most definitive miRNA annotation, yet this miRNA list is a work in progress with additional miRNAs still unidentified. Compounding the problem of missing data, there is no consensus about how miRNA gene families (as well as sub-families and superfamilies) should be defined. Current miRNA classifications recognize distinct miR-15, miR-16, miR-103, and miR-107 families 26; 27; 28; 29; 30; 31.
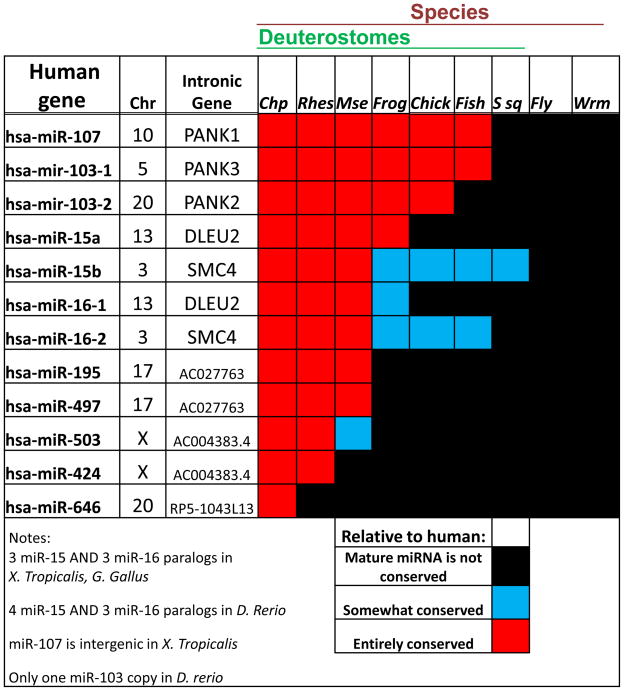
The known phylogenetic distribution of AGCx2 miRNAs suggests that they are unique to deuterostomes (a superphylum of animals that includes chordates but not insects or worms), and the number of AGCx2 genes has increased dramatically within the mammals32; 33; 34. No such miRNA has been described in a plant or non-chordate animal (Figure 2). Seven of the 12 AGCx2 miRNAs expressed in humans are widespread within the vertebrata, having been identified in other primates, rodents, birds, amphibians, and bony fishes. The other five are apparently mammalian specific, and miR-646 appears in humans and chimpanzees but has not been found in Rhesus monkeys. MiR-322 has only been identified in mammals other than humans, so will not be discussed here. Note that all twelve of the human AGCx2 miRNA genes are associated with protein-coding genes, or transcribed pseudogenes, compared to ~50% of human miRNA genes that reside in intergenic regions35; 36. As can happen with paralogs that arise via gene duplication, some genes are paired genomically in tandem: miR-15 paralogs with miR-16 paralogs, miR-424 with miR-503, and miR-497 with miR-195; these syntenies are evolutionarily conserved.
Figure 2.
Human genomic context including chromosome (Chr) and evolutionary conservation for the miR-15/107 gene group between human and Pan troglodytes (Chimpanzee; Chp), Macacca mulatta (Rhesus; Rhes), Mus musculus (Mouse; Mse), Xenopus tropicalis (Frog), Gallus gallus (Chicken; Chick), Danio rerio (Zebrafish; Fish), Ciona intestinalis (Sea squirt; Ssq), Drosophila melanogaster (Fruitfly; Fly), and Caenorhabditis elegans (Worm; Wrm). MiR-15/107 gene group members are only expressed in chordates, and tend to be located within or near a protein-coding gene or transcribed pseudogene. Mir-103 and miR-107 are entirely conserved in vertebrates whereas miR-15 and miR-16 show more variability. A number of miR-15/107 gene group members are mammalian specific and miR-646 is only known to be shared between humans and chimpanzees. Source: miRBase miRNA registry 13.
According to a recent phylogenetic analysis, miR-103 and miR-107 originated in the common ancestor of deuterostomes; Peterson and co-workers define a miR-103 superfamily that includes these two genes along with miR-201330. MiR-103 and miR-107 paralogs have been stable across vertebrate genomes, residing almost always in pantothenate kinase (PANK) gene introns. One exception: in the frog, Xenopus tropicalis, miR-103 is in a PANK gene intron but miR-107 is located intergenically. PANK genes encode polypeptides that are important in metabolism-related cell functions37 (see below). Intronic miRNAs tend to be co-expressed with their protein-coding ‘mother’ genes although this is not always the case38; 39; 40.
A miR-15 superfamily, comprising miR-15a, miR-15b, miR-16, miR-195, miR-322, miR-424, miR-457, and miR-497 are hypothesized to have evolved in the common ancestor of vertebrates 32; 33; 34. There are two miR-15/16 clusters in mammals. Many vertebrate species have three or more paralogs of miR-15 and miR-16 as shown in Figure 2. The location of miR-15a/16 in the DLEU2 (“deleted in leukemia 2”, a non-coding transcript that affects cell proliferation41; 42) gene seems to have evolved more recently. It is tempting to speculate that the physical location of miR-15/16 (located within chromatin/DNA and cell proliferation-related genes SMC4 and DLEU2) and miR-103/107 (located within cellular metabolism-related PANK genes) along with their known overlap in targets (see below), provide clues that these genes serve functional roles linked to cross-talk between the key cellular functions of cell division and metabolism.
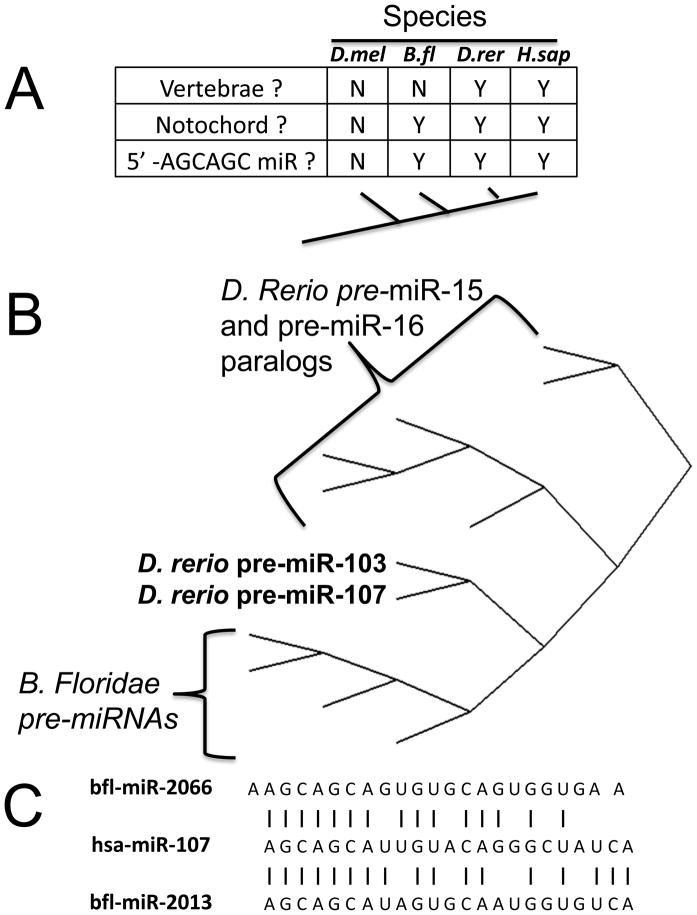
Cephalochordates are generally regarded as the sister group of vertebrates 6, and the model cephalochordate, Branchiostoma floridae, expresses four miRNAs with 5′-end AGCAGC motifs6; 43 (Figure 3). The B. floridae mir-2013 is most similar to human miR-107, with 17/23 (74%) of the mature miRNAs’ nts matching exactly, including the 5′-end eight nts. A comparison between the different pre-miRNAs in Danio rerio and B. floridae show that the zebrafish miR-103 and miR-107 cluster in an intermediate position between the B.floridae pre-miRNAs and the zebrafish miR-15/16 pre-miRNAs 44.
Figure 3.
The miR-15/107 gene group is chordate-specific but exact phylogenetic relationships are elusive A. Table compares some features of Drosophila melanogaster (Fruitfly; D.mel), the amphioxus cephalochordate Branchiostoma floridae (Lancelet; B.fl), Danio rerio (Zebrafish; D.rer), and Homo sapiens (H.sap). Note B. floridae, a relatively close evolutionary relative of vertebrates, does express four miRNAs with 5′AGCAGC sequence. B. A phylogenetic tree was created to evaluate the similarities in the sequences of the ~70 nucleotide (nt) pre-miRNA sequences of B. floridae and D.rerio, the former a relatively late-evolving pre-vertebrate species and the latter a relatively early-evolving vertebrate species. This phylogenetic tree shows that the miR-103 and miR-107 genes have sequences with intermediate similarity between the B. floridae pre-miRNAs and the other members of the gene group expressed in D.rerio. C. In addition to the apparent homology of the pre-miRNAs, the sequence of the mature miR-107 has the closest similarity to mature B. floridae miRNAs. Note that between vertebrates’ miR-107 and B. floridae bfl-mir-2013 17/23 (74%) of the mature miRNAs’ nts matching exactly.
Urochordates are generally regarded as the sister group to vertebrates plus cephalochordates. The sea squirt Ciona intestinalis appears to have lost (or never expressed) miR-103 and miR-107 genes. C. intestinalis has only been shown to express a single AGCx2 miRNA, which resembles miR-15 and resides in an intron of an SMC4 (“structural maintenance of chromosomes 4”) ortholog. The genomic location in C. intestinalis SMC4 is significant because as stated above, a miR-15 ortholog also is expressed in a SMC4 intron in vertebrates. SMC4 is an essential, evolutionarily-conserved ATPase that participates in DNA/chromatin dynamics45; 46. More distantly-related species such as Drosophila and Caenorhabditis express no AGCx2 miRNAs.
We conclude that the origins of the miR-15 and miR-107 genes remain an unsolved puzzle for now. We cannot reject definitively the possibility that the miR-15 superfamily members had an independent origin in an early vertebrate ancestor, and they have converged in some respects with a separately-evolved miR-107 superfamily. However, it also is possible that the miR-15 superfamily and the miR-107 superfamily evolved from a common ancestor possessing a 5′-end AGCAGC sequence. If so, these miRNAs may exhibit functions that have been retained from their common ancestor and it would be appropriate to recognize the “miR-15/107 gene superfamily.” The existence of functional overlap between human miR-15/-16 and miR-103/-107 genes lends credibility to this hypothesis (see below). In either case, insights into the biology of the ancestral member of this group of genes could be gleaned from functional analyses of miRNAs in non-vertebrate deuterostomes. Other insights can be gained by studying the function and expression of these intriguing miRNAs in mammals.
Expression of the miR-15/107 group in mammalian tissues
MiRNA profiling experiments have shown that miR-15/107 group miRNAs are expressed at moderate-to-high levels in many mammalian tissues. Here we are distilling a large, sometimes contradictory literature and readers are encouraged to refer to the primary sources for more detailed information. There are technical challenges in assessing these genes’ expression levels, and a wide range of methods were used in preparing and profiling the miRNAs in the various studies. Further, in many miRNA profiling platforms there is cross-reactions in terms of closely-paralogous mature miRNAs such as miR-103 and miR-107.
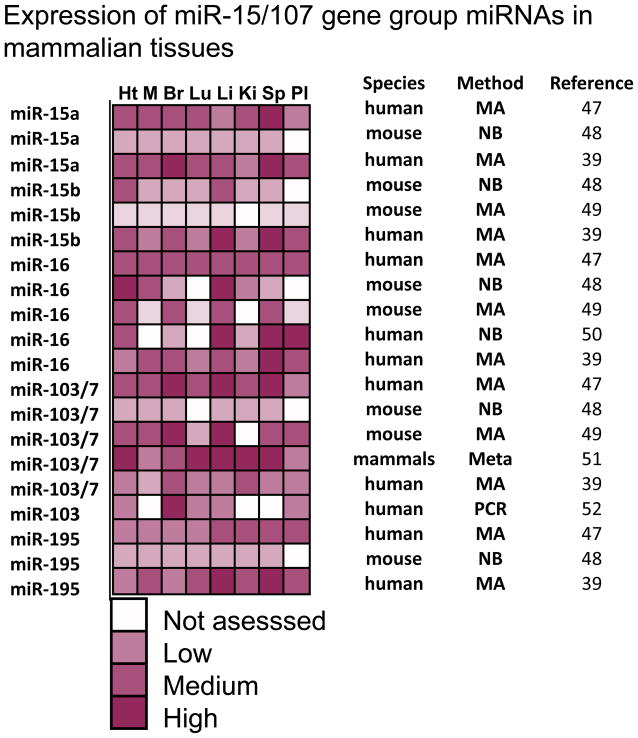
Perhaps due to the abovementioned technical factors, there has been inconsistency among prior studies in terms of miR-15/107 gene group member expression profiling in mammals. Figure 4 shows results from a subset of prior studies that evaluated miRNA expression in multiple mammalian tissues including heart, skeletal muscle, brain, lung, liver, kidney, spleen, and placenta39; 47; 48; 49; 50; 51; 52. There may be similar expression profiles among some group members across human tissues (miR-15, miR-16, and miR-195) 39, but this is not always seen. Some but not all studies have indicated that miR-15/miR-16 expression is relatively high in hematopoietic cells including T lymphocytes47; 48; 51; 53; 54; 55; 56. Published data is also discrepant as to whether miR-103/7 levels are highest in brain39; 48; 49; 52; 56; 57 and whether miR-195 is expressed at highest levels in lung47; 56. Nor is there a clear-cut consensus for the miR-15/107 group member(s) that tend to have highest expression. The complexity of this situation is illustrated by Ason et al58 who used in situ hybridization to study embryos from different species, finding that miR-107 is essentially brain-specific in the Japanese killifish, highly expressed in almost all tissues in chicken, but neither pattern is observed in mouse. Bargaje et al51 identified a subset of 18 miRNAs that are “constitutively” expressed in all mammalian tissues. From the miR-15/107 group, only miR-103 and miR-107 were on this list, which may indicate that the other miRNAs have more specialized functions. Distilling all these observations, the miR-15/107 gene group of miRNAs can be described as ubiquitous in that as far as we know there has not been a human cell type described that lacks expression of at least one miR-15/107 gene group member.
Figure 4.
Expression profiling of selected miR-15/107 gene group members from prior studies in human and mouse tissues. Profiling was performed from heart (Ht), skeletal muscle (M), brain (Br), liver (Li), lung (Lu), kidney (Ki), spleen (Sp), and placenta (Pl). The amount of miRNAs expressed is displayed in a semi-quantitative way from negligible/nondetected to very high expression. White squares indicate that the miRNAs were not evaluated in that tissue in the cited study. A variety of different profiling platforms is represented: microarray (MA), northern blots (NB), PCR, and a meta-analysis of other profiling studies (Meta). The semi-quantitative scoring reflects a subjective interpretation from the primary sources cited in the right-hand column. The results could not be mapped in a purely quantitative manner due to the nature of the data (for example, NBs yield no quantitative readout), known differences in dynamic ranges, and differential specificities to individual miRNAs. Note that there is no obvious tissue-specific pattern of expression that has been found across the different studies. However, the multiple miR-15/107 gene group members are consistently found to be expressed in multiple mammalian tissues.
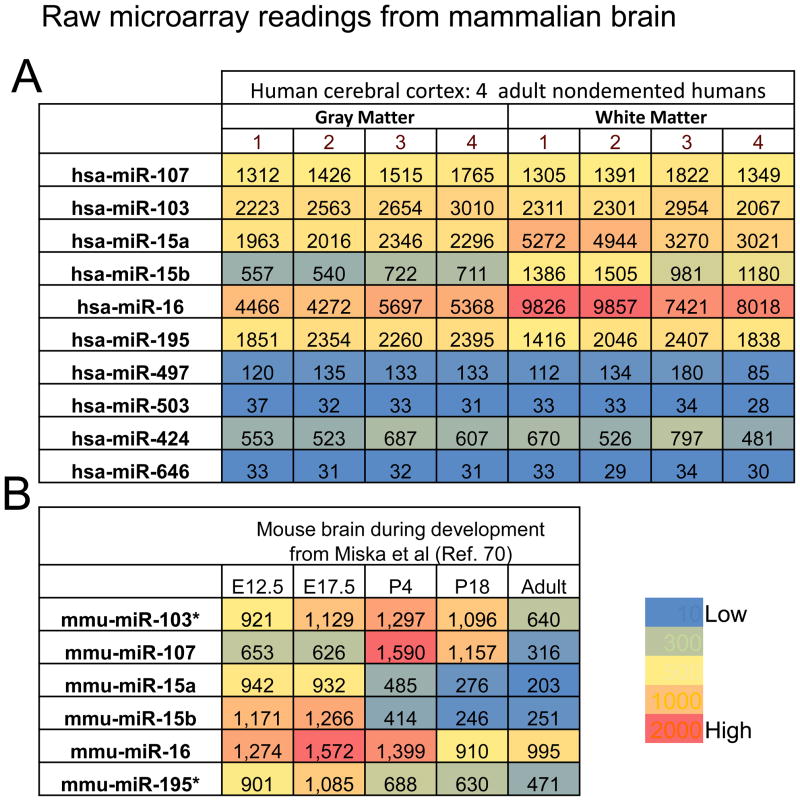
In order to include some of the more recently annotated miR-15/107 group genes in our discussion, the results of recent expression profiling of miR-15/107 group members in human brain samples is displayed in Figure 5 (superior and middle temporal gyri, N=4 each from nondemented individuals older than age 80). These were evaluated using a locked nucleic acid (LNA)-based microarray and here raw data numbers from the microarrays are described. The results were different for gray matter and white matter -- the latter contains more oligodendrocytes and fewer neurons. Note that miR-16 is the highest-expressing miRNA according to this platform and is expressed at highest levels in white matter. Using a different microarray platform we found that miR-107 was the highest expressing group member in similarly processed brain tissue 59; this discrepancy is not surprising. Researchers from many laboratories have now found that miRNA profiling results obtained using different platforms can vary dramatically for individual miRNAs despite good overall inter-platform correlation60; 61; 62; 63; 64. Notwithstanding these technical issues, an interesting correlation is observed: the miRNAs apparently expressed at highest levels in human tissues (miR-15a, miR-15b, miR-16, miR-103, miR-107, and miR-195) include a 7-nt common sequence: AGCAGCA (see Figure 1).
Figure 5.
Raw microarray data from mammalian brain experiments help illustrate particular aspects of miR-15/107 gene group member expression. A. Results of a new experiment using the LNA-microarray including the relatively recently characterized genes miR-497, miR-503, miR-424, and miR-646. These data include readings from four aged nondemented persons and the RNA was isolated from the human superior and middle temporal gyri (Brodmann Areas 21 and 22). White matter and gray matter were dissected and analyzed separately. Note that all the highly-expressed miRNAs in mammals include the sequence AGCAGCA at the 5′ end (see Figure 1). Also note that the gray matter and white matter have different expression for some miRNAs (especially miR-15a, miR-15b, miR-16 which are more highly expressed in white matter). B. Microarray results at different developmental time points from the microarray study by Miska et al 70. Mouse brains were studied at embryonic day 12.5 and 17.5, postnatal days 4 and 18, and adults. Note that most members of the miR-15/107 gene group had highest expression at some pre-adult developmental stage. The data for miR-103 and miR-195 (as shown with *) are averages of two different results.
In addition to tissue- and cell-specific factors, developmental stages constitute another layer of gene expression complexity. Recall that animal miRNAs were discovered in the context of stage-specific expression during worm development65; 66; 67. There are altered expressions of miR-15/107 group members in certain stages of mammalian development also. For example, miR-15 and miR-16 expression correlates to particular stages in erythropoiesis26; 68. Assessing archival human brains that had been formalin-fixed and paraffin-embedded we found that miR-16 expression is highly expressed in early human brain development 69. Figure 5B shows the results from Miska et al70, which provides compelling evidence that multiple miR-15/107 group members have increased expression levels during early mouse brain development.
We conclude the following about the expression of miR-15/107 group members in mammalian tissues:
Expression of multiple miR-15/107 group miRNAs are moderate-to-high across many tissues including cells derived from mesoderm, ectoderm, and endoderm;
Gene expression among group members tends not to be purely specific to particular tissue or cell types;
MiR-15/107 group gene expression is relatively high in some early developmental stages;
In humans the highest-expressing miRNAs tend to be miR-15a, miR-15b, miR-16, miR-103, and miR-107 (all of which contain the sequence AGCAGCA), with the caveat that some miRNAs were not assessed in earlier studies. Thus, there are important similarities between the highly-expressed miR-15/107 group members.
MiR-15/107 group miRNA processing and mRNA targeting
Clues related to the evolution and gene expression of miR-15/107 group members support the idea of a homologous gene group, but what about the functions of the mature miRNAs? There is compelling evidence that different members of the miR-15/107 group regulate overlapping lists of mRNA targets. This is presumably because the AGCAGC(A) sequence near the miRNAs’ 5′ end controls the mRNA target specificity. Some studies have focused on the mRNA targeting overlap between the “miR-15/16 group” paralogs such as miR-15a, miR-15b, miR-16, miR-195, miR-424, and miR-50371; 72; 73. Yet there is also functional overlap even in the more dissimilar group members, miR-103 and miR-10774; 75. In a rigorous and systematic analysis, Linsley et al27 performed mRNA-detecting microarray studies after miRNA transfections in cultured cancer cell lines. They concluded that “(miR-15a, miR-16, and miR-103) are members of a single miRNA group”. Linsley et al27 also note that transfections with “miR-15a, miR-16, and miR-195 also gave nearly identical expression profiles, but miR-103 gave an overlapping but distinct profile compared to other members of this group, consistent with the 1-nt offset of the miR-103 seed sequence.” More specifically: “Only a subset of [consensus miR-16 responsive] transcripts was regulated by miR-103 and miR-107. These transcripts were not regulated in cells transfected with miR-16 duplexes having mismatches at positions 2 and 3 or 4 and 5, but their regulation was unaltered in cells transfected with duplexes with mismatches at positions 18 and 19 and 19 and 20.” 27
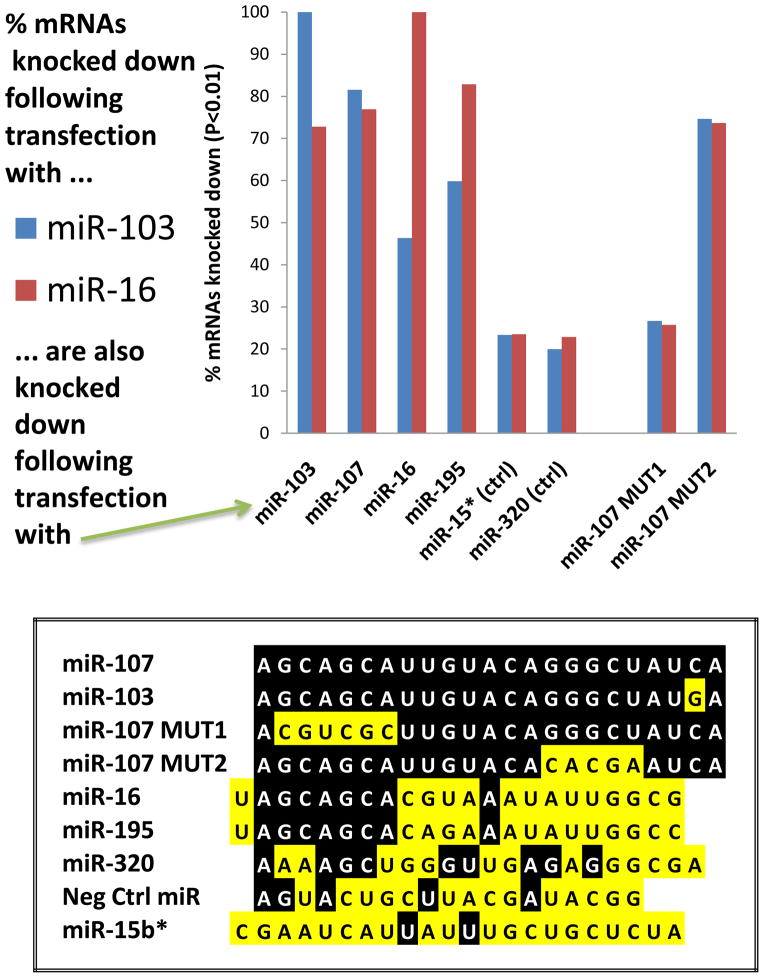
We compared experimentally the effects of transfections using different miR-15/107 group members in cultured cancer cells (Figure 6). Studies were performed using H4 “glioneuronal” cells using methods that we recently described in detail76; 77. For these experiments, an individual mRNA was considered to be a target of miRNA X if the level of that mRNA was significantly reduced following a miRNA transfection with miRNA X, relative to the level of that mRNA after transfection with a Negative control miRNA (sequence shown in Figure 6). Three transfections were performed for each miRNA. We found that mRNAs that are knocked down after transfection by miR-107, miR-103, and miR-16 tend to overlap substantially. Note that in this experiment the correlation was best (in terms of mRNAs knocked down) between close paralogs miR-103 and miR-107, but even between miR-103 and miR-195 the correlation was better than to controls. MiR-15b* was used as a control miRNA because its sequence (roughly antisense to miR-15b) has similar G/C content. Two additional non-physiological controls (miR-107MUT1 and miR-107MUT2; see Figure 6) were used that have been described before75. Inclusion of these controls helped to strongly support the importance of the miRNA 5′ seed portion in terms of dictating mRNA targeting specificity, because the miR-107MUT2, lacking the 5′ seed sequence, does not impact the same mRNAs as miR-103 or miR-16. It should be emphasized that these data describe mRNAs downregulated in lysates following miRNA transfections, rather than co-immunoprecipitated definitive “targets”, so some of these downregulated mRNAs may represent downstream effects. However, these results underscore the appreciable overlap between different miR-15/107 group members in terms of their biochemical impact.
Figure 6.
There is considerable overlap in the mRNAs targeted by various miR-15/107 gene group members in cultured H4 glioneuronal cancer cells. Transfections with “RIP-Chip” experimental design were performed as previously described in detail 75; 77. Briefly, H4 cells (American Type Culture Collection, Manassas, VA), were cultured under the vendor’s recommended conditions and plated at a density of 2.5×106/10cm plate a day before transfections. Cells were transfected with 25 nM of exogenous RNA duplexes (Ambion, Austin, TX) with sequences shown on bottom, using RNAiMAX (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions. Transfections with “miR-107MUT” artificial miRNAs was recently described75. Cells were harvested 48 hr after transfection, the RNA was isolated, converted to cDNA and profiled using Affymetrix 1.0 ST Gene microarrays according to manufacturer’s instructions. A total of 21,898 annotated genes were profiled. Genes were evaluated in which mRNA levels were lower after miR-103 transfection (N=1484 genes; blue bars) or after miR-16 transfections (N= 945 genes; red bars) relative to the “Neg control” miRNA transfections (N=3 biological replicates for each transfection). Comparisons were performed using unpaired Student’s t-test with a P<0.01 cutoff. Note that a majority of genes knocked down by miR-103 were also knocked down by close paralog mir-107 and the miR-107MUT2 reagents, and also miR-16 and miR-195. Similarly, genes knocked down by miR-16 tended to also knocked down after miR-195 and somewhat less so with miR-103, miR-107, and miR-107MUT2 but not controls. These results strongly support the importance of the 5′ AGCAGC sequences in determining which mRNAs are targeted by genes in the miR-15/107 gene group.
Before discussing biological pathways and individual targets affected by miR-15/107 group genes, we note that miRNAs work through multiple distinct mechanisms78; 79; 80; 81; 82. There are special considerations related to mRNA targeting by genes in the miR-15/107 group. For example, miR-107 may act preferentially by systematically targeting sequences in mRNAs’ open reading frame, as opposed to their 3′ untranslated region12. It has been established that miRNAs may target the protein-coding sequence of target mRNAs83, with ~25% of miRNA targets in mouse brain mRNA corresponding to protein-coding sequences according to high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) data84. However, miR-107 is the first example of an individual miRNA that may recognize protein-coding portions of target mRNAs systematically. The phenomenon (a focus of ongoing work in the Nelson laboratory) was discovered using RIP-Chip technique in H4 cells and will require confirmatory studies in other cells, tissues, and organisms.
Beyond mRNA targeting, there are additional levels of complexity that distinguish the miR-15/107 gene group. MiR-107 causes feedback inhibition of the entire miRNA processing machinery by repressing expression of Argonaute proteins with profound global changes in miRNA processing 77. Martello et al reported that miR-107 also targets Dicer, the exonuclease that processes precursor miRNAs85. The interplay between miR-107 and the miRNA-processing machinery is additionally complicated, because the miR-107 gene contains a specific sequence motif that controls how the precursor miRNA molecule is processed by Dicer, and thus how the mature miRNA is expressed. This novel mechanism, with special relevance to miR-107 and only a few other miRNAs, is associated with the uridylyl transferase protein “TUTase 4” (TUT4)86. It is currently unknown whether these phenomena relate to the abovementioned tendency of miR-107 to target mRNAs’ protein-coding sequences. We are also only beginning to understand the complex mechanisms regulating transcription of genes in the miR-15/107 group42; 87; 88; 89. In sum, the many unanswered questions pertaining to miR-15/107 group members’ biological chemistry are an important consideration as we begin to assess the kaleidoscopic extant data about the hypothesized functions of miR-15/107 gene group members.
The biochemical impact of a well-expressed mature miRNA is thought to relate to the “sum of its targets”--the aggregate mRNAs targeted weighted by the strength of each interaction. Computational predictive methods may provide information but these methods entail some shortcomings; computational algorithms differ amongst themselves and each is anchored in prior knowledge90; 91. For a bench researcher wishing to prove that a particular miRNA targets a specific mRNA, the experimental strategies usually involve a combination of the following:
miRNA transfection or over-expression→ down-regulation of target (mRNA and/or protein);
In vivo or cell culture miRNA knockout/knockdown→ up-regulation of target (mRNA and/or protein);
reporter assays with subcloned 3′UTR sequences help assess sequence specificity and confirm direct interaction;
and target(s) co-purify with anti-Argonaute co-immunoprecipitation with or without RNA cross-linking.
Each of these methods entails both benefits and difficulties. In terms of potential pitfalls, the physiological relevance and specificity of cell culture miRNA perturbation techniques bears critical scrutiny due to the intrinsic artificiality of the experiments, and reporter assays tend to depend on mechanisms that are not identical to the target mRNAs’ biochemical contexts91; 92; 93; 94. Co-immunoprecipitation methods entail other assumptions (such as the stability of the miRNA:mRNA particles) and may only capture a subset of miRNA:target interactions91. High-throughput methods such as HITS-CLIP and PAR-CLIP84; 95; 96 may be more indicative of the wide web of miRNA effects than lower-throughput tools because the -CLIP methods identify miRNA recognition sites transcriptome-wide. However, every current technique for miRNA target identification is at least somewhat biased by current dogma and there are probably many interactions yet to be identified. In sum, each and every experimental method – as with each computational method – includes potential limitations. These caveats are not mentioned out of pessimism but as a reminder that the field of miRNA biology is still very much in flux with many exciting discoveries ahead. In that spirit, we provide updates of various “works in progress” pertaining to miR-15/107 group members.
Biological functions of the miR-15/107 group
A. Cell division
Cellular pathways related to mitosis in vertebrate species are partly regulated by miRNAs in the miR-15/107 gene group. Most studies have focused on the impact of miR-15 and miR-16 paralogs on cell division, but other miR-15/107 group members have also been studied in this context. Linsley et al27 described this group of miRNAs as the “miR-16 group” and noted that miR-15a and miR-16 (and to a lesser extent miR-103) target a highly disproportionate number of cell cycle genes and transfection with these miRNAs leads to G0/G1 arrest. Specifically, for miR-103, miR-15a, and miR-16, the percentage of targets that were involved in cell cycle regulation were 12%, 14%, and 22% respectively as determined using microarray data after miRNA transfection with correlation to GO process terms, and these percentages were much higher than would have been expected by chance27. Other labs have confirmed that miR-15/107 group members (again usually focusing on miR-15 and miR-16) target cell cycle-related genes (and see below) 27; 28; 42; 97; 98; 99. MiR-107 transfection also leads to decreased rates of cell division with cell cycle arrest 100. The biological significance of miR-15/107 gene group-mediated cell cycle arrest is still not known but is an area of active enquiry partly because of the obvious implications in cancer biology as discussed below.
B. Cellular metabolism and stress
Numerous lines of evidence indicate that miR-15/107 group genes participate in pathways involved in cellular metabolism. Tang et al101 studied miRNA expression changes in response to glucose, using a cDNA array to test the effects of 20 mM pyruvate, 1 or 25 mM glucose in the cell medium of pancreatic β-cell line MIN6. A handful of miRNAs were up-regulated following increased extracellular glucose, including miR-107. We subsequently confirmed this effect of glucose on miR-107 expression using a nearly identical experimental design but with cultured H4 cells75. MiR-107 is also up-regulated in cells exposed to folate deficiency102, and dietary intake of various lipids altered specifically miR-107 expression in the colonic epithelium and cancerous tissue from mice103. Further, mice forced to perform endurance exercise had over 50% increased miR-107 expression in quadriceps femoris muscle tissue relative to a sedentary control group (this was the largest increase of any miRNA tested)104.
There are other hints that the miR-15/107 group genes are involved in metabolic pathways. In vertebrate species, miR-103 and miR-107 paralogs reside in introns of genes that also encode PANK. PANK enzymes catalyze the universal rate-determining step in the formation of Coenzyme A (CoA). CoA participates in many metabolic reactions105; 106. MiR-103/107 paralogs and PANK genes may be coordinately transcribed gene products with synergistic effects in regulating cellular Acetyl-CoA levels37. Bioinformatics predict that miR-103/107 paralogs exhibit an exceptional tendency to target mRNAs in a pattern suggestive of synergy with PANK function. Like PANKs, miR-103/7 may function to increase acetyl-CoA levels available for the Krebs Cycle, and miR-103/7 may also inhibit the synthesis and metabolism of cellular lipids37. Although the research to date cannot be fully harmonized into a unitary framework, miR-15/107 group members clearly are both responsive to, and impactful on, metabolic pathways in human cells.
It may be artificial to consider biological mechanisms separately from each other because in a cellular context these pathways interact dynamically. For example, perturbations in cellular metabolism can cause cell stress and vice versa. Multiple experimental stress paradigms are associated with changed expression of miR-15/107 group members. In cultured cells, miR-16 expression increases after ultraviolet light damage and then seems to participate in regulating the DNA-damage response and cell division machinery107. It has also been shown that exposure to mice of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), a common environmental toxicological contaminant, led to decreased miR-15 and miR-16 expression108. Members of the miR-15/107 group have not been shown to be aberrantly regulated in heat stress109 which may indicate a more focused cellular role. MiR-107 expression is down-regulated following rodent traumatic brain injury (TBI)110. We have found that TBI-related miR-107 down-regulation may trigger increased GRN expression in mouse hippocampi after TBI75, which may be an adaptive response in that context. The miR-15/107 group may also help regulate cells’ response to ischemic stress: miR-107 and miR-424 are downregulated in hypoxia111; 112, and miR-15a and miR-497 may help cells respond to oxygen deprivation in vivo and cell culture models113; 114. It remains to be seen how these cell stress pathways relate to the other physiological pathways miR-15/107 group members participate in.
C. Angiogenesis
An exciting recent discovery is the connection between miR-107 and angiogenesis. Angiogenesis, the dynamic process of growing new small blood vessels from existing vessels, is directly relevant to development, neoplasia, neurobiology, and various healthy responses to stress. Yamakuchi et al88 reported that miR-107 is regulated transcriptionally by P53 protein and in turn miR-107 inhibits HIF-1beta (also known as aryl hydrocarbon receptor nuclear translocator, ARNT). Through this pathway, miR-107 regulates colon cancer tumor growth through a mechanism related to angiogenesis. This study provides insights into upstream regulation of miR-107: agents were identified that up-regulate miR-107 expression through specific p53-responsive promoter regions. Thus, a small non-coding RNA neither necessary nor sufficient to subserve any cellular function is hypothesized to represent a key regulatory intermediary connecting p53 expression, hypoxia, angiogenesis, and the clinical-biological behavior of colon cancer. These changes may be synergistic with other miR-15/107 group members. For example, it has been shown that miR-16 can regulate VEGF strongly in angiogenesis115. As in other important biological processes, it can be assumed that miRNAs in addition to those of the miR-15/107 group play roles in angiogenesis. Specifically, miR-17-92 cluster, miR-378, miR-143, miR-145, miR-93a, miR-27b, miR-130a, let-7f, miR-221, and miR-222 have been shown to participate in blood vessel development in normal and/or tumor tissues116; 117.
In summary, the ubiquitously expressed miR-15/107 gene group may help orchestrate gene expression adaptively in boundary areas between important human cellular pathways (e.g., cross-talk between hypoxia with angiogenesis, and linking cell proliferation with metabolic needs). The known functions of this miRNA gene group dovetail intriguingly with the hypothesis that miRNAs provide cells with a “safeguard against turmoil”118 by regulating the stability and translation of mRNA. However, any adaptive mechanism can go awry. Thus it comes as no surprise that dysregulation of miR-15/107 group gene expression may contribute to, or even cause, human diseases.
Human diseases
A. Neoplasia
An important role for genes in the miR-15/107 gene group has been described in human cancers. Multiple miRNAs from this group have been implicated, spanning a broad range of tumor types. Table 1 summarizes miR-15/107 group gene expression changes that have been found in human neoplastic conditions. In seminal studies, Calin et al119; 120; 121 established that the miR-15a and miR-16 tandem gene locus was the important genomic susceptibility factor within the minimal deletion region that determines susceptibility to chronic lymphocytic leukemia (CLL), as had been hinted at by Lagos-Quintana et al20. Subsequent studies have produced a compelling corpus of experimental evidence to suggest that miR-15a/miR-16 constitute key tumor suppressors whose deletion contribute to cancer. Genes with mRNAs targeted by miR-15a and miR-16, and thought to have clinico-biological impact for CLL patients, include BCL-2, CCND3, CCNE1, CDK6, CAPRIN1, and HMGA127; 28; 42; 97; 98; 99. This is an active area of ongoing research and recent reviews are available122; 123; 124.
Table 1.
MiR-15/107 gene group may be potential cancer prognostic biomarkers, either up-regulated (“oncomiRs”) or down-regulated (tumor suppressors) in tumor cells
| miRNA expression is a possible prognostic biomarker | Reference(s) | |
|---|---|---|
| miR-16, miR-195 | Esophageal carcinoma | 159 |
| miR-15a, miR-16 | Myelodysplastic syndrome | 160 |
| miR-16 | Acute lymphoblastic leukemia | 161 |
| miR-103/7 | Esophageal carcinoma | 162 |
| miR-195 | Adrenocortical carcinoma | 163 |
| miR-15b | Hepatocellular carcinoma | 164 |
| miRNA expression increased in tumor vs. normal tissues | ||
| miR-16 | Head and neck carcinoma | 165 |
| miR-16 | Renal cell carcinoma | 166 |
| miR-15b, miR-16 | Cervical carcinoma | 167 |
| miR-503 | Retinoblastoma | 168 |
| miR-424 | Colonic carcinoma | 169 |
| miR-15b | Colonic carcinoma | 170 |
| miR-15b | Malignant melanoma | 171 |
| miRNA decreased in tumor vs. normal tissues | ||
| miR-15a, miR-16 | Prostatic carcinoma | 172; 173; 174 |
| miR-15a, miR-16 | Chronic lymphocytic leukemia | 120; 175 |
| miR-15a, miR-16 | Non-small cell lung carcinoma | 97 |
| miR-15a, miR-16 | Pituitary adenomas | 176; 177 |
| miR-15a, miR-16 | Ovarian carcinoma | 178; 179; 180 |
| miR-107 | Colonic carcinoma | 103 |
| miR-107 | Head and neck carcinoma | 181 |
| miR-107 | Acute promyelocytic leukemia | 182 |
| miR-107 | Pancreatic carcinoma | 89; 183 |
| miR-107 | Tongue squamous cell carcinoma | 184 |
| miR-195 | Hepatocellular carcinoma | 185 |
| miR-195 | Bladder carcinoma | 186 |
| miR-195 | Gastric carcinoma | 187 |
| miR-195 | Chronic lymphocytic leukemia | 188 |
| mir-195, miR-397 | Primary peritoneal carcinoma | 189 |
| miR-497 | Male breast carcinoma | 190 |
| miR-503 | Parathyroid carcinoma | 191 |
The roles of miRNAs in cancer can provide new insights into disease mechanisms and the rich complexity of mammalian gene regulation. Lee et al reported that promoter methylation may induce down-regulation of miR-107 in pancreatic carcinomas89. We also note that granulin (GRN), which is regulated by miR-15/107 gene group members, is an active mitogen and growth factor relevant to many cancers 125; 126; 127; 128; 129; 130. It remains to be seen if the impact of miR-107 in pancreatic and other cancers is mediated partly through GRN. Unlike the case for CLL, pancreatic carcinoma, and other cancers, miRNAs in the miR-15/107 group are up-regulated in some other cancers such as cervical carcinoma (Table 1). This is compatible with the hypothesis that miRNAs can act as either tumor suppressors or oncomirs131; 132; 133. Providing yet another layer of complexity in cancers as in other conditions, the degree of functional redundancy among the different members of the miR-15/107 group is an important, unresolved, and deceptively complex issue.
B. Neurodegenerative diseases
MiRNAs may have direct relevance to human neurodegenerative diseases including Alzheimer’s disease (AD), Parkinson’s disease, and frontotemporal dementias (FTD)134; 135; 136. Members of the miR-15/107 group have been shown to be downregulated in AD by at least three different laboratories 59; 137; 138. We studied the cerebral cortical gray matter of aged humans and found that miR-107 is downregulated very early in AD pathogenesis59. This finding was recently validated using a different group of brain samples139. MiR-107 targets the beta-amyloid cleavage enzyme 1 (BACE1) gene so the miRNA down-regulation may have important pathogenetic consequences59; 103. Yet the physiological significance of the miR-107 downregulation in AD brains has not been systematically tested. Noren Hooten et al compared miRNA expression in young versus old individuals’ blood mononuclear cells by real-time RT-PCR analysis to test aging-related differences, and found that two of the nine miRNAs significantly decreased in aged individuals were miR-103 and miR-107140. The link between decreased miR-103/107 expression and aging merits further study because age is the most significant risk factor for AD. As noted above, miR-107 and other group members strongly regulate the expression of the GRN gene75. This targeting depends on elements in the open reading frame of GRN mRNA. GRN is a susceptibility gene for FTD141; 142 and we found that the regulation may also be relevant to neuroinflammation and the brain response to traumatic brain injury. Two laboratories independently reported that miR-15a is downregulated in AD although these studies employed different technical parameters137; 138. Members of the miR-15/107 group are also predicted to target the neurodegeneration-related genes such as the β-amyloid precursor protein and α-synuclein136; 143. In addition, miR-15/107 group members seem involved in the fine-tuning of tau phosphorylation (Hébert et al., unpublished data). The findings in AD may tie in with studies that have suggested that miR-15/107 group member miR-195 may regulate the important brain growth factor BDNF with relevance to schizophrenia72; 144; 145. We note that miRNAs outside the miR-15/107 gene group, including miR-29a/b, miR-128, miR-146a, and others have also been shown to have possible relevance to AD pathogenesis134; 138; 146; 147; 148; 149; 150; 151; 152; 153; 154. As with other conditions, it is not known how targeting by different miRNAs overlaps or interacts with the function of individual miR-15/107 group members in the context of neurodegenerative diseases.
In addition to the miRNA expression down-regulation in AD brains, there are other connections between the miR-15/107 gene group and neurodegenerative diseases. PANK2, which harbors an intronic miR-103 gene, is the susceptibility gene for a separate neurodegenerative disease, pantothenate kinase-associated neurodegeneration (PKAN)155. PKAN is characterized neuropathologically by progressive brain atrophy with iron pigment deposition and axonal spheroids mostly in subcortical structures. Intriguingly, the brains of some PKAN patients harbor neurofibrillary tangles similar to those seen in AD patients’ brains156; 157. We conclude that it is unclear whether miR-15/107 group members participate causally in neurodegenerative diseases; however, there are already numerous intriguing connections between the expression of the miR-15/107 gene group members and the manifestations of neurological diseases.
C. Heart disease
MiR-15/107 group members are among a subset of human-expressed miRNAs that have been implicated in cardiovascular disease. Most of this research has focused on miR-195. MiR-195 is up-regulated during cardiac hypertrophy and conversely over-expressing miR-195 in murine cardiomyocytes led to fatal dilated cardiomyopathy158. An recent review on the roles of miRNAs in cardiovascular disease is available30.
Summary and Conclusions
Research on the miR-15/107 gene group is less than a decade old but there is compelling evidence that these genes play essential roles in human biology (Figure 7). MiR-15, miR-16, miR-103, and miR-107 are stably situated in the genomes of vertebrates; however, the evolutionary biology of these miRNAs is still incompletely understood. Moderate-to-high expression of multiple miR-15/107 gene group members has been observed in most human tissues, with increased expression in particular tissues and at earlier stages of development. The miR-15/107 group genes that are relatively highly expressed in humans include the sequence AGCAGCA at or near the 5′ seed region of the mature miRNAs, and this sequence appears to be a strong determinant of mRNA target selection. Functionally, miRNAs in this gene group serve fundamental roles in human tissues. Some of these biological functions occur via novel mechanisms including systematic targeting of mRNA target protein coding sequences. Laboratory data dovetail with evidence from evolutionary biology suggesting that miR-15/107 group members could provide some sort of link between the mechanisms of cell division and metabolism. MiR-15, miR-107, and their paralogs are dysregulated in some human diseases and provide potential therapeutic targets. Although there has been substantial recent scientific progress, much more work remains to be done in studying these genetic regulators whose complexity and impact are enhanced by functional synergy between different miRNA genes.
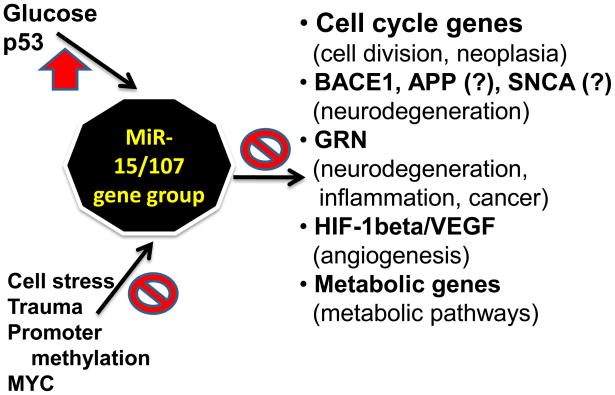
Figure 7.
Summary cartoon regarding the regulation and function of the miR-15/107 gene group. Upstream positive regulation has been documented for glucose (through unknown transcription factors) and P53 with the caveat that each miRNA is transcriptionally activated via differing mechanisms. Suppression of miR-15/107 gene group member expression can occur in cell stress and trauma, and secondary to promoter methylation or MYC. The normal functions of miR-15/107 gene group members seem to include roles in cell division, angiogenesis, and metabolic pathways. Dysfunction in miR-107 expression may contribute to neoplasia, neurodegeneration, cardiovascular dysfunction, and other diseases.
Acknowledgments
We apologize to those researchers whose work could not be cited because of space limitations. We thank Ms Willa Huang for technical and collegial assistance in the project. We are deeply grateful to study participants at the University of Kentucky Alzheimer’s Disease Center. This research was supported by grants R01 NS061933, K08 NS050110, and P30-AG028383, from the NIH, Bethesda, MD and NIRG-89917 from the Alzheimer Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Liu X, Fortin K, Mourelatos Z. MicroRNAs: biogenesis and molecular functions. Brain Pathol. 2008;18:113–21. doi: 10.1111/j.1750-3639.2007.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saito T, Saetrom P. MicroRNAs-targeting and target prediction. N Biotechnol. doi: 10.1016/j.nbt.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 5.Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. The microRNA world: small is mighty. Trends Biochem Sci. 2003;28:534–40. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Dai Z, Chen Z, Ye H, Zhou L, Cao L, Wang Y, Peng S, Chen L. Characterization of microRNAs in cephalochordates reveals a correlation between microRNA repertoire homology and morphological similarity in chordate evolution. Evol Dev. 2009;11:41–9. doi: 10.1111/j.1525-142X.2008.00301.x. [DOI] [PubMed] [Google Scholar]
- 7.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–21. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 8.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–78. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–87. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang WX, Wilfred BR, Xie K, Jennings MH, Hu Y, Stromberg AJ, Nelson PT. Individual microRNAs (miRNAs) display distinct mRNA targeting “rules”. RNA Biol. :7. doi: 10.4161/rna.7.3.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–11. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 15.Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–8. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scherer WF, Syverton JT, Gey GO. Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med. 1953;97:695–710. doi: 10.1084/jem.97.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasashima K, Nakamura Y, Kozu T. Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem Biophys Res Commun. 2004;322:403–10. doi: 10.1016/j.bbrc.2004.07.130. [DOI] [PubMed] [Google Scholar]
- 18.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, Raymond CK, Roberts BS, Juhl H, Kinzler KW, Vogelstein B, Velculescu VE. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–92. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 20.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–9. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–70. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 22.Sewer A, Paul N, Landgraf P, Aravin A, Pfeffer S, Brownstein MJ, Tuschl T, van Nimwegen E, Zavolan M. Identification of clustered microRNAs using an ab initio prediction method. BMC Bioinformatics. 2005;6:267. doi: 10.1186/1471-2105-6-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thornton JW, DeSalle R. Gene family evolution and homology: genomics meets phylogenetics. Annu Rev Genomics Hum Genet. 2000;1:41–73. doi: 10.1146/annurev.genom.1.1.41. [DOI] [PubMed] [Google Scholar]
- 24.Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW. The evolution of mammalian gene families. PLoS ONE. 2006;1:e85. doi: 10.1371/journal.pone.0000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choong ML, Yang HH, McNiece I. MicroRNA expression profiling during human cord blood-derived CD34 cell erythropoiesis. Exp Hematol. 2007;35:551–64. doi: 10.1016/j.exphem.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M, Lim L. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–52. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–9. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 30.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 121:1022–32. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. 2009 doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson KJ, Dietrich MR, McPeek MA. MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion. Bioessays. 2009 doi: 10.1002/bies.200900033. [DOI] [PubMed] [Google Scholar]
- 33.Sempere LF, Cole CN, McPeek MA, Peterson KJ. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J Exp Zoolog B Mol Dev Evol. 2006;306:575–88. doi: 10.1002/jez.b.21118. [DOI] [PubMed] [Google Scholar]
- 34.Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci U S A. 2008;105:2946–50. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci U S A. 2007;104:17719–24. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab. 2007;91:209–17. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. Structure and activity of putative intronic miRNA promoters. RNA. 16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–7. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang YP, Li KB. Correlation of expression profiles between microRNAs and mRNA targets using NCI-60 data. BMC Genomics. 2009;10:218. doi: 10.1186/1471-2164-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, Dalla-Favera R. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 42.Lerner M, Harada M, Loven J, Castro J, Davis Z, Oscier D, Henriksson M, Sangfelt O, Grander D, Corcoran MM. DLEU2, frequently deleted in malignancy, functions as a critical host gene of the cell cycle inhibitory microRNAs miR-15a and miR-16-1. Exp Cell Res. 2009;315:2941–52. doi: 10.1016/j.yexcr.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Li Q, Wang J, Guo X, Jiang X, Ren Z, Weng C, Sun G, Wang X, Liu Y, Ma L, Chen JY, Zen K, Zhang J, Zhang CY. Identification and characterization of novel amphioxus microRNAs by Solexa sequencing. Genome Biol. 2009;10:R78. doi: 10.1186/gb-2009-10-7-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, Sheridan R, John B, Marks DS, Gaidatzis D, Sander C, Zavolan M, Tuschl T. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005;19:1288–93. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Losada A, Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 2005;19:1269–87. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- 46.Strunnikov AV, Jessberger R. Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions. Eur J Biochem. 1999;263:6–13. doi: 10.1046/j.1432-1327.1999.00509.x. [DOI] [PubMed] [Google Scholar]
- 47.Shingara J, Keiger K, Shelton J, Laosinchai-Wolf W, Powers P, Conrad R, Brown D, Labourier E. An optimized isolation and labeling platform for accurate microRNA expression profiling. RNA. 2005;11:1461–70. doi: 10.1261/rna.2610405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–9. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu CG, Spizzo R, Calin GA, Croce CM. Expression profiling of microRNA using oligo DNA arrays. Methods. 2008;44:22–30. doi: 10.1016/j.ymeth.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bargaje R, Hariharan M, Scaria V, Pillai B. Consensus miRNA expression profiles derived from interplatform normalization of microarray data. RNA. 16:16–25. doi: 10.1261/rna.1688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–52. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA profiling of naive, effector and memory CD8 T cells. PLoS ONE. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur J Haematol. 2008;81:304–10. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 55.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, Negrini M, Croce CM. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A. 2004;101:9740–4. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, Weng T, Gou D, Chen Z, Chintagari NR, Liu L. Identification of rat lung-specific microRNAs by micoRNA microarray: valuable discoveries for the facilitation of lung research. BMC Genomics. 2007;8:29. doi: 10.1186/1471-2164-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soares AR, Pereira PM, Santos B, Egas C, Gomes AC, Arrais J, Oliveira JL, Moura GR, Santos MA. Parallel DNA pyrosequencing unveils new zebrafish microRNAs. BMC Genomics. 2009;10:195. doi: 10.1186/1471-2164-10-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RH. Differences in vertebrate microRNA expression. Proc Natl Acad Sci U S A. 2006;103:14385–9. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–23. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang WX, Wilfred BR, Baldwin DA, Isett RB, Ren N, Stromberg A, Nelson PT. Focus on RNA isolation: obtaining RNA for microRNA (miRNA) expression profiling analyses of neural tissue. Biochim Biophys Acta. 2008;1779:749–57. doi: 10.1016/j.bbagrm.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Git A, Dvinge H, Salmon-Divon M, Osborne M, Kutter C, Hadfield J, Bertone P, Caldas C. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA. doi: 10.1261/rna.1947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Y, Gelfond JA, McManus LM, Shireman PK. Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics. 2009;10:407. doi: 10.1186/1471-2164-10-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ach RA, Wang H, Curry B. Measuring microRNAs: comparisons of microarray and quantitative PCR measurements, and of different total RNA prep methods. BMC Biotechnol. 2008;8:69. doi: 10.1186/1472-6750-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Willenbrock H, Salomon J, Sokilde R, Barken KB, Hansen TN, Nielsen FC, Moller S, Litman T. Quantitative miRNA expression analysis: comparing microarrays with next-generation sequencing. RNA. 2009;15:2028–34. doi: 10.1261/rna.1699809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 66.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 67.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–69. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 68.Bruchova H, Yoon D, Agarwal AM, Mendell J, Prchal JT. Regulated expression of microRNAs in normal and polycythemia vera erythropoiesis. Exp Hematol. 2007;35:1657–67. doi: 10.1016/j.exphem.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–91. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Forrest AR, Kanamori-Katayama M, Tomaru Y, Lassmann T, Ninomiya N, Takahashi Y, de Hoon MJ, Kubosaki A, Kaiho A, Suzuki M, Yasuda J, Kawai J, Hayashizaki Y, Hume DA, Suzuki H. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia. 24:460–6. doi: 10.1038/leu.2009.246. [DOI] [PubMed] [Google Scholar]
- 72.Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17:3030–42. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joglekar MV, Parekh VS, Mehta S, Bhonde RR, Hardikar AA. MicroRNA profiling of developing and regenerating pancreas reveal post-transcriptional regulation of neurogenin3. Dev Biol. 2007;311:603–12. doi: 10.1016/j.ydbio.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 74.Pallasch CP, Patz M, Park YJ, Hagist S, Eggle D, Claus R, Debey-Pascher S, Schulz A, Frenzel LP, Claasen J, Kutsch N, Krause G, Mayr C, Rosenwald A, Plass C, Schultze JL, Hallek M, Wendtner CM. miRNA deregulation by epigenetic silencing disrupts suppression of the oncogene PLAG1 in chronic lymphocytic leukemia. Blood. 2009;114:3255–64. doi: 10.1182/blood-2009-06-229898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang W-X, Wilfred BR, Madathil SK, Tang G, Hu Y, Dimayuga J, Stromberg AJ, Huang Q, Saatman KE, Nelson PT. MiR-107 regulates Granulin/Progranulin with implicatins for traumatic brain injury and neurodegenerative disease. Am J Pathol. 2010 doi: 10.2353/ajpath.2010.091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arnstein P, Taylor DO, Nelson-Rees WA, Huebner RJ, Lennette EH. Propagation of human tumors in antithymocyte serum-treated mice. J Natl Cancer Inst. 1974;52:71–84. doi: 10.1093/jnci/52.1.71. [DOI] [PubMed] [Google Scholar]
- 77.Wang WX, Wilfred BR, Hu Y, Stromberg AJ, Nelson PT. Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. RNA. 2010;16:394–404. doi: 10.1261/rna.1905910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–98. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008;7:2643–6. doi: 10.4161/cc.7.17.6597. [DOI] [PubMed] [Google Scholar]
- 81.Iwasaki S, Tomari Y. Argonaute-mediated translational repression (and activation) Fly (Austin) 2009;3:204–6. [PubMed] [Google Scholar]
- 82.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–60. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 83.Rigoutsos I. New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res. 2009;69:3245–8. doi: 10.1158/0008-5472.CAN-09-0352. [DOI] [PubMed] [Google Scholar]
- 84.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, Parenti AR, Daidone MG, Bicciato S, Piccolo S. A MicroRNA targeting dicer for metastasis control. Cell. 141:1195–207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 86.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 87.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–33. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 88.Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, Mendell JT, Huso D, Lowenstein CJ. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci U S A. 107:6334–9. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee KH, Lotterman C, Karikari C, Omura N, Feldmann G, Habbe N, Goggins MG, Mendell JT, Maitra A. Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology. 2009;9:293–301. doi: 10.1159/000186051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yue D, Liu H, Huang Y. Survey of Computational Algorithms for MicroRNA Target Prediction. Curr Genomics. 2009;10:478–92. doi: 10.2174/138920209789208219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nelson PT, Kiriakidou M, Mourelatos Z, Tan GS, Jennings M, Xie K, Wang WX. High-throughput experimental studies to identify miRNA targets directly, with special focus on the mammalian brain. Brain Res. doi: 10.1016/j.brainres.2010.03.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tamura Y, Yoshida M, Ohnishi Y, Hohjoh H. Variation of gene silencing involving endogenous microRNA in mammalian cells. Mol Biol Rep. 2009;36:1413–20. doi: 10.1007/s11033-008-9330-4. [DOI] [PubMed] [Google Scholar]
- 93.Lennox KA, Behlke MA. A Direct Comparison of Anti-microRNA Oligonucleotide Potency. Pharm Res. doi: 10.1007/s11095-010-0156-0. [DOI] [PubMed] [Google Scholar]
- 94.Koshiol J, Wang E, Zhao Y, Marincola F, Landi MT. Strengths and limitations of laboratory procedures for microRNA detection. Cancer Epidemiol Biomarkers Prev. 19:907–11. doi: 10.1158/1055-9965.EPI-10-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–9. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, Kappeler A, Brunner T, Vassella E. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553–9. doi: 10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- 98.Chen RW, Bemis LT, Amato CM, Myint H, Tran H, Birks DK, Eckhardt SG, Robinson WA. Truncation in CCND1 mRNA alters miR-16-1 regulation in mantle cell lymphoma. Blood. 2008;112:822–9. doi: 10.1182/blood-2008-03-142182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang F, Fu XD, Zhou Y, Zhang Y. Down-regulation of the cyclin E1 oncogene expression by microRNA-16-1 induces cell cycle arrest in human cancer cells. BMB Rep. 2009;42:725–30. doi: 10.5483/bmbrep.2009.42.11.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takahashi Y, Forrest AR, Maeno E, Hashimoto T, Daub CO, Yasuda J. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS ONE. 2009;4:e6677. doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tang X, Muniappan L, Tang G, Ozcan S. Identification of glucose-regulated miRNAs from pancreatic {beta} cells reveals a role for miR-30d in insulin transcription. RNA. 2009;15:287–93. doi: 10.1261/rna.1211209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–8. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 103.Davidson LA, Wang N, Shah MS, Lupton JR, Ivanov I, Chapkin RS. n-3 Polyunsaturated fatty acids modulate carcinogen-directed non-coding microRNA signatures in rat colon. Carcinogenesis. 2009;30:2077–84. doi: 10.1093/carcin/bgp245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Safdar A, Abadi A, Akhtar M, Hettinga BP, Tarnopolsky MA. miRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57Bl/6J male mice. PLoS ONE. 2009;4:e5610. doi: 10.1371/journal.pone.0005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Leonardi R, Zhang YM, Rock CO, Jackowski S. Coenzyme A: back in action. Prog Lipid Res. 2005;44:125–53. doi: 10.1016/j.plipres.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 106.Jackowski S, Rock CO. Regulation of coenzyme A biosynthesis. J Bacteriol. 1981;148:926–32. doi: 10.1128/jb.148.3.926-932.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pothof J, Verkaik NS, van IW, Wiemer EA, Ta VT, van der Horst GT, Jaspers NG, van Gent DC, Hoeijmakers JH, Persengiev SP. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J. 2009;28:2090–9. doi: 10.1038/emboj.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang B, Pan X. RDX induces aberrant expression of microRNAs in mouse brain and liver. Environ Health Perspect. 2009;117:231–40. doi: 10.1289/ehp.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wilmink GJ, Roth CL, Ibey BL, Ketchum N, Bernhard J, Cerna CZ, Roach WP. Identification of microRNAs associated with hyperthermia-induced cellular stress response. Cell Stress Chaperones. doi: 10.1007/s12192-010-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Redell JB, Liu Y, Dash PK. Traumatic brain injury alters expression of hippocampal microRNAs: potential regulators of multiple pathophysiological processes. J Neurosci Res. 2009;87:1435–48. doi: 10.1002/jnr.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, Ivan M. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–67. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Donker RB, Mouillet JF, Nelson DM, Sadovsky Y. The expression of Argonaute2 and related microRNA biogenesis proteins in normal and hypoxic trophoblasts. Mol Hum Reprod. 2007;13:273–9. doi: 10.1093/molehr/gam006. [DOI] [PubMed] [Google Scholar]
- 113.Yin C, Salloum FN, Kukreja RC. A novel role of microRNA in late preconditioning: upregulation of endothelial nitric oxide synthase and heat shock protein 70. Circ Res. 2009;104:572–5. doi: 10.1161/CIRCRESAHA.108.193250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yin KJ, Deng Z, Hamblin M, Xiang Y, Huang H, Zhang J, Jiang X, Wang Y, Chen YE. Peroxisome proliferator-activated receptor delta regulation of miR-15a in ischemia-induced cerebral vascular endothelial injury. J Neurosci. 30:6398–408. doi: 10.1523/JNEUROSCI.0780-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Karaa ZS, Iacovoni JS, Bastide A, Lacazette E, Touriol C, Prats H. The VEGF IRESes are differentially susceptible to translation inhibition by miR-16. RNA. 2009;15:249–54. doi: 10.1261/rna.1301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–8. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 117.Daubman S. MicroRNAs in angiogenesis and vascular smooth muscle cell function. Circ Res. 106:423–5. doi: 10.1161/RES.0b013e3181d61a0d. [DOI] [PubMed] [Google Scholar]
- 118.Leung AK, Sharp PA. microRNAs: a safeguard against turmoil? Cell. 2007;130:581–5. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 119.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 121.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell’Aquila ML, Alder H, Rassenti L, Kipps TJ, Bullrich F, Negrini M, Croce CM. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101:11755–60. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 17:215–20. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 123.Mraz M, Pospisilova S, Malinova K, Slapak I, Mayer J. MicroRNAs in chronic lymphocytic leukemia pathogenesis and disease subtypes. Leuk Lymphoma. 2009;50:506–9. doi: 10.1080/10428190902763517. [DOI] [PubMed] [Google Scholar]
- 124.Scaglione BJ, Salerno E, Balan M, Coffman F, Landgraf P, Abbasi F, Kotenko S, Marti GE, Raveche ES. Murine models of chronic lymphocytic leukaemia: role of microRNA-16 in the New Zealand Black mouse model. Br J Haematol. 2007;139:645–57. doi: 10.1111/j.1365-2141.2007.06851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bateman A, Bennett HP. The granulin gene family: from cancer to dementia. Bioessays. 2009 doi: 10.1002/bies.200900086. [DOI] [PubMed] [Google Scholar]
- 126.Cheung ST, Wong SY, Leung KL, Chen X, So S, Ng IO, Fan ST. Granulin-epithelin precursor overexpression promotes growth and invasion of hepatocellular carcinoma. Clin Cancer Res. 2004;10:7629–36. doi: 10.1158/1078-0432.CCR-04-0960. [DOI] [PubMed] [Google Scholar]
- 127.Davidson B, Alejandro E, Florenes VA, Goderstad JM, Risberg B, Kristensen GB, Trope CG, Kohn EC. Granulin-epithelin precursor is a novel prognostic marker in epithelial ovarian carcinoma. Cancer. 2004;100:2139–47. doi: 10.1002/cncr.20219. [DOI] [PubMed] [Google Scholar]
- 128.Donald CD, Laddu A, Chandham P, Lim SD, Cohen C, Amin M, Gerton GL, Marshall FF, Petros JA. Expression of progranulin and the epithelin/granulin precursor acrogranin correlates with neoplastic state in renal epithelium. Anticancer Res. 2001;21:3739–42. [PubMed] [Google Scholar]
- 129.Jones MB, Michener CM, Blanchette JO, Kuznetsov VA, Raffeld M, Serrero G, Emmert-Buck MR, Petricoin EF, Krizman DB, Liotta LA, Kohn EC. The granulin-epithelin precursor/PC-cell-derived growth factor is a growth factor for epithelial ovarian cancer. Clin Cancer Res. 2003;9:44–51. [PubMed] [Google Scholar]
- 130.Zanocco-Marani T, Bateman A, Romano G, Valentinis B, He ZH, Baserga R. Biological activities and signaling pathways of the granulin/epithelin precursor. Cancer Res. 1999;59:5331–40. [PubMed] [Google Scholar]
- 131.Lee YS, Dutta A. MicroRNAs: small but potent oncogenes or tumor suppressors. Curr Opin Investig Drugs. 2006;7:560–4. [PubMed] [Google Scholar]
- 132.Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 134.Hebert SS, De Strooper B. Molecular biology. miRNAs in neurodegeneration. Science. 2007;317:1179–80. doi: 10.1126/science.1148530. [DOI] [PubMed] [Google Scholar]
- 135.Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 136.Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–8. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nunez-Iglesias J, Liu CC, Morgan TE, Finch CE, Zhou XJ. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PLoS ONE. 5:e8898. doi: 10.1371/journal.pone.0008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–20. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nelson PT, Wang WX. MiR-107 is Reduced in Alzheimer’s Disease Brain Neocortex: Validation Study. J Alzheimers Dis. doi: 10.3233/JAD-2010-091603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Noren Hooten N, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, Evans MK. MicroRNA expression patterns reveal differential expression of target genes with age. PLoS ONE. 5:e10724. doi: 10.1371/journal.pone.0010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ahmed Z, Mackenzie IR, Hutton ML, Dickson DW. Progranulin in frontotemporal lobar degeneration and neuroinflammation. J Neuroinflammation. 2007;4:7. doi: 10.1186/1742-2094-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Eriksen JL, Mackenzie IR. Progranulin: normal function and role in neurodegeneration. J Neurochem. 2008;104:287–97. doi: 10.1111/j.1471-4159.2007.04968.x. [DOI] [PubMed] [Google Scholar]
- 143.Liu W, Liu C, Zhu J, Shu P, Yin B, Gong Y, Qiang B, Yuan J, Peng X. MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer’s-associated pathogenesis in SAMP8 mice. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 144.Guo AY, Sun J, Jia P, Zhao Z. A novel microRNA and transcription factor mediated regulatory network in schizophrenia. BMC Syst Biol. 4:10. doi: 10.1186/1752-0509-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mellios N, Huang HS, Baker SP, Galdzicka M, Ginns E, Akbarian S. Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiatry. 2009;65:1006–14. doi: 10.1016/j.biopsych.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 146.Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci Lett. 2009;459:100–4. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 147.Patel N, Hoang D, Miller N, Ansaloni S, Huang Q, Rogers JT, Lee JC, Saunders AJ. MicroRNAs can regulate human APP levels. Mol Neurodegener. 2008;3:10. doi: 10.1186/1750-1326-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Provost P. Interpretation and applicability of microRNA data to the context of Alzheimer’s and age-related diseases. Aging (Albany NY) 2 doi: 10.18632/aging.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 150.Lukiw WJ, Pogue AI. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J Inorg Biochem. 2007;101:1265–9. doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shioya M, Obayashi S, Tabunoki H, Arima K, Saitoh Y, Ishida T, Satoh J. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neuron navigator-3. Neuropathol Appl Neurobiol. doi: 10.1111/j.1365-2990.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- 152.Hebert SS, Horre K, Nicolai L, Bergmans B, Papadopoulou AS, Delacourte A, De Strooper B. MicroRNA regulation of Alzheimer’s Amyloid precursor protein expression. Neurobiol Dis. 2009;33:422–8. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 153.Carrettiero DC, Hernandez I, Neveu P, Papagiannakopoulos T, Kosik KS. The cochaperone BAG2 sweeps paired helical filament- insoluble tau from the microtubule. J Neurosci. 2009;29:2151–61. doi: 10.1523/JNEUROSCI.4660-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 155.Zhou B, Westaway SK, Levinson B, Johnson MA, Gitschier J, Hayflick SJ. A novel pantothenate kinase gene (PANK2) is defective in Hallervorden-Spatz syndrome. Nat Genet. 2001;28:345–9. doi: 10.1038/ng572. [DOI] [PubMed] [Google Scholar]
- 156.Eidelberg D, Sotrel A, Joachim C, Selkoe D, Forman A, Pendlebury WW, Perl DP. Adult onset Hallervorden-Spatz disease with neurofibrillary pathology. A discrete clinicopathological entity. Brain. 1987;110 (Pt 4):993–1013. doi: 10.1093/brain/110.4.993. [DOI] [PubMed] [Google Scholar]
- 157.Wakabayashi K, Fukushima T, Koide R, Horikawa Y, Hasegawa M, Watanabe Y, Noda T, Eguchi I, Morita T, Yoshimoto M, Iwatsubo T, Takahashi H. Juvenile-onset generalized neuroaxonal dystrophy (Hallervorden-Spatz disease) with diffuse neurofibrillary and lewy body pathology. Acta Neuropathol. 2000;99:331–6. doi: 10.1007/s004010050049. [DOI] [PubMed] [Google Scholar]
- 158.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hu Y, Correa AM, Hoque A, Guan B, Ye F, Huang J, Swisher SG, Wu TT, Ajani JA, Xu XC. Prognostic significance of differentially expressed miRNAs in esophageal cancer. Int J Cancer. doi: 10.1002/ijc.25330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pons A, Nomdedeu B, Navarro A, Gaya A, Gel B, Diaz T, Valera S, Rozman M, Belkaid M, Montserrat E, Monzo M. Hematopoiesis-related microRNA expression in myelodysplastic syndromes. Leuk Lymphoma. 2009;50:1854–9. doi: 10.3109/10428190903147645. [DOI] [PubMed] [Google Scholar]
- 161.Kaddar T, Chien WW, Bertrand Y, Pages MP, Rouault JP, Salles G, Ffrench M, Magaud JP. Prognostic value of miR-16 expression in childhood acute lymphoblastic leukemia relationships to normal and malignant lymphocyte proliferation. Leuk Res. 2009;33:1217–23. doi: 10.1016/j.leukres.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 162.Guo Y, Chen Z, Zhang L, Zhou F, Shi S, Feng X, Li B, Meng X, Ma X, Luo M, Shao K, Li N, Qiu B, Mitchelson K, Cheng J, He J. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 163.Tombol Z, Szabo PM, Molnar V, Wiener Z, Tolgyesi G, Horanyi J, Riesz P, Reismann P, Patocs A, Liko I, Gaillard RC, Falus A, Racz K, Igaz P. Integrative molecular bioinformatics study of human adrenocortical tumors: microRNA, tissue-specific target prediction, and pathway analysis. Endocr Relat Cancer. 2009;16:895–906. doi: 10.1677/ERC-09-0096. [DOI] [PubMed] [Google Scholar]
- 164.Chung GE, Yoon JH, Myung SJ, Lee JH, Lee SH, Lee SM, Kim SJ, Hwang SY, Lee HS, Kim CY. High expression of microRNA-15b predicts a low risk of tumor recurrence following curative resection of hepatocellular carcinoma. Oncol Rep. 23:113–9. [PubMed] [Google Scholar]
- 165.Hui AB, Lenarduzzi M, Krushel T, Waldron L, Pintilie M, Shi W, Perez-Ordonez B, Jurisica I, O’Sullivan B, Waldron J, Gullane P, Cummings B, Liu FF. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 16:1129–39. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- 166.Jung M, Mollenkopf HJ, Grimm C, Wagner I, Albrecht M, Waller T, Pilarsky C, Johannsen M, Stephan C, Lehrach H, Nietfeld W, Rudel T, Jung K, Kristiansen G. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med. 2009;13:3918–28. doi: 10.1111/j.1582-4934.2009.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhao JJ, Yang J, Lin J, Yao N, Zhu Y, Zheng J, Xu J, Cheng JQ, Lin JY, Ma X. Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Childs Nerv Syst. 2009;25:13–20. doi: 10.1007/s00381-008-0701-x. [DOI] [PubMed] [Google Scholar]
- 169.Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen XM, Gao HJ. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J Dig Dis. 11:50–4. doi: 10.1111/j.1751-2980.2009.00413.x. [DOI] [PubMed] [Google Scholar]
- 170.Xi Y, Formentini A, Chien M, Weir DB, Russo JJ, Ju J, Kornmann M. Prognostic Values of microRNAs in Colorectal Cancer. Biomark Insights. 2006;2:113–121. [PMC free article] [PubMed] [Google Scholar]
- 171.Satzger I, Mattern A, Kuettler U, Weinspach D, Voelker B, Kapp A, Gutzmer R. MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int J Cancer. 126:2553–62. doi: 10.1002/ijc.24960. [DOI] [PubMed] [Google Scholar]
- 172.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De Maria R. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–7. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 173.Yoon S, De Micheli G. Prediction of regulatory modules comprising microRNAs and target genes. Bioinformatics. 2005;21(Suppl 2):ii93–100. doi: 10.1093/bioinformatics/bti1116. [DOI] [PubMed] [Google Scholar]
- 174.Takeshita F, Patrawala L, Osaki M, Takahashi RU, Yamamoto Y, Kosaka N, Kawamata M, Kelnar K, Bader AG, Brown D, Ochiya T. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol Ther. 18:181–7. doi: 10.1038/mt.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Amaral FC, Torres N, Saggioro F, Neder L, Machado HR, Silva WA, Jr, Moreira AC, Castro M. MicroRNAs differentially expressed in ACTH-secreting pituitary tumors. J Clin Endocrinol Metab. 2009;94:320–3. doi: 10.1210/jc.2008-1451. [DOI] [PubMed] [Google Scholar]
- 177.Bottoni A, Piccin D, Tagliati F, Luchin A, Zatelli MC, degli Uberti EC. miR-15a and miR-16-1 down-regulation in pituitary adenomas. J Cell Physiol. 2005;204:280–5. doi: 10.1002/jcp.20282. [DOI] [PubMed] [Google Scholar]
- 178.Bhattacharya R, Nicoloso M, Arvizo R, Wang E, Cortez A, Rossi S, Calin GA, Mukherjee P. MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer. Cancer Res. 2009;69:9090–5. doi: 10.1158/0008-5472.CAN-09-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, Kim JW, Kim S. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–5. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 180.Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, Johnstone CN, Megraw MS, Adams S, Lassus H, Huang J, Kaur S, Liang S, Sethupathy P, Leminen A, Simossis VA, Sandaltzopoulos R, Naomoto Y, Katsaros D, Gimotty PA, DeMichele A, Huang Q, Butzow R, Rustgi AK, Weber BL, Birrer MJ, Hatzigeorgiou AG, Croce CM, Coukos G. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:7004–9. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu X, Chen Z, Yu J, Xia J, Zhou X. MicroRNA Profiling and Head and Neck Cancer. Comp Funct Genomics. 2009:837514. doi: 10.1155/2009/837514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Careccia S, Mainardi S, Pelosi A, Gurtner A, Diverio D, Riccioni R, Testa U, Pelosi E, Piaggio G, Sacchi A, Lavorgna S, Lo-Coco F, Blandino G, Levrero M, Rizzo MG. A restricted signature of miRNAs distinguishes APL blasts from normal promyelocytes. Oncogene. 2009;28:4034–40. doi: 10.1038/onc.2009.255. [DOI] [PubMed] [Google Scholar]
- 183.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, Croce CM. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–84. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 184.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin Cancer Res. 2008;14:2588–92. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 185.Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50:113–21. doi: 10.1002/hep.22919. [DOI] [PubMed] [Google Scholar]
- 186.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K, Tsujimoto G, Nakagawa M, Seki N. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–52. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 187.Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–7. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 188.Zanette DL, Rivadavia F, Molfetta GA, Barbuzano FG, Proto-Siqueira R, Silva WA, Jr, Falcao RP, Zago MA. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz J Med Biol Res. 2007;40:1435–40. doi: 10.1590/s0100-879x2007001100003. [DOI] [PubMed] [Google Scholar]
- 189.Flavin RJ, Smyth PC, Laios A, O’Toole SA, Barrett C, Finn SP, Russell S, Ring M, Denning KM, Li J, Aherne ST, Sammarae DA, Aziz NA, Alhadi A, Sheppard BL, Lao K, Sheils OM, O’Leary JJ. Potentially important microRNA cluster on chromosome 17p13.1 in primary peritoneal carcinoma. Mod Pathol. 2009;22:197–205. doi: 10.1038/modpathol.2008.135. [DOI] [PubMed] [Google Scholar]
- 190.Lehmann U, Streichert T, Otto B, Albat C, Hasemeier B, Christgen H, Schipper E, Hille U, Kreipe HH, Langer F. Identification of differentially expressed microRNAs in human male breast cancer. BMC Cancer. 10:109. doi: 10.1186/1471-2407-10-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Corbetta S, Vaira V, Guarnieri V, Scillitani A, Eller-Vainicher C, Ferrero S, Vicentini L, Chiodini I, Bisceglia M, Beck-Peccoz P, Bosari S, Spada A. Differential expression of microRNAs in human parathyroid carcinomas compared with normal parathyroid tissue. Endocr Relat Cancer. 17:135–46. doi: 10.1677/ERC-09-0134. [DOI] [PubMed] [Google Scholar]