Abstract
Summary: The program package CopyMap identifies copy number variation from oligo-hybridization and CGH data. Using a time-dependent hidden Markov model to combine evidence of copy number variants (CNVs) across multiple carriers, CopyMap is substantially more accurate than standard hidden Markov methods in identifying CNVs and calling CNV-carriers. Moreover, CopyMap provides more precise estimates of CNV-boundaries.
Availability: The C-source code and detailed documentation for the program CopyMap is available on the Internet at http://www.sph.umich.edu/csg/szoellner/
Contact: szoellne@umich.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Copy number variants (CNVs) are segments of the genome that exist in different copy numbers in the population. About 90% of CNVs have two allelic states (McCarroll et al., 2008), the remaining 10% have multiple states. As CNVs encompass genes as well as non-coding DNA, they are good candidates for functional variation (Conrad et al., 2010). Commonly used method for identifying copy number variable sites are Representational Oligonucleotide Microarray Analysis (Lucito et al., 2003), Agilent competitive genomic hybridization (CGH) (Barrett et al., 2004) and tiling arrays (Geigl et al., 2009). These methods interrogate DNA using a dense set of probes covering the non-repetitive regions of the genome. However, the resolution of each individual probe is usually low. As CNVs often extend over multiple probes, resolution is improved by combining information across neighboring probes. Hidden Markov model (HMM) methods are among the most commonly applied tools to analyze such data. First proposed by Fridlyand et al. (2004), these methods exploit the local correlation of trait status and are computationally highly efficient. More complex versions of these algorithms specifically designed for CNVs have since been developed (Yin et al., 2010) and applied to large datasets (Conrad et al., 2010). However, most HMM algorithms analyze datasets one individual at a time. Thus, CNVs that are shared among individuals are only identified after comparing CNV calls between individuals. Conceptually, this approach has two disadvantages: first, no detection power is gained by the fact that multiple individuals carry the same CNV. In most regions of the genome, CNVs are rare or absent and therefore models assign a low probability of transitioning into the CNV state. However, at a locus known to have variable copy number in one individual, the probability that another individual has a variant copy number at the same locus should be higher. Jointly analyzing individuals results in a higher detection rate. Secondly, each time a CNV is identified, its borders will be estimated independently of all other individuals. Jointly estimating the borders from all carriers results in a more efficient use of information. This idea has been applied in several recent methods for mapping CNVs in selected regions from genotyping data (Korn et al., 2008; Zöllner and Teslovich, 2009; Zöllner et al., 2009).
I present the program CopyMap, an alternative HMM method for CNV detection and CNV calling that jointly analyzes genome-wide data for all individuals in a sample. CopyMap is specifically designed for hybridization arrays, but it can also be applied to single nucleotide polymorphism genotyping arrays.
2 DESCRIPTION
CopyMap models a hidden Markov process, where the copy number at each probe has one of three hidden states (baseline, duplication and deletion) and the hybridization intensity of that probe is the observed signal. The program uses an expectation maximization-algorithm similar to the classical Baum–Welch algorithm to infer the location of CNVs. CopyMap has two novel features.
Location-dependent transition rates: uniform transition rates used by classic HMMs model each locus in the genome to be equally likely as a starting and end point of a CNV. CopyMap models the process of generating CNVs in the sample as a time-dependent Markov process and each individual in the sample as a realization of this process. The rate of transitioning into CNV state is estimated between every consecutive pair of probes jointly from all individuals and can be considered as the population frequency of a CNV starting at that locus. Hence, regions that show evidence for CNVs in some individuals will be identified by a higher transition rate from the baseline to the CNV state, combining the evidence for that region to be copy variable across all individuals.
Markov-k process: individual probes of most arrays do not provide sufficient information to call CNVs and short inferred CNVs are often false positives. Therefore, it is a common practice to filter all called CNVs, removing CNVs spanning less than k probes, with k between 5 and 10. These filters are usually applied post hoc after all modeling parameters were estimated. Hence, parameter estimates are including likely false signals. In contrast, I allow the user to initially set a minimum length k for CNV. Implementing a Markov-k model, the transition probabilities depend on the last k states rather than just on the present state. By restricting the transition matrix, the computation time for this process is designed to be linear with k, rather than quadratic as typical Markov-k models.
The program implements two methods for calling CNVs. Either a Viterbi algorithm or a heuristic to identify regions with high overall evidence for variable copy number and to then estimate the posterior probability for copy number for each individual in that region. Technical details of this algorithm are described elsewhere (Henrichsen et al., 2009; She et al., 2008).
3 EVALUATION
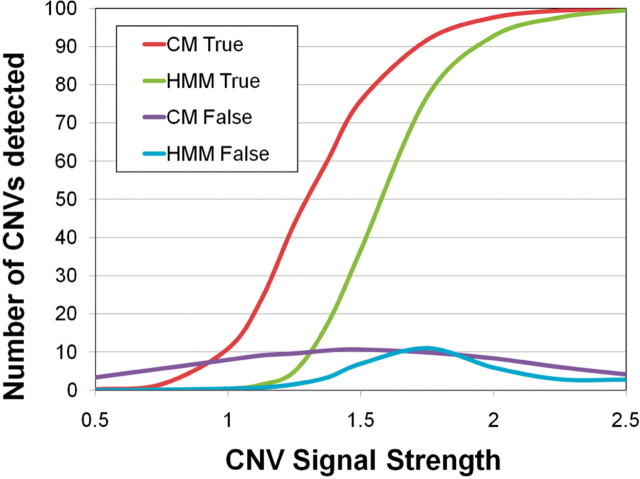
CopyMap has been applied to several large studies, reporting error rates between 95% (She et al., 2008) and 67% (Henrichsen et al., 2009). To highlight the conceptual advantages of the algorithm, I compared the performance of CopyMap with our implementation of an HMM as proposed by Fridlyand et al. (2004) by simulating a dataset of 10 CNVs with minor allele frequency 0.1 in a sample of 100 individuals; each CNV was equally likely to be a deletion or a duplication. Individuals were assessed by 10 000 consecutive probes. I assumed the intensity of probes was normally distributed and the mean intensity of probes covered by a CNV was shifted by δ = 0.5 − 2.5 SDs. I further assumed that the distributions of intensity were known, so these results represent a best-case scenario (See Supplementary online appendix for simulation details). Analyzing each dataset required ∼1 min with HMM and ∼8.5 min with CopyMap on a single 2.33 GHz processor. For intermediate values of delta between 1 and 2, CopyMap is able to detect substantially more CNVs than HMM (Fig. 1). For δ = 1.5 CopyMap detects 76 of the 100 simulated CNVs, compared to 37 detected by HMM. Both algorithms have comparable numbers of false discoveries; CopyMap has 3–11 false discoveries while HMM has 0–11.
Fig. 1.
Comparison of CopyMap and classical HMM algorithms. The horizontal axis shows the difference in signal strength δ between the CNV-probes and probes with baseline copy number. I simulated a total of 100 CNVs. The vertical axis displays the number of CNVs detected using CopyMap (CM, red line) and using a classic HMM (HMM, orange line). It also shows the number of false CNVs detected (blue and grey line).
I compared the inferred boundaries of the CNV with the true boundaries by calculating the boundary error. A boundary error of 1 indicates that either the start or the endpoint of the CNV was miscalled by one probe. CopyMap generates substantially more precise boundary calls for all values of δ (Supplementary Fig. 1S); for δ > 1.25 the average boundary error is <1. Even for signal intensities where both HMM and CopyMap have high power to detect CNVs (δ ≥ 2), CopyMap provides more precise estimates of each CNV's position.
4 CONCLUSIONS
CopyMap is a computationally efficient command line-driven C-program that can be easily run on modern parallel cluster architecture. By combining evidence across multiple individuals, it has increased power to detect CNVs and it has higher resolution to infer CNV boundaries. The source code and extensive documentation is available at http://www.sph.umich.edu/csg/zollner/software/.
Supplementary Material
ACKNOWLEDGEMENTS
I would like to thank Doug Hom for help with the CopyMap manual.
Funding: National Institute of Health grant (R01GM073822 to J.B.).
Conflict of Interest: none declared.
REFERENCES
- Barrett MT, et al. Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA. Proc. Natl Acad. Sci. USA. 2004;101:17765–17770. doi: 10.1073/pnas.0407979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridlyand J, et al. Hidden Markov models approach to the analysis of array CGH data. J. Multivariate Anal. 2004;90:132–153. [Google Scholar]
- Geigl JB, et al. Identification of small gains and losses in single cells after whole genome amplification on tiling oligo arrays. Nucleic Acids Res. 2009;37:e105. doi: 10.1093/nar/gkp526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichsen CN, et al. Segmental copy number variation shapes tissue transcriptomes. Nat. Genet. 2009;41:424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- Korn JM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucito R, et al. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- She X, et al. Extensive copy number variation of mouse segmental duplications. Nat. Genet. 2008;40:909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XL, et al. Detecting copy number variations from array CGH data based on a conditional random field model. J. Bioinform. Comput. Biol. 2010;8:295–314. doi: 10.1142/s021972001000480x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöllner S, Teslovich TM. Using GWAS data to identify copy number variants contributing to common complex diseases. Stat. Sci. 2009;24:530–546. [Google Scholar]
- Zöllner S, et al. Bayesian EM algorithm for scoring polymorphic deletions from SNP data and application to a common CNV on 8q24. Genet. Epidemiol. 2009;33:357–368. doi: 10.1002/gepi.20391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.