Summary
Here, we report that MSH2/MSH3 maintains lesion specificity for small loops by a distinctly different mechanism than does MHSH2/MSH6 for single base mismatches. ADP and ATP have no preference for the subunits of hMSH2/MSH3. Upon lesion binding, however, hMSH2/MSH3 adopts a single “nucleotide signature” in which one ADP binds within the hMSH2 subunit and the hMSH3 subunit is empty. On the lesion, ADP-hMSH2/MSH3-empty binds and hydrolyzes ATP in the empty hMSH3 subunit, which reduces ADP affinity and increases ATP affinity for the hMSH2 subunit. ADP/ATP exchange converts (CA)4-loop-bound ADP-MSH2/MSH3-ATP into an ATP-hMSH2/MSH3-ADP intermediate in which ATP hydrolysis is inhibited in the hMSH2 subunit. We propose a model in which lesion binding converts hMSH2/MSH3 into a distinct nucleotide-bound form, and poises it to be a molecular sensor for lesion specificity.
Keywords: Mismatch repair, DNA repair, MSH2/MSH3, MSH2/MSH6
The Mismatch Repair (MMR) system binds and corrects mispaired or extrahelical base lesions within DNA. MMR supports the intrinsic proofreading properties of DNA polymerases during replication and decreases the mutation rate of newly replicated DNA by nearly 1,000-fold1–3. MMR proteins also correct heteroduplex DNA after recombination events, and have been linked functionally to base and nucleotide excision repair pathways4,5. In mammals, loss of MMR function confers a mutator phenotype and contributes to the development of cancers, such as hereditary nonpolyposis colorectal cancer (HNPCC) and sporadic solid tumors6–8.
Many steps of MMR are not well understood, but the structural and functional basis for lesion recognition has been derived from prokaryotic MutS9,10, and eukaryotic MSH2/MSH611. For both the MutS homodimer and the MHS2/MSH6 heterodimer, DNA binding is asymmetric with one subunit in direct contact with a mispaired base, and the other making non-specific contacts with the phosphodiester backbone9,10. Both MMR complexes contain a conserved Phe-X-Glu motif in which the phenylalanine residue stacks onto the mispaired DNA base, and is essential for efficient repair1–3,9,10,12. MutS and MSH2/MSH6 each bind and hydrolyze ATP13–18 within a conserved Walker-type nucleotide-binding domain in the carboxy- terminus of each subunit9–14.
In eukaryotes, a second MMR complex, MSH2/MSH31–3, can carry out lesion recognition. Although they share a common MSH2 subunit, MSH3 and MSH6 have little sequence homology in their DNA binding domains and different lesion specificities1–3,9–12. MSH2/MSH6 has a high affinity for single mismatched or an unpaired base as measured by both DNA binding and ATPase activity19–21. Recent data have suggested that MSH2/MSH3 can repair some base-base mispairs22. However, MSH2/MSH3 has a higher apparent affinity and specificity for insertion/deletion DNA loops (IDL) composed of 2–13 bases1–3,18, 23,24.
The underlying basis for lesion specificity between the two respective MMR complexes is poorly understood, but is likely to reside in MSH3- and MSH6-specific sequences. Changes in any of the Phe-X-Glu residues of MSH6 result in a strong mutator phenotype suggesting that these residues are essential for effective repair of single base mismatches12. However, at the analogous position of Phe-X-Glu in MSH6, MSH3 from yeast or human has a lysine instead of a Phe, and Lys or Arg instead of a Glu1–3,22. Thus, repair by MSH2/MSH3 and MSH2/MSH6, even for the same lesion, is unlikely to occur by the same mechanism12,24. For human MSH2/MSH6, the hMSH2 subunit makes no contact with DNA even within the phosphodiester backbone11. Furthermore, when the mispair-binding domain (MBD) of yMSH6 is switched for the DNA-binding domain of yMSH3, the resulting chimera assumes the lesion specificity of yMSH3, and complements an yMSH3 deletion mutant in vivo25. The effect is not reversible. A chimeric hMSH3 containing a yMSH6 MBD does not complement a yMSH6 deletion mutant25. Furthermore, yeast strains bearing a deletion in Domain 1 of yMSH2 are nearly wild-type in MSH2/MSH6-mediated MMR and recombination, but these strains are defective in MSH2/MSH3-mediated MMR and recombination functions26. These data suggest that the MSH3 and MSH6 subunits are structurally and functionally different.
Despite extensive differences in their properties, initiation of repair by both MSH2/MSH3 and MSH2/MSH6 require coupling of DNA binding and ATP hydrolysis13–18. Recent mapping studies in yeast indicate that the yMSH6 subunit of yMSH2/MSH6 binds ATP with high apparent affinity, while yMSH2 exclusively binds ADP18. Interestingly, DNA binding does not alter either the nucleotide binding properties18 or the conformation11 of yMSH2/MSH6 with a mismatched DNA substrate. A model has been proposed in which both MSH2 and the MSH6 subunit of yMSH2/MSH6 are simultaneously occupied with ATP in order to slide off the DNA, and interact with MLH1/PMS2 in the absence of ATP hydrolysis15–18. Differential nucleotide occupancy, therefore, is a major determinant for MSH2/MSH6 to initiate repair of single base mismatches1–3,18,27–29.
MSH2/MSH3 recognizes structurally distinct lesions, and differs significantly in its DNA binding domain, yet the biochemical properties of MSH2/MSH3 have been largely inferred from those of bacterial MutS or its eukaryotic homologue, MSH2/MSH6. Although small heteroduplex loops are among the most frequent lesions in DNA, the mechanism by which MSH2/MSH3 might recognize and repair them is not understood, and the nucleotide binding properties have not been measured. To begin addressing this issue, we have purified the human MSH2/MSH330 and MSH2/MSH6 complexes, measured the nucleotide binding affinities in each complex, and mapped the nucleotide subunit occupancy during lesion recognition prior to ATP hydrolysis. Based on our findings, we propose a novel model for the recognition and repair of small loops by MSH2/MSH3 that differs substantially from recognition and repair of single base mismatches by MSH2/MSH6.
RESULTS
hMSH2/MSH3 has nucleotide-binding profiles distinct from those of hMSH2/MSH6
The apparent affinity and subunit specificity for ADP and ATP has been recently reported for yeast MSH2/MSH6 (yMSH2/MSH6)18. Therefore, as a reference point for evaluating human MSH2/MSH3 (hMSH2/MSH3), we first determined whether there were significant differences in nucleotide binding profiles between the yeast and human MSH2/MSH6 protein. To map the subunit occupancy and nucleotide affinity, we measured uv-crosslinking (x-linking) of [α32P]-labeled ADP or [α32P]-labeled ATP(−Mg2+) to hMSH2/MSH6, and resolved the x-linking products on SDS-PAGE gels (Fig. 1). Because covalent attachment depends on the geometry of the binding sites and is irreversible, x-linking can over estimate affinity if there is non-specific binding, or underestimate affinity if x-linking is inefficient. To avoid potential artifacts, we also measured equilibrium binding of fluorescent Bo-Dipy (BD)-labeled nucleotides in solution using fluorescence anisotropy (FA). A summary of the measured affinities by both methods is listed (Supplemental Fig. 1). In all experiments, we confirmed that contaminants within an ADP, ATP or AMP-PNP preparation were less than 1% (Supplemental Fig. 2a) or, in the case of ATP, an ATP-regeneration system was included in the reaction (Supplemental Fig. 2b). Preparations of purified hMSH2/MSH3 were checked with a sensitive luciferase assay28 to insure that any ADP pre-bound nucleotide had been removed.
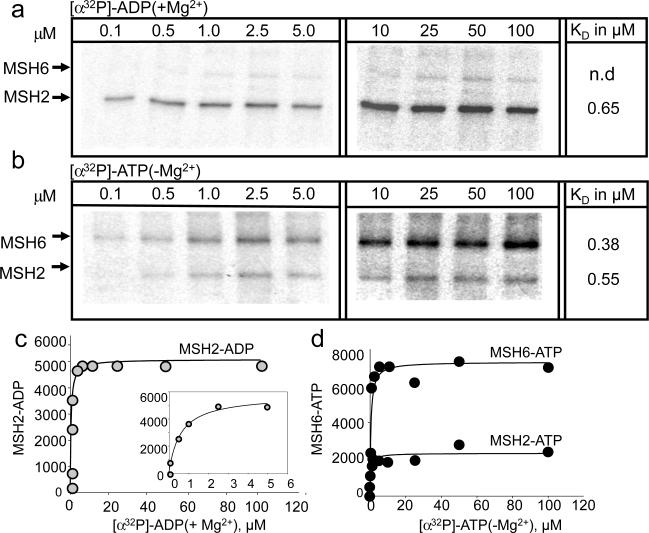
Fig. 1. Human MSH2/MSH6 binds ADP and ATP in a subunit-specific manner.
(a) Representative PAGE gel of cross-linked (x-linked) products of (a) [α32P]-ADP(+Mg2+) or (b) [α32P]-ATP(−Mg2+) bound to either the hMSH2 or the hMSH6 subunits of hMSH2/MSH6, as indicated. Reactions in (a) and (b) were performed using 100nM hMSH2/MSH6 and 0.1–5.0μM [α32P]-ADP(+Mg2+) (a, left) or [α32P]-ATP(−Mg2+)(b, left) or 500nM hMSH2/MSH6 and 10–10μM [α32P]-ADP(+Mg2+) (a, right) or [α32P]-ATP(−Mg2+)(b, right). Unless otherwise specified, Mg2+ was 5mM and reactions were preformed in 10μl volumes. Bands corresponding to hMSH2 and hMSH6 are indicated. (c, d) Quantified band intensities from (a) and (b). (c) Plot of ADP-bound (in hMSH2 subunit) versus [α32P]-ADP(+Mg2+) concentration (gray circles) and the single ligand binding fit from quantified band intensities. The inset illustrating the binding curve between 1–5μM nucleotide, (d) Plot of ATP-hMSH2 and ATP-hMSH6 versus ATP concentration (−Mg2+, black circles) and single ligand binding fit from quantified band intensities. The KDs (see text) are indicated and summarized in Supplemental Fig. 1a. Incubation time of nucleotide and protein was 20 minutes at room temperature, followed by x-linking for 20 minutes in ice.
The nucleotide binding properties of hMSH2/MSH6 were similar to those of yMSH2/MSH6 (Figure 1a,c, Supplemental Fig. 1a). For ADP, the high affinity-binding site resided in the human MSH2 subunit (hMSH2) (KD = 650 nM), while the human MSH6 subunit (hMSH6) did not show quantifiable cross-linking at concentrations up to 100 μM (Fig. 1a,c and Supplemental Fig. 3a). The binding experiments were repeated in solution using equilibrium binding of BD-ADP(+Mg2+) with similar results (Supplemental Fig. 1b). Together, these data indicated that ADP had a preference for the MSH2 subunit in both hMSH2/MSH6 and yMSH2/MSH618,27.
The nucleotide binding properties of ATP for hMSH2/MSH6 and yMSH2/MSH6 were also similar (Fig. 1b,d). ATP can be hydrolyzed during the course of binding reactions, therefore, we measured the ATP binding reaction under non-hydrolyzing conditions using either [α32P]-ATP(−Mg2+) or a non-hydrolyzable ATP analogue, AMP-PNP(+Mg2+.). As reported for yMSH2/MSH6, ATP had two binding sites, with a high affinity site residing in the hMSH6 subunit (Fig. 1b,d, Supplemental 1a and 3a). X-linking of [α32P] -ATP(−Mg2+) to the hMSH2 subunit was only a fraction (about 20%) of that covalently attached to hMSH6 (Fig. 1b,d, Supplementary Fig. 1a). However, x-linking of ATP to hMSH2 was saturable (Fig. 1 b,d), and, in solution, the binding affinity for BD-ATP(−Mg2+) and BD-AMP-PNP(+Mg2+) was 390 and 230 nM, respectively (Supplemental Fig. 1b). To verify that the BD tag did not affect the BD-ATP(−Mg2+) binding affinity (KD), we measured the ability of unlabeled ATP(−Mg2+) to inhibit binding of BD-ATP(−Mg2+) (the Ki) to hMSH2/MSH6. If unlabeled ADP directly competes with pre-bound BD-ADP, then the binding and dissociation reflect the same reaction. Indeed, we found that the Ki for ATP(−Mg2+) inhibition (Supplemental 1a,c) was similar to the KD for BD-ATP(−Mg2+) binding (Supplemental 1a,c). Thus, the fluorophore had no measurable effect on the nucleotide binding properties of ATP to hMSH2/MSH6. Thus, x-linking of [α32P] -ATP(−Mg2+) to the hMSH2 subunit of hMSH2/MSH6 was apparently inefficient. As a whole, however, the nucleotide binding properties of ADP and ATP to hMSH2/MSH6 and to yMSH2/MSH6 were comparable. In both species, the hMSH2 subunit was a high affinity-binding site for ADP, and hMSH6 was a high affinity-binding site for ATP.
The subunits of hMSH2/MSH3 bind ADP and ATP stochastically
The nucleotide binding properties of hMHS2/MSH3 were distinct from those of hMSH2/MSH6 (Fig. 2). Neither ADP(+Mg2+) nor ATP(−Mg2+) had a preference for the subunits of hMSH2/MSH3. X-linking with [α32P]-ADP(+Mg2+) or [α32P]-ATP(−Mg2+) indicated that both subunits bound either nucleotide with high affinity, as quantified by the gel scans (Fig. 2a–c, Supplemental Fig. 3a, Supplemental Fig. 1a). Anisotropy measurements cannot distinguish between the subunits. However, we found that the KD for BD-ADP (+Mg2+) binding to hMSH2/MSH3 (Supplemental Fig. 1b) reflected the average affinity for the individual hMSH2 and hMSH3 subunits measured by x-linking (Supplemental Fig. 1a and b). Thus, binding of ADP and ATP to either subunit of hMSH2/MSH3 was stochastic (Fig. 2d–f, and Supplemental Fig. 1a).
Fig. 2. Human MSH2/MSH3 binds ATP and ADP with equal apparent affinity in both subunits.
(a) Representative PAGE gel of x-linked products of [α32P]-ADP(+Mg2+), bound to either the hMSH2 or the hMSH3 subunits of hMSH2/MSH3, as indicated. The x-linking reactions and band quantification were performed as described in Fig. 1. (b, c) Quantified band intensities and single ligand binding fits of data in (a). (b) Plot of ADP-bound in the MSH2 subunit versus [α32P]-ADP(+Mg2+) concentration, (c) same as b for the hMSH3 subunit. The KD was determined as described in Fig. 1, and summarized in (d) Representative PAGE gel of resolved x-linked products in the presence of increasing concentrations of [α32P]-ATP(−Mg2+), as described in Fig. 1. (e,f) Plots of binding data for (d) (black circles) for the hMSH2 (e) and the hMSH3 (f) subunits of hMSH2/MSH3. Insets are binding data from 1–5μM [α32P]-ATP(−Mg2+). The KD for [α32P]-ATP(−Mg2+) was determined as described in Fig. 1, are indicated, and summarized in Supplemental Fig. 1a. (g, left) Representative PAGE gel of x-linked products of 0 to 5.0μM [α32P]-ADP(+Mg2+) in the presence of increasing concentrations of unlabeled AMP-PNP(+Mg2+) added simultaneously. (g, right) Same as (g, left) except that unlabeled AMP-PNP(+Mg2+) was added at 5μM simultaneously with increasing concentrations of [α32P]-ADP(+Mg2+).
As a second test for stochastic binding of nucleotides, we simultaneously added a constant amount (5 μM) of unlabeled AMP-PNP(+Mg2+) and a specified (increasing) concentration of labeled ADP(+Mg2+) to MSH2MSH3 (Fig. 2g, left panel). If nucleotide binding was stochastic, then ADP and ATP, when added together, should compete equally well for binding to each subunit of hMSH2/MSH3. Indeed, as the concentration of the unlabeled AMP-PNP(+Mg2+) approached the concentration of the labeled ADP(+Mg2+), cross-linking to each subunit was reduced to the same extent (Fig. 2g, left panel). In a complementary experiment, we simultaneously added a constant concentration of unlabeled AMP-PNP(+Mg2+) (5μM) with increasing concentrations of labeled [α32P]-ADP(+Mg2+) (Fig. 2g, right panel). The results of the two experiments were equivalent. As the concentration of the labeled [α32P]-ADP(+Mg2+) approached that of unlabeled AMP-PNP(+Mg2+), cross-linking to each subunit increased to the same extent. Thus, the nucleotide-binding properties of hMSH2/MSH3 were significantly different from those of hMSH2/MSH6. ADP and ATP did not discriminate between the subunits of hMSH2/MSH3.
Binding of nucleotides in one subunit of hMSH2/MSH3 excludes binding in the other
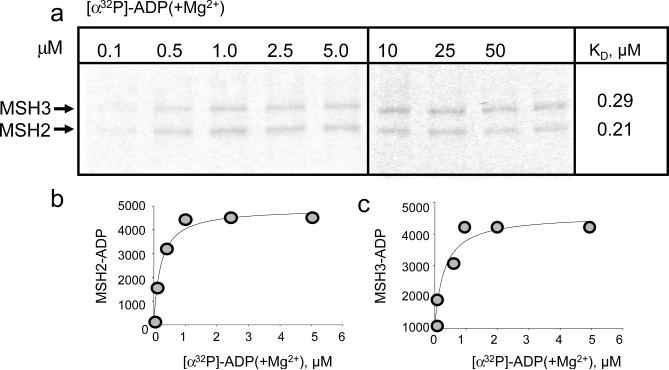
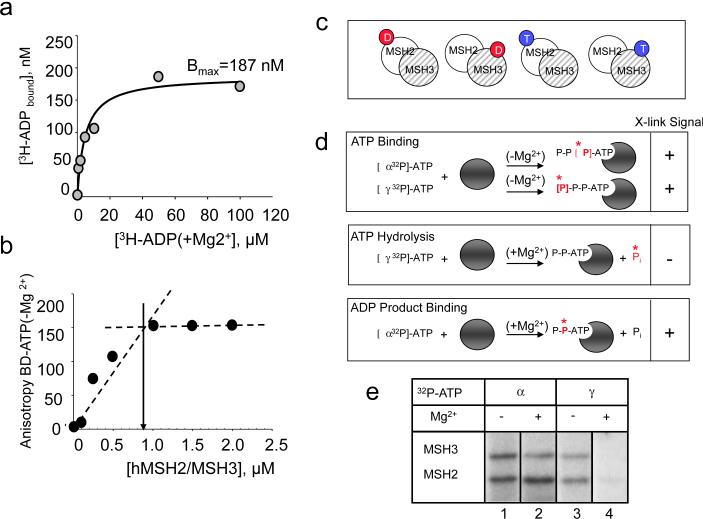
Two Walker-type ATP binding sites are present in all MutS homologues9–11. Since ADP and ATP showed no preference for the subunits of hMSH2/MSH3, we tested whether the same nucleotide could occupy both sites simultaneously. To measure the stoichiometry of hMSH2/MSH3 to ADP(+Mg2+), we used a filter-binding assay with 3H-ADP(+Mg2+) (Fig. 3a). If both nucleotide-binding sites were occupied, then the ratio of labeled 3H-ADP(+Mg2+) to the heterodimeric hMSH2/MSH3 should be 2, while an equimolar ratio would indicate that only one site was occupied. Indeed, we observed that 3H-ADP(+Mg2+) binding was equimolar and occupied 1 of the available nucleotide-binding sites (187 nM bound versus 400 nM total sites, Fig. 3a). Similar results were obtained for ATP (Fig. 3b). In this FA experiment, we observed that binding of BD-ATP(−Mg2+) reached saturation at 0.9 moles of bound per mole of hMSH2/MSH3 heterodimer (Fig. 3b). ADP or ATP could occupy only one Walker sites at a time. Thus, four distinct nucleotide-bound complexes of hMSH2/MSH3 were possible. In the absence of hydrolysis, half of the respective nucleotide was bound to hMSH2 subunit, and half to the hMSH3 subunit (Fig. 3c).
Fig. 3. Binding of either ADP or ATP to one subunit of hMSH2/MSH3 excludes binding in the other.
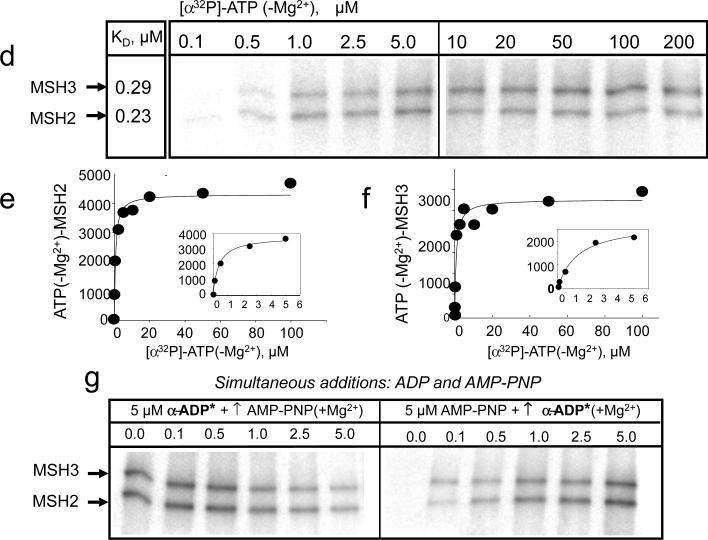
(a) Plot of the concentration of bound [3H]-ADP(+Mg2) versus concentration of hMSH2/MSH3 as measured by filter binding. hMSH2/MSH3 at a concentration of 200 nMolar (400 nMolar total nucleotide binding sites) was titrated with increasing amounts of [3H]-ADP(+Mg2) as indicated (gray circles). Maximal binding (Bmax) was derived from a fit of the binding data to a single site model. Bmax was 187 nMolar and, therefore, bound to 46.8% of total available sites. (b) Plot of BD-ATP(−Mg2)(1.0μM) versus concentration of hMSH2/MSH3 determined by FA (black circles). Dotted lines indicate the intersection of initial slope and anisotropy signal at saturation. The arrow indicates the concentration of hMSH2/MSH3 at saturation (0.9μM). (c) Stochastic and exclusive binding generates 4 unique nucleotide-bound species of hMSH2/MSH3. (d) Schematic of possible outcomes for x-linking of radiolabeled ATPs. Red and star indicates that 32P-labled phosphate, and grey balls indicate a generic hMSH2/MSH3 subunit. (ATP binding) Retention (+) of the [α32P]-ATP(−Mg2+) or [γ 32P]-ATP(−Mg2+) x-linking signal in the absence of magnesium (−Mg2+) indicates ATP binding; (ATP hydrolysis) Loss (−) of the [γ 32P]-ATP(+Mg2+) x-linking signal in the presence of magnesium (+Mg2+) indicates hydrolysis; (ADP product binding) Retention (+) of the [α32P]-ATP(+Mg2+) x-linking signal in the presence of magnesium (+Mg2+) indicates ADP binding. (e) Representative PAGE gel of x-linked products of hMSH2/MSH3 bound with equimolar [α32P]-ATP(−Mg2+) at 1.0 μM under non-hydrolytic (−Mg2+)(lane 1) and hydrolytic (+Mg2+)(lane 2) conditions; x-linked products of hMSH2/MSH3 with [γ 32P]-ATP under non-hydrolytic (−Mg2+)(lane 3) and hydrolytic (+Mg2+)(lane 4) conditions. The hMSH2 or the hMSH3 subunits of hMSH2/MSH3 is indicated.
To evaluate which of the four forms might be available for DNA binding, we tested whether the proportion of ATP- and ADP-bound species of hMSH2/MSH3 changed under hydrolytic conditions. hMSH2/MSH3 was incubated with [α32P]-ATP or [γ 32P]-ATP in the presence or absence of magnesium, and the products were evaluated after x-linking (see Fig. 3d,e). The retention or loss of the labeled phosphate was used to determine whether ATP bound to the respective subunits, and whether the bound ATP was hydrolyzed (see Fig. 3d for schematic of outcomes). [α32P]-ATP x-linked to a similar extent to both subunits of hMSH2/MSH3 under hydrolyzing and under non-hydrolyzing conditions (Fig. 3e, lanes 1 and 2). In the presence of magnesium, however, the [γ 32P]-ATP(+Mg2+) signal was lost (Fig. 3e, compare lanes 3 and 4). Thus, both subunits of hMSH2/MSH3 bound ATP, hydrolyzed it, and retained the ADP product.
ADP-bound forms of hMHS2/MSH3 are stable in solution
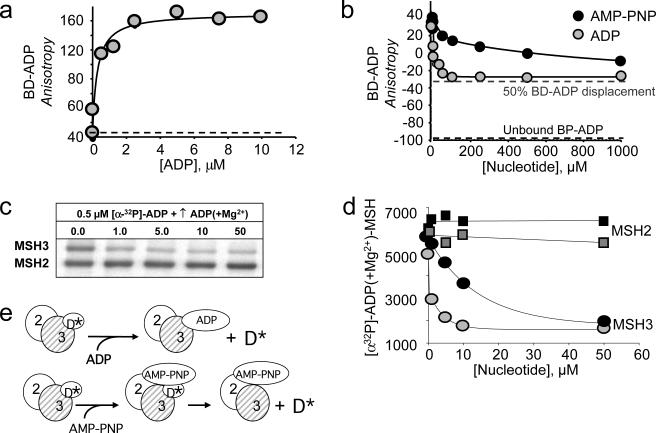
Stochastic binding predicted that the ADP in the hMSH2/MSH3 subunits, once formed, would freely exchange ADP or ATP in solution. Given the excess of ATP in the cell, we anticipated that the population of ATP-bound forms of hMSH2/MSH3 would be substantial. However, ADP not only formed within the subunits of hMSH2/MSH3, but the ADP-bound forms appeared to be stable. To test this hypothesis, we measured the ability of AMP-PNP(+Mg2+) or ADP(+Mg2+) to displace pre-bound BD-ADP(+Mg2+) within the subunits of hMSH2/MSH3. Since ADP binds equally well to either subunit BD-ADP-hMSH2/MSH3-empty and empty-hMSH2/MSH3-BD-ADP were present in equal amounts at the beginning of the experiment (Fig. 4a, dotted line is unbound nucleotide). However, competition with either AMP-PNP(+Mg2+) or ADP(+Mg2+) failed to displace half of the pre-bound BD-ADP (Fig. 4b; lower dashed line represents the anisotropy of unbound nucleotide and upped dashed line the anisotropy at half saturation). Indeed, one of the hMSH2/MSH3 subunits apparently did not exchange ADP for other nucleotides in solution.
Fig. 4. ADP-bound forms of hMHS2/MSH3 are stable in solution.
(a) BD- ADP(+Mg2+) binding to hMSH2/MSH3. Plot of BD-ADP(+Mg2+) anisotropy versus concentration of hMSH2/MSH3. The dashed line indicates baseline for unbound BD-ADP. (b) Plot of BD-ADP(+Mg2+) anisotropy versus added competitor, either unlabeled ADP(+Mg2+) (grey circles) or unlabeled AMP-PNP (+Mg2+)(black circles). The bottom dashed line indicates the anisotropy signal of unbound BD-ADP, and the upper dashed line indicates half-bound. (c) Representative PAGE gel resolving x-linked products of [α32P]-ADP(+Mg2+)(0.5μM) to either the MSH2 or the MSH3 subunits of hMSH2/MSH3 after competition with an increasing concentration of unlabeled ADP(+Mg2+). (d) Quantified band intensities from data in (c). [α32P]-ADP(+Mg2+) x-linking to the hMSH2 and the hMSH3 subunits after addition of either unlabeled ADP(+Mg2+)(gray squares and circles) or AMP-PNP (+Mg2+)(black squares and circles). The apparent Ki values derived from plots are reported in the text, and in Supplemental Fig. 1c. (e,f) Schematic diagram illustrating 2 possible mechanisms by which AMP-PNP(+Mg2+) could dissociate pre-bound ADP. D* indicates labeled ADP, either BD-ADP(+Mg2+) or [α32P]-ADP(+Mg2+) in different experiments. (e) Direct competition model: ADP(+Mg2+) directly displaces D* (+Mg2+) in the same subunit. hMSH2/MSH3 is not dually occupied with nucleotide, (f) Allosteric model: AMP-PNP(+Mg2+) binds to the empty subunit resulting in dissociation of pre-bound D* from the occupied subunit. In this case, hMSH2/MSH3 assumes a dually occupied intermediate. Data, as reported in text, are consistent with model presented in (f).
To determine which complex retained ADP, we repeated the experiment using pre-bound [α32P]-ADP(+Mg2+), and x-linked it during the competition. Even at 100-fold molar excess of AMP-PNP(+Mg2+) or ADP(+Mg2+), hMSH2 subunit did not completely release its bound [α32P]-ADP(+Mg2+) during the crosslinking period (Fig. 4c,d). In fact, when 500 μM of unlabeled AMP-PNP(+Mg2+) or ADP(+Mg2+) was added to a BD-ADP-bound hMSH2/MSH3 complex, dissociation of the BD-ADP(+Mg2+) was not complete even after four hours (Supplementary Fig. 4).
ADP binding to the hMSH3 subunit of hMSH2/MSH3 was more labile, and pre-bound ADP exchanged with added ADP or ATP in solution (Fig. 4c,d). Indeed, dissociation of the pre-bound ADP by added ADP was a reversible reaction (compare KD and Ki for ADP in Supplemental Fig. 1a,c), and occurred by simple competition (Fig. 4e). However, the dissociation of the hMSH3-bound ADP by added ATP occurred by a different pathway. BD-ADP(+Mg2+) and BD-AMP-PNP(+Mg2+) bound to hMSH3 with similar affinity (Supplemental Fig. 1b), however, the KI for AMP-PNP(+Mg2+)-induced dissociation of BD-ADP(+Mg2+) was 40-fold higher (11.3 μM) (Fig. 4c,d and Supplemental Fig. 1c). The results were consistent with a model in which AMP-PNP bound first to the empty hMSH2 subunit and displaced ADP from the hMSH3 subunit (Fig. 4f). Altogether, the considerable communication between the nucleotide-bound subunits of hMSH2/MSH3 indicated that ADP-bound forms would predominate in solution, even when ATP was in excess. MSH2 did not release its bound ADP, while bound ATP in either subunit was converted to ADP (Fig. 3e).
DNA-binding binding determines the nucleotide-bound state of hMSH2/MSH3
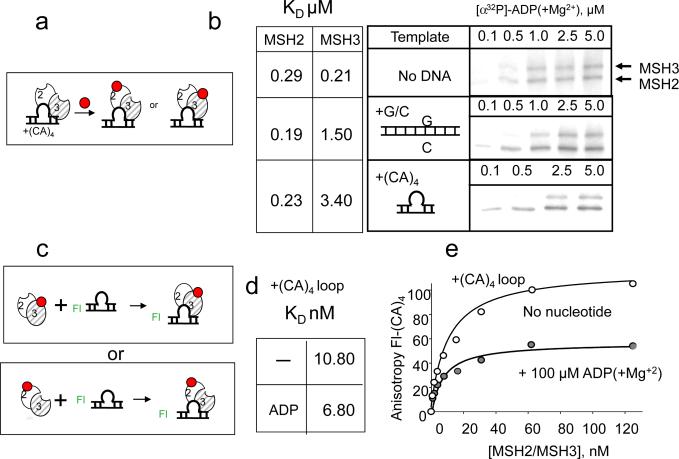
The major function for MutS-related complexes is lesion recognition, and two forms, ATP-hMSH2/MSh3-empty and empty-hMSH2/MSH3-ADP, were prominent in solution. Thus, we tested whether one or both bound to DNA, and whether DNA binding altered the nucleotide affinity (Fig. 5). Apo-hMSH2/MSH3 was added to DNA to form stoichiometric DNA/protein complexes. Nucleotide affinity was measured by x-linking of added [α32P]-ADP(+Mg2+), [α32P]-ATP(+Mg2+), or [α32P]-AMP-PNP(+Mg2+)(Fig. 5a). For ADP(+Mg2+), neither perfectly matched nor mismatched DNA had any significant effects on the binding to the hMSH2 subunit of hMSH2/MSH3 (Fig. 5b, Supplemental Fig. 1a). However, DNA binding reduced the affinity of ADP to the hMSH3 subunit 5-fold when bound to homoduplex DNA, and 10-fold when bound to a small heteroduplex loop comprising four CA residues ((CA)4-loop DNA) (Fig. 5b, Supplemental Fig. 5). Thus, DNA binding significantly inhibited ADP occupation of the hMSH3 subunit, and suggested that empty-MSH2/MSH3-ADP might not stably bind to the lesion. To test this possibility, (FL)-labeled DNA was added to stoichiometric amounts of ADP(+Mg2+)-bound hMSH2/MSH3 (Fig. 5c), and the KD for binding to the (CA)4-loop DNA was measured using FA (Fig. 5d,e). Indeed, the (CA)4-loop DNA bound only half of the added BD-ADP (Fig. 5e). Since ADP primarily occupied the hMSH2 subunit of hMSH2/MSH3 when bound to DNA (see Fig. 5b, panel 2, 3), ADP-hMSH2/MSH3-empty, but not empty-hMSH2/MSH3-ADP was competent for DNA binding.
Fig. 5. ADP-MSH2/MSH3-empty stably binds DNA.
DNA binding of hMSH2/MSH3 strongly reduces the apparent affinity for nucleotides in the hMSH3 subunit of hMSH2/MSH3. (a) Schematic of experimental design for measuring apparent ADP affinity for the hMSH2 or for the hMSH3 subunits of DNA-bound hMSH2/MSH3. Red balls indicate ADP. [α32P]-ADP(−Mg2+) is added to apo-hMSH2/MSH3 pre-bound to DNA. (b) Representative gels resolving the [α32P]-ADP(+Mg2+) x-linked products in the absence of DNA (No DNA), pre-bound to homoduplex DNA (+G/C), or pre-bound to the (CA)4 heteroduplex loop (+(CA)4). The concentration of nucleotides (μM) is indicated. Cross-linking reactions are as described in Fig. 1. The KD (μM) are indicated. (c) Schematic diagram of experimental design for measuring DNA binding affinity. Two ADP-bound complexes are available for DNA binding. Apo-nucleotide hMSH2/MSH3 or ADP-hMSH2/MSH3 (100μM) is added to fluorescein (FL, green)-labeled (CA)4-loop DNA, and DNA binding is measured as increasing FL anisotropy versus hMSH2/MSH3 concentration. ADP is denoted as red balls. The KD (nM) are indicated and summarized in Supplemental Fig. 1d: (−) is apo-MSH2/MSH3 and (ADP) is ADP-bound hMSH2/MSH3 binding to DNA. (e) Plots of anisotropy versus hMSH2/MSH3 concentration. Shown are the best fits for a single-ligand binding model. The change in anisotropy for ADP-(hMSH2/MSH3) (gray circles) complex binding to DNA is half that compared to apo-nucleotide hMSH2/MSH3 (open circles). Only one of the two ADP-bound hMSH2/MSH3 complexes illustrated in (c) can bind DNA. (f) Schematic of experimental design of ATP x-linking experiment. Same as (a) except for ATP or AMP-PNP is the added nucleotide. (g,h) Same as (b) for [α32P]-ATP(−Mg2+) (g) and [α32P]-AMP-PNP(+Mg2+)(h). The KDs (μM) are indicated, and are summarized in Supplemental Fig. 1a.
ATP had little affinity for DNA-bound hMSH2/MSH3 (Fig. 5f–h, Supplemental Fig. 6). For the hMSH2 subunit, ATP(−Mg2+) binding affinity decreased 5-fold when bound to homoduplex DNA, and 10-fold when bound to the (CA)4-loop (Fig. 5g, Supplemental Fig. 1a). The impact of DNA binding on the hMSH3 subunit was more significant. The apparent affinity of ATP to the hMSH3 subunit decreased by 100-fold with homoduplex, and nearly 200-fold when bound to the (CA)4-loop DNA, and rendered the hMSH3 subunit an essentially “empty” site (Fig. 5g, Supplemental Fig. 1a). Results for x-linking with AMP-PNP displayed the same trend (Fig. 5h and Supplemental Fig. 1a). Thus, only ADP-hMSH2/MSH3-empty bound stably to the lesion.
ATP hydrolysis enhances ADP to ATP exchange in the hMSH2 subunit of (CA)4-loop bound hMSH2/MSH3
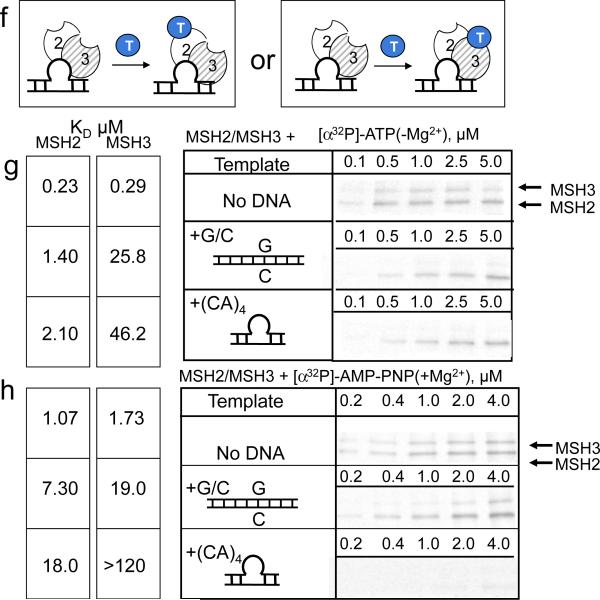
MutSα and MSH2/MSH6 become doubly occupied when bound to single base mismatched DNA, and ATP hydrolysis is used to initiate repair1–3. Therefore, we tested (1) whether DNA-bound hMSH2/MSH3 was doubly occupied with nucleotide, and (2) whether nucleotide occupancy was influenced by ATP hydrolysis (Fig. 6). ADP was added to stiochiometric complexes of (CA)4-loop bound-hMSH2/MSH3. Thus, at the beginning of the experiments, only the hMSH2 subunit of the DNA-protein complex was bound with ADP (Fig. 6h). Nucleotide occupancy was detected in the hMSH2 and hMSH3 subunits by x-linking after addition of [α32P]-ATP(+Mg2+), [γ 32P]-ATP(+Mg2+) or AMP-PNP(+Mg2+) competitors, representing both hydrolyzing and non-hydrolyzing conditions (Fig. 6).
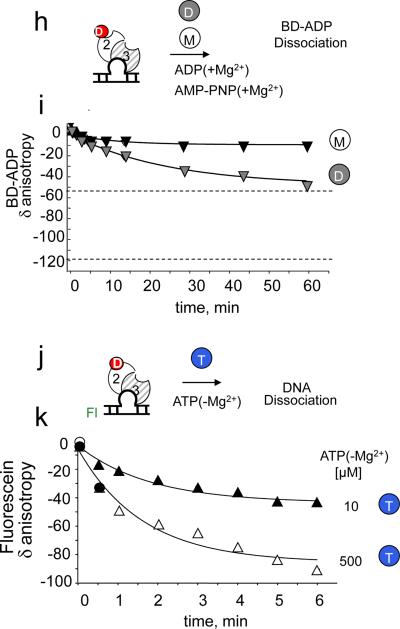
Figure 6. ATP hydrolysis enhances ADP to ATP exchange in the hMSH2 subunit of (CA)4-loop bound hMSH2/MSH3 and promotes a hydrolysis independent state.
Schematic diagram for the hMSH2/MSH3 complexes: DNA-free complex (panel 1), (CA)4 loop-bound hMSH2/MSH3 (panel 2), and homoduplex-bound hMSH2/MSH3 (panel 3), (a,b) Representative PAGE gel of [α32P]-ADP(+Mg2+) x-linked products upon competition with unlabeled ATP(+Mg2+)(a), or unlabled AMP-PNP(+Mg+2)(b), (c,d) Representative PAGE gel of the increase in x-linked [α32P]-ATP(+Mg2+)(c) or [γ 32P]-ATP(+Mg2+)(d) as competitors for unlabeled ADP pre-bound to hMSH2/MSH3 (panel 1) or to (CA)4 loop-bound hMSH2/MSH3 (panel 2). Concentrations (μM) of the competing nucleotide are indicated. Labeled nucleotide denoted by star (*) and bold letters. Each radionucleotide has a different specific activity, and the x-linking intensities cannot be compared between panels of different labels. The [α32P]-ADP(+Mg2+) x-linking reactions with hMSH2/MSH3 were performed as described in Fig. 1. (e) Quantification of x-linking results for the hMSH2 and hMSH3 subunits in (b); (f) Quantification of x-linking results for the hMSH2 and hMSH3 subunits in (a); (g) Quantification of x-linking results for (c). (h) Schematic of experimental design to measure loss of BD-ADP(+Mg+2) with time upon competition with ADP(+Mg+2) or AMP-PNP(+Mg+2). Unlabeled ADP(+Mg+2) (white ball, D) or unlabeled AMP-PNP(+Mg+2) (gray ball, M) is added to BD-ADP-(hMSH2/MSH3-(CA)4 complex. (i) Results of (h) for (CA)4-loop-bound hMSH2/MSH3 (triangles) after competition with either 500μM unlabeled ADP(+Mg2+) (gray) or 500μM unlabeled AMP-PNP(+Mg2+) (black). Dissociation of BD-ADP is followed as the loss of anisotropy with time. Since only the MSH2 subunit in an hMSH2/MSH3-(CA)4-loop DNA complex bound the BD-ADP, 50% of the added labeled ADP was free. The upper dashed line marks the 50% bound BD-ADP(+Mg2+) from MSH2/MSH3-(CA)4, while the lower dashed line marks unbound BD-ADP. (j) Schematic of experimental design for DNA dissociation of fluorescein-lableled (FL, in green) (CA)4loop complexes and ADP-hMSH2/MSH3. (k) Results for DNA dissociation by the loss of FL anisotropy with time after addition of ATP(−Mg+2)(T-blue circle) at the indicated concentrations: 10μM (black symbols), 500 μM (open symbols).
For the DNA-free hMSH2/MSH3, addition of AMP-PNP(+Mg2+) had little effect on [α32P]-ADP(+Mg2+) occupancy of the hMSH2 subunit. (Fig. 6b, panels 1 and 2; Fig 4c,d). A modest amount of ADP occurred in the hMSH2 subunit of DNA-free protein under hydrolyzing conditions (Fig. 6a, panel 1), which was mirrored by a comparable gain in ATP binding (Fig. 6c, panel 1). ATP bound in either subunit was hydrolyzed to ADP (compare Fig. 6c and d, panel 1)
Addition of AMP-PNP(+Mg2+) also had little effect on the ADP occupancy of the (CA)4-loop-bound hMSH2/MSH3 (compare Fig. 6a,b, panel 2). However, hydrolytic conditions not only promoted double occupancy of CA)4-loop-bound ADP-hMSH2/MSH3-empty, but also resulted in three key effects relative to the DNA-free complex (Fig. 6, quantified results in Fig. e–g). First, release of ADP in the hMSH2 subunit of (CA)4-loop-bound ADP-hMSH2/MSH3-empty under required ATP binding and hydrolysis in the hMSH3 subunit (compare Fig. 6a,b, panel 2). The loss of the [α32P]-ADP(+Mg2+) x-linking signal occurred only when ATP(+Mg2+) was the competitor (Compare Fig. 6a and b, panel 2). Second, binding of [α32P]-ATP(+Mg2+) in the hMSH3 subunit of (CA)4-loop-bound ADP-hMSH2/MSH3-empty increased the affinity of ATP for the hMSH2 subunit, resulting in ADP to ATP exchange (Compare Fig. 6b and 6c, panel 2). Third, after exchange, ATP hydrolysis in the hMSH2 subunit was inhibited (Fig. 6c and d, panel 2). Thus, under hydrolytic conditions, binding of ATP(+Mg2+) in the hMSH3 subunit of (CA)4-loop-bound hMSH2/MSH3 led to conversion of (CA)4-loop-bound ADP-MSH2/MSH3-empty into a hydrolysis independent ATP-hMSH2/MSH3-ADP intermediate.
Binding to the (CA)4 loop suppresses ADP/ATP exchange relative to homoduplex DNA
To test if the nucleotide dynamics of hMSH2/MSH3 depended on binding to the heteroduplex loop, we prebound hMSH2/MSH3 to excess homoduplex DNA and repeated the x-linking experiments. Under those conditions, ATP was an even better competitor for pre-bound [α32P]-ADP(+Mg2+) compared to (CA)4 loop-bound or unbound hMSH2/MSH3 (Fig. 6a,b, panel 3, and 6f). Thus, ADP-hMSH2/MSH3 binding to the (CA)4 heteroduplex loop suppressed ADP/ATP exchange in the hMSH2 subunit relative to perfectly paired DNA.
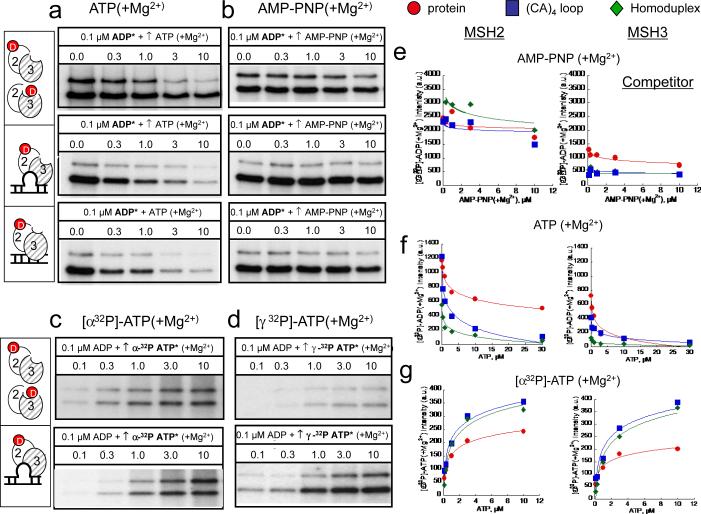
ATP hydrolysis suppresses ADP/ATP exchange in hMSH2 subunit, and promotes DNA dissociation of ADP-hMSH2/MSH3-empty
We further investigated the consequence on double occupancy of (CA)4 loop-bound ADP-hMSH2/MSH3. In these kinetic experiments, as before, the fluorescent analogue, BD-ADP was added in a 1:1 ratio to pre-formed stoichiometric complexes of (CA)4-loop DNA and hMSH2/MSH3. Thus, the hMSH2/MSH3 heterodimers were bound to DNA and only the hMSH2 subunit was bound with BD-ADP (Fig. 6h). Unlabeled ADP(+Mg2+) or AMP-PNP(+Mg2+) was added as a competitor (500 μM), and BD-ADP dissociation was followed for one hour by FA (Fig. 6h).
Unlabeled ADP(+Mg2+) competed directly for and displaced nearly all of the pre-bound BD-ADP(+Mg2+) in the (CA)4 loop-bound ADP-MSH2/MSH3-empty. Since ADP occupied only one site, the BD-ADP released was half (Fig, 6i, upper dashed line) that of the total added BD-ADP(+Mg2+) (Fig. 6i, lower dashed line). In contrast, under non-hydrolyzable conditions, AMP-PNP(+Mg2+) displaced little of the BD-ADP(+Mg2+) in the hMSH2 subunit of (CA)4-loop-ADP-hMSH2/MSH3 within the one hour observation time (Fig. 6i). However, (CA)4 loop-bound ADP-hMSH2/MSH3-ATP intermediate dissociated from DNA within minutes (Fig. 6j,k). To measure the kinetics of DNA dissociation in the absence of hydrolysis, we incubated FL-labeled (CA)4-loop DNA with ADP-hMSH2/MSH3-empty, and followed loss of the FL anisotropic signal upon addition of ATP(−Mg2+) (Fig. 6j). The ADP-hMSH2/MSH3-ATP was not stably retained on DNA under these conditions. FL-(CA)4-loop dissociation was ATP concentration dependent, and FL-(CA)4-loop dissociation from ADP-hMSH2/MSH3-ATP was nearly complete by six minutes after addition of 500 μM ATP (Fig. 6k). Thus, under non-hydrolyzing conditions, the hMSH2 subunit failed to undergo ADP to ATP exchange, and the ADP-hMSH2/MSH3-ATP intermediate, dissociated from DNA with ADP still bound.
DISCUSSION
The nucleotide-binding properties in hMSH2/MSH3 are DNA-dependent
The biochemical properties of MSH2/MSH3 have been largely inferred from those of bacterial MutS or eukaryotic MSH2/MSH6. However, we report here that the nucleotide binding properties of hMSH2/MSH3 and hMSH2/MSH6 are remarkably different. Both hMSH2/MSH3 and hMSH2/MSH6 maintain a subunit asymmetry for binding nucleotides, but the mechanism is distinct for each protein. For hMSH2/MSH6 and for yMSH2/MSH6, the apparent affinity for each nucleotide determines the subunit occupancy18. ATP binds with high affinity to the MSH6 subunit, ADP binds with high affinity to hMSH2. Thus, ADP binding to the hMSH2 subunit arises from rapid hydrolysis of ATP and subsequent release of ADP by MSH6 followed by stable binding of this ADP by MSH2. The high affinity binding of ATP and ADP are minimally affected by lesion binding (Supplemental Fig. 7).
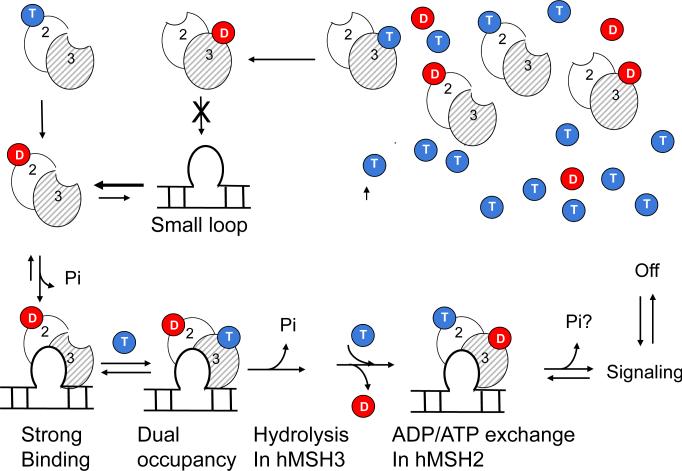
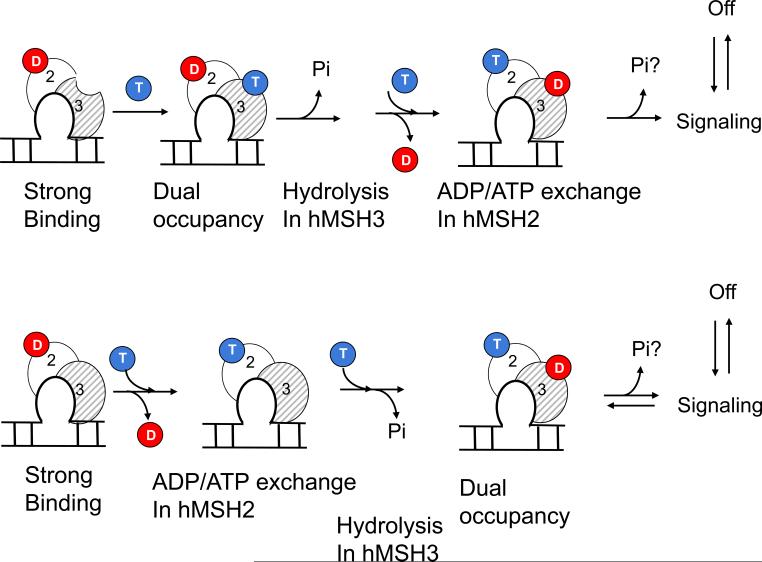
In contrast, ADP and ATP bind stochastically to either subunit of hMSH2/MSH3, and DNA binding determines the nucleotide bound state (Fig. 7). ATP hydrolysis, in the absence of DNA, results in two equal populations of ADP-MSH2/MSH3-empty and empty-hMSH2/MSH3-ADP in solution. However, binding of hMSH2/MSH3 to its canonical (CA)4 loop substrate excludes or substantially weakens nucleotide occupation of the hMSH3 subunit (Fig. 7, strong binding). Thus, DNA binding creates a “nucleotide signature” in which one ADP binds within the hMSH2 subunit and the hMSH3 subunit is empty.
Fig. 7. Allosteric model for loop repair by hMSH2/MSH3.
The concentration of ATP is 10-fold higher than ADP in the resting cellular environment33 (ATP depicted as blues balls and red depicts ADP), but both subunits of hMSH2/MSH3 hydrolyze ATP and retain ADP. Moreover, we have shown previously that DNA binding by MSH2/MSH3 increases the ATP hydrolysis rate by 10-fold23,30. Thus, the initial state of hMSH2/MSH3 before encountering DNA is likely to be one that is ADP-bound. Only ADP-MSH2/MSH3-empty stably binds to its canonical DNA substrate (Strong Binding). Loop binding shifts the equilibrium towards the doubly bound state in which ADP occupies the hMSH2 subunit and ATP occupies the hMSH3 subunit (Dual occupancy). ATP hydrolysis in the hMSH3 subunit (Hydrolysis) promotes ADP to ATP exchange in the hMSH2 subunit by decreasing the apparent affinity of ADP and increasing the apparent affinity ATP for both subunits (affinity change: ADP/ATP exchange). ATP hydrolysis is suppressed in the hMSH2 subunit relative to hMSH3. It is possible that a downstream co-factor binding or interaction with an MutL-homologue (MHL) complex triggers a second round of ATP hydrolysis in the hMSH2 subunit (Pi question mark). Sliding and/or dissociation occur subsequently, and return the MSH2/MSH3 to the pool of hMSH2/MSH3. Three complexes are potential DNA binding complexes nucleotide-bound precursors (hMSH3 is likely to be hydrolyzed to hMSH3-ADP). A second “rogue complex”, (empty)-MSH2/MSH3-(ADP), does not bind a small loop DNA substrate, but is present in solution. The DNA binding properties or a role for this complex has not been characterized.
The ability for lesion binding to convert hMSH2/MSH3 into a distinct nucleotide-bound state, poises it to be a molecular sensor for lesion specificity. Indeed, this prediction is consistent with the finding that replacement of yMSH3 protein with the yMSH6 MBD does not complement the mutational profile of a yMSH6(−/−)26. However, some aspects of downstream signaling are also likely to be shared since a chimeric yMSH6 protein with the yMSH3 MBD is functional for repair26. Our data provide a biochemical basis for these recent genetic observations, and underscore fundamental differences in the mechanisms of hMSH2/MSH6 and hMSH2/MSH3 recognition.
A model for lesion recognition by hMSH2/MSH3
For MutSα and MSH2/MSH6, binding to mispaired DNA causes a shift to the doubly occupied state that is required for downstream MMR events. Recent x-linking analysis indicates that, in yeast, empty-yMSH2/MSH6-empty or ADP-yMSH2/MSH6-empty binds to mispaired DNA, which stabilizes ATP binding and inhibits its hydrolysis in the MSH6 subunit. ADP/ATP and ADP/ADP doubly occupied states of MutS or MSH2/MSH6 have been observed16,27,32. However, in yeast18, stable ATP binding in hMSH6 destabilizes ADP occupation of the yMSH2 subunit, which is subsequently replaced by ATP. ADP/ATP exchange leads to formation of an ATP-yMSH2/MSH6-ATP “hydrolysis independent” intermediate, which is capable of end-dependent dissociation18. ATP hydrolysis presumably requires additional factors and occurs downstream.
The mechanism for downstream signaling by hMSH2/MSH3 differs significantly. Like yMSH2/MSH6, binding of ADP-hMSH2/MSH3-empty to the heteroduplex loop shifts the equilibrium towards a doubly bound state (Fig. 7, dual occupancy). However, on the lesion, ADP-hMSH2/MSH3-empty binds and hydrolyzes ATP in the empty hMSH3 subunit (Fig. 7, hydrolysis), which both reduces ADP affinity and increases ATP affinity for the hMSH2 subunit. ADP/ATP exchange converts (CA)4-loop-bound ADP-MSH2/MSH3-ATP into an ATP-hMSH2/MSH3-ADP intermediate in which ATP hydrolysis is inhibited in the hMSH2 subunit (Fig. 7, ADP/ATP exchange).
Both yMSH2/MSH6 and hMSH2/MSH3 undergo ADP to ATP exchange in the MSH2 subunit in response to ATP binding, but their requirement for ATP hydrolysis in the exchange reaction is distinct. ADP release from DNA-bound yMSH2/MSH6 requires ATP stabilization in the hMSH6 subunit, while ADP release from DNA-bound hMSH2/MSH3 requires ATP hydrolysis in the hMSH3 subunit (Figure 7, hydrolysis). The latter finding implies that ADP would occupy the hMSH3 subunit before binding of ATP in the hMSH2 subunit of DNA-bound hMSH2/MSH3. Thus, hMSH2/MSH3 ultimately adopts an ATP-hMSH2/MSH3-ADP intermediate, in which ADP binding stabilizes ATP in, at last a partially, “hydrolysis independent” state (Fig. 7, ADP/ATP exchange). Depending on the relative rates of nucleotide binding and ATP hydrolysis, it remains possible that an ADP-hMSH2/MSH3-ADP or even an ATP-hMSH2/MSH3-ATP intermediate briefly forms. However, we have not resolved either in the present measurements.
Whether the ADP-hMSH2/MSH3-ATP or ATP-hMSH2/MSH3-ADP intermediates act as sliding clamps is not yet known. Both subunits of hMSH2/MSH3 bind either nucleotide, and hydrolyze ATP at different rates. Thus, sliding and/or dissociation will require a detailed kinetic analysis of nucleotide binding dynamics in the hMSH2 and hMSH3 subunits, the differential rate of hydrolysis in the respective subunits, and the DNA association during coordination of each of these events. Nonetheless, our data indicate that interaction of hMSH2/MSH3 with the (CA)4 loop inhibits ADP release in the hMSH2 subunit relative to the homoduplex DNA, indicating that the loop is necessary for the suppression. At the same time, ADP release depends on ATP binding and hydrolysis in the hMSH3 subunit. Thus, at a minimum, the ADP-hMSH2/MSH3-empty to ATP-hMSH2/MSH3-ADP conversion is likely to occur on the (CA)4 loop. The resulting ATP-hMSH2/MSH3-ADP intermediate is likely to bind there, at least transiently, to verify the mismatch32 or before moving away from the mismatch (Fig. 7).
Based on these data, we propose a model for lesion recognition by hMSH2/MSH3, in which ATP hydrolysis and heteroduplex loop binding have opposing effects. Interaction of hMSH2/MSH3 with the heteroduplex loop suppresses ADP release from the hMSH2 subunit relative to homoduplex DNA, while ATP hydrolysis in the hMSH3 subunit promotes ADP release. Together, these functions regulate ATP/ADP exchange and provide a balance that maximizes the retention of the doubly occupied state on the heteroduplex loop. The inability of hMSH3 to hydrolyze ATP shifts the ADP/ATP equilibrium to the left, and hMSH2/MSH3 leaves the site before converting to the appropriate signaling intermediate. The homoduplex DNA differs from heteroduplex DNA in that the equilibrium for ADP/ATP exchange is shifted to the right, and DNA binding of doubly occupied hMSH2/MSH3 is short-lived.
Nucleotide dynamics of hMSH2/MSH3 at other lesions
A conformational change in the nucleotide-bound state of hMSH2/MSH3, and adoption of a “nucleotide signature” at its canonical (CA)4 loop substrate provides a plausible starting point to direct repair through a canonical pathway. The nucleotide binding properties of hMSH2/MSH3 at other lesions have not yet been determined. However, conceivably, there could be alternative nucleotide “signatures” in hMSH2/MSH3 that are lesion dependent. Consistent with this type of model, our data reveal that hMSH2/MSH3 forms a “rogue” complex (hMSH2/MSH3-ADP), which is stable in solution but does not bind to the (CA)4 loop. Empty-hMSH2/MSH3-ADP is not predicted to have an hMSH2/MSH6 counterpart based on current data18. Thus, one or more nucleotide-bound forms of hMSH2/MSH3 present in solution may sense a different spectrum of non-canonical lesions.
Indeed, lesion-dependent mutations that specifically implicate a role for hMSH2/MSH3 have been reported4,30,31. hMSH2/MSH3 is a causative factor in trinucleotide DNA expansion of CAG or CTG sequences in vivo30,31. hMSH2/MSH6 does not appear to be involved in trinucleotide-repeat expansion in vivo in spite of its capacity to bind synthetic CAG hairpins with high apparent affinity in vitro (Owen, unpublished). We have shown previously that binding of a CAG-repeat synthetic DNA folded into a hairpin compromises both nucleotide apparent affinity and ATPase activity in vitro30. The overall ATPase activity of hMSH2/MSH3 bound to a CAG-hairpin DNA is inhibited relative to binding of repair-competent substrates. Thus, the hairpin can bind with high apparent affinity to hMSH2/MSH3, but its ATPase activity is compromised upon interaction with the heteroduplex loop30. In light of our findings, binding to a complex lesion such as a CAG-hairpin may fail to specify the nucleotide bound state required for canonical loop repair by inhibiting ATPase activity in at least one of the hMSH2/MSH3 subunits. Alternatively, inhibition of ATPase activity may allow hMSH2/MSH3 to direct repair to alternative, non-canonical pathway that is competent to carry out error-prone repair. In an alternative pathway, DNA-hMSH2/MSH3 complexes may associate with different cofactors or different downstream signaling molecules in order to complete repair of the site. Whatever the differences, lesion-specific interactions will ultimately shape and determine whether the biological outcome is repair, mutation, or apoptosis.
EXPERIMENTAL PROCEDURES
Protein Purification
Human MSH3 and His-tagged MSH2 were over-expressed in SF9 insect cells using a pFastBac dual expression vector (GIBCO-BRL) that had been sub-cloned from pFastBac1 plasmids expressing untagged human MSH2 and MSH3 graciously obtained from Josef Jiricny. SF9 insect cell pellets expressing the recombinant proteins were obtained under contract with the University of Colorado Cancer Center's Cell Culture Core Facility (Denver, CO). Proteins were purified and stored as described30. Detailed description is provided in supplemental methods.
DNA Substrates and Nucleotides
Synthetic DNA substrates were prepared as described30 with an unlabeled 5' strand and either an unlabeled 3' strand or labeled with fluorescein as indicated. All complexes were purified by non-denaturing PAGE and stored at 4° C. Unlabeled ADP, ATP, and AMP-PNP were purchased from Sigma and purity was assessed by chromatography (Supplemental methods) using a mono-Q (5/5) column (Amersham/GE Healthcare). [α32P]-ATP was purchased from Amersham/GE Healthcare, and [α32P]-ADP was derived by incubation of [α32P]-ATP with hexokinase. Hexokinase, at 112 μg/ml was added to [α32P]-ATP in a buffer containing 1 mM glucose, 20 mM TEA, pH 7.0, 200 mM KCl, 0.5 mM EDTA, 0.5 mM DDT, 10 mM MgCl2, and 10% glycerol. Conversion of ATP to ADP was assessed by thin layer chromatographic (TLC) analysis as described previously30 (Supplemental Fig. 8). [α32P]-AMP-PNP was purchased from Apparent affinity Labeling Technologies. All preparations of nucleotides used contained less than 1% contamination of other nucleotides, except AMP-PNP which contained about 20% of a previously recognized but uncharacterized contaminant (arrow 4 minutes, Supplemental 2a, bottom chromatogram). This species underwent further degradation over time to a component that eluted at the same place as AMP when analyzed by MonoQ chromatography (Supplemental Fig. 2a, arrow 3 minutes, bottom chromatogram). The concentration of AMP-PNP was adjusted for the presence of this contaminant.
Anisotropy
Anisotropy was measured using a Safire fluorescent plate reader (Tecan Group Ltd.) at ambient temperature. Purified proteins were added at concentrations as indicated and binding was monitored to either bodipy-labeled nucleotides (ADP, ATP, or AMP-PNP) or fluorescein-labeled DNA. Protein-nucleotide or protein-DNA complexes were pre-incubated at RT for 30 minutes prior to measurements. G-factor was determined by measuring 1:1 complexes of each protein assayed bound to its' respective fluor-labeled substrate at the highest protein concentration and polarization calculations were adjusted accordingly using the instrument's software (XFluor). All anisotropy measurements for KD determinations were performed in triplicates and comparable measurements were performed using proteins from different purifications.
Nucleotide binding affinity on DNA-bound complexes
To prepare the DNA-bound complexes for these experiments, we utilized conditions under which the majority of the DNA-bound complex was primarily ADP-hMSH2/MSH3-empty. Specifically, hMSH2/MSH3 was incubated with either the (CA)4 loop DNA or homoduplex DNA at a 1:10 protein to DNA ratio before addition of nucleotides, and [α32P]-ADP(+Mg2+) was added in 1:1 ratio to the protein. In parallel, we added [α32P]-ADP(+Mg2+) in stoichiometric ratio to hMSH2/MSH in the absence of DNA. Each sample was x-linked for 7.5 minutes, and the x-linking profiles of DNA-bound-[α32P]-ADP-hMSH2/MSH3-(empty) and DNA-free ADP-hMSH2/MSH3-empty were directly compared under hydrolyzing (ATP(+Mg2+)) and non-hydrolyzing (AMP-PNP(+Mg2+)) conditions.
Filter Binding
Filter binding of [3H]-ADP to human MSH2/MSH3 was performed on ice using 100nM protein (200nM nucleotide binding-sites) and increasing concentrations of labeled-ADP was added, as indicated. (HAWP filters, Millipore). The amount of 3H-ADP bound to MSH2/MSH3 was determined from a standard curve constructed from known concentrations of 3H-ADP spotted on the filters and processed identically. Triplicates at each nucleotide concentration were assayed for both standard curve and protein-binding reactions.
Cross-linking
Cross linking (Stratalinker) reactions, with respect to time and temperature, were done essentially as described previously18. However, we used, for experiments with both MSH2/MSH6 and MSH2/MSH3, buffer conditions that were ideal for MSH2/MSH3 stability and activity: 25 mM Hepes, pH 8.1, 110 mM NaCl, 1.0 mM dithiothreitol, 100 μg/ml BSA, and 10% glycerol, and 5.0 mM MgCl2 was added where indicated. Experiments were performed at least three times and with different lots of protein and nucleotide preparations, except for [α32P]-AMP-PNP, where only one preparation was available. Non-linear regression fits of the data were performed using SigmaPlot (Systat Software, Inc.)
Competitive Inhibition measurements by reverse anisotropy (Ki)
Reversibility of binding for Bodipy-labeled nucleotides was measured by competition of pre-bound Bodipy-nucleotide-MSH protein complexes. 2.5μM protein (heterodimer) was mixed with 1.0 μM Bodipy-labeled nucleotide and incubated at RT for 30 minutes. Available binding sites in the protein were in excess relative to the added nucleotide to ensure complete binding, as unbound Bodipy-nucleotide would complicate the measurement. Increasing concentrations of unlabeled nucleotide was added and nucleotide concentration was plotted versus loss of anisotropic signal. The data were fit to a 3-parameter hyperbolic decay curve using Sigmaplot. The resulting EC50 was used to calculate the Ki using the following equation: The same method was used for determining the binding apparent affinity of homoduplex (G/C) DNA, with the following exceptions: a 10 nanomolar complex was formed between MSH2/MSH3 and (CA)4-loop DNA labeled on the 3' strand with fluorescein and increasing concentrations of unlabeled G/C DNA was added up to 1000μM.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Richard Weinshilboum, Thomas C. Wood and Louis J. (Jim) Maher, III for providing generous access to critical equipment; Irina Kovtun and Jania Trushina for helpful comments. This work was supported by the Mayo Foundation, the National Institutes of Health grants NS40738 (CTM), GM 066359 (CTM), the CA092584 (CTM).
REFERENCES
- 1.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–23. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 2.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 3.Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 4.Surtees JA, Alani E. Mismatch repair factor MSH2/MSH3 binds and alters the conformation of branched DNA structures predicted to form during genetic recombination. J Mol Biol. 2006;360:523–36. doi: 10.1016/j.jmb.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 5.Kovtun IV, McMurray CT. Crosstalk of DNA glycosylases with pathways other than base excision repair. DNA Repair (Amst) 2007;6:517–29. doi: 10.1016/j.dnarep.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 6.Fishel R. The selection for mismatch repair defects in hereditary nonpolyposis colorectal cancer: revising the mutator hypothesis. Cancer Res. 2001;61:7369–74. [PubMed] [Google Scholar]
- 7.Peltomaki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet. 2001;10:735–40. doi: 10.1093/hmg/10.7.735. [DOI] [PubMed] [Google Scholar]
- 8.Wei K, Kucherlapati R, Edelmann W. Mouse models for human DNA mismatch-repair gene defects. Trends Mol Med. 2002;8:346–53. doi: 10.1016/s1471-4914(02)02359-6. [DOI] [PubMed] [Google Scholar]
- 9.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–10. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 10.Lamers MH, et al. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature. 2000;407:711–7. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 11.Warren JJ, et al. Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell. 2007;26:579–92. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 12.Alani E, et al. Crystal structure and biochemical analysis of the MutS·ADP·beryllium fluoride complex suggest a conserved mechanism for ATP interactions in mismatch repair. J Biol Chem. 2003;278:16068–16094. doi: 10.1074/jbc.M213193200. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto A, Schofield MJ, Biswas I, Hsieh P. Requirement for Phe36 for DNA binding and mismatch repair by Escherichia coli MutS protein. Nucleic Acids Res. 2000;28:3564–9. doi: 10.1093/nar/28.18.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blackwell LJ, Bjornson KP, Allen DJ, Modrich P. Distinct MutS DNA-binding modes that are differentially modulated by ATP binding and hydrolysis. J Biol Chem. 2001;276:34339–47. doi: 10.1074/jbc.M104256200. [DOI] [PubMed] [Google Scholar]
- 15.Iaccarino I, Marra G, Palombo F, Jiricny J. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSalpha. Embo J. 1998;17:2677–86. doi: 10.1093/emboj/17.9.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gradia S, et al. hMSH2/MSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 17.Mendillo ML, Mazur DJ, Kolodner RD. Analysis of the interaction between the Saccharomyces cervisiae MSH2/MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J Biol Chem. 2005;280:22245–22257. doi: 10.1074/jbc.M407545200. [DOI] [PubMed] [Google Scholar]
- 18.Mazur DJ, Mendillo ML, Kolodner RD. Inhibition of MSH6 ATPase activity by mispaired DNA induces a MSH2(ATP)-MSH6(ATP) state capable of hydrolysis-independent movement along DNA. Mol Cell. 2006;22:39–49. doi: 10.1016/j.molcel.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Acharya S, et al. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci U S A. 1996;93:13629–34. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gradia S, Acharya S, Fishel R. The role of mismatched nucleotides in activating the hMSH2-hMSH6 molecular switch. J Biol Chem. 2000;275:3922–30. doi: 10.1074/jbc.275.6.3922. [DOI] [PubMed] [Google Scholar]
- 21.Marsischky GT, Kolodner RD. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2/MSH6 complex and mispaired bases in DNA. J Biol Chem. 1999;274:26668–82. doi: 10.1074/jbc.274.38.26668. [DOI] [PubMed] [Google Scholar]
- 22.Harrington JM, Kolodner RD. Saccharomyces cerevisiae MSH2/MSH3 acts in repair of base-base mispairs. Mol Cell Biol. 2007;27:6546–54. doi: 10.1128/MCB.00855-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson T, Guerrette S, Fishel R. Dissociation of mismatch recognition and ATPase activity by hMSH2-hMSH3. J Biol Chem. 1999;274:21659–64. doi: 10.1074/jbc.274.31.21659. [DOI] [PubMed] [Google Scholar]
- 24.Palombo F, et al. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6:1181–4. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 25.Shell SS, Putnam CD, Kolodner RD. Chimeric Saccharomyces cerevisiae MSH6 protein with an MSH3 mispair-binding domain combines properties of both proteins. Proc Natl Aca)d Sci U S A. 2007;104:10956–61. doi: 10.1073/pnas.0704148104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SD, Surtees JA, Alani E. Saccharomyces cerevisiae MSH2/MSH3 and MSH2/MSH6 complexes display distinct requirements for DNA binding domain I in mismatch recognition. J Mol Biol. 2007;366:53–66. doi: 10.1016/j.jmb.2006.10.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martik D, Baitinger C, Modrich P. Differential specificities and simultaneous occupancy of human MutS□ nucleotide binding sites. J Biol Chem. 2004;279:28402–10. doi: 10.1074/jbc.M312108200. [DOI] [PubMed] [Google Scholar]
- 28.Lamers MH, Winterwerp HHK, Sixma TK. The alternating ATPase domains of MutS control DNA mismatch repair. EmboJ. 2003;22:746–756. doi: 10.1093/emboj/cdg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bjornson KP, Modrich P. Differential and simultaneous adenosine di- and triphosphate binding by MutS. J Biol Chem. 2003;278:18557–62. doi: 10.1074/jbc.M301101200. [DOI] [PubMed] [Google Scholar]
- 30.Owen BA, et al. (CAG)(n)-hairpin DNA binds to MSH2/MSH3 and changes properties of mismatch recognition. Nat Struct Mol Biol. 2005;12:663–70. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 31.van den Broek WJ, et al. Somatic expansion behavior of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by MSH3 and MSH6 mismatch-repair proteins. Hum Mol Genet. 2002;11:191–8. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 32.Junop M, Obmolova G, Rausch K, Hsieh P, Yang W. Composite Active Site of an ABC ATPaseMutS Uses ATP to Verify Mismatch Recognition and Authorize DNA Repair. Mol. Cell. 7:1–12. doi: 10.1016/s1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- 33.Bradbury DA, Simmons TD, Slater KJ, Crouch SP. Measurement of the ADP:ATP ratio in human leukaemic cell lines (CA)n be used as an indicator of cell viability, necrosis and apoptosis. J Immunol Methods. 2000;240:79–92. doi: 10.1016/s0022-1759(00)00178-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.